- BHF Centre for Cardiovascular Science, The Queen’s Medical Research Institute, University of Edinburgh, Edinburgh, UK
The tropical freshwater zebrafish has recently emerged as a valuable model organism for the study of adipose tissue biology and obesity-related disease. The strengths of the zebrafish model system are its wealth of genetic mutants, transgenic tools, and amenability to high-resolution imaging of cell dynamics within live animals. However, zebrafish adipose research is at a nascent stage and many gaps exist in our understanding of zebrafish adipose physiology and metabolism. By contrast, adipose research within other, closely related, teleost species has a rich and extensive history, owing to the economic importance of these fish as a food source. Here, we compare and contrast knowledge on peroxisome proliferator-activated receptor gamma (PPARG)-mediated adipogenesis derived from both biomedical and aquaculture literatures. We first concentrate on the biomedical literature to (i) briefly review PPARG-mediated adipogenesis in mammals, before (ii) reviewing Pparg-mediated adipogenesis in zebrafish. Finally, we (iii) mine the aquaculture literature to compare and contrast Pparg-mediated adipogenesis in aquaculturally relevant teleosts. Our goal is to highlight evolutionary similarities and differences in adipose biology that will inform our understanding of the role of adipose tissue in obesity and related disease.
Adipogenesis—the process of progenitor cell differentiation to generate mature, lipid-laden adipocytes (fat cells) is central to physiological homeostasis. Dysregulation of adipogenesis and a reduced capacity to sequester lipid within cytoplasmic lipid droplets (LDs) of adipocytes leads to lipodystrophy, ectopic lipid deposition, systemic metabolic dysfunction, and increased risk for developing diabetes and cardiovascular disease (1–3). Members of the peroxisome proliferator-activated receptor (PPAR) family of nuclear receptors have paramount roles in lipid metabolism; and, in particular, PPAR gamma (PPARG) is critical for adipogenesis. Much is known on PPARG-mediated adipogenesis in mammalian model systems; however, extensive research has also been conducted on adipogenesis in fish species relevant to the aquaculture industry. The aim of this mini-review is to integrate findings on Pparg-mediated adipogenesis from the aquaculture industry into the larger biomedical-centered literature. This review is focused on adipogenesis in white adipose tissue (WAT); however, adipogenesis in brown adipose has also recently been reviewed (4).
PPARG: A Master Regulator of Mammalian Adipogenesis
Peroxisome proliferator-activated receptor gamma is both necessary and sufficient for WAT adipogenesis in mammals, and is considered a “master regulator” of adipogenesis. In mouse, Pparg plays an important role in placental vascularization, monocyte differentiation, and cardiac development (5, 6); however, Pparg is also required for adipogenesis both in vitro (7) and in vivo (5, 7, 8). Naturally occurring mutations within the PPARG coding sequence can lead to PPARG loss-of-function (LOF), severe lipodystrophy, insulin resistance, and diabetes in humans (2, 3, 9). Further, adipocyte-specific deletion of Pparg in mouse results in the complete absence of WAT (8). Strikingly, expression of Pparg, together with provision of an activating ligand, is sufficient to initiate an adipogenic program and maintain an adipocyte phenotype in previously non-adipogenic cells (10, 11). Therefore, PPARG has a central role in mammalian adipogenesis, typified by PPARG LOF in humans, which is associated with severe lipodystrophy, and metabolic dysfunction and disease.
In mammals, PPARG exists as two isoforms, G1 (γ1) and G2 (γ2), derived from a single gene, and transcribed by distinct promoters (12, 13). PPARG2 contains additional 30 amino acids at the N-terminal of PPARG1 and is specific to WAT—whereas, PPARG1 can be expressed at low levels in non-WAT tissues (12, 13). Both γ1 and γ2 isoforms can instruct a similar adipogenic gene expression program; however, PPARG2 exhibits a quantitatively greater adipogenic ability (14). Structurally, PPARG contains six protein domains (domains A–F) (Figure 1A): the N-terminal A/B-domain contains the ligand-independent transactivation function 1 (AF-1); the C-domain is a highly conserved DNA-binding domain (DBD), consisting of two type II zinc fingers; the D-domain is a flexible hinge region; the E-domain contains the AF-2 ligand-binding domain (LBD); and at the C-terminus, a small F-domain has been shown to interact with cofactors (15).
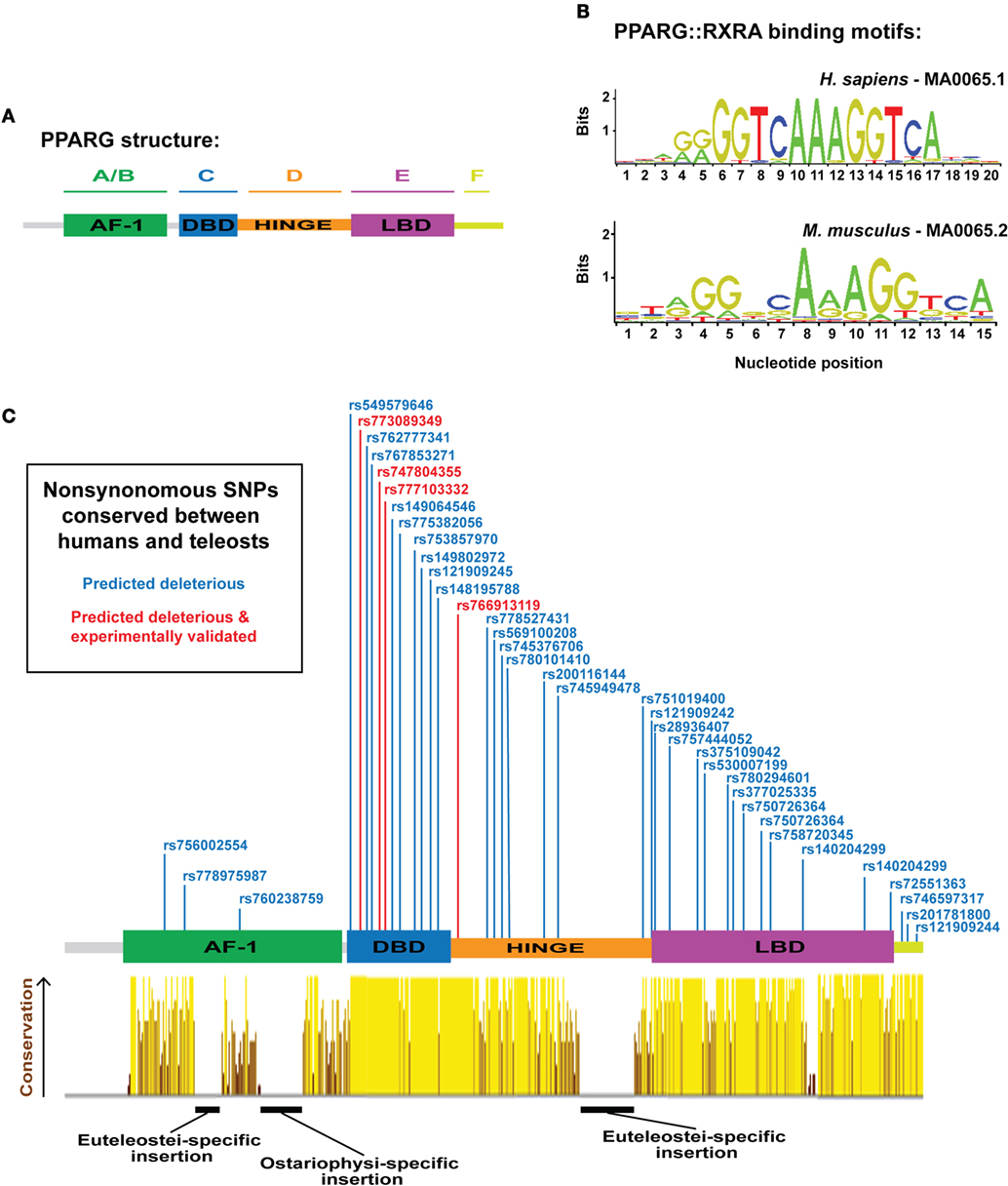
Figure 1. Overview of peroxisome proliferator-activated receptor gamma (PPARG) structure, DNA-binding specificity, and identification of human PPARG genetic variation conserved to fish. (A) Schematic illustrating the domain organization of human PPARG. (B) PPARG:RXRA-binding motifs for human (upper motif) and mouse (lower motif). Motifs are derived from the JASPAR database (http://jaspar.genereg.net/). (C) PPARG domain structure with dbSNPS predicted to be deleterious using SIFT and Polyphen, and conserved to zebrafish, Nile tilapia, and fugu. Red single nucleotide polymorphisms (SNPs) indicate functional verification (9). Yellow–brown histogram indicates the degree of conservation in PPARG between human, mouse, coelacanth, spotted gar, zebrafish, fugu, and Nile tilapia. Height and color indicate the degree of conservation.
