- 1Graduate School of Medicine, Korea University, Seoul, South Korea
- 2INSERM U982, University of Rouen, Mont-Saint-Aignan, France
Kisspeptin (KISS) plays a key role in regulating reproduction by binding to its receptor, GPR54. Because of the Arg-Phe (RF) sequence at its carboxyl terminus, KISS has been proposed to be a member of the RF-amide peptide family consisting of neuropeptide FF (NPFF), neuropeptide VF (NPVF), pyroglutamylated RF-amide peptide (QRFP), and prolactin-releasing hormone (PRLH). Evolutionary relationships of protein families can be determined through phylogenetic analysis. However, phylogenetic analysis among related peptide families often fails to provide sufficient information because only short mature peptide sequences from full preprohormone sequences are conserved. Considering the concept of the coevolution of peptide ligands and their cognate receptors, evolutionary relationships among related receptor families provide clues to explore relationships between their peptides. Although receptors for NPFF, NPVF, and QRFP are phylogenetically clustered together, receptors for PRLH and KISS are on different branches of the phylogenetic tree. In particular, KISS has been proposed to be a member of the KISS/galanin/spexin family based on synteny analysis and the phylogenetic relationship between their receptors. This article discusses the evolutionary history of the receptors for the proposed RF-amide peptide family and proposes that, from an evolutionary aspect, KISS has emerged from an ancestor, which is distinct from those of the other RF-amide peptides, and so should be classed separately.
Introduction
The RF-amide peptides that harbor the Arg-Phe-amide sequence in their carboxyl (C)-termini were first discovered in a species of mollusk, Macrocallista nimbosa (1). Following this discovery, various RF-amide peptides have been identified in other invertebrate species (2, 3). In vertebrates, neuropeptide FF (NPFF) along with its receptor GPR74 (NPFFR2) was the first to be identified in the central nervous system of mammals (4). Fifteen years later, paralogous peptides, gonadotropin-inhibitory hormone (GnIH) in quail brain (5), and neuropeptide VF (NPVF) in mammals (6, 7) were identified. Currently, GnIH and NPVF are found to be orthologous to each other and activate the receptor GPR147 (NPFFR1) (6–10). Another paralogous peptide, pyroglutamylated RF-amide peptide (QRFP or 43RF-amide peptide), and its receptor GPR103 (QRFPR) have been identified in the hypothalamus and spinal cord of mammals (11–13). The receptors for these three vertebrate RF-amide peptides, NPFFR1, NPFFR2, and QRFPR, are phylogenetically very close to each other (Figure 1), indicating an evolutionarily common origin for these receptors. The other RF-amide-related peptides in vertebrates are prolactin-releasing hormone (PRLH or PrRP) and kisspeptin (KISS). Interestingly, the receptors for PRLH (PRLHR) and KISS (KISSR) are phylogenetically distant from NPFFR1, NPFFR2, and QRFPR. They are more closely related to neuropeptide Y (NPY) receptors (14, 15) and galanin (GAL) receptors (16), respectively (Figure 1).
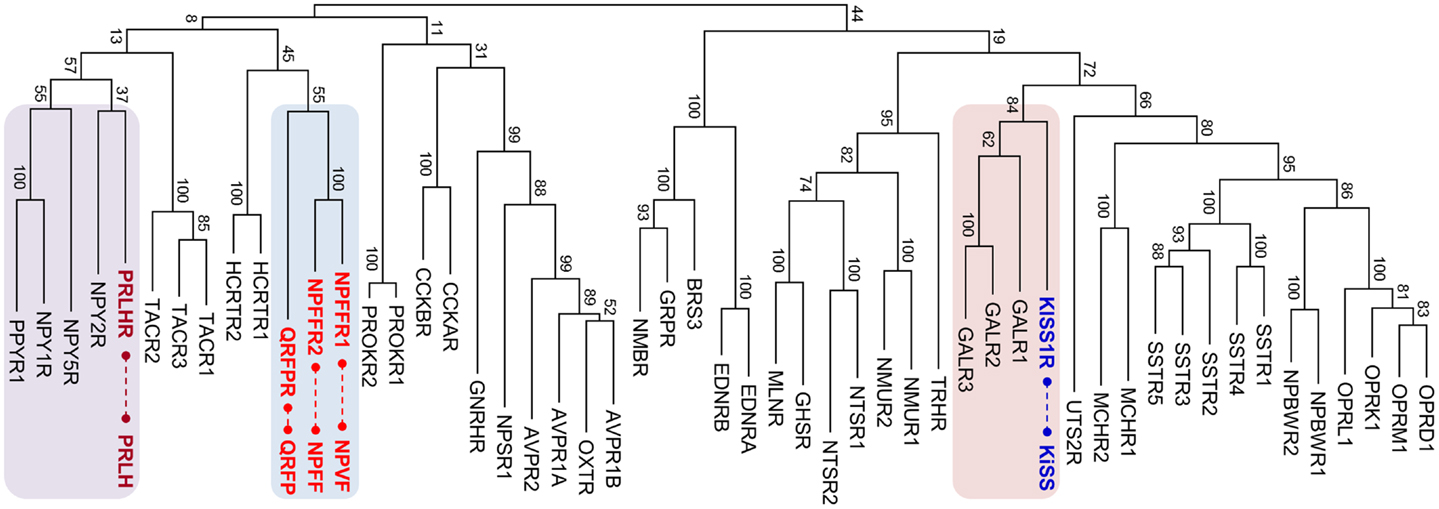
Figure 1. Maximum likelihood phylogenetic tree for human rhodopsin-like neuropeptide GPCRs. The amino acid sequences of the receptors were aligned using MUSCLE, and the tree was constructed by MEGA 5.1. The RF-amide peptide receptor groups are indicated with colored boxes and corresponding peptides are connected by dashed lines. Bootstrap values represent 100 replicates. MLNR, motilin receptor; GHSR, ghrelin receptor; NTSR, neurotensin receptor; NMUR, neuromedin U repceptor; TRHR, thyrotropin-releasing hormone receptor; NMBR, neuromedin B receptor; GRPR, gastrin-releasing peptide receptor; BRS, bombesin receptor; EDNR, endothelin receptor; PRLHR, prolactin-releasing hormone receptor; NPYR, neuropeptide Y receptor; PROKR, prokineticin receptor; TACR, tachykinin receptor; CCKR, cholecystokinin receptor; HCRTR, hypocretin (orexin) receptor; QRFPR, pyroglutamylated RFamide peptide receptor; NPFFR, neuropeptide FF receptor; AVPR, arginine vasopressin receptor; OXTR, oxytocin receptor; NPSR, neuropeptide S receptor; GnRHR, gonadotropin-releasing hormone receptor; UTSR, urotensin receptor; GALR, galanin receptor; KISSR, kisspeptin receptor; MCHR, melanin-concentrating hormone receptor; SSTR, somatostatin receptor; NPBWR, neuropeptides B/W receptor; OPR, opioid receptor.
