- Department of Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, MA, United States
The CDKN2A/B genomic locus is associated with risk of human cancers and metabolic disease. Although the locus contains several important protein-coding genes, studies suggest disease roles for a lesser-known antisense lncRNA encoded at this locus, called ANRIL. ANRIL is a complex gene containing at least 21 exons in simians, with many reported linear and circular isoforms. Like other genes, abundance of ANRIL is regulated by epigenetics, classic transcription regulation, splicing, and post-transcriptional influences such as RNA stability and microRNAs. Known molecular functions of ANRIL include in cis and in trans gene regulation through chromatin modification complexes, and influence over microRNA signaling networks. Polymorphisms at the ANRIL gene are linked to risk for many different cancers, as well as risk of atherosclerotic cardiovascular disease, bone mass, obesity and type 2 diabetes. A broad array of variable reported impacts of polymorphisms on ANRIL abundance, splicing and function suggests that ANRIL has cell-type and context-dependent regulation and actions. In cancer cells, ANRIL gain of function increases proliferation, metastasis, cell survival and epithelial-mesenchymal transformation, whereas ANRIL loss of function decreases tumor size and growth, invasion and metastasis, and increases apoptosis and senescence. In metabolic disease, polymorphisms at the ANRIL gene are linked to risk of type 2 diabetes, coronary artery disease, coronary artery calcium score, myocardial infarction, and stroke. Intriguingly, with the exception of one polymorphism in exon 2 of ANRIL, the single nucleotide polymorphisms (SNPs) associated with atherosclerosis and diabetes are non-overlapping. Evidence suggests that ANRIL gain of function increases atherosclerosis; in diabetes, a risk-SNP reduced the pancreatic beta cell proliferation index. Studies are limited by the uncertain relevance of rodent models to ANRIL studies, since most ANRIL exons do not exist in mouse. Diverse cell-type-dependent results suggest it is necessary to perform studies in the relevant primary human tissue for each disease. Much remains to be learned about the biology of ANRIL in human health and disease; this research area may lead to insight into disease mechanisms and therapeutic approaches.
Introduction
The discovery of functional noncoding RNAs has opened a kaleidoscopic world of unanticipated mechanisms extending far beyond the DNA-RNA-protein paradigm; noncoding RNAs may in fact outnumber coding RNAs (1). Long noncoding RNAs (lncRNAs) have been discovered throughout the genome; scientists are working to explore their functions in health and disease. The ANRIL lncRNA was first identified in a melanoma kindred with a large (403 kb) deletion at the CDKN2A/B locus (2). ANRIL has attracted broad attention because it is located at a genomic hotspot for disease heritability, the CDKN2A/B locus. Although protein coding genes at this locus have important well-studied roles in cell cycle regulation, data suggest that some locus disease-associated single nucleotide polymorphisms (SNPs) act through effects on ANRIL itself. Intriguingly, studies suggest ANRIL not only impacts the biology of cancer, but also has cell-type-specific roles in metabolic disease. Although ANRIL has been reviewed in the past (3, 4), knowledge has exponentially increased in recent years. Here we review advances in ANRIL SNPs, gene regulation, cell biology, and disease roles of ANRIL.
The CDKN2A/B Locus
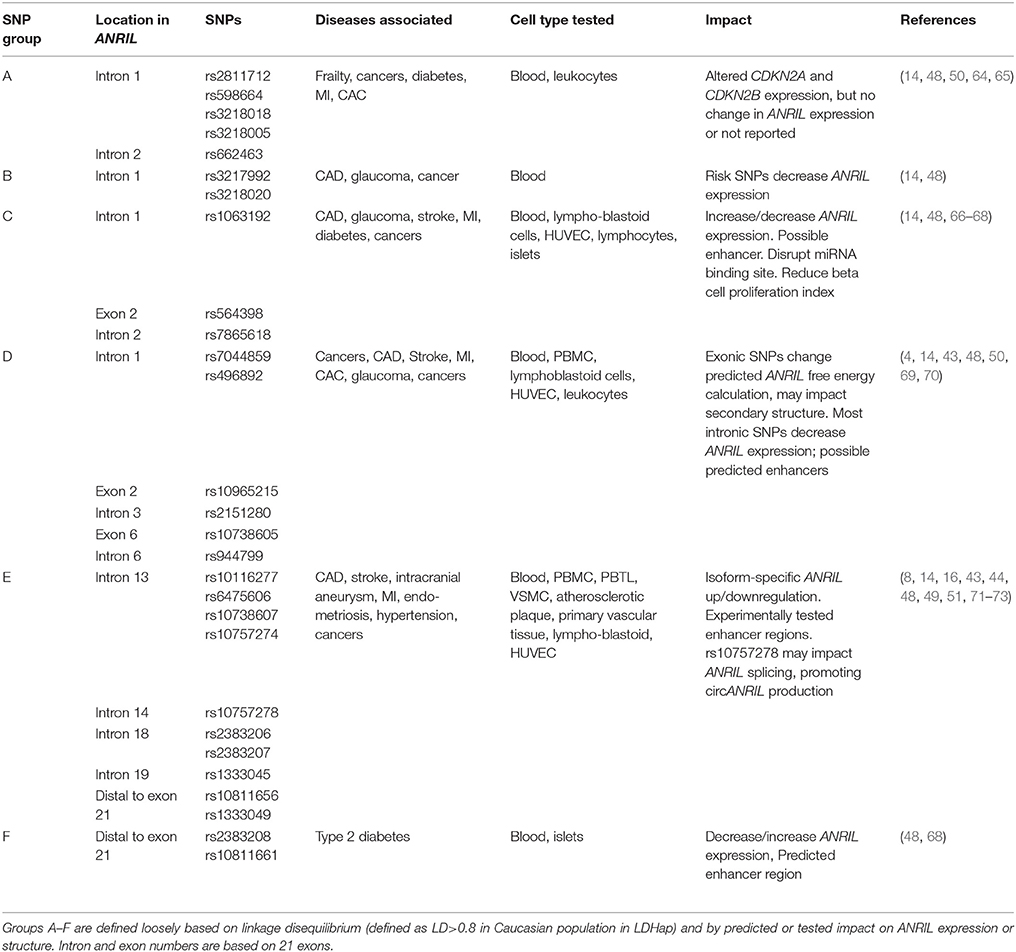
ANRIL, or CDKN2B-AS1, is located at the human CDKN2A/B locus at 9p21.3. This gene cluster, extending over a nearly 350 kb genomic region housed within a single topologically associated domain (TAD) (5), contains three protein coding genes and, antisense to them, the ANRIL lncRNA (Figure 1). The protein coding genes include S-methyl-5′-thioadenosine phosphorylase (MTAP), CDKN2A, which encodes splice variants p16INK4A and p14ARF, and CDKN2B, which encodes p15INK4B (9, 10). MTAP lies at one end of the locus, 192 kb telomeric to the 5′ start of ANRIL. At the centromeric end of the locus, the ANRIL gene contains 19–21 reported exons over a 126 kb region. CDKN2A lies between MTAP and ANRIL, near the first exon of ANRIL; CDKN2B is located within the first intron of ANRIL, in an antisense direction. The proteins encoded by CDKN2A and CDKN2B are tumor suppressors with well-established roles in cell proliferation, apoptosis, senescence and aging (11, 12). p16INK4A and p15INK4B are cyclin dependent kinase (CDK) inhibitors, inhibiting retinoblastoma phosphorylation by CDK4/6. The p14ARF protein, a splice variant of CDKN2A which due to a frame shift has no amino acid homology to the principal other CDKN2A splice variant, p16INK4A, modulates p53 activity. ANRIL is transcribed by RNA polymerase II and spliced into multiple linear and circular isoforms in a tissue-specific manner. In general, ANRIL roles, explored in detail below, include gene regulation in cis and in trans through interaction with polycomb repressive complex (PRC) histone modifiers, as well as RNA-RNA interactions such as microRNA (miRNA) sponge activity (3, 13). Known biological impact of ANRIL activities include modulation of proliferation, apoptosis and cellular adhesion pathways (14).
Figure 1. CDKN2A/B locus and ANRIL isoforms. (A) At the CDKN2A/B locus, the ANRIL lncRNA is antisense to the protein coding genes. The p15INK4B gene is contained within intron 1 of ANRIL. (B) To date, ANRIL has 21 reported exons. (C,D) Numerous linear (C) and circular (D) exons have been reported. Due to the discovery of additional exons, distal exons in some circular isoforms have been renumbered based on the current 21 reported exons. *Exons 15-16 refer to exons 14-15 in Holdt et al. (6). **Exon 16 refers to exon 14 in Sarkar et al. (7) and Burd et al. (8).
