- 1College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 2Fujian Collaborative Innovation Center for Exploitation and Utilization of Marine Biological Resources, Xiamen, China
Neuropeptides, in addition to their classical role in the nervous system, act on intraovarian factors to regulate reproductive functions in vertebrates. However, this function of neuropeptides has not been characterized in crustaceans. Short neuropeptide F (sNPF), a highly conserved invertebrate neuropeptide, has been reported to be involved in feeding, metabolism, and in differentiation processes including reproduction. Although sNPF and its receptor (sNPFR) have been detected in the ovary in different species, ovarian colocalization of sNPF/sNPFR has not been investigated. In this study, we identified Scylla paramamosain (mud crab) sNPF (Sp-sNPF) as an endogenous ligand for the S. paramamosain orphan G protein-coupled receptor NPY2R in mammalian cell line HEK293T. We designated this receptor as Sp-sNPFR. RNA in situ hybridization in pre-vitellogenic ovary and reverse transcription-PCR on isolated denuded oocytes and follicle layers showed that Sp-sNPF was exclusively localized to the follicle cells, whereas Sp-sNPFR was detected in both follicle cells and oocytes. We also found that Sp-sNPF partly suppressed spontaneous maturation of denuded oocytes and caused intracellular cAMP accumulation and Ca2+ mobilization. Moreover, injection of synthetic Sp-sNPF peptides inhibited the expression of vitellogenin and vitellogenin receptor genes in vivo. These combined results suggest for the first time that Sp-sNPF may have inhibitory functions in vitellogenesis and oocyte maturation possibly via the autocrine/paracrine pathway.
Introduction
Neuropeptides are widespread in the nervous system of animals (1). Neuropeptides are secreted from neural cells and act as neuromodulators or neurotransmitters in the nervous system (2). Furthermore, neuropeptides are secreted from neuroendocrine cells, transported through blood to peripheral organs, and function as neurohormones (3). They also play a pivotal role in regulating reproduction (4). One such example is that of gonadotropin-releasing hormone (GnRH), a key decapeptide hormone involved in the regulation of reproduction which is secreted by the hypothalamus and transported via the hypophysial portal system to stimulate the production of luteinizing hormone (LH) and follicle-stimulating hormone (FSH), which further act on the ovary (5, 6). In addition to its involvement in this classical hypothalamic-pituitary-gonadal (HPG) reproduction axis in vertebrates, GnRH also plays a role as a neuroendocrine factor by directly binding to its receptor (GnRHR) in the ovary (7, 8). Mostly, neuropeptides are released by the nervous system and regulate reproduction directly or indirectly (8, 9). However, neuropeptides can also act as intraovarian regulatory factors. Intraovarian GnRH/GnRHR system has been suggested to play an autocrine/paracrine role in gonadal steroidogenesis, cell proliferation, and apoptosis (8, 10–13). In addition to GnRH/GnRHR, two key upstream regulation neuropeptide systems, gonadotrophin-inhibitory hormone (GnIH)/GnIH receptor (GnIHR) and kisspeptin/kisspeptin receptor, in the vertebrate reproduction axis also exist in ovary (14–23). The regulation of intraovarian neuropeptides has been well studied in vertebrates (7, 8, 18, 24, 25). However, a detailed knowledge about the intraovarian neuropeptides acting on ovary in invertebrates has not been reported.
Short neuropeptide F (sNPF), a homolog of vertebrate neuropeptide Y (NPY), is a C-terminally conserved neuropeptide in invertebrates. sNPF belongs to FMRFa-like peptides (FLPs) with the RFamide conserved motif (26) and has been widely studied in insects, especially in Drosophila (27). The first sNPF isolated from the midgut of American cockroach, Periplaneta americana, was postulated to play a role in digestion (28). Studies have mainly focused on the role of sNPF in regulation of feeding and metabolism (29–34). However, a role for sNPF in reproduction has also been identified. For instance Led-NPF-1, sNPF peptide from Leptinotarsa decemlineata (Colorado potato beetle) and Scg-NPF, a Locusta migratoria sNPF peptide, were shown to accelerate egg development in L. migratoria (34–37). However, their regulatory role in signaling is not known. Drosophila sNPF and its receptor (sNPFR) have been considered as upstream regulators of insulin signaling pathway (30). In Drosophila, sNPF and sNPFR1 proteins were detected in the larval brain where sNPFR1 was localized to the brain insulin producing cells (IPCs) and sNPFnergic neurons were found adjacent to these IPCs (30, 38). sNPF peptides stimulate insulin-like peptides (ILPs) synthesis in IPCs and affect insulin signaling (30). In insects, insulin signaling pathway regulates developmental and differentiation functions such as reproduction (39, 40). Juvenile hormone (JH) and 20-hydroxyecdysone (20E) were found to act as the insect gonadotropic hormones and to be regulated by insulin signaling by a complex physiological regulatory network (39, 40). sNPF peptides can directly regulate the production of JH. In the silkworm, Bombyx mori, sNPF peptides localize to the corpora cardiaca and inhibit JH biosynthesis in the corpora allata (41, 42). Moreover, sNPF was speculated as a neurohormone directly functioning on the ovary of red imported fire ant, Solenopsis invicta Buren as the S. invicta sNPF receptor (Si-sNPFR) protein was detected in oocytes by immunolocalization (43). These findings suggest that sNPF may serve as neuro-regulator in reproduction. Whether or not sNPF directly stimulates ovary is not known.
Recently, with the rapid development of methodologies, such as next-generation sequencing and mass spectrometry, many sNPF peptides have been identified in various crustacean species including the mud crab (Scylla paramamosain) (44–50). However, the reproductive function of sNPF in crustaceans is not known. S. paramamosain sNPF (Sp-sNPF) transcript was identified to encode a 126 amino acids (aa) precursor peptide with a 25 aa signal peptide followed by three mature peptides, each 9–12 aa in length and each containing a XPXRLRFamide (X represent variable residues) conserved motif (44). Interestingly, in our previous study, we found that Sp-sNPF transcript was highly expressed in the ovary in addition to the nervous system (44). We hypothesized that sNPF may act as intraovarian regulatory factor similar to vertebrate neuropeptides. In this study, we investigated the potential intraovarian autocrine/paracrine role of Sp-sNPF by receptor identification, ligand-receptor localization, signal transduction, and by biological relevance.
Materials and Methods
Animals
Crabs were purchased in the eighth market in Xiamen, Fujian Province, China. According to histological characteristics (51) and gonadosomatic index (GSI) (52), ovarian development was classified into five stages: undeveloped stage (stage I, GSI = 0.57 ± 0.47), pre-vitellogenic stage (stage II, GSI = 2.19 ± 0.21), early vitellogenic stage (stage III, GSI = 3.68 ± 0.20), late vitellogenic stage (stage IV, GSI = 7.81 ± 0.94), and mature stage (stage V, GSI = 10.49 ± 0.49). Anesthesia on ice was administered before dissections. The study was approved by Xiamen University animal care committee.
Molecular Cloning and Plasmid Construction
Total RNA from the crab ovary was isolated with the Trizol Reagent (Invitrogen) according to the manufacturer's instructions. The cDNA was synthetized using the RevertAid First Strand cDNA Synthesis Kit (Thermo) as per manufacturer's instructions. To clone the entire coding region of Sp-sNPFR gene, primers (Table 1) were designed based on the sequence in the transcriptome databases of brain (SRA accession: SRP068003) and muscle (SRA accession: SRP111448) from S. paramamosain. PCR product was sequenced prior to inserting into the HindIII and BamHI sites of the pEGFP-N1 vector. The construct was sequenced to validate the sequence and orientation.