The RF-amide peptides do not exhibit any sequence similarity to each other, other than the presence of the common RF-amide at their C-termini (17). As most neuropeptides have coevolved with their cognate receptors (18, 19), phylogenetic analysis of the related receptor families may mirror the evolutionary history of the peptide families (20, 21). Phylogeny of the RF-amide peptide receptor family, however, revealed distant relationships between PRLHR, KISSR, and the other RF-amide receptors (15, 16). This suggests that PRLH and KISS, at least, are likely to have originated from ancestors different from that of NPFF, NPVF, and QRFP. Therefore, it is timely to redefine the RF-amide family group members according to the evolutionary histories of individual RF-amide peptides. This article reviews the evolutionary relationships between KISS, and the other RF-amide peptides along with their paired receptors, and proposes that KISS is independent from the RF-amide peptide family.
General Mechanism for the Evolution of Neuropeptides and Their Receptors
Neuropeptide and receptor families have expanded through whole-genome duplications (WGD) and local tandem gene duplications before and after WGD during vertebrate evolution (14, 16, 20, 22–26). To date, synteny analyses of vertebrate genome fragments and comparison of entire chromosomes of evolutionarily distinct taxa support two rounds (2R) of WGD during early vertebrate evolution. These events produced four paralogous chromosomal regions (paralogons) sharing similar sets of genes (27–29). A third round (3R) of WGD during an early phase of bony fish emergence resulted in octupled paralogons in teleosts (30).
The evolutionary history of a gene family can be traced by phylogenetic analysis. However, in the case of peptide gene families, phylogenetic analysis often does not provide sufficient information to conclude the evolutionary relationship among related peptide gene families (20, 31). In general, signal peptide sequences are not conserved and propeptide sequences, other than the mature peptide, are highly variable because they are free from evolutionary selective pressure (32). Sequence comparison of only a short, conserved mature peptide is not enough to extrapolate correct relational information. Furthermore, paralogous peptide genes that arose by local gene duplication before 2R exhibit considerable variation even in the mature peptide sequences (20, 31). For instance, the mature peptide sequences of the ligand genes for class B (secretin-like) G-protein-coupled receptors (GPCR) are highly variable (20, 31) while maintaining conserved three-dimensional structure to a general extent (33). Thus, alternative methods need to be applied to explore the evolutionary relationships among peptide gene families.
In contrast to peptide genes, evolutionary relationships among related receptor families can be readily assessed by phylogenetic analysis since the transmembrane domains of the receptors are relatively conserved across vertebrate species and the amino acid sequences of receptors are long enough to generate a convincing phylogenetic tree. Thus, tracing relationships among related receptor families can provide clues to understand the evolutionary history of their corresponding peptide genes. For instance, the evolutionary histories of receptors for secretin family peptides, including corticotropin-releasing hormone, calcitonin, parathyroid hormone, glucagon, and secretin subfamilies may allow us to speculate about possible evolutionary histories for the cognate peptide genes, for which the sequence similarities are not well preserved (20, 25, 31). In addition, locating related peptide genes on the reconstructed pre-2R ancestral chromosomes (or linkage group) is an alternative tool to explore the relationships among related gene families (20, 34). Paralogous genes that emerged through local duplications before 2R reside in the same vicinity on the pre-2R linkage group. For instance, many secretin family peptide genes are located on the same pre-2R linkage group, indicating close evolutionary relationships among the peptide genes (20). The gene families of the neuropeptides, KISS, GAL, and spexin (SPX), are in the vicinity of the same pre-2R linkage group (16). This possibility is further supported by phylogenetic relationships among the receptors of these peptides (Figure 1) (16). The neuropeptide Y (NPY) gene and its paralogous genes, including peptide YY (PYY) and pancreatic polypeptide (PPY), reside on the same chromosome or on paralogons (35, 36). Likewise, the NPY receptor (NPYR) family seems to have been generated by local gene duplications followed by 2R WGD (14, 15, 37).
