
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Endocrinol. , 13 December 2017
Sec. Obesity
Volume 8 - 2017 | https://doi.org/10.3389/fendo.2017.00349
This article is part of the Research Topic Understanding the Role of Gut Microbiota in Obesity View all 6 articles
The developing infant gut microbiome affects metabolism, maturation of the gastrointestinal tract, immune system function, and brain development. Initial seeding of the neonatal microbiota occurs through maternal and environmental contact. Maternal diet, antibiotic use, and cesarean section alter the offspring microbiota composition, at least temporarily. Nutrients are thought to regulate initial perinatal microbial colonization, a paradigm known as the “Restaurant” hypothesis. This hypothesis proposes that early nutritional stresses alter both the initial colonizing bacteria and the development of signaling pathways controlled by microbial mediators. These stresses fine-tune the immune system and metabolic homeostasis in early life, potentially setting the stage for long-term metabolic and immune health. Dysbiosis, an imbalance or a maladaptation in the microbiota, can be caused by several factors including dietary alterations and antibiotics. Dysbiosis can alter biological processes in the gut and in tissues and organs throughout the body. Misregulated development and activity of both the innate and adaptive immune systems, driven by early dysbiosis, could have long-lasting pathologic consequences such as increased autoimmunity, increased adiposity, and non-alcoholic fatty liver disease (NAFLD). This review will focus on factors during pregnancy and the neonatal period that impact a neonate’s gut microbiome, as well as the mechanisms and possible links from early infancy that can drive increased risk for diseases including obesity and NAFLD. The complex pathways that connect diet, the microbiota, immune system development, and metabolism, particularly in early life, present exciting new frontiers for biomedical research.
Microbes introduced through the mother can serve as regulators of the infant immune system by contributing to and altering the composition and diversity of the infant gut microbiota. Because neonates have a limited capacity to initiate an immune response, the blooming of “pioneering” microbes in the neonate exerts critically important effects on postnatal immune responses that can be more persistent than those resulting from microbiota disruption during adult life, highlighting the neonatal period as a critical developmental window. While many factors alter the microbiome throughout life, the early infant pattern of microbiome development can have life-long implications for disease risk (1, 2) and the timing and dynamics of bacterial colonization later in development. In the first 1,000 days of life, many biological systems are established and are developmentally driven in part by environmental stimuli. The developmental origins of health and disease paradigm suggests that nutrient deficiency as well as nutrient excess in utero and in early infancy increases susceptibility to metabolic disease later in life (3, 4) and across generations (5). Maternal obesity and poor diet can influence the types and abundance of pioneering microbes. Changes in the early microbiome are associated with inflammatory diseases, such as asthma, allergies, and increased obesity risk (1, 6, 7). Early microbes affect the liver and other organs through direct communication via the portal system, through alterations in metabolite production or gut barrier integrity, and the hematopoietic immune cell axis (Figure 1). In rodents, non-human primates, and in human infants born to obese mothers or those consuming a high-fat diet (HFD), infant dysbiosis increases risk for both obesity and non-alcoholic fatty liver disease (NAFLD), a prevalent and harmful disease in children and adults with limited treatment options. Improper “training” of the innate immune system by microbial crosstalk with the immune system has been implicated in obesity and NAFLD (8, 9). Given that inoculation of obese human or obese mouse microbiota to germ-free (GF) mice alters the assembly of their intestinal microbiome in a manner that favors adipogenesis and inflammation (10–16), modifications to the structure of the infant intestinal microbiome by maternal obesity is plausible and might carry similar metabolic risk to the infant.
