- 1State Key Laboratory of Biocontrol, Institute of Aquatic Economic Animals, Guangdong Provincial Key Laboratory for Aquatic Economic Animals, College of Life Sciences, Sun Yat-Sen University, Guangzhou, China
- 2Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China
- 3Department of Experimental Research, Sun Yat-Sen University Cancer Center, Guangzhou, China
Interferon gamma (IFNγ) is a Th1 cytokine that is critical for innate and adaptive immunity. Toll-like receptors (TLRs) signaling pathways are critical in early host defense against invading pathogens. miR-146a has been reported to participate in the regulation of host immunity. The known mechanisms of integrations between the IFNγ and TLR signaling pathways are incompletely understood, especially in teleosts. In this study, orange-spotted grouper (Epinephelus coioides) IFNγ1 and IFNγ2, their biological activities, especially their involvements in TLR pathway, were explored. We identified and cloned two IFNγ genes of E. coioides, namely EcIFNγ1 and EcIFNγ2. The produced recombinant E. coioides IFNγ1 (rEcIFNγ1) and IFNγ2 (rEcIFNγ2) proteins showed functions, which are similar to those of other bony fishes, such as enhancing nitric oxide responses and respiratory burst response. rEcIFNγ2 could regulate TLR pathway by enhancing the promoter activity of miR-146a upstream sequence and thus increasing the expression level of miR-146a, which possibly targets TNF receptor-associated factor 6 (TRAF6), a key adapter molecule in TLR signaling pathway. Taken together, these findings unravel a novel regulatory mechanism of anti-inflammatory response by IFNγ2, which could mediate TLR pathway through IFNγ2–miR-146a–TRAF6 negative regulation loop. It is suggested that IFNγ2 may provide a promising therapeutic, which may help to fine tune the immune response.
Introduction
Interferon gamma (IFNγ) is a Th1 cytokine that is critical for almost all phases of immune and inflammatory responses. Only one single IFNγ gene is identified in mammalian (1), avian (2), and amphibian (3) species, while many fishes possess two IFNγ genes, namely IFNγ1 and IFNγ2. Like mammalian IFNγ, teleost IFNγs mediate their protective effects as an activator of macrophages, through the enhancing of respiratory burst activity, nitric oxide production, and bacterial phagocytosis (4–8), inducing of the expression of pro-inflammatory cytokines (4, 5, 9) and the typical antiviral genes (8, 10). Respiratory burst and nitric oxide are well known as potent antimicrobials (11).
Orange-spotted grouper (Epinephelus coioides) is one of the most commercially important species in Southeast Asian. However, along with the rapid development of aquaculture industry, diseases caused by viruses, bacteria, and parasites emerge more and more frequently and have led to great number of economic loss. Injection of Poly(I:C), an interferon inducer, or recombinant IFNa1 protein could provide significant protection against viruses in sevenband grouper (Epinephelus septemfasciatus) (12, 13). In addition, inactivated Singapore grouper iridovirus vaccine, which showed high efficiency in orange-spotted grouper, could induce the expression of type I interferon-stimulated genes, suggesting that type I interferon system may be involved in the antivirus immune responses (14). Taken together, it was suggested that interferons might play important roles in the immune system of groupers, while the function of IFNγ and its potential applications in grouper farming are still unclear and worth investigating.
Toll-like receptors (TLRs), a family of evolutionarily conserved receptors, play crucial roles in early host defense against invading pathogens. IFNγ was produced by natural killer and T cells during the recognition of pathogens by TLRs (15). Like mammalian IFNγ (16), teleost IFNγs also play various roles in response to bacterial or viral infection (8, 9, 17, 18). IFNγ and TLR signaling pathways are important for both innate and adaptive immune responses. However, the cross talk between IFNγ and TLR signal pathways is incompletely understood.
MicroRNAs (miRNAs) are conserved small non-coding RNAs that function as posttranscriptional regulators of gene expression by binding to the 3ʹ untranslated region (UTR) of target mRNAs (19–22). The genesis of miRNAs was mediated by a two-step processing pathway, in which long primary miRNAs are first processed to approximately 60-bp hairpin precursor miRNAs (pre-miRNAs), then, these pre-miRNAs are cleaved to generate mature miRNAs (23, 24). miR-146a has been reported to participate in the regulation of host immunity (25–28). miR-146a, whose transcription is controlled by NF-κB and is induced by TLR activation, negatively regulates the TLR pathway by targeting TNF receptor-associated factor 6 (TRAF6) and interleukin-1 receptor-associated kinase 1 (IRAK1), which are key adapter molecules in TLR signaling cascades, mediating activation of NF-κB pathway (29–34).
In the present study, we report the cloning, expression profiles of E. coioides IFNγ1 and IFNγ2 and their potential functions in regulation of immune response. First, the functional recombinant E. coioides IFNγ1 (rEcIFNγ1) and recombinant E. coioides IFNγ2 (rEcIFNγ2) proteins were obtained. We detected their ability of enhancing respiratory burst activity, nitric oxide production. Then the potential cross talk between E. coioides IFNγs and TLR pathway was explored. Our data demonstrated that rEcIFNγ2 could regulate TLR pathway by enhancing the miR-146a upstream sequence transcription activity and thus increasing miR-146a expression, while miR-146a may target TRAF6. These findings unravel a novel regulatory mechanism of anti-inflammatory response by IFNγ2, which may function as novel negative regulator and promising therapeutic that help to fine tune the immune response.
Materials and Methods
Ethics Statement
All animal experiments were conducted in accordance with the guidelines and approval of the Animal Research and Ethics Committees of Sun Yat-Sen University. All efforts were made to minimize suffering.
Fish
Healthy E. coioides weighing approximately 500 g were purchased from the Guangdong Daya Bay Fishery Development Center (Huizhou, Guangdong, P. R. China). The fish were maintained in a recirculating seawater system, with a 12 h light/12 h dark cycle, at 25–30°C for 7 days before use. The fish were fed with commercial pellets twice daily. Food was withheld from the fish for 24 h prior to sample collection. All of the fish used in this study appeared to be healthy before the experiments were conducted. Before sample collection, the fish were anesthetized with MS-222 in dechlorinated water for 2 min.
Molecular Cloning
The head kidney was dissected from healthy E. coioides, and total RNA was extracted by TRIzol reagent (Invitrogen, USA). The first strand of cDNA was synthesized with ReverTra Ace qPCR RT Kit (TOYOBO, Japan) according to the manufacturer’s instructions.
The open reading frame (ORF) regions of IFNγ1 and IFNγ2 were amplified with the primers listed in Table 1. PCR amplification were performed at 94°C for 5 min, followed by 40 cycles at 94°C for 20 s, 55°C for 20 s, and 72°C for 45 s, with 72°C for a final 10 min at the end of the last cycle. All PCR products were ligated into the pTZ57R/T Vector (Fermentas, USA) after analyzing on 1.5% agarose gels, finally sequenced by Invitrogen Bioengineering Corporation, Guangdong, China.
The upstream sequence of pre-miR-146a was cloned from E. coioides genomic DNA using the primers of miR-146a F/R (Table 1). The pre-miR-146a upstream fragment was ligated into pGL4.10[luc2] vector (Promega, USA). Mutated versions of these constructs were obtained by site-directed mutagenesis using Mut Express II Fast Mutagenesis Kit (Vazyme, China).
TNF receptor-associated factor 6 3ʹUTR was cloned from head kidney cDNA by PCR with the primers of TRAF6 F/R (Table 1). To create 3ʹUTR luciferase reporter constructs, fragments of 3ʹUTR of TRAF6 gene were sub-cloned downstream of CMV-driven firefly luciferase cassette in pMIR-REPORT vector (Ambion, USA).
