- Department of Molecular, Cellular and Developmental Biology, University of Michigan, Ann Arbor, MI, United States
Insulin-like growth factors (IGFs) are key growth-promoting peptides that act as both endocrine hormones and autocrine/paracrine growth factors. In the bloodstream and in local tissues, most IGF molecules are bound by one of the members of the IGF-binding protein (IGFBP) family, of which six distinct types exist. These proteins bind to IGF with an equal or greater affinity than the IGF1 receptor and are thus in a key position to regulate IGF signaling globally and locally. Binding to an IGFBP increases the half-life of IGF in the circulation and blocks its potential binding to the insulin receptor. In addition to these classical roles, IGFBPs have been shown to modulate IGF signaling locally under various conditions. Although members of the IGFBP family share significant sequence homology, they each have unique structural features and play distinct roles. These IGFBP genes also have different modes of regulation and distinct expression patterns. Some IGFBPs have been found to bind to their own receptors or to translocate into the interior compartments of cells where they may execute IGF-independent actions. In spite of this functional and regulatory diversity, it has been puzzling that loss-of-function studies have yielded relatively little information about the physiological functions of IGFBPs. In this review, we suggest that evolution has tended to retain an array of IGFBPs in order to facilitate fine-tuning of IGF signaling. We explore the emerging explanation that many IGFBP functions have evolved to allow the targeted adjustment of IGF signaling under stressful or irregular conditions, which would likely not be revealed in a standard laboratory setting.
Introduction
The insulin and insulin-like growth factor (IGF) signaling pathway is highly conserved among the metazoans. Many invertebrates have large numbers of insulin-like peptides (ILPs); for instance, the Caenorhabditis elegans genome contains around 40 (http://wormbase.org), and the Drosophila melanogaster genome contains 8 (http://flybase.org). In vertebrates, the ancestral insulin-like gene has diverged into insulin, IGFs-1 and -2, and several ILPs including relaxin and relaxin-like peptide (1). Insulin primarily acts in an endocrine fashion to regulate metabolism, whereas IGFs have a variety of roles as endocrine, paracrine and autocrine factors that promote cell growth, proliferation, differentiation, survival, etc. Both IGF-1 and IGF-2 bind to the IGF-1 receptor (IGF1R), which is expressed in almost all cells, with hepatocytes being an important exception in mammals. The liver secretes IGF-1 into the circulation in response to growth hormone (GH) stimulation (2, 3). Local tissues also secrete IGF-1 in response to GH, and this paracrine/autocrine IGF-1 acts together with the endocrine IGF-1 (mostly liver derived) to mediate the global growth promoting effects of GH (4). In addition to their role in regulating fetal, neonatal, and postnatal growth, IGFs are also involved in diverse processes including wound healing (5), development of CNS and other tissues (6), regulation of protein, carbohydrate, and lipid metabolism (7), neuroprotection (8), aging (9), etc.
The diverse functions of this central hormonal pathway require that robust regulatory mechanisms be in place to avoid inappropriate regulation and/or dysfunction in different tissues and at different times. We now understand that IGF signaling is regulated by a family of specific IGF-binding proteins (IGFBPs) of which there are six distinct types in vertebrates. These IGFBPs share significant sequence homology and they are capable of binding IGFs with equal or greater affinity than the IGF1R. In fact, in both the circulation and in local tissues, most IGFs can be found bound to an IGFBP (10–13). In this review, we discuss the complex interplay of both overlapping and unique functions by which IGFBPs influence IGF signaling.
The IGFBP Family
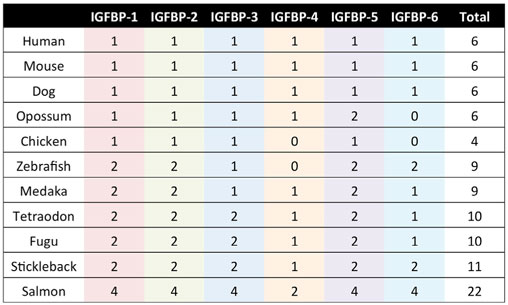
The IGFBP family is evolutionarily ancient and highly conserved in vertebrates (11, 14–16). The six types of IGFBPs have been designated IGFBP-1 through IGFBP-6. Mammals generally possess one gene that belongs to each of the six types, and humans follow this pattern (Table 1). Some vertebrate species occasionally lack one or more of the types, and others have more than one IGFBP gene that can be classified within one type (Table 1). It is believed that the IGFBP family evolved via successive rounds of whole genome duplications. Notably, many teleost fish possess two copies of each of the six types of IGFBPs (Table 1), which is attributable to the third round of whole genome duplication that they are believed to have undergone following their divergence from the other vertebrates (16–21). Salmonid fish experienced an additional round of genome wide duplication and can have four copies of each IGFBP (22–24). We discuss the evolution of the IGFBP family in more detail in a later section.
All IGFBPs generally have approximately 200–300 amino acids and share a conserved structure consisting of a highly cysteine-rich N-terminal domain that is highly conserved among the IGFBP family and across species, a linker domain whose sequence is variable, and a cysteine-rich C-terminal domain that is also evolutionarily conserved (Figure 1A). The N- and C-terminal domains are globular and are structurally stabilized by multiple disulfide bonds between the conserved cysteine residues. Both of these domains participate in forming the IGF-binding site. The central linker domain is unstructured and serves to tether the N- and C-terminal domains together but also provides a location for functional motifs (10, 25).
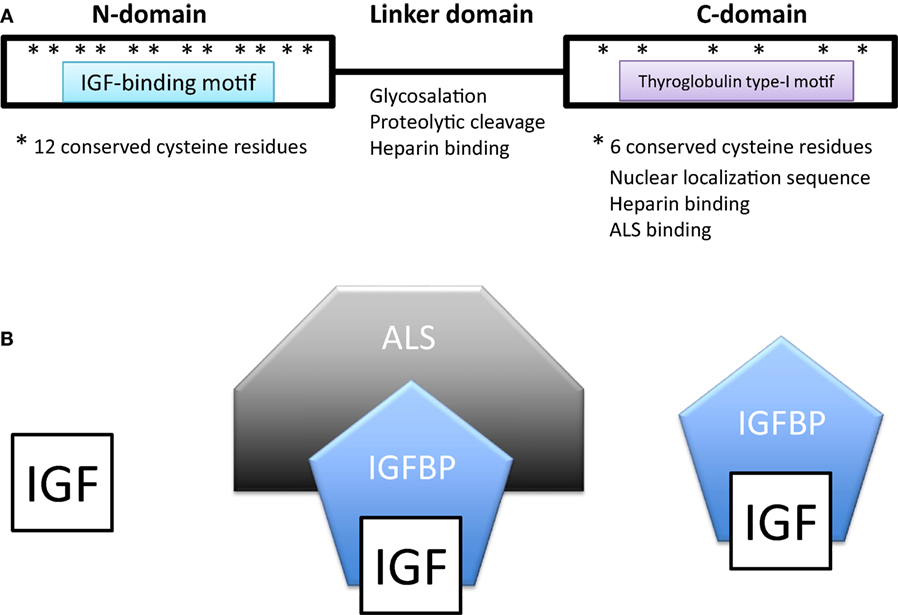
Figure 1. (A) Domain structure of insulin-like growth factor-binding proteins (IGFBPs). IGFBPs contain conserved N- and C-terminal domains and a variable linker domain between them. The N-domain contains an insulin-like growth factor (IGF)-binding motif and the C-domain contains a thyroglobulin type-I repeat. The N-domain usually contains 12 conserved cysteine residues and the C-domain contains 6. (B) In extracellular environments, most IGFs are bound with IGFBPs, either in a binary complex containing one IGF and one IGFBP or a ternary complex consisting of an IGF, IGFBP-3 (or less often IGFBP-5), and a glycoprotein called acid labile subunit (ALS).
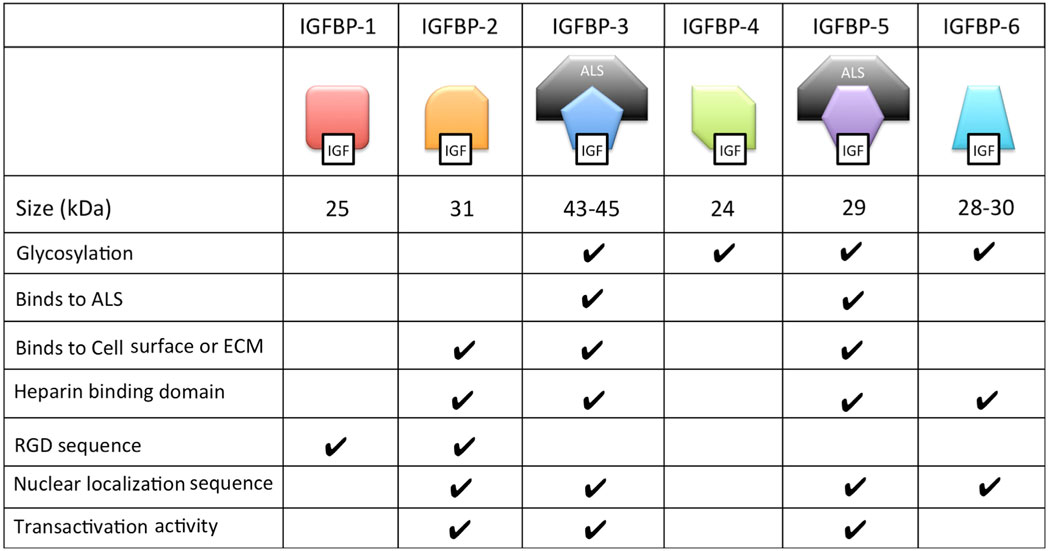
Many of the functions of different IGFBPs are made possible by their unique collection of functional motifs (Table 2). These functional motifs include binding sites for heparin, components of the extracellular matrix and cell surface proteoglycans; proteolytic cleavage sites; sites of post-translational modifications including glycosylation, etc. In addition, IGFBP-2, -3, -5, and -6 all contain functional nuclear localization sequences by which they are imported into the cell nucleus in certain cell types as we discuss below.