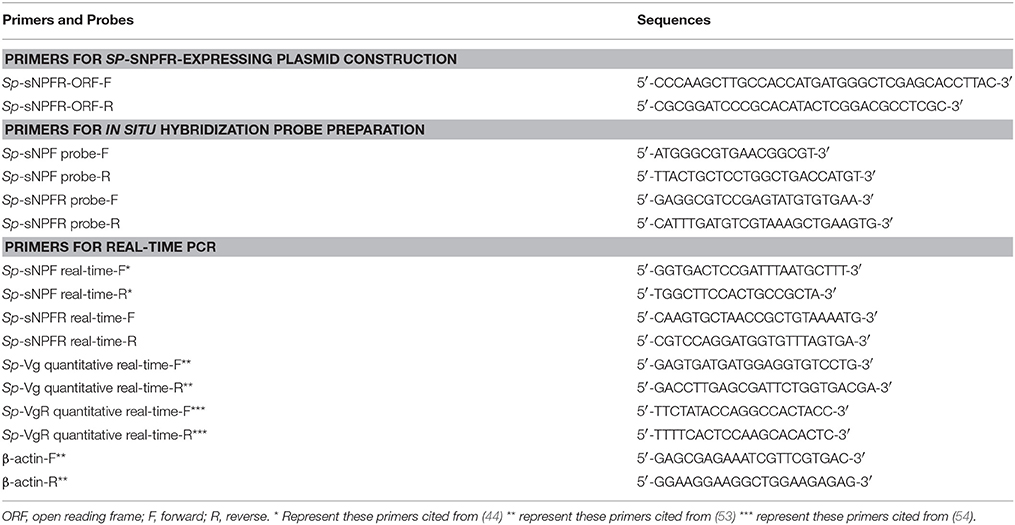
Table 1. Primers used for plasmid construction, real-time PCR and in situ hybridization probe preparation.
Cell Culture and Transient Transfection
Human embryonic kidney cell lines (HEK293T) were maintained in high glucose DMEM (Hyclone) supplemented with 10% fetal bovine serum (Gibco), 100 U/mL penicillin and 100 μg/mL streptomycin (Hyclone) at 37°C in a humidified incubator (Thermo) containing 5% CO2. Cells were seeded overnight in 6-well plates and transiently co-transfected with 1 μg of the Sp-sNPFR/pEGFP-N1 plasmid, 0.8 μg of the reporter gene pCRE-luc, and 0.4 μg of internal control gene pRL-TK using 5 μL Lipofectamine 2000 (Invitrogen) reagent following the manufacturer's protocols. Empty pEGFP-N1 plasmid was used for mock transfection.
Confocal Microscopy
For detection of sNPFR expression on the cell surface, HEK293T transiently transfected with Sp-sNPFR/pEGFP-N1 plasmid for 24 h were stained with the cell membrane probe DiI (Beyotime) at 37°C for 10 min and fixed with 4% paraformaldehyde (PFA) for 15 min. Cells were further incubated with a nuclear dye DAPI (Invitrogen) for 10 min. Stained cells were mounted in 50% glycerol and imaged using a two-photon laser confocal fluorescence microscopy (LSM 780 NLO, Zeiss).
Activation of Signaling
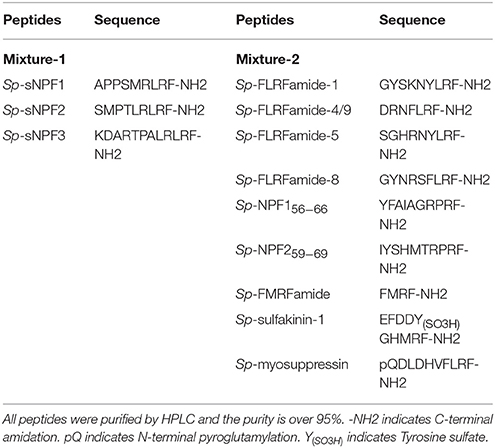
HEK293T transiently co-transfected with Sp-sNPFR/pEGFP-N1 and dual-luciferase reporter system vectors were grown to 90% confluency in a 24-well plate. Cells were first treated with synthetic Sp-sNPF peptide mixture and the mixture containing 9 synthetic FLPs (GL Biochem, Table 2) at 1 μM each. Further, various concentrations of individual peptides were used to record dose-dependent curves. After incubation for 6 h, ligand-induced changes in luciferase activity were measured by GloMax® 20/20 Luminometer (Promega) with Dual-Luciferase® Reporter Assay System Kit (Promega).
In situ Hybridization
Crab ovaries in a pre-vitellogenic stage were dissected, fixed in 4% PFA, and prepared for in situ hybridization as described (53). Primers, listed in Table 1, spanning 381 and 250 bp nucleotides of Sp-sNPF and Sp-sNPFR cDNA, respectively, were used for the preparation digoxigenin-labeled cRNA riboprobes using a DIG-RNA Labeling Kit (Roche). Hybridization was performed at 57°C for 16 h according to the manufacturer's instructions.
Isolation and Incubation of Crab Ovary
Ovaries (late vitellogenic stage) with fully grown (FG) oocytes were removed from female crabs and placed in 10 cm culture dish containing calcium-free crab saline (440 mM NaCl, 11.3 mM KCl, 26 mM MgCl2, 10 mM Hepes, 10 mM Glucose, pH 7.4 adjusted with NaOH) (55). The follicle layers were carefully separated with fine forceps under stereoscope (Leica). To determine the expression of Sp-sNPF and Sp-sNPFR gene, total RNA from intact ovary, denuded oocytes, and follicle layers were extracted with the Trizol Reagent (Invitrogen). cDNA was synthesized using TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix kit (TransGen Biotech) according to the manufacturer's protocols. The cDNA templates were used for PCR amplification using Sp-sNPF and Sp-sNPFR primers (Table 1). PCR products were analyzed by 1.5% agarose gel electrophoresis.
Intracellular Calcium Measurement
The denuded oocytes were washed thrice after they were separated from the ovaries with FG oocytes and were loaded with 1 μM Fluo-4 AM (Molecular Probes) for 40 min in 25°C. Oocytes were washed thrice, resuspended in calcium-free crab saline, and incubated for 30 min at 25°C. Calcium flux was measured in oocytes stimulated with Sp-sNPF peptides using excitation at 486 and emission at 526 nm in a fluorescence spectrometer (Infinite 200Pro, Tecan).
Effect of Sp-sNPFs on Oocyte Maturation
To obtain spontaneously matured oocytes, ovaries with the oocytes in germinal vesicle (GV) stage were placed in a glass dish with crab serum (the status of GV stage oocytes was maintained better in crab serum than that in crab saline, data not shown). After the follicle layers were removed, the denuded oocytes were isolated from ovary and incubated in crab serum containing 5 μM Sp-sNPF peptides. The observation of oocyte germinal vesicle breakdown (GVBD) was carried out as described (56). The GVBD was counted in intact ovary as well as in the denuded oocytes following incubation with in Sp-sNPF peptides for 0, 5, 15, 30, and 60 min.
Measurement of the Accumulation of cAMP
The measurement of cAMP accumulation and the calculation of GVBD rate were performed at the same time. Oocytes were lysed and the concentration of cAMP was determined using a competitive binding technique based on enzyme-linked immunosorbent assay (ELISA) (cAMP EIA kit, Cayman) according to the manufacturer's protocols.