The function of each PPARG domain has been extensively studied. The N-terminal AF-1 domain regulates the transcriptional activity of PPARG by (i) influencing Pparg ubiquitination and receptor turnover (16), (ii) controlling localization of Pparg to distinct cellular compartments (17, 18), (iii) facilitating communication with the LBD and enhancing ligand-dependent transcription (19), and (iv) recruitment of coactivators (20, 21) and corepressors (22). Importantly, many AF-1 focused regulatory mechanisms rely on posttranscriptional modifications of PPARG and can be both ligand-dependent or ligand-independent (23). Accordingly, inhibiting phosphorylation of serine 112 (S112) of Pparg2 in mouse results in improved insulin sensitivity when fed a high-fat diet (24). In addition, humans carrying a mutation blocking phosphorylation of an equivalent serine residue also have improved insulin sensitivity (18, 25). Together, these studies show that multiple diverse mechanisms converge on the AF-1 domain to regulate the transcriptional activity, and insulin sensitizing potential, of PPARG.
The transcriptional activity of PPARG is highly dependent on its DBD. Mutations within the DBD of human PPARG inhibit the transcriptional potential of PPARG and patients carrying such mutations exhibit severe insulin resistance and an increased risk for diabetes (3, 9, 26). The core DBD is highly conserved between different nuclear receptors; both within the PPAR family, and between distinct nuclear receptor families (27). Indeed, some nuclear receptors bind identical DNA motifs (28) and, in support, Pparg retains the ability to conduct an adipogenic program even when fused to alternative DBDs (29). These data suggest that the specificity of PPARG-mediated gene activation is not entirely contained within the DBD. Pparg generally binds DNA as obligate heterodimers with members of the retinoid X receptor (RXR) family of nuclear receptors (30), although some evidence suggests Pparg can also function as a homodimer (31). Strikingly, mutations within RXR DBDs have severe consequences for the transcriptional activity of PPARG:RXR heterodimers, suggesting the DNA-binding activity of RXR is also central to PPARG function (32). PPARG/RXR heterodimers bind to cis-acting peroxisome proliferator response elements (PPREs) containing direct repeats of 5′-AGGTCA-3′ separated by n nucleotides (DRn) (Figure 1B) (33–35). Along with an AAACT flanking sequence situated immediately 5′ to the core DRn motif, which helps guide selective PPAR binding (36–38). ChIP-Seq analyses for Pparg binding have identified DR1 as the canonical motif for PPARG binding (33, 39, 40), and binding is dependent on the sequence, and affinity, of specific DR1 motifs (40). Wider chromatin organization and accessibility also appear key for PPARG-mediated adipogenesis, as extensive chromatin remodeling occurs early in adipogenesis, prior to Pparg binding, and creates “hotspots” primed for future Pparg binding (41).
The LBD of PPARG, Known Ligands, and Modulation of Transcriptional Activity
Ligand binding regulates the transcriptional activity of PPARG and, as such, the LBD is central to the ability of PPARG to direct adipogenesis and regulate insulin sensitivity. Numerous lipid metabolites have been identified as PPARG ligands; including, polyunsaturated fatty acids (PUFAs), such as docosahexaenoic acid and linoleic acid, eicosanoids, and 15-deoxy-Δ12,14-prostanglandin J2 [PGJ(2)] (10, 42–47). Many of these ligands bind PPARG with low affinity and are unlikely to be present at concentrations required to activate PPARG in vivo (48). However, derivatives of linoleic acid have been shown to potently bind Pparg, and may represent an endogenous ligand for PPARG (49, 50). Intriguingly, a cAMP-induced, transient Pparg ligand is produced by 3T3-L1 adipocytes during the early stages of adipogenesis (51) and drives Pparg-mediated progenitor differentiation. Furthermore, this transient Pparg ligand is suggestive of a positive feedback loop, which is autonomous to adipocytes and acts in a paracrine manner. Synthetic ligands also bind and regulate PPARG activity. Most prominently, thiazolidinediones (TZDs) are potent PPARG agonists that lower hyperglycemia, decrease plasma triacylglycerides and free fatty acids, and increase insulin sensitivity (51). As such, TZDs have incredible potential to improve insulin sensitivity and glucose homeostasis in diabetic patients. However, many TZDs have been withdrawn from clinical use, or are under extensive review, owing to toxic side effects (52, 53). In particular, TZDs induce adipogenesis in patients and can lead to increased weight gain (51, 54, 55).
Zebrafish as a Model to Study Pparg-Mediated Adipogenesis
As a complement to mammalian model systems, the zebrafish has recently emerged as a tractable model for studying adipogenesis in vivo. Zebrafish possess adipose tissue that is morphologically similar to mammalian WAT (56–58), and which is deposited in anatomically homologous regions to mammalian WAT (56, 58, 59). Further, zebrafish adipose responds to nutritional manipulation, suggesting a conserved role for WAT as an energy store or supply during periods of caloric excess or restriction (56, 60, 61). Zebrafish possess a single pparg ortholog on chromosome 11 (47), which exhibits 67% overall similarity to human PPARG (47). The LBD and DBD of zebrafish pparg show especially high conservation to human PPARG (80.5 and 94.3% of amino acids are identical in LDB and DBD, respectively) (Figure 1C) (47, 62). The N-terminal AF-1 domain shows much less conservation between zebrafish and human (Figure 1C) (47, 62); however, this is unsurprising as the AF-1 domain is well known to exhibit low similarity even between more closely related species (23). Interestingly, zebrafish pparg contain multiple regions with amino acid insertions not present in mammalian PPARG, suggesting the potential for neo-functionalization of fish Pparg (Figure 1C) (47). Importantly, zebrafish pparg mRNA is detected in adipocytes (56, 58, 60). Moreover, many compounds known to stimulate mammalian Pparg also modulate zebrafish pparg mRNA; including, organotin compounds such as tributyltin (63, 64), halogenated analogs of bisphenol A (65), and PGJ(2) (66). Construction of a zebrafish transgenic line expressing the human PPARG LBD fused to a Gal4 DBD exhibits increased transcriptional activity after treatment with TZDs including rosiglitazone, pioglitazone, or troglitazone (67), thus suggesting that ligand-dependent coactivators of Pparg are conserved and functional in zebrafish. Intriguingly, recent work showed that treatment of zebrafish with the TZD rosiglitazone increased adiposity, suggesting that the role of Pparg in stimulating zebrafish adipogenesis may also be conserved to mammals (68).