It should be noted that a number of proteins belonging to the CCN (cyr61, ctgt, and Nov) protein family have been reported to contain sequences similar to the IGFBP N-domain and were once named IGFBP 7–12. It was later recognized that these were not IGFBPs and were renamed as IGFBP-related proteins and classified as part of a broader superfamily with the IGFBPs (26). The latter nomenclature has been questioned because these CCN family proteins not only lack high-affinity IGF-binding abilities but are also structurally no more related to IGFBPs than to von Willebrand factor, thrombospondin, or growth factor cysteine knots (27). These CCN family proteins will not be discussed in this review.
IGFBP Biological Actions
In this section, we discuss a selection of the vast literature on the many reported biological actions/activities of IGFBPs that have been reported in gain-of-function studies in vivo or in vitro. Each species has its own standard nomenclature for gene and protein names. In this article, we deal with a large number of species. To increase the readability, we will use the same symbol for each IGFBP name. Whenever required, the species name is added to avoid confusion.
Endocrine Actions of IGFBPs
In extracellular environments, most IGFs are bound with IGFBPs, either in a binary complex or a ternary complex (Figure 1B). The vast majority of IGFs in the serum are bound to an IGFBP. IGFBP-3 is the most prevalent in adult serum with a concentration of around 100 nM/L, while all of the other IGFBPs are present at concentrations of less than 20 nM/L (25, 28). About 75–80% of serum IGFs were found in a ternary complex of about 150 kDa consisting of an IGF, IGFBP-3 (or less often IGFBP-5) and a glycoprotein called acid labile subunit (ALS). The remaining 20–25% of IGFs were complexed with one of the other IGFBPs (25, 28). Unbound IGFs have a half-life of less than 10 min (29). Binding to an IGFBP increases IGF half-life in the circulation to around 25 min, but the binary complexes are able to rapidly leave the circulation (29). Most of the circulating IGFs are present in the IGF-IGFBP3/5-ALS ternary complex (30, 31). The addition of ALS increases the molecular size of the complex and this has the effect of preventing the bound IGF from leaving the capillaries thereby confining it within the circulation (32). The ternary complex thereby greatly prolongs the half-life of bound IGFs to about 16 h or more, forming a long-lasting reservoir of IGFs in the circulation (28). Deletion of the ALS gene in mice results in a 60% reduction in circulating IGFs and a 15–20% reduction in postnatal growth (33).
Insulin-like growth factors are sufficiently structurally similar to insulin that they can cross react with the insulin receptor (IR) (34). Another important function of circulating IGFBPs is to prevent the potential interaction of IGFs with the IR, which is crucial since IGF concentrations are high enough in the serum to cause hypoglycemic effects even given their lower affinity for the IR (25, 35).
Insulin-like growth factor-binding protein-3 is produced in the liver and in other tissues and secreted into the serum, and its hepatic expression level is regulated by GH (36). This ensures that as the amount of secreted IGF-1 increases in response to GH stimulation, there will be an increased quantity of IGFBP-3 to absorb it in the circulation. IGFBP-1 is also synthesized in the liver and its expression and secretion are highly regulated by catabolic factors and hormones. For example, hepatic IGFBP-1 expression level is highly induced by starvation, hypoxia, and stress (37, 38). Its expression is reduced by insulin and increased by glucocorticoids (39, 40). These regulatory mechanisms serve to promote IGFBP-1 expression in response to starvation and catabolic conditions, including amino acid shortages and hypoxia (41, 42). The functional role of IGFBP-1 in these conditions is to reduce the rate of development and growth by binding to IGFs and inhibiting IGF activity (37, 38).
Local Actions of IGFBPs
While the bulk of circulating IGFBP-3 and IGFBP-1 are produced in the liver, IGFBP-3 and other IGFBPs are also expressed in many peripheral tissues (43, 44). The importance of local IGF-1 is supported by the finding that deletion of IGF-1 specifically in the liver resulted in an 80% reduction in circulating endocrine IGF-1 but no change in postnatal growth (45). Biochemical and cell culture studies suggest that IGFBPs generally bind IGFs with equal or higher affinity than the IGF1R and can inhibit IGF signaling by sequestration of the ligands (12, 13, 25, 46) (see Figure 2A). An example of this behavior is found in vascular smooth muscle cells (VSMCs) where IGFBP-4 acts to block IGF-1 from interacting with the IGF1R and thereby inhibits IGF-1-stimulated DNA synthesis (47). When IGFBP-4 was overexpressed in various tissues in mice, it resulted in hypoplasia of the affected tissue, suggesting that this may be a common action in different cell types (48).
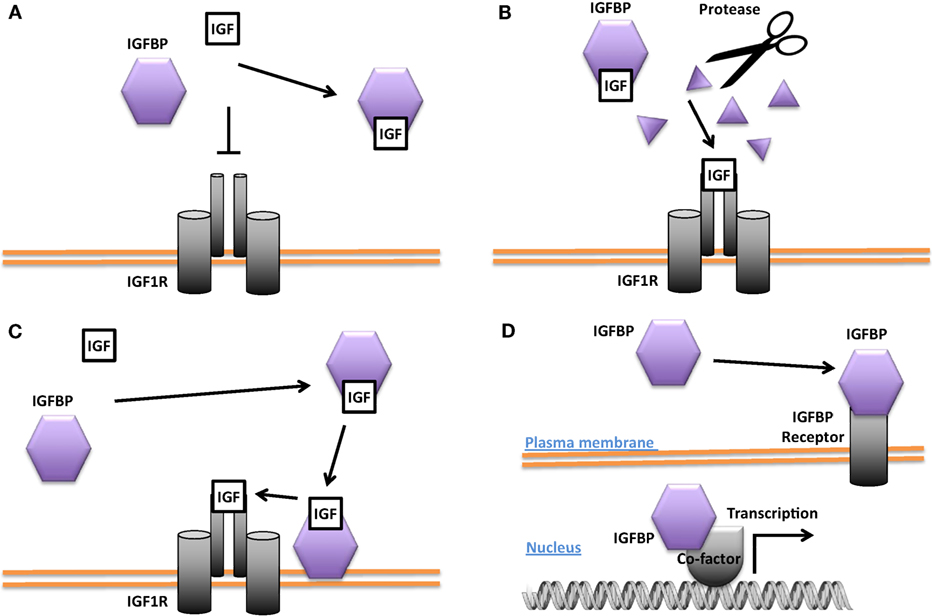
Figure 2. Different modes of Insulin-like growth factor-binding protein (IGFBP) actions. (A) Inhibition of insulin-like growth factor (IGF) signaling by sequestering IGFs away from the IGF-1 receptor (IGF1R). (B) Promotion of IGF signaling by proteolytic cleavage of the IGFBP and liberation of IGFs from the IGF/IGFBP complex for binding to the IGF1R. (C) Enhancement of IGF signaling by concentrating IGF locally and increasing IGF availability for binding to the IGF1R. (D) IGF-independent actions. Some IGFBPs have been shown to be capable of translocating into the nucleus in certain cells and may affect gene transcription directly or indirectly. Some IGFBPs have been found to bind to cell surface proteins that may act as IGFBP receptors.
Some IGFBPs have been shown to potentiate IGF signaling. Several proteases are known to cleave IGFBPs, and the resulting proteolytic fragments have greatly reduced binding affinity for IGFs. This leads to the liberation of IGFs from the IGF/IGFBP complex and increases the amount of IGFs available for IGF1R binding, thereby converting the inhibition of IGF signaling into an enhancement (Figure 2B). The proteases pregnancy-associated plasma protein A (PAPP-A) and PAPP-A2 are specific IGFBP proteases (49). IGFBP-4 is cleaved by PAPP-A when bound to IGF. This results in the release of the IGF ligand from the complex and a consequent increase in IGF ligand available for binding to the IGF-1R (50). PAPP-A knockout mice were about 40% smaller than wild-type littermates, which is consistent with the idea that PAPP-A cleaves inhibitory IGFBPs and thereby promotes IGF action (51). IGFBP-4 knockout mice were paradoxically slightly smaller than wild-type littermates, but mice null for both IGFBP-4 and PAPP-A were not smaller than IGFBP-4 knockout mice, indicating that the growth-promoting effects of PAPP-A likely result from the cleavage of IGFBP-4 (52). A number of other proteases have also been found to cleave various IGFBPs (53–55).
