
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Endocrinol. , 25 April 2017
Sec. Endocrinology of Aging
Volume 8 - 2017 | https://doi.org/10.3389/fendo.2017.00090
This article is part of the Research Topic Non-coding RNAs: Entwining Metabolism and Aging View all 7 articles
One of the long-standing principles of molecular biology has been that DNA functions as a template for transcription of messenger RNAs, which are eventually translated into a protein. Thus, proteins were seen as the main mediators of nearly all aspects of cell and tissue function. However, this perception started changing rapidly when high-throughput sequencing platforms became available, unraveling that more than two-thirds of the human genome are transcribed into RNA but only <2% of transcripts encode proteins (1, 2). Thus, the majority of the transcriptome falls into the category of non-coding RNAs (ncRNAs). These include long-known and well-characterized classes of ncRNAs with basic cellular housekeeping functions such as translation (transfer RNAs and ribosomal RNAs), splicing (small nuclear RNAs), or RNA editing (small nucleolar RNAs) (3, 4). Furthermore, short regulatory ncRNAs (20–30 nt in length) including microRNAs, piwi-associated RNAs, or endogenous short-interfering RNAs are highly conserved among species and have been proven to be crucial regulators of gene expression (5–7). Apart from these rather well-studied ncRNAs, the more recently identified class of lncRNAs has gained increasing scientific interest over the past years, and we are only beginning to appreciate their significance in a multitude of cellular processes and their complex modes of action (8, 9).
The original classification of lncRNAs is based on a length of at least 200 nt and lack of protein-coding potential. lncRNAs can be spliced, capped, and/or polyadenylated and localize either to the nucleus or the cytoplasm of the cell (1, 10). Interestingly, several lncRNAs were recently shown to act as templates for small peptides, and a number of mRNAs appear to adopt additional non-coding functionality (11–16). These observations suggest that classification of RNAs based on protein-coding potential might not in all cases be sufficiently exhaustive.
In contrast to mRNAs, lncRNAs generally show less primary sequence conservation among species, contain fewer but longer exons, and exhibit an intriguingly cell type-specific expression (8, 9). In addition, lncRNAs have been proven essential for processes such as cellular differentiation and progenitor cell regulation, epigenetic imprinting, X-chromosome inactivation, promoter-specific gene regulation, and nuclear import (17–25). Moreover, aberrant lncRNA expression has been linked to several diseases, including many types of cancer, highlighting their functional relevance during these diverse processes and rendering lncRNAs a captivating and novel research field (26–30). The frequently observed high level of complexity and diversity of gene loci, however, can significantly complicate functional characterization of lncRNAs. Hence, careful analysis of lncRNA function should start with a close characterization of its genomic locus, especially if the lncRNA is not yet characterized or its gene locus not well annotated, to lower the chance of drawing wrong conclusions and dissipating time and money. Below, we will illustrate mechanisms of lncRNA isoform generation using selected examples, introduce several approaches for lncRNA locus studies, and discuss potential pitfalls in investigating lncRNA loci.
With the lncRNA field still being in its infancy, novel lncRNAs are detected in human cells and tissues on a regular basis, resulting in several thousand predicted human lncRNAs to date (8, 31, 32). Studies focusing on the complexity within lncRNA loci revealed up to 40 different isoforms for the lncRNA PCBP1-AS1, with an average of 2.3–3.9 different isoforms per locus, accentuating the necessity to complement the functional analysis of an lncRNA with a thorough characterization of its gene locus (8, 9, 33). Scattered all over the genome, lncRNA genes can be found far away from other annotated genes, or lncRNAs can emerge in the opposite direction of a neighboring gene locus (divergent). In addition, several lncRNAs were found to reside within an intron (intronic) or being the antisense transcript of a protein-coding gene, thus sharing the same gene locus (see Figure 1) (34).