Evolutionary History of NPFF/NPVF, QRFP, and PRLH
Neuropeptide NPFF was the first RF-amide peptide characterized in the central nervous system of mammals (4, 8, 38, 39). NPFF is known to be involved in morphine tolerance, adipogenesis, and anorectic activity (40–45). In 2000, Tsutsui et al. found a new RF-amide peptide, GnIH, in quail brain (5, 46). This turned out to be an ortholog of NPVF in humans (9, 47). Albeit with some cross-reactivity, NPFF has a high affinity to NPFFR2 (4) while NPVF more selectively activates NPFFR1 (6, 48, 49).
The neuropeptides NPFF and NPVF seem to be 2R-generated paralogs (ohnologs) as their genes are located on two 2R-generated paralogons of human chromosomes. NPFF is located in the vicinity of the neuropeptide tachykinin 3 (TAC3) gene on human chromosome 12 while NPVF is on human chromosome 7, which also contains the TAC1 gene. Likewise, the receptors NPFFR1 and NPFFR2 are ohnologous to each other since NPFFR1/TACR2 and NPFFR2/TACR3 pairs are on paralogons of human chromosomes 10 and 4, respectively (6, 15, 50). It is of interest to note that human chromosome 10 also harbors PRLHR and PPYR (a receptor for NPY family peptides) and that human chromosome 4 has QRFPR and three NPYRs, NPY1R, NPY2R, and NPY5R (14, 15, 51). The phylogenetic tree in Figure 1 also shows that these receptors are clustered together. These observations suggest that NPFFRs, QRFPR, TACRs, NPYRs, and PRLHR may have emerged through local duplications from a common ancestor before 2R and expanded their members via 2R. Although the hypocretin (orexin) receptor (HCRTR) is shown in that branch, the HCRTR gene is not shown in the paralogons. This may be due to translocation of the gene to other chromosomal regions during evolution (20). Similarly, many peptide genes for these receptors are clustered on 2R-generated paralogons. For instance, the HCRT, PYY, PPY, and TAC4 genes are closely located on a region of human chromosome 17, which is likely another paralogon of NPFF/TAC-containing regions of human chromosomes 7 and 12 (15). Thus, it seems likely that these peptide genes arose during vertebrate evolution in a manner similar to that of receptor genes. It is noteworthy that the chicken C-RF-amide peptide (PRLH2), an ohnolog of PRLH resides near NPY, TAC1, and NPVF on the chromosomes of chickens, medaka, and tetraodon (52, 53). These findings suggest the presence of evolutionary relationships between the NPFF family and the other RF-amide peptides QRFP and PRLH, along with non-RF-amide peptides such as NPY, TAC, and HCRT.
Pyroglutamylated RF-amide peptide (43RF-amide), a long form of 26RF-amide, has been discovered in the hypothalamus and spinal cord of humans (11–13, 54). Intravenous or intracerebroventricular administration of QRFP increased plasma aldosterone levels and food intake in rats (11, 12, 55). While humans have only one QRFP receptor (12, 13), rodents have two receptors QRFPR1 and QRFPR2 (56, 57). QRFPR2 appears to be an ohnolog of QRFPR1. A region of mouse chromosome 6 has QRFPR2 and TACR1, indicating that this region is another paralogon for the QRFPR/NPFFR/TACR-containing chromosomal regions as described previously. As the phylogenetic tree reveals a close relationship between QRFPR and NPFFRs, an evolutionarily close relationship between QRFR and NPFF/NPVF can be postulated.
Prolactin-releasing hormone was first identified by a reverse pharmacological approach in 1998 (58). Physiological roles of PRLH include regulation of stress response (59–61), reduced appetite (62–64), and stimulation of luteinizing hormone and follicle stimulating hormone (65). The orphan receptor GPR10 was identified as the receptor for PRLH (58, 66, 67). PRLHR exhibits a close relationship in its sequence identity with the NPYR family (14, 15). NPY4R, a member of the NPYR family is positioned together with PRLHR1 on the same chromosome of humans and chickens (15). There are also some structural similarities between PRLH and NPY family peptides. Both PRLH and NPY family genes have two coding exons. The first coding exon contains a signal peptide sequence followed by the N-terminal region of the mature peptide sequence. The second coding exon contains the C-terminal region of the mature peptide with conserved Arg-Phe-Gly (RFG) or Arg-Tyr-Gly (RYG) residues followed by a dibasic cleavage site. Like PRLH, some NPY peptide family members such as bovine PYY and PPY have RF-amide sequences, while NPY, PPY, and PYY from most vertebrates contain the Arg-Tyr-amide (RY-amide) sequence (15, 68, 69). It is also interesting to note that the NPY peptide is able to activate PRLHR1 at micromolar levels (15). These observations together with phylogenetic analysis of neuropeptide receptors suggest that the PRLH/NPY family genes and their receptor genes emerged by local gene duplications during early vertebrate evolution (14, 15). These local duplication events are likely to have occurred after the genes split from their common ancestors into the PRLH/NPY and NPFF/QRFP systems.