The potentiating action of some IGFBPs can occur when the IGFBP binds to the target cell’s surface proteoglycans and/or extracellular matrix components, resulting in a concentration of local IGF that can then be released to the IGF1R (Figure 2C). It has been reported that IGFBP-5 undergoes a reduction of affinity for IGFs when it binds to certain extracellular matrix components, allowing it to deliver and then release IGF ligands at target sites (56, 57). Differentiating myoblasts provide one example of the interplay between a locally produced IGFBP and autocrine/paracrine IGF signaling. During myogenesis, IGF-2 is produced locally at high levels and this is required for myoblast differentiation (58). Prior to the onset of IGF-2 secretion, there is an increase in the expression and secretion of IGFBP-5. The secreted IGFBP-5 potentiates IGF-2 signaling and increases myoblast cell differentiation by binding to IGF-2 and promoting its interaction with the IGF-1R (59).
Insulin-like growth factor-binding proteins have also been reported to act locally under certain pathological conditions. A good example is the role of IGFBP-5 in the progression of atherosclerosis. IGFBP-5 is normally produced and secreted by VSMCs but its expression is upregulated in the VSMCs found within atherosclerotic plaques (60). Immunostaining for IGFBP-5 protein was dense within atherosclerotic plaques and especially around calcified areas (61). Locally secreted IGFs have been suggested to play an important role in atherogenesis by promoting the VSMC proliferation and their migration into the area of the arterial wall known as the intima (62, 63). These actions are promoted by local IGFBP-5 (57). Interestingly, a protease resistant IGFBP-5 mutant actually inhibited VSMC proliferation and migration, suggesting a mechanism by which IGFBP-5 is normally cleaved in order to present the IGF ligands to the IGF-1R on the surface of VSMCs (56). IGFBP-5 also binds to certain extracellular matrix proteins that are enriched in atherosclerotic lesions. It was reported that these interactions enhanced the mitogenic effects of IGFs on VSMCs (53, 55, 64). These studies support a model in which local IGFBP-5 is concentrated within atherosclerotic lesions (by both increased local expression and secretion and by binding to locally enriched ECM components), where it then acts to concentrate and deliver IGFs to the IGF1R on local VSMCs.
Studies suggest that IGFBP-4 may be involved in inhibiting atherosclerosis. A protease-resistant IGFBP-4 mutant was able to inhibit atherosclerotic lesion development in hypercholesterolemic pigs (65). When the PAPP-A protease that cleaves IGFBP-4 was knocked down in a mouse model of atherosclerosis (ApoE KO), there was decreased formation of atherosclerotic lesions (66, 67). When PAPP-A expression was transgenically increased locally within VSMCs in artery walls in ApoE KO mice, there was a substantial increase in atherosclerotic plaque formation that was associated with an increase in local IGF-1 availability (68). Targeting PAPP-A in ApoE KO mice with a monoclonal antibody that inhibited its proteolytic activity resulted in a 70% reduction in aortic plaque burden (69). These results suggest that the proteolysis of IGFBP-4 by PAPP-A releases IGF-1 that acts locally to promote atherosclerotic plaque formation.
IGF-Independent Actions
Several IGFBPs have been reported to have cellular actions that are independent of their IGF binding (Figure 2D). Some IGF-independent actions are mediated by binding to cell surface proteins. For example, the integrin-binding RGD motif found in IGFBPs 1 and 2 allows them to promote cell migration and influence cell adhesion, respectively (70, 71). IGFBP-5 and -3 possess functional nuclear localization sequences and can enter the nucleus (72, 73). The nuclear localization of IGFBP-3 and -5 was found to be mediated by importin beta (72). Locally produced IGFBP-5 was found to stimulate porcine VSMC migration by an IGF-independent mechanism (74). IGFBP-5 was shown to possess transactivation activity (73). This transactivation activity was mapped to the N-domain and was also demonstrated in the N-domains of IGFBPs-2 and 3 (75). Nuclear localization and transactivation activity are also present in zebrafish IGFBP-3 and -5 (21, 76). In cephalochordate amphioxus, which diverged from the vertebrates approximately 520 million years ago, there is a single IGFBP-like gene. The amphioxus IGFBPs contains a functional nuclear localization signal and a transactivation domain (77). The lamprey IGFBP3, a jawless agnathan vertebrate, has been reported to possess both IGF-dependent action and the transactivation activity. The conservation of IGFBP transactivation activity across eons of evolution suggests that it likely has an important function. Along this line, several studies have found roles for nuclear IGFBPs in altering transcription in cancer cells (78, 79), but the physiological role(s) of the endogenous IGFBPs in the nucleus remain unclear.
Other IGF-independent actions have been reported that do not apparently involve nuclear localization. Paracrine IGFBP-4 was shown to promote differentiation of cardiomyocytes by inhibiting Wnt signaling in an IGF-independent manner (80). The physiological relevance of this effect was supported by the fact that knockdown of IGFBP-4 in Xenopus embryos resulted in cardiac defects attributable to impaired cardiomyogenesis (80). On the other hand, IGFBP-4 knockout mice have no cardiac phenotype (81). The lack of phenotype may be due to genetic redundancy and/or compensation by other IGFBPs. Another example is the antagonization of bone morphogenic protein signaling by IGFBP-3 in zebrafish (76). It has been reported that human IGFBP-6 has antiangiogenic activity when tested using in vitro assays. This action is independent from IGF binding because an IGFBP-6 mutant with 10,000-fold lower binding affinity for IGFs was as potent as the wild-type human IGFBP-6 in inhibiting angiogenesis (82). Interestingly, IGFBP-6 was found to be able to bind vascular endothelial growth factor (VEGF) and coincubation with IGFBP-6 abolished VEGF-stimulated angiogenesis. This antiangiogenic action of IGFBP-6 was demonstrated in vivo in a tumor model by transplanting human Rh30 rhabdomyosarcoma cells stably transfected with IGFBP-6 into BALB/c nude mice (82). Expression of zebrafish IGFBP-6b had similar effects, indicating that this antiangiogenic activity is evolutionarily conserved (82).
Some IGFBPs may have cell surface receptor-mediated IGF-independent actions (Figure 2D). Exogenous IGFBP-3 was reported to inhibit cultured cell growth by an IGF-independent mechanism (83, 84). This effect was shown to be related to the binding of IGFBP-3 to the type 5 transforming growth factor β (TFGβ) receptor (85). This receptor was then shown to be identical to the low-density lipoprotein receptor-related protein-1 (LRP-1) (86). LRP-1 is known to be responsible for the uptake and clearance of various molecules from the circulation (87). The downstream mechanisms by which the interaction of IGFBP-3 with LRP-1 may lead to growth inhibition remain unclear. IGFBP-2 has been shown to bind to a receptor called receptor protein tyrosine phosphatase β (RPTPβ), which triggers a signal transduction cascade that leads to reduced PTEN phosphatase activity and a consequent enhancement of IGF-1-induced Akt pathway activation (88). This interaction between IGFBP-2 and RPTPβ was shown to be responsible for the ability of IGFBP-2 to trigger osteoblast differentiation (89). This role of IGFBP-2 was independent of IGF-binding and a 13-residue peptide corresponding of IGFBP-2’s heparin-binding domain 1 was shown to mediate its binding to RPTPβ (88, 89).
Loss-of-Function Studies
Given the numerous biological actions of IGFBPs found in gain-of-function studies, it was surprising that little or no phenotypic change was observed when individual IGFBP genes were deleted in mice (81, 90–93). IGFBP-1 knockout mice were indistinguishable from their wild-type littermates and no embryonic lethality was observed (91). IGFBP-2 knockout mice were phenotypically normal with the exception of minor gender specific changes in bone structure and minor changes in the weights of spleen and liver in adult males (90, 92). IGFBP-3 knockout mice were also normal (81). Deletion of the IGFBP-4 gene in mice resulted in a mild 10–15% reduction in prenatal growth, which is somewhat paradoxical given that overexpression of IGFBP-4 also reduces growth (81). IGFBP-5 knockout mice were also phenotypically normal (81). Genetic deletion of IGFBP genes using CRISPR-Cas9 or TALEN in zebrafish have also resulted in little or no alteration in phenotype. Zebrafish IGFBP-3 knockout fish are morphologically normal and their growth rate and developmental speed are comparable to their siblings. Likewise, IGFBP-5a and -5b knockout zebrafish are morphologically indistinguishable from their wild-type siblings when kept under optimized lab conditions (unpublished data).
