- 1 Department of Developmental Biology, Harvard School of Dental Medicine, Boston, MA, USA
- 2 Department of Oncology Biomarker Development, Genentech Inc., South San Francisco, CA, USA
- 3 Department of Microbiology, Harbin Medical University, Harbin, China
Despite early detection and reduced risk of death, prostate cancer still remains the second leading cause of cancer death in American men. There is currently no cure for advanced prostate cancer. The multistage, stochastic and highly heterogeneous nature of prostate cancer, coupled with genetic and epigenetic alterations that occur during disease progression and response to therapy, represent fundamental challenges in our quest to understand and control this complex and prevalent disease. Recent advances in drug