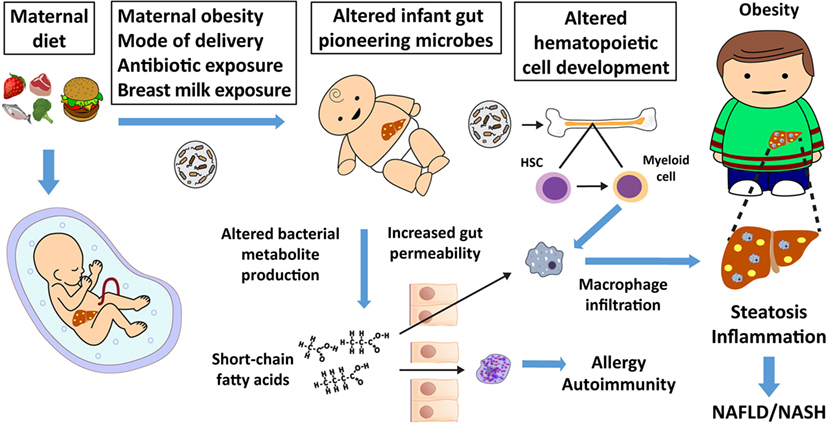
Figure 1. Maternal nutrition affects the fetal liver, microbiome, and offspring immunity, thereby increasing non-alcoholic fatty liver disease (NAFLD) and obesity risk. Maternal diet, mode of delivery, early offspring diet, and antibiotic exposure alter the infant’s pioneering microbiota and metabolite production. Changes in short-chain fatty acid production by the microbiota influence gut permeability, which can allow short-chain fatty acids and bacterial metabolites to directly influence adaptive and innate immune cell function and development. Altered adaptive immunity can lead to atopy, allergy, and autoimmunity. Microbes or their products alter hematopoietic stem cell (HSC) differentiation to macrophage progenitor cells in bone marrow and other tissues (17), which influences innate immunity. Altered innate immunity predisposes offspring to dysbiosis, macrophage activation, inappropriate macrophage infiltration, childhood obesity, and NAFLD, which often presents as non-alcoholic steatohepatitis (NASH) in obese youth.
Current data suggest that the microbiome might begin to assemble earlier in development than previously thought. Some have even suggested that the infant gut microbiome is partially established during pregnancy. Supporting this idea are reports that bacterial DNA is detectable in placental tissue (18) and amniotic fluid (19). Additionally, microbes have been detected in infant meconium (20, 21) wherein the bacterial composition of meconium is about 61% shared with the bacterial composition of amniotic fluid, further suggesting bacterial colonization in utero (20). These findings challenge the idea that the infant develops in a sterile environment. However, functional studies of the microbiota inhabiting these tissues are lacking, as many studies have only documented the presence of bacterial DNA, rather than viable bacteria, in embryonic tissues (22). Whether bacterial DNA detected during an uneventful, healthy pregnancy represents a transient exposure to microbes or microbial products (akin to transient bacteremia occurring in healthy adults), a potentially pathogenic phenomenon, or nascent assembly and succession of a microbial community is unknown.
The concept that the first nutrient availability dictates the community structure of the intestine suggested a hypothesis, first termed the “Restaurant” hypothesis (23, 24), which proposes that nutritional stress can alter the original colonizing bacteria, particularly Escherichia coli, and signaling pathways controlled by microbial mediators. Since bacterial species grow at different rates under nutrient-limiting conditions, the rate of growth of each species during colonization can be modulated by competitive metabolic interactions within the intestine (25). E. coli normally represents only a small fraction of the human adult gut ecosystem; however, in the neonatal gut, its prevalence is higher (26). Under healthy conditions, vaginally delivered infants born to normal-weight mothers are initially colonized by mostly facultative anaerobic bacteria including species of Escherichia, Staphylococcus, and Streptococcus (21). E. coli is metabolically flexible and has a short doubling time, making this bacterium highly adaptable and allowing it to flourish in the presence of oxygen and nitrogen. Due to these attributes, its presence might explain significant functional variability in the human gut microbiome (27). Presumably, by consuming oxygen, altering the pH, lowering the redox potential, and producing carbon dioxide and nutrients, E. coli make the habitat in the gut suitable for colonization by strict anaerobes.
Although certain early-colonizing gut bacteria are thought to have evolved mechanisms that influence the growth of other microbes and maturation of the host immune system, nutrients can influence the gut microbial community by regulating the host gut epithelium (28). This creates specific niches for the early gut microbiota and for other microorganisms with similar nutrient biochemical capacity. A recent study reported that the microbiota is fundamentally shaped by the availability of nitrogen and microbial strategies for obtaining nitrogen in the gut (29), suggesting that the competitive advantage of certain bacteria in obtaining nitrogen can shape microbiota composition. This result reinforces the restaurant concept; what feeds the developing microbiota determines its composition, and hence its function.