When IGFBP-3, -4, and -5 were knockout together in mice, there was a 25% reduction in body growth, decreased fat accumulation and quadriceps muscle mass, expanded pancreatic islets, and enhanced glucose homeostasis (81). These triple mutant mice were viable (81). Considering that knockout of IGF-1 itself results in a 60% reduction in prenatal growth followed by perinatal lethality for over 95% of mutant pups (94), the phenotype of the triple IGFBP-3/4/5 knockout mice can be viewed as relatively moderate.
The lack of substantial phenotypes in these IGFBP mutant mice and the finding that these animals can survive without three out of the six IGFBPs suggests a high degree of functional redundancy and/or genetic compensatory mechanisms. Indeed, elevated levels of IGFBPs-1, -3, and -4 were found in the IGFBP-2 knockout mice, supporting the notion that the lack of IGFBP-2 may be compensated for by upregulation of other IGFBPs (90).
Genetic redundancy among paralogous genes is a widespread phenomenon and can result in the masking of phenotypes in loss-of-function studies (95, 96). One study of the Drosophila genome suggested that when gene duplications occur, only 4% of the resulting paralogs survive (97). One explanation for the stable retention of redundant paralogous genes is that genes with redundant functions may also acquire functions that are unique to themselves. This can result in the coselection of the redundant functions with the unique functions in a model referred to as the “piggyback” mechanism (98, 99). In this model, whenever it is the case that most mutations tend to inactivate both the redundant and non-redundant functions simultaneously, redundant functions can then be retained in both gene duplicates. Unique functions could be obtained by gain-of-function mutations, but it is more common for complementary inactivating mutations to cause ancestral functions to be partitioned between the duplicates in the process of subfunctionalization (100). Redundant functions can be maintained in both duplicates when at least one unique function is maintained in each duplicate (100).
Why are there so many IGFBPs?
Why has evolution favored the retention of so many IGFBP genes? One potential explanation is that, given the crucial importance of the IGF pathway in determining central life history traits such as body size and longevity, it may be that even relatively minor fine-tuning of IGF signaling levels would be strongly selected for. A possible example comes from the zebrafish IGFBP genes. In zebrafish, there are two IGFBP-1 genes, being paralogs of mammalian IGFBP-1 (17). Zebrafish IGFBP-1a and -1b have similar expression patterns and regulatory responses, but IGFBP-1a has a higher affinity for IGFs than IGFBP-1b, which may allow more graded inhibition of IGF signaling during catabolic conditions than was possible with only a single IGFBP-1 gene (17). The zebrafish genome also contains two IGFBP-2 genes. In this case, the IGFBP-2a and -2b proteins have similar biological activities (18, 101). However, these two paralogous genes exhibit distinct spatiotemporal expression patterns. During embryogenesis, IGFBP-2a mRNA is found in the lens and the brain boundary vasculature; it subsequently becomes highly expressed in the liver. IGFBP-2b is detected initially in all tissues at low levels, but later becomes abundant in the liver (18). In the adult stage, liver has the highest levels of IGFBP-2a mRNA, followed by the brain. IGFBP-2b mRNA, on the other hand, is only detected in the liver (18, 101). The two zebrafish IGFBP-5 genes have diverged both in gene expression patterns and protein functions. Zebrafish IGFBP-5a and -5b are expressed in spatially restricted, mostly non-overlapping domains during early development (21). The IGF-binding site is conserved in both zebrafish IGFBP-5a and -5b, and they are both secreted and capable of IGF binding (21). While zebrafish IGFBP-5b has transactivation activity, no such activity is found with IGFBP-5a (21). Given their divergence in both expression patterns and cellular actions, zebrafish IGFBP-5a and 5b may regulate IGF-signaling within their respective domains in subtly differing ways. This may provide enhanced fine-tuning of IGF signaling as compared with a single IGFBP-5 gene.
A second possible explanation is that genetic compensation is responsible for masking what would otherwise be more significant phenotypes. It has been recognized recently that permanent genetic deletions (knockouts), often result in a less severe phenotype than transient reductions in expression (knockdowns) (102). The mechanisms responsible for this phenomenon remain unclear but a number of hypotheses have been proposed, including the idea that related or unrelated genes could be upregulated in the permanent mutants (102). When zebrafish IGFBP-3 was deleted, for example, no phenotypes were detected. However, when zebrafish IGFBP-3 was knocked down using antisense morpholinos, it resulted in defects in the development of the pharyngeal skeleton and inner ear (103).
Another possible explanation is that, in addition to their somewhat overlapping functions of transporting and protecting IGFs in the circulation, the individual IGFBPs are also involved context-dependent regulation of IGF signaling in specific cell types and under specific stressful or aberrant conditions. Flexible and versatile modes of regulation such as these would be highly advantageous for organisms in the wild and would be strongly selected for, despite being unlikely to produce observable phenotypes under optimized laboratory conditions. One example is the role of IGFBP-1 in responding to catabolic conditions by throttling back growth and developmental rate in order to conserve scarce resources (38, 42, 104). Another example is the specific role of IGFBP-1 in liver regeneration. IGFBP-1 knockout mice exhibited normal growth but were found to have impaired liver regeneration (91). Their liver cells were highly sensitive to induction of apoptosis by treatment with Fas agonist. This effect could be ameliorated by pretreatment with IGFBP-1 (105), suggesting that IGFBP-1 has a crucial but conditional role in protecting the liver when facing injury and healing. The role of IGFBP-5 in mammary gland remodeling is a further example. The IGFBP-5 knockout mice had normal body growth and normal mammary gland development under standard laboratory conditions. However, these mutant mice exhibited delayed mammary gland involution and enhanced alveolar bud formation after ovariectomy and estradiol/progesterone treatment (106). Another example is provided by zebrafish IGFBP5a, which is specifically expressed in a specific type of epithelial cell (ionocytes) on the larval yolk sac skin that are responsible for transporting Ca2+ ions. When wild type larvae are raised in embryo solution containing a very low calcium concentration, these ionocytes rapidly proliferate via a mechanism that requires the activation of IGF signaling in these cells (107). This allows increased calcium import and is necessary for survival under these conditions. This proliferation is blunted in the IGFBP5a knockout fish larvae, causing lethality. However, under optomized and calcium-rich conditions, these mutant fish are indistinguisable from their wild type siblings. This suggests that IGFBP-5a is critical for calcium ionocytes to activate a conditional proliferation program in order to maintain calcium homeostasis.
As always in biology, the question of why there are so many IGFBPs can only be fully understood in the context of evolutionary history. Based on analyses of phylogenetic relationships, the surrounding chromosomal regions in which modern IGFBPs sit, and IGFBP sequences from a large number of species, the evolution of the IGFBP family in early vertebrate ancestors has been reconstructed (16). An ancestral IGFBP sequence in an ancient early chordate was duplicated resulting in two adjacent IGFBP sequences in a chromosomal region that also bore the homeobox (HOX) genes. These two original genes were then duplicated along with the entire genome in the two successive rounds of tetraploidization that occurred in early vertebrates (108), resulting in eight IGFBP genes. It is thought that two of them were lost, resulting in the six types of IGFBPs seen in mammals and most other vertebrate classes (Figure 3). In many teleost fish, another round of tetraploidization occurred, resulting in a further doubling of IGFBP genes (16, 109). Some of these additional duplicates were subsequently retained in modern fish. Indeed, there is substantial variation in numbers of IGFBP genes between fish species (Table 1). The salmonid fish, whose common ancestor underwent a fourth round of whole genome duplication, exhibit the largest known repertoire of 22 IGFBP genes (22, 23). The preservation and evolutionary conservation of most of the IGFBP gene duplicates implies that these genes might have aquired unique evolutionarily adaptive roles, either by developing new functions opportunistically (neofunctionalization) or by retention of a subset of the parent gene’s original functions in each duplicate (subfunctionalization). This is in agreement with the idea that fine tuning of IGF signaling is strongly adaptive to the extent that perhaps even small changes in the regulation of IGF signaling would be sufficient to account for the conservation of additional IGFBP genes to provide these regulatory advantages.
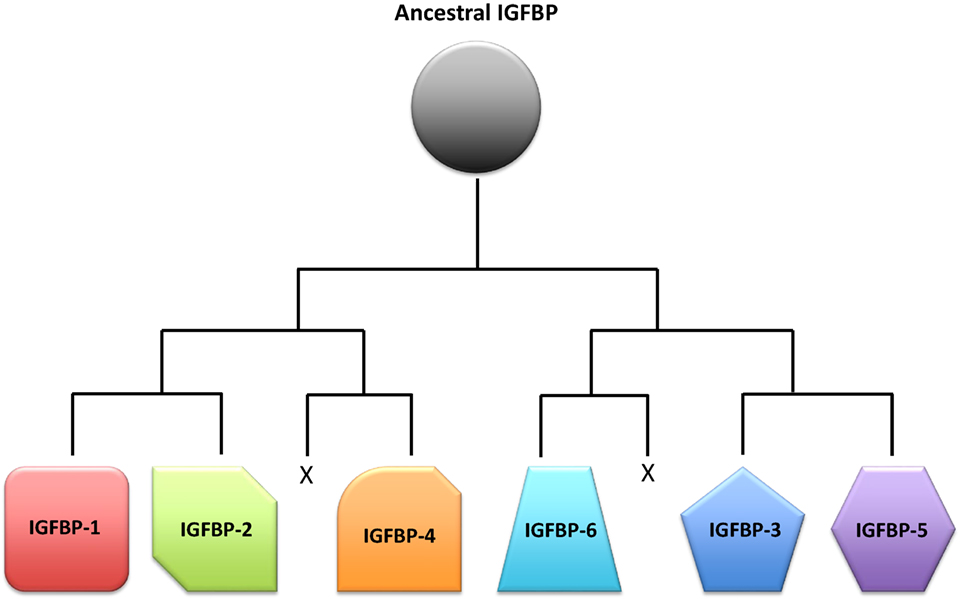
Figure 3. Schematic representation of a proposed scenario of the insulin-like growth factor-binding protein (IGFBP) family evolution. A single ancestral IGFBP gene was duplicated in an early chordate. This duplication was followed by two successive rounds of chromosomal duplications or tetraploidization events in early vertebrates. Of the eight IGFBPs that resulted from this process, two were subsequently lost, leaving six types of IGFBPs that are seen in modern vertebrates.