Bioinformatics
BLAST was used for identifying cDNA and deducing amino acid sequences in the NCBI.1 Prediction of ORF were performed in the DNAssist 2.0 software. Multiple-sequence alignment of the EcIFNγ with other vertebrate IFNγs were performed with the Clustal Omega.2 A vertebrata IFNγ phylogenetic tree was constructed with MEGA6 software using the maximum likelihood (ML) method, with a bootstrap of 100 times to verify its credibility. The SignalP program3 was applied to search for the signal peptide of vertebrata IFNγ. N-glycosylation sites were predicted by NetNGlyc4 and nuclear localization sequences (NLS) were predicted by Brameier et al. (35). Target prediction between miR-146a and the 3ʹUTR region of TRAF6 was performed using FINDTAR3. Sequences of mature miR-146a were obtained from our lab by whole genome sequencing. The pre-miR-14a was confirmed by structure prediction using RNAfold WebServer.5
Expression Study
Twelve tissues including the thymus, head kidney, trunk kidney, spleen, heart, gill, eye, skin, intestine, stomach, skin, and liver were aseptically dissected from three independent individuals. The tissue expression profiles were detected by real-time PCR performed with SYBR Green PCR Master Mix (Life Technologies, USA) and the primers of IFNγ1 F/R or IFNγ2 F/R, respectively. E. coioides 18S rRNA (18S-F/R) was amplified as an internal control (Table 1).
Production and Purification of rEcIFNγ Proteins
The putative mature peptides of E. coioides IFNγ1 and IFNγ2 were predicted by the SignalP program. Then, the cDNA fragments encoding the putative mature peptide with deletion of the the signal peptide from the N terminus were amplified by PCR using the primers of IFNγ1 NdeI-F/IFNγ1 XhoI-R and IFNγ2 EcoRI-F/IFNγ2 XhoI-R (Table 1), respectively. The fragments were separated on a 1.5% agarose gel and purified by Qiagen gel extraction kit (Qiagen, Germany). After subcloning into pTZ57R/T vector for sequencing, the fragments were digested with restriction enzymes and then inserted into the pET22b expression vector (Novagen, USA). The E. coli BL21 (DE3) cells transformed with IFNγ1 recombinant construct and E. coli Rosetta (DE3) cells transformed with IFNγ2 recombinant construct were induced by different concentrations of IPTG. The cells were collected by centrifugation and the resultant recombinant proteins named rEcIFNγ1 and rEcIFNγ2 were purified using His·Bind Column (Novagen) according to the manufacturer’s protocol. The purity of rEcIFNγs was checked on SDS-PAGE gel stained with coomassie brilliant blue R-250 (Sigma, USA), and the size of target proteins was measured by comparing the protein band location with a standard protein (Fermentas). Both proteins were detected by western blotting using a primary anti-His-tag monoclonal antibody (Novagen) and a secondary goat anti-mouse IgG (Amersham Biosciences, UK).
Isolation of Blood Lymphocytes and Head Kidney Monocytes
The whole blood of fish was directly obtained from three fish (n = 3) using 5 mL syringe. Blood lymphocytes were isolated from whole blood using lymphocyte separation medium. The primary head kidney cells were obtained as previously described (36). Head kidney monocytes were isolated from primary head kidney cells using monocyte isolating kit (TBDscience, China). The isolated cells were washed and enumerated on a hemocytometer with trypan blue and resuspended at a concentration of 106 cells/mL in tissue culture medium (TCM). The TCM was prepared from RPMI-1640 medium by adding 10% fetal bovine serum (Life Technologies), 2 mM l-glutamine (Sigma), and penicillin/streptomycin (Sigma).
Nitric Oxide Assay
Primary blood lymphocytes were distributed into 96-well plates at a density of 106 cells/mL. rEcIFNγ1 or rEcIFNγ2 were respectively added to the culture medium to reach a working concentration of 1, 10, and 100 ng/mL, and the control group was treated with TCM. These cells were incubated at 28°C in 5% CO2 for 72 h. Nitrite production was determined based on the Griess reaction with a NO determination kit (Beyotime Institute of Biotechnology, China).
Respiratory Burst Assay
Primary blood lymphocytes cultivation and in vitro stimulation were performed as described in Nitric oxide assay. The respiratory burst assay was performed as previously described (8). These cells were incubated at 28°C in 5% CO2 for 18 h. Then, NBT (2 mg/mL, Sigma) and PMA (final concentration, 100 ng/mL, Sigma) were added to the cell cultures at room temperature. Absolute methanol was applied to fix the pelleted cells, and 70% methanol was applied to remove the non-reduced NBT. After air drying, the reduced NBT was dissolved using 2 M KOH and the blue crystals in the cytoplasm were dissolved by DMSO. Finally, the OD values were detected at 630 nm.
Western Blot
Primary head kidney cells were distributed into 6-well plates at a density of 106 cells/mL. rEcIFNγ1 or rEcIFNγ2 were, respectively, added to the culture medium to reach a working concentration of 1, 10, and 100 ng/mL, and the control group was treated with TCM. After incubation for 3 h at 28°C in 5% CO2, these cells were washed with PBS and lysed in a lysis buffer (Beyotime), which contained protease inhibitors (Sigma) and phosphatase inhibitors (Sigma). The protein lysates were separated by SDS-PAGE, then transferred onto nitrocellulose membranes. The membranes were blocked in 5% BSA in TBST for 1 h at room temperature followed by incubations with primary antibodies and the relevant HRP-conjugated secondary antibodies. The membranes were processed for ECL Western Blotting Detection Reagents (Pierce). Meanwhile, the TRAF6 expression levels in primary head monocytes after rEcIFNγ1 or rEcIFNγ2 treatment were also detected by western blot. Antibodies used in the study were TRAF6 (Santa Cruz, CA, USA) and β-actin (Proteintech, USA). The bands were analyzed semiquantitively by densitometry (grayscale analysis) with ImageJ software and normalized to their controls.
miR-146a Expression Levels Detection
After treatment with rEcIFNγ1 or rEcIFNγ2, head kidney monocytes were collected. Total RNA was extracted by TRIzol reagent (Invitrogen), then reverse-transcribed and amplified with the Hairpin-it miRNAs RT-PCR Quantitation kit (GenePharma, China). E. coioides U6 snRNA served as control. The relative level of miR-146a expression was calculated using the comparative threshold (2−ΔΔCt) method (37).
Detection of the Relationship between miR-146a and TRAF6
In order to confirm whether miR-146a could directly target the mRNA of TRAF6, HEK-293T cells were transfected with 40 nM miR-146a mimics (GenePharma) or negative control (nc) (GenePharma) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. 24 h after transfection, luciferase activity in 293T cells was measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. The luciferase data were normalized by dividing the firefly luciferase activity by the activity of Renilla luciferase.
Primary head kidney monocytes were also transfected with miR-146a mimics or nc. The cells were lysed and the TRAF6 protein levels in the lysates were detected with western-blot.
Pre-miR-146a Upstream Sequence Activity Assay
Mutant or wild-type pre-miR-146a upstream-reporter vectors were transfected into 293T cell using Lipofectamine 2000. After 6 h, the cells were stimulated with rEcIFNγ1 or rEcIFNγ2 and luciferase activities were measured.
Statistical Analyses
All data were expressed as mean values ± SEM. Statistical analysis was carried out by one-way analysis of variance (ANOVA) or t-test. Differences were considered significant with a p-value less than 0.05 and were marked with asterisks.
Results
Cloning and Sequence Characterization of EcIFNγ1 and EcIFNγ2 Genes
The ORF of the EcIFNγ1 and EcIFNγ2 genes transcript was obtained (Figure 1). EcIFNγ1 ORF was 567 bp in length and translated into a 188-aa precursor molecule with a 19-aa signal peptide, while EcIFNγ2 ORF was 603 bp in length and encoded a 200-aa putative protein with a 19-aa signal peptide. The predicted mature EcIFNγ1 peptides contain two potential N-glycosylation sites (NTS and NVT), while EcIFNγ2 have one N-glycosylation site (NRT). Furthermore, EcIFNγ1 and EcIFNγ2 both possess an IFNγ signature sequence ([I/V]-Q-X-[K/Q]-A-X2-E-[L/F]-X2-[I/V]) at the C-terminus, which was conserved among known IFNγ molecules. In particular, a conserved motif RRRRRR similar to the nuclear localization signal of the known IFNγ molecules is found in EcIFNγ2 at its C-terminal tail, which was absent in EcIFNγ1.

Figure 1. The nucleotide and predicted peptide sequences of Epinephelus coioides IFNγ1 (A) and IFNγ2 (B). Start (ATG) and stop (TAG) codons are indicated in boldface type. The predicted signal sequence is underlined and glycosylation sites are double underlined. IFNγ signature sequence is boxed. The nuclear localization sequence (RRRR) is bolded.
