
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 12 March 2018
Sec. Experimental Endocrinology
Volume 9 - 2018 | https://doi.org/10.3389/fendo.2018.00080
This article is part of the Research Topic Current Perspectives on Insulin-like Growth Factor Binding Protein (IGFBP) Research View all 16 articles
The insulin-like growth factor (Igf) binding protein (Igfbp) family has a broad range of physiological functions and a fascinating evolutionary history. This review focuses on the Igfbps of teleost fishes, where genome duplication events have diversified gene repertoire, function, and physiological regulation—with six core Igfbps expanded into a family of over twenty genes in some lineages. In addition to briefly summarizing the current state of knowledge on teleost Igfbp evolution, function, and expression-level regulation, we highlight gaps in our understanding and promising areas for future work.
The insulin-like growth factor (Igf) binding protein (Igfbps) are highly studied, especially in mammals, and a vast literature has emerged on their roles as mediators of Igf signaling events, and diverse functions that extend beyond Igf regulation. This review focuses on the Igfbp family of teleost fishes, which remains poorly understood compared to the mammalian system. Our goal is to summarize the status of knowledge on teleost Igfbps in an evolutionary context, considering work on gene function and physiological regulation, in addition to phylogenetics and genomics. Our coverage of literature is non-encompassing, and we point the reader to additional reviews. The scope of the review is exclusive to the “true” Igfbps, which each bind Igfs with high affinity, rather than the broader proposed superfamily containing Igfbp-related proteins [reviewed in Ref. (1)], that are distantly related in both sequence and function (2). We also assume that the reader has prior knowledge of the core genetic components of the Igf system, where comprehensive reviews with a non-mammalian focus already exist (3–6).
Gene duplication and subsequent divergence is central to the evolutionary “narrative” of the Igfbp family. While it is long-established that many vertebrates possess six ancestral subtypes (Igfbp-1, -2, -3, -4, -5, and -6), with the primary cDNAs first reported over 25 years ago [e.g., Ref. (7, 8)], their evolutionary origins were elucidated more recently. An important study reported in 2011 (9), built on past work revealing linkage between Igfbp genes and Hox clusters [e.g., Ref. (10); Hox clusters being well-established markers of genome duplication events], to present a realistic scenario for the origin and expansion of core Igfbp subtypes. The hypothesis is that an ancestral Igfbp gene was duplicated in tandem during an early stage of vertebrate evolution to produce a pair of Igfbp genes (9, 10). Subsequently, two genome duplication events in the ancestor to extant vertebrates (11) led one gene to give rise to Igfbp-1, -2, and -4, and the other to Igfbp-3, -5, and -6. A single Igfbp is present in amphioxus, a chordate that did not undergo the same duplications, and this molecule failed to bind Igf-I or Igf-II, indicating that Igf-binding is either a vertebrate-specific function (12), or was secondarily lost. The same study confirmed that Igf-independent functions had evolved before vertebrates (12).
It is also important to remember that the diversification of the core vertebrate Igfbp system occurred alongside expansions in other key gene families within the Igf system, including both hormones (13) and receptors (14). It now seems certain that the early vertebrate genome duplication events were crucial for the evolution of distinct insulin and Igf systems [e.g., Ref. (15)]. For the remainder of this review, we focus on the Igfbp system of teleosts, where additional genetic expansions—some dramatic—have been recently characterized.
A further round of genome duplication occurred in the ancestor to extant teleost fishes (i.e., around half of known vertebrate species) 300–350 million years ago (11). This led to retention of duplicated copies (paralogs) for all the core Igfbp subtypes barring Igfbp-4, where one paralog was lost early (9, 16). In different lineages that have not experienced further genome duplication events, the number of Igfbps retained is variable, but always higher than mammals and most non-teleosts. For example, zebrafish (Danio rerio), the most studied teleost in terms of Igfbp function, retains nine unique genes. This includes paralog pairs for Igfbp-1 (17), -2 (18), -5 (19), and -6 (20), along with a single Igfbp-3 copy and no Igfbp-4 gene, owing to lineage-specific losses (9, 16). The phylogenetic relationships of teleost Igfbp paralogs have been established using robust methods (9, 16). An “-a”/“-b” nomenclature common to different teleosts is preferred (e.g., “Igfbp-1a” and “-1b”) (16), as it acknowledges a common ancestral origin from the same duplication event, while accommodating zebrafish nomenclature [e.g., Ref. (17–20)].
Several teleost lineages experienced additional rounds of genome duplication. This includes a well-studied event ~95 million years ago in the salmonid ancestor (21, 22) that caused dramatic genetic expansions within the Igf system (summarized in Figure 1). For example, we reported in 2013 that salmonids retain at least 19 unique Igfbp genes, with salmonid-specific paralogs of igfbp-1a, -1b, -2b, -3a, -3b, -5b, -6a, and -6b (16). We proposed a nomenclature with either “1” or “2” after the “a” and “b” teleost symbols (e.g., “igfbp-1a1” and “-1a2”). Several of these Igfbp pairs are highly divergent compared to the genome-wide average for paralogs retained from the salmonid genome duplication event (Figure 1; e.g., sharing <80% amino acid identity, compared to an average of ~93% across thousands of paralog pairs) (23). This points to functional divergence at the protein level that remains entirely unexplored.
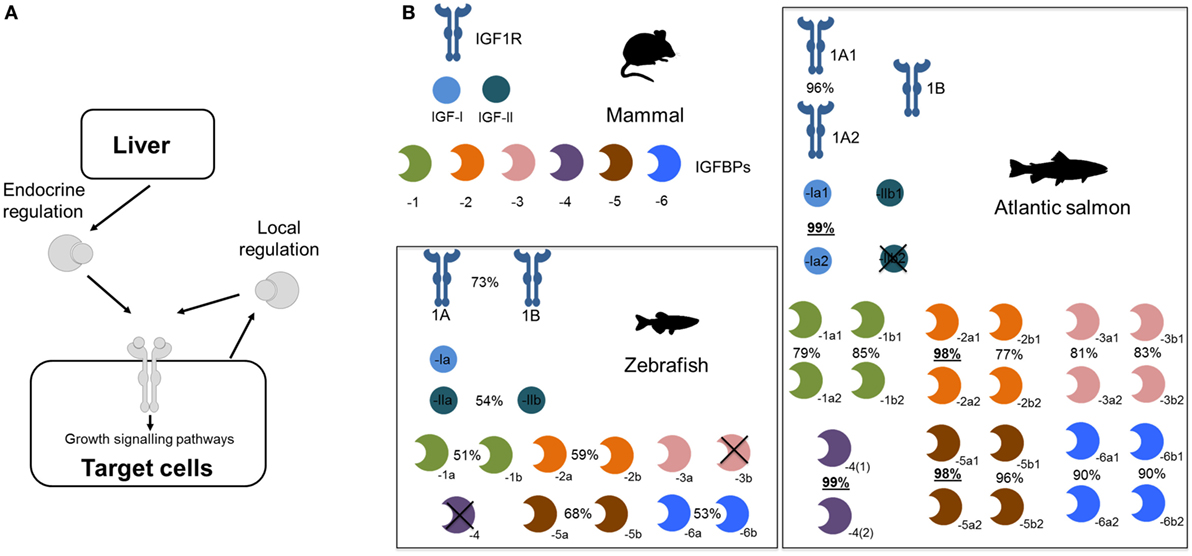
Figure 1. Expansion of the core insulin-like growth factor (Igf) system—including Igfbps—during teleost evolution. (A) Simplified depiction of Igf system. (B) Core Igfbp system components (i.e., proteins encoded by distinct genes) in different vertebrate groups, contrasting a typical mammalian system with that of two teleost lineages. For teleosts, % identity is shown for paralogous amino acid sequences. For Atlantic salmon, the underlined % identities highlight paralog pairs residing in regions of the genome that experienced a delay in cytological rediploidization after genome duplication (24), a process required for paralogs to diverge in sequence on distinct chromosomes—hence, these genes have had less evolutionary time to diverge, leading to extremely high identity. Phylogenetic relationships of the Igfbp families from these different lineages, along with another group of teleosts that experienced a separate lineage-specific genome duplication event, are depicted in Figure 2.
More recently, an improved understanding of the complexities of genome evolution following the salmonid genome duplication, which was a spontaneous genome doubling event (or “autotetraploidization”) (21, 24), led to the discovery of salmonid Igfbp paralog pairs for igfbp-4, igfbp-2a, and igfbp-5a (24), which share extremely similar sequences and were previously unrecognized or ignored as alleles (see Figure 1; legend contains additional information). Thus, some salmonid species, including the commercially important Atlantic salmon (Salmo salar), possess 22 unique igfbp genes, with 11 paralog pairs (Figure 1), some of which may have evolved adaptively (25). Remarkably, every possible Igfbp paralog generated from the salmonid-specific genome duplication was evidently maintained, despite the genome-wide paralog retention rate being around 50% in the same species (21). We and others have also identified expansions to other core gene components of the Igf system due to the salmonid genome duplication, including Igf-I [e.g., Ref. (24, 26)], Igf-II (25), and Igf-1R (27). These paralogs remain of substantial interest, but we are at an early stage of understanding their roles in salmonid biology.