The acquisition of IGF-independent actions of IGFBPs presents an intriguing question. One possible explanation is that they were present in the ancestral IGFBP gene. A comparative study suggested that the single amphioxus IGFBP has a functional nuclear localization sequence and transactivation activity but lacks the ability to bind modern IGFs (77). Both IGF-dependent and IGF-independent actions appear to have been present in the earliest vertebrates as indicated by the fact that an IGFBP from sea lamprey exhibited both IGF-dependent and -independent actions (110). Therefore, the IGF-binding function of IGFBPs may have been acquired later in evolution.
Concluding Remarks and Prospects
We propose that IGFBPs provide a set of tools with which evolution has acted to increase the flexibility and versatility in the regulation of the IGF signaling system. An ancestral IGFBP gene has diversified into a number of IGFBP genes, which have both overlapping and unique expression patterns and functions. These IGFBPs can be viewed as different tools that all apply leverage but also provide individual context specific advantages. A number of attributes of IGFBPs may help to give rise to the increased flexibility and versatility in their abilities to regulate IGF actions. These include: (1) distinct spatiotemporal expression patterns of these IGFBP genes, (2) differences in their ligand-binding affinity and selectivity, (3) different roles in the circulation including formation of the ternary complex with ALS, (4) different abilities to interact with cell surface proteins, extracellular proteins, and other growth factors, (5) different subcellular localization, and (6) various IGF-independent activities (Figure 4). The existence of multiple IGFBPs can contribute to the fine-tuning of IGF signaling both globally and locally, and under various physiological and pathological conditions. The involvement of IGFBPs in mammary gland growth, liver regeneration, and atherogenesis, and the adaptive proliferation of calcium ionocytes in zebrafish are all examples of this sort of process. It is plausible that more IGFBPs will be found to participate in other roles of this type. A great deal of work has identified many roles for IGFBPs in cancer cells despite the fact that IGFBPs are not commonly mutated in human cancers (12). Given the involvement of IGFBPs in tissue remodeling and conditional proliferation of certain cell types, it is not surprising that their physiological actions would be coopted by cancer cells in order to facilitate the needs of tumor growth.
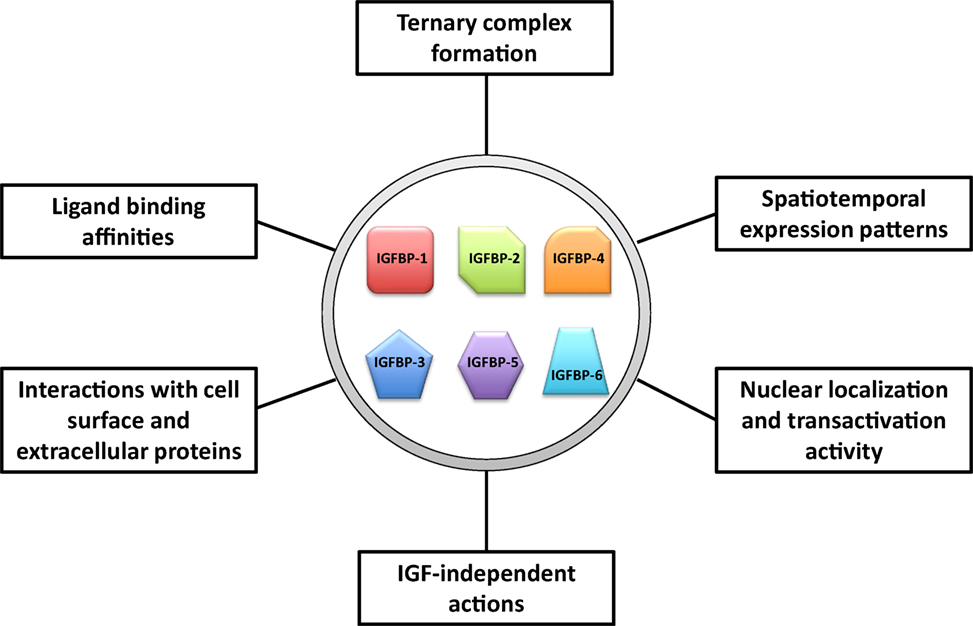
Figure 4. Major attributes of insulin-like growth factor-binding proteins (IGFBPs) that may help to give rise to the increased flexibility and versatility in their abilities to regulate insulin-like growth factor (IGF) actions.
Much has been learned in recent decades about the cell type-specific actions of IGFBPs but many questions remain unanswered. One major question is, why do several of the IGFBPs have the ability to enter the cell nucleus? Although certain IGFBPs have a functional nuclear localization motif and a transactivation domain that are both evolutionarily conserved, the physiological functions of the nuclear IGFBPs remain unknown. Another area of inquiry for future research will be to identify additional stressful conditions that IGFBPs have evolved to respond to. It will also be of great interest to identify pathological processes that depend on the misregulation of IGFBP(s) to increase or decrease IGF signaling, or on inappropriate activation of their IGF-independent actions. We also have much more to learn regarding the evolutionary history of the IGFBPs in early vertebrates and the nature of its IGF-independent functions. This may shed light on the complex biology of modern IGFBPs.
CRISPR/Cas9-based genetic editing technology will allow the generation of mutant animals whose endogenous IGFBP genes are directly mutated to disrupt individual functionalities such as IGF-binding, nuclear translocation, or interaction with cell surface proteins, to allow assessing the roles of those capabilities individually or collectively under physiological conditions in vivo. The CRISPR-Cas9-based approaches will allow the physiological roles of redundant paralogs to be determined much more readily by enabling the generation of multiple knockouts at reasonable cost. Increasing our understanding of IGFBPs will yield insights into the array of biological processes to which IGF signaling is linked, including many that are crucial for human health and diseases.
Author Contributions
CD conceived the article. JA and CD wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the many colleagues in the field of IGFBP biology whose work we were unable to cite due to the narrow focus of this article. This work was supported by NSF grant IOS-1557850 and MCube2.0 Project U0496246 to CD.
References
1. Wood AW, Duan C, Bern HA. Insulin-like growth factor signaling in fish. Int Rev Cytol (2005) 243:215–85. doi:10.1016/S0074-7696(05)43004-1
2. Liu JL, LeRoith D. Insulin-like growth factor I is essential for postnatal growth in response to growth hormone. Endocrinology (1999) 140(11):5178–84. doi:10.1210/endo.140.11.7151
3. Duan C, Ren H, Gao S. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation. Gen Comp Endocrinol (2010) 167(3):344–51. doi:10.1016/j.ygcen.2010.04.009
4. Ohlsson C, Mohan S, Sjogren K, Tivesten A, Isgaard J, Isaksson O, et al. The role of liver-derived insulin-like growth factor-I. Endocr Rev (2009) 30(5):494–535. doi:10.1210/er.2009-0010
5. Emmerson E, Campbell L, Davies FC, Ross NL, Ashcroft GS, Krust A, et al. Insulin-like growth factor-1 promotes wound healing in estrogen-deprived mice: new insights into cutaneous IGF-1R/ERalpha cross talk. J Invest Dermatol (2012) 132(12):2838–48. doi:10.1038/jid.2012.228
6. Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell (1993) 75(1):59–72. doi:10.1016/S0092-8674(05)80084-4
7. Rajpathak SN, Gunter MJ, Wylie-Rosett J, Ho GY, Kaplan RC, Muzumdar R, et al. The role of insulin-like growth factor-I and its binding proteins in glucose homeostasis and type 2 diabetes. Diabetes Metab Res Rev (2009) 25(1):3–12. doi:10.1002/dmrr.919
8. Lioutas VA, Alfaro-Martinez F, Bedoya F, Chung CC, Pimentel DA, Novak V. Intranasal insulin and insulin-like growth factor 1 as neuroprotectants in acute ischemic stroke. Transl Stroke Res (2015) 6(4):264–75. doi:10.1007/s12975-015-0409-7
9. Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis, and life span. Annu Rev Physiol (2008) 70:191–212. doi:10.1146/annurev.physiol.70.113006.100533
10. Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev (2002) 23(6):824–54. doi:10.1210/er.2001-0033
11. Duan C, Xu Q. Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen Comp Endocrinol (2005) 142(1–2):44–52. doi:10.1016/j.ygcen.2004.12.022
12. Baxter RC. IGF binding proteins in cancer: mechanistic and clinical insights. Nat Rev Cancer (2014) 14(5):329–41. doi:10.1038/nrc3720
13. Clemmons DR. Role of IGF binding proteins in regulating metabolism. Trends Endocrinol Metab (2016) 27(6):375–91. doi:10.1016/j.tem.2016.03.019
14. Upton Z, Chan SJ, Steiner DF, Wallace JC, Ballard FJ. Evolution of insulin-like growth factor binding proteins. Growth Regul (1993) 3(1):29–32.