Homology Alignment and Phylogenetic Analysis
Both EcIFNγ1 and EcIFNγ2 sequences and the IFNγs from other species were subjected to multiple alignment (Figure 2). The alignment confirms that EcIFNγ1 and EcIFNγ2 have relatively low sequence similarity compared with their vertebrate counterparts, respectively. EcIFNγ1 has 18.42–44.89% amino acid identity with other known fish IFNγ1, which shared the highest degree with tetraodon (Tetraodon nigroviridis) (44.89%). EcIFNγ2 shared a higher similarity with teleosts IFNγ2 sequences varied from 25.15 to 61.86% than with other vertebrates (12.41–22.02%), being most similar to IFNγ from Japanese flounder (Paralichthys olivaceus) (61.86%).

Figure 2. Multiple alignments of Epinephelus coioides IFNγ1 (A) and IFNγ2 (B). A multiple alignment of the deduced amino acid sequences of E. coioides IFNγ1 and IFNγ2 with those of other mammalian and piscine sequences was created using the ClustalX program. The putative signal peptides are indicated by single underlines. The signature motif ([IV]-Q-X-[KQ]-A-X2-E-[LF]-X2-[IV]) is boxed. The N-glycosylation sites are shaded in gray. The nuclear localization sequences sequence is in bold. Identical amino acids among all sequences are indicated by “*,” whereas those with high or low similarity are indicated by “:” and “.”. Amino acids were collected form NCBI were listed as follows: NP_000610.2, Homo sapiens IFNγ; NP_032363.1, Mus musculus IFNγ; NP_620235.1, Rattus norvegicus IFNγ; NP_990480.1, Gallus gallus IFNγ; XP_002188959.1, Taeniopygia guttata IFNγ; NP_999113.1, Sus scrofa IFNγ; XP_007664509.1, Ornithorhynchus anatinus IFNγ; XP_019398803.1, Crocodylus porosus IFNγ; XP_002938555.1, Xenopus tropicalis IFNγ; NP_001153975.1, Oncorhynchus mykiss IFNγ1; NP_001153976.1, O. mykiss IFNγ2; BAD72865.1, Danio rerio IFNγ1-1; NP_998029.1, Danio rerio IFNγ1-2; AHZ62713.1, Tetraodon nigroviridis IFNγ1; AHZ62714.1, T. nigroviridis IFNγ2; BAG50577.1, Paralichthys olivaceus IFNγ; CAJ98867.1, Cyprinus carpio IFNγ1; CAJ51089.1, C. carpio IFNγ2b; CAJ51088.1, C. arpio IFNγ2a; AAZ40504.1, Ictalurus punctatus IFNγ1; AAZ40505.1, I. punctatus IFNγ2a; AAZ40506.1, I. punctatus IFNγ2b.
A phylogenetic tree was also constructed based on the alignments of EcIFNγ1 and EcIFNγ2 sequences with that of other species (Figure 3). The EcIFNγ1 was branched with T. nigroviridis. EcIFNγ2 was most closely related to those of P. olivaceus and T. nigroviridis, all of which belonged to marine teleost IFNγ2 clade. The previously reported rainbow trout (Oncorhynchus mykiss) IFNγ1 might also belong to the IFNγ2 family based on the phylogenetic tree and protein alignments.
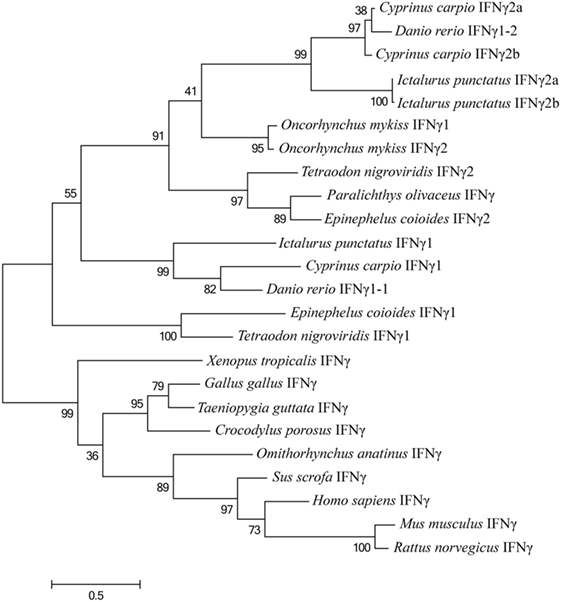
Figure 3. Phylogenetic tree, comparing the amino acid sequences of vertebrate interferon gamma (IFNγ) genes. This tree was generated with MEGA6 software using the maximum likelihood method, with a bootstrap of 100 times to verify its credibility. Values in percentage are indicated at branch nodes. All the amino acids sequences have already been applied in the multiple alignments in Figure 2.
Expression Profile of EcIFNγ1 and EcIFNγ2
Quantitative expression analysis of EcIFNγ1 or EcIFNγ2 in tissues of healthy fish revealed that EcIFNγ1 and EcIFNγ2 mRNA were expressed in different patterns. The highest mRNA levels of EcIFNγ1 were in the thymus and intestine, while the expression was relatively low in stomach, heart, eye, and spleen (Figure 4A). EcIFNγ2 was globally expressed in almost all tissues, and its expression levels were higher in gills and spleen, but lower in intestine, head kidney and eye (Figure 4B).
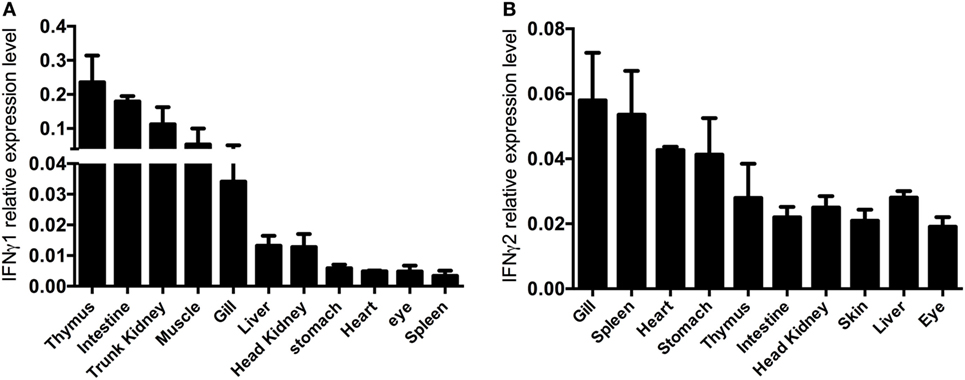
Figure 4. Constitutive interferon gamma (IFNγ) expression. cDNA of different organs of four control fish were used as template for quantitative real-time PCR. The IFNγ1 (A) and IFNγ2 (B) mRNA expression data are shown relative to the 18S rRNA.
Prokaryotic Production of rIFNγ1 and rIFNγ2
For further study of the biological activities of EcIFNγ1 and EcIFNγ2, the two putative mature peptides were expressed as C-terminal 6 His-tagged fusion protein in BL21 (DE3) and Rosetta (DE3) cells, respectively, followed by purification with affinity chromatography. SDS-PAGE analysis (Figures 5A,B) indicated that the purified sample exhibited a single protein band after induction by IPTG, and the molecular weights were close to the predicted size of the 6-His fused rEcIFNγ1 and rEcIFNγ2 peptides, respectively. Besides, both rEcIFNγ1 and rEcIFNγ2 were confirmed by Western blot assay with monoclonal antibody against the His-tag, and a single band was detected for the each of fused proteins (Figure 5C).