Coevolution of the Spexin/Galanin/Kisspeptin Family
The KISS gene was first identified as a tumor suppressor gene expressed in human melanoma and breast cancer cells (70, 71). Later, the KISS gene was found to produce a functional peptide with an RF-amide sequence (designated as kisspeptin or metastin) that activates an orphan GPCR, GPR54 (KISSR) (72–74). KISS and KISSR are involved in the onset of puberty and the control of reproduction and food consumption (75–80). After the discoveries of mammalian KISS and KISSR, two paralogs of the KISS gene (KISS2 and KISS3) and three paralogs of the KISSR gene (KISSR2, KISSR3, and KISSR4) have been identified in a variety of vertebrate species (32, 81–83). All these paralogs seem to be ohnologs as each of them is located on 2R-generated paralogons (32, 82, 83).
Although KISS has been acknowledged as a member of the RF-amide peptide family, no supporting evidence, other than the presence of the RF-amide sequence in its C-terminus, has been provided. KISSR was originally reported to have considerably high degree of sequence similarity with galanin receptors (GALR) (84). This result raises the possibility that KISS/KISSR and GAL/GALR pairs have diverged from a common ancestor and not from the RF-amide peptide/receptor ancestors. Recently, it was shown that the novel neuropeptides spexin1 (SPX1) and spexin2 (SPX2) are functional ligands for GALR2 and GALR3 but not GALR1 (16, 85). In particular, SPXs are more potent than GAL in activation of GALR3, while they show potencies to GALR2 similar to that of GAL (16). Synteny analysis and relocation of the gene families on the reconstructed vertebrate ancestral linkage groups show that SPX, GAL, and KISS family genes are distributed among 2R-generated 4 paralogons (4 linkage groups). Three linkage groups contain SPX1 and KISS2, SPX2 and GAL, and KISS3 and galanin-like peptide (GALP), respectively. The fourth linkage group has KISS1 alone (16, 86). This study proposed that ancestral forms of KISS, GAL, and SPX arose by tandem local duplications before 2R and expanded their family members through 2R (16). Likewise, KISSRs and GALRs are likely to have emerged through local duplications before 2R, producing three subgroups in vertebrates: KISSRs (4 KISSRs), GALR1 (GALR1a and GALR1b), and GALR2/3 (GALR2a, GALR2b, and GALR3) (Figure 2). Altogether, these results suggest that the evolutionary origin of the KISS/SPX/GAL family is far distant from those of the NPFF/QRFP and PRLH/NPY families.
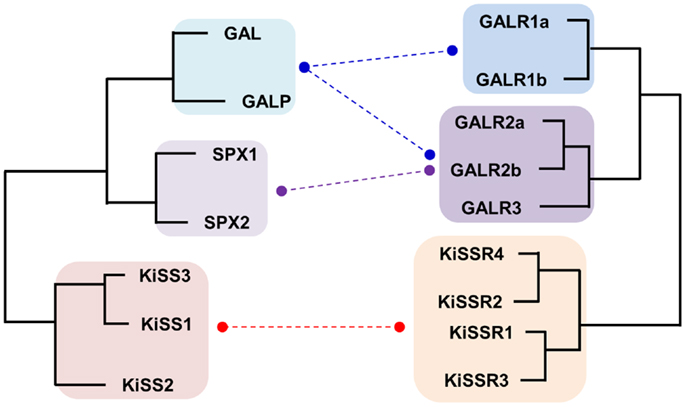
Figure 2. Schematic diagram for the evolutionary relationship between KISS/GAL/SPX family peptides and their corresponding receptors in vertebrates. Paralogs for each peptide and receptor found in vertebrates are included. Functional interaction between the ligands and receptors are indicated by dashed lines. The KISS peptides are able to activate all types of KISSRs with some differences in their ligand potencies but they do not activate any type of GALRs. GALs exhibit high potencies toward GALR1 and GALR2 family receptors but low potencies toward GALR3. SPXs activate GALR2 and GALR3 with high affinities but do not activate GALR1.
Conclusion
Rapid accumulation of genomic sequence information from various invertebrate and vertebrate species and the development of bioinformatic tools, including phylogenetic analysis, small scale genome comparison to identify orthologous and paralogous relationships of genes, and reconstruction of ancient chromosomes, have facilitated exploration of the relationships and origins of peptide and receptor gene families (20, 28, 32, 34, 87, 88). Based on the phylogenetic analysis of neuropeptide receptors and the syntenic relationships of neuropeptide genes, we suggest that the proposed RF-amid peptide family arose from three different ancestors. NPFF and NPVF are likely 2R-generated paralogs and have a close evolutionary relationship with QRFP. PRLH is evolutionarily closer to the NPY family than the NPFF/NPVF/QRFP group. KISS is likely a member of the KISS/GAL/SPX peptide family and their evolutionary origin is far distant from those of the other RF-amide peptides. This study may provide an insight into the mechanism for coevolution of neuropeptides and their receptors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant (2013R1A2A2A01068295) from the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning.