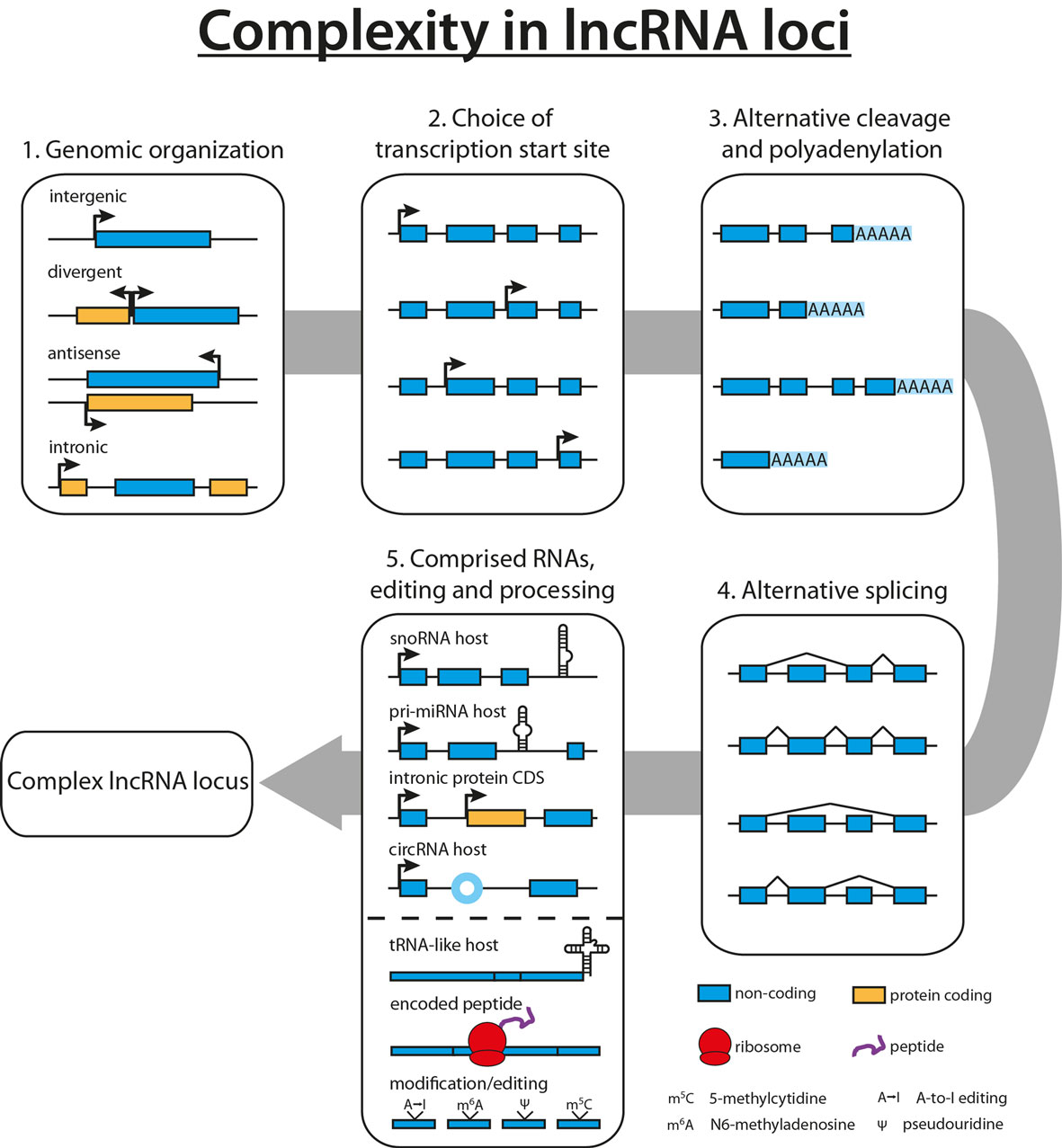
Figure 1. Complexity in long non-coding RNA (lncRNA) loci. Diversity in lncRNA loci originates from the genomic organization of the lncRNA (1). A plethora of lncRNA isoforms arises from the combination of multiple transcription start sites (2), alternative cleavage and polyadenylation sites (3), as well as alternative splicing events (4). Finally, lncRNAs have been shown to harbor other non-coding RNAs such as snoRNAs, miRNAs, or tRNAs or to contain intronic protein-coding genes, increasing the potential complexity of lncRNA loci (5). In addition to diverse loci, lncRNAs can give rise to tRNA-like molecules, encode small peptides, or are subject to RNA modification and editing events (5).