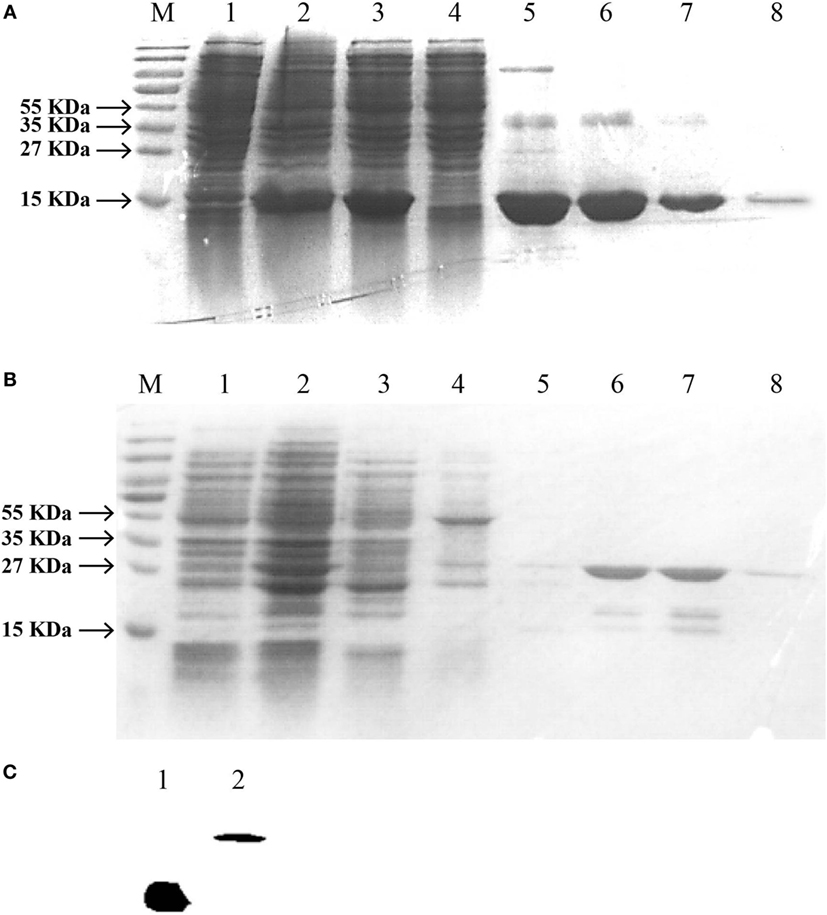
Figure 5. SDS-PAGE and western blotting analysis of Epinephelus coioides interferon gamma (IFNγ). IFNγ1 (A) or IFNγ2 (B) proteins was expressed using the pET22b expression vector. (A) Lane M: protein size marker; Lane 1: the total lysates from non-induced cells; Lane 2: the total lysates from IPTG-induced cells; Lane 3: soluble IFNγ1 protein; Lane 4–7: IFNγ1 protein by washing buffer with different imidazole concentration; Lane 8: purified recombinant protein with a His-tag; (B) Lane M: protein size marker; Lane 1: the total lysates from non-induced cells; Lane 2: the total lysates from IPTG-induced cells; Lane 3: soluble IFNγ2 protein; Lane 4–8: IFNγ2 protein by washing buffer with pH6.3, pH5.9, pH5.4, pH5.0, and pH4.5. (C) Verification of recombinant IFNγ1 and IFNγ2 by Western blot. Lane 1: IFNγ1; Lane 2: IFNγ2; they were analyzed on a 15% SDS-PAGE gel, followed by western blotting analysis using a mAb against the His-tag. The molecular weight of the lane 1 IFNγ1 was the same to that of the lane 8 of Figure 5A. Similarly, that of lane 2 IFNγ2 was also the same to the lane 8 of Figure 5B.
rEcIFNγ1 and rEcIFNγ2 Activated Nitric Oxide Responses and Respiratory Burst Response
Blood lymphocytes were exposed to different concentrations of rEcIFNγ1 or rEcIFNγ2, followed by detection of the respiratory burst or nitric oxide levels. Both rEcIFNγ1 and rEcIFNγ2 at concentrations of 1 and 10 ng/mL induced nitric oxide responses in blood lymphocytes that were significantly higher than that of none recombinant protein treatment (Figure 6A). In addition, rEcIFNγ1 or rEcIFNγ2 at 1 ng/mL induced an obvious respiratory burst response in blood lymphocytes (Figure 6B).
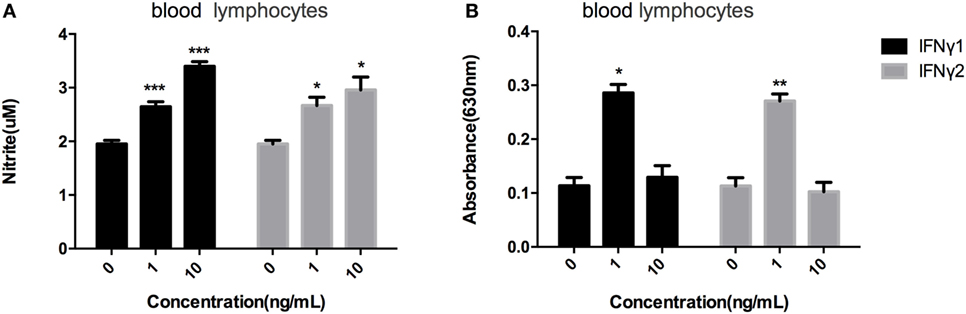
Figure 6. rEcIFNγ1 and rEcIFNγ2 activated nitric oxide responses and respiratory burst response. (A) Nitric oxide production of Epinephelus coioides primary blood lymphocytes after stimulation with various concentrations of rEcIFNγ1 or rEcIFNγ2 for 72 h was determined using the Griess reaction and nitrite concentration was determined using a nitrite standard curve. (B) Respiratory burst activity of E. coioides primary blood lymphocytes after stimulation with various concentrations of rEcIFNγ1 or rEcIFNγ2 for 18 h was detected. Each bar indicates the mean ± SEM (n = 6). Statistical analysis was done using t-test. *, **, and ***: significant differences from control cells at the p < 0.05, p < 0.01, and p < 0.001 levels, respectively.
rEcIFNγ1 and rEcIFNγ2 Induced miR-146a Expression Level by Enhancing the miR-146a Upstream Region Activity
The miR-146a expression level was detected by real-time PCR. Compared to the control group, the expression level of miR-146a in group with rEcIFNγ1 or rEcIFNγ2 treatment was significantly increased at 0.5 and 2 h (Figure 7A). miR-146a expression level reached a peak (increased approximately 11-fold) at about 2 h after rEcIFNγ2 treatment. The rapid induction of miR-146a in response to rEcIFNγ1 or rEcIFNγ2 suggested that miR-146a might be involved in response to IFNγ.
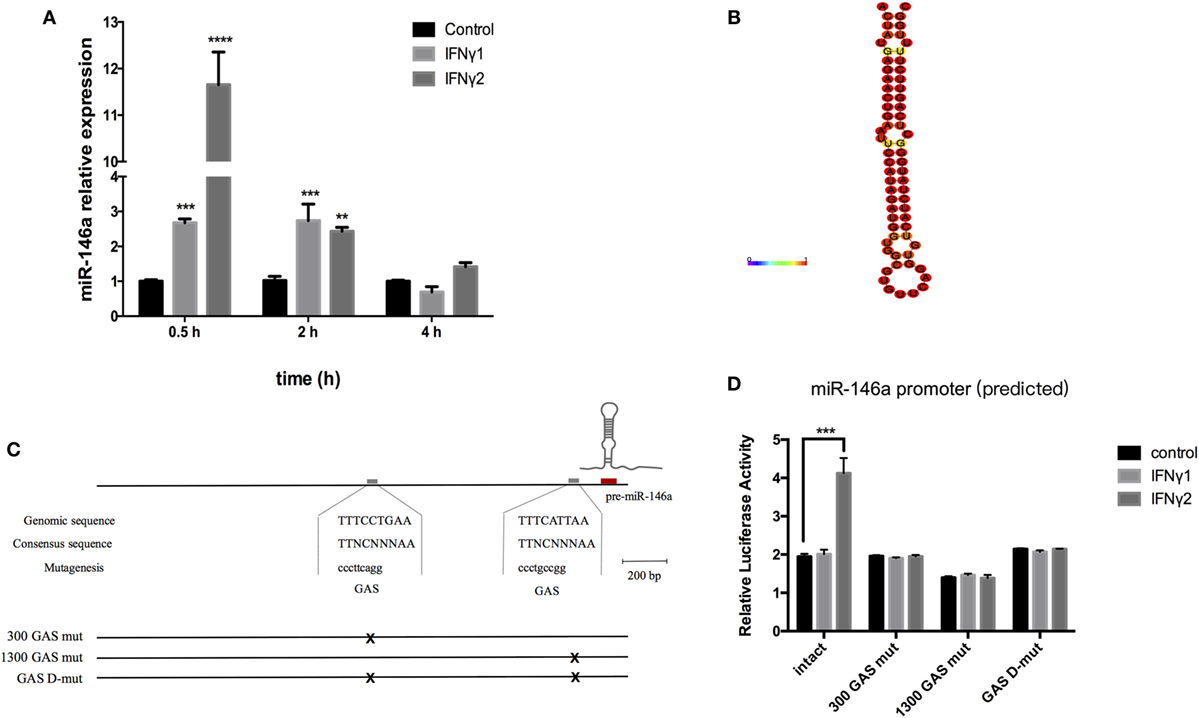
Figure 7. rEcIFNγ1 and rEcIFNγ2 induced the expression level of miR-146a. (A) Primary monocytes of head kidney were stimulated with 10 ng/mL rEcIFNγ1 or rEcIFNγ2 for different hours. The amount of miR-146a was normalized to that of U6 snRNA and was presented as the relative fold changes using the comparative threshold (2−ΔΔCt) method. (B) The stem loop of Epinephelus coioides pre-miR-146a was identified by RNA fold. (C) The approximately 2.5 kb genomic region upstream of mature miR-146a was analyzed. Two gamma-activated sequence (GAS, TTNCTTTAA) were found, namely 300 GAS and 1300 GAS. (D) The miR-146a upstream approximately 2.5 kb fragment was inserted into pGL4.10 vector. Dual-luciferase report assay was conducted after transfection with wild-type or GAS mutant pGL4-miR-146a vectors followed by rEcIFNγ1 or rEcIFNγ2 stimulation. The miR-146a upstream region luciferase activity significant increased after rEcIFNγ2 treatment. Upon mutant of either GAS, rEcIFNγ2 had no effect on luciferase activity of miR-146a upstream region. Each bar indicates the mean ± SEM (n = 6). Statistical analysis was performed using one-way analysis of variance. *, **, and ***: significant differences from control cells at the p < 0.05, p < 0.01, and p < 0.001 levels, respectively.