Adipogenesis and the Aquaculture Industry
The use of zebrafish as a biomedical model system to study adipogenesis is at a nascent stage, and many gaps exist in our understanding. However, the aquaculture industry has conducted extensive investigation into adipogenesis in closely related fish species, owing to the fact that adipogenesis affects meat quality, animal health, and harvest yields (69). Aquaculture is defined as the farming of aquatic organisms; including fish, crustaceans, mollusks, and plants. The aquaculture industry contributes ~50% of the world’s aquatic food source (70); thus representing a significant proportion of all food consumed worldwide (71). For this review, we focus on teleost species most closely related to zebrafish. For a comprehensive review of teleost phylogeny, we refer you to the following articles (72, 73). The teleost lineage is divided into three branches; clupeocephalans (including the majority of teleosts); and the relatively minor elopomorpha (including eels and tarpons), and osteoglossomorpha (fish possessing toothed or bony tongues) (72, 73). For this review, we only consider clupeocephalans, which belong to two main lineages; ostariophysi and euteleostei. In 2010, freshwater fish production was dominated by ostariophysi such as silver carp (Hypophthalmichthys molitrix), grass carp (Ctenopharyngodon idella), common carp (Cyprinus carpio), and the euteleostei, Nile tilapia (Oreochromis niloticus). The euteleostei Atlantic salmon (Salmo salar) was the most farmed saltwater fish (70). Extensive regional differences exist in the species of fish farmed; for example, Asian countries primarily farm ostariophysi carp species, accounting for 89% of world aquaculture (70). By contrast, Mediterranean countries farm euteleostei species including gilthead sea bream (Sparus aurata) (74), European sea bass (Dicentrarchus labrax), and flathead gray mullet (Mugil cephalus) (70). Northern European countries, primarily farm euteleostei salmonid species such as Atlantic salmon and rainbow trout (Oncorhynchus mykiss) (70). A characteristic of teleosts is a teleost-specific third whole genome duplication (Ts3R), which is estimated to have occurred ~225–333 million years ago (72, 75). Recent genome sequencing projects have revealed that, in addition to Ts3R, certain teleost lineages have undergone further extensive genome duplication; including, salmonids (76, 77) and common carp (78). Genome duplications are hypothesized to underlie the dramatic radiation of teleosts and often lead to multiple gene copies, under reduced selective constraint, and thus receptive to neo-, non-, and sub-functionalization of the ancestral gene role (73).
Extensive Syntenic Conservation at Teleost pparg Loci
As PPARG exerts such a central role in mammalian adipogenesis, we first wished to assess whether duplicated teleost pparg paralogs have been retained. Only a single pparg ortholog was identified in 9 (of 10) teleost fish species with genome data on Ensembl (79). These data are striking, as the other members of the PPAR family (ppara and ppard) have been extensively duplicated, with paralogs retained, in teleosts (Ensembl Gene Tree: ENSGT00870000136388) (47). Teleost species with a single pparg ortholog include, ostariophysi such as zebrafish (cyprinidae); and euteleostei such as, Atlantic cod (gadiformes), pufferfish (tetradontiformes, both fugu and tetraodon), stickleback (gasterodae), and Nile tilapia (cichlidae) (Ensembl Gene Tree: ENSGT00870000136388) (47). The single teleost species with a retained pparg paralog is the ostariophysi blind cavefish (Astyanax mexicanus) of the chariciformes order (Figure 2). The striking loss of duplicated pparg genes in the majority of teleosts suggest stringent selective pressures for retaining Pparg copy number and function. To construct a predicted ancestral pparg locus, we examined synteny at the Pparg locus in tetrapods (mouse and human), a basal sarcopterygian (coelacanth), an actinopterygian holosteian basal to teleosts (spotted gar), and a chondrichthyan, cartilaginous fish (elephant shark) (Figure 2). Following the Ts3R, two pparg loci can be identified which each share extensive synteny to the predicted ancestral locus (Figure 2). Remarkably, in both ostariophysi and euteleostei, pparg appeared to be retained at a specific single locus (locus 1) (Figure 2), with the exception being Atlantic salmon, which retained pparg at locus 2 (Figure 2). Strikingly, in all euteleostei species examined, the region downstream of pparg contained multiple new genes not found in other species (iffo2b, akr7a3, mrto4, megf6b), suggesting an euteleostei-specific recombination event that completely changed the sequence downstream of pparg (Figure 2).
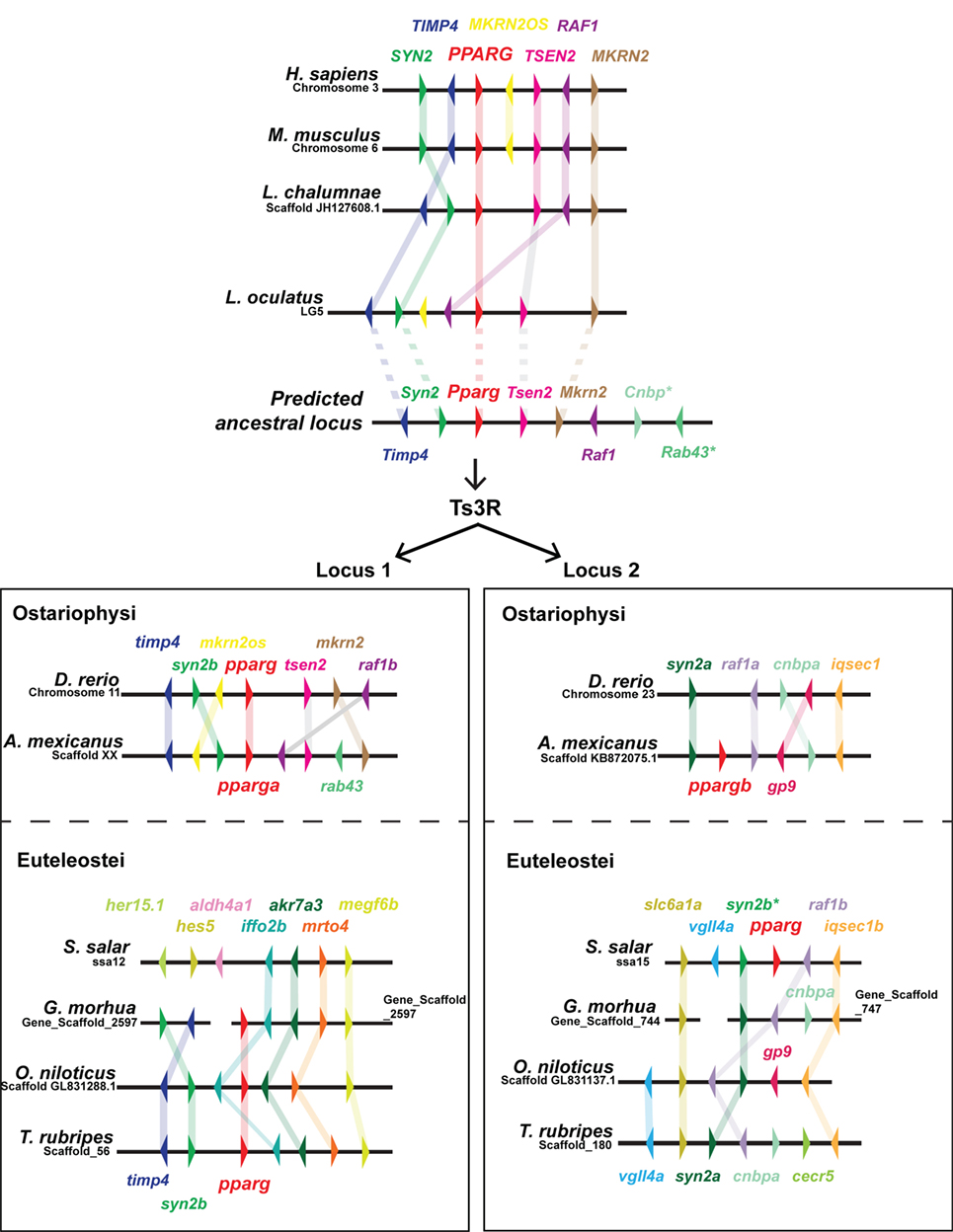
Figure 2. Extensive shared synteny between mammals and fish at the Peroxisome proliferator-activated receptor (PPARG) locus. The predicted ancestral locus was inferred from comparing the loci documented in the figure, together with the chondrichthyan elephant shark (Callorhinchus milii) locus (not shown). The C. milii locus contained Cenp and Rab43 genes (indicated with an asterisk). Ts3R indicates the teleost-specific genome duplication. Note the inversion of TIMP4 and SYN2 upstream of PPARG in the mammalian lineage. Duplicated raf1 paralogs (raf1a and raf1b) are only retained in zebrafish and cavefish.