Evolution of the ANRIL Gene
The evolutionary development of the human ANRIL gene has been studied by comparative analysis of the genomes of 27 organisms including non-mammalian vertebrates, non-placental mammals, non-primate placental mammals, and primates (15). ANRIL originated in ancestors of the Eutherian (placental) mammalian clade. Initially the gene contained only a few exons; over time, ANRIL underwent clade-specific evolution, adding exons in many mammals but losing exons in rodents. The full 21 exon gene is present only in simians. ANRIL genes contain many repeat elements, both intronic, and exonic; evidence suggests that transposon activity has mediated many of the observed evolutionary changes in exon presence or absence, location, sequence, conservation, and structure, as well as introduction of splice sites (15).
Early ANRIL variants were likely not transcribed or functional (15). One hallmark of functional RNAs is splice signals at intron/exon boundaries. In simians, 191 intron/exon boundaries contained canonical splice signals, while 20 did not. In lower mammals, however, only about half of intron/exon junctions contained identifiable splice signals (15). This finding suggests that as ANRIL gained exons, and exon sequences became more conserved across species, it also increased the number of splice signals and gained functionality. Taken together, data suggest that ANRIL may be functional only in simians, and that functionality may have been introduced by transposon activity (15).
ANRIL Isoforms and Structure
With at least 21 exons (new exons discovered as recently as 2017 (7)), the ANRIL gene can potentially generate a large number of splice variants. In fact, many ANRIL isoforms have been reported (Figure 1) (16). Exon numbering has changed over time as new exons were discovered. Studies observe multiple isoforms in any given cell type, mostly at low abundance. A different range of isoforms may be identified from one cell type to another, but tissue-dependent isoform expression in primary cells or tissues has not yet been comprehensively quantified using the same reagents and techniques. Intriguingly, many studies have now identified both linear and circular ANRIL isoforms (6–8). The longest open reading frame identified in any ANRIL variant is 86 codons, supporting the concept that functionality of this gene is through RNA activity (16).
Linear and Circular Isoforms
Many conventional linear polyadenylated ANRIL isoforms are detected in different cell types. Circular ANRIL (circANRIL) isoforms, without polyadenylation, have also been described. Circular RNAs, which are formed by “back-splicing” in which a downstream splice donor site is joined to an upstream splice acceptor site, were discovered in 2012 to be a broadly occurring phenomenon across developmental stages and tissues, arising from at least 14% of human transcribed genes (17, 18). CircRNAs enjoy distinct properties from linear RNAs, including, in general, enhanced stability and longevity, cytoplasmic localization, and lack of translation (although if an IRES is engineered, circRNAs can support translation) (19). Traditional PCR using antisense-oriented primers cannot distinguish between linear and circular isoforms; other methodology, such as PCR using “outward-facing” primers directed away from each other, detection of specific exon-exon junctions, or protection from RNAse R digestion can quantify circular RNAs.
Careful examination of melanoma cell lines showed that the abundance of individual ANRIL exons is non-uniform, supporting the presence of different isoforms (7). In both transformed cell lines and in human brain derived cells, abundance of ANRIL exons was lower than exons from locus protein-coding genes CDKN2A and CDKN2B (8). Linear isoforms tend to include proximal exons (1-2), whereas isoforms with only central exons (4-16) are more likely to be circular (8). In melanoma cells, proximal exons (exon 1 and exon 5-6) were more highly expressed than distal exons. This suggests that short isoforms of ANRIL, which tend to include proximal exons, are more abundant than longer isoforms in this cell type (7). In human peripheral blood mononuclear cells and a monocyte cell line, four major groups of ANRIL transcripts were found, all with common proximal exons including exons 1, 5, and 6 but with different distal exons, of various lengths (8). Multiple circular ANRIL isoforms have been detected. A circANRIL isoform with an exon 14-5 head-to-tail junction was reported to be the predominant form in both an immortalized fibroblast cell line (8) and in a majority of melanoma cell lines (7). Other non-canonical back-spliced junctions observed in melanoma samples included exon 14-5, 7-4, 10-5, and 14-4 (7). The exons most commonly observed in circANRIL in melanoma cells were 4, 5, 6, 7, 10, 13, and 14; in varied human cell types, the majority of circANRIL species were exon 5-6-7 containing (6). ANRIL exons 1, 2, 3, 8, 9, 11, and 12 were rarely included in circular RNA products (8). In melanoma lines, no correlation was observed between abundance of linear and circANRIL (7). However, circANRIL expression was inversely correlated with linear ANRIL expression in peripheral blood mononuclear cells in a cardiovascular cohort (6). CircANRIL was found to be resistant to RNAse R digestion compared with linear ANRIL, and an actinomycin D time course confirmed enhanced stability of the 14-5 circANRIL isoform compared with linear isoforms (7, 8).
Secondary Structure
Structure and function of lncRNAs is of high interest in the scientific community, given the increasing recognition of lncRNA roles in cancer and the normal biology of higher organisms. As such, prediction of lncRNA structures is an important computational challenge. One approach is to identify structural elements through comparison of related lncRNAs. The MONSTER tool was used to compare ANRIL to two lncRNAs with similar biological function: HOTAIR and COLDAIR (20). MONSTER identifies sequence-predicted secondary structure, such as regions likely to be single stranded RNA, double stranded RNA, hairpin loops, interior loops and bulges. Comparing predictions of two lncRNAs with similar functions is proposed as a mechanism to identify structural motifs. When HOTAIR, COLDAIR, and ANRIL were compared, several common structures were identified, putative structural motifs related to their common function in epigenetic regulation, which could lead to a molecular understanding of mechanism of action in future studies (20). Another study identified the region of ANRIL that interacts with CBX7, a polycomb repressor component; secondary structure analysis revealed hairpin structural motifs with significant binding affinity to CBX7. Fluorescence anisotropy suggested a ternary complex between a particular loop of ANRIL, CBX7 and a H3K27me3 methylated histone peptide (21).
Cellular Localization
RNA localization impacts function. In melanoma cells, linear ANRIL species containing proximal (exon 1) and distal (exons 13b, 19) exons were predominantly found in the nucleus. However, middle exons (exons 5, 6, and 7), which are found in both linear and circANRIL, were observed in cytoplasmic fractions, suggesting that circANRIL species may be predominantly cytoplasmic (7). Nuclear localization suggests linear isoforms may be responsible for the known ANRIL function of regulating gene transcription via chromatin modulation (see below). Conversely, cytoplasmic localization suggests circANRIL forms may participate in post-transcriptional functions. In gastric (22), prostate (21), and urothelial (23) cancer cells, ANRIL was predominantly nuclear. In a beautiful high-resolution analysis of single-molecule lncRNA localization, ANRIL was found to be mostly localized to cell nuclei, in one or several bright foci. Like other lncRNAs analyzed, ANRIL nuclei foci were lost in mitotic cells (24). Physiological stimuli that change ANRIL localization may provide clues as to ANRIL functions. Intriguingly, in a retinal cell line ANRIL isoforms were observed by fluorescence in situ hybridization to localize to the peri-nuclear cytoplasmic space. ANRIL abundance was induced by glucose, but ANRIL localization did not change with high glucose exposure (25). On the other hand, a study in HUVEC cells, using primers predicted to detect both linear and circular isoforms, found ANRIL to be mostly nuclear; nuclear ANRIL was increased after exposure to TNF-α (26). At least one study has used ANRIL as a nuclear positive control to test localization of other transcripts (27). Future cell type specific studies of ANRIL localization under basal, stimulated, and stress conditions may lead to clues as to ANRIL roles in tissue health and disease.
Regulation of ANRIL Abundance
Abundance of ANRIL species is determined by promoter transcriptional activity, splicing decisions, and RNA stability (Figure 2). Like other genes, ANRIL promoter activity is influenced by epigenetic control and transcription factor occupancy. Intriguingly, epidemiological findings suggest that epigenetic regulation of ANRIL, through promoter methylation, has important long-lasting consequences for tissue function (29–31). As such, ANRIL regulation is one mediator of the impact of early life environmental signals on adult human health.