We first identified the stem loop of E. coioides pre-miR-146a (Figure 7B). Then, approximately 2.5 kb genomic region upstream of mature miR-146a was analyzed. Two gamma-activated sequence (GAS, TTNCTTTAA) were found, namely 300 GAS (−198 to −190 bp) and 1300 GAS (−1,136 to −1,128 bp) (Figure 7C). To explore the relationship between GAS and E. coioides IFNγs, the miR-146a upstream approximately 2.5 kb fragment was inserted into pGL4.10 vector. Dual-luciferase report assay was conducted after transfection with wild-type or mutant pGL4-miR-146a vectors followed by rEcIFNγ1 or rEcIFNγ2 stimulation. The miR-146a upstream region luciferase activity significant increased after rEcIFNγ2 treatment (Figure 7D). Upon mutantion of either GAS, rEcIFNγ2 had no effect on luciferase activity of miR-146a upstream region.
miR-146a May Target TRAF6
To further investigate the effect of miR-146a induction, we predicted its mRNA target sites. 3ʹUTR of TRAF6 mRNA was found to contain miR-146a target sequences (Figure 8A). In mammals, TRAF6 are key adapter molecules in TLR receptor signaling cascades, mediating activation of NF-κB pathway. To test the possibility that the TRAF6 expression may be regulated posttranscriptionally by miR-146a, we constructed a reporter vector that contained the firefly luciferase gene fused to 900 bp of the 3ʹUTR region of TRAF6 containing putative miR-146 target site. The reporter construct was transiently transfected into 293T cells together with miR-146a mimics. Compared to nc, a significant downregulative effect in the relative luciferase activity was observed when cells were transfected with miR-146a mimics (Figure 8B). We further examined the effect of miR-146a on TRAF6 protein expression in primary head kidney monocytes of E. coioides. After miR-146a mimic treatment, miR-146a mRNA expression was found to increase, which suggested that miR-146a mimics was successfully transfected into the primary head kidney monocytes (Figure 8C). The western blot results showed that TRAF6 protein level decreased after miR-146a mimics transfection (Figure 8D). These data suggested that the TRAF6 gene might be the target for posttranscriptional repression by miR-146a.
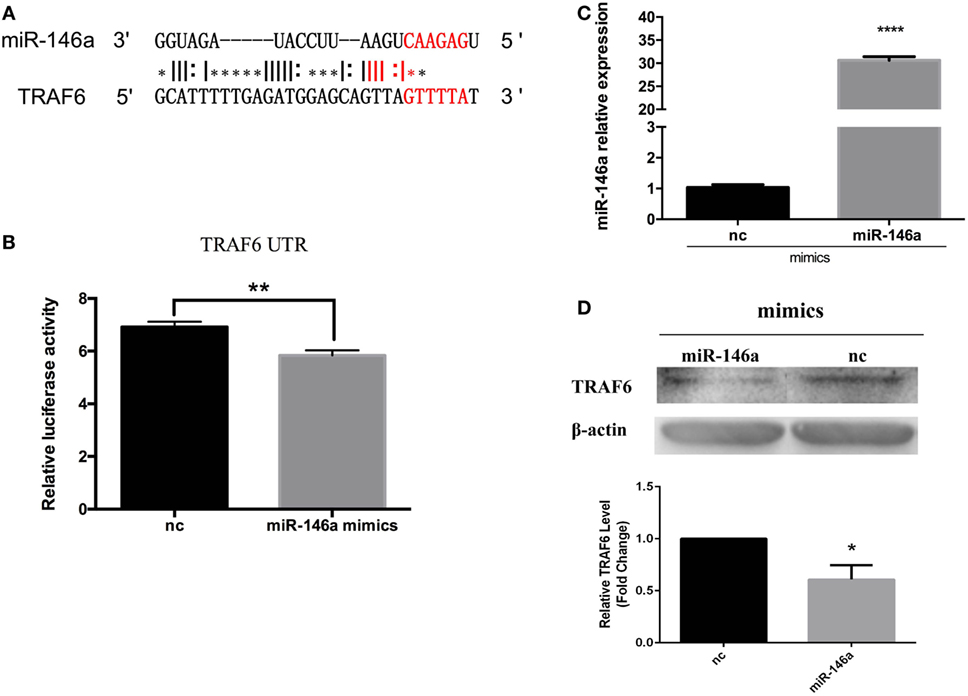
Figure 8. Epinephelus coioides miR-146a may target TNF receptor-associated factor 6 (TRAF6). (A) The target site in TRAF6 was predicted by FINDTAR3. (B) A reporter construct that contain the firefly luciferase gene fused to 900 bp of the 3ʹ untranslated region from TRAF6 containing putative miR-146 target site was obtained. miR-146a mimics induced the obvious downregulation of TRAF6 luciferase activity. (C) The E. coioides monocytes of head kidney were treated miR-146a mimics or negative control (nc). Then, the fold expression of miR-146a was calculated by the 2−ΔΔCt method using U6 as the reference gene, respectively. Compared with the nc, the expression of miR-146a significantly increased after miR-146a mimic treatment. (D) Treatment with miR-146a mimics triggered the weakening of the TRAF6 protein. The quantified graph was shown below the blots. Each bar indicates the mean ± SEM (n = 6). Statistical analysis was performed using one-way analysis of variance. *, **, and ***: significant differences from control cells at the p < 0.05, p < 0.01, and p < 0.001 levels, respectively.
rEcIFNγ1 and rEcIFNγ2 Regulated TRAF6 Expression
Compared to the control group, both rEcIFNγ1 and rEcIFNγ2 treatments obviously induced TRAF6 protein expression from 0.5 to 2 h. After 4 h, rEcIFNγ1 treatment slightly increased the TRAF6 protein expression, while rEcIFNγ2 treatment resulted in slight decrease (Figure 9).
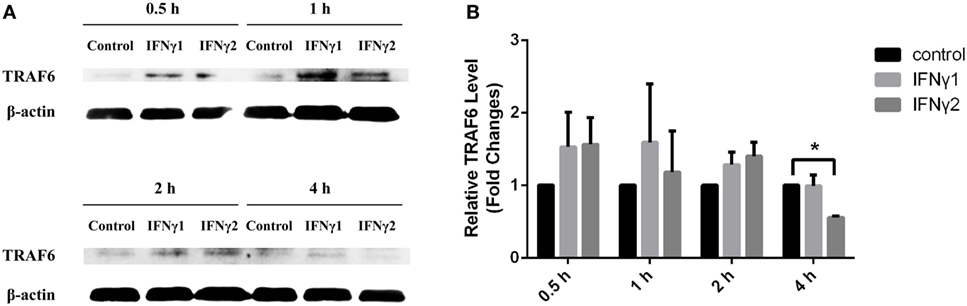
Figure 9. rEcIFNγ1 and rEcIFNγ2 regulated TNF receptor-associated factor 6 (TRAF6) expression. (A) Primary monocytes of head kidney were stimulated with 10 ng/mL rEcIFNγ1 or rEcIFNγ2 for different hours. The expression of TRAF6 protein was detected using western blot. The β-actin was used as an internal control. (B) Grayscale analysis was performed to semi-quantify the blots. *: significant difference from control blots at the p < 0.05 levels (n = 3).