In vivo Effect of Sp-sNPF Peptides on Sp-Vg and Sp-VgR Expression
Female crabs in pre-vitellogenic stage (weighing 45–56 g) were reared in the laboratory and were fed a shrimp diet. The initial control crabs were sacrificed on first day of the injection assay. Ten microliter of 1 mM Sp-sNPF peptides (final concentration in hemolymph about 5 μM) were injected into the arthrodial membrane at the base of last walking leg every 2 days while concurrent control crabs received the same volume of carrier. After 10 days, crabs were dissected and RNA extracted. cDNA was synthesized from hepatopancreas and ovaries and used in quantitative real-time PCR (qRT-PCR) to detect the expression level of vitellogenin (Sp-Vg) and vitellogenin receptor (Sp-VgR). The qRT-PCR was performed in the 7500 Fast Real-Time PCR system (Applied Biosystems) with SYBR Premix Ex Taq (TaKaRa) as following protocol: 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s.
Data Analysis
All values are expressed as mean ± SD. Multiple sequence alignment of sNPFR sequences was performed with ClustalX and embellished by LaTEX TexShade (57). Data of ligand-receptor assays were analyzed using nonlinear curve fitting (GraphPad Prism 6.0) to obtain activation curves and EC50 values. The mRNA levels of Sp-Vg and Sp-VgR gene were normalized against the internal control β-actin, and the relative expression was calculated using the 2−ΔΔCt method. Statistical significance was determined using one-way ANOVA followed by Duncan's multiple range tests. All statistical analyses were performed using IBM SPSS Statistics 20.0. Pictures were drawn by GraphPad Prism 6.0 and modified with Adobe Photoshop CS5.
Results
Cloning and Expression of Sp-sNPFR in HEK239T Cells
Two partial sequences annotated as neuropeptide Y receptor type 2 (NPY2R) were mined from two transcriptome databases of S. paramamosain. The predicted Sp-sNPFR gene sequence (GenBank accession number: MH382826) encoding 458 amino acids was obtained when the two partial sequences were assembled. The protein sequence showed 60.1% identity to the mosquito, Aedes aegypti sNPF receptor, 61.1% identity to the African malaria mosquito, Anopheles gambiae sNPF receptor, 58.7% identity to B. mori sNPF receptor, 56.4% identity to S. invicta sNPF receptor, 61.9% identity to the tsetse fly, Glossina morsitans morsitans sNPF receptor, 52.4% identity to the fruit fly, Drosophila melanogaster sNPF receptor, and 54.4% identity to the Desert Locust, Schistocerca gregaria sNPF receptor (Figure 1). Enhanced green fluorescent protein (EGFP) fused to the C-terminus of Sp-sNPFR was transiently expressed in HEK293T. Confocal microscopy showed that Sp-sNPFR-EGFP mainly localized to the cell membrane as demonstrated (Figure 2).

Figure 1. Comparative sequence alignment of sNPF receptors in different animals. Protein sequence alignment of sNPF receptors for S. paramamosain with Aedes aegypti, Anopheles gambiae, Bombyx mori, Solenopsis invicta, Glossina morsitans morsitans, Drosophila melanogaster and Schistocerca gregaria. The transmembrane helices are indicated by TM. Sequence logo is shown above alignments.
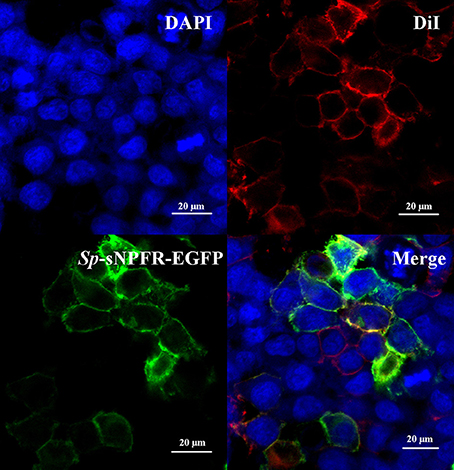
Figure 2. Expression of Sp-sNPFR in HEK293T. HEK293T were transiently co-transfected with Sp-sNPFR/pEGFP-N1, pCRE-luc and pRL-TK. Cells expressing Sp-sNPFR-EGFP fusion protein were stained with a nuclei probe (DAPI) and a membrane plasma probe (DiI).
Activation of Sp-sNPFR by Sp-sNPFs
We found that in cells transfected with Sp-sNPFR and dual-luciferase reporter, Sp-sNPFR was exclusively activated by Sp-sNPF peptides mixture, while all other FLPs with the RFamide C-terminus did not activate Sp-sNPFR at 1 μM concentration (Figure 3A). Furthermore, Sp-sNPFR was found to be activated by all the three Sp-sNPF peptides in a dose-dependent manner. The EC50 values were found to be 1.2, 3.8, and 8.5 μM for Sp-sNPF1, Sp-sNPF2, and Sp-sNPF3, respectively (Figure 3B).
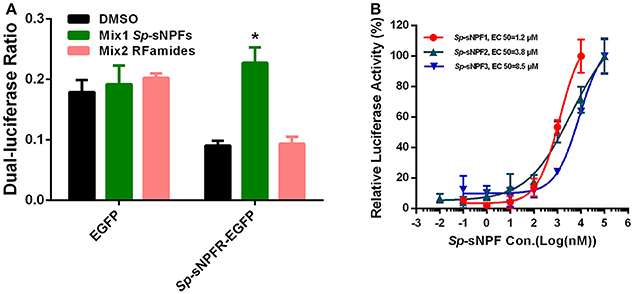
Figure 3. Ligand-receptor activation of signaling. (A) Relative luciferase activity of Sp-sNPFR-EGFP expressing cells after exposure to two peptide mixutures with a fixed concentration of 1 μM each peptide (*P < 0.05). (B) HEK293T transiently co-transfected with Sp-sNPFR/pEGFP-N1, pCRE-luc, and pRL-TK were treated with different doses of Sp-sNPF1, Sp-sNPF2, and Sp-sNPF3 and the concentration of cAMP was determined. Values are plotted as mean ± SD from three biological replicates.
Localization of Sp-sNPF and Sp-sNPFR mRNA in the Ovary
In the mud crab, ovary consists of oocytes and follicle layers surrounding the oocytes (Figure 4). Histological study showed that oocytes were larger in size than the follicle cells at pre-vitellogenic stage (Figures 5A,B). In situ hybridization showed that the positive signals of Sp-sNPF mRNA were located on the follicle cells (Figure 5C) and Sp-sNPFR was detected in both oocytes and follicle layer cells (Figure 5E). No signal was detected when the sense riboprobes of Sp-sNPF (Figure 5D) and Sp-sNPFR (Figure 5F) were used. Results of RT-PCR matched the findings of in situ hybridization (Figure 6).