In addition to being part of another transcriptional unit, lncRNAs themselves can harbor protein-coding genes or other ncRNAs such as circular RNAs, tRNAs, miRNAs, and snoRNAs (8, 35–39). A prime example for this complexity is the lncRNA GAS5, which hosts 10 C/D box snoRNAs, five of which can be further processed to piRNAs (40, 41).
Despite their classification as long “non-coding” RNAs, several studies showed that lncRNAs can be translated into small peptides and are associated with ribosomes, further increasing the complexity within lncRNA loci (14, 42, 43). In addition, mRNAs can harbor regulatory RNA functions such as miRNA sponges, transcription elongation, or translational control (12, 13, 16). One of the earliest observations of these bifunctional RNAs is that both SRA1 and its protein product SRAP can act as transcriptional coactivators of nuclear receptors (11, 44). More recently, the peptide DWORF was identified in the lncRNA LOC100507537, and many additional putative peptides are predicted to arise from lncRNAs (15, 42, 43). On the other hand, Bánfai et al. correlated tandem mass spectrometry data with RNA sequencing (RNA-seq) data (both generated in two different cell lines by ENCODE) and found over 90% of GENCODE lncRNAs to be unlikely to encode a peptide. This is in accordance to a similar approach by Gascoigne et al., suggesting that the majority of lncRNAs likely is truly non-coding (45, 46). Thus, experimental validation of a bioinformatically predicted small peptide is needed to verify translation, stability, and functional relevance.
Diversity within a lncRNA locus is not solely reflected by overlapping or embedded transcripts within a lncRNA but also the transcription initiation and termination sites can vary. Correspondingly, almost two different 3′ ends can be found for each transcriptional start of a given lncRNA (47, 48). One cause for alternative 3′ ends is alternative cleavage and polyadenylation (APA). Roughly, 70% of human and mouse genes undergo APA, and many lncRNAs exhibit alternative polyadenylation sites upstream of the most 3′ exon, whereas for mRNAs, alternative polyadenylation sites are often located within the last exon (49, 50). Interestingly, 15–45% of the conserved elements in lncRNAs are located behind the first polyadenylation site, suggesting a switch in lncRNA function regulated by APA (49). Thus, it is not surprising that lncRNAs can be guided to different cellular compartments by alternative cleavage and polyadenylation as reported for CCAT-1 (51).
Besides alternative polyadenylation, 3′ processing of lncRNAs has been described, further expanding the potential diversity of lncRNA isoforms generated within the cell. As an example, the lncRNAs MALAT1 and NEAT1 feature a tRNA-like structure at their 3′ end, which is subject to RNAse P cleavage, resulting in a stable RNA triple-helix at the 3′ end of both lncRNAs, which serve as compensatory poly-A tails (52–54).
Similar to mRNAs, lncRNAs exploit the usage of alternative transcription start sites (55, 56). For the lncRNA Tsix, which is involved in the process of X-chromosome inactivation, two different transcription start sites have been identified. Correspondingly, the gene locus of the lncRNA SOX2OT has at least two promoter regions (57, 58).
By using both alternative polyadenylation and transcription start sites, a multiplicity of lncRNAs originating from the DM1-AS locus has been reported, which is even further increased through alternative splicing events (59).