Discussion
IFNγ1 and IFNγ2 are already identified in various teleosts. However, their immune functions, especially their involvement in TLR pathway, remained unclear. In this research, the biological activities of rEcIFNγ1 and rEcIFNγ2 were preliminarily characterized. We demonstrated that E. coioides IFNγ1 and IFNγ2 not only functioned similarly to those of other bony fishes but could also mediate TLR pathway through IFNγ2-miR-146a-TRAF6 negative regulation loop.
EcIFNγ1 shares highest sequence similarity with T. nigroviridis (44.89%) IFNγ1 molecules and lower similarity with IFNγ2 proteins. In bony fish, IFNγ2 shows higher sequence similarity with marine fishes generally than freshwater fishes and shares highest identity with P. olivaceus (61.86%). Based on the relatively low similarity between species, it might be suggested that IFNγs of different species play different roles. However, the protein tertiary structure of the whole IFNγs family as six α-helices remained conserved. Besides, a conserved motif ([IV]-Q-X-[KQ]-A-X2-E-[LF]-X2-[IV]) could be found in EcIFNγ1, EcIFNγ2 and all the other known IFNγs. The C-terminal cationic motif RRRRRR is found in EcIFNγ2, which is similar to its mammalian counterparts. This motif is considered as the NLS region that plays indispensable role in IFNγ functioning. Loss of NLS resulted in a failure of the inducing ability of rIFNγ on IP-10 expression in O. mykiss (7).
EcIFNγ1 and EcIFNγ2 present ubiquitous and constitutive expression in many tissues of teleost fish but show different expression patterns. In Atlantic cod (Gadus morhua) and Atlantic halibut (Hippoglossus hippoglossus L.), IFNγ2 is mainly expressed in gills and spleen, while its expression levels are low in stomach and liver (18, 38). In common carp (Cyprinus carpio L.) and goldfish, IFNγ2 is ubiquitously expressed in almost all tissues, while the highest expression levels could be found in immune organs as well (5, 39). In channel catfish (Ictalurus punctatus), IFNγ1 was found to be highly expressed in the thymus and intestines, while IFNγ2 was found to be only expressed in head kidney (40). In contrast, in rainbow trout, IFNγ1 mRNA transcription profile is similar to IFNγ2, both of whom were highly expressed in gills and spleen, with lower expression in head kidney and skin (7), which further suggested that the rainbow trout IFNγ1 might belong to IFNγ2 family.
In this study, EcIFNγ1 and EcIFNγ2 gene showed broad expression in all sampled tissues. EcIFNγ1 was expressed at highest level in thymus and intestines while EcIFNγ2 in spleen and gills, suggesting the two IFNγ genes might participate in signaling pathways of both immune and in non-immune tissues to resist pathogens in different manner probably.
Although recombinant proteins produced from E. coli generally lack glycosylation, whether the functions of interferons rely on glycosylation or not remains unclear. In black carp, it was reported that the un-glycosylated mutation form of IFNα could still be secreted and showed the similar antiviral ability as that of normal IFNα (41). For IFNγ, it was found that the unglycosylated IFNγ suffered from shorter half-life and lower protease resistance, while its function seemed to be unchanged (42). Functional in vitro assays exhibited that rEcIFNγ1 and rEcIFNγ2 could induce nitric oxide response, which effects are similar with the previous researches from mammals, goldfish, tetraodon, and common carp, in which, IFNγ were found to induce a nitric oxide response (5, 6, 8, 39, 43). Meanwhile, both rEcIFNγ1 and rEcIFNγ2 significantly enhanced the respiratory burst response, which effects were also observed in mammalian, goldfish and rainbow trout IFNγs (5–7, 44). These results suggested that the rEcIFNγ1 and rEcIFNγ2 obtained from E. coli have normal biological activities similar to those of the type II interferons of other teleosts.
After rEcIFNγ1 or rEcIFNγ2 treatment, the expression of TRAF6 protein was first upregulated, which lasted for 2 h and then gradually decreased to the basic level. It suggested that there might also exist a cross talk between IFNγ and TLR signaling pathway in teleost. In mammals, IFNγ regulates TLR signaling components (45) by including positive TLR signaling components, such as TLR itself, signaling adaptors MyD88 and IRAK1 (46, 47), and inhibitory TLR signaling components, such as SOCS1 and SOCS3 (48). IFNγ also could modulate the expression of ICSBP expression, which may participate more directly in the TLR signaling pathway, via interaction with TRAF6 (49). Our results indicated a tight interplay between E. coioides IFNγs and TLR signaling pathways through induction of TRAF6 proteins.
miR-146a is one of the most important and well-characterized miRNAs (26, 27, 50) and is reported as a key regulator of the immune response through mediating TLR signaling. In mammals, the TLR4 signaling pathway can activate the expression of miR-146a mediated by NF-κB (30). Besides, miR-146a negatively regulates NF-κB activation through targeted inhibition of the signaling proteins of innate immune responses, such as NF-κB inducers TRAF6 and IRAK1 (51). In shellfish, miR-146a was identified in Pinctada martensii and represented a critical role in inflammatory response (52). In consistent with our results of the rEcIFNγ1 and rEcIFNγ2 induction of miR-146a expression, miR-146a expression level was also upregulated when A375 cells were stimulated with IFNγ in a short time (less than 12 h) (53). It raised a question whether IFNγ was involved in the regulation of miR-146a expression. What is more, the luciferase reporter assay further suggested that rEcIFNγ2 enhanced miR-146a upstream sequence luciferase activity, which was mediated by GAS site in miR-146a upstream region. Meanwhile, as the findings of previous researches in mammals, E. coioides miR-146a may target TRAF6, which is an adaptor in TLR pathway. Thus, we proposed that an IFNγ2-miR-146a-TRAF6 negative loop may exist in teleost. In addition, overexpression of NF-κB p65 increased the miR-146a upstream sequence luciferase reporter activity, which is mediated by binding to NF-κB binding site in miR-146a upstream region (data not shown). These results also showed that the IFNγ2-induced transcription of miR-146a in E. coioides relied on NF-κB as that of mammals.
Our previous study in T. nigroviridis indicated that although T. nigroviridis IFNγs were not able to protect T. nigroviridis from Vibrio parahaemolyticus infection, T. nigroviridis IFNγ1 may promote an overwhelming inflammatory response, whereas T. nigroviridis IFNγ2 suppressed the inflammatory reaction promoted by V. parahaemolyticus infection (8). These further supported our speculation that rEcIFNγ2 would regulate immune response by IFNγ2-miR-146a-TRAF6 negative regulation loop (Figure 10). A model of pathogen infection of E. coioides is needed to be established to further explore and characterize the E. coioides IFNγs function in vivo during immune response.
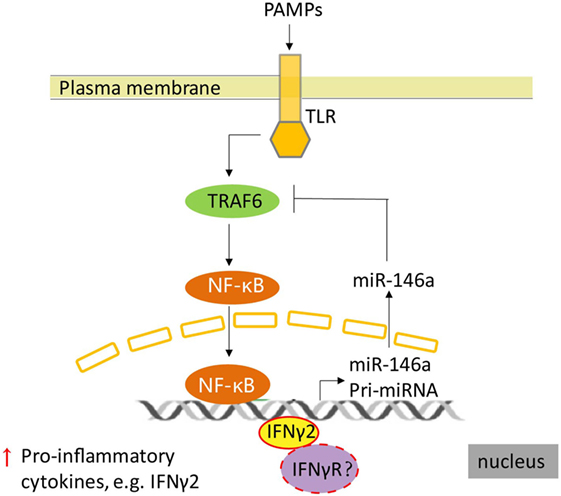
Figure 10. A supposed schematic of regulation between IFNγ2 and toll-like receptor (TLR) pathway in Epinephelus coioides. TLRs recognized the components of bacterial, then triggered the inflammatory response mediated by NF-κB signal, which induced the expression of pro-inflammatory factors, including IFNγ2. In E. coioides, subsequently, the expression of miR-146a was activated by NF-κB (the data were not shown) and IFNγ2. Upregulation of miR-146a reduced the expression of TNF receptor-associated factor 6, which is a signal transduction component in the TLR signaling pathway. Then the NF-κB signal was regulated. Meanwhile, how does E. coioides IFNγ2 activate the transcription of miR-146a and whether IFNγ receptor is involved in this regulation still remain unclear in E. coioides.