Additional lineage-specific genome duplication events have occurred in several teleost groups, including the ancestor to goldfish (Carassius auratus) and common carp (Cyprinus carpio). This event is younger than the salmonid-specific genome duplication event, occurring around 8–12 million years ago (28, 29). It also involved a distinct mechanism (“allotetraploidization”), where two species hybridized before genome duplication (28). This event created a large set of paralogs (28), some of which are known to have experienced functional divergence [e.g., Ref. (29, 30)]. However, no accompanying expansions to Igfbp repertoire are yet reported. To explore this knowledge gap, we performed a bioinformatic and phylogenetic analysis, revealing that common carp retains at least 17 unique igfbp genes, including paralog pairs for igfbp-1a, -1b, -2b, -3a, -5a, -5b, -6a, and -6b (see Figure 2; methods provided therein). To avoid confusion with the salmonid-specific paralogs, we suggest “α”/“β” is added to the existing teleost nomenclature when these duplicates are studied in the future (e.g., igfbp-1aα/-1aβ) (Figure 2). The results confirm that salmonids are not unique among teleosts in retaining a highly expanded Igfbp repertoire. In fact, as many vertebrate groups have experienced lineage-specific genome duplication events, both fishes and non-mammalian groups, including anuran frogs [e.g., Ref. (31)], it seems likely that many other species possess expanded Igfbp repertoires, contributing additional complexity to their growth regulation.
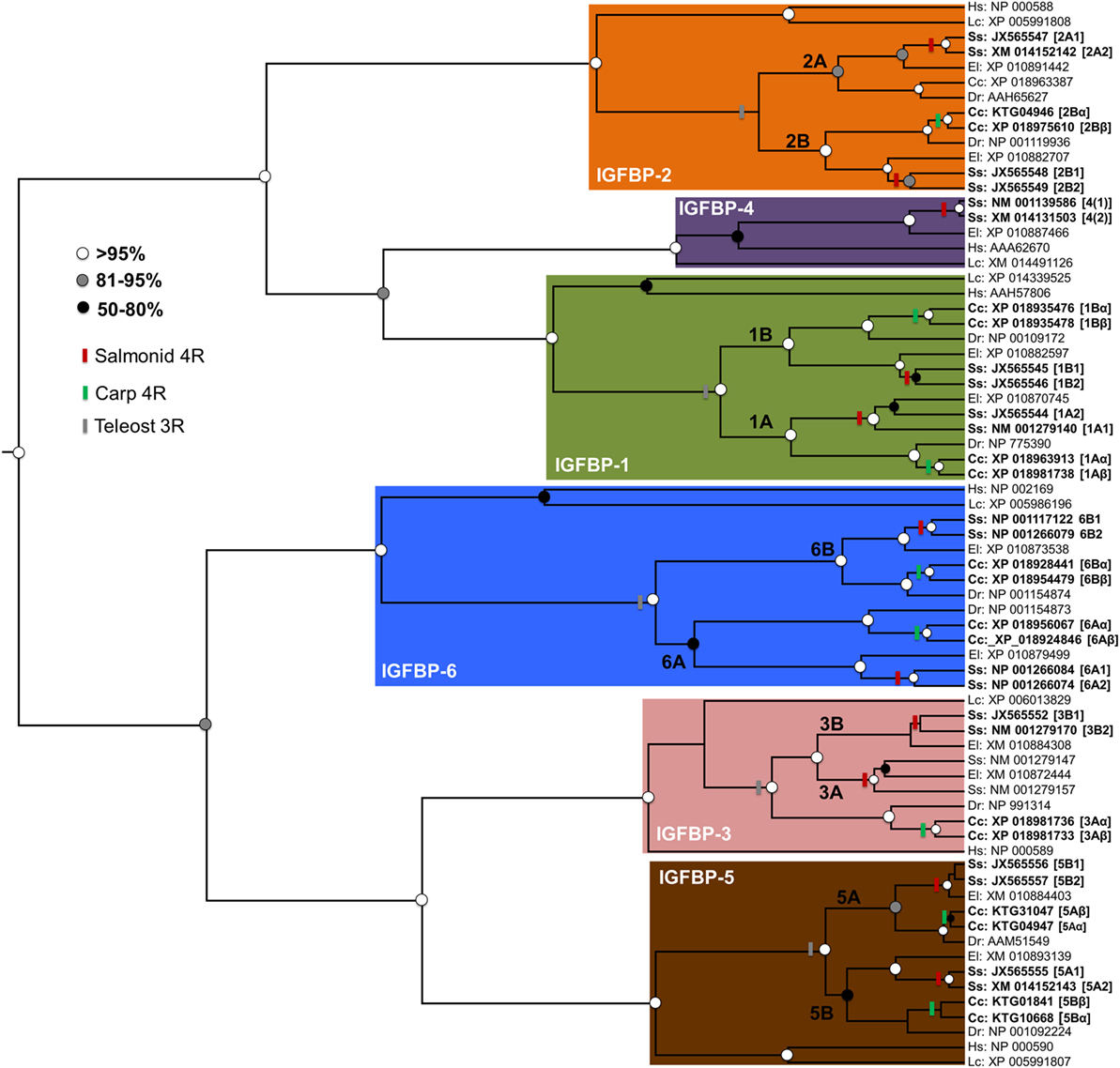
Figure 2. Independent evolutionary expansions to the Igfbp family of teleosts. A phylogenetic analysis was performed, including 71 unique Igfbp amino acid sequences from a standardized set of taxa: Human Homo sapiens (“Hs”), coelacanth Latimeria chalumnae (“Lc”), zebrafish Danio rerio (“Dr’”), common carp Cyprinus carpio (“Cc”), Atlantic salmon Salmo salar (“Ss”), and northern pike Esox lucius (“El,” from a sister lineage to salmonids that did not undergo the salmonid-specific genome duplication). Accession numbers are given for all sequences, which were gathered from the NCBI RefSeq database, facilitated by BLAST analyses (32). The sequences were aligned using Mafft V.7 (33) with default settings. Alignment quality filtering was done using the Guidance2 algorithm (34) to remove the least confidently aligned regions. This led to a high-confidence alignment of 212 amino acids positions (SI file 1). The alignment was used in Bayesian phylogenetic analysis, using methods published elsewhere (35). Briefly, this was done in BEAST v. 1.8 (36) using the best-fitting amino acid substitution model (JTT+G+I), determined by maximum likelihood via the IQ-tree server (37), along with a relaxed molecular clock model (38), allowing probabilistic estimation of the trees root [consistent with previous studies (9, 16)]. The tree is annotated to show genome duplication events in teleost evolutionary history, including in the teleost ancestor (“3R”) and additional events in the salmonid and carp lineages. The nomenclature for salmonid and carp paralogs is given as described in the text. Branch support values (posterior probability) are highlighted by circles placed on nodes, with colors matching a legend. Minor inconsistencies in branching patterns in some Igfbp clades (e.g., relating to the 3R or salmonid 4R) compared to other studies (16), can be explained by the short alignment length. Nonetheless, the tree demonstrates independent expansions within the salmonid and carp Igfbp repertoire, additional to paralogs retained in many other teleosts.
Many studies have investigated the physiological roles of Igfbp genes in the teleost lineage. Barring a few model species (e.g., zebrafish), it has been historically challenging to perform functional analyses in most teleosts, although this is changing in light of emerging genome-editing methods (see Perspectives and Future Work). Hence, while in mammals, Igf-dependent and Igf-independent functions have been widely demonstrated, the majority of studies in teleosts have failed to reach similar levels of functional insight. In fact, most work has focused on expression-level regulation of igfbp genes or proteins under a diverse set of experimental stimuli. It is also important to note that most teleost Igfbp research has focused on aquaculture species of high commercial value, including the salmonid, perciform (perch-like fish), pleuronectiform (flatfish), cypriniform (carp spp. and relatives), and siluriform (catfish) groups. This has led to a bias toward physiological processes relevant to commercial production, especially growth, muscle development, stress, and disease resistance. In this section, we briefly summarize the literature on teleost Igfbp function and regulation, considering the core vertebrate subtypes separately. We make attempts to distinguish Igfbp paralogs according to the evolutionary histories and nomenclature described above, although this is often not possible as many studies failed to distinguish paralogs, especially for the most recently discovered genes.
In mammals, Igfbp-1 is mainly produced in the liver and secreted to circulation, where it acts to limit Igf signaling in catabolic contexts, such as fasting, stress, and hypoxia (39). It is widely considered a negative regulator of somatic growth, reproduction, and development (4, 40); and interacts with cell surface integrins to stimulate cellular motility (41). It has Igf-dependent and Igf-independent functions, along with important roles in the regulation of metabolism [reviewed in Ref. (42)].
In salmonids, Igfbp-1a and Igfbp-1b are two of the three major circulatory Igfbps (43), first identified by molecular weight (20–15 and 28–32 kDa, respectively) (4, 44). It is likely that similar molecular weight Igfbps detected in others teleosts plasma are Igfbp-1 orthologs (45–49). Igfbp-1 encoding genes, as in mammals, are mainly expressed in teleost liver (16, 50–55). In zebrafish embryos, igfbp-1a mRNA is expressed during early development; while igfbp-1b is expressed later, after which time both paralogs become restricted to liver (17, 50). At the functional level, both Igfbp-1a and -1b of zebrafish can bind to Igf-I and Igf-II, but Igfbp-1b had a lower affinity for each hormone, and a lesser ability to downregulate Igf-I signaling (17). In other species, it has been reported that igfbp-1a genes are expressed in non-hepatic tissues, but typically at lower levels than in liver (16, 51, 52, 54–58). This supports the hypothesis that Igfbp-1a evolved more localized functions than Igfbp-1b (6).