Evidence that maternal diet is a significant driver of the infant microbiota composition comes from studies in non-human primates demonstrating that maternal HFD, independent of body habitus, results in the loss of key bacteria in offspring including decreased Campylobacter, Helicobacter, and Bacteroidetes, as well as decreased overall bacterial diversity when compared with control diet-fed mothers (2). This dysbiosis was not fully corrected by weaning offspring onto a control diet at a time when the microbiome is thought to be stabilized (2). Evidence based on diet recall suggests that HFD in human mothers affects the neonatal microbiota (30); while in a different study (31), feeding pregnant mice an HFD altered microbiota composition in the offspring, including an increase in Lachnospiraceae and Clostridiales, despite offspring being weaned onto a control diet. In addition to exhibiting dysbiosis, these offspring also had worse outcomes in models of infection, autoimmunity, and allergic sensitization. This has important clinical relevance as the early-colonizing bacteria, driven by maternal diet, can have a long-lasting effect on the commensal microbe population in offspring, setting the stage for increased risk of immunologic and metabolic disease. It is also important to note that maternal diet impacts multiple metabolic systems that can contribute to obesity, such as behavior and appetite (32).
With regard to specific gut bacteria in infants born to obese mothers, we reported that vaginally delivered, exclusively breastfed infants with no neonatal or postnatal exposure to antibiotics born to mothers with obesity had a significant 50% reduction in Gammaproteobacteria at 2 weeks of age compared with infants born to normal-weight mothers (33). This relative depletion in Proteobacteria was also reported in 2-day-old infants delivered vaginally, but not by cesarean section, to overweight/obese mothers (34). These results suggest that differences in the relative abundance of Proteobacteria might stem from vertical transmission of microbiota from the mother, but this requires confirmation of the mothers’ bacteria. Additional studies (35, 36) have suggested that maternal obesity can alter early gut microbes in the offspring, although factors such as antibiotic use, cesarean section delivery, and weight gain during pregnancy might confound maternal obesity as a factor impacting early offspring microbiota composition. Human milk oligosaccharides (37) and secretory IgA production (38) are involved in the selective suppression of Proteobacteria during establishment of the early gut microbiota. A delayed or reduced Proteobacteria content might have important consequences on lipopolysaccharide (LPS) production that is critical for immune education as discussed below.
Maternal obesity is frequently associated with cesarean delivery, and this mode of delivery carries increased risk of pediatric obesity (39). In addition, early antibiotic exposure and neonatal/infant diet influence the initial gut microbial community, and these changes are at least somewhat predictive of higher risks of allergy, atopy, and obesity (28, 29, 40, 41). Breastfeeding alters the composition of Bifidobacteriaceae and Lactobacillaceae in the early infant gut and promotes growth of important pro-inflammatory bacteria when compared with formula feeding (42, 43). Besides being a potential source of bacteria, human milk contains human milk oligosaccharides, a group of unconjugated glycans resistant to human enzymatic digestion which act as prebiotics for certain infant gut bacteria such as Bifidobacterium. Microbiota in breast milk from mothers with obesity have been shown to have less community diversity, with increased Staphylococcus and decreased Bifidobacterium, than that of normal-weight mothers (44). However, whether this is a true microbial community with a low-biomass relative to the gut, or whether these findings represent contamination by maternal skin or other body sites needs clarification. An urgent question that remains unanswered is whether maternal phenotype (i.e., maternal obesity or diabetes, or maternal diet) can also modify disease risk by altering the glycobiome of human milk.