Ethics Statement
All animal experiments were conducted in accordance with the guidelines and approval of the Animal Research and Ethics Committees of Sun Yat-Sen University. All efforts were made to minimize suffering.
Author Contributions
WP and D-QL conceived and designed the experiments; WP and YS performed most of the experiments; L-GH, XY, and R-ZL performed some experiments; Y-SL and XD contributed reagents/materials. YZ and H-RL provided scientific advice; WP analyzed the data and wrote the manuscript. D-QL and G-FL revised and edited the manuscript. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jia-Nan He for his excellent technical assistance.
Funding
This work was supported by the National Natural Science Foundation of China (No.31001126), Guangdong Natural Science Foundation (No. 2014A030313214, 2015A030313069, 2015A030313409), Science and Technology Planning Project of Guangzhou (No.201607020014 and 201607010043), Modern Agriculture Talents Support Program (2016-2020), the Special Fund for Fisheries-Scientific Research of Guangdong Province (A201501A03, A201600A02) and the Fundamental Research Funds for the Central Universities (161gzd14, 161gpy35).
Abbreviations
IFNγ, interferon gamma; TLR, toll-like receptor; rEcIFNγ1, recombinant E. coioides IFNγ1; rEcIFNγ2, recombinant E. coioides IFNγ2; TRAF6, TNF receptor-associated factor 6; IRAK1, interleukin-1 receptor-associated kinase 1; UTR, untranslated region; ORF, open reading frame; NLS, nuclear localization sequences; TCM, tissue culture medium; PAMPs, pathogen-associated molecular patterns; IFNγR, IFNγ receptor.
Footnotes
References
1. Zimonjic DB, Rezanka LJ, Evans CH, Polymeropoulos MH, Trent JM, Popescu NC. Mapping of the immune interferon gamma gene (IFNG) to chromosome band 12q14 by fluorescence in situ hybridization. Cytogenet Cell Genet (1995) 71:247–8. doi:10.1159/000134119
2. Kaiser P, Wain HM, Rothwell L. Structure of the chicken interferon-gamma gene, and comparison to mammalian homologues. Gene (1998) 207:25–32. doi:10.1016/S0378-1119(97)00600-8
3. Qi Z, Nie P, Secombes CJ, Zou J. Intron-containing type I and type III IFN coexist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J Immunol (2010) 184:5038–46. doi:10.4049/jimmunol.0903374
4. Arts JAJ, Tijhaar EJ, Chadzinska M, Savelkoul HFJ, Kemenade BMLV. Analysis of carp interferon-γ: evolutionary conservation of classical phagocyte activation. Fish Shellfish Immunol (2010) 29:793–802. doi:10.1016/j.fsi.2010.07.010
5. Grayfer L, Belosevic M. Molecular characterization, expression and functional analysis of goldfish (Carassius aurutus L.) interferon gamma. Dev Comp Immunol (2009) 33:235–46. doi:10.1016/j.dci.2008.09.001
6. Grayfer L, Garcia EG, Belosevic M. Comparison of macrophage antimicrobial responses induced by type II interferons of the goldfish (Carassius auratus L.). J Biol Chem (2010) 285:23537–47. doi:10.1074/jbc.M109.096925
7. Zou J, Carrington A, Collet B, Dijkstra JM, Yoshiura Y, Bols N, et al. Identification and bioactivities of IFN-gamma in rainbow trout Oncorhynchus mykiss: the first Th1-type cytokine characterized functionally in fish. J Immunol (2005) 175:2484–94. doi:10.4049/jimmunol.175.4.2484
8. Peng W, Lu D-Q, Li G-F, Zhang X, Yao M, Zhang Y, et al. Two distinct interferon-γ genes in Tetraodon nigroviridis: functional analysis during Vibrio parahaemolyticus infection. Mol Immunol (2016) 70:34–46. doi:10.1016/j.molimm.2015.12.004
9. Jung CY, Hikima J, Ohtani M, Jang HB, del Castillo CS, Nho SW, et al. Fish Shellfish Immunol (2012) 33:197–203. doi:10.1016/j.fsi.2012.04.015
10. Sun B, Skjaeveland I, Svingerud T, Zou J, Jørgensen J, Robertsen B. Antiviral activity of salmonid gamma interferon against infectious pancreatic necrosis virus and salmonid alphavirus and its dependency on type I interferon. J Virol (2011) 85(17):9188–98. doi:10.1128/JVI.00319-11
11. Urban CF, Lourido S, Zychlinsky A. How do microbes evade neutrophil killing? Cell Microbiol (2006) 8:1687–96. doi:10.1111/j.1462-5822.2006.00792.x
12. Ohta T, Ueda Y, Ito K, Miura C, Yamashita H, Miura T, et al. Anti-viral effects of interferon administration on sevenband grouper, Epinephelus septemfasciatus. Fish Shellfish Immunol (2011) 30:1064–71. doi:10.1016/j.fsi.2011.02.003
13. Nishizawa T, Takami I, Kokawa Y, Yoshimizu M. Fish immunization using a synthetic double-stranded RNA Poly(I:C), an interferon inducer, offers protection against RGNNV, a fish nodavirus. Dis Aquat Organ (2009) 83:115–22. doi:10.3354/dao02001
14. Ou-yang Z, Wang P, Huang X, Cai J, Huang Y, Wei S, et al. Immunogenicity and protective effects of inactivated Singapore grouper iridovirus (SGIV) vaccines in orange-spotted grouper, Epinephelus coioides. Dev Comp Immunol (2012) 38:254–61. doi:10.1016/j.dci.2012.07.004
15. Sturge CR, Benson A, Raetz M, Wilhelm CL, Mirpuri J, Vitetta ES, et al. TLR-independent neutrophil-derived IFN-γ is important for host resistance to intracellular pathogens. Proc Natl Acad Sci U S A (2013) 110:10711–6. doi:10.1073/pnas.1307868110
16. Scharton-Kersten TM, Wynn TA, Denkers EY, Bala S, Grunvald E, Hieny S, et al. In the absence of endogenous IFN-gamma, mice develop unimpaired IL-12 responses to Toxoplasma gondii while failing to control acute infection. J Immunol (1996) 157:4045–54.