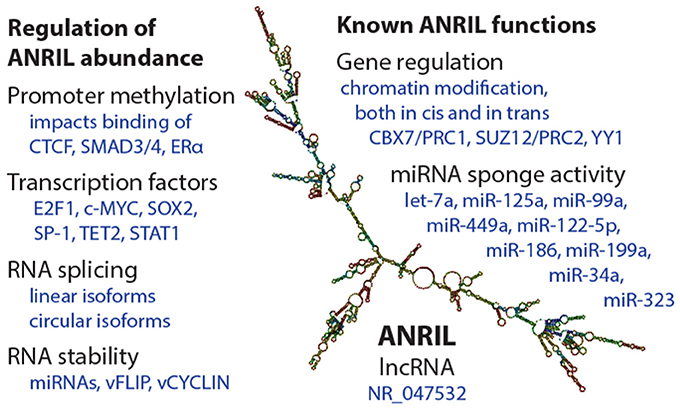
Figure 2. Summary of regulation and functions of the ANRIL lncRNA. (Left) Some of the known mechanisms by which ANRIL abundance is regulated, at the transcriptional and post-transcriptional levels. On the (Right), a selection of known ANRIL cellular functions are depicted. We apologize for observations not included in this summary image. The ANRIL structural prediction in the center is of a common long-isoform of ANRIL, and was generated from Gruber et al. (28).
ANRIL Promoter Methylation
Epidemiological and experimental findings demonstrate that methylation of the ANRIL promoter region regulates ANRIL gene expression and has functional importance. The first exons of ANRIL and p14ARF are separated by only 300 bp, in head-to-head antisense orientation; the intergenic region between them is a bidirectional promoter (4, 32, 33). In silico analysis of ENCODE ChromHMM data (34) revealed that this region is enriched for both promoter and enhancer activity, and DNAse I hypersensitivity, across multiple cell types, suggesting this is a regulatory region. This region is bound by CTCF, usually considered to be a transcriptional repressor, insulating promoters from enhancer activity. Oddly, CTCF binding at the ANRIL promoter was associated with active-chromatin mark histone H3K4 trimethylation (35). CTCF binding, and ANRIL and p14ARF expression, were inhibited by methylation of local CpG islands and increased by demethylation. Knockdown of CTCF prevented the demethylation-induced expression of ANRIL and p14ARF, confirming that CTCF is a methylation-sensitive positive regulator of ANRIL promoter activity (35).
Additional evidence supports the functional importance of CpG sites for ANRIL promoter activity, locus gene expression and transcription factor binding. Mutagenesis of the CpG sites affects both ANRIL and p14ARF promoter activity (29, 30). Methylation status of several CpG differentially methylated regions at ANRIL in umbilical cord tissues was positively associated with abundance of linear but not circular ANRIL, but inversely associated with p14ARF and p16INK4a expression (29). CpG methylation also affects other transcription factors binding at the ANRIL promoter to regulate downstream gene expression, such as interferon gamma, SMAD3/4 and ERα (29, 30). Methylation status of CpG islands around the p16INK4A transcription start site was also shown to coordinate transcription of ANRIL and p16INK4A in human cells (36). Given the multiple protein complexes binding across different CpG sites, and variable quantitative impact of individual CpG region mutagenesis on ANRIL isoforms and locus gene expression, regulation appears to be complex.
Transcription Factors Regulate ANRIL Production
ANRIL expression is influenced by cellular processes such as genotoxic stress, tumorigenesis, senescence, and inflammation. Activity at the bidirectional promoter region upstream of both ANRIL and p14ARF genes is influenced by the critical cell cycle regulator E2F1 (32, 33). In response to the genotoxic stress of DNA damage, E2F1 transcriptionally activates ANRIL in an ATM-dependent manner (33, 37). In this case, ANRIL is thought to promote cell growth by suppressing locus INK-family inhibitors after DNA repair is complete, allowing re-entry into cell cycling (33). The ANRIL promoter was also responsive to E2F1 in cancer cells (32). In addition to E2F1, several known potent oncogenes regulate ANRIL expression in various cancers. In lung cancer, c-MYC binds to an E-box in the ANRIL promoter and induces ANRIL expression (38). In nasopharyngeal carcinoma, transcription factor SOX2 was shown to bind directly to the ANRIL promoter and activate transcription of ANRIL and its downstream effector β-catenin (39). In liver cancer, SP1 binds the ANRIL promoter and positively regulates ANRIL transcription (40). On the other hand, TET2, a tumor suppressor in human gastric cancer, binds to the promoter region of ANRIL and regulates expression of ANRIL as well as p16INK4a, p15INK4b, and p14ARF (41). Transcription regulation of ANRIL is involved not only in cell DNA damage and oncogenesis, but also in disparate processes such as cell senescence and inflammation. In senescence, oncogenic Ras was found to reduce expression of ANRIL (13, 21, 42). In inflammation, STAT1 activates the ANRIL locus in vascular endothelial cells has been reported; CAD-associated ANRIL SNP rs10757278, located in a known downstream enhancer region, disrupts the STAT1 binding site and modulates IFN-γ induced ANRIL expression via stimulation (43). Intriguingly, the binding of STAT1 at this enhancer exerts cell-type specific regulation of ANRIL expression: repression in lymphoblastoid cells lines, but activation in HUVEC cells (43). In sum, data support an important role for cell-type specific transcriptional regulation of the ANRIL lncRNA in a range of cellular processes and outcomes.
Regulation of ANRIL Splicing
Cell type dependent variation in abundance of different ANRIL isoforms suggests that splicing may be a point of regulation (8, 44). Almost nothing is known about ANRIL splicing decisions. Disease-associated ANRIL gene polymorphisms have shed light on this process. In lymphocytes, the coronary artery disease (CAD) associated SNP rs10757278 (intron 12) correlates with abundance of certain circular (14-5 and 4-6) and linear (exon 1-2, but not 18-19, containing) isoforms (8). The rs10757278 A allele was found to inhibit skipping of exon 15, promoting circANRIL species ending in exon 14 (8). Mechanisms regulating ANRIL splicing require further study.
Post-Transcriptional Regulation
Determinants of ANRIL transcript longevity and stability remain uncertain, but miRNAs can participate. ANRIL, downregulated following Kaposi's sarcoma associated herpesvirus (KSHV) infection, contains multiple seed matches for KSHV miRNAs. Forced miRNA expression decreased ANRIL abundance, and miRNA pull-down experiments confirmed a direct interaction. In addition, KSHV latency associated proteins vFLIP and vCyclin also decreased ANRIL abundance, suggesting post-transcriptional miRNA-dependent and independent regulation (45).
Functions of the ANRIL lncRNA
Transcription Regulation via Chromatin Modifying Complexes
Many studies show that ANRIL functions in cells to regulate gene expression via chromatin modification. Acting in cis, ANRIL interacts with both PRC-1 and−2 to mediate epigenetic transcriptional repression of neighboring genes CDKN2A and CDKN2B, through mechanisms involving histone modification and chromatin remodeling (13, 21, 33). ANRIL interacts with PRC1 component CBX7 to recruit PRC1 to the p14ARF and p16INK4A loci, silencing the CDKN2A locus by H3K27-trimethylation (21). At CDKN2B, ANRIL was shown to recruit SUZ12, a subunit of the PRC2 (13). ANRIL also interacts with PRC-associated protein YY1 (46). Intriguingly, the structural conformation of the methyl-lysine binding pocket in the chromodomain of CBX7, which interacts with H3K27-trimethylation to cause chromatin compaction, is influenced by allosteric RNA-protein binding with ANRIL (47). However, despite this well-documented repression of other locus genes by ANRIL, a positive correlation between ANRIL (both short and long isoforms), CDKN2A and CDKN2B RNA abundance has been frequently reported, suggesting transcriptional co-regulation of these genes predominates in many tissues (8, 10, 14, 16, 44, 48–51).
ANRIL also acts in a PRC1/2 dependent mechanism to repress distant genes in trans (32, 46). Trans regulation by ANRIL may be dependent on Alu motifs, which are found both in ANRIL transcripts and in the promoters of ANRIL target genes (46). This mechanism was shown to regulate the CARD8 gene in endothelial cells (52). Polycomb group proteins, which are highly enriched near Alu motifs across the genome, are recruited to target gene promoters upon ANRIL over-expression. In support of this concept, silencing ANRIL impacts expression of a large number of genes across the genome (14). Separate from chromatin modification, ANRIL is reported to regulate Wnt signaling by binding to SOX2, increasing transcriptional activity of the WNT/β-catenin pathway (39).
miRNA Abundance and Activity
ANRIL also influences gene expression via miRNA networks. ANRIL regulates miRNAs both at the epigenetic level, through regulation of miRNA transcription, and through direct binding to miRNAs, acting as a miRNA “sponge.” In gastric cancer cells, ANRIL epigenetically silences miR-99a/miR-449a through a PRC2 mechanism (22). In general, expression of ANRIL and its target miRNAs are negatively correlated in tissues and cell lines (22, 53–57). ANRIL has been described as having pro-oncogenic effects by sponging miRNAs (see below for more details). On the other hand, circANRIL containing exons 5-6-7 was found to lack miRNA sponge activity (6). Inhibition of miRNAs can reverse the effects of ANRIL knockdown.