Despite functioning in the regulation of RNA splicing, lncRNAs too can be alternatively spliced, which presumably alters their function within the cell (60, 61). One notable example is the lncRNA GNG12-AS1. Splicing of GNG12-AS1 results in a total of 38 different isoforms with up to 10 exons. Furthermore, cohesin has been identified as a splice regulator of GNG12-AS1, evoking the idea of tight splicing regulation to be crucial for maintaining the isoform-specific functions of GNG12-AS1 (39). Another example for the complexity of the human transcriptome through alternative splicing can be found for the lncRNA HOTAIR, which can act as a molecular scaffold: the 5′ end of HOTAIR binds the polycomb repressive complex 2 (PRC2), and the 3′ end interacts with the histone demethylase LSD1 (62). By bringing these two chromatin modifying complexes in close proximity and guiding them to target chromatin, HOTAIR mediates epigenetic silencing of the HOXD locus, thus leading to increased cancer invasiveness and metastasis (19, 20). Through alternative splicing, the PRC2-binding domain of HOTAIR can be removed, potentially changing the functionality of this lncRNA.
Along these lines, another study focused on the transcriptome of hepatocellular carcinoma (HCC) patients. As a result, Zhang et al. found that in addition to differential expression, lncRNAs also displayed alternative splicing in HCC specimens compared to controls, suggesting a potential role for those splice variants as biomarkers and therapeutic targets for HCC (63). Taken together, alternative lncRNA splicing may alter the function of a given lncRNA.
Given the examples above, which illustrate multiple means of RNA isoform generation, we can assume that our current knowledge of diversity within gene loci in general is far from being complete, and current annotations are not always exhaustive. Accordingly, comprehensive analysis of gene loci might in many cases be necessary to enable accurate functional and mechanistic investigation of the resulting isoforms. Below, we will discuss approaches to identify expressed lncRNA isoforms and further explore a given lncRNA locus.
The gold standard in elucidating lncRNA expression and isoform discovery on a large scale is RNA-seq. In general, this sensitive method requires significant bioinformatical expertise, especially when investigating lncRNA isoforms or alternative splicing [for a review, see Ref. (64, 65)]. Nevertheless, there are some useful tools that require only limited bioinformatical knowledge and are publicly available, which can be seen as a starting point for isoform prediction. Employing genome browsers such as UCSC allows direct uploading of aligned RNA-seq data from the cell or tissue type of interest. This enables locus-specific mapping and comparison with potentially annotated lncRNA isoforms or publically available histone mark occupancy data as well as further expression tracks to support predictions of lncRNA isoforms from genomic areas devoid of any annotation (66). When inspecting RNA-seq reads, one should keep in mind that many lncRNAs are rather low expressed and exhibit a tissue-specific expression pattern, so adequate sequencing depth is required for isoform analysis (8, 9). Attention should also be attributed to the employed library preparation technique. Using oligo-dT-based enrichment strategies will result in loss of the non-polyadenylated lncRNA population within the transcriptome. Usage of non-poly-A selective library preparation methods can circumvent this problem, but at the cost of sequencing depth and the requirement for additional means of rRNA removal (67). Furthermore, strand-specific library preparation protocols offer the possibility to distinguish between sense and antisense transcripts (68).
Even though RNA-seq is very sensitive and bioinformatical tools are constantly improving, the experimental validation of potential isoforms employing methods such as northern blot or rapid amplification of cDNA ends (RACE) is still required to complement the bioinformatic predictions. With RACE, the RNA ends can be deciphered; however, most approaches utilize a 3′ poly-A tail, circumventing the detection of non-polyadenylated transcripts (69). For 5′ RACE, protocols exploiting the 5′ cap have been established, ensuring only detection of intact transcripts rather than also picking up potential degradation products (70). Identification of uncapped transcripts on the other hand requires classic 5′ RACE approaches, which might result in an overestimation of isoforms and transcription start sites (70). Supplementing RACE, cap analysis gene expression (CAGE) may unravel the 5′ end of capped RNAs, and recent modifications to the original protocol such as nanoCAGE or nAnT-iCAGE have been developed to work with minimal starting material and exclude bias from PCR amplification or tag cleavage (71, 72). Before performing own CAGE analysis, the recently published FANTOM5 data set can be mined for the occurrence of 5′ start sites for a given locus (73).