Sequence Homology of Teleost pparg Genes
Extensive synteny at teleost and mammalian PPARG suggest the locus is under considerable selective constraint; therefore, we next assessed whether the PPARG amino acid sequence was equally conserved. We aligned PPARG sequences from representative tetrapods, a basal sarcopterygian, a holosteian, an ostariophysan, and euteleostians (Figure 1C). As expected, not all elements of the mammalian PPARG sequence were conserved from mammals to fish (Figure 1C). Strikingly, the DBD, hinge region, and LDB exhibited high levels of conservation from mammals to fish (Figure 1C). However, we found a large insertion into the hinge region specific to the euteleosts, fugu, and Nile tilapia (Figure 1C). We further found euteleost-specific and zebrafish-specific insertions into the AF-1 domain (Figure 1C). Aside from these three regions inserted into teleost Pparg, conservation was extensive (Figure 1C). Previous studies have identified that the LDB of fish Pparg (red sea bream and Nile tilapia) often contains additional amino acids compared to human PPARG (80, 81). However, the DBD of PPARG is well conserved between fish and mammals (81). These distinct patterns of conservation have been suggested to reflect the fact that PPARG target genes are well conserved, while there may be greater diversity in ligands, which activate PPARG (81) and may explain why some human PPARG agonists are unable to stimulate pparg expression across teleost species (81, 82).
Conservation of Pparg Amino Acids Affected by Disease-Associated Genetic Variation in Humans
The considerable sequence conservation between mammalian and teleost PPARG suggest that residues affected by naturally occurring, disease-associated, mutations in human PPARG may also be conserved in teleosts. To address this, we (i) collected all known human PPARG single nucleotide polymorphisms (SNPs) from dbSNP (347 SNPs), (ii) filtered these SNPs to identify 73 SNPs predicted to have a highly deleterious effect on PPARG function, (iii) identified amino acids altered by the deleterious SNPs, which were conserved to teleosts (39 SNPs/amino acids), (iv) filtered the conserved deleterious SNPs to ones that had been experimentally verified to have an effect on adipogenesis and PPARG function in humans (4 SNPs) (9). The resulting collection of SNPs (Figure 1C) represent ideal initial targets for modeling Pparg function during teleost adipogenesis and highlight the highly conserved nature of PPARG from mammals to fish.
Expression Dynamics of pparg in Farmed Fish Species
Although little is known regarding the expression of pparg in zebrafish, extensive experiments have been undertaken in farmed fish species to determine the dynamics of pparg during adipogenesis. In grass carp (ostariophysi), gilthead sea bream, large yellow croaker, and Atlantic salmon (all euteleostei), Pparg/pparg appeared coincident with early stages of adipocyte differentiation and increased gradually throughout adipogenesis (69, 83–87). These dynamics mirror those observed in mammalian 3T3-L1 cells, where Pparg mRNA is present at low levels in adipocyte progenitors, and increases upon stimulation of adipogenesis (10, 88). Similar to 3T3-L1 cells, cebpb mRNA was also induced prior to pparg during differentiation of adipocyte progenitors in the euteleostei, cobia (Rachycentron canadum), and in Atlantic Salmon (89). By contrast, in red sea bream (Pagrus major) (another euteleostei), pparg mRNA appeared to remain stable during a 10-day preadipocyte culture; however, isolated cells were maintained for 4 days prior to induction of adipogenesis; therefore, it remains possible that fluctuations in pparg mRNA expression occurred prior to analysis (80). However, by this method, accumulation of LDs appeared late and was not robust (80). Furthermore, in gilthead sea bream (euteleostei), pparg mRNA decreased in preadipocytes upon the addition of an adipogenic cocktail (83). The experimental reasons for differences in teleost pparg expression dynamics is unclear; however, in most fish species, the induction and maintenance of pparg mRNA during adipogenesis appears largely conserved to mammals. Furthermore, multiple pparg isoforms have been found in Nile tilapia and Atlantic salmon (both long and short isoforms) (81, 85, 90, 91), suggesting that teleost pparg is alternatively spliced similar to mammalian PPARG.
The Functional Role of Pparg-Mediated Adipogenesis in Farmed Fish Species
In addition to expression dynamics, extensive experiments on Pparg-mediated adipogenesis have been conducted in aquaculturally relevant fish species. Much of the evidence for Pparg-mediated adipogenesis in fish species derive from primary adipocyte progenitor, or “preadipocyte,” cell culture systems. Primary preadipocyte cultures have been established in multiple species, including; Atlantic salmon (84), red sea bream (92), rainbow trout (93), grass carp (94), large yellow croaker (69), gilthead sea bream (74), and cobia (89). In all of these systems, primary stromal-vascular cells were isolated from visceral adipose tissue (VAT) [the VAT source most likely equates to the pancreatic VAT and abdominal VAT deposits described in zebrafish (59)]. The preadipocyte culture methods closely follow established methods for the growth and differentiation of mammalian 3T3-L1 cells (95) and enable the incubation of preadipocytes, and differentiated adipocytes, with a range of pharmacological and biological agents to study potential roles during Pparg-mediated adipogenesis.