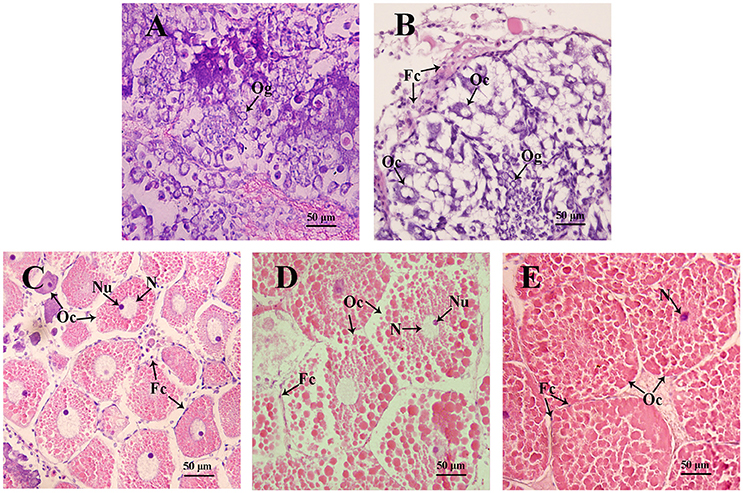
Figure 4. Sections of ovaries from various stages of ovarian development in S. paramamosain. (A) undeveloped stage; (B) pre-vitellogenic stage; (C) early vitellogenic stage; (D) late vitellogenic stage; (E) mature stage. Fc, follicle cell; Oc, oocyte; Og, oogonium; N, nuclear; Nu, nucleolus. The arrows indicate different types of cells from various stages of ovaries.
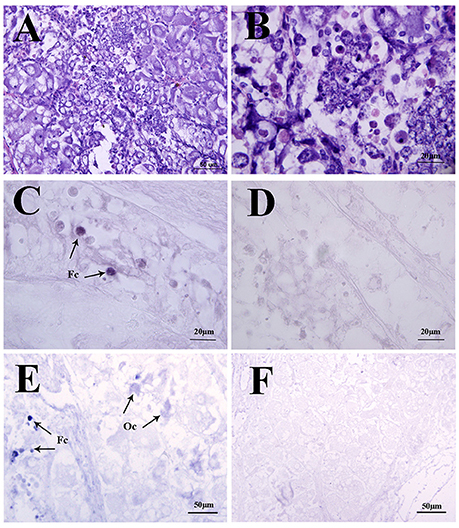
Figure 5. Localization of Sp-sNPF and Sp-sNPFR mRNA by in situ hybridization in the mud crab ovaries at the pre-vitellogenic stage. (A, B) Hematoxylin and eosin (HE) staining in the ovaries. Arrows indicate the specific Sp-sNPF (C) and Sp-sNPFR (E) mRNA signals with the antisense riboprobes in ovaries. Sense riboprobes of Sp-sNPF (D) and Sp-sNPFR (F) were used as the negative control. Fc, follicle cell; Oc, oocyte.
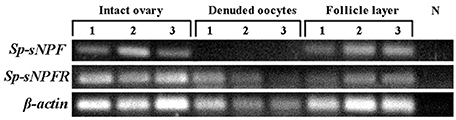
Figure 6. Intraovarian distribution of Sp-sNPF and Sp-sNPFR mRNA in a full-grown ovary. RT-PCR detection of Sp-sNPF and Sp-sNPFR gene expression in different compartments of full-grown ovaries (follicle layer and denuded oocyte). Sp-sNPF was exclusively detected in follicle layer, whereas Sp-sNPFR was expressed in both denuded oocytes and follicle layer. β-actin as the housekeeping gene.
Oocytes Were Directly Activated by Sp-sNPFs
Based on our finding that Sp-sNPFR mRNA was detected in denuded oocytes (Figure 6), we employed Ca2+ mobilization technology to investigate whether oocytes were activated by Sp-sNPFs. As shown in Figure 7, oocytes exhibited a dose-dependent response in the intracellular Ca2+ mobilization when challenged with different concentrations of Sp-sNPF peptides.
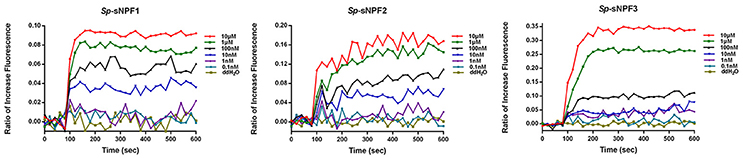
Figure 7. Intracellular Ca2+ mobilization of oocytes was induced by Sp-sNPF peptides. Sp-sNPF peptides evoked intracellular Ca2+ flux in a ligand concentration-dependent manner in oocytes. All events were performed in at least three independent experiments.
Effect of Sp-sNPFs on Oocyte Maturation
To explore the potential role of Sp-sNPF in oocyte maturation, denuded oocytes (GV stage) which were separated from the ovaries were chosen. As indicated in Figure 8A, denuded oocytes underwent GVBD fast and over 90% oocytes exhibited GVBD after 15 min in the control group. However, the time of GVBD ratio was delayed to 30 min in oocytes treated with 5 μM Sp-sNPF peptides (Figure 8A). Moreover, the GVBD ratio of oocytes was decreased in oocytes incubated with Sp-sNPF1, Sp-sNPF2, and Sp-sNPF3 for 5 min by 18, 33, and 21%, respectively. GVBD ratio of oocytes was decreased by 14, 12, and 14%, respectively, when Sp-sNPF1, Sp-sNPF2, and Sp-sNPF3 were used for incubation at 15 min (Figure 8A). We also observed a higher level of cAMP in oocytes incubated with 5 μM Sp-sNPF peptides (Figure 8B). This was in contrast to control oocytes where the cAMP level was reduced during the GVBD process (Figure 8B).
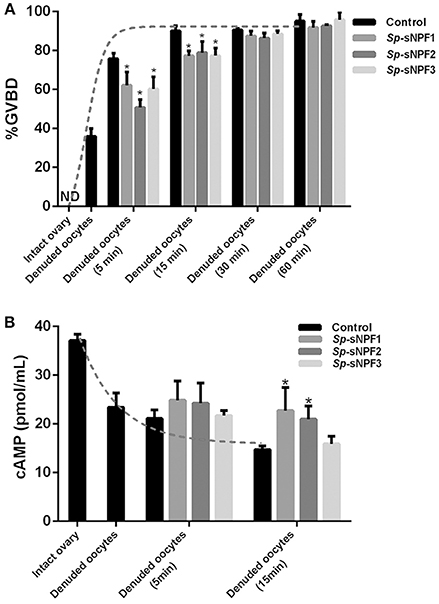
Figure 8. Inverse effects of Sp-sNPF peptides on maturation of denuded oocytes. (A) Sp-sNPF peptides suppressed GVBD rate in denuded oocytes. Denuded oocytes were incubated in the presence or absence of Sp-sNPF peptides (5 μM) for 0, 5, 15, 30, and 60 min before scoring GVBD oocytes. GVBD was inhibited by Sp-sNPF peptides at 5 and 15 min of incubation. The dotted line represents the GVBD rate trend of denuded oocytes which were isolated from an intact ovary in the control group. (B) cAMP accumulation was measured with ELISA. The dotted line represents the cAMP accumulation trend of denuded oocytes, which were isolated from intact ovary, in the control group. All values are presented as mean ± SD (n = 3) from three independent experiments (*P < 0.05).
Effect of Sp-sNPFs on Expression of Sp-Vg and Sp-VgR In Vivo
Our previous study demonstrated that the expression of Sp-sNPF decreased continuously during vitellogenesis (44). In this study, we investigated the potential role of sNPF in vitellogenesis by analyzing the effect of injected synthetic Sp-sNPF peptides on the expression of Sp-Vg and Sp-VgR in hepatopancreas and ovaries derived from pre-vitellogenic females. We found that injection of Sp-sNPF1, Sp-sNPF2, and Sp-sNPF3 significantly inhibited Sp-Vg expression in hepatopancreas by 78, 59, and 83%, respectively. The expression level of Sp-VgR in ovaries was decreased by 74%, 56%, and 68%, in the presence of Sp-sNPF1, Sp-sNPF2, and Sp-sNPF3 respectively (Figure 9).