Several teleost studies have reported protein or transcript level upregulation of Igfbp-1 genes during catabolic a process, which probably serves to downregulate growth by sequestering Igfs from Igf-1Rs, allowing allocation of resources to metabolic processes essential for survival. Consistent with these findings, overexpression of igfbp-1a (and igfbp-1a in zebrafish) in cypriniform embryos (17, 54, 59) caused growth and developmental retardation. Nutrient deprivation has been shown to increase circulatory Igfbp-1 proteins and igfbp-1 gene expression in liver (for both teleost paralogs, when distinguished) and skeletal muscle igfbp-1a expression, which is reversed by a return to anabolic conditions (50, 51, 55, 60–65). It has also been shown that dietary amino acid deficiency can upregulate igfbp-1 gene expression through a not well-described mechanism (64). Teleost igfbp-1 genes are also negatively regulated by growth hormone (Gh) and Igf-I, consistent with a negative role in growth regulation (44, 46, 48, 53, 57, 62).
Igfbp-1 expression in teleosts is also modulated by hormones others than the Igfs. For instance, igfbp-1a evidently plays a role in zebrafish sexual maturation, in a way that seems to differ from its classical anti-proliferative role. Specifically, igfbp-1a expression increases in response to T3 and the follicle stimulating (Fsh) hormones (well-known for stimulating spermatogonia proliferation) (58). The female sex hormone 17β-estradiol also increased Igfbp-1 secretion in striped bass liver explants (46).
Stress is also known to induce igfbp-1 gene expression in teleosts. Cortisol, the main stress hormone in vertebrates (66, 67), and other synthetic glucocorticoids such as dexamethasone increases both Igfbp-1a and Igfbp-1b circulatory levels, as well as gene expression in liver and cultured myotubes (43, 57, 62, 68, 69). Environmental stressors, such as hypoxia, confinement, temperature, heavy metals, and salinity, were also found to increase igfbp-1 expression (17, 49–53, 56, 61, 70, 71). It is also possible that igfbp-1 upregulation in response to food deprivation (see above) is related to a rise in cortisol, as some studies have found increasing levels of circulatory glucocorticoids in response to fasting (70, 72, 73). There is also emerging evidence from salmonids that igfbp-1a1 upregulation during infection serves a role in linking growth to innate immunity, potentially promoting downregulation of growth in favor of a more effective immune response (74). This expression response represents an example of salmonid-specific divergence in the regulation of Igfbp paralogs, as igfbp-1a2 was unaltered by infection in the same study (74).
In mammals, Igfbp-2 is highly expressed during embryonic stages, and more lowly expressed in adult tissues, with highest levels in liver, adipocytes, the central nervous and reproductive systems, heart, and kidney (75). Mice embryos overexpressing Igfbp-2 show a reduced growth rate, likely through reduced Igf availability (76). However, Igfbp-2 deletion in mice embryos does not have any significant effect on growth or development (77), which may indicate compensatory effects with other Igfbps. While the functional roles of Igfbp-2 remain relatively poorly established, recent studies have linked it to several pathological states. For example, Igfbp-2 may act as a tumor promoter (78) by suppressing epidermis growth factor receptor nuclear signaling (79). There is also increasing evidence that Igfbp-2 plays a role in mammalian bone formation (80).
Igfbp-2b is the third main circulatory Igfbp in salmonids and probably other teleost species (41 kDa form) and the main Igf carrier (6, 81, 82). For a long time, Igfbp-2b was wrongly considered to be Igfbp-3 (83) due to its similar physiological regulation to Igfbp-3 in mammals [e.g., Ref. (83)]. Teleost igfbp-2 genes are expressed in a range of tissues [e.g., Ref. (16, 84–86)] with zebrafish igfbp-2a and igfbp-2b having different spatiotemporal patterns during early development, and each being expressed in liver in adults (18). In adult salmon, igfbp-2a was expressed across multiple tissues, with especially high abundance in liver, whereas igfbp-2b1 and igfbp-2b1 were liver-restricted (16). Overexpression of igfbp-2a and igfbp-2b causes a reduction in growth and developmental rate in early-stage zebrafish (18, 87), suggesting an equivalent role to that observed in mammals. Similar to Igfbp-1, past work has suggested a role for Igfbp-2 in teleost sexual maturation, with igfbp-2 mRNA being expressed in pre-ovulatory ovaries of rainbow trout, and upregulated in response to female sex hormones (83).
Mixed results exist on the regulation of teleost igfbp-2 genes by nutritional status. For example, some past studies showed that igfbp-2 genes are downregulated or unchanged in liver and skeletal muscle of fasted fish (55, 88–90), which does not support an obvious role in growth inhibition. By contrast, a significant increase of igfbp-2 expression was observed in fasted zebrafish (91), although this same effect was not clearly observed in a later study of zebrafish that distinguished igfbp-2a and -2b (65). In Atlantic salmon, a significant decrease in igfbp-2a (formerly “igfbp-2.1”) expression was reported in skeletal muscle during refeeding after a period of restricted food intake, suggesting an inhibitory role on growth (90, 92). Similarly, all three tested igfbp-2 paralogs (igfbp-2a, -2b1, and -2b1) were downregulated in Atlantic salmon liver upon post-fasting refeeding, again suggesting an inhibitory role on growth (16). However, such data have not been replicated in vitro where neither amino acid deprivation nor addition of Igf-I and amino acids to Atlantic salmon cultured myotubes modified the expression of the same paralogs (57).
Moreover, the regulation of igfbp-2 expression by Gh does not clearly support a growth inhibitory role common to teleosts. While a study in zebrafish embryos reported that Gh inhibits igfbp-2 expression (91), work in Atlantic salmon demonstrated an increase in circulating Igfbp-2b in response to Gh (48, 81, 93). By contrast, treatment with dexamethasone, known to enhance catabolism, led to an increase in igfbp-2a expression in salmon skeletal muscle myotubes (57). Despite not distinguishing teleost paralogs, recent work revealed upregulation of skeletal muscle igfbp-2 expression in fine flounder (Paralichthys adspersus), concomitant to a rise in blood cortisol (94). Differences in Igfbp-2 expression across studies suggest a complex role for this Igfbp family member in teleost growth, dependent on both physiological and species context.
Igfbp-3 is the main carrier of circulating Igf in mammals, forming a tertiary structure with the acid-labile subunit (ALS) that increases Igf half-life and regulates Igf bioavailability (95). Igfbp-3 has anti-proliferative effects in many mammalian cell types, preventing the interaction of Igf-I and Igf-II with Igf-1R, and it also has Igf-independent roles (96). In this respect, Igfbp-3 directly interacts with two-cell surface receptors independently of Igf-I, Lrp1, and Tmem29, which mediates its anti-proliferative effects (97, 98). However, mammalian Igfbp-3 can also enhance cellular proliferation in some conditions, through both Igf-dependent and Igf-independent mechanisms (99, 100).
In contrast to mammals, teleost Igfbp-3 proteins are not considered major circulatory Igfbps (6). In fact, there exists no known association between Igfbp-3 proteins—or indeed any teleost Igfbp subtype—and ALS (6), highlighting fundamental differences in the way Igfs are regulated in circulation. In zebrafish, the single igfbp-3a (16) gene has important roles in early development, showing dorsalizing effects in embryos through an Igf-independent interaction with bone morphogenic protein 2b (101), one of few studies demonstrating an Igf-independent role for a teleost Igfbp. The four distinct igfbp-3 paralogs of salmonids (igfbp-3a1, -3a2, -3b1, and -3b2) were lowly expressed in 11 tested adult Atlantic salmon tissues (and each absent in liver), although igfbp-3a1 was among the most abundant of all Igfbp family member genes in heart (16) and the only detected igfbp-3 gene in primary myotube culture (57). Conversely, igfbp-3b of adult fine flounder was reported as more highly expressed in liver (the main route for Igfbp to circulation) than several other tested tissues, while igfbp-3a was not considered in the same study (88).
Studies in zebrafish, flounder, and yellowtail reported a significant increase in the expression of igfbp-3 genes in liver and/or muscle in response to fasting (55, 88, 102), which may act to restrict Igf signaling. However, on the other hand, studies in salmonids have reported no changes in muscle igfbp-3 gene expression in response to food deprivation (57, 89) with an increase in igfbp-3a1 expression in liver during post-fasting refeeding, more consistent with a growth-promoting function (16). Similarly, in coho salmon (Oncorhynchus kisutch), igfbp-3a1 muscle expression was increased by Gh transgenesis (103), again supporting a growth-promoting role. However, stress caused a downregulation of igfbp-3b gene expression in skeletal muscle of fine flounder (94), which is inconsistent with a role in growth inhibition.
Overall, there is a relatively limited body of research on teleost Igfbp-3 genes, leaving their roles unclear in many species, with the available evidence suggesting functional divergence among different lineages.
In mammals, Igfbp-4 is expressed in many cell types and tissues, where it is often considered to inhibit Igf action (104, 105). However, it is also considered to have growth-promoting roles during early embryogenesis, where it enhances Igf-II activity (106). Some studies have reported Igf-independent actions for Igfbp-4, including in relation to the inhibition of apoptosis (104, 105) and cardiogenesis (107).