How the early microbiome impacts the immune system in the short- and long-term remains a critical concern. The microbiome has been implicated in altering macrophage development and polarization and an extensive area of research focuses on how bacterial metabolites, such as short-chain fatty acids, regulate macrophage function (45). A recent study showed that monocytes derived from newborn infants born to obese mothers displayed reduced LPS responsiveness, associated with altered cytokine promoter methylation (46), suggestive of early programming of the innate immune system by maternal obesity. An important role for early nutrition in the control of immune function is the fermentation of dietary polysaccharides to short-chain fatty acids by the microbiota. Acetate, proprionate, and butyrate have been shown to remodel regulatory T cell expansion in mice by inhibition of histone deacetylase activity in the FoxP3 promoter (47), as well as by causing histone deacetylase inhibition in murine macrophages that dampens immunity (48), and enhancing chemotaxis in neutrophils from rats (49), thereby skewing inflammation. In a murine model, oral administration of acetate during pregnancy was sufficient for the priming of FoxP3+ regulatory T cells and preventing allergic airway inflammation in the adult offspring (50), suggesting that in utero exposure to maternal gut microbial metabolites contributes to the development of immune function in the offspring. Although the mechanisms are poorly understood, the maternal microbiota has the ability to directly induce and/or regulate the offspring immune system (51). Using a reversible colonization model, microbial constituents, such as aryl hydrocarbon ligands, were shown to induce major transcriptional changes in the fetal gut and enhance the cellularity of the innate immune system (52). Fetal murine intestinal macrophages are constitutively non-responsive to LPS (53), whereas shortly after birth, acquisition of LPS resistance coincides with spontaneous activation of intestinal epithelial cells. This activation occurred in mice born vaginally, but not in offspring delivered by cesarean section or in toll-like receptor 4-deficient mice. These findings, coupled with local epithelial endotoxin measurements, identified LPS as a stimulatory agent (53) and suggest that epithelial activation shortly after birth is critical to maintain intestinal homeostasis.
The significant increase in common myeloid progenitor cells derived from bone marrow in diet-induced obese mice is associated with an altered microbiota composition (45). A key study has shown that the addition of microbes to gestating GF mice increases the prevalence of certain groups of intestinal innate immune system cells in offspring and also alters these cells’ transcriptional output (54), suggesting that maternal microbes alter function of the offspring immune system. A recent study has also shown that GF mice have compromised hematopoiesis and a decrease in several lines of myeloid cells in bone marrow and other tissues (17). The decrease in monocyte and granulocyte cells in bone marrow was reversible upon restoring the bacterial community via gavage in conjunction with heat killed E. coli, but not in conjunction with short-chain fatty acids (17). In a murine model, maternal HFD has also been shown to restrict the expansion and renewal of fetal hematopoietic stem cells by altering the transcriptional output of genes regulating metabolism, stress response, proliferation, and other functions (55). Importantly, these findings suggest that early microbes or maternal diet might alter immune function through alterations in hematopoietic cell development.
Gut microbes and inflammation play an active role in promoting the progression, and possibly the initiation, of NAFLD in children and adults (56–58). NAFLD describes a spectrum of liver abnormalities, ranging from simple hepatic steatosis to the more severe non-alcoholic steatohepatitis (NASH), with varying degrees of inflammation and fibrosis that can lead to cirrhosis. NAFLD affects 40% of all adults in the USA and well over 80% of adults with obesity (59). Importantly, NAFLD also affects approximately 34% of obese children ages 3–18 in North America and half have already progressed to NASH at the time of diagnosis (60–62). In addition, NAFLD is one of the fastest growing causes for liver cancer in the USA (63).
Intestinal dysbiosis has been correlated with NAFLD in children and adults; however, whether early life microbial composition influences hepatic fat accumulation and immunity before the disease occurs in humans is unclear (61, 64, 65). Our human studies (66), and those of others (67), using MRI/MRS, have documented that maternal BMI predicts newborn intrahepatocellular lipid storage, suggesting risk factors for pediatric obesity/NAFLD begin in early life (even in utero) and might permanently change the body’s structure, physiology, immune system, and metabolism, leading to an increased lifetime disease risk (68). A murine study showed that maternal HFD promoted increased liver adiposity, increased pro-fibrinogenic gene expression, and epigenetic changes in many genes in their offspring compared with offspring born from control diet-fed dams (69). This study also found that maternal HFD changed the microbial profile in offspring, significantly lowering the microbial biodiversity within a sample (69).
Highlighting the role of microbiota in the development of NAFLD is a recent study in mice showing that HFD-induced NASH can be attenuated by fecal transplant from chow-fed donors, as evidenced by a decrease in liver adiposity, and decreased inflammatory cytokines (70). Many other studies in rodent models similarly showed that probiotic treatment can slow NAFLD progression and help reverse the disease (71, 72). An increase in blood microbial DNA and a decrease in microbial biodiversity in patients with obesity was associated with liver fibrosis compared with patients without fibrosis (73). Therefore, there seems to be a distinct microbial community indicative of NAFLD rather than solely of obesity, which might suggest more specific microbial mechanisms that promote NASH.