References
1. Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science (1977) 197:670–1. doi: 10.1126/science.877582
2. Walker RJ. Neuroactive peptides with an RFamide or Famide carboxyl terminal. Comp Biochem Physiol C (1992) 102:213–22. doi:10.1016/0742-8413(92)90104-F
3. Walker RJ, Papaioannou S, Holden-Dye L. A review of FMRFamide- and RFamide-like peptides in metazoa. Invert Neurosci (2009) 9:111–53. doi:10.1007/s10158-010-0097-7
4. Yang HY, Fratta W, Majane EA, Costa E. Isolation, sequencing, synthesis, and pharmacological characterization of two brain neuropeptides that modulate the action of morphine. Proc Natl Acad Sci U S A (1985) 82:7757–61. doi:10.1073/pnas.82.22.7757
5. Tsutsui K, Saigoh E, Ukena K, Teranishi H, Fujisawa Y, Kikuchi M, et al. A novel avian hypothalamic peptide inhibiting gonadotropin release. Biochem Biophys Res Commun (2000) 275:661–7. doi:10.1006/bbrc.2000.3350
6. Bonini JA, Jones KA, Adham N, Forray C, Artymyshyn R, Durkin MM, et al. Identification and characterization of two G protein-coupled receptors for neuropeptide FF. J Biol Chem (2000) 275:39324–31. doi:10.1074/jbc.M004385200
7. Elshourbagy NA, Ames RS, Fitzgerald LR, Foley JJ, Chambers JK, Szekeres PG, et al. Receptor for the pain modulatory neuropeptides FF and AF is an orphan G protein-coupled receptor. J Biol Chem (2000) 275:25965–71. doi:10.1074/jbc.M004515200
8. Perry SJ, Yi-Kung Huang E, Cronk D, Bagust J, Sharma R, Walker RJ, et al. A human gene encoding morphine modulating peptides related to NPFF and FMRFamide. FEBS Lett (1997) 409:426–30. doi:10.1016/S0014-5793(97)00557-7
9. Hinuma S, Shintani Y, Fukusumi S, Iijima N, Matsumoto Y, Hosoya M, et al. New neuropeptides containing carboxy-terminal RFamide and their receptor in mammals. Nat Cell Biol (2000) 2:703–8. doi:10.1038/35036326
10. Ubuka T, Morgan K, Pawson AJ, Osugi T, Chowdhury VS, Minakata H, et al. Identification of human GnIH homologs, RFRP-1 and RFRP-3, and the cognate receptor, GPR147 in the human hypothalamic pituitary axis. PLoS One (2009) 4:e8400. doi:10.1371/journal.pone.0008400
11. Chartrel N, Dujardin C, Anouar Y, Leprince J, Decker A, Clerens S, et al. Identification of 26RFa, a hypothalamic neuropeptide of the RFamide peptide family with orexigenic activity. Proc Natl Acad Sci U S A (2003) 100:15247–52. doi:10.1073/pnas.2434676100
12. Fukusumi S, Yoshida H, Fujii R, Maruyama M, Komatsu H, Habata Y, et al. A new peptidic ligand and its receptor regulating adrenal function in rats. J Biol Chem (2003) 278:46387–95. doi:10.1074/jbc.M305270200
13. Jiang Y, Luo L, Gustafson EL, Yadav D, Laverty M, Murgolo N, et al. Identification and characterization of a novel RF-amide peptide ligand for orphan G-protein-coupled receptor SP9155. J Biol Chem (2003) 278:27652–7. doi:10.1074/jbc.M302945200
14. Larhammar D, Salaneck E. Molecular evolution of NPY receptor subtypes. Neuropeptides (2004) 38:141–51. doi:10.1016/j.npep.2004.06.002
15. Lagerstrom MC, Fredriksson R, Bjarnadottir TK, Fridmanis D, Holmquist T, Andersson J, et al. Origin of the prolactin-releasing hormone (PRLH) receptors: evidence of coevolution between PRLH and a redundant neuropeptide Y receptor during vertebrate evolution. Genomics (2005) 85:688–703. doi:10.1016/j.ygeno.2005.02.007
16. Kim DK, Yun S, Son GH, Hwang JI, Park CR, Kim JI, et al. Coevolution of the spexin/galanin/kisspeptin family: spexin activates galanin receptor type II and III. Endocrinology (2014) 155:1864–73. doi:10.1210/en.2013-2106
17. Tsutsui K, Bentley GE, Kriegsfeld LJ, Osugi T, Seong JY, Vaudry H. Discovery and evolutionary history of GnIH and kisspeptin: new key neuropeptides controlling reproduction. J Neuroendocrinology (2010) 22:716–27. doi:10.1111/j.1365-2826.2010.02018.x
18. Acharjee S, Do-Rego JL, Oh DY, Moon JS, Ahn RS, Lee K, et al. Molecular cloning, pharmacological characterization, and histochemical distribution of frog vasotocin and mesotocin receptors. J Mol Endocrinol (2004) 33:293–313. doi:10.1677/jme.0.0330293
19. Cho HJ, Acharjee S, Moon MJ, Oh DY, Vaudry H, Kwon HB, et al. Molecular evolution of neuropeptide receptors with regard to maintaining high affinity to their authentic ligands. Gen Comp Endocrinol (2007) 153:98–107. doi:10.1016/j.ygcen.2006.12.013
20. Hwang JI, Moon MJ, Park S, Kim DK, Cho EB, Ha N, et al. Expansion of secretin-like G protein-coupled receptors and their peptide ligands via local duplications before and after two rounds of whole-genome duplication. Mol Biol Evol (2013) 30:1119–30. doi:10.1093/molbev/mst031
21. Mirabeau O, Joly JS. Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci U S A (2013) 110:E2028–37. doi:10.1073/pnas.1219956110
23. Lundin LG. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics (1993) 16:1–19. doi:10.1006/geno.1993.1133
24. Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl (1994):125–33.