To our knowledge, no functional genetic data on the role of Pparg in farmed fish species is currently published. However, extensive data exist on pharmacological manipulation of Pparg and adipogenesis. Troglitazone, an insulin sensitizing TZD, potently stimulates preadipocyte differentiation in porcine and human preadipocytes (96, 97); and co-incubation with insulin induced preadipocyte differentiation in rainbow trout (98). A second TZD tested in teleosts, ciglitazone, induced pparg expression in preadipocytes of red sea bream (80), suggesting that TZDs induce both pparg and adipogenesis in teleosts. The role of several pro- and anti-adipogenic factors have also been studied in fish. Insulin has potent stimulatory effects on Pparg mRNA levels, and the proliferation and differentiation of mammalian preadipocytes, acting through IRS1 and the MAPK pathway (99, 100). In large yellow croaker (Percomorpha), insulin increased pparg mRNA, along with stimulating preadipocyte proliferation and differentiation (69). In accordance with mammalian data, Insulin inhibited lipolysis in differentiated adipocytes of rainbow trout (69, 101). Similarly, insulin also stimulated the differentiation of adipocyte progenitors and lipid accumulation in red sea bream (92). These findings suggest that insulin has a conserved role in stimulating pparg expression and promoting adipogenesis. Insulin-mediated induction of pparg and adipogenesis is also potentially conserved to other fish species, as the insulin-IRS1-MAPK signaling axis is also functional in rainbow trout adipocytes (102). However, unlike in rainbow trout, insulin had no effect on adipocyte lipolysis in gilthead sea bream (103). Tumor necrosis factor alpha (TNFA) is secreted from mammalian adipocytes and inhibits adipogenesis (104). Treating large yellow croaker preadipocytes with human TNFA reduced pparg mRNA levels, suppressed proliferation and differentiation, and stimulated lipolysis in differentiated adipocytes (69). An anti-adipogenic role for TNFA was also found in rainbow trout adipocytes at both RNA and protein levels (93, 105). PUFAs inhibit the proliferation and differentiation of mammalian preadipocytes (106, 107). DHA, an omega-3 fatty acid, was used in the treatment of large yellow croaker preadipocytes and led to decreased pparg mRNA levels and reductions in cell proliferation (69). It has further been shown that DHA stimulates lipolysis in 3T3-L1 preadipocytes (108); however, DHA did not exert a positive effect on lipolysis within large yellow croaker adipocytes and was actually observed to have an anti-lipolytic effect (69). Interestingly, DHA reduced lipid accumulation in Atlantic salmon adipocytes, although a mechanisms by which this occurred was not identified (85). Conversely, an analog of the saturated fatty acid palmitate, 2-bromopalmitate, increased pparg mRNA (red sea bream) (80). pparg cooperates with rxra to transcribe fabp4 suggesting that fish Pparg also functions as an obligate heterodimer with Rxr proteins to guide adipogenic gene expression (Nile tilapia) (81). In Atlantic salmon, pparg mRNA was induced after addition of liver X receptor (lxr) agonists (109), suggesting Pparg:Lxr coordinate gene expression in teleosts as they do in mammals (110).
Conclusion and Future Directions
Peroxisome proliferator-activated receptor gamma is a master regulator of adipogenesis in mammals, and mutations deleterious to PPARG function lead to increased susceptibility to diabetes and cardiovascular disease. In this review, we assessed the literature on Pparg-mediated adipogenesis in teleost fish species, including the biomedical model system, zebrafish, and multiple aquaculturally relevant farmed fish species. We found a high degree of synteny and conservation at/in pparg in teleost fish, along with evidence of conserved expression, regulation, and function derived from primary preadipocyte culture studies. Altogether, information on the role of Pparg gleaned from aquaculturally relevant species is likely to be highly informative for future zebrafish and mammalian biomedical studies on adipogenesis.
Author Contributions
Background literature research, writing, and review were conducted by RW, PT, and JM. Sequence and locus analysis was conducted by JM.
Conflict of Interest Statement
These authors declare no commercial or financial activities that may act as potential conflicts of interest.
Funding
This mini review was funded by a British Heart Foundation and University of Edinburgh Fellowship to JM, and a Diabetes UK Early Career Small Grant to JM (16/0005494).
References
1. Luo L, Liu M. Adipose tissue in control of metabolism. J Endocrinol (2016) 231:R77–99. doi: 10.1530/JOE-16-0211
2. Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature (1999) 402:880–3.
3. Agostini M, Schoenmakers E, Mitchell C, Szatmari I, Savage D, Smith A, et al. Non-DNA binding, dominant-negative, human PPARgamma mutations cause lipodystrophic insulin resistance. Cell Metab (2006) 4:303–11. doi:10.1016/j.cmet.2006.09.003
4. Nakagami H. The mechanism of white and brown adipocyte differentiation. Diabetes Metab J (2013) 37:85–90. doi:10.4093/dmj.2013.37.2.85
5. Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell (1999) 4:585–95. doi:10.1016/S1097-2765(00)80209-9
6. Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell (1998) 93:241–52. doi:10.1016/S0092-8674(00)81575-5
7. Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell (1999) 4:611–7. doi:10.1016/S1097-2765(00)80211-7
8. Wang F, Mullican SE, DiSpirito JR, Peed LC, Lazar MA. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARgamma. Proc Natl Acad Sci U S A (2013) 110:18656–61. doi:10.1073/pnas.1314863110
9. Majithia AR, Flannick J, Shahinian P, Guo M, Bray MA, Fontanillas P, et al. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci U S A (2014) 111:13127–32. doi:10.1073/pnas.1410428111
10. Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell (1994) 79:1147–56. doi:10.1016/0092-8674(94)90006-X
11. Hu E, Tontonoz P, Spiegelman BM. Transdifferentiation of myoblasts by the adipogenic transcription factors PPAR gamma and C/EBP alpha. Proc Natl Acad Sci U S A (1995) 92:9856–60. doi:10.1073/pnas.92.21.9856
12. Zhu Y, Qi C, Korenberg JR, Chen XN, Noya D, Rao MS, et al. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc Natl Acad Sci U S A (1995) 92:7921–5. doi:10.1073/pnas.92.17.7921
13. Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev (1994) 8:1224–34. doi:10.1101/gad.8.10.1224
14. Mueller E, Drori S, Aiyer A, Yie J, Sarraf P, Chen H, et al. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor gamma isoforms. J Biol Chem (2002) 277:41925–30. doi:10.1074/jbc.M206950200
15. Qi C, Zhu Y, Reddy JK. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem Biophys (2000) 32(Spring):187–204. doi:10.1385/CBB:32:1-3:187