Cellular Outcomes of ANRIL Activity
ANRIL has broad impacts on cell biology, including influence over proliferation, senescence, apoptosis, extracellular matrix remodeling, and inflammation (14). In cancer, ANRIL-miRNA interactions regulate networks of downstream targets of miRNAs, promoting an oncogenic role for ANRIL in cell proliferation, metastasis, invasion, radio-resistance, drug-induced cytotoxicity and apoptosis, involving many different signaling pathways (22, 53–57). Specifically, repression of cell cycle inhibitors p14ARF, p15INK4B, and p16INK4A increases proliferation, decreases senescence, and contributes to the DNA damage response (13, 21, 33). PRC-mediated epigenetic repression of Kruppel-like factor 2 (KLF2) influences proliferation and apoptosis (40, 58). Cooperation between ANRIL and PRC-associated YY1 increases TNF-alpha dependent inflammatory mediators (IL-6, IL-8) through NF-kB (26). ANRIL influences the cellular response to oxidative stress through a miR-125a regulation of MCL-1 (59). Circular ANRIL species were found to regulate ribosome biogenesis in vascular smooth muscle cells (6).
The ANRIL Gene Is Associated With Human Disease
A primary driver of interest in ANRIL is the large body of genomic data linking the ANRIL gene with risk of human disease. Genome-wide association studies (GWAS) have identified many disease-associated SNPs in or near the ANRIL gene (60). The CDKN2A/B locus is remarkable for the large number of associated diseases, ranging from aging and frailty to cancer to metabolic disease. Perhaps surprisingly given the validated importance of the products of the CDKN2A and CDKN2B genes in cell biology, in some cases ANRIL expression shows stronger phenotype association than protein-coding CDKN2A/B locus genes (4, 48), linking ANRIL itself to a range of important human diseases.
ANRIL SNPs and Disease Risk
Studies indicate that SNPs in the ANRIL gene can impact ANRIL expression and function. The CDKN2A/B locus is associated with risk of cancer, atherosclerotic disease, type 2 diabetes, stroke, aneurysm, periodontitis, Alzheimer's disease, aging, frailty, glaucoma, endometriosis, multiple sclerosis, hypertension (10, 61). Reviewed here are only SNPs within or downstream of the ANRIL gene; broader CDKN2A/B locus disease associations have been reviewed previously (10, 62). Integrating information from published observations and the NCBI linkage disequilibrium database (63), we find that disease-associated SNPs in the ANRIL gene that modulate locus gene expression fall into approximately six groups (Table 1) defined loosely by linkage block and reported effects. Exceptions outnumber the rules, however; for nearly all groups there are reports of SNPs with different or even opposite effects. Summarized here is a generalized synopsis of the majority of reports. Group A SNPs, while located in ANRIL introns, generally impact CDKN2A/B but not ANRIL biology (48, 50, 64). All other SNP groups have reported impacts on ANRIL itself, but reports often describe conflicting direction of change. Some SNPs are reported to fall in enhancer regions (43, 49, 64) or to impact ANRIL splicing (8) or secondary structure (69, 74). The data are incomplete. A particular weakness of the field is that although tissue-specific effects are likely to determine how polymorphisms impact disease risk, in many cases the relevant primary tissue has not been tested.
Disease-Associated SNPs May Influence ANRIL Abundance
There is no consistent global pattern with respect to SNP impact on ANRIL abundance. For most SNP groups, risk-SNPs are reported that both increase and decrease ANRIL levels in different studies. Variability may be related to differences in technique used to detect ANRIL that favor one isoform over others, cell type studied, acute and chronic biology and genetic origin of the cellular material studied, and of course the individual biology of each polymorphism. Most ANRIL SNPs fall in large linkage blocks, which are variable among different human genetic groups; in many cases the SNP tested may not be the causative SNP in the linkage block, and published linkage blocks may not apply to the material tested if not carefully matched by origin. It is entirely possible that all conflicting results are correct; for example, a CAD risk-SNP could increase pro-proliferative ANRIL isoforms in endothelial, macrophage or vascular smooth muscle cells to drive atherosclerosis, whereas a diabetes risk-SNP at the same position could decrease proliferative ANRIL isoforms in beta cells to limit beta cell mass. The complexity of the human system necessitates testing the relevant ANRIL isoforms in the relevant cell type, preferably in primary cells, in tissue- and disease-specific manner.
A comprehensive review of all SNP effects is beyond the scope of this review. Some ANRIL located disease-associated SNPs impact both ANRIL expression and CDKN2A/CDKN2B expression (14, 48); others impact ANRIL but not CDKN2A or CDKN2B (14, 48, 60), and still others impact CDKN2A/CDKN2B but not ANRIL (48, 50, 64). Some SNPs are located within predicted or proven enhancer regions (10, 43, 48, 49, 64, 75) or miRNA binding sites (65, 66), providing possible mechanisms of cell type specific gene regulation.
Disease-Associated SNPs May Influence ANRIL Structure or Function
Beyond regulation of ANRIL transcription, polymorphisms could impact ANRIL function by influencing relative abundance of different isoforms through RNA splicing or stability, or through altering the secondary structure or interactions of any given isoform. Several studies have identified ANRIL isoform-specific effects (50, 53, 67, 71, 72); for example, four SNPs forming an atherosclerosis risk haplotype were associated with increased expression of some, but not all, ANRIL isoforms (44). SNPs may influence the relative abundance of linear compared to circular isoforms (8). Several SNPs are reported to impact ANRIL free energy of folding, resulting in a predicted change in secondary structure, with implications for function and stability (48, 69, 74).
ANRIL in Cancer
ANRIL was initially identified in a kindred of familial melanoma-neural system tumor with a germ-line deletion of the entire CDKN2A/B locus (2). Although the CDKN2A/B locus is deleted or silenced in approximately 40% of human cancers, related to the tumor suppressive actions of CDKN2A and CDKN2B (76), ANRIL itself has pro-oncogenic properties. ANRIL is implicated in many malignancies, including cancers of the bladder (77), ovary (78, 79), lung (38, 58, 80–82), liver (40, 54, 83), stomach (22), breast (57, 84, 85), esophagus (86), nasopharyngeal cavity (39, 87, 88), thyroid (89), bone (90), cervix (91), colon (92), prostate (21, 56), glioma (55), and others (76). High tissue abundance of ANRIL in cancers is associated with aggressive clinicopathologic features such as high histological grade tumor size, advanced tumor-node-metastasis stage, and poor overall survival (22, 38, 40, 58, 78, 79, 83, 87, 89, 91–93). Certain SNPs within the ANRIL gene are associated with ANRIL and CDKN2A/B locus gene expression and clinical parameters (4, 48, 70, 94–96). ANRIL may be useful as a prognostic biomarker and a therapeutic target for clinical cancer management.
Molecular Mechanisms of ANRIL in Cancer
Accumulating evidence suggests that ANRIL participates in tumorigenesis by influencing cell proliferation, apoptosis and metastasis. Depletion or overexpression of ANRIL changes expression levels of many genes involved in proliferation, cellular adhesion and apoptosis (14, 32, 46). ANRIL overexpression promotes proliferation, migration, invasion, and epithelial-mesenchymal transformation but inhibits cell apoptosis; ANRIL loss-of-function represses tumor size and growth rate, cell proliferation, migration, invasion, metastasis, and enhances apoptosis and senescence (22, 38, 55, 56, 58, 77, 80, 81, 84, 89–92). Suppression of ANRIL is required for Ras-induced senescence (13, 21, 42). High ANRIL levels are associated with resistance to chemotherapy, and ANRIL knockdown may promote chemosensitivity (37, 79, 88, 97–99). On the other hand, ANRIL mediated anti-oncogenic effects of phospholipase D in lung cancers (82).