15. Duan C, Ding J, Li Q, Tsai W, Pozios K. Insulin-like growth factor binding protein 2 is a growth inhibitory protein conserved in zebrafish. Proc Natl Acad Sci U S A (1999) 96(26):15274–9. doi:10.1073/pnas.96.26.15274
16. Daza DO, Sundstrom G, Bergqvist CA, Duan C, Larhammar D. Evolution of the insulin-like growth factor binding protein (IGFBP) family. Endocrinology (2011) 152(6):2278–89. doi:10.1210/en.2011-0047
17. Kamei H, Lu L, Jiao S, Li Y, Gyrup C, Laursen LS, et al. Duplication and diversification of the hypoxia-inducible IGFBP-1 gene in zebrafish. PLoS One (2008) 3(8):e3091. doi:10.1371/journal.pone.0003091
18. Zhou J, Li W, Kamei H, Duan C. Duplication of the IGFBP-2 gene in teleost fish: protein structure and functionality conservation and gene expression divergence. PLoS One (2008) 3(12):e3926. doi:10.1371/journal.pone.0003926
19. Li M, Li Y, Lu L, Wang X, Gong Q, Duan C. Structural, gene expression, and functional analysis of the fugu (Takifugu rubripes) insulin-like growth factor binding protein-4 gene. Am J Physiol Regul Integr Comp Physiol (2009) 296(3):R558–66. doi:10.1152/ajpregu.90439.2008
20. Wang X, Lu L, Li Y, Li M, Chen C, Feng Q, et al. Molecular and functional characterization of two distinct IGF binding protein-6 genes in zebrafish. Am J Physiol Regul Integr Comp Physiol (2009) 296(5):R1348–57. doi:10.1152/ajpregu.90969.2008
21. Dai W, Kamei H, Zhao Y, Ding J, Du Z, Duan C. Duplicated zebrafish insulin-like growth factor binding protein-5 genes with split functional domains: evidence for evolutionarily conserved IGF binding, nuclear localization, and transactivation activity. FASEB J (2010) 24(6):2020–9. doi:10.1096/fj.09-149435
22. Macqueen DJ, Garcia de la Serrana D, Johnston IA. Evolution of ancient functions in the vertebrate insulin-like growth factor system uncovered by study of duplicated salmonid fish genomes. Mol Biol Evol (2013) 30(5):1060–76. doi:10.1093/molbev/mst017
23. Shimizu M, Dickhoff WW. Circulating insulin-like growth factor binding proteins in fish: their identities and physiological regulation. Gen Comp Endocrinol (2017) 252:150–61. doi:10.1016/j.ygcen.2017.08.002
24. Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, et al. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol (2017) 18(1):111. doi:10.1186/s13059-017-1241-z
25. Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev (1995) 16:3–34. doi:10.1210/edrv-16-1-3
26. Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev (1999) 20(6):761–87. doi:10.1210/edrv.20.6.0382
27. Grotendorst GR, Lau LF, Perbal B. CCN proteins are distinct from and should not be considered members of the insulin-like growth factor-binding protein superfamily. Endocrinology (2000) 141(6):2254–6. doi:10.1210/endo.141.6.7485
28. Rajaram S, Baylink DJ, Mohan S. Insulin-like growth factor-binding proteins in serum and other biological fluids: regulation and functions. Endocr Rev (1997) 18(6):801–31. doi:10.1210/edrv.18.6.0321
29. Guler HP, Zapf J, Schmid C, Froesch ER. Insulin-like growth factors I and II in healthy man. Estimations of half-lives and production rates. Acta Endocrinol (Copenh) (1989) 121(6):753–8.
30. Baxter RC, Martin JL. Structure of the Mr 140,000 growth hormone-dependent insulin-like growth factor binding protein complex: determination by reconstitution and affinity-labeling. Proc Natl Acad Sci U S A (1989) 86(18):6898–902. doi:10.1073/pnas.86.18.6898
31. Leong SR, Baxter RC, Camerato T, Dai J, Wood WI. Structure and functional expression of the acid-labile subunit of the insulin-like growth factor-binding protein complex. Mol Endocrinol (1992) 6(6):870–6. doi:10.1210/mend.6.6.1379671
32. Lewitt MS, Saunders H, Phuyal JL, Baxter RC. Complex formation by human insulin-like growth factor-binding protein-3 and human acid-labile subunit in growth hormone-deficient rats. Endocrinology (1994) 134(6):2404–9. doi:10.1210/endo.134.6.7514998
33. Yakar S, Rosen CJ, Bouxsein ML, Sun H, Mejia W, Kawashima Y, et al. Serum complexes of insulin-like growth factor-1 modulate skeletal integrity and carbohydrate metabolism. FASEB J (2009) 23(3):709–19. doi:10.1096/fj.08-118976
34. Nakae J, Kido Y, Accili D. Distinct and overlapping functions of insulin and IGF-I receptors. Endocr Rev (2001) 22(6):818–35. doi:10.1210/edrv.22.6.0452
35. Daughaday WH, Rotwein P. Insulin-like growth factors I and II. Peptide, messenger ribonucleic acid and gene structures, serum, and tissue concentrations. Endocr Rev (1989) 10(1):68–91. doi:10.1210/edrv-10-1-68
36. Blum WF, Albertsson-Wikland K, Rosberg S, Ranke MB. Serum levels of insulin-like growth factor I (IGF-I) and IGF binding protein 3 reflect spontaneous growth hormone secretion. J Clin Endocrinol Metab (1993) 76(6):1610–6. doi:10.1210/jcem.76.6.7684744
37. Maures TJ, Duan C. Structure, developmental expression, and physiological regulation of zebrafish IGF binding protein-1. Endocrinology (2002) 143(7):2722–31. doi:10.1210/endo.143.7.8905
38. Kajimura S, Aida K, Duan C. Understanding hypoxia-induced gene expression in early development: in vitro and in vivo analysis of hypoxia-inducible factor 1-regulated zebra fish insulin-like growth factor binding protein 1 gene expression. Mol Cell Biol (2006) 26(3):1142–55. doi:10.1128/MCB.26.3.1142-1155.2006
39. Suwanichkul A, Allander SV, Morris SL, Powell DR. Glucocorticoids and insulin regulate expression of the human gene for insulin-like growth factor-binding protein-1 through proximal promoter elements. J Biol Chem (1994) 269(49):30835–41.