In teleosts, igfbp-4 was expressed in most tissues for each species investigated, including Atlantic salmon (16), tiger pufferfish (Takifugu rubripes) (108) and fine flounder (88). Moreover, in Atlantic salmon, igfbp-4 was described as showing high abundance in gill (108). Atlantic salmon was recently shown to retain two highly similar Igfbp-4 paralogs (see Figures 1 and 2), which show conserved regulation across tissues (24). In tiger pufferfish, fasting caused upregulation of igfbp-4 expression in several tissues, consistent with an inhibitory role on growth (108). In addition, the expression of recombinant pufferfish Igfbp-4 in zebrafish embryos resulted in significant growth retardation (108). However, these findings contrast studies of several species (including salmonids and fine flounder), where igfbp-4 expression in response to nutritional status manipulation suggested a growth-promoting role. Such work revealed no change in igfbp-4 expression during fasting (57, 65, 88–90, 92), but upregulation in response to subsequent refeeding (57, 65, 88–90, 92, 109–111). A study of Arctic charr (Salvelinus alpinus) showed that dwarf populations with highly restricted growth had lower constitutive igfbp-4 expression in muscle than populations reaching larger body size (112). A growth-promoting role for igfbp-4 in salmonids was also supported by a strong positive correlation between igfbp-4 and several pro-myogenic gene markers during in vitro myogenesis in Atlantic salmon (110). Studies of Igfbp-4 expression in response to stress also suggest a growth-promoting role. For instance, addition of dexamethasone to Atlantic salmon myotubes (57), and stress confinement in fine flounder (94) induced a significant reduction in igfbp-4 expression. Conversely, an increase in igfbp-4 expression was reported in skeletal muscle during maturation-induced atrophy in rainbow trout (113).
Overall, the available evidence suggests that the physiological role of Igfbp-4, when conserved, differs across species and physiological contexts, though for some lineages, particularly salmonids, a growth-promoting function is implicated.
Igfbp-5 is the most conserved Igfbp family member. In mammals, it forms a ternary complex with ALS, similar to Igfbp-3, although much more circulating Igf is carried by Igfbp-3-ALS (114). Igfbp-5 represents an essential regulator of many processes in mammalian bone, kidney, mammary gland, and skeletal muscle (114) and can assert both stimulatory and inhibitory effects (depending on cell type) through Igf-dependent or Igf-independent routes. For instance, it has growth factor-like actions, stimulating bone growth in Igf-I knockout mice (115), and smooth muscle cell migration (116). There is also evidence that Igfbp-5 can translocate into the nucleus (117) and have nuclear functions (118). It is thought that Igfbp-5 cellular internalization is achieved by interaction with membrane proteins such as heparin sulfate proteoglycans, and that the Igfbp-5 N-terminal region has an Igf-independent transcriptional activity (118). Furthermore, Igfbp-5 can interact with transcription co-activators such as the four and half Lim domains 2 (119).
In zebrafish and grass carp embryos, igfbp-5a and igfbp-5b have distinctive patterns of expression during early development, suggesting evolutionary divergence in regulation (19, 120), which has also been demonstrated at the functional level (19). In adult zebrafish, igfbp-5a was expressed at high levels in brain and gill, and lower levels in several other tissues, but was absent in liver or skeletal muscle; while igfbp-5b was ubiquitously expressed. Similarly, in other studied teleost species igfbp-5 genes were reported to show a broad tissue distribution, with differences noted among species and paralogs (51, 52, 55, 63, 120), including three paralogs in Atlantic salmon (16, 24).
The importance of igfbp-5 genes for muscle differentiation and growth in teleosts is well established. Igfbp-5 expression has been studied across in vitro myogenesis, with both teleost paralogs (when distinguished) detected from early stages (i.e., myogenic progenitor cells) through to fully differentiated myotubes (57, 110, 111). In Atlantic salmon, both igfbp-5a (formerly: igfbp5.1) and igfbp-5b (formerly: igfbp5.2) showed highest expression in early-stage myoblasts, which decreased during myogenic differentiation (110). Using the same in vitro models, it was observed that pro-growth stimuli such as amino acids and Igfs increase igfbp-5 gene expression (111, 113), including both igfbp-5a and igfbp-5b paralogs distinguished in salmonids (57, 110). However, igfbp-5 paralogs appear to have different patterns of expression in response to catabolic signals. For instance, while amino acid deprivation had no effect on the regulation of any igfbp-5 paralog in Atlantic salmon myotubes (57, 113), dexamethasone reduced igfbp-5a expression, while simultaneously increasing igfbp-5b1 (57). A past study of rainbow trout skeletal muscle recorded no change in igfbp-5 gene expression in response to fasting or re-feeding (89), though it was unclear which paralog was measured. Similarly, igfbp-5a and igfbp-5b muscle expression did not change in response to short- or long-term fasting in Atlantic salmon (90, 92). However, in grass carp, igfbp-5a and igfbp-5b expression decreased in skeletal muscle during fasting, while both paralogs were upregulated in liver, and upon injection of Gh in both tissues (120). In Gh transgenic coho salmon, igfbp-5b1 was significantly upregulated (103).
There is also emerging evidence that igfbp-5 genes play a role in ionic homeostasis and Igf regulation in zebrafish (121), stickleback [e.g., Ref. (122)] and Atlantic salmon gills (123) with Igfbp-5a acting to regulate calcium influx in zebrafish gills (121) and being differentially expressed and under divergent selective pressures in marine vs. freshwater sticklebacks (122, 124).
To sum up, the available evidence suggests that Igfbp-5 genes play conserved functions in multiple aspects of teleost biology, with roles most clearly demonstrated in myogenesis, muscle growth, and gill function. There is also considerable evidence that both teleost and salmonid-specific Igfbp-5 paralogs have evolved divergent roles.
Igfbp-6 represents a special case among the Igfbp family. In mammals, it has a 50-fold binding preference for Igf-II over Igf-I (125, 126) (a unique feature among Igfbps), but also shows differences in key protein motifs, with three disulfide bonds in the N-terminal region instead of the 4 found in Igfbp-1 to 5 (127). Igfbp-6 is a relatively specific inhibitor of Igf-II actions and, therefore, regulates processes where Igf-II is involved such as proliferation, survival, migration, and differentiation (125, 126). Igfbp-6 also has known Igf-independent actions (125, 128), including the inhibition of fibroblast proliferation (129), cancer cell migration (130), and apoptosis (131). The gene has a broad tissue expression distribution in mammals, including lung, liver, and the gastrointestinal tract.
While differences in the affinity of Igfbp-6 proteins for Igf-II and Igf-I are yet to be confirmed in teleosts, the main underlying structural features are conserved (16). In zebrafish adults, igfbp-6a was highly expressed in muscle, and almost undetectable in other tissues, while igfbp-6b was only abundant in brain, heart, and muscle (20). In adult fine flounder, igfbp-6b was most highly expressed in heart, gills, and the gastrointestinal tract (88). In adult Atlantic salmon, neither igfbp-6a1 nor -6a2 were notably expressed across a panel of 11 tissues, while igfbp-6b1 and 6b2 were each broadly expressed, with the latter being especially highly expressed in gill, brain, and spleen (16). Both igfbp-6b1 and 6b2 were also reported as being highly expressed in Atlantic salmon gills, where they were dynamically regulated during smoltification (123).
The overexpression of either zebrafish igfbp-6 paralog caused a significant reduction of embryonic growth (20), suggesting a role in growth inhibition. Studies of igfbp-6 gene regulation in skeletal muscle support a similar role in other species, though some conflicting data exist. For example, a study in Atlantic salmon reported no change in igfbp-6b expression in response to food intake manipulation (92), while another reported downregulation of igfbp-6b in tilapia skeletal muscle in response to feeding after a period of fasting (53). Similar results were observed in fine flounder skeletal muscle, where igfbp-6b expression decreased in response to feeding immediately post-fasting, although expression then increased during long-term refeeding (88). However, igfbp-6b was repressed in fine flounder skeletal muscle in response to stress (94), which is less consistent with a negative role in growth. Conversely, in Atlantic salmon myotubes treated with dexamethasone, igfbp-6a1 was downregulated, while igfbp-6b2 was upregulated, highlighting complex functions that cannot be easily interpreted without functional data (57).
Recent work also implies a novel role for igfbp-6 genes in linking growth and immune regulation in teleosts. Alzaid et al. observed a significant increase of igfbp-6a2 in primary immune tissues of rainbow trout following a bacterial infection, and provided evidence that immune-responsive igfbp-6a2 upregulation was stimulated by immune signaling pathways driven by pro-inflammatory cytokines (27). Past work in salmonids has also shown that pro-inflammatory cytokines can promote the expression of igfbp-6 genes in skeletal muscle cell cultures (132) and in vivo (103), which may be linked to the balancing of energetic allocation toward effective immune function.
In summary, Igfbp-6 genes of teleosts are rather understudied, and it is difficult to draw overarching conclusions about their roles and functions at this time.
Our current understanding of the Igfbp repertoire of different teleosts has benefited greatly from recent expansions to genomic resources. We can now be confident in the existence of many teleost paralogs, which are expressed and presumably functional. However, our understanding of the functions and regulatory control of these genes remains highly fragmented across teleosts as a group and remains highly underdeveloped compared to mammals. It is becoming increasingly clear—perhaps with the exception of Igfbp-1—that teleost and mammalian Igfbp orthologs have evolved distinct expression-level regulation. This points to distinct functional roles in the regulation of growth in teleosts compared to mammals, which may be related to differences in growth dynamics, for example, indeterminate growth in teleosts. Moreover, there is also evidence that Igfbp orthologs from different species have evolved distinct regulation and hence, potentially functions, during teleost evolution. This can be speculatively linked to the additional functional flexibility or redundancy linked to Igfbp duplication events, which has allowed divergent regulation of paralogs to evolve under different physiological contexts.