ANRIL may promote carcinogenesis through a number of mechanisms. Canonical ANRIL transcriptional mechanisms may play a role, such as by in cis suppression of the CDKN2A/CDKN2B tumor suppressor genes (80, 81, 100), or through PRC-mediated in trans gene regulation (40, 58, 80). ANRIL miRNA regulation has been implicated in cancers as well, including mechanisms involving let-7a and miR-125a in nasopharyngeal and oral carcinoma (56, 88, 101), miR-99a/miR-449a in gastric cancer (22), miR-122-5p in hepatocellular carcinoma (54), miR-186 in cervical cancer (91), and miR-199a in breast cancer (57), miR-34a in glioma (55), and miR-323 in pediatric medulloblastoma (102). Transcription factors affected by ANRIL in cancers include KLF2 (40, 58), SMAD (56, 86, 89) and β-catenin. ANRIL interacts with signal transduction pathways in cancers such as PI3K/AKT, p38 MAPK, TGF-β, ATM-E2F1, and MTOR (33, 55, 56, 86, 89, 99, 103). ANRIL can also drive cancer progression by increasing glucose uptake for glycolysis (87), through lymphangiogenesis via LYVE-1, VEFG-C, and VEGFR-3 (92), and through invasion and metastasis via MET and MMP3 (78). An intriguing but mostly unexplored phenomenon is breakpoint fusion transcripts including exons from ANRIL fused with exons from MTAP, a neighboring protein-coding gene, which were identified in 20% of screened melanoma cell lines (104).
ANRIL in Metabolic Disease
In addition to cancer, genome-wide association studies have repeatedly and confidently identified links between the genomic region containing ANRIL and risk of developing cardiometabolic disease, including type 2 diabetes and manifestations of atherosclerosis such as CAD and stroke (10, 62). This locus influences risk not only of classic type 2 (obesity-related) diabetes, but also with related syndromes such as gestational diabetes, transplant-associated diabetes, and cystic fibrosis related diabetes, but not risk of type 1 (autoimmune) diabetes (10). Although diabetes is a clinical risk factor for atherosclerosis, the genetic influence for these conditions at the ANRIL locus is mostly non-overlapping, with atherosclerosis SNPs located throughout the ANRIL gene, and T2D SNPs located distal to the last ANRIL exon (7). One exception is a SNP located in ANRIL exon 2, rs564398, which is associated with both T2D and CAD (105). Since CDKN2A/B locus genes are known for their roles in cell cycle regulation and cancer, and not metabolism, many questions remain as to how this locus impacts metabolic disease.
ANRIL and Atherosclerotic Disease
Since ANRIL locus SNPs influence risk of atherosclerosis, many studies have now tested whether ANRIL gene expression is related to atherosclerosis-associated diseases. In subjects with angiographically confirmed CAD in the Leipzig heart study, specific ANRIL isoforms were positively correlated with CAD risk SNP haplotype in PBMCs, whole blood, and atherosclerotic plaque tissue (44). In the Framingham heart study, ANRIL SNPs were associated with multiple CAD-related outcomes, and showed isoform-specific ANRIL correlation in leukocytes, with short isoforms predicted to contribute to CAD pathogenesis (50). CAD risk-SNPs may regulate the relative abundance of linear and circular ANRIL isoforms (8). Intriguingly, abundance of ANRIL in circulating plasma was positively correlated with in-stent restenosis (53), but in PBMCs harvested at the time of angioplasty/reperfusion, ANRIL levels were lower in subjects with myocardial infarction, but higher in subjects with older age, diabetes, hypertension. In this cohort, ANRIL levels in PBMCs improved model prediction of subsequent left ventricular dysfunction (106). ANRIL promoter methylation may mediate an epigenetic influence on future cardiac risk; higher CpG methylation at birth was associated with higher pulse wave velocity, a marker for increased arterial stiffness indicating greater cardiovascular risk, at 9 years of age (31).
Mechanisms by which ANRIL impacts atherosclerotic disease remain debated. In aortic smooth muscle cells, knockdown of ANRIL using siRNA targeting exon 1 or exon 19 revealed altered gene expression networks impacting cell proliferation, apoptosis, extracellular matrix, and inflammation (14). Atherogenic gene expression networks were regulated by ANRIL via the Alu mechanism, in which Alu motifs target ANRIL to particular gene locations, recruiting PRC complexes and altering gene methylation status (46). ANRIL may impact risk of ischemic stroke by regulating the Caspase recruitment domain 8 (CARD8) gene in endothelial cells (52). A known CAD-associated miRNA, miR-92a, may mediate some ANRIL effects; ANRIL targets GATA2, MAP1B, and ARG1 were found to require miR-92a, placing this miRNA downstream of ANRIL for some atherogenic effects (69). Finally, ANRIL is related to inflammation: ANRIL is increased by pro-inflammatory factors NF-κ B and TNF-α in endothelial cells, and ANRIL was found to bind directly to the YY1 transcription factor to mediate TNF-a induction of cytokines IL-6 and IL-8 (26).
ANRIL and Obesity, Bone Mass, and Estrogen Signaling
Although GWAS studies do not suggest a link between CDKN2A/B locus SNPs and obesity risk in adult populations, intriguingly, ANRIL may be a genomic site of environmental epigenetic influence on obesity. The ANRIL promoter contains CpG methylation sites that are differentially regulated across samples. In human tissues taken at birth, lower CpG methylation in infancy predicted higher fat mass at 6 years of age, as well as increased bone size, mineralization and density (29, 30). ANRIL promoter methylation was also negatively correlated with BMI in contemporaneous samples of peripheral blood from adolescents and in adipose tissue from adults (29). Methylation of these CpG sites increased tissue abundance of ANRIL RNA, in a mechanism that might include increased activity of an estrogen response element. Functional studies in a liposarcoma cell line showed that transcription factor binding to an adjacent ERE was enhanced by methylation, and estradiol increased ANRIL expression (29).
ANRIL and Type 2 Diabetes
Multiple SNPs in different linkage blocks at the CDKN2A/B locus are associated with T2D risk; evidence in human populations suggests these SNPs impact pancreatic islet mass or function (10). Despite the fact that the T2D risk SNPs are located in or near the ANRIL gene, the field has largely assumed the effect was mediated by the protein coding genes at the locus, due to extensive published work implicating p16INK4A in the regulation of beta cell mass (10). However, although studies have found no association between CDKN2A/B T2D SNPs and transcript level of p14ARF, p15INK4B, or p16INK4A in human islets (10, 107), an age-dependent positive association was identified between distal T2D risk-SNPs (group F in Table 1) and ANRIL expression (68). On the other hand, a T2D risk-SNP in ANRIL exon 2 (group C in Table 1) was associated with reduced ANRIL expression, again with no change in p14ARF, p15INK4B, or p16INK4A expression (14, 48); however, these studies were carried out in blood rather than islets. In human islets, this exon 2 SNP was shown to remove a CpG methylation site; risk allele was associated with reduced islet insulin content but no change in locus gene expression (108). Risk allele at this SNP was associated with impaired beta cell proliferation response to high glucose (68). In a study relevant to diabetic retinopathy, high glucose exposure increased ANRIL expression in human retinal epithelial cells (25). In ANRIL was found to increase expression of VEGF, a critical element of the neovascularization that is central to damage from retinopathy, via a mechanism involving PRC2 and miR200b (25, 109).
Summary
Studies suggest the ANRIL lncRNA influences risk of a number of diseases, including many types of cancer as well as metabolic disease. Current understanding of ANRIL biology indicates the primary function of this lncRNA is to regulate gene expression, both locally at CDKN2A/B as well as across the genome, via mechanisms including chromatin modulation, transcription factor binding, and miRNA regulation. Knowledge concerning ANRIL function in cancers is more solid and advanced than for metabolic tissues. Mechanisms by which SNPs influence ANRIL abundance remain uncertain and require more study; how DNA methylation regulates ANRIL in cancers also will benefit from more study. Much remains to be learned about the structural complexity of ANRIL; how the various identified linear and circular isoforms impact tissue biology to modulate disease risk is mostly unknown. There is an urgent need for deeper understanding of how ANRIL isoforms modulate cellular function in human organs and tissues, and to explore the differing roles of ANRIL in cancer and metabolic disease. Given the advent of RNA therapeutics, and the broad disease relevance of ANRIL, it is possible that these studies may lead to future disease prevention and treatment.
Author Contributions
YK, C-HH, and LA wrote and revised the manuscript.
Funding
This work was supported by NIH/NIDDK: R01DK114686 and NIH/NIDDK DK095140 (LA), the American Diabetes Association grant #1-18-IBS-233 (LA) in collaboration with the Order of the Amaranth, and the George F. and Sybil H. Fuller Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the Beta Cell Biology Group at the University of Massachusetts Medical School for helpful discussions.