40. O’Brien RM, Noisin EL, Suwanichkul A, Yamasaki T, Lucas PC, Wang JC, et al. Hepatic nuclear factor 3- and hormone-regulated expression of the phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein 1 genes. Mol Cell Biol (1995) 15(3):1747–58. doi:10.1128/MCB.15.3.1747
41. McLellan KC, Hooper SB, Bocking AD, Delhanty PJ, Phillips ID, Hill DJ, et al. Prolonged hypoxia induced by the reduction of maternal uterine blood flow alters insulin-like growth factor-binding protein-1 (IGFBP-1) and IGFBP-2 gene expression in the ovine fetus. Endocrinology (1992) 131(4):1619–28. doi:10.1210/endo.131.4.1382958
42. Kajimura S, Duan C. Insulin-like growth factor-binding protein-1: an evolutionarily conserved fine tuner of insulin-like growth factor action under catabolic and stressful conditions. J Fish Biol (2007) 71:309–25. doi:10.1111/j.1095-8649.2007.01606.x
43. Schneider MR, Wolf E, Hoeflich A, Lahm H. IGF-binding protein-5: flexible player in the IGF system and effector on its own. J Endocrinol (2002) 172(3):423–40. doi:10.1677/joe.0.1720423
44. Zhou R, Diehl D, Hoeflich A, Lahm H, Wolf E. IGF-binding protein-4: biochemical characteristics and functional consequences. J Endocrinol (2003) 178(2):177–93. doi:10.1677/joe.0.1780177
45. Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, et al. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci U S A (1999) 96(13):7324–9. doi:10.1073/pnas.96.13.7324
46. Sitar T, Popowicz GM, Siwanowicz I, Huber R, Holak TA. Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins. Proc Natl Acad Sci U S A (2006) 103(35):13028–33. doi:10.1073/pnas.0605652103
47. Duan C, Clemmons DR. Differential expression and biological effects of insulin-like growth factor-binding protein-4 and -5 in vascular smooth muscle cells. J Biol Chem (1998) 273(27):16836–42. doi:10.1074/jbc.273.27.16836
48. Schneider MR, Lahm H, Wu M, Hoeflich A, Wolf E. Transgenic mouse models for studying the functions of insulin-like growth factor-binding proteins. FASEB J (2000) 14(5):629–40. doi:10.1096/fasebj.14.5.629
49. Oxvig C. The role of PAPP-A in the IGF system: location, location, location. J Cell Commun Signal (2015) 9(2):177–87. doi:10.1007/s12079-015-0259-9
50. Lawrence JB, Oxvig C, Overgaard MT, Sottrup-Jensen L, Gleich GJ, Hays LG, et al. The insulin-like growth factor (IGF)-dependent IGF binding protein-4 protease secreted by human fibroblasts is pregnancy-associated plasma protein-A. Proc Natl Acad Sci U S A (1999) 96(6):3149–53. doi:10.1073/pnas.96.6.3149
51. Conover CA, Bale LK, Overgaard MT, Johnstone EW, Laursen UH, Fuchtbauer EM, et al. Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development. Development (2004) 131(5):1187–94. doi:10.1242/dev.00997
52. Ning Y, Schuller AG, Conover CA, Pintar JE. Insulin-like growth factor (IGF) binding protein-4 is both a positive and negative regulator of IGF activity in vivo. Mol Endocrinol (2008) 22(5):1213–25. doi:10.1210/me.2007-0536
53. Besnard N, Pisselet C, Monniaux D, Monget P. Proteolytic activity degrading insulin-like growth factor-binding protein-2, -3, -4, and -5 in healthy growing and atretic follicles in the pig ovary. Biol Reprod (1997) 56(4):1050–8. doi:10.1095/biolreprod56.4.1050
54. Busby WH Jr, Nam TJ, Moralez A, Smith C, Jennings M, Clemmons DR. The complement component C1s is the protease that accounts for cleavage of insulin-like growth factor-binding protein-5 in fibroblast medium. J Biol Chem (2000) 275(48):37638–44. doi:10.1074/jbc.M006107200
55. Shi Z, Xu W, Loechel F, Wewer UM, Murphy LJ. ADAM 12, a disintegrin metalloprotease, interacts with insulin-like growth factor-binding protein-3. J Biol Chem (2000) 275(24):18574–80. doi:10.1074/jbc.M002172200
56. Imai Y, Busby WH Jr, Smith CE, Clarke JB, Garmong AJ, Horwitz GD, et al. Protease-resistant form of insulin-like growth factor-binding protein 5 is an inhibitor of insulin-like growth factor-I actions on porcine smooth muscle cells in culture. J Clin Invest (1997) 100(10):2596–605. doi:10.1172/JCI119803
57. Yin P, Xu Q, Duan C. Paradoxical actions of endogenous and exogenous insulin-like growth factor-binding protein-5 revealed by RNA interference analysis. J Biol Chem (2004) 279(31):32660–6. doi:10.1074/jbc.M401378200
58. Florini JR, Magri KA, Ewton DZ, James PL, Grindstaff K, Rotwein PS. “Spontaneous” differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J Biol Chem (1991) 266(24):15917–23.
59. Ren H, Yin P, Duan C. IGFBP-5 regulates muscle cell differentiation by binding to IGF-II and switching on the IGF-II auto-regulation loop. J Cell Biol (2008) 182(5):979–91. doi:10.1083/jcb.200712110
60. Zheng B, Duan C, Clemmons DR. The effect of extracellular matrix proteins on porcine smooth muscle cell insulin-like growth factor (IGF) binding protein-5 synthesis and responsiveness to IGF-I. J Biol Chem (1998) 273(15):8994–9000. doi:10.1074/jbc.273.15.8994
61. Kim KS, Seu YB, Baek SH, Kim MJ, Kim KJ, Kim JH, et al. Induction of cellular senescence by insulin-like growth factor binding protein-5 through a p53-dependent mechanism. Mol Biol Cell (2007) 18(11):4543–52. doi:10.1091/mbc.E07-03-0280
62. Duan C, Bauchat JR, Hsieh T. Phosphatidylinositol 3-kinase is required for insulin-like growth factor-I-induced vascular smooth muscle cell proliferation and migration. Circ Res (2000) 86(1):15–23. doi:10.1161/01.RES.86.1.15
63. Zaina S, Pettersson L, Ahren B, Branen L, Hassan AB, Lindholm M, et al. Insulin-like growth factor II plays a central role in atherosclerosis in a mouse model. J Biol Chem (2002) 277(6):4505–11. doi:10.1074/jbc.M108061200
64. Nam TJ, Busby WH Jr, Rees C, Clemmons DR. Thrombospondin and osteopontin bind to insulin-like growth factor (IGF)-binding protein-5 leading to an alteration in IGF-I-stimulated cell growth. Endocrinology (2000) 141(3):1100–6. doi:10.1210/endo.141.3.7386
65. Nichols TC, Busby WH Jr, Merricks E, Sipos J, Rowland M, Sitko K, et al. Protease-resistant insulin-like growth factor (IGF)-binding protein-4 inhibits IGF-I actions and neointimal expansion in a porcine model of neointimal hyperplasia. Endocrinology (2007) 148(10):5002–10. doi:10.1210/en.2007-0571
66. Resch ZT, Simari RD, Conover CA. Targeted disruption of the pregnancy-associated plasma protein-A gene is associated with diminished smooth muscle cell response to insulin-like growth factor-I and resistance to neointimal hyperplasia after vascular injury. Endocrinology (2006) 147(12):5634–40. doi:10.1210/en.2006-0493
67. Harrington SC, Simari RD, Conover CA. Genetic deletion of pregnancy-associated plasma protein-A is associated with resistance to atherosclerotic lesion development in apolipoprotein E-deficient mice challenged with a high-fat diet. Circ Res (2007) 100(12):1696–702. doi:10.1161/CIRCRESAHA.106.146183
68. Conover CA, Mason MA, Bale LK, Harrington SC, Nyegaard M, Oxvig C, et al. Transgenic overexpression of pregnancy-associated plasma protein-A in murine arterial smooth muscle accelerates atherosclerotic lesion development. Am J Physiol Heart Circ Physiol (2010) 299(2):H284–91. doi:10.1152/ajpheart.00904.2009
69. Conover CA, Bale LK, Oxvig C. Targeted inhibition of pregnancy-associated plasma protein-A activity reduces atherosclerotic plaque burden in mice. J Cardiovasc Transl Res (2016) 9(1):77–9. doi:10.1007/s12265-015-9666-9
70. Jones JI, Gockerman A, Busby WH Jr, Wright G, Clemmons DR. Insulin-like growth factor binding protein 1 stimulates cell migration and binds to the alpha 5 beta 1 integrin by means of its Arg-Gly-Asp sequence. Proc Natl Acad Sci U S A (1993) 90(22):10553–7. doi:10.1073/pnas.90.22.10553
71. Feng N, Zhang Z, Wang Z, Zheng H, Qu F, He X, et al. Insulin-like growth factor binding protein-2 promotes adhesion of endothelial progenitor cells to endothelial cells via integrin alpha5beta1. J Mol Neurosci (2015) 57(3):426–34. doi:10.1007/s12031-015-0589-3
72. Schedlich LJ, Le Page SL, Firth SM, Briggs LJ, Jans DA, Baxter RC. Nuclear import of insulin-like growth factor-binding protein-3 and -5 is mediated by the importin beta subunit. J Biol Chem (2000) 275(31):23462–70. doi:10.1074/jbc.M002208200
73. Xu Q, Li S, Zhao Y, Maures TJ, Yin P, Duan C. Evidence that IGF binding protein-5 functions as a ligand-independent transcriptional regulator in vascular smooth muscle cells. Circ Res (2004) 94(5):E46–54. doi:10.1161/01.RES.0000124761.62846.DF
74. Hsieh T, Gordon RE, Clemmons DR, Busby WH Jr, Duan C. Regulation of vascular smooth muscle cell responses to insulin-like growth factor (IGF)-I by local IGF-binding proteins. J Biol Chem (2003) 278(44):42886–92. doi:10.1074/jbc.M303835200
75. Zhao Y, Yin P, Bach LA, Duan C. Several acidic amino acids in the N-domain of insulin-like growth factor-binding protein-5 are important for its transactivation activity. J Biol Chem (2006) 281(20):14184–91. doi:10.1074/jbc.M506941200
76. Zhong Y, Lu L, Zhou J, Li Y, Liu Y, Clemmons DR, et al. IGF binding protein 3 exerts its ligand-independent action by antagonizing BMP in zebrafish embryos. J Cell Sci (2011) 124(Pt 11):1925–35. doi:10.1242/jcs.082644
77. Zhou J, Xiang J, Zhang S, Duan C. Structural and functional analysis of the amphioxus IGFBP gene uncovers ancient origin of IGF-independent functions. Endocrinology (2013) 154(10):3753–63. doi:10.1210/en.2013-1201
78. Schedlich LJ, O’Han MK, Leong GM, Baxter RC. Insulin-like growth factor binding protein-3 prevents retinoid receptor heterodimerization: implications for retinoic acid-sensitivity in human breast cancer cells. Biochem Biophys Res Commun (2004) 314(1):83–8. doi:10.1016/j.bbrc.2003.12.049
79. Azar WJ, Azar SH, Higgins S, Hu JF, Hoffman AR, Newgreen DF, et al. IGFBP-2 enhances VEGF gene promoter activity and consequent promotion of angiogenesis by neuroblastoma cells. Endocrinology (2011) 152(9):3332–42. doi:10.1210/en.2011-1121
80. Zhu W, Shiojima I, Ito Y, Li Z, Ikeda H, Yoshida M, et al. IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature (2008) 454(7202):345–9. doi:10.1038/nature07027
81. Ning Y, Schuller AG, Bradshaw S, Rotwein P, Ludwig T, Frystyk J, et al. Diminished growth and enhanced glucose metabolism in triple knockout mice containing mutations of insulin-like growth factor binding protein-3, -4, and -5. Mol Endocrinol (2006) 20(9):2173–86. doi:10.1210/me.2005-0196
82. Zhang C, Lu L, Li Y, Wang X, Zhou J, Liu Y, et al. IGF binding protein-6 expression in vascular endothelial cells is induced by hypoxia and plays a negative role in tumor angiogenesis. Int J Cancer (2012) 130(9):2003–12. doi:10.1002/ijc.26201
83. Oh Y, Muller HL, Lamson G, Rosenfeld RG. Insulin-like growth factor (IGF)-independent action of IGF-binding protein-3 in Hs578T human breast cancer cells. Cell surface binding and growth inhibition. J Biol Chem (1993) 268(20):14964–71.