Supporting the idea that prebiotics can be therapeutic is a study showing that bovine milk oligosaccharides prevented HFD-associated increases in gut permeability, dysbiosis, and obesity in mice (74). Furthermore, addition of the oligosaccharides decreased macrophage infiltration into adipose tissue, even in the setting of an HFD (74). A recent murine study reported that reversal of dysbiosis induced by maternal HFD and resulting pathologies might be possible by bacterial supplementation of defined microbial species in the offspring early in development, but not later in life (75). The study demonstrated not only that a maternal HFD causes dysbiosis, social problems, and defects in synaptic plasticity in offspring, but also that these defects can be reversed by administering Lactobacillus reuteri to the dysbiotic offspring who lacked this bacterium during weaning (75). This suggests that if definitive effects of the loss or the presence of certain bacteria can be identified, the negative effects of dysbiosis might be correctable with the relatively simple treatment of a probiotic in the early stages of life. While evidence suggests that microbe composition is alterable later in life by changes in the diet (76), the microbiota have different impacts on physiology during adulthood that make changes in composition less functionally relevant than changes in early life.
Despite considerable intra- and inter-personal variations in the infant microbiota, keystone species in the neonatal microbial community can be patterned by diet and can have life-long effects on immunity and disease pathways. Specifically, early life bacterial species composition might be driven by nutrient availability, which supports the “Restaurant” hypothesis. This concept is clinically relevant because bacteria and their metabolites seem to influence development of the immune system, which impacts many aspects of physiology. Microbial-derived factors shaping macrophage metabolism, transcription, and polarization toward functional phenotypes are needed in the gut and other tissues to ensure homeostasis. Remarkably, animal models fed an HFD during pregnancy are programmed for inflammation, even if the offspring are switched to a normal diet at weaning. Legacy effects of maternal obesity or diet exposure might direct the development of the infant microbiota and innate immunity, and underlie common disorders including obesity and NAFLD. Delineating the links between microbiota function and composition in early life will help draw mechanistic connections between altered microbiota composition and disease risk. Thus, advancing our understanding of early life contributors to the microbiota and metabolic and inflammatory diseases is a critical and unmet need, and should motivate future studies.
Both MN and JF participated in drafting, writing, and editing the manuscript. DF participated in writing and editing the manuscript.
JF is a consultant and recipient of a grant from Janssen Pharmaceuticals. MN and DF declared no conflict of interests.
The authors thank Rachel Janssen for her expert assistance editing the manuscript.
The work was supported in part by NIH R24-DK090964-08 and NIH P30-DK048520-23 (JF).
1. Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep (2012) 13:440–7. doi:10.1038/embor.2012.32
2. Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun (2014) 5:3889. doi:10.1038/ncomms4889
3. Dabelea D, Hanson RL, Lindsay RS, Pettitt DJ, Imperatore G, Gabir MM, et al. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes (2000) 49:2208–11. doi:10.2337/diabetes.49.12.2208
4. Friedman JE. Obesity and gestational diabetes mellitus pathways for programming in mouse, monkey, and man-where do we go next? The 2014 Norbert Freinkel award lecture. Diabetes Care (2015) 38:1402–11. doi:10.2337/dc15-0628
5. Derraik JG, Ahlsson F, Diderholm B, Lundgren M. Obesity rates in two generations of Swedish women entering pregnancy, and associated obesity risk among adult daughters. Sci Rep (2015) 5:16692. doi:10.1038/srep16692
6. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med (2015) 7:307ra152. doi:10.1126/scitranslmed.aab2271
7. Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature (2012) 488:621–6. doi:10.1038/nature11400
8. Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science (2010) 328:228–31. doi:10.1126/science.1179721
9. Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature (2008) 455:1109–13. doi:10.1038/nature07336
10. Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature (2016) 534:213–7. doi:10.1038/nature18309
11. Ellekilde M, Selfjord E, Larsen CS, Jakesevic M, Rune I, Tranberg B, et al. Transfer of gut microbiota from lean and obese mice to antibiotic-treated mice. Sci Rep (2014) 4:5922. doi:10.1038/srep05922
12. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science (2013) 341:1241214. doi:10.1126/science.1241214
13. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444:1027–31. doi:10.1038/nature05414
14. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A (2005) 102:11070–5. doi:10.1073/pnas.0504978102
15. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature (2006) 444:1022–3. doi:10.1038/4441022a
16. Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med (2009) 1:6ra14. doi:10.1126/scitranslmed.3000322
17. Khosravi A, Yáñez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promotes hematopoiesis tocontrol bacterial infection. Cell Host Microbe (2014) 15:374–81. doi:10.1016/j.chom.2014.02.006
18. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med (2014) 6:237ra65. doi:10.1126/scitranslmed.3008599
19. DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One (2008) 3:e3056. doi:10.1371/journal.pone.0003056
20. Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One (2014) 9:e90784. doi:10.1371/journal.pone.0090784
21. Brumbaugh DE, Arruda J, Robbins K, Ir D, Santorico SA, Robertson CE, et al. Mode of delivery determines neonatal pharyngeal bacterial composition and early intestinal colonization. J Pediatr Gastroenterol Nutr (2016) 63:320–8. doi:10.1097/MPG.0000000000001124
22. Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome (2017) 5:48. doi:10.1186/s40168-017-0268-4
23. Leatham-Jensen MP, Frimodt-Moller J, Adediran J, Mokszycki ME, Banner ME, Caughron JE, et al. The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infect Immun (2012) 80:1716–27. doi:10.1128/IAI.06193-11
24. Conway T, Cohen PS. Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol Spectr (2015) 3. doi:10.1128/microbiolspec.MBP-0006-2014
25. Freter R, Brickner H, Fekete J, Vickerman MM, Carey KE. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun (1983) 39:686–703.
26. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe (2015) 17:690–703. doi:10.1016/j.chom.2015.04.004
27. Bradley PH, Pollard KS. Proteobacteria explain significant functional variability in the human gut microbiome. Microbiome (2017) 5:36. doi:10.1186/s40168-017-0244-z
28. Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe (2013) 14:559–70. doi:10.1016/j.chom.2013.10.004
29. Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med (2016) 8:343ra82. doi:10.1126/scitranslmed.aad7121
30. Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, et al. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med (2016) 8:77. doi:10.1186/s13073-016-0330-z
31. Myles IA, Fontecilla NM, Janelsins BM, Vithayathil PJ, Segre JA, Datta SK. Parental dietary fat intake alters offspring microbiome and immunity. J Immunol (2013) 191:3200–9. doi:10.4049/jimmunol.1301057
32. Thompson JR, Valleau JC, Barling AN, Franco JG, DeCapo M, Bagley JL, et al. Exposure to a high-fat diet during early development programs behavior and impairs the central serotonergic system in juvenile non-human primates. Front Endocrinol (2017) 8:164. doi:10.3389/fendo.2017.00164
33. Lemas DJ, Young BE, Baker PR II, Tomczik AC, Soderborg TK, Hernandez TL, et al. Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome. Am J Clin Nutr (2016) 103:1291–300. doi:10.3945/ajcn.115.126375
34. Mueller NT, Shin H, Pizoni A, Werlang IC, Matte U, Goldani MZ, et al. Birth mode-dependent association between pre-pregnancy maternal weight status and the neonatal intestinal microbiome. Sci Rep (2016) 6:23133. doi:10.1038/srep23133
35. Cerdo T, Ruiz A, Jauregui R, Azaryah H, Torres-Espinola FJ, Garcia-Valdes L, et al. Maternal obesity is associated with gut microbial metabolic potential in offspring during infancy. J Physiol Biochem (2017). doi:10.1007/s13105-017-0577-x
36. Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS One (2014) 9:e113026. doi:10.1371/journal.pone.0113026
37. De Leoz ML, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, et al. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res (2015) 14:491–502. doi:10.1021/pr500759e
38. Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes (2014) 5:28–39. doi:10.4161/gmic.26489
39. Yuan C, Gaskins AJ, Blaine AI, Zhang C, Gillman MW, Missmer SA, et al. Association between cesarean birth and risk of obesity in offspring in childhood, adolescence, and early adulthood. JAMA Pediatr (2016) 170:e162385. doi:10.1001/jamapediatrics.2016.2385
40. Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell (2014) 158:705–21. doi:10.1016/j.cell.2014.05.052
41. O’Sullivan A, He X, McNiven EM, Haggarty NW, Lonnerdal B, Slupsky CM. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res (2013) 12:2833–45. doi:10.1021/pr4001702
42. O’Sullivan A, Farver M, Smilowitz JT. The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr Metab Insights (2015) 8:1–9. doi:10.4137/NMI.S29530
43. Madan JC, Hoen AG, Lundgren SN, Farzan SF, Cottingham KL, Morrison HG, et al. Association of cesarean delivery and formula supplementation with the intestinal microbiome of 6-week-old infants. JAMA Pediatr (2016) 170:212–9. doi:10.1001/jamapediatrics.2015.3732
44. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr (2012) 96:544–51. doi:10.3945/ajcn.112.037382
45. Luo Y, Chen GL, Hannemann N, Ipseiz N, Kronke G, Bauerle T, et al. Microbiota from obese mice regulate hematopoietic stem cell differentiation by altering the bone niche. Cell Metab (2015) 22:886–94. doi:10.1016/j.cmet.2015.08.020
46. Sureshchandra S, Wilson RM, Rais M, Marshall NE, Purnell JQ, Thornburg KL, et al. Maternal pregravid obesity remodels the DNA methylation landscape of cord blood monocytes disrupting their inflammatory program. J Immunol (2017) 199:2729–44. doi:10.4049/jimmunol.1700434
47. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol (2015) 8:80–93. doi:10.1038/mi.2014.44
48. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A (2014) 111:2247–52. doi:10.1073/pnas.1322269111
49. Vinolo MA, Rodrigues HG, Hatanaka E, Hebeda CB, Farsky SH, Curi R. Short-chain fatty acids stimulate the migration of neutrophils to inflammatory sites. Clin Sci (Lond) (2009) 117:331–8. doi:10.1042/CS20080642
50. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun (2015) 6:7320. doi:10.1038/ncomms8320
51. Macpherson AJ, de Aguero MG, Ganal-Vonarburg SC. How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol (2017) 17:508–17. doi:10.1038/nri.2017.58
52. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature (2016) 535:65–74. doi:10.1038/nature18847
53. Lotz M, Gutle D, Walther S, Menard S, Bogdan C, Hornef MW. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med (2006) 203:973–84. doi:10.1084/jem.20050625
54. Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science (2016) 351:1296–302. doi:10.1126/science.aad2571
55. Kamimae-Lanning AN, Krasnow SM, Goloviznina NA, Zhu X, Roth-Carter QR, Levasseur PR, et al. Maternal high-fat diet and obesity compromise fetal hematopoiesis. Mol Metab (2015) 4:25–38. doi:10.1016/j.molmet.2014.11.001
56. Wieland A, Frank DN, Harnke B, Bambha K. Systematic review: microbial dysbiosis and nonalcoholic fatty liver disease. Aliment Pharmacol Ther (2015) 42:1051–63. doi:10.1111/apt.13376
57. Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology (2016) 63:764–75. doi:10.1002/hep.28356
58. Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology (2014) 146:1513–24. doi:10.1053/j.gastro.2014.01.020
59. Cholankeril G, Perumpail RB, Pham EA, Ahmed A, Harrison SA. Nonalcoholic fatty liver disease: epidemiology, natural history, and diagnostic challenges. Hepatology (2016) 64:954. doi:10.1002/hep.28719
60. Anderson EL, Howe LD, Jones HE, Higgins JP, Lawlor DA, Fraser A. The Prevalence of non-alcoholic fatty liver disease in children and adolescents: a systematic review and meta-analysis. PLoS One (2015) 10:e0140908. doi:10.1371/journal.pone.0140908