It is also clear that differences in the expression of homologous Igfbp genes across teleost species are often difficult to interpret. In many cases, this may be linked to the historic ignorance of paralogous genes, either by considering one paralog in pair, or detecting signals from both paralogs in gene expression analyses. Hence, a fuller understanding of Igfbp genes will be possible in the presence of high-quality reference genomes, where all genes are properly annotated and can then be distinguished experimentally. The evidence for divergent regulation of Igfbp paralog expression is overwhelming, even for genes with very similar coding sequences (24), suggesting gene expression studies should make every effort to distinguish Igfbp paralogs.
An additional priority for future research should be to characterize the individual protein-level functions of all teleost Igfbp paralogs in multiple species extending beyond model organisms. While it has classically been challenging to perform functional analyses in non-model teleosts, the research landscape is rapidly changing. For example, genome editing using engineered CRIPSR/Cas9 systems has been demonstrated in vivo for various large commercial species, including salmonids (133) and catfishes (134), as well as in teleost cell culture (135). Hence, while even 5 years ago, the full repertoire of Igfbp genes was not even recognized in many teleosts, we can look forward to a future where every paralog within a species has its function cataloged by such approaches, even in lineages with hugely expanded Igfbp gene families. This will open the door for associating protein-level functional divergence in Igfbp paralogs with evolutionary changes in gene expression regulation, generating a fuller picture of the biological roles of this fascinating gene family in teleosts.
DGS and DJM wrote the manuscript and prepared the figures. DM built the phylogenetic tree showed in the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
DGS would like to thank the Marine Alliance Science Technology Scotland (MASTS) for their support. MASTS is funded by the Scottish Funding Council (grant reference HR09011) and contributing institutions.
1. Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev (1999) 20(6):761–87. doi:10.1210/er.20.6.761
2. Rodgers BD, Roalson EH, Thompson C. Phylogenetic analysis of the insulin-like growth factor binding protein (IGFBP) and IGFBP-related protein gene families. Gen Comp Endocrinol (2008) 155(1):201–7. doi:10.1016/j.ygcen.2007.04.013
3. Kelley KM, Schmidt KE, Berg L, Sak K, Galima MM, Gillespie C, et al. Comparative endocrinology of the insulin-like growth factor-binding protein. J Endocrinol (2002) 175(1):3–18. doi:10.1677/joe.0.1750003
4. Wood AW, Duan C, Bern HA. Insulin-like growth factor signaling in fish. Int Rev Cytol (2005) 243:215–85. doi:10.1016/S0074-7696(05)43004-1
5. Reindl KM, Sheirdan MA. Peripheral regulation of the growth hormone-insulin-like growth factor system in fish and other vertebrates. Comp Biochem Physiol A Mol Integr Physiol (2012) 163(3–4):231–45. doi:10.1016/j.cbpa.2012.08.003
6. Shimizu M, Dickhoff WW. Circulating insulin-like growth factor binding proteins in fish: their identities and physiological regulation. Gen Comp Endocrinol (2017) 252:150–61. doi:10.1016/j.ygcen.2017.08.002
7. Shimasaki S, Ling N. Identification and molecular characterization of insulin-like growth factors binding proteins (IGFBP-1, -2, -3, -4, -5 and -6). Prog Growth Factor Res (1991) 3(4):243–66. doi:10.1016/0955-2235(91)90003-M
8. Shimasaki S, Shimonaka M, Zhang H, Ling N. In: Spencer E, editor. Modern Concepts on Insulin-Like Growth Factors. New York: Elsevier (1991). p. 343–58.
9. Ocampo Daza D, Sundström G, Bergqvist CA, Duan C, Larhammar D. Evolution of the insulin-like growth factor binding protein (IGFBP) family. Endocrinology (2011) 152(6):2278–89. doi:10.1210/en.2011-0047
10. Sundström G, Larsson TA, Larhammar D. Phylogenetic and chromosomal analyses of multiple gene families syntenic with vertebrate Hox clusters. BMC Evol Biol (2008) 8:254. doi:10.1186/1471-2148-8-254
11. Van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy. Nat Rev Genet (2017) 18(7):411–24. doi:10.1038/nrg.2017.26
12. Zhou J, Xiang J, Zhang S, Duan C. Structural and functional analysis of the amphioxus IGFBP gene uncovers ancient origin of IGF-independent functions. Endocrinology (2013) 154(10):3753–63. doi:10.1210/en.2013-1201
13. Chan SJ, Cao QP, Steiner DF. Evolution of the insulin superfamily: cloning of a hybrid insulin/insulin-like growth factor cDNA from amphioxus. Proc Natl Acad Sci U S A (1990) 87(23):9319–23. doi:10.1073/pnas.87.23.9319
14. Pashmforoush M, Chan SJ, Steiner DF. Structure and expression of the insulin-like peptide receptor from amphioxus. Mol Endocrinol (1996) 10(7):857–66. doi:10.1210/mend.10.7.8813726
15. Holland PW, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl (1994):125–33.
16. Macqueen DJ, Garcia de la serrana D, Johnston IA. Evolution of ancient functions in the vertebrate insulin-like growth factor system uncovered by study of duplicated salmonid fish genomes. Mol Biol Evol (2013) 30(5):1060–76. doi:10.1093/molbev/mst017
17. Kamei H, Lu L, Jiao S, Li Y, Gyrup C, Laursen LS, et al. Duplication and diversification of the hypoxia-inducible IGFBP-1 gene in zebrafish. PLoS One (2008) 28(3):e3091. doi:10.1371/journal.pone.0003091
18. Zhou J, Li W, Kamei H, Duan C. Duplication of the IGFBP-2 gene in teleost fish: protein structure and functionality conservation and gene expression divergence. PLoS One (2008) 3(12):e3926. doi:10.1371/journal.pone.0003926
19. Dai W, Kamei H, Zhao Y, Ding J, Du Z, Duan C. Duplicated zebrafish insulin-like growth factor binding protein-5 genes with split functional domains: evidence for evolutionarily conserved IGF binding, nuclear localization, and transactivation activity. FASEB J (2010) 24(6):2020–9. doi:10.1096/fj.09-149435
20. Wang X, Lu L, Li Y, Li M, Chen C, Feng Q, et al. Molecular and functional characterization of two distinct IGF binding protein-6 genes in zebrafish. Am J Physiol Regul Integr Comp Physiol (2009) 296(5):1348–57. doi:10.1152/ajpregu.90969.2008
21. Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, et al. The Atlantic salmon genome provides insights into rediploidization. Nature (2016) 533(7602):200–5. doi:10.1038/nature17164
22. Macqueen DJ, Johnston IA. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc Biol Sci (2014) 281(1778):20132881. doi:10.1098/rspb.2013.2881
23. Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noël B, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun (2014) 5:3657. doi:10.1038/ncomms4657
24. Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, et al. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol (2017) 18(1):111. doi:10.1186/s13059-017-1241-z
25. Lappin FM, Shaw RL, Macqueen DJ. Targeted sequencing for high-resolution evolutionary analyses following genome duplication in salmonid fish: proof of concept for key components of the insulin-like growth factor axis. Mar Genomics (2016) 30:15–26. doi:10.1016/j.margen.2016.06.003
26. Wallis AE, Devlin RH. Duplicate insulin-like growth factor-I genes in salmon display alternative splicing pathways. Mol Endocrinol (1993) 7(3):409–22. doi:10.1210/me.7.3.409
27. Alzaid A, Martin SA, Macqueen DJ. The complete salmonid IGF-IR gene repertoire and its transcriptional response to disease. Sci Rep (2016) 6:34806. doi:10.1038/srep34806
28. Xu P, Zhang X, Wang X, Li J, Kuang Y, Xu J, et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genet (2014) 46(11):1212–9. doi:10.1038/ng.3098
29. David L, Blum S, Feldman MW, Lavi U, Hillel J. Recent duplication of the common carp (Cyprinus carpio L.) genome as revealed by analyses of microsatellite loci. Mol Biol Evol (2003) 20(9):1425–34. doi:10.1093/molbev/msg173