16. Fujimoto Y, Shiraki T, Horiuchi Y, Waku T, Shigenaga A, Otaka A, et al. Proline cis/trans-isomerase Pin1 regulates peroxisome proliferator-activated receptor gamma activity through the direct binding to the activation function-1 domain. J Biol Chem (2010) 285:3126–32. doi:10.1074/jbc.M109.055095
17. Burgermeister E, Chuderland D, Hanoch T, Meyer M, Liscovitch M, Seger R. Interaction with MEK causes nuclear export and downregulation of peroxisome proliferator-activated receptor gamma. Mol Cell Biol (2007) 27:803–17. doi:10.1128/MCB.00601-06
18. von Knethen A, Tzieply N, Jennewein C, Brune B. Casein-kinase-II-dependent phosphorylation of PPARgamma provokes CRM1-mediated shuttling of PPARgamma from the nucleus to the cytosol. J Cell Sci (2010) 123:192–201. doi:10.1242/jcs.055475
19. Shao D, Rangwala SM, Bailey ST, Krakow SL, Reginato MJ, Lazar MA. Interdomain communication regulating ligand binding by PPAR-gamma. Nature (1998) 396:377–80. doi:10.1038/24634
20. Bugge A, Grontved L, Aagaard MM, Borup R, Mandrup S. The PPARgamma2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment. Mol Endocrinol (2009) 23:794–808. doi:10.1210/me.2008-0236
21. Gelman L, Zhou G, Fajas L, Raspé E, Fruchart JC, Auwerx J. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. J Biol Chem (1999) 274:7681–8. doi:10.1074/jbc.274.12.7681
22. Takahashi Y, Ohoka N, Hayashi H, Sato R. TRB3 suppresses adipocyte differentiation by negatively regulating PPARgamma transcriptional activity. J Lipid Res (2008) 49:880–92. doi:10.1194/jlr.M700545-JLR200
23. Bugge A, Mandrup S. Molecular mechanisms and genome-wide aspects of PPAR subtype specific transactivation. PPAR Res (2010) 2010. doi:10.1155/2010/169506
24. Rangwala SM, Rhoades B, Shapiro JS, Rich AS, Kim JK, Shulman GI, et al. Genetic modulation of PPARgamma phosphorylation regulates insulin sensitivity. Dev Cell (2003) 5:657–63. doi:10.1016/S1534-5807(03)00274-0
25. Ristow M, Muller-Wieland D, Pfeiffer A, Krone W, Kahn CR. Obesity associated with a mutation in a genetic regulator of adipocyte differentiation. N Engl J Med (1998) 339:953–9. doi:10.1056/NEJM199810013391403
26. Savage DB, Agostini M, Barroso I, Gurnell M, Luan J, Meirhaeghe A, et al. Digenic inheritance of severe insulin resistance in a human pedigree. Nat Genet (2002) 31:379–84. doi:10.1038/ng926
27. Aagaard MM, Siersbaek R, Mandrup S. Molecular basis for gene-specific transactivation by nuclear receptors. Biochim Biophys Acta (2011) 1812:824–35. doi:10.1016/j.bbadis.2010.12.018
28. Umesono K, Giguere V, Glass CK, Rosenfeld MG, Evans RM. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature (1988) 336:262–5. doi:10.1038/336262a0
29. Hummasti S, Tontonoz P. The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis. Mol Endocrinol (2006) 20:1261–75. doi:10.1210/me.2006-0025
30. Tontonoz P, Graves RA, Budavari AI, Erdjument-Bromage H, Lui M, Hu E, et al. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPAR gamma and RXR alpha. Nucleic Acids Res (1994) 22:5628–34. doi:10.1093/nar/22.25.5628
31. Okuno M, Arimoto E, Ikenobu Y, Nishihara T, Imagawa M. Dual DNA-binding specificity of peroxisome-proliferator-activated receptor gamma controlled by heterodimer formation with retinoid X receptor alpha. Biochem J (2001) 353:193–8. doi:10.1042/bj3530193
32. Temple KA, Cohen RN, Wondisford SR, Yu C, Deplewski D, Wondisford FE. An intact DNA-binding domain is not required for peroxisome proliferator-activated receptor gamma (PPARgamma) binding and activation on some PPAR response elements. J Biol Chem (2005) 280:3529–40. doi:10.1074/jbc.M411422200
33. Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev (2008) 22:2953–67. doi:10.1101/gad.501108
34. Nakachi Y, Yagi K, Nikaido I, Bono H, Tonouchi M, Schönbach C, et al. Identification of novel PPARgamma target genes by integrated analysis of ChIP-on-chip and microarray expression data during adipocyte differentiation. Biochem Biophys Res Commun (2008) 372:362–6. doi:10.1016/j.bbrc.2008.05.037
35. Hamza MS, Pott S, Vega VB, Thomsen JS, Kandhadayar GS, Ng PW, et al. De-novo identification of PPARgamma/RXR binding sites and direct targets during adipogenesis. PLoS One (2009) 4:e4907. doi:10.1371/journal.pone.0004907
36. Juge-Aubry C, Pernin A, Favez T, Burger AG, Wahli W, Meier CA, et al. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. Importance of the 5'-flanking region. J Biol Chem (1997) 272:25252–9. doi:10.1074/jbc.272.40.25252
37. Palmer CN, Hsu MH, Griffin HJ, Johnson EF. Novel sequence determinants in peroxisome proliferator signaling. J Biol Chem (1995) 270:16114–21. doi:10.1074/jbc.270.27.16114
38. Isakova A, Berset Y, Hatzimanikatis V, Deplancke B. Quantification of cooperativity in heterodimer-DNA binding improves the accuracy of binding specificity models. J Biol Chem (2016) 291:10293–306. doi:10.1074/jbc.M115.691154
39. Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev (2008) 22:2941–52. doi:10.1101/gad.1709008
40. Grossman SR, Zhang X, Wang L, Engreitz J, Melnikov A, Rogov P, et al. Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc Natl Acad Sci U S A (2017) 114:E1291–300. doi:10.1073/pnas.1621150114
41. Siersbæk R, Nielsen R, John S, Sung MH, Baek S, Loft A, et al. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J (2011) 30:1459–72. doi:10.1038/emboj.2011.65
42. Schupp M, Lazar MA. Endogenous ligands for nuclear receptors: digging deeper. J Biol Chem (2010) 285:40409–15. doi:10.1074/jbc.R110.182451
43. Yu K, Bayona W, Kallen CB, Harding HP, Ravera CP, McMahon G, et al. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J Biol Chem (1995) 270:23975–83. doi:10.1074/jbc.270.41.23975
44. Kliewer SA, Lenhard JM, Willson TM, Patel I, Morris DC, Lehmann JM. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell (1995) 83:813–9. doi:10.1016/0092-8674(95)90194-9
45. Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell (1995) 83:803–12. doi:10.1016/0092-8674(95)90193-0
46. Lehmann JM, Lenhard JM, Oliver BB, Ringold GM, Kliewer SA. Peroxisome proliferator-activated receptors alpha and gamma are activated by indomethacin and other non-steroidal anti-inflammatory drugs. J Biol Chem (1997) 272:3406–10. doi:10.1074/jbc.272.6.3406
47. Den Broeder MJ, Kopylova VA, Kamminga LM, Legler J. Zebrafish as a model to study the role of peroxisome proliferating-activated receptors in adipogenesis and obesity. PPAR Res (2015) 2015:358029. doi:10.1155/2015/358029
48. Bell-Parikh LC, Ide T, Lawson JA, McNamara P, Reilly M, FitzGerald GA. Biosynthesis of 15-deoxy-delta12,14-PGJ2 and the ligation of PPARgamma. J Clin Invest (2003) 112:945–55. doi:10.1172/JCI200318012
49. Schopfer FJ, Lin Y, Baker PR, Cui T, Garcia-Barrio M, Zhang J, et al. Nitrolinoleic acid: an endogenous peroxisome proliferator-activated receptor gamma ligand. Proc Natl Acad Sci U S A (2005) 102:2340–5. doi:10.1073/pnas.0408384102