25. Hwang JI, Yun S, Moon MJ, Park CR, Seong JY. Molecular evolution of GPCRs: GLP1/GLP1 receptors. J Mol Endocrinol (2014) 52:T15–27. doi:10.1530/JME-13-0137
26. Sefideh FA, Moon MJ, Yun S, Hong SI, Hwang JI, Seong JY. Local duplication of gonadotropin-releasing hormone (GnRH) receptor before two rounds of whole genome duplication and origin of the mammalian GnRH receptor. PLoS One (2014) 9:e87901. doi:10.1371/journal.pone.0087901
27. Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol (2005) 3:e314. doi:10.1371/journal.pbio.0030314
28. Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res (2007) 17:1254–65. doi:10.1101/gr.6316407
29. Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature (2008) 453:1064–71. doi:10.1038/nature06967
30. Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays (2005) 27:937–45. doi:10.1002/bies.20293
31. Cardoso JC, Vieira FA, Gomes AS, Power DM. The serendipitous origin of chordate secretin peptide family members. BMC Evol Biol (2010) 10:135. doi:10.1186/1471-2148-10-135
32. Lee YR, Tsunekawa K, Moon MJ, Um HN, Hwang JI, Osugi T, et al. Molecular evolution of multiple forms of kisspeptins and GPR54 receptors in vertebrates. Endocrinology (2009) 150:2837–46. doi:10.1210/en.2008-1679
33. Neumann JM, Couvineau A, Murail S, Lacapere JJ, Jamin N, Laburthe M. Class-B GPCR activation: is ligand helix-capping the key? Trends Biochem Sci (2008) 33:314–9. doi:10.1016/j.tibs.2008.05.001
34. Yegorov S, Good S. Using paleogenomics to study the evolution of gene families: origin and duplication history of the relaxin family hormones and their receptors. PLoS One (2012) 7:e32923. doi:10.1371/journal.pone.0032923
35. Cerdá-Reverter JM, Larhammar D. Neuropeptide Y family of peptides: structure, anatomical expression, function, and molecular evolution. Biochem Cell Biol (2000) 78:371–92. doi:10.1139/o00-004
36. Larhammar D, Fredriksson R, Larson ET, Salaneck E. Phylogeny of NPY-family peptides and their receptors. In: Michel MC, editor. Neuropepide Y and Related Peptides. Berlin-Heidelberg: Springer-Verlag (2004). p. 75–100.
37. Larhammar D, Bergqvist CA. Ancient grandeur of the vertebrate neuropeptide Y system shown by the coelacanth Latimeria chalumnae. Front Neurosci (2013) 7:27. doi:10.3389/fnins.2013.00027
38. Yang HY, Martin RM. Isolation and characterization of a neuropeptide FF-like peptide from brain and spinal cord of rat. Soc Neurosci Abstr (1995) 21:760.
39. Bonnard E, Burlet-Schiltz O, Monsarrat B, Girard JP, Zajac JM. Identification of proNeuropeptide FFA peptides processed in neuronal and non-neuronal cells and in nervous tissue. Eur J Biochem (2003) 270:4187–99. doi:10.1046/j.1432-1033.2003.03816.x
40. Murase T, Arima H, Kondo K, Oiso Y. Neuropeptide FF reduces food intake in rats. Peptides (1996) 17:353–4. doi:10.1016/0196-9781(95)02137-X
41. Sunter D, Hewson AK, Lynam S, Dickson SL. Intracerebroventricular injection of neuropeptide FF, an opioid modulating neuropeptide, acutely reduces food intake and stimulates water intake in the rat. Neurosci Lett (2001) 313:145–8. doi:10.1016/S0304-3940(01)02267-4
42. Herrera-Herrera ML, Salazar-Olivo LA. RFamide neuropeptides inhibit murine and human adipose differentiation. Biochem Biophys Res Commun (2008) 377:29–34. doi:10.1016/j.bbrc.2008.09.071
43. Mouledous L, Frances B, Zajac JM. Modulation of basal and morphine-induced neuronal activity by a NPFF(2) selective agonist measured by c-Fos mapping of the mouse brain. Synapse (2010) 64:672–81. doi:10.1002/syn.20774
44. Wu CH, Tao PL, Huang EY. Distribution of neuropeptide FF (NPFF) receptors in correlation with morphine-induced reward in the rat brain. Peptides (2010) 31:1374–82. doi:10.1016/j.peptides.2010.03.036
45. Jhamandas JH, Goncharuk V. Role of neuropeptide FF in central cardiovascular and neuroendocrine regulation. Front Endocrinol (Lausanne) (2013) 4:8. doi:10.3389/fendo.2013.00008
46. Ukena K, Tsutsui K. A new member of the hypothalamic RF-amide peptide family, LPXRF-amide peptides: structure, localization, and function. Mass Spectrom Rev (2005) 24:469–86. doi:10.1002/mas.20031
47. Liu Q, Guan XM, Martin WJ, McDonald TP, Clements MK, Jiang Q, et al. Identification and characterization of novel mammalian neuropeptide FF-like peptides that attenuate morphine-induced antinociception. J Biol Chem (2001) 276:36961–9. doi:10.1074/jbc.M105308200
48. Mollereau C, Mazarguil H, Marcus D, Quelven I, Kotani M, Lannoy V, et al. Pharmacological characterization of human NPFF(1) and NPFF(2) receptors expressed in CHO cells by using NPY Y(1) receptor antagonists. Eur J Pharmacol (2002) 451:245–56. doi:10.1016/S0014-2999(02)02224-0
49. Findeisen M, Rathmann D, Beck-Sickinger AG. Structure-activity studies of RFamide peptides reveal subtype-selective activation of neuropeptide FF1 and FF2 receptors. ChemMedChem (2011) 6:1081–93. doi:10.1002/cmdc.201100089