30. Helbo S, Dewilde S, Williams DR, Berghmans H, Berenbrink M, Cossins AR, et al. Functional differentiation of myoglobin isoforms in hypoxia-tolerant carp indicates tissue-specific protective roles. Am J Physiol Regul Integr Comp Physiol (2012) 302(6):693–701. doi:10.1152/ajpregu.00501.2011
31. Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature (2016) 538(7625):336–43. doi:10.1038/nature19840
32. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol (1990) 214:403–10. doi:10.1016/S0022-2836(05)80360-2
33. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol (2013) 30(4):772–80. doi:10.1093/molbev/mst010
34. Sela I, Ashkenazy H, Katoh K, Pupko T. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res (2015) 43(W1):W7–14. doi:10.1093/nar/gkv318
35. Macqueen DJ, Wilcox AH. Characterization of the definitive classical calpain family of vertebrates using phylogenetic, evolutionary and expression analyses. Open Biol (2014) 4:130219. doi:10.1098/rsob.130219
36. Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol (2012) 29(8):1969–73. doi:10.1093/molbev/mss075
37. Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res (2016) 44(W1):W232–5. doi:10.1093/nar/gkw256
38. Drummond AJ, Ho SYW, Philips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol (2006) 4(5):e88. doi:10.1371/journal.pbio.0040088
39. Kajimura S, Duan C. Insulin-like growth factor-binding protein-1: an evolutionarily conserved fine tuner of insulin-like growth factor action under catabolic and stressful conditions. J Fish Biol (2007) 71:309–25. doi:10.1111/j.1095-8649.2007.01606.x
40. Lee PD, Giudice LC, Conover CA, Powell DR. Insulin-like growth factor binding protein-1: recent finding and new directions. Proc Soc Exp Biol Med (1997) 216(3):319–57. doi:10.3181/00379727-216-44182
41. Jones JI, Gockerman A, Busby WH, Wright G, Clemmons DR. Insulin-like growth factor binding protein 1 stimulates cell migration and binds to the alpha 5 beta 1 integrin by means of its Arg-Gly-Asp sequence. Proc Natl Acad Sci U S A (1993) 90(22):10553–7. doi:10.1073/pnas.90.22.10553
42. Wheatcroft SB, Kearney MT. IGF-dependent and IGF-independent actions of IGF-binding protein-1 and -2: implications for metabolic homeostasis. Trends Endocrinol Metab (2009) 20(4):153–62. doi:10.1016/j.tem.2009.01.002
43. Shimizu M, Kishimoto K, Yamaguchi T, Nakano Y, Hara A, Dickhoff WW. Circulating salmon 28- and 22-kDa insulin-like growth factor binding proteins (IGFBPs) are co-orthologs of IGFBP-1. Gen Comp Endocrinol (2011) 174(2):97–106. doi:10.1016/j.ygcen.2011.08.005
44. Shimizu M, Cooper KA, Dickhoff WW, Beckman BR. Postprandial changes in plasma growth hormone, insulin, insulin-like growth factor (IGF)-I, and IGF-binding proteins in coho salmon fasted for varying periods. Am J Physiol Regul Integr Comp Physiol (2009) 297(2):352–61. doi:10.1152/ajpregu.90939.2008
45. Johnston J, Silverstein J, Wolters WR, Shimizu M, Dickhoff WW, Shepherd BS. Disparate regulation of insulin-like growth factor-binding proteins in a primitive, ictalurid, teleost (Ictalurus punctatus). Gen Comp Endocrinol (2003) 134(2):122–30. doi:10.1016/S0016-6480(03)00244-2
46. Fukazawa Y, Siharath K, Iguchi T, Bern HA. In vitro secretion of insulin-like growth factor-binding proteins from liver of striped bass, Morone saxatilis. Gen Comp Endocrinol (1995) 99(2):239–47. doi:10.1006/gcen.1995.1107
47. Gómez-Requeni P, Calduch-Giner J, Vega-Rubín de Celis S, Médale F, Sadasivam JK, Pérez-Sánchez J. Regulation of the somatotropic axis by dietary factors in rainbow trout (Oncorhynchus mykiss). Br J Nutr (2005) 94(3):353–61. doi:10.1079/BJN20051521
48. Park R, Shepherd BS, Nishioka RS, Grau EG, Bern HA. Effects of homologous pituitary hormone treatment on serum insulin-like growth-factor-binding proteins (IGFBPs) in hypophysectomized tilapia, Oreochromis mossambicus with special reference to a novel 20-kDa IGFBP. Gen Comp Endocrinol (2000) 117(3):404–12. doi:10.1006/gcen.1999.7421
49. Davis BK, Peterson BC. The effect of temperature, stress, and cortisol on plasma IGF-I and IGFBPs in sunshine bass. Gen Comp Endocrinol (2006) 149(3):219–25. doi:10.1016/j.ygcen.2006.05.009
50. Maures TJ, Duan C. Structure, developmental expression, and physiological regulation of zebrafish IGF binding protein-1. Endocrinology (2002) 143(7):2722–31. doi:10.1210/endo.143.7.8905
51. Rahman MS, Thomas P. Characterization of three IGFBP mRNAs in Atlantic croaker and their regulation during hypoxic stress: potential mechanisms of their upregulation by hypoxia. Am J Physiol Endocrinol Metab (2011) 301(4):637–48. doi:10.1152/ajpendo.00168.2011
52. Rahman MS, Thomas P. Molecular and biochemical responses of hypoxia exposure in Atlantic croaker collected from hypoxic regions in the northern Gulf of Mexico. PLoS One (2017) 12(9):e0184341. doi:10.1371/journal.pone.0184341
53. Breves JP, Tipsmark CK, Stough BA, Seale AP, Flack BR, Moorman BP, et al. Nutritional status and growth hormone regulate insulin-like growth factor binding protein (igfbp) transcripts in Mozambique tilapia. Gen Comp Endocrinol (2014) 207:66–73. doi:10.1016/j.ygcen.2014.04.034
54. Tian YM, Chen J, Tao Y, Jiang XY, Zou SM. Molecular cloning and function analysis of insulin-like growth factor-binding protein 1a in blunt snout bream (Megalobrama amblycephala). Dongwuxue Yanjiu (2014) 35(4):300–6. doi:10.13918/j.issn.2095-8137.2014.4.300
55. Pedroso FL, Fukada H, Masumoto T. Molecular characterization, tissue distribution patterns and nutritional regulation of IGFBP-1, -2, -3 and -5 in, Seriola quinqueradiata. Gen Comp Endocrinol (2009) 161(3):344–53. doi:10.1016/j.ygcen.2009.01.010
56. Chen W, Zhang Z, Dong H, Yan F. Insulin-like growth factor-binding protein-1 (IGFBP-1) in goldfish, Carassius auratus: molecular cloning, tissue expression, and mRNA expression responses to periprandial changes and cadmium exposure. Fish Physiol Biochem (2016) 42(3):1043–52. doi:10.1007/s10695-015-0195-x
57. Garcia de la serrana D, Fuentes EN, Martin SAM, Johnston IA, Macqueen DJ. Divergent regulation of insulin-like growth factor binding protein genes in cultured Atlantic salmon myotubes under different models of catabolism and anabolism. Gen Comp Endocrinol (2017) 247:53–65. doi:10.1016/j.ygcen.2017.01.017
58. Safian D, Morais RD, Bogerd J, Schulz RW. Igf binding proteins protect undifferentiated spermatogonia in the zebrafish testis against excessive differentiation. Endocrinology (2016) 157(11):4423–33. doi:10.1210/en.2016-1315
59. Kajimura S, Aida K, Duan C. Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc Natl Acad Sci U S A (2005) 102(4):1240–5. doi:10.1073/pnas.0407443102
60. Shimizu M, Beckman BR, Hara A, Dickhoff WW. Measurement of circulating salmon IGF binding protein-1: assay development, response to feeding ration and temperature, and relation to growth parameters. J Endocrinol (2006) 188(1):101–10. doi:10.1677/joe.1.06475
61. Waagbø R, Jørgensen SM, Timmerhaus G, Breck O, Olsvik PA. Short-term starvation at low temperature prior to harvest does not impact the health and acute stress response of adult Atlantic salmon. PeerJ (2017) 5:e3273. doi:10.7717/peerj.3273
62. Pierce AL, Shimizu M, Felli L, Swanson P, Dickhoff WW. Metabolic hormones regulate insulin-like growth factor binding protein-1 mRNA levels in primary cultured salmon hepatocytes; lack of inhibition by insulin. J Endocrinol (2006) 191(2):379–86. doi:10.1677/joe.1.06986
63. Breves JP, Phipps-Costin SK, Fujimoto CK, Einarsdottir IE, Regish AM, Bjornsson BT, et al. Hepatic insulin-like growth-factor binding protein (igfbp) responses to food restriction in Atlantic salmon smolts. Gen Comp Endocrinol (2016) 233:79–87. doi:10.1016/j.ygcen.2016.05.015