50. Baker PR, Lin Y, Schopfer FJ, Woodcock SR, Groeger AL, Batthyany C, et al. Fatty acid transduction of nitric oxide signaling: multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J Biol Chem (2005) 280:42464–75. doi:10.1074/jbc.M504212200
51. Larsen TM, Toubro S, Astrup A. PPARgamma agonists in the treatment of type II diabetes: is increased fatness commensurate with long-term efficacy? Int J Obes Relat Metab Disord (2003) 27:147–61. doi:10.1038/sj.ijo.802223
52. Hampp C, Pippins J. Pioglitazone and bladder cancer: FDA’s assessment. Pharmacoepidemiol Drug Saf (2017) 26:117–8. doi:10.1002/pds.4154
53. Derosa G, Sahebkar A, Maffioli P. The role of various peroxisome proliferator-activated receptors and their ligands in clinical practice. J Cell Physiol (2017). doi:10.1002/jcp.25804
54. Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). J Biol Chem (1995) 270:12953–6. doi:10.1074/jbc.270.22.12953
55. Giannini S, Serio M, Galli A. Pleiotropic effects of thiazolidinediones: taking a look beyond antidiabetic activity. J Endocrinol Invest (2004) 27:982–91. doi:10.1007/BF03347546
56. Flynn EJ III, Trent CM, Rawls JF. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio). J Lipid Res (2009) 50:1641–52. doi:10.1194/jlr.M800590-JLR200
57. Song Y, Cone RD. Creation of a genetic model of obesity in a teleost. FASEB J (2007) 21:2042–9. doi:10.1096/fj.06-7503com
58. Imrie D, Sadler KC. White adipose tissue development in zebrafish is regulated by both developmental time and fish size. Dev Dyn (2010) 239:3013–23. doi:10.1002/dvdy.22443
59. Minchin JE, Rawls JF. In vivo imaging and quantification of regional adiposity in zebrafish. Methods Cell Biol (2017) 138:3–27. doi:10.1016/bs.mcb.2016.11.010
60. Minchin JE, Dahlman I, Harvey CJ, Mejhert N, Singh MK, Epstein JA, et al. Plexin D1 determines body fat distribution by regulating the type V collagen microenvironment in visceral adipose tissue. Proc Natl Acad Sci U S A (2015) 112:4363–8. doi:10.1073/pnas.1416412112
61. McMenamin SK, Minchin JE, Gordon TN, Rawls JF, Parichy DM. Dwarfism and increased adiposity in the gh1 mutant zebrafish vizzini. Endocrinology (2013) 154:1476–87. doi:10.1210/en.2012-1734
62. Zhao Y, Zhang K, Giesy JP, Hu J. Families of nuclear receptors in vertebrate models: characteristic and comparative toxicological perspective. Sci Rep (2015) 5:8554. doi:10.1038/srep08554
63. Lima D, Castro LF, Coelho I, Lacerda R, Gesto M, Soares J, et al. Effects of tributyltin and other retinoid receptor agonists in reproductive-related endpoints in the zebrafish (Danio rerio). J Toxicol Environ Health A (2015) 78:747–60. doi:10.1080/15287394.2015.1028301
64. Boyer IJ. Toxicity of dibutyltin, tributyltin and other organotin compounds to humans and to experimental animals. Toxicology (1989) 55:253–98. doi:10.1016/0300-483X(89)90018-8
65. Riu A, Grimaldi M, le Maire A, Bey G, Phillips K, Boulahtouf A, et al. Peroxisome proliferator-activated receptor gamma is a target for halogenated analogs of bisphenol A. Environ Health Perspect (2011) 119:1227–32. doi:10.1289/ehp.1003328
66. Ibabe A, Herrero A, Cajaraville MP. Modulation of peroxisome proliferator-activated receptors (PPARs) by PPAR(alpha)- and PPAR(gamma)-specific ligands and by 17beta-estradiol in isolated zebrafish hepatocytes. Toxicol In Vitro (2005) 19:725–35. doi:10.1016/j.tiv.2005.03.019
67. Tiefenbach J, Moll PR, Nelson MR, Hu C, Baev L, Kislinger T, et al. A live zebrafish-based screening system for human nuclear receptor ligand and cofactor discovery. PLoS One (2010) 5:e9797. doi:10.1371/journal.pone.0009797
68. Tingaud-Sequeira A, Ouadah N, Babin PJ. Zebrafish obesogenic test: a tool for screening molecules that target adiposity. J Lipid Res (2011) 52:1765–72. doi:10.1194/jlr.D017012
69. Wang X, Huang M, Wang Y. The effect of insulin, TNFalpha and DHA on the proliferation, differentiation and lipolysis of preadipocytes isolated from large yellow croaker (Pseudosciaena Crocea R.). PLoS One (2012) 7:e48069. doi:10.1371/journal.pone.0048069
70. Bostock J, McAndrew B, Richards R, Jauncey K, Telfer T, Lorenzen K, et al. Aquaculture: global status and trends. Philos Trans R Soc Lond B Biol Sci (2010) 365:2897–912. doi:10.1098/rstb.2010.0170
71. Food and Agriculture Organization of the United Nations (FAO). The State of World Fisheries and Aquaculture 2016. Contributing to Food Security and Nutrition for All. Rome (2016). 200 p.
72. Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, et al. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A (2012) 109:13698–703. doi:10.1073/pnas.1206625109
73. Braasch I, Peterson SM, Desvignes T, McCluskey BM, Batzel P, Postlethwait JH. A new model army: emerging fish models to study the genomics of vertebrate Evo-Devo. J Exp Zool B Mol Dev Evol (2015) 324:316–41. doi:10.1002/jez.b.22589
74. Salmeron C, Acerete L, Gutierrez J, Navarro I, Capilla E. Characterization and endocrine regulation of proliferation and differentiation of primary cultured preadipocytes from gilthead sea bream (Sparus aurata). Domest Anim Endocrinol (2013) 45:1–10. doi:10.1016/j.domaniend.2013.02.002
75. Santini F, Harmon LJ, Carnevale G, Alfaro ME. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol Biol (2009) 9:194. doi:10.1186/1471-2148-9-194
76. Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noël B, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun (2014) 5:3657. doi:10.1038/ncomms4657
77. Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, et al. The Atlantic salmon genome provides insights into rediploidization. Nature (2016) 533:200–5. doi:10.1038/nature17164
78. Xu P, Zhang X, Wang X, Li J, Liu G, Kuang Y, et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genet (2014) 46:1212–9. doi:10.1038/ng.3098
79. Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, et al. Ensembl 2017. Nucleic Acids Res (2017) 45:D635–42. doi:10.1093/nar/gkw1104
80. Oku H, Umino T. Molecular characterization of peroxisome proliferator-activated receptors (PPARs) and their gene expression in the differentiating adipocytes of red sea bream Pagrus major. Comp Biochem Physiol B Biochem Mol Biol (2008) 151:268–77. doi:10.1016/j.cbpb.2008.07.007
81. He AY, Liu CZ, Chen LQ, Ning LJ, Qin JG, Li JM, et al. Molecular characterization, transcriptional activity and nutritional regulation of peroxisome proliferator activated receptor gamma in Nile tilapia (Oreochromis niloticus). Gen Comp Endocrinol (2015) 223:139–47. doi:10.1016/j.ygcen.2015.05.008
82. Leaver MJ, Boukouvala E, Antonopoulou E, Diez A, Favre-Krey L, Ezaz MT, et al. Three peroxisome proliferator-activated receptor isotypes from each of two species of marine fish. Endocrinology (2005) 146:3150–62. doi:10.1210/en.2004-1638
83. Salmeron C, Riera-Heredia N, Gutierrez J, Navarro I, Capilla E. Adipogenic gene expression in Gilthead Sea Bream mesenchymal stem cells from different origin. Front Endocrinol (2016) 7:113. doi:10.3389/fendo.2016.00113