Abbreviations
CAD, coronary artery disease; CAC, coronary artery calcium; CDK, cyclin dependent kinase; circANRIL, circular ANRIL; circRNA, circular RNA; GWAS, genome-wide association study; lncRNA, long noncoding RNA; miRNA, microRNA; MI, myocardial infarction; MTAP, S-methyl-5′-thioadenosine phosphorylase; PBMC, peripheral blood mononuclear cell; PBTL, peripheral blood T-lymphocytes; PRC, polycomb repressive complex; SNP, single nucleotide polymorphism; VSMC, vascular smooth muscle cells.
References
1. Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci. (2015) 16:3251–66. doi: 10.3390/ijms16023251
2. Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. (2007) 67:3963–9. doi: 10.1158/0008-5472.CAN-06-2004
3. Congrains A, Kamide K, Ohishi M, Rakugi H. ANRIL: molecular mechanisms and implications in human health. Int J Mol Sci (2013) 14:1278–92. doi: 10.3390/ijms14011278
4. Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J Off Publ Fed Am Soc Exp Biol. (2011) 25:444–8. doi: 10.1096/fj.10-172452
5. Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell (2014) 159:1665–80. doi: 10.1016/j.cell.2014.11.021
6. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. (2016) 7:12429. doi: 10.1038/ncomms12429
7. Sarkar D, Oghabian A, Bodiyabadu PK, Joseph WR, Leung EY, Finlay GJ, et al. Multiple isoforms of ANRIL in melanoma cells: structural complexity suggests variations in processing. Int J Mol Sci. (2017) 18:E1378. doi: 10.3390/ijms18071378
8. Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet (2010) 6:e1001233. doi: 10.1371/journal.pgen.1001233
9. Holdt LM, Sass K, Gäbel G, Bergert H, Thiery J, Teupser D. Expression of Chr9p21 genes CDKN2B (p15INK4b), CDKN2A (p16INK4a, p14ARF) and MTAP in human atherosclerotic plaque. Atherosclerosis (2011) 214:264–70. doi: 10.1016/j.atherosclerosis.2010.06.029
10. Kong Y, Sharma RB, Nwosu BU, Alonso LC. Islet biology, the CDKN2A/B locus and type 2 diabetes risk. Diabetologia (2016) 59:1579–93. doi: 10.1007/s00125-016-3967-7
11. Matheu A, Maraver A, Collado M, Garcia-Cao I, Cañamero M, Borras C, et al. Anti-aging activity of the Ink4/Arf locus. Aging Cell (2009) 8:152–61. doi: 10.1111/j.1474-9726.2009.00458.x
12. Cánepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, et al. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life (2007) 59:419–26. doi: 10.1080/15216540701488358
13. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 INK4B tumor suppressor gene. Oncogene (2011) 30:1956–62. doi: 10.1038/onc.2010.568
14. Congrains A, Kamide K, Katsuya T, Yasuda O, Oguro R, Yamamoto K, et al. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem Biophys Res Commun (2012) 419:612–6. doi: 10.1016/j.bbrc.2012.02.050
15. He S, Gu W, Li Y, Zhu H. ANRIL/CDKN2B-AS shows two-stage clade-specific evolution and becomes conserved after transposon insertions in simians. BMC Evol Biol (2013) 13:247. doi: 10.1186/1471-2148-13-247
16. Folkersen L, Kyriakou T, Goel A, Peden J, Mälarstig A, Paulsson-Berne G, et al. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PLoS ONE (2009) 4:e7677. doi: 10.1371/journal.pone.0007677
17. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE (2012) 7:e30733. doi: 10.1371/journal.pone.0030733
18. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (2013) 19:141–57. doi: 10.1261/rna.035667.112
19. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Dev Camb Engl. (2016) 143:1838–47. doi: 10.1242/dev.128074
20. Fiscon G, Paci P, Iannello G. MONSTER v1.1: a tool to extract and search for RNA non-branching structures. BMC Genomics (2015) 16:S1. doi: 10.1186/1471-2164-16-S6-S1
21. Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular Interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by Polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. (2010) 38:662–74. doi: 10.1016/j.molcel.2010.03.021
22. Zhang E, Kong R, Yin D, You L, Sun M, Han L, Xu T, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget (2014) 5:2276–92. doi: 10.18632/oncotarget.1902
23. Hoffmann MJ, Dehn J, Droop J, Niegisch G, Niedworok C, Szarvas T, Schulz WA. Truncated Isoforms of lncRNA ANRIL are overexpressed in bladder cancer, but do not contribute to repression of INK4 tumor suppressors. Non Coding RNA (2015) 1:266–84. doi: 10.3390/ncrna1030266
24. Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. (2015) 16:20. doi: 10.1186/s13059-015-0586-4
25. Thomas AA, Feng B, Chakrabarti S. ANRIL: a regulator of VEGF in diabetic retinopathy. Invest Ophthalmol Vis Sci. (2017) 58:470–80. doi: 10.1167/iovs.16-20569
26. Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X, et al. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. (2016) 13:98–108. doi: 10.1080/15476286.2015.1122164
27. Cao S, Moss W, O'Grady T, Concha M, Strong MJ, Wang X, et al. New noncoding lytic transcripts derived from the epstein-barr virus latency origin of replication, oriP, are hyperedited, bind the paraspeckle protein, NONO/p54nrb, and support viral lytic transcription. J Virol. (2015) 89:7120–32. doi: 10.1128/JVI.00608-15
28. Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. (2008) 36:W70–4. doi: 10.1093/nar/gkn188
29. Lillycrop K, Murray R, Cheong C, Teh AL, Clarke-Harris R, Barton S, et al. ANRIL promoter DNA methylation: a perinatal marker for later adiposity. EBioMedicine (2017) 19:60–72. doi: 10.1016/j.ebiom.2017.03.037
30. Curtis EM, Murray R, Titcombe P, Cook E, Clarke-Harris R, Costello P, et al. Perinatal DNA methylation at CDKN2A is associated with offspring bone mass: findings from the southampton Women's survey. J Bone Miner Res Off J Am Soc Bone Miner Res. (2017) 32:2030–40. doi: 10.1002/jbmr.3153
31. Murray R, Bryant J, Titcombe P, Barton SJ, Inskip H, Harvey NC, et al. DNA methylation at birth within the promoter of ANRIL predicts markers of cardiovascular risk at 9 years. Clin Epigenetics (2016) 8:90. doi: 10.1186/s13148-016-0259-5
32. Sato K, Nakagawa H, Tajima A, Yoshida K, Inoue I. ANRIL is implicated in the regulation of nucleus and potential transcriptional target of E2F1. Oncol Rep. (2010) 24:701–7. doi: 10.3892/or_00000910
33. Wan G, Mathur R, Hu X, Liu Y, Zhang X, Peng G, et al. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. (2013) 25:1086–95. doi: 10.1016/j.cellsig.2013.02.006
34. Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature (2011) 473:43–9. doi: 10.1038/nature09906
35. Rodriguez C, Borgel J, Court F, Cathala G, Forné T, Piette J. CTCF is a DNA methylation-sensitive positive regulator of the INK/ARF locus. Biochem Biophys Res Commun. (2010) 392:129–34. doi: 10.1016/j.bbrc.2009.12.159
36. Gan Y, Ma W, Wang X, Qiao J, Zhang B, Cui C, et al. Coordinated transcription of ANRIL and P16 genes is silenced by P16 DNA methylation. Chin J Cancer Res. (2018) 30:93–103. doi: 10.21147/j.issn.1000-9604.2018.01.10
37. Özgür E, Mert U, Isin M, Okutan M, Dalay N, Gezer U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin Exp Med. (2013) 13:119–26. doi: 10.1007/s10238-012-0181-x
38. Lu Y, Zhou X, Xu L, Rong C, Shen C, Bian W. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. OncoTargets Ther. (2016) 9:3077–84. doi: 10.2147/OTT.S102658
39. Wu JH, Tang JM, Li J, Li XW. Upregulation of SOX2-activated lncRNA ANRIL promotes nasopharyngeal carcinoma cell growth. Sci Rep. (2018) 8:3333. doi: 10.1038/s41598-018-21708-z
40. Huang M, Chen W, Qi F, Xia R, Sun M, Xu T, et al. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J Hematol Oncol. (2015) 8:50. doi: 10.1186/s13045-015-0146-0
41. Deng W, Wang J, Zhang J, Cai J, Bai Z, Zhang Z. TET2 regulates LncRNA-ANRIL expression and inhibits the growth of human gastric cancer cells. IUBMB Life (2016) 68:355–64. doi: 10.1002/iub.1490