64. Rolland M, Dalsgaard J, Holm J, Gómez-Requeni P, Skov PV. Dietary methionine level affects growth performance and hepatic gene expression of GH–IGF system and protein turnover regulators in rainbow trout (Oncorhynchus mykiss) fed plant protein-based diets. Comp Biochem Physiol B Biochem Mol Biol (2015) 181:33–41. doi:10.1016/j.cbpb.2014.11.009
65. Amaral IP, Johnston IA. Insulin-like growth factor (IGF) signalling and genome-wide transcriptional regulation in fast muscle of zebrafish following a single-satiating meal. J Exp Biol (2011) 214(Pt 13):2125–39. doi:10.1242/jeb.053298
66. Haase CG, Long AK, Gillooly JF. Energetics of stress: linking plasma cortisol levels to metabolic rate in mammals. Biol Lett (2016) 12(1):20150867. doi:10.1098/rsbl.2015.0867
67. Fürtbauer I, Heistermann M. Cortisol coregulation in fish. Sci Rep (2016) 6:30334. doi:10.1038/srep30334
68. Kajimura S, Hirano T, Visitacion N, Moriyama S, Aida K, Grau EG. Dual mode of cortisol action on GH/IGF-I/IGF binding proteins in the tilapia, Oreochromis mossambicus. J Endocrinol (2003) 178(1):91–9. doi:10.1677/joe.0.1780091
69. Peterson BC, Small BC. Effects of exogenous cortisol on the GH/IGF-I/IGFBP network in channel catfish. Domest Anim Endocrinol (2005) 28(4):391–404. doi:10.1016/j.domaniend.2005.01.003
70. Kelley KM, Haigwood JT, Perez M, Galima MM. Serum insulin-like growth factor binding proteins (IGFBPs) as markers for anabolic/catabolic condition in fishes. Comp Biochem Physiol B Biochem Mol Biol (2001) 129(2–3):229–36. doi:10.1016/S1096-4959(01)00314-1
71. Shepherd BS, Johnson JK, Silverstein JT, Parhar IS, Vijayan MM, McGuire A, et al. Endocrine and orexigenic actions of growth hormone secretagogues in rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol A Mol Integr Physiol (2007) 146(3):390–9. doi:10.1016/j.cbpa.2006.11.004
72. Peterson BC, Small BC. Effects of fasting on circulating IGF-binding proteins, glucose, and cortisol in channel catfish (Ictalurus punctatus). Domest Anim Endocrinol (2004) 26(3):231–40. doi:10.1016/j.domaniend.2003.10.005
73. Davis KB, Peterson BC. Comparison of insulin-like growth factor-I and insulin-like growth factor binding protein concentrations of the palmetto and sunshine bass and the effects of gender and stress. J World Aquacult Soc (2005) 36(3):384–92. doi:10.1111/j.1749-7345.2005.tb00342.x
74. Alzaid A, Castro R, Wang T, Secombes CJ, Boudinot P, Macqueen DJ, et al. Cross talk between growth and immunity: coupling of the IGF axis to conserved cytokine pathways in rainbow trout. Endocrinology (2016) 157(5):1942–55. doi:10.1210/en.2015-2024
75. Shin M, Kang HS, Park JH, Bae JH, Song DK, Im SS. Recent insights into insulin-like growth factor binding protein 2 transcriptional regulation. Endocrinol Metab (Seoul) (2017) 32(1):11–7. doi:10.3803/EnM.2017.32.1.11
76. Hoeflich A, Wu M, Mohan S, Föll J, Wanke R, Froehlich T, et al. Overexpression of insulin-like growth factor-binding protein-2 in transgenic mice reduces postnatal body weight gain. Endocrinology (1999) 140(12):5488–96. doi:10.1210/endo.140.12.7169
77. Wood TL, Rogler LE, Czick ME, Schuller AG, Pintar JE. Selective alterations in organ sizes in mice with a targeted disruption of the insulin-like growth factor binding protein-2 gene. Mol Endocrinol (2000) 14(9):1472–82. doi:10.1210/mend.14.9.0517
78. Schutt BS, Langkapm M, Rauschnabel U, Ranke MB, Elmlinger MW. Integrin-mediated action of insulin-like growth factor binding protein-2 in tumor cells. J Mol Endocrinol (2004) 32(3):859–68. doi:10.1677/jme.0.0320859
79. Chua CY, Liu Y, Granberg KJ, Hu L, Haapasalo H, Annala MJ, et al. IGFBP2 potentiates nuclear EGFR-STAT3 signaling. Oncogene (2016) 35(6):738–47. doi:10.1038/onc.2015.131
80. Amin S, Riggs BL, Melton LJ III, Achenbach SJ, Atkison EJ, Khosla S. High serum IGFBP-2 is predictive of increased bone turnover in aging men and women. J Bone Miner Res (2007) 22(6):799–807. doi:10.1359/jbmr.070306
81. Shimizu M, Swanson P, Hara A, Dickhoff WW. Purification of a 41-kDa insulin-like growth factor binding protein from serum of chinook salmon, Oncorhynchus tshawytscha. Gen Comp Endocrinol (2003) 132(1):103–11. doi:10.1016/S0016-6480(03)00052-2
82. Shimizu M, Suzuki S, Horikoshi M, Hara A, Dickhoff WW. Circulating salmon 41-kDa insulin-like growth factor binding protein (IGFBP) is not IGFBP-3 but an IGFBP-2 subtype. Gen Comp Endocrinol (2011) 171(3):326–31. doi:10.1016/j.ygcen.2011.02.013
83. Kamangar BB, Gabillard JC, Bobe J. Insulin-like growth factor-binding protein (IGFBP)-1, -2, -3, -4, -5, and -6 and IGFBP-related protein 1 during rainbow trout postvitellogenesis and oocyte maturation: molecular characterization, expression profiles, and hormonal regulation. Endocrinology (2006) 147(5):2399–410. doi:10.1210/en.2005-1570
84. Radaelli G, Domeneghini C, Arrighi S, Bosi G, Patruno M, Funkenstein B. Localization of IGF-I, IGF-I receptor, and IGFBP-2 in developing Umbrina cirrosa (Pisces: Osteichthyes). Gen Comp Endocrinol (2003) 130(3):232–44. doi:10.1016/S0016-6480(02)00609-3
85. Funkenstein B, Tsai W, Maures T, Duan C. Ontogeny, tissue distribution, and hormonal regulation of insulin-like growth factor binding protein-2 (IGFBP-2) in a marine fish, Sparus aurata. Gen Comp Endocrinol (2002) 128(2):112–22. doi:10.1016/S0016-6480(02)00059-X
86. Chen W, Li W, Lin H. Common carp (Cyprinus carpio) insulin-like growth factor binding protein-2 (IGFBP-2): molecular cloning, expression profiles, and hormonal regulation in hepatocytes. Gen Comp Endocrinol (2009) 161(3):390–9. doi:10.1016/j.ygcen.2009.02.004
87. Wood WA, Schlueter PJ, Duan C. Targeted knockdown of insulin-like growth factor binding protein-2 disrupts cardiovascular development in zebrafish embryos. Mol Endocrinol (2005) 19(4):1024–34. doi:10.1210/me.2004-0392
88. Safian D, Fuentes EN, Valdes JA, Molina A. Dynamic transcriptional regulation of autocrine/paracrine igfbp1, 2, 3, 4, 5, and 6 in the skeletal muscle of the fine flounder during different nutritional statuses. J Endocrinol (2012) 214(1):95–108. doi:10.1530/JOE-12-0057
89. Gabillard JC, Kamangar BB, Montserrat N. Coordinated regulation of the GH/IGF system genes during refeeding in rainbow trout (Oncorhynchus mykiss). J Endocrinol (2006) 191(1):15–24. doi:10.1677/joe.1.06869
90. Valente LM, Bower NI, Johnston IA. Postprandial expression of growth-related genes in Atlantic salmon (Salmo salar L.) juveniles fasted for 1 week and fed a single meal to satiation. Br J Nutr (2012) 108(12):2148–57. doi:10.1017/S0007114512000396
91. Duan C, Ding J, Li Q, Tsai W, Pozios K. Insulin-like growth factor binding protein 2 is a growth inhibitory protein conserved in zebrafish. Proc Natl Acad Sci U S A (1999) 96(26):15274–9. doi:10.1073/pnas.96.26.15274
92. Bower NI, Li X, Taylor R, Johnston IA. Switching to fast growth: the insulin-like growth factor (IGF) system in skeletal muscle of Atlantic salmon. J Exp Biol (2008) 211(Pt 24):3859–70. doi:10.1242/jeb.024117
93. Shimizu M, Swanson P, Dickhoff WW. Free and protein-bound insulin-like growth factor-I (IGF-I) and IGF-binding proteins of coho salmon, Oncorhynchus kisutch. Gen Comp Endocrinol (1999) 115(3):398–405. doi:10.1006/gcen.1999.7328
94. Valenzuela CA, Zuloaga R, Mercado L, Einarsdottir IE, Bjornsson BT, et al. Chronic stress inhibits growth and induces proteolytic mechanisms through two different non-overlapping pathways in the skeletal muscle of a teleost fish. Am J Physiol Regul Integr Comp Physiol (2018) 314(1):R102–13. doi:10.1152/ajpregu.00009.2017
95. Baxter RC, Martin JL, Beniac VA. High molecular weight insulin-like growth factor binding protein complex. Purification and properties of the acid-labile subunit from human serum. J Biol Chem (1989) 264(20):11843–8.