17. Sieger D, Stein C, Neifer D, van der Sar AM, Leptin M. The role of gamma interferon in innate immunity in the zebrafish embryo. Dis Model Mech (2009) 2:571–81. doi:10.1242/dmm.003509
18. Furnes C, Seppola M, Robertsen B. Molecular characterisation and expression analysis of interferon gamma in Atlantic cod (Gadus morhua). Fish Shellfish Immunol (2009) 26:285–92. doi:10.1016/j.fsi.2008.12.002
19. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function genomics: the miRNA genes. Cell (2004) 116:281–97. doi:10.1016/S0092-8674(04)00045-5
20. Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE (2007) 2007:re1. doi:10.1126/stke.3672007re1
21. O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of toll-like receptor signalling. Nat Rev Immunol (2011) 11:163–75. doi:10.1038/nri2957
22. Bartel DP. Review microRNAs: target recognition and regulatory functions. Cell (2009) 136:215–33. doi:10.1016/j.cell.2009.01.002
23. Gregory RI, Yan K, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The microprocessor complex mediates the genesis of microRNAs. Nature (2004) 208:2053–6. doi:10.1038/nature03120
24. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature (2005) 436:740–4. doi:10.1038/nature03868
25. Park H, Huang X, Lu C, Cairo MS, Zhou X. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J Biol Chem (2015) 290:2831–41. doi:10.1074/jbc.M114.591420
26. Saba R, Sorensen DL, Booth SA. MicroRNA-146a: a dominant, negative regulator of the innate immune response. Front Immunol (2014) 5:1–11. doi:10.3389/fimmu.2014.00578
27. Roos J, Enlund E, Jan-bernd F, Tews D, Holzmann K, Debatin K, et al. miR-146a-mediated suppression of the inflammatory response in human adipocytes. Sci Rep (2016) 6:38339. doi:10.1038/srep38339
28. Li M, Wang J, Fang Y, Gong S, Li M, Wu M, et al. microRNA-146a promotes mycobacterial survival in macrophages through suppressing nitric oxide production. Sci Rep (2016) 6:23351. doi:10.1038/srep23351
29. Cameron JE, Yin Q, Fewell C, Lacey M, Mcbride J, Wang X, et al. Epstein-Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol (2008) 82:1946–58. doi:10.1128/JVI.02136-07
30. Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A (2006) 103:12481–6. doi:10.1073/pnas.0605298103
31. Li Y, Shi X. MicroRNAs in the regulation of TLR and RIG-I pathways. Cell Mol Immunol (2013) 10:65–71. doi:10.1038/cmi.2012.55
32. Pacifico F, Crescenzi E, Mellone S, Iannetti A, Porrino N, Liguoro D, et al. Nuclear factor-κB contributes to anaplastic thyroid carcinomas through up-regulation of miR-146a. J Clin Endocrinol Metab (2010) 95:1421–30. doi:10.1210/jc.2009-1128
33. Li S, Yue Y, Xu W, Xiong S. MicroRNA-146a represses mycobacteria-induced inflammatory response and facilitates bacterial replication via targeting IRAK-1 and TRAF-6. PLoS One (2013) 8:81483-81438. doi:10.1371/journal.pone.0081438
34. Curtale G, Mirolo M, Renzi TA, Rossato M, Bazzoni F, Locati M. Negative regulation of toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci U S A (2013) 110:11499–504. doi:10.1073/pnas.1219852110
35. Brameier M, Krings A, MacCallum RM. NucPred–predicting nuclear localization of proteins. Bioinformatics (2007) 23(9):1159–60. doi:10.1093/bioinformatics/btm066
36. Yi S, Lu D, Peng W, Wang T, Zhang Y, Lin H. Differential expression profiling of spleen microRNAs in response to two distinct type II interferons in Tetraodon nigroviridis. PLoS One (2014) 9:1–12. doi:10.1371/journal.pone.0096336
37. Ma T, Jiang H, Gao Y, Zhao Y, Dai L, Xiong Q, et al. Microarray analysis of differentially expressed microRNAs in non-regressed and regressed bovine corpus luteum tissue; microRNA-378 may suppress luteal cell apoptosis by targeting the interferon gamma receptor 1 gene. J Appl Genet (2011) 52:481–6. doi:10.1007/s13353-011-0055-z
38. Øverga AC, Nepstad I, Nerland AH, Patel S. Characterisation and expression analysis of the Atlantic halibut (Hippoglossus hippoglossus L.) cytokines: IL-1 b, IL-6, IL-11, IL-12 b and IFNγ. Mol Biol Rep (2012) 39:2201–13. doi:10.1007/s11033-011-0969-x
39. Stolte EH, Savelkoul HFJ, Wiegertjes G, Flik G, Lidy Verburg-van Kemenade BM. Differsential expression of two interferon-gamma genes in common carp (Cyprinus carpio L.). Dev Comp Immunol (2008) 32:1467–81. doi:10.1016/j.dci.2008.06.012
40. Milev-Milovanovic I, Long S, Wilson M, Bengten E, Miller NW, Chinchar VG. Identification and expression analysis of interferon gamma genes in channel catfish. Immunogenetics (2006) 58:70–80. doi:10.1007/s00251-006-0081-x
41. Huang Z, Chen S, Liu J, Xiao J, Yan J, Feng H. IFNa of black carp is an antiviral cytokine modified with N-linked glycosylation. Fish Shellfish Immunol (2015) 46:477–85. doi:10.1016/j.fsi.2015.07.020
42. Razaghi A, Owens L, Heimann K. Review of the recombinant human interferon gamma as an immunotherapeutic: Impacts of production platforms and glycosylation. J Biotechnol (2016) 240:48–60. doi:10.1016/j.jbiotec.2016.10.022
43. Martin E, Nathan C, Xie QW. Role of interferon regulatory factor 1 in induction of nitric oxide synthase. J Exp Med (1994) 180:977–84. doi:10.1084/jem.180.3.977
44. Cassatella MA, Bazzoni F, Flynn RM, Dusi S, Trinchieri G, Rossi F. Molecular basis of interferon-gamma and lipopolysaccharide enhancement of phagocyte respiratory burst capability. Studies on the gene expression of several NADPH oxidase components. J Biol Chem (1990) 265:20241–6.
45. Schroder K, Sweet MJ, Hume DA. Signal integration between IFNγ and TLR signalling pathways in macrophages. Immunobiology (2006) 211:511–24. doi:10.1016/j.imbio.2006.05.007
46. Adib-Conquy M, Cavaillon JM. Gamma interferon and granulocyte/monocyte colony-stimulating factor prevent endotoxin tolerance in human monocytes by promoting interleukin-1 receptor-associated kinase expression and its association to MyD88 and not by modulating TLR4 expression. J Biol Chem (2002) 277:27927–34. doi:10.1074/jbc.M200705200
47. Bosisio D, Polentarutti N, Sironi M, Bernasconi S, Miyake K, Webb GR, et al. Stimulation of toll-like receptor 4 expression in human mononuclear phagocytes by interferon-gamma: a molecular basis for priming and synergism with bacterial lipopolysaccharide. Blood (2002) 99:3427–31. doi:10.1182/blood.V99.9.3427
48. Dalpke AH, Eckerle S, Frey M, Heeg K. Triggering of toll-like receptors modulates IFN signalling: Involvement of serine 727 STAT1 phosphorylation and suppressors of cytokine signaling. Eur J Immunol (2003) 33:1776–87. doi:10.1002/eji.200323621
49. Zhao J, Kong HJ, Li H, Huang B, Yang M, Zhu C, et al. IRF-8/ interferon (IFN) consensus sequence-binding protein is involved in toll-like receptor (TLR) signaling and contributes to the cross-talk between TLR and IFN-γ signaling pathways. J Biol Chem (2006) 281:10073–80. doi:10.1074/jbc.M507788200
50. Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell (2010) 142:914–29. doi:10.1016/j.cell.2010.08.012
51. Liu R, Liu C, Chen D, Yang W, Liu X, Liu G, et al. FOXP3 controls an miR-146/NFκB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res (2016) 75:1703–13. doi:10.1158/0008-5472.CAN-14-2108.FOXP3
52. Zheng Z, Liang J, Huang R, Du X, Wang Q, Deng Y, et al. Identification of a novel miR-146a from Pinctada martensii involved in the regulation of the inflammatory response. Fish Shellfish Immunol (2016) 54:40–5. doi:10.1016/j.fsi.2016.03.025
Keywords: Epinephelus coioides, IFNγ1, IFNγ2, miR-146a, TNF receptor-associated factor 6
Citation: Peng W, Sun Y, Li G-F, He L-G, Li R-Z, Liang Y-S, Ding X, Yu X, Zhang Y, Lin H-R and Lu D-Q (2018) Two Distinct Interferon-γ in the Orange-Spotted Grouper (Epinephelus coioides): Molecular Cloning, Functional Characterization, and Regulation in Toll-Like Receptor Pathway by Induction of miR-146a. Front. Endocrinol. 9:41. doi: 10.3389/fendo.2018.00041
Received: 02 October 2017; Accepted: 31 January 2018;
Published: 26 February 2018
Edited by:
Yan-ling Wang, Institute of Zoology (CAS), ChinaReviewed by:
Takashi Yazawa, Asahikawa Medical College, JapanTsubasa Sakai, Suntory Foundation for Life Sciences, Japan
Copyright: © 2018 Peng, Sun, Li, He, Li, Liang, Ding, Yu, Zhang, Lin and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan-Qi Lu, bHVkYW5xaUBtYWlsLnN5c3UuZWR1LmNu
 Wan Peng
Wan Peng Yan Sun1
Yan Sun1 Hao-Ran Lin
Hao-Ran Lin Dan-Qi Lu
Dan-Qi Lu