Once a pool of potential isoforms has been established, northern blots can be used to verify their presence and predicted length (74). Moreover, the abundance of approved isoforms can be determined by (q)RT-PCR using isoform-specific primer sets (75). Overall, none of the mentioned techniques alone might be sufficient for verification and mapping of multiple isoforms because each technique not only has certain strengths but also has weaknesses. However, using these methods as complementary approaches and compiling insights from all analyses may allow isoform prediction as well as isoform verification.
With new lncRNAs being continuously identified, this exciting research field is rapidly growing. In the hunt for new functions and mechanisms, close attention has to be paid to the wealth of lncRNA isoforms and their potential to being processed to other ncRNAs or translated into small peptides to discover new facets of lncRNAs. Recently, the presence of lncRNA modifications and lncRNA editing has been reported and associated with structural and functional changes, increasing the variety of lncRNAs (52, 53, 76–78). For example, m6A on position 2,577 of MALAT1 was found to alter its secondary structure, resulting in tighter binding of heterogeneous nuclear ribonucleoprotein C (77, 78).
Within this article, we highlighted the complexity of lncRNA isoform generation and outlined approaches for lncRNA isoform detection and their drawbacks, which should be rather seen as impulses than an exhausting discussion and motivate researchers to move forward into this very intriguing and challenging field of study.
CZ and MK drafted and revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Our research is supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 960 project B9 to MK).
1. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature (2012) 489:101–8. doi:10.1038/nature11233
2. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature (2004) 431:931–45. doi:10.1038/nature03001
3. Noller HF. Ribosomal RNA and translation. Annu Rev Biochem (1991) 60:191–227. doi:10.1146/annurev.bi.60.070191.001203
4. Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol (2007) 8:209–20. doi:10.1038/nrm2124
5. Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol (2008) 9:673–8. doi:10.1038/nrm2479
6. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell (2009) 136:215–33. doi:10.1016/j.cell.2009.01.002
7. Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development (2008) 135:1201–14. doi:10.1242/dev.005629
8. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res (2012) 22:1775–89. doi:10.1101/gr.132159.111
9. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev (2011) 25:1915–27. doi:10.1101/gad.17446611
10. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet (2013) 15:7–21. doi:10.1038/nrg3606
11. Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem J (2003) 369:163–71. doi:10.1042/bj20020743
12. Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, et al. p53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol (2008) 10:1098–105. doi:10.1038/ncb1770
13. Rutnam ZJ, Yang BB. The non-coding 3’ UTR of CD44 induces metastasis by regulating extracellular matrix functions. J Cell Sci (2012) 125:2075–85. doi:10.1242/jcs100818
14. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell (2015) 160:595–606. doi:10.1016/j.cell.2015.01.009
15. Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science (2016) 351:271–5. doi:10.1126/science.aad4076
16. Young TM, Tsai M, Tian B, Mathews MB, Pe’ery T. Cellular mRNA activates transcription elongation by displacing 7SK RNA. PLoS One (2007) 2:e1010. doi:10.1371/journal.pone.0001010
17. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature (2012) 493:231–5. doi:10.1038/nature11661
18. Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev (2012) 26:338–43. doi:10.1101/gad.182121.111
19. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell (2007) 129:1311–23. doi:10.1016/j.cell.2007.05.022
20. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature (2010) 464:1071–6. doi:10.1038/nature08975
21. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science (2010) 329:689–93. doi:10.1126/science.1192002
22. Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature (2007) 445:666–70. doi:10.1038/nature05519
23. Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature (1991) 351:153–5. doi:10.1038/351153a0
24. Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science (2005) 309:1570–3. doi:10.1126/science.1115901
25. Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature (1991) 349:38–44. doi:10.1038/349038a0
26. Hombach S, Kretz M. The non-coding skin: exploring the roles of long non-coding RNAs in epidermal homeostasis and disease: review essay. Bioessays (2013) 35:1093–100. doi:10.1002/bies.201300068
27. Zhang Z. Long non-coding RNAs in Alzheimer’s disease. Curr Top Med Chem (2016) 16:511–9. doi:10.2174/1568026615666150813142956
28. Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell (2013) 152:727–42. doi:10.1016/j.cell.2013.01.034
29. Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med (2016) 22:861–8. doi:10.1038/nm.4135
30. Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev (2016) 30:34–51. doi:10.1101/gad.270959.115
31. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet (2015) 47:199–208. doi:10.1038/ng.3192
32. Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res (2015) 43:D174–80. doi:10.1093/nar/gku1060
33. Kornienko AE, Dotter CP, Guenzl PM, Gisslinger H, Gisslinger B, Cleary C, et al. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol (2016) 17:14. doi:10.1186/s13059-016-0873-8
34. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. doi:10.1146/annurev-biochem-051410-092902
35. Stein JM. The effect of adrenaline and of alpha- and beta-adrenergic blocking agents on ATP concentration and on incorporation of 32Pi into ATP in rat fat cells. Biochem Pharmacol (1975) 24:1659–62. doi:10.1016/0006-2952(75)90002-7
36. Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, et al. Long noncoding RNAs with snoRNA ends. Mol Cell (2012) 48:219–30. doi:10.1016/j.molcel.2012.07.033
37. Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA (2009) 15:2013–27. doi:10.1261/rna.1705309
38. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet (2015) 17:47–62. doi:10.1038/nrg.2015.10
39. Niemczyk M, Ito Y, Huddleston J, Git A, Abu-Amero S, Caldas C, et al. Imprinted chromatin around DIRAS3 regulates alternative splicing of GNG12-AS1, a long noncoding RNA. Am J Hum Genet (2013) 93:224–35. doi:10.1016/j.ajhg.2013.06.010
40. Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5’-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol (1998) 18:6897–909. doi:10.1128/MCB.18.12.6897
41. He X, Chen X, Zhang X, Duan X, Pan T, Hu Q, et al. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res (2015) 43:3712–25. doi:10.1093/nar/gkv214
42. Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell (2011) 147:789–802. doi:10.1016/j.cell.2011.10.002
43. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature (2011) 477:295–300. doi:10.1038/nature10398
44. Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell (1999) 97:17–27. doi:10.1016/S0092-8674(00)80711-4
45. Gascoigne DK, Cheetham SW, Cattenoz PB, Clark MB, Amaral PP, Taft RJ, et al. Pinstripe: a suite of programs for integrating transcriptomic and proteomic datasets identifies novel proteins and improves differentiation of protein-coding and non-coding genes. Bioinformatics (2012) 28:3042–50. doi:10.1093/bioinformatics/bts582
46. Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res (2012) 22:1646–57. doi:10.1101/gr.134767.111
47. Kawaji H, Severin J, Lizio M, Waterhouse A, Katayama S, Irvine KM, et al. The FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Genome Biol (2009) 10:R40. doi:10.1186/gb-2009-10-4-r40
48. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science (2005) 309:1559–63. doi:10.1126/science.1112014
49. Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, et al. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods (2012) 10:133–9. doi:10.1038/nmeth.2288
50. Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res (2012) 22:1173–83. doi:10.1101/gr.132563.111
51. Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res (2014) 24:513–31. doi:10.1038/cr.2014.35
52. Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell (2008) 135:919–32. doi:10.1016/j.cell.2008.10.012
53. Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res (2008) 19:347–59. doi:10.1101/gr.087775.108
54. Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev (2012) 26:2392–407. doi:10.1101/gad.204438.112
55. Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A (2003) 100:15776–81. doi:10.1073/pnas.2136655100
56. de Klerk E, ‘t Hoen PAC. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet (2015) 31:128–39. doi:10.1016/j.tig.2015.01.001
57. Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development (2001) 128:1275–86.
58. Saghaeian Jazi M, Samaei NM, Ghanei M, Shadmehr MB, Mowla SJ. Identification of new SOX2OT transcript variants highly expressed in human cancer cell lines and down regulated in stem cell differentiation. Mol Biol Rep (2016) 43:65–72. doi:10.1007/s11033-015-3939-x
59. Gudde AE, van Heeringen SJ, de Oude AI, van Kessel ID, Estabrook J, Wang ET, et al. Antisense transcription of the myotonic dystrophy locus yields low-abundant RNAs with and without (CAG)n repeat. RNA Biol (2017). doi:10.1080/15476286.2017.1279787
60. Gonzalez I, Munita R, Agirre E, Dittmer TA, Gysling K, Misteli T, et al. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol (2015) 22(5):370–6. doi:10.1038/nsmb.3005
61. Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry (2014) 19:486–94. doi:10.1038/mp.2013.45
62. Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol (2011) 30:99–104. doi:10.1038/nbt.2024
63. Zhang L, Liu X, Zhang X, Chen R. Identification of important long non-coding RNAs and highly recurrent aberrant alternative splicing events in hepatocellular carcinoma through integrative analysis of multiple RNA-seq datasets. Mol Genet Genomics (2016) 291:1035–51. doi:10.1007/s00438-015-1163-y
64. Li W, Dai C, Kang S, Zhou XJ. Integrative analysis of many RNA-seq datasets to study alternative splicing. Methods (2014) 67:313–24. doi:10.1016/j.ymeth.2014.02.024
65. Signal B, Gloss BS, Dinger ME. Computational approaches for functional prediction and characterisation of long noncoding RNAs. Trends Genet (2016) 32:620–37. doi:10.1016/j.tig.2016.08.004
66. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res (2002) 12:996–1006. doi:10.1101/gr.229102
67. Hrdlickova R, Toloue M, Tian B. RNA-seq methods for transcriptome analysis: RNA-seq. Wiley Interdiscip Rev RNA (2017) 8:e1364. doi:10.1002/wrna.1364
68. Borodina T, Adjaye J, Sultan M. Methods in Enzymology. (Vol. 500). London: Elsevier (2011). p. 79–98.
69. Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A (1988) 85:8998–9002. doi:10.1073/pnas.85.23.8998
70. Schaefer BC. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem (1995) 227:255–73. doi:10.1006/abio.1995.1279
71. Plessy C, Bertin N, Takahashi H, Simone R, Salimullah M, Lassmann T, et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat Methods (2010) 7:528–34. doi:10.1038/nmeth.1470
72. Murata M, Nishiyori-Sueki H, Kojima-Ishiyama M, Carninci P, Hayashizaki Y, Itoh M. Detecting expressed genes using CAGE. Methods Mol Biol (2014) 1164:67–85. doi:10.1007/978-1-4939-0805-9_7
73. Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature (2017) 543:199–204. doi:10.1038/nature21374
74. Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet (2006) 2:e37. doi:10.1371/journal.pgen.0020037
75. Vandenbroucke II, Vandesompele J, Paepe AD, Messiaen L. Quantification of splice variants using real-time PCR. Nucleic Acids Res (2001) 29:E68–68. doi:10.1093/nar/29.13.e68
76. Gong J, Liu C, Liu W, Xiang Y, Diao L, Guo AY, et al. LNCediting: a database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res (2017) 45:D79–84. doi:10.1093/nar/gkw835
77. Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, et al. N6-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J Mol Biol (2016) 428:822–33. doi:10.1016/j.jmb.2015.08.021
Keywords: long non-coding RNAs, non-coding RNA, alternative splicing, alternative polyadenylation, bifunctional RNA
Citation: Ziegler C and Kretz M (2017) The More the Merrier—Complexity in Long Non-Coding RNA Loci. Front. Endocrinol. 8:90. doi: 10.3389/fendo.2017.00090
Received: 01 February 2017; Accepted: 06 April 2017;
Published: 25 April 2017
Edited by:
Jan-Wilhelm Kornfeld, Max-Planck Institute for Metabolism Research, GermanyReviewed by:
Eleonora Leucci, vib-KU Leuven, BelgiumCopyright: © 2017 Ziegler and Kretz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Markus Kretz, bWFya3VzLmtyZXR6QHZrbC51bmktcmVnZW5zYnVyZy5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.