96. Yamada PM, Lee KW. Perspectives in mammalian IGFBP-3 biology: local vs. systemic action. Am J Physiol Cell Physiol (2009) 296(5):C954–76. doi:10.1152/ajpcell.00598.2008
97. Huang SS, Ling TY, Tseng WF, Huang YH, Tang FM, Leal SM, et al. Cellular growth inhibition by IGFBP-3 and TGF-beta1 requires LRP-1. FASEB J (2003) 17(14):2068–81. doi:10.1096/fj.03-0256com
98. Ingermann AR, Yang YF, Han J, Mikami A, Garza AE, Mohanraj L, et al. Identification of a novel cell death receptor mediating IGFBP-3-induced anti-tumor effects in breast and prostate cancer. J Biol Chem (2010) 285(39):30233–46. doi:10.1074/jbc.M110.122226
99. Baxter RC. Insulin-like growth factor binding protein-3 (IGFBP-3): novel ligands mediate unexpected functions. J Cell Commun Signal (2013) 7(3):179–89. doi:10.1007/s12079-013-0203-9
100. Conover CA, Clarkson JT, Bale LK. Factors regulating insulin-like growth factor-binding protein-3 binding, processing, and potentiation of insulin-like growth factor action. Endocrinology (1996) 137(6):2286–92. doi:10.1210/endo.137.6.8641177
101. Zhong Y, Lu L, Zhou J, Li Y, Liu Y, Clemmons DR, et al. IGF binding protein 3 exerts its ligand-independent action by antagonizing BMP in zebrafish embryos. J Cell Sci (2011) 124(Pt 11):1925–35. doi:10.1242/jcs.082644
102. Chen JY, Chen JC, Huang WT, Liu CW, Hui CF, Chen TT, et al. Molecular cloning and tissue-specific, developmental-stage-specific, and hormonal regulation of IGFBP3 gene in zebrafish. Mar Biotechnol (NY) (2004) 6(1):1–7. doi:10.1007/s10126-002-0115-9
103. Alzaid A, Kim JH, Devlin RH, Martin SAM, Macqueen DJ. Growth hormone transgenesis disrupts immune function in muscle of coho salmon (Oncorhynchus kisutch) impacting cross-talk with growth systems. bioRxiv (2017) 210104. doi:10.1101/210104
104. Zhou R, Diehl D, Hoeflich A, Lahm H, Wolf E. IGF-binding protein-4: biochemical characteristics and functional consequences. J Endocrinol (2003) 178(2):177–93. doi:10.1677/joe.0.1780177
105. Durai R, Davies M, Yang W, Yang SY, Seifalian A, Goldspink G, et al. Biology of insulin-like growth factor binding protein-4 and its role in cancer (review). Int J Oncol (2006) 28(6):1317–25. doi:10.3892/rjo.28.6.1317
106. Ning Y, Schuller AG, Conover CA, Pintar JE. Insulin-like growth factor (IGF) binding protein-4 is both a positive and negative regulator of IGF activity in vivo. Mol Endocrinol (2008) 22(5):1213–25. doi:10.1210/me.2007-0536
107. Zhu W, Shiojima I, Ito Y, Li Z, Ikeda H, Yoshida M, et al. IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature (2008) 454(7202):345–9. doi:10.1038/nature07027
108. Li M, Li Y, Lu L, Wang X, Gong Q, Duan C. Structural, gene expression, and functional analysis of the fugu (Takifugu rubripes) insulin-like growth factor binding protein-4 gene. Am J Physiol Regul Integr Comp Physiol (2009) 296(3):558–66. doi:10.1152/ajpregu.90439.2008
109. Garcia de la serrana D, Vieira VL, Andree KB, Daria M, Estevez A, Gisbert E, et al. Development temperature has persistent effects on muscle growth responses in gilthead sea bream. PLoS One (2012) 7(12):e51884. doi:10.1371/journal.pone.0051884
110. Bower NI, Johnston IA. Transcriptional regulation of the IGF signaling pathway by amino acids and insulin-like growth factors during myogenesis in Atlantic salmon. PLoS One (2010) 5(6):e11100. doi:10.1371/journal.pone.0011100
111. Azizi S, Nematollahi MA, Mojazi Amiri B, Velez EJ, Lutfi E, Navarro I, et al. Lysine and leucine deficiencies affect myocytes development and IGF signaling in gilthead sea bream (Sparus aurata). PLoS One (2016) 11(1):e0147618. doi:10.1371/journal.pone.0147618
112. Macqueen DJ, Kristjansson BK, Paxton CG, Vieira VL, Johnston IA. The parallel evolution of dwarfism in Arctic charr is accompanied by adaptive divergence in mTOR-pathway gene expression. Mol Ecol (2011) 20(15):3167–84. doi:10.1111/j.1365-294X.2011.05172.x
113. Salem M, Kenney PB, Rexroad CE III, Yao J. Proteomic signature of muscle atrophy in rainbow trout. J Proteomics (2010) 73(4):778–89. doi:10.1016/j.jprot.2009.10.014
114. Schneider MR, Wolf E, Hoeflich A, Lahm H. IGF-binding protein-5: flexible player in the IGF system and effector on its own. J Endocrinol (2002) 172(3):423–40. doi:10.1677/joe.0.1720423
115. Miyakoshi N, Richman C, Kasukawa Y, Linkhart TA, Baylink DJ, Mohan S. Evidence that IGF-binding protein-5 functions as a growth factor. J Clin Invest (2001) 107(1):73–81. doi:10.1172/JCI10459
116. Hsieh T, Gordon RE, Clemmons DR, Busby WH, Duan C. Regulation of vascular smooth muscle cell responses to insulin-like growth factor (IGF)-I by local IGF-binding proteins. J Biol Chem (2003) 278(44):42886–92. doi:10.1074/jbc.M303835200
117. Sun M, Long J, Yi Y, Xia W. Importin α-importin β complex mediated nuclear translocation of insulin-like growth factor binding protein-5. Endocr J (2017) 64(10):963–75. doi:10.1507/endocrj.EJ17-0156
118. Xu Q, Li S, Zhao Y, Maures TJ, Yin P, Duan C. Evidence that IGF binding protein-5 functions as a ligand-independent transcriptional regulator in vascular smooth muscle cells. Circ Res (2004) 94(5):46–54. doi:10.1161/01.RES.0000124761.62846.DF
119. Amaar YG, Thompson GR, Linkhart TA, Chen ST, Baylink DJ, Mohan S. Insulin-like growth factor-binding protein 5 (IGFBP-5) interacts with a four and a half LIM protein 2 (FHL2). J Biol Chem (2002) 277(14):12053–60. doi:10.1074/jbc.M110872200
120. Zeng GD, Zhou CX, Lin ST, Chen J, Jian XY, Zou SM. Two grass carp (Ctenopharyngodon idella) insulin-like growth factor-binding protein 5 genes exhibit different yet conserved functions in development and growth. Comp Biochem Physiol B Biochem Mol Biol (2016) 204:69–76. doi:10.1016/j.cbpb.2016.11.008
121. Dai W, Bai Y, Zhong X, Hebda L, Liu J, Kao J, et al. Calcium deficiency-induced and TRPV channel-regulated IGF-PI3K-Akt signaling stimulates abnormal epithelial proliferation. Cell Death Differ (2014) 21(4):568–81. doi:10.1038/cdd.2013.177
122. Kusakabe M, Ishikawa A, Ravinet M, Yoshida K, Makino T, Toyoda A, et al. Genetic basis for variation in salinity tolerance between stickleback ecotypes. Mol Ecol (2017) 26(1):304–19. doi:10.1111/mec.13875
123. Breves JP, Fujimoto CK, Phipps-Costin SK, Einarsdottir IE, Bjornsson BT, McCormick SD. Variation in branchial expression among insulin-like growth-factor binding proteins (igfbps) during Atlantic salmon smoltification and seawater exposure. BMC Physiol (2017) 17(1):2. doi:10.1186/s12899-017-0028-5
124. Pellisier T, Al Nafea H, Good SV. Divergence of insulin superfamily ligands, receptors and Igf binding proteins in marine versus freshwater stickleback: evidence of selection in known and novel genes. Comp Biochem Physiol Part D Genomics Proteomics (2017) 25:53–61. doi:10.1016/j.cbd.2017.10.006
125. Bach LA. Recent insights into the actions of IGFBP-6. J Cell Commun Signal (2015) 9(2):189–200. doi:10.1007/s12079-015-0288-4
127. Neumann GM, Bach LA. The N-terminal disulfide linkages of human insulin-like growth factor binding protein-6 (hIGFBP-6) and hIGFBP-1 are different as determined by mass spectrometry. J Biol Chem (1999) 274(21):14587–94. doi:10.1074/jbc.274.21.14587
128. Zeng C, Feng X, Wang W, Lv L, Fang C, Chi L, et al. Decreased expression of insulin-like growth factor binding protein 6 is associated with gastric adenocarcinoma prognosis. Oncol Lett (2017) 13(6):4161–41–68. doi:10.3892/ol.2017.5993
129. Raykha C, Crawford J, Gan BS, Fu LA, Bach LA, O’Gorman DB. IGF-II and IGFBP-6 regulate cellular contractility and proliferation in Dupuytren’s disease. Biochim Biophys Acta (2013) 1832(10):1511–9. doi:10.1016/j.bbadis.2013.04.018
130. Fu P, Yang Z, Bach LA. Prohibitin-2 binding modulates insulin-like growth factor binding protein-6 (IGFBP-6)-induced rhabdomyosarcoma cell migration. J Biol Chem (2013) 288(41):29890–900. doi:10.1074/jbc.M113.510826
131. Sueoka N, Lee HY, Wiehle S, Cristiano RJ, Fang B, Ji L, et al. Insulin-like growth factor binding protein-6 activates programmed cell death in non-small cell lung cancer cells. Oncogene (2000) 19(38):4432–6. doi:10.1038/sj.onc.1203813
132. Pooley NJ, Tacchi L, Secombes CJ, Martin SA. Inflammatory responses in primary muscle cell cultures in Atlantic salmon (Salmo salar). BMC Genomics (2013) 14:747. doi:10.1186/1471-2164-14-747
133. Edvardsen RB, Leininger S, Kleppe L, Skaftnesmo KO, Wargelius A. Targeted mutagenesis in Atlantic salmon (Salmo salar L.) using the CRISPR/Cas9 system induces complete knockout individuals in the F0 generation. PLoS One (2014) 9(9):e108622. doi:10.1371/journal.pone.0108622
134. Khalil K, Elayat M, Khalifa E, Daghash S, Elaswad A, Miller M, et al. Generation of myostatin gene-edited channel catfish (Ictalurus punctatus) via zygote injection of CRISPR/Cas9 System. Sci Rep (2017) 7(1):7301. doi:10.1038/s41598-017-07223-7
Keywords: insulin-like growth factor binding protein, teleost fish, genome duplication, physiology, comparative biology, gene family evolution
Citation: Garcia de la Serrana D and Macqueen DJ (2018) Insulin-Like Growth Factor-Binding Proteins of Teleost Fishes. Front. Endocrinol. 9:80. doi: 10.3389/fendo.2018.00080
Received: 11 January 2018; Accepted: 22 February 2018;
Published: 12 March 2018
Edited by:
Briony Forbes, Flinders University, AustraliaReviewed by:
Gustavo M. Somoza, Instituto de Investigaciones Biotecnológicas (IIB-INTECH), ArgentinaCopyright: © 2018 Garcia de la Serrana and Macqueen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel Garcia de la Serrana, ZGdkbHNjQHN0LWFuZHJld3MuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.