61. Goyal NP, Schwimmer JB. The progression and natural history of pediatric nonalcoholic fatty liver disease. Clin Liver Dis (2016) 20:325–38. doi:10.1016/j.cld.2015.10.003
62. Brumbaugh DE, Friedman JE. Developmental origins of nonalcoholic fatty liver disease. Pediatr Res (2014) 75:140–7. doi:10.1038/pr.2013.193
63. Mittal S, El-Serag HB, Sada YH, Kanwal F, Duan Z, Temple S, et al. Hepatocellular carcinoma in the absence of cirrhosis in United States veterans is associated with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol (2016) 14:124–31.e1. doi:10.1016/j.cgh.2015.07.019
64. Ayonrinde OT, Oddy WH, Adams LA, Mori TA, Beilin LJ, de Klerk N, et al. Infant nutrition and maternal obesity influence the risk of non-alcoholic fatty liver disease in adolescents. J Hepatol (2017) 67(3):568–76. doi:10.1016/j.jhep.2017.03.029
65. Nobili V, Alkhouri N, Alisi A, Della Corte C, Fitzpatrick E, Raponi M, et al. Nonalcoholic fatty liver disease: a challenge for pediatricians. JAMA Pediatr (2015) 169:170–6. doi:10.1001/jamapediatrics.2014.2702
66. Brumbaugh DE, Tearse P, Cree-Green M, Fenton LZ, Brown M, Scherzinger A, et al. Intrahepatic fat is increased in the neonatal offspring of obese women with gestational diabetes. J Pediatr (2013) 162:930–6.e1. doi:10.1016/j.jpeds.2012.11.017
67. Modi N, Murgasova D, Ruager-Martin R, Thomas EL, Hyde MJ, Gale C, et al. The influence of maternal body mass index on infant adiposity and hepatic lipid content. Pediatr Res (2011) 70:287–91. doi:10.1203/PDR.0b013e318225f9b1
68. Wesolowski SR, Hay WW Jr. Role of placental insufficiency and intrauterine growth restriction on the activation of fetal hepatic glucose production. Mol Cell Endocrinol (2016) 435:61–8. doi:10.1016/j.mce.2015.12.016
69. Wankhade UD, Zhong Y, Kang P, Alfaro M, Chintapalli SV, Thakali KM, et al. Enhanced offspring predisposition to steatohepatitis with maternal high-fat diet is associated with epigenetic and microbiome alterations. PLoS One (2017) 12:e0175675. doi:10.1371/journal.pone.0175675
70. Zhou D, Pan Q, Shen F, Cao HX, Ding WJ, Chen YW, et al. Total fecal microbiota transplantation alleviates high-fat diet-induced steatohepatitis in mice via beneficial regulation of gut microbiota. Sci Rep (2017) 7:1529. doi:10.1038/s41598-017-01751-y
71. Kim DH, Kim H, Jeong D, Kang IB, Chon JW, Kim HS, et al. Kefir alleviates obesity and hepatic steatosis in high-fat diet-fed mice by modulation of gut microbiota and mycobiota: targeted and untargeted community analysis with correlation of biomarkers. J Nutr Biochem (2017) 44:35–43. doi:10.1016/j.jnutbio.2017.02.014
72. Xue L, He J, Gao N, Lu X, Li M, Wu X, et al. Probiotics may delay the progression of nonalcoholic fatty liver disease by restoring the gut microbiota structure and improving intestinal endotoxemia. Sci Rep (2017) 7:45176. doi:10.1038/srep45176
73. Lelouvier B, Servant F, Paisse S, Brunet AC, Benyahya S, Serino M, et al. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: a pilot analysis. Hepatology (2016) 64:2015–27. doi:10.1002/hep.28829
74. Hamilton MK, Ronveaux CC, Rust BM, Newman JW, Hawley M, Barile D, et al. Prebiotic milk oligosaccharides prevent development of obese phenotype, impairment of gut permeability, and microbial dysbiosis in high fat-fed mice. Am J Physiol Gastrointest Liver Physiol (2017) 312:G474–87. doi:10.1152/ajpgi.00427.2016
75. Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell (2016) 165:1762–75. doi:10.1016/j.cell.2016.06.001
Keywords: pregnancy, innate immunity, microbiome, non-alcoholic fatty liver disease, proteobacteria
Citation: Nash MJ, Frank DN and Friedman JE (2017) Early Microbes Modify Immune System Development and Metabolic Homeostasis—The “Restaurant” Hypothesis Revisited. Front. Endocrinol. 8:349. doi: 10.3389/fendo.2017.00349
Received: 24 August 2017; Accepted: 28 November 2017;
Published: 13 December 2017
Edited by:
Margaret Morris, University of New South Wales, AustraliaReviewed by:
Amanda Cox, Griffith University, AustraliaCopyright: © 2017 Nash, Frank and Friedman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacob E. Friedman, amVkLmZyaWVkbWFuQHVjZGVudmVyLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.