42. Kotake Y, Naemura M, Kitagawa K, Niida H, Tsunoda T, Shirasawa S, et al. Oncogenic Ras influences the expression of multiple lncRNAs. Cytotechnology (2016) 68:1591–6. doi: 10.1007/s10616-014-9834-9
43. Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature (2011) 470:264–68. doi: 10.1038/nature09753
44. Holdt LM, Beutner F, Scholz M, Gielen S, Gäbel G, Bergert H, et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. (2010) 30:620–7. doi: 10.1161/ATVBAHA.109.196832
45. Sethuraman S, Gay LA, Jain V, Haecker I, Renne R. microRNA dependent and independent deregulation of long non-coding RNAs by an oncogenic herpesvirus. PLoS Pathog (2017) 13:e1006508. doi: 10.1371/journal.ppat.1006508
46. Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. (2013) 9:e1003588. doi: 10.1371/journal.pgen.1003588
47. Ren C, Smith SG, Yap K, Li S, Li J, Mezei M, et al. Structure-Guided Discovery of Selective Antagonists for the Chromodomain of Polycomb Repressive Protein CBX7. ACS Med Chem Lett. (2016) 7:601–5. doi: 10.1021/acsmedchemlett.6b00042
48. Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet (2010) 6:e1000899. doi: 10.1371/journal.pgen.1000899
49. Jarinova O, Stewart AFR, Roberts R, Wells G, Lau P, Naing T, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. (2009) 29:1671–7. doi: 10.1161/ATVBAHA.109.189522
50. Johnson AD, Hwang SJ, Voorman A, Morrison A, Peloso GM, Hsu YH, et al. Resequencing and clinical associations of the 9p21.3 region: a comprehensive investigation in the Framingham heart study. Circulation (2013) 127:799–810. doi: 10.1161/CIRCULATIONAHA.112.111559
51. Motterle A, Pu X, Wood H, Xiao Q, Gor S, Liang Ng F, et al. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. Hum Mol Genet. (2012) 21:4021–9. doi: 10.1093/hmg/dds224
52. Bai Y, Nie S, Jiang G, Zhou Y, Zhou M, Zhao Y, et al. Regulation of CARD8 expression by ANRIL and association of CARD8 single nucleotide polymorphism rs2043211 (p.C10X) with ischemic stroke. Stroke (2014) 45:383–8. doi: 10.1161/STROKEAHA.113.003393
53. Wang F, Su X, Liu C, Wu M, Li B. Prognostic value of plasma long noncoding RNA ANRIL for in-stent restenosis. Med Sci Monit Int Med J Exp Clin Res. (2017) 23:4733–9. doi: 10.12659/MSM.904352
54. Ma J, Li T, Han X, Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. (2018) 144:205–14. doi: 10.1007/s00432-017-2543-y
55. Dong X, Jin Z, Chen Y, Xu H, Ma C, Hong X, et al. Knockdown of long non-coding RNA ANRIL inhibits proliferation, migration, and invasion but promotes apoptosis of human glioma cells by upregulation of miR-34a. J Cell Biochem. (2018) 119:2708–18. doi: 10.1002/jcb.26437
56. Zhao B, Lu YL, Yang Y, Hu LB, Bai Y, Li RQ, et al. Overexpression of lncRNA ANRIL promoted the proliferation and migration of prostate cancer cells via regulating let-7a/TGF-β1/ Smad signaling pathway. Cancer Biomark Sect Dis Markers (2018) 21:613–20. doi: 10.3233/CBM-170683
57. Xu ST, Xu JH, Zheng ZR, Zhao QQ, Zeng XS, Cheng SX, et al. Long non-coding RNA ANRIL promotes carcinogenesis via sponging miR-199a in triple-negative breast cancer. Biomed Pharmacother Biomedecine Pharmacother. (2017) 96:14–21. doi: 10.1016/j.biopha.2017.09.107
58. Nie F, Sun M, Yang J, Xie M, Xu T, Xia R, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. (2015) 14:268–77. doi: 10.1158/1535-7163.MCT-14-0492
59. Li R, Yin F, Guo YY, Zhao KC, Ruan Q, Qi YM. Knockdown of ANRIL aggravates H2O2-induced injury in PC-12 cells by targeting microRNA-125a. Biomed Pharmacother Biomedecine Pharmacother. (2017) 92:952–61. doi: 10.1016/j.biopha.2017.05.122
60. Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. (2008) 17:806–14. doi: 10.1093/hmg/ddm352
61. Rezazadeh M, Gharesouran J, Moradi M, Noroozi R, Omrani MD, Taheri M, et al. Association study of ANRIL genetic variants and multiple sclerosis. J Mol Neurosci. (2018) 65:54–9 doi: 10.1007/s12031-018-1069-3
62. Hannou SA, Wouters K, Paumelle R, Staels B. Functional genomics of the CDKN2A/B locus in cardiovascular and metabolic disease: what have we learned from GWASs? Trends Endocrinol Metab. (2015) 26:176–84. doi: 10.1016/j.tem.2015.01.008
63. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. (2016) 44:D877–81. doi: 10.1093/nar/gkv1340
64. Hungate EA, Vora SR, Gamazon ER, Moriyama T, Best T, Hulur I, et al. A variant at 9p21.3 functionally implicates CDKN2B in paediatric B-cell precursor acute lymphoblastic leukaemia aetiology. Nat Commun. (2016) 7:10635. doi: 10.1038/ncomms10635
65. Gong J, Liu C, Liu W, Wu Y, Ma Z, Chen H, et al. An update of miRNASNP database for better SNP selection by GWAS data, miRNA expression and online tools. Database (2015) 2015:bav029. doi: 10.1093/database/bav029
66. Wang X, Li W, Ma L, Gao J, Liu J, Ping F, et al. Association study of the miRNA-binding site polymorphisms of CDKN2A/B genes with gestational diabetes mellitus susceptibility. Acta Diabetol. (2015) 52:951–8. doi: 10.1007/s00592-015-0768-2
67. Zhao W, Smith JA, Mao G, Fornage M, Peyser PA, Sun YV, et al. The cis and trans effects of the risk variants of coronary artery disease in the Chr9p21 region. BMC Med Genomics (2015) 8:21. doi: 10.1186/s12920-015-0094-0
68. Kong Y, Sharma RB, Ly S, Stamateris RE, Jesdale WM, Alonso LC. CDKN2A/B T2D genome-wide association study risk SNPs impact locus gene expression and proliferation in human islets. Diabetes (2018) 67:872–84. doi: 10.2337/db17-1055
69. Cheng M, An S, Li J. CDKN2B-AS may indirectly regulate coronary artery disease-associated genes via targeting miR-92a. Gene (2017) 629:101–7. doi: 10.1016/j.gene.2017.07.070
70. Poi MJ, Li J, Sborov DW, VanGundy Z, Cho YK, Lamprecht M, et al. Polymorphism in ANRIL is associated with relapse in patients with multiple myeloma after autologous stem cell transplant. Mol Carcinog. (2017) 56:1722–32. doi: 10.1002/mc.22626
71. Zhuang J, Peng W, Li H, Wang W, Wei Y, Li W, et al. Methylation of p15INK4b and expression of ANRIL on chromosome 9p21 are associated with coronary artery disease. PLoS ONE (2012) 7:e47193. doi: 10.1371/journal.pone.0047193
72. Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS ONE (2009) 4:e5027. doi: 10.1371/journal.pone.0005027
73. Zhang W, Chen Y, Liu P, Chen J, Song L, Tang Y, et al. Variants on chromosome 9p21.3 correlated with ANRIL expression contribute to stroke risk and recurrence in a large prospective stroke population. Stroke (2012) 43:14–21. doi: 10.1161/STROKEAHA.111.625442
74. Gong W, Peng J, Yin J, Li X, Zheng W, Xiao L, et al. Association between well-characterized lung cancer lncRNA polymorphisms and platinum-based chemotherapy toxicity in Chinese patients with lung cancer. Acta Pharmacol Sin. (2017) 38:581–90. doi: 10.1038/aps.2016.164
75. Nakaoka H, Gurumurthy A, Hayano T, Ahmadloo S, Omer WH, Yoshihara K, et al. Allelic Imbalance in Regulation of ANRIL through chromatin interaction at 9p21 endometriosis risk locus. PLOS Genet. (2016) 12:e1005893. doi: 10.1371/journal.pgen.1005893
76. Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. (2012) 3:219. doi: 10.3389/fgene.2012.00219
77. Zhu H, Li X, Song Y, Zhang P, Xiao Y, Xing Y. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem Biophys Res Commun. (2015) 467:223–8. doi: 10.1016/j.bbrc.2015.10.002
78. Qiu JJ, Lin YY, Ding JX, Feng WW, Jin HY, Hua KQ. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. (2015) 46:2497–505. doi: 10.3892/ijo.2015.2943
79. Hu X, Bao J, Wang Z, Zhang Z, Gu P, Tao F, et al. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumour Biol J Int Soc Oncodevelopmental Biol Med. (2016) 37:3497–504. doi: 10.1007/s13277-015-4023-9
80. Naemura M, Murasaki C, Inoue Y, Okamoto H, Kotake Y. Long noncoding RNA ANRIL regulates proliferation of non-small cell lung cancer and cervical cancer cells. Anticancer Res. (2015) 35:5377–82.