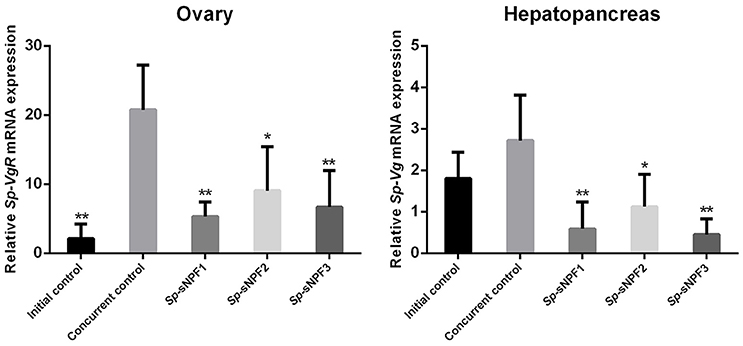
Figure 9. Inhibitory effects of Sp-sNPF peptides on the expression of Sp-Vg and Sp-VgR in vivo. Hepatopancreas and ovaries were sampled at 10 days post-injection of Sp-sNPF peptides for detecting the expression levels of Sp-Vg and Sp-VgR. The relative expression levels were normalized to β-actin. All data are presented as mean ± SD (n = 4) from a representative experiment. *P < 0.05, **P < 0.01 vs. concurrent control.
Discussion
sNPFs have been identified from a broad range of arthropods (27). Since the discovery of the first sNPFR, a GPCR with structural similarity to NPY2R, in D. melanogaster (58), some sNPFRs have been identified in insects (33, 41, 59–68). Although a large number of sNPFs have been characterized in different crustacean species (44–50), no sNPFR has been identified in crustaceans. In the present study, analyses of sequence similarity in transcriptome database revealed a candidate sNPFR annotated as a NPY-like receptor (NPY2R) in S. paramamosain. In addition, the ligand-receptor activation of signaling demonstrated that the candidate sNPFR was activated by three synthetic S. paramamosain sNPF peptides, Sp-sNPF1, Sp-sNPF2, and Sp-sNPF3, in a dose-dependent manner, with EC50 of 1.2, 3.8, and 8.5 μM respectively and which resulted in cAMP accumulation. All other tested peptides with the RFamide C-terminus did not elicit a cAMP response. Therefore, we designated the candidate sNPFR as the S. paramamosain sNPF receptor (Sp-sNPFR). To our knowledge, this receptor is the first crustacean sNPF receptor that has been characterized in this study.
To date, several insect sNPF receptors have been cloned and characterized (33, 41, 59–68). Among these receptors, the sNPFRs from D. melanogaster (67, 68), A. gambiae (60), B. mori (41, 61), S. gregaria (33), A. aegypti (62), S. invicta (63), G. m. morsitans (64), and the oriental fruit fly, Bactrocera dorsalis (Hendel) (65) have been deorphanized by ligand-receptor activation of signaling in a heterologous cell expression system, with EC50 values in the nanomolar range. Compared to the high sensitivity of insect sNPFRs, Sp-sNPFR in our study had a relatively higher half-maximal effective concentration (EC50) in the HEK293T expression system. Likewise, the intracellular Ca2+ concentration of S. paramamosain oocytes were mobilized by relatively high concentrations of synthetic Sp-sNPF peptides in vitro. Nonetheless, our findings are similar to a study which showed that a short neuropeptide F-related receptor from the mollusk, Crassostrea gigas exhibited EC50 value in the micromolar range, similar to our results (69).
Studies in insect sNPFRs revealed that Si-sNPFR is localized in the ovary (43) suggesting that sNPF may directly act on ovary. As S. invicta sNPF was detected in the nervous system, authors speculated that sNPF may act as a neurohormone on the ovaries (43). In our previous study (44), we found a higher level of Sp-sNPF transcript in the ovary of S. paramamosain suggesting that Sp-sNPF may function as an autocrine/paracrine factor within the ovary.
Our results showed that Sp-sNPF mRNA was exclusively localized in the follicle cells, whereas Sp-sNPFR was present in both follicle cells and oocytes. These results implied that Sp-sNPF may bind Sp-sNPFR via an autocrine/paracrine way. The localization of Sp-sNPF and Sp-sNPFR is consistent with the intraovarian expression pattern of S. paramamosain bone morphogenetic protein (Sp-BMP) and its receptors (54). BMP belongs to the transforming growth factor β (TGF-β) superfamily, classical ovarian local autocrines/paracrines factors (70, 71). In vertebrates, BMPs and their receptors have a different localization, and are speculated to have an autocrine/paracrine regulatory mechanism in the ovary (72–75). The distribution of Sp-sNPF and Sp-sNPFR suggested that there may be a similar autocrine/paracrine regulatory mechanism mediated by Sp-sNPF in the ovary for follicle cells to signal the follicle cells/oocytes. Finally, the finding that the Sp-sNPF peptides elicited a dramatic increase in intracellular Ca2+ is consistent with its proposed role as an intraovarian regulatory factor through an autocrine/paracrine regulatory mechanism.
When FG oocytes were separated from the follicle cells, they underwent spontaneous maturation with almost all oocytes exhibiting GVBD in 15 min. Meanwhile, the cAMP level in the oocytes decreased significantly. In contrast, when Sp-sNPF peptides were added to the culture medium, GVBD rate of oocytes was significantly reduced, while the cAMP levels were partly reversed. This phenomenon is in agreement with the previous reports in zebrafish where removal of the follicle layers significantly increased in spontaneous maturation of the oocytes (76, 77). These findings suggest that surrounding follicle layer cells may play a similar role in both vertebrates and invertebrates in inhibiting oocyte maturation. The secretory factors that may be responsible for this inhibitory effect of follicle cells on oocyte maturation have not been fully explored. Estradiol (E2) and Pituitary Adenylate Cyclase Activating Polypeptide (PACAP) were identified as two such factors where E2 partly inhibited the spontaneous maturation of the denuded oocytes via GPER, a membrane receptor located in the oocytes (76), and PACAP had a similar inhibitory effect on oocyte maturation (77). Sp-sNPF may be one such factor that suppresses oocyte maturation by binding to Sp-sNPFR to promote cAMP accumulation. However, like E2 and PACAP, the inhibition of Sp-sNPF was incomplete suggesting that other factors could be involved in this regulatory mechanism.
To provide further clues to elucidate the importance of Sp-sNPF in S. paramamosain ovary, we performed in vivo experiments. In contrast to the previous reports in L. migratoria (35, 36), injection of Sp-sNPF peptides significant suppressed the expression of Sp-VgR in the pre-vitellogenic stage ovary in our study. VgR, located in the oocytes, has been regarded as a key receptor in the ovary which mediates the release of vitellogenin from hepatopancreas in crabs (78). Considering the correlation of Vg/VgR system, we also examined the expression level of Sp-Vg in hepatopancreas when the crabs were injected with Sp-sNPF peptides. In agreement with the expression pattern of Sp-VgR, Sp-sNPF peptides exhibited a similar inhibitory effect on Sp-Vg mRNA expression. The results suggest that Sp-sNPF may be an inhibitor of vitellogenesis.