84. Valentinis B, Bhala A, DeAngelis T, Baserga R, Cohen P. The human insulin-like growth factor (IGF) binding protein-3 inhibits the growth of fibroblasts with a targeted disruption of the IGF-I receptor gene. Mol Endocrinol (1995) 9(3):361–7. doi:10.1210/mend.9.3.7539889
85. Leal SM, Liu Q, Huang SS, Huang JS. The type V transforming growth factor beta receptor is the putative insulin-like growth factor-binding protein 3 receptor. J Biol Chem (1997) 272(33):20572–6. doi:10.1074/jbc.272.33.20572
86. Huang SS, Ling TY, Tseng WF, Huang YH, Tang FM, Leal SM, et al. Cellular growth inhibition by IGFBP-3 and TGF-beta1 requires LRP-1. FASEB J (2003) 17(14):2068–81. doi:10.1096/fj.03-0256com
87. Strickland DK, Gonias SL, Argraves WS. Diverse roles for the LDL receptor family. Trends Endocrinol Metab (2002) 13(2):66–74. doi:10.1016/S1043-2760(01)00526-4
88. Shen X, Xi G, Maile LA, Wai C, Rosen CJ, Clemmons DR. Insulin-like growth factor (IGF) binding protein 2 functions coordinately with receptor protein tyrosine phosphatase beta and the IGF-I receptor to regulate IGF-I-stimulated signaling. Mol Cell Biol (2012) 32(20):4116–30. doi:10.1128/MCB.01011-12
89. Xi G, Wai C, DeMambro V, Rosen CJ, Clemmons DR. IGFBP-2 directly stimulates osteoblast differentiation. J Bone Miner Res (2014) 29(11):2427–38. doi:10.1002/jbmr.2282
90. Wood TL, Rogler LE, Czick ME, Schuller AG, Pintar JE. Selective alterations in organ sizes in mice with a targeted disruption of the insulin-like growth factor binding protein-2 gene. Mol Endocrinol (2000) 14(9):1472–82. doi:10.1210/mend.14.9.0517
91. Leu JI, Crissey MA, Craig LE, Taub R. Impaired hepatocyte DNA synthetic response posthepatectomy in insulin-like growth factor binding protein 1-deficient mice with defects in C/EBP beta and mitogen-activated protein kinase/extracellular signal-regulated kinase regulation. Mol Cell Biol (2003) 23(4):1251–9. doi:10.1128/MCB.23.4.1251-1259.2003
92. DeMambro VE, Clemmons DR, Horton LG, Bouxsein ML, Wood TL, Beamer WG, et al. Gender-specific changes in bone turnover and skeletal architecture in igfbp-2-null mice. Endocrinology (2008) 149(5):2051–61. doi:10.1210/en.2007-1068
93. Gray A, Aronson WJ, Barnard RJ, Mehta H, Wan J, Said J, et al. Global Igfbp1 deletion does not affect prostate cancer development in a c-Myc transgenic mouse model. J Endocrinol (2011) 211(3):297–304. doi:10.1530/JOE-11-0240
94. Powell-Braxton L, Hollingshead P, Warburton C, Dowd M, Pitts-Meek S, Dalton D, et al. IGF-I is required for normal embryonic growth in mice. Genes Dev (1993) 7(12B):2609–17. doi:10.1101/gad.7.12b.2609
95. Dean EJ, Davis JC, Davis RW, Petrov DA. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet (2008) 4(7):e1000113. doi:10.1371/journal.pgen.1000113
96. Ewen-Campen B, Mohr SE, Hu Y, Perrimon N. Accessing the phenotype gap: enabling systematic investigation of paralog functional complexity with CRISPR. Dev Cell (2017) 43(1):6–9. doi:10.1016/j.devcel.2017.09.020
97. Rogers RL, Bedford T, Hartl DL. Formation and longevity of chimeric and duplicate genes in Drosophila melanogaster. Genetics (2009) 181(1):313–22. doi:10.1534/genetics.108.091538
98. Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature (1997) 388(6638):167–71. doi:10.1038/40618
99. Vavouri T, Semple JI, Lehner B. Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet (2008) 24(10):485–8. doi:10.1016/j.tig.2008.08.005
100. Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics (1999) 151(4):1531–45.
101. Wood AW, Schlueter PJ, Duan C. Targeted knockdown of insulin-like growth factor binding protein-2 disrupts cardiovascular development in zebrafish embryos. Mol Endocrinol (2005) 19:1024–34. doi:10.1210/me.2004-0392
102. El-Brolosy MA, Stainier DYR. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet (2017) 13(7):e1006780. doi:10.1371/journal.pgen.1006780
103. Li Y, Xiang J, Duan C. Insulin-like growth factor-binding protein-3 plays an important role in regulating pharyngeal skeleton and inner ear formation and differentiation. J Biol Chem (2005) 280(5):3613–20. doi:10.1074/jbc.M411479200
104. Kajimura S, Aida K, Duan C. Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc Natl Acad Sci U S A (2005) 102(4):1240–5. doi:10.1073/pnas.0407443102
105. Leu JI, Crissey MA, Taub R. Massive hepatic apoptosis associated with TGF-beta1 activation after Fas ligand treatment of IGF binding protein-1-deficient mice. J Clin Invest (2003) 111(1):129–39. doi:10.1172/JCI16712
106. Ning Y, Hoang B, Schuller AG, Cominski TP, Hsu MS, Wood TL, et al. Delayed mammary gland involution in mice with mutation of the insulin-like growth factor binding protein 5 gene. Endocrinology (2007) 148(5):2138–47. doi:10.1210/en.2006-0041
107. Dai W, Bai Y, Hebda L, Zhong X, Liu J, Kao J, et al. Calcium deficiency-induced and TRP channel-regulated IGF1R-PI3K-Akt signaling regulates abnormal epithelial cell proliferation. Cell Death Differ (2014) 21(4):568–81. doi:10.1038/cdd.2013.177
108. Lundin LG, Larhammar D, Hallbook F. Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. J Struct Funct Genomics (2003) 3(1–4):53–63. doi:10.1023/A:1022600813840
109. Sundstrom G, Larsson TA, Larhammar D. Phylogenetic and chromosomal analyses of multiple gene families syntenic with vertebrate Hox clusters. BMC Evol Biol (2008) 8:254. doi:10.1186/1471-2148-8-254
Keywords: insulin-like growth factor, insulin-like growth factor-binding protein, insulin-like growth factor 1 receptor, insulin-like growth factor signaling, evolution
Citation: Allard JB and Duan C (2018) IGF-Binding Proteins: Why Do They Exist and Why Are There So Many? Front. Endocrinol. 9:117. doi: 10.3389/fendo.2018.00117
Received: 11 January 2018; Accepted: 08 March 2018;
Published: 09 April 2018
Edited by:
Andreas Hoeflich, Leibniz-Institut für Nutztierbiologie (FBN), GermanyReviewed by:
Taisen Iguchi, National Institute for Basic Biology, JapanTom Ole Nilsen, University of Bergen, Norway
Isabel Navarro, Universitat de Barcelona, Spain
Copyright: © 2018 Allard and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cunming Duan, Y2R1YW5AdW1pY2guZWR1
 John B. Allard
John B. Allard Cunming Duan
Cunming Duan