84. Vegusdal A, Sundvold H, Gjoen T, Ruyter B. An in vitro method for studying the proliferation and differentiation of Atlantic salmon preadipocytes. Lipids (2003) 38:289–96. doi:10.1007/s11745-003-1063-3
85. Todorcević M, Vegusdal A, Gjøen T, Sundvold H, Torstensen BE, Kjaer MA, et al. Changes in fatty acids metabolism during differentiation of Atlantic salmon preadipocytes; effects of n-3 and n-9 fatty acids. Biochim Biophys Acta (2008) 1781:326–35. doi:10.1016/j.bbalip.2008.04.014
86. Liu P, Ji H, Li C, Tian J, Wang Y, Yu P, et al. Ontogenetic development of adipose tissue in grass carp (Ctenopharyngodon idellus). Fish Physiol Biochem (2015) 41:867–78. doi:10.1007/s10695-015-0053-x
87. Todorcevic M, Skugor S, Krasnov A, Ruyter B. Gene expression profiles in Atlantic salmon adipose-derived stromo-vascular fraction during differentiation into adipocytes. BMC Genomics (2010) 11:39. doi:10.1186/1471-2164-11-39
88. Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology (1994) 135:798–800. doi:10.1210/endo.135.2.8033830
89. Cheng Y, Chen H. Effects of different fatty acids on cell differentiation and lipid accumulation in preadipocytes of warm water fish cobia (Rachycentron canadum Linnaeus, 1766). Aquac Res (2015) 46:590–601. doi:10.1111/are.12204
90. Andersen O, Eijsink VG, Thomassen M. Multiple variants of the peroxisome proliferator-activated receptor (PPAR) gamma are expressed in the liver of atlantic salmon (Salmo salar). Gene (2000) 255:411–8. doi:10.1016/S0378-1119(00)00350-4
91. Sundvold H, Ruyter B, Ostbye TK, Moen T. Identification of a novel allele of peroxisome proliferator-activated receptor gamma (PPARG) and its association with resistance to Aeromonas salmonicida in Atlantic salmon (Salmo salar). Fish Shellfish Immunol (2010) 28:394–400. doi:10.1016/j.fsi.2009.11.023
92. Oku H, Tokuda M, Okumura T, Umino T. Effects of insulin, triiodothyronine and fat soluble vitamins on adipocyte differentiation and LPL gene expression in the stromal-vascular cells of red sea bream, Pagrus major. Comp Biochem Physiol B Biochem Mol Biol (2006) 144:326–33. doi:10.1016/j.cbpb.2006.03.008
93. Bouraoui L, Gutierrez J, Navarro I. Regulation of proliferation and differentiation of adipocyte precursor cells in rainbow trout (Oncorhynchus mykiss). J Endocrinol (2008) 198:459–69. doi:10.1677/JOE-08-0264
94. Li Y. Establishment and evaluation of a new model for studying lipogenesis in grass carp (Ctenopharyngodon idella) preadipocytes. In Vitro Cell Dev Biol Anim (2012) 48:37–42. doi:10.1007/s11626-011-9474-8
95. Ruiz-Ojeda FJ, Ruperez AI, Gomez-Llorente C, Gil A, Aguilera CM. Cell models and their application for studying adipogenic differentiation in relation to obesity: a review. Int J Mol Sci (2016) 17. doi:10.3390/ijms17071040
96. Poulos SP, Hausman GJ. A comparison of thiazolidinedione-induced adipogenesis and myogenesis in stromal-vascular cells from subcutaneous adipose tissue or semitendinosus muscle of postnatal pigs. J Anim Sci (2006) 84:1076–82. doi:10.2527/2006.8451076x
97. Brown JM, Halvorsen YD, Lea-Currie YR, Geigerman C, McIntosh M. Trans-10, cis-12, but not cis-9, trans-11, conjugated linoleic acid attenuates lipogenesis in primary cultures of stromal vascular cells from human adipose tissue. J Nutr (2001) 131:2316–21.
98. Bouraoui L, Cruz-Garcia L, Gutierrez J, Capilla E, Navarro I. Regulation of lipoprotein lipase gene expression by insulin and troglitazone in rainbow trout (Oncorhynchus mykiss) adipocyte cells in culture. Comp Biochem Physiol A Mol Integr Physiol (2012) 161:83–8. doi:10.1016/j.cbpa.2011.09.008
99. Boney CM, Gruppuso PA, Faris RA, Frackelton AR Jr. The critical role of Shc in insulin-like growth factor-I-mediated mitogenesis and differentiation in 3T3-L1 preadipocytes. Mol Endocrinol (2000) 14:805–13. doi:10.1210/mend.14.6.0487
100. Rieusset J, Andreelli F, Auboeuf D, Roques M, Vallier P, Riou JP, et al. Insulin acutely regulates the expression of the peroxisome proliferator-activated receptor-gamma in human adipocytes. Diabetes (1999) 48:699–705. doi:10.2337/diabetes.48.4.699
101. Albalat A, Gutierrez J, Navarro I. Regulation of lipolysis in isolated adipocytes of rainbow trout (Oncorhynchus mykiss): the role of insulin and glucagon. Comp Biochem Physiol A Mol Integr Physiol (2005) 142:347–54. doi:10.1016/j.cbpa.2005.08.006
102. Bouraoui L, Capilla E, Gutierrez J, Navarro I. Insulin and insulin-like growth factor I signaling pathways in rainbow trout (Oncorhynchus mykiss) during adipogenesis and their implication in glucose uptake. Am J Physiol Regul Integr Comp Physiol (2010) 299:R33–41. doi:10.1152/ajpregu.00457.2009
103. Albalat A, Gómez-Requeni P, Rojas P, Médale F, Kaushik S, Vianen GJ, et al. Nutritional and hormonal control of lipolysis in isolated gilthead seabream (Sparus aurata) adipocytes. Am J Physiol Regul Integr Comp Physiol (2005) 289:R259–65. doi:10.1152/ajpregu.00574.2004
104. Cawthorn WP, Sethi JK. TNF-alpha and adipocyte biology. FEBS Lett (2008) 582:117–31. doi:10.1016/j.febslet.2007.11.051
105. Bou M, Todorčević M, Rodríguez J, Capilla E, Gutiérrez J, Navarro I. Interplay of adiponectin, TNFalpha and insulin on gene expression, glucose uptake and PPARgamma, AKT and TOR pathways in rainbow trout cultured adipocytes. Gen Comp Endocrinol (2014) 205:218–25. doi:10.1016/j.ygcen.2014.05.005
106. Awad AB, Begdache LA, Fink CS. Effect of sterols and fatty acids on growth and triglyceride accumulation in 3T3-L1 cells. J Nutr Biochem (2000) 11:153–8. doi:10.1016/S0955-2863(99)00087-X
107. Hensler M, Bardova K, Jilkova ZM, Wahli W, Meztger D, Chambon P, et al. The inhibition of fat cell proliferation by n-3 fatty acids in dietary obese mice. Lipids Health Dis (2011) 10:128. doi:10.1186/1476-511X-10-128
108. Kim HK, Della-Fera M, Lin J, Baile CA. Docosahexaenoic acid inhibits adipocyte differentiation and induces apoptosis in 3T3-L1 preadipocytes. J Nutr (2006) 136:2965–9.
109. Carmona-Antonanzas G, Tocher DR, Martinez-Rubio L, Leaver MJ. Conservation of lipid metabolic gene transcriptional regulatory networks in fish and mammals. Gene (2014) 534:1–9. doi:10.1016/j.gene.2013.10.040
Keywords: adipose, zebrafish, adipogenesis, pparg, aquaculture
Citation: Wafer R, Tandon P and Minchin JEN (2017) The Role of Peroxisome Proliferator-Activated Receptor Gamma (PPARG) in Adipogenesis: Applying Knowledge from the Fish Aquaculture Industry to Biomedical Research. Front. Endocrinol. 8:102. doi: 10.3389/fendo.2017.00102
Received: 09 March 2017; Accepted: 01 May 2017;
Published: 22 May 2017
Edited by:
Encarnación Capilla, University of Barcelona, SpainReviewed by:
Marta Bou, Nofima, NorwayMercedes Blazquez, Consejo Superior de Investigaciones Científicas (CSIC), Spain
Copyright: © 2017 Wafer, Tandon and Minchin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James E. N. Minchin, amFtZXMubWluY2hpbkBlZC5hYy51aw==
 Rebecca Wafer
Rebecca Wafer James E. N. Minchin
James E. N. Minchin