81. Qiu J, Wang Y, Liu Y, Zhang Y, Ding J, Hua K. The long non-coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget (2016) 7:32478–92. doi: 10.18632/oncotarget.8744
82. Kang YH, Kim D, Jin EJ. Down-regulation of phospholipase D stimulates death of lung cancer cells involving up-regulation of the Long ncRNA ANRIL. Anticancer Res. (2015) 35:2795–2803.
83. Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ, Ma WL. High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. (2015) 8:3076–82.
84. Liu M, Xing LQ, Liu YJ. A three-long noncoding RNA signature as a diagnostic biomarker for differentiating between triple-negative and non-triple-negative breast cancers. Medicine (2017) 96:e6222. doi: 10.1097/MD.0000000000006222
85. Meseure D, Vacher S, Alsibai KD, Nicolas A, Chemlali W, Caly M, et al. Expression of ANRIL-polycomb complexes-CDKN2A/B/ARF genes in breast tumors: identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol Cancer Res. (2016) 14:623–33. doi: 10.1158/1541-7786.MCR-15-0418
86. Chen D, Zhang Z, Mao C, Zhou Y, Yu L, Yin Y, et al. ANRIL inhibits p15(INK4b) through the TGFβ1 signaling pathway in human esophageal squamous cell carcinoma. Cell Immunol. (2014) 289:91–6. doi: 10.1016/j.cellimm.2014.03.015
87. Zou ZW, Ma C, Medoro L, Chen L, Wang B, Gupta R, et al. LncRNA ANRIL is up-regulated in nasopharyngeal carcinoma and promotes the cancer progression via increasing proliferation, reprograming cell glucose metabolism and inducing side-population stem-like cancer cells. Oncotarget (2016) 7:61741–54. doi: 10.18632/oncotarget.11437
88. Wang Y, Cheng N, Luo J. Downregulation of lncRNA ANRIL represses tumorigenicity and enhances cisplatin-induced cytotoxicity via regulating microRNA let-7a in nasopharyngeal carcinoma. J Biochem Mol Toxicol. (2017) 31:e21904 doi: 10.1002/jbt.21904
89. Zhao JJ, Hao S, Wang LL, Hu CY, Zhang S, Guo LJ, et al. Long non-coding RNA ANRIL promotes the invasion and metastasis of thyroid cancer cells through TGF-β/Smad signaling pathway. Oncotarget (2016) 7:57903–18. doi: 10.18632/oncotarget.11087
90. Cheng S, Huang T, Li P, Zhang W, Wang Z, Chen Y. Long non-coding RNA ANRIL promotes the proliferation, migration and invasion of human osteosarcoma cells. Exp Ther Med. (2017) 14:5121–5. doi: 10.3892/etm.2017.5123
91. Zhang JJ, Wang DD, Du CX, Wang Y. Long noncoding RNA ANRIL promotes cervical cancer development by acting as a sponge of miR-186. Oncol Res. (2017) 26:345–52. doi: 10.3727/096504017X14953948675449
92. Sun Z, Ou C, Ren W, Xie X, Li X, Li G. Downregulation of long non-coding RNA ANRIL suppresses lymphangiogenesis and lymphatic metastasis in colorectal cancer. Oncotarget (2016) 7:47536–55. doi: 10.18632/oncotarget.9868
93. Wang H, Liu Y, Zhong J, Wu C, Zhong Y, Yang G, et al. Long noncoding RNA ANRIL as a novel biomarker of lymph node metastasis and prognosis in human cancer: a meta-analysis. Oncotarget (2018) 9:14608–18. doi: 10.18632/oncotarget.21825
94. Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. (2009) 41:909–14. doi: 10.1038/ng.412
95. Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. (2009) 41:920–5. doi: 10.1038/ng.411
96. Gu F, Pfeiffer RM, Bhattacharjee S, Han SS, Taylor PR, Berndt S, et al. Common genetic variants in the 9p21 region and their associations with multiple tumours. Br J Cancer (2013) 108:1378–86. doi: 10.1038/bjc.2013.7
97. Xu R, Mao Y, Chen K, He W, Shi W, Han Y. The long noncoding RNA ANRIL acts as an oncogene and contributes to paclitaxel resistance of lung adenocarcinoma A549 cells. Oncotarget (2017) 8:39177–84. doi: 10.18632/oncotarget.16640
98. Lan WG, Xu DH, Xu C, Ding CL, Ning FL, Zhou YL, et al. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol Rep. (2016) 36:263–70. doi: 10.3892/or.2016.4771
99. Zhang D, Ding L, Li Y, Ren J, Shi G, Wang Y, et al. Midkine derived from cancer-associated fibroblasts promotes cisplatin-resistance via up-regulation of the expression of lncRNA ANRIL in tumour cells. Sci Rep. (2017) 7:16231. doi: 10.1038/s41598-017-13431-y
100. Xu S, Wang H, Pan H, Shi Y, Li T, Ge S, et al. ANRIL lncRNA triggers efficient therapeutic efficacy by reprogramming the aberrant INK4-hub in melanoma. Cancer Lett. (2016) 381:41–8. doi: 10.1016/j.canlet.2016.07.024
101. Chai L, Yuan Y, Chen C, Zhou J, Wu Y. The role of long non-coding RNA ANRIL in the carcinogenesis of oral cancer by targeting miR-125a. Biomed Pharmacother. (2018) 103:38–45. doi: 10.1016/j.biopha.2018.01.105
102. Zhang H, Wang X, Chen X. Potential role of long non-coding RNA ANRIL in pediatric medulloblastoma through promotion on proliferation and migration by targeting miR-323. J Cell Biochem. (2017) 118:4735–44. doi: 10.1002/jcb.26141
103. Yu G, Liu G, Yuan D, Dai J, Cui Y, Tang X. Long non-coding RNA ANRIL is associated with a poor prognosis of osteosarcoma and promotes tumorigenesis via PI3K/Akt pathway. J Bone Oncol. (2018) 11:51–5. doi: 10.1016/j.jbo.2018.02.002
104. Xie H, Rachakonda PS, Heidenreich B, Nagore E, Sucker A, Hemminki K, et al. Mapping of deletion breakpoints at the CDKN2A locus in melanoma: detection of MTAP-ANRIL fusion transcripts. Oncotarget (2016) 7:16490–504. doi: 10.18632/oncotarget.7503
105. Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science (2007) 316:1336–41. doi: 10.1126/science.1142364
106. Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res (2014) 115:668–77. doi: 10.1161/CIRCRESAHA.115.303836
107. Taneera J, Lang S, Sharma A, Fadista J, Zhou Y, Ahlqvist E, et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. (2012) 16:122–34. doi: 10.1016/j.cmet.2012.06.006
108. Dayeh TA, Olsson AH, Volkov P, Almgren P, Rönn T, Ling C. Identification of CpG-SNPs associated with type 2 diabetes and differential DNA methylation in human pancreatic islets. Diabetologia (2013) 56:1036–46. doi: 10.1007/s00125-012-2815-7
Keywords: ANRIL, CDKN2A, CDKN2B, long noncoding RNA, diabetes, pancreatic islet, cancer, metabolic disease
Citation: Kong Y, Hsieh C-H and Alonso LC (2018) ANRIL: A lncRNA at the CDKN2A/B Locus With Roles in Cancer and Metabolic Disease. Front. Endocrinol. 9:405. doi: 10.3389/fendo.2018.00405
Received: 07 May 2018; Accepted: 29 June 2018;
Published: 24 July 2018.
Edited by:
Che-Pei Kung, School of Medicine, Washington University in St. Louis, United StatesReviewed by:
Subrata Chakrabarti, University of Western Ontario, CanadaTania Lee Slatter, University of Otago, New Zealand
Copyright © 2018 Kong, Hsieh and Alonso. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura C. Alonso, TGF1cmEuQWxvbnNvQHVtYXNzbWVkLmVkdQ==
 Yahui Kong
Yahui Kong Chih-Heng Hsieh
Chih-Heng Hsieh Laura C. Alonso
Laura C. Alonso