50. Zhou W, Li S, Liu Y, Qi X, Chen H, Cheng CH, et al. The evolution of tachykinin/tachykinin receptor (TAC/TACR) in vertebrates and molecular identification of the TAC3/TACR3 system in zebrafish (Danio rerio). Mol Cell Endocrinol (2012) 361:202–12. doi:10.1016/j.mce.2012.04.007
51. Ukena K, Osugi T, Leprince J, Vaudry H, Tsutsui K. Molecular evolution of GPCRs: 26Rfa/GPR103. J Mol Endocrinol (2014) 52:T119–31. doi:10.1530/JME-13-0207
52. Lagerstrom MC, Fredriksson R, Bjarnadottir TK, Schioth HB. The ancestry of the prolactin-releasing hormone precursor. Ann N Y Acad Sci (2005) 1040:368–70. doi:10.1196/annals.1327.064
53. Wang Y, Wang CY, Wu Y, Huang G, Li J, Leung FC. Identification of the receptors for prolactin-releasing peptide (PrRP) and Carassius RFamide peptide (C-RFa) in chickens. Endocrinology (2012) 153:1861–74. doi:10.1210/en.2011-1719
54. Bruzzone F, Lectez B, Tollemer H, Leprince J, Dujardin C, Rachidi W, et al. Anatomical distribution and biochemical characterization of the novel RFamide peptide 26RFa in the human hypothalamus and spinal cord. J Neurochem (2006) 99:616–27. doi:10.1111/j.1471-4159.2006.04090.x
55. Do Rego JC, Leprince J, Chartrel N, Vaudry H, Costentin J. Behavioral effects of 26RFa and related peptides. Peptides (2006) 27:2715–21. doi:10.1016/j.peptides.2006.04.017
56. Kampe J, Wiedmer P, Pfluger PT, Castaneda TR, Burget L, Mondala H, et al. Effect of central administration of QRFP(26) peptide on energy balance and characterization of a second QRFP receptor in rat. Brain Res (2006) 1119:133–49. doi:10.1016/j.brainres.2006.08.055
57. Takayasu S, Sakurai T, Iwasaki S, Teranishi H, Yamanaka A, Williams SC, et al. A neuropeptide ligand of the G protein-coupled receptor GPR103 regulates feeding, behavioral arousal, and blood pressure in mice. Proc Natl Acad Sci U S A (2006) 103:7438–43. doi:10.1073/pnas.0602371103
58. Hinuma S, Habata Y, Fujii R, Kawamata Y, Hosoya M, Fukusumi S, et al. A prolactin-releasing peptide in the brain. Nature (1998) 393:272–6. doi:10.1038/30515
59. Maruyama M, Matsumoto H, Fujiwara K, Noguchi J, Kitada C, Fujino M, et al. Prolactin-releasing peptide as a novel stress mediator in the central nervous system. Endocrinology (2001) 142:2032–8. doi:10.1210/en.142.5.2032
60. Lin SH, Leslie FM, Civelli O. Neurochemical properties of the prolactin releasing peptide (PrRP) receptor expressing neurons: evidence for a role of PrRP as a regulator of stress and nociception. Brain Res (2002) 952:15–30. doi:10.1016/S0006-8993(02)03183-9
61. Dodd GT, Luckman SM. Physiological roles of GPR10 and PrRP signaling. Front Endocrinol (Lausanne) (2013) 4:20. doi:10.3389/fendo.2013.00020
62. Lawrence CB, Celsi F, Brennand J, Luckman SM. Alternative role for prolactin-releasing peptide in the regulation of food intake. Nat Neurosci (2000) 3:645–6. doi:10.1038/76597
63. Seal LJ, Small CJ, Dhillo WS, Stanley SA, Abbott CR, Ghatei MA, et al. PRL-releasing peptide inhibits food intake in male rats via the dorsomedial hypothalamic nucleus and not the paraventricular hypothalamic nucleus. Endocrinology (2001) 142:4236–43. doi:10.1210/endo.142.10.8419
64. Takayanagi Y, Matsumoto H, Nakata M, Mera T, Fukusumi S, Hinuma S, et al. Endogenous prolactin-releasing peptide regulates food intake in rodents. J Clin Invest (2008) 118:4014–24. doi:10.1172/JCI34682
65. Seal LJ, Small CJ, Kim MS, Stanley SA, Taheri S, Ghatei MA, et al. Prolactin releasing peptide (PrRP) stimulates luteinizing hormone (LH) and follicle stimulating hormone (FSH) via a hypothalamic mechanism in male rats. Endocrinology (2000) 141:1909–12. doi:10.1210/endo.141.5.7528
66. Fujii R, Fukusumi S, Hosoya M, Kawamata Y, Habata Y, Hinuma S, et al. Tissue distribution of prolactin-releasing peptide (PrRP) and its receptor. Regul Pept (1999) 83:1–10. doi:10.1016/S0167-0115(99)00028-2
67. Langmead CJ, Szekeres PG, Chambers JK, Ratcliffe SJ, Jones DN, Hirst WD, et al. Characterization of the binding of [(125)I]-human prolactin releasing peptide (PrRP) to GPR10, a novel G protein coupled receptor. Br J Pharmacol (2000) 131:683–8. doi:10.1038/sj.bjp.0703617
68. Conlon JM. The origin and evolution of peptide YY (PYY) and pancreatic polypeptide (PP). Peptides (2002) 23:269–78. doi:10.1016/S0196-9781(01)00608-8
69. Sundstrom G, Larsson TA, Brenner S, Venkatesh B, Larhammar D. Evolution of the neuropeptide Y family: new genes by chromosome duplications in early vertebrates and in teleost fishes. Gen Comp Endocrinol (2008) 155:705–16. doi:10.1016/j.ygcen.2007.08.016
70. Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE, et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst (1996) 88:1731–7. doi:10.1093/jnci/88.23.1731