In summary, this study shows the S. paramamosain orphan receptor NPY2R is a receptor for the neuropeptide Sp-sNPF (Sp-sNPFR). To our knowledge, this is the first sNPFR reported in crustaceans. Furthermore, the tissue and cellular localization of Sp-sNPF/Sp-sNPFR and the signal transduction are consistent with its proposed function as an intraovarian regulatory factor inhibiting vitellogenesis and oocyte maturation through an autocrine/paracrine regulatory mechanism. This research provides a new insight into the sNPF functions in reproduction.
Author Contributions
CB: Conceptualization, data curation, software, methodology, writing-original draft, project administration; YY: Conceptualization, data curation, methodology, writing-review, and editing; HH: Supervision; HY: Conceptualization, supervision, funding acquisition.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos. 41476119 and 31772827). We wish to thank Mr. Yuting Zhang for his technical support.
References
1. Grimmelikhuijzen C, Leviev IK, Carstensen K. Peptides in the nervous systems of cnidarians: structure, function, and biosynthesis. Int Rev Cytol. (1996) 167:37–89. doi: 10.1016/S0074-7696(08)61345-5
2. Boonen K, Landuyt B, Baggerman G, Husson SJ, Huybrechts J, Schoofs L. Peptidomics: the integrated approach of MS, hyphenated techniques and bioinformatics for neuropeptide analysis. J Sep Sci. (2008) 31:427–45. doi: 10.1002/jssc.200700450
3. Fricker LD. Neuropeptides and other bioactive peptides: from discovery to function. In: Devi L, Fricker LD, editors. Colloquium Series on Neuropeptides. California, CA: Morgan & Claypool Publishers (2012). p. 1–122.
4. Hartenstein V. The neuroendocrine system of invertebrates: a developmental and evolutionary perspective. J Endocrinol. (2006) 190:555–70. doi: 10.1677/joe.1.06964
5. Seeburg PH, Mason AJ, Stewart TA, Nikolics K. The mammalian GnRH gene and its pivotal role in reproduction. In: Proceedings of the 1986 Laurentian Hormone Conference. Quebéc (1987). p. 69–98.
6. Millar RP, Newton CL. Current and future applications of GnRH, kisspeptin and neurokinin B analogues. Nat Rev Endocrinol. (2013) 9:451–66. doi: 10.1038/nrendo.2013.120
7. Millar RP, Lu ZL, Pawson AJ, Flanagan CA, Morgan K, Maudsley SR. Gonadotropin-releasing hormone receptors. Endocr Rev. (2004) 25:235–75. doi: 10.1210/er.2003-0002
8. Metallinou C, Asimakopoulos B, Schröer A, Nikolettos N. Gonadotropin-releasing hormone in the ovary. Reprod Sci. (2007) 14:737–49. doi: 10.1177/1933719107310707
9. Cheung LW, Wong AS. Gonadotropin-releasing hormone: GnRH receptor signaling in extrapituitary tissues. FEBS J. (2008) 275:5479–95. doi: 10.1111/j.1742-4658.2008.06677.x
10. Shahed A, Young KA. Intraovarian expression of GnRH-1 and gonadotropin mRNA and protein levels in Siberian hamsters during the estrus cycle and photoperiod induced regression/recrudescence. Gen Comp Endocrinol. (2011) 170:356–64. doi: 10.1016/j.ygcen.2010.10.008
11. Hong IS, Cheung AP, Leung PC. Gonadotropin-releasing hormones I and II induce apoptosis in human granulosa cells. J Clin Endocrinol Metab. (2008) 93:3179–85. doi: 10.1210/jc.2008-0127
12. Leung PC, Cheng CK, Zhu XM. Multi-factorial role of GnRH-I and GnRH-II in the human ovary. Mol Cell Endocrinol. (2003) 202:145–53. doi: 10.1016/S0303-7207(03)00076-5
13. Ramakrishnappa N, Rajamahendran R, Lin YM, Leung PCK. GnRH in non-hypothalamic reproductive tissues. Anim Reprod Sci. (2005) 88:95–113. doi: 10.1016/j.anireprosci.2005.05.009
14. Bentley GE, Ubuka T, McGuire NL, Chowdhury VS, Morita Y, Yano T, et al. Gonadotropin-inhibitory hormone and its receptor in the avian reproductive system. Gen Comp Endocrinol. (2008) 156:34–43. doi: 10.1016/j.ygcen.2007.10.003
15. Maddineni SR, Ocon-Grove OM, Krzysik-Walker SM, Hendricks GL, Ramachandran R. Gonadotropin-inhibitory hormone (GnIH) receptor gene is expressed in the chicken ovary: potential role of GnIH in follicular maturation. Reproduction (2008) 135:267–74. doi: 10.1530/REP-07-0369
16. Li X, Su J, Lei Z, Zhao Y, Jin M, Fang R, et al. Gonadotropin-inhibitory hormone (GnIH) and its receptor in the female pig: cDNA cloning, expression in tissues and expression pattern in the reproductive axis during the estrous cycle. Peptides (2012) 36:176–85. doi: 10.1016/j.peptides.2012.05.008
17. Oishi H, Klausen C, Bentley GE, Osugi T, Tsutsui K, Gilks CB, et al. The human gonadotropin-inhibitory hormone ortholog RFamide-related peptide-3 suppresses gonadotropin-induced progesterone production in human granulosa cells. Endocrinology (2012) 153:3435–45. doi: 10.1210/en.2012-1066
18. Ubuka T, Son YL, Bentley GE, Millar RP, Tsutsui K. Gonadotropin-inhibitory hormone (GnIH), GnIH receptor and cell signaling. Gen Comp Endocrinol. (2013) 190:10–7. doi: 10.1016/j.ygcen.2013.02.030
19. Gaytan F, Garcia-Galiano D, Dorfman MD, Manfredi-Lozano M, Castellano JM, Dissen GA, et al. Kisspeptin receptor haplo-insufficiency causes premature ovarian failure despite preserved gonadotropin secretion. Endocrinology (2014) 155:3088–97. doi: 10.1210/en.2014-1110
20. Shahed A, Young KA. Differential ovarian expression of KiSS-1 and GPR-54 during the estrous cycle and photoperiod induced recrudescence in Siberian hamsters (Phodopus sungorus). Mol Reprod Dev. (2009) 76:444–52. doi: 10.1002/mrd.20972
21. Castellano JM, Gaytan M, Roa J, Vigo E, Navarro VM, Bellido C, et al. Expression of KiSS-1 in rat ovary: putative local regulator of ovulation? Endocrinology (2006) 147:4852–62. doi: 10.1210/en.2006-0117
22. Gaytán F, Gaytán M, Castellano JM, Romero M, Roa J, Aparicio B, et al. KiSS-1 in the mammalian ovary: distribution of kisspeptin in human and marmoset and alterations in KiSS-1 mRNA levels in a rat model of ovulatory dysfunction. Am J Physiol Endocrinol Metab. (2009) 296:E520–31. doi: 10.1152/ajpendo.90895.2008
23. Cielesh ME, McGrath BM, Scott CJ, Norman ST, Stephen CP. The localization of kisspeptin and kisspeptin receptor in the canine ovary during different stages of the reproductive cycle. Reprod Domest Anim. (2016) 52:24–8. doi: 10.1111/rda.12841
24. Törnell J, Carlsson B, Hillensjö T. Vasoactive intestinal peptide stimulates oocyte maturation, steroidogenesis, and cyclic adenosine 3',5'-monophosphate production in isolated preovulatory rat follicles. Biol Reprod. (1988) 39:213–20. doi: 10.1095/biolreprod39.2.213
25. Canipari R, Di Paolo V, Barberi M, Cecconi S. PACAP in the reproductive system. In: Reglodi D, Tamas A, editors. Pituitary Adenylate Cyclase Activating Polypeptide-PACAP. Cham: Springer International Publishing (2016). p. 405–20.
26. Walker RJ, Papaioannou S, Holden-Dye L. A review of FMRFamide- and RFamide-like peptides in Metazoa. Invert Neurosci. (2009) 9:111–53. doi: 10.1007/s10158-010-0097-7
27. Dick RN, Christian W. A comparative review of short and long neuropeptide F signaling in invertebrates: any similarities to vertebrate neuropeptide Y signaling. Peptides (2011) 32:1335. doi: 10.1016/j.peptides.2011.03.013
28. Veenstra JA, Lambrou G. Isolation of a novel RFamide peptide from the midgut of the American cockroach, Periplaneta americana. Biochem Biophys Res Commun. (1995) 213:519–24. doi: 10.1006/bbrc.1995.2162
29. Lee KS, You KH, Choo JK, Han YM, Yu K. Drosophila short neuropeptide F regulates food intake and body size. J Biol Chem. (2004) 279:50781–9. doi: 10.1074/jbc.M407842200
30. Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, et al. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol. (2008) 10:468–75. doi: 10.1038/ncb1710
31. Lee KS, Hong SH, Kim AK, Ju SK, Kwon OY, Yu K. Processed short neuropeptide F peptides regulate growth through the ERK insulin pathway in Drosophila melanogaster. FEBS Lett. (2009) 583:2573–7. doi: 10.1016/j.febslet.2009.07.024
32. Dillen S, Verdonck R, Zels S, van Wielendaele P, vanden Broeck J. Identification of the short neuropeptide F precursor in the desert locust: evidence for an inhibitory role of sNPF in the control of feeding. Peptides (2014) 53:134–9. doi: 10.1016/j.peptides.2013.09.018
33. Dillen S, Zels S, Verlinden H, Spit J, van Wielendaele P, VandenBroeck J. Functional characterization of the short neuropeptide F receptor in the desert locust, Schistocerca gregaria. PLoS ONE (2013) 8:e53604. doi: 10.1371/journal.pone.0053604
34. Root CM, Ko KI, Jafari A, Wang JW. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell (2011) 145:133–44. doi: 10.1016/j.cell.2011.02.008
35. Cerstiaens ANJA, Benfekih L, Zouiten H, Verhaert P, De Loof A, Schoofs L. Led-NPF-1 stimulates ovarian development in locusts. Peptides (1999) 20:39–44. doi: 10.1016/S0196-9781(98)00152-1
36. Schoofs L, Clynen E, Cerstiaens A, Baggerman G, Wei Z, Vercammen T, et al. Newly discovered functions for some myotropic neuropeptides in locusts. Peptides (2001) 22:219–27. doi: 10.1016/S0196-9781(00)00385-5
37. De Loof A, Baggerman G, Breuer M, Claeys I, Cerstiaens A, Clynen E, et al. Gonadotropins in insects: an overview. Arch Insect Biochem Physiol. (2001) 47:129–38. doi: 10.1002/arch.1044
38. Kapan N, Lushchak V, Luo J, Nässel DR. Identified peptidergic neurons in the Drosophila brain regulate insulin-producing cells, stress responses and metabolism by coexpressed short neuropeptide F and corazonin. Cell Mol Life Sci. (2012) 69:4051–66. doi: 10.1007/s00018-012-1097-z
39. Badisco L, Van Wielendaele P, Broeck JV. Eat to reproduce: a key role for the insulin signaling pathway in adult insects. Front Physiol. (2013) 4:202. doi: 10.3389/fphys.2013.00202
40. Nässel DR, Broeck JV. Insulin/IGF signaling in Drosophila and other insects: factors that regulate production, release and post-release action of the insulin-like peptides. Cell Mol Life Sci. (2016) 73:271–90. doi: 10.1007/s00018-015-2063-3
41. Yamanaka N, Yamamoto S, Žitnan D, Watanabe K, Kawada T, Satake H, et al. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS ONE (2008) 3:e3048. doi: 10.1371/journal.pone.0003048
42. Kaneko Y, Hiruma K. Short neuropeptide F (sNPF) is a stage-specific suppressor for juvenile hormone biosynthesis by corpora allata, and a critical factor for the initiation of insect metamorphosis. Dev Biol. (2014) 393:312–9. doi: 10.1016/j.ydbio.2014.07.014
43. Lu HL, Pietrantonio PV. Immunolocalization of the short neuropeptide F receptor in queen brains and ovaries of the red imported fire ant (Solenopsis invicta Buren). BMC Neurosci. (2011) 12:57. doi: 10.1186/1471-2202-12-57
44. Bao C, Yang Y, Huang H, Ye H. Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: transcriptomic analysis and expression profiles during vitellogenesis. Sci Rep. (2015) 5:17055. doi: 10.1038/srep17055
45. Christie AE. Expansion of the Litopenaeus vannamei and Penaeus monodon peptidomes using transcriptome shotgun assembly sequence data. Gen Comp Endocrinol. (2014) 206:235–54. doi: 10.1016/j.ygcen.2014.04.015
46. Christie AE. Prediction of the peptidomes of Tigriopus californicus and Lepeophtheirus salmonis (Copepoda, Crustacea). Gen Comp Endocrinol. (2014) 201:87–106. doi: 10.1016/j.ygcen.2014.02.015
47. Christie AE. Prediction of Scylla olivacea (Crustacea; Brachyura) peptide hormones using publicly accessible transcriptome shotgun assembly (TSA) sequences. Gen Comp Endocrinol. (2016) 230:1–16. doi: 10.1016/j.ygcen.2016.03.008
48. Christie AE, Pascual MG. Peptidergic signaling in the crab Cancer borealis: Tapping the power of transcriptomics for neuropeptidome expansion. Gen Comp Endocrinol. (2016) 237:53–67. doi: 10.1016/j.ygcen.2016.08.002
49. Christie AE, Stemmler EA, Dickinson PS. Crustacean neuropeptides. Cell Mol Life Sci. (2010) 67:4135–69. doi: 10.1007/s00018-010-0482-8
50. Veenstra JA. Similarities between decapod and insect neuropeptidomes. Peer J. (2016) 4:e2043. doi: 10.7717/peerj.2043
51. Huang X, Ye H, Huang H, Yang Y, Gong J. An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen Comp Endocrinol. (2014) 204:229–38. doi: 10.1016/j.ygcen.2014.06.002
52. Han K, Dai Y, Zou Z, Fu M, Wang Y, Zhang Z. Molecular characterization and expression profiles of cdc2 and cyclin B during oogenesis and spermatogenesis in green mud crab (Scylla paramamosain). Comp Biochem Physiol B Biochem Mol Biol. (2012) 163:292–302. doi: 10.1016/j.cbpb.2012.07.001
53. Gong J, Ye H, Xie Y, Yang Y, Huang H, Li S, et al. Ecdysone receptor in the mud crab Scylla paramamosain: a possible role in promoting ovarian development. J Endocrinol. (2015) 224:273–87. doi: 10.1530/JOE-14-0526
54. Shu L, Yang Y, Huang H, Ye H. A bone morphogenetic protein ligand and receptors in mud crab: A potential role in the ovarian development. Mol Cell Endocrinol. (2016) 434:99–107. doi: 10.1016/j.mce.2016.06.023
55. Zeng H, Bao C, Huang H, Ye H, Li S. The mechanism of regulation of ovarian maturation by red pigment concentrating hormone in the mud crab Scylla paramamosain. Anim Reprod Sci. (2016) 164:152–61. doi: 10.1016/j.anireprosci.2015.11.025
56. Song YN, Shi LL, Liu ZQ, Qiu GF. Global analysis of the ovarian microRNA transcriptome: implication for miR-2 and miR-133 regulation of oocyte meiosis in the Chinese mitten crab, Eriocheir sinensis (Crustacea: Decapoda). BMC Genomics (2014) 15:47. doi: 10.1186/1471-2164-15-547
57. Beitz E. TeXshade: shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics (2000) 16:135–9. doi: 10.1093/bioinformatics/16.2.135
58. Mertens I, Meeusen T, Huybrechts R, De Loof A, Schoofs L. Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochem Biophys Res Commun. (2002) 297:1140–8. doi: 10.1016/S0006-291X(02)02351-3
59. Feng G, Reale V, Chatwin H, Kennedy K, Venard R, Ericsson C, et al. Functional characterization of a neuropeptide F-like receptor from Drosophila melanogaster. Eur J Neurosci. (2003) 18:227–38. doi: 10.1046/j.1460-9568.2003.02719.x
60. Garczynski SF, Crim JW, Brown MR. Characterization of neuropeptide F and its receptor from the African malaria mosquito, Anopheles gambiae. Peptides (2005) 26:99–107. doi: 10.1016/j.peptides.2004.07.014
61. Ma Q, Cao Z, Yu Y, Yan L, Zhang W, Shi Y, et al. Bombyx neuropeptide G protein-coupled receptor A7 is the third cognate receptor for short neuropeptide F from silk worm. J Biol Chem. (2017) 292:20599–612. doi: 10.1074/jbc.M117.815191
62. Liesch J, Bellani LL, Vosshall LB. Functional and genetic characterization of neuropeptide Y-like receptors in Aedes aegypti. PLoS Negl Trop Dis. (2013) 7:e2486. doi: 10.1371/journal.pntd.0002486
63. Bajracharya P, Lu HL, Pietrantonio PV. The red imported fire ant (Solenopsis invicta Buren) kept Y not F: predicted sNPY endogenous ligands deorphanize the short NPF (sNPF) receptor. PloS ONE (2014) 9:e109590. doi: 10.1371/journal.pone.0109590
64. Caers J, Peymen K, Van Hiel MB, Van Rompay L, Van Den Abbeele J, Schoofs L, et al. Molecular characterization of a short neuropeptide F signaling system in the tsetse fly, Glossina morsitans morsitans. Gen Comp Endocrinol. (2016) 235:142–9. doi: 10.1016/j.ygcen.2016.06.005
65. Jiang HB, Gui SH, Xu L, Pei YX, Smagghe G, Wang JJ. The short neuropeptide F modulates olfactory sensitivity of Bactrocera dorsalis upon starvation. J Insect Physiol. (2017) 99:78–85. doi: 10.1016/j.jinsphys.2017.03.012
66. Chen ME, Pietrantonio PV. The short neuropeptide F-like receptor from the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae). Arch Insect Biochem Physiol. (2006) 61:195–208. doi: 10.1002/arch.20103
67. Hauser F, Cazzamali G, Williamson M, Park Y, Li B, Tanaka Y, et al. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front Neuroendocrinol. (2008) 29:142–65. doi: 10.1016/j.yfrne.2007.10.003
68. Veenstra JA, Rombauts S, Grbić M. In silico cloning of genes encoding neuropeptides, neurohormones and their putative G-protein coupled receptors in a spider mite. Insect Biochem Mol Biol. (2012) 42:277–95. doi: 10.1016/j.ibmb.2011.12.009
69. Bigot L, Beets I, Dubos MP, Boudry P, Schoofs L, Favrel P. Functional characterization of a short neuropeptide F-related receptor in a lophotrochozoan, the mollusk Crassostrea gigas. J Exp Biol. (2014) 217:2974–82. doi: 10.1242/jeb.104067
70. Elvin JA, Clark AT, Wang P, Wolfman NM, Matzuk MM. Paracrine actions of growth differentiation factor-9 in the mammalian ovary. Mol Endocrinol. (1999) 13:1035–48. doi: 10.1210/mend.13.6.0310
71. Elvin JA, Yan C, Matzuk MM. Oocyte-expressed TGF-β superfamily members in female fertility. Mol Cell Endocrinol. (2000) 159:1–5. doi: 10.1016/S0303-7207(99)00185-9
72. Clelland E, Peng C. Endocrine/paracrine control of zebrafish ovarian development. Mol Cell Endocrinol. (2009) 312:42–52. doi: 10.1016/j.mce.2009.04.009
73. Ge W. Intrafollicular paracrine communication in the zebrafish ovary: the state of the art of an emerging model for the study of vertebrate folliculogenesis. Mol Cell Endocrinol. (2005) 237:1–10. doi: 10.1016/j.mce.2005.03.012
74. Li CW, Ge W. Spatiotemporal expression of bone morphogenetic protein family ligands and receptors in the zebrafish ovary: a potential paracrine-signaling mechanism for oocyte-follicle cell communication. Bio Reprod. (2011) 85:977–86. doi: 10.1095/biolreprod.111.092239
75. Li CW, Zhou R, Ge W. Differential regulation of gonadotropin receptors by bone morphogenetic proteins in the zebrafish ovary. Gen Comp Endocrinol. (2012) 176:420–5. doi: 10.1016/j.ygcen.2011.12.032
76. Pang Y, Thomas P. Role of G protein-coupled estrogen receptor 1, GPER, in inhibition of oocyte maturation by endogenous estrogens in zebrafish. Dev Biol. (2010) 342:194–206. doi: 10.1016/j.ydbio.2010.03.027
77. Zhou R, Tsang AH, Lau SW, Ge W. Pituitary adenylate cyclase-activating polypeptide (PACAP) and its receptors in the zebrafish ovary: evidence for potentially dual roles of pacap in controlling final oocyte maturation. Bio Reprod. (2011) 85:615–25. doi: 10.1095/biolreprod.111.091884
Keywords: short neuropeptide F, ovary, oocyte maturation, vitellogenesis, crustacean
Citation: Bao C, Yang Y, Huang H and Ye H (2018) Inhibitory Role of the Mud Crab Short Neuropeptide F in Vitellogenesis and Oocyte Maturation via Autocrine/Paracrine Signaling. Front. Endocrinol. 9:390. doi: 10.3389/fendo.2018.00390
Received: 17 April 2018; Accepted: 25 June 2018;
Published: 13 July 2018.
Edited by:
Takayoshi Ubuka, Monash University Malaysia, MalaysiaReviewed by:
Honoo Satake, Suntory Foundation for Life Sciences, JapanTsuyoshi Ohira, Kanagawa University, Japan
Copyright © 2018 Bao, Yang, Huang and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haihui Ye, aGFpaHVpeWVAeG11LmVkdS5jbg==
 Chenchang Bao
Chenchang Bao Yanan Yang1
Yanan Yang1 Haihui Ye
Haihui Ye