71. Lee JH, Welch DR. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res (1997) 57:2384–7.
72. Kotani M, Detheux M, Vandenbogaerde A, Communi D, Vanderwinden JM, Le Poul E, et al. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem (2001) 276:34631–6. doi:10.1074/jbc.M104847200
73. Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature (2001) 411:613–7. doi:10.1038/35079135
74. Muir AI, Chamberlain L, Elshourbagy NA, Michalovich D, Moore DJ, Calamari A, et al. AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem (2001) 276:28969–75. doi:10.1074/jbc.M102743200
75. de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A (2003) 100:10972–6. doi:10.1073/pnas.1834399100
76. Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr, Shagoury JK, et al. The GPR54 gene as a regulator of puberty. N Engl J Med (2003) 349:1614–27. doi:10.1056/NEJMoa035322
77. Messager S, Chatzidaki EE, Ma D, Hendrick AG, Zahn D, Dixon J, et al. Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54. Proc Natl Acad Sci U S A (2005) 102:1761–6. doi:10.1073/pnas.0409330102
78. Roa J, Aguilar E, Dieguez C, Pinilla L, Tena-Sempere M. New frontiers in kisspeptin/GPR54 physiology as fundamental gatekeepers of reproductive function. Front Neuroendocrinol (2008) 29:48–69. doi:10.1016/j.yfrne.2007.07.002
79. Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev (2009) 30:713–43. doi:10.1210/er.2009-0005
80. Choe HK, Kim HD, Park SH, Lee HW, Park JY, Seong JY, et al. Synchronous activation of gonadotropin-releasing hormone gene transcription and secretion by pulsatile kisspeptin stimulation. Proc Natl Acad Sci U S A (2013) 110:5677–82. doi:10.1073/pnas.1213594110
81. Kitahashi T, Ogawa S, Parhar IS. Cloning and expression of kiss2 in the zebrafish and medaka. Endocrinology (2009) 150:821–31. doi:10.1210/en.2008-0940
82. Um HN, Han JM, Hwang JI, Hong SI, Vaudry H, Seong JY. Molecular coevolution of kisspeptins and their receptors from fish to mammals. Ann N Y Acad Sci (2010) 1200:67–74. doi:10.1111/j.1749-6632.2010.05508.x
83. Pasquier J, Lafont AG, Tostivint H, Vaudry H, Rousseau K, Dufour S. Comparative evolutionary histories of kisspeptins and kisspeptin receptors in vertebrates reveal both parallel and divergent features. Front Endocrinol (Lausanne) (2012) 3:173. doi:10.3389/fendo.2012.00173
84. Lee DK, Nguyen T, O’Neill GP, Cheng R, Liu Y, Howard AD, et al. Discovery of a receptor related to the galanin receptors. FEBS Lett (1999) 446:103–7. doi:10.1016/S0014-5793(99)00009-5
85. Leroy X. Neuropeptide q as Modulator of GPCR galr2 and Uses of Thereof. WO2012042455 A1 (2012). Available from: http://www.google.com/patents/WO2012042455A1?cl=en
86. Liu Y, Li S, Qi X, Zhou W, Liu X, Lin H, et al. A novel neuropeptide in suppressing luteinizing hormone release in goldfish, Carassius auratus. Mol Cell Endocrinol (2013) 374:65–72. doi:10.1016/j.mce.2013.04.008
87. Kim DK, Cho EB, Moon MJ, Park S, Hwang JI, Kah O, et al. Revisiting the evolution of gonadotropin-releasing hormones and their receptors in vertebrates: secrets hidden in genomes. Gen Comp Endocrinol (2011) 170:68–78. doi:10.1016/j.ygcen.2010.10.018
Keywords: kisspeptin, RF-amide, spexin, galanin, coevolution, gene duplication, evolutionary history
Citation: Yun S, Kim D-K, Furlong M, Hwang J-I, Vaudry H and Seong JY (2014) Does kisspeptin belong to the proposed RF-amide peptide family? Front. Endocrinol. 5:134. doi: 10.3389/fendo.2014.00134
Received: 23 May 2014; Paper pending published: 03 July 2014;
Accepted: 30 July 2014; Published online: 13 August 2014.
Edited by:
Sylvie Dufour, Muséum National d’Histoire Naturelle, FranceReviewed by:
Yoshitaka Oka, University of Tokyo, JapanGustavo M. Somoza, Instituto de Investigaciones Biotecnológicas-Instituto Tecnológico de Chascomús, Argentina
Copyright: © 2014 Yun, Kim, Furlong, Hwang, Vaudry and Seong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae Young Seong, Graduate School of Medicine, Korea University, Seoul 136-705, South Korea e-mail:anlzZW9uZ0Brb3JlYS5hYy5rcg==
 Seongsik Yun1
Seongsik Yun1 Jong-Ik Hwang
Jong-Ik Hwang Hubert Vaudry
Hubert Vaudry Jae Young Seong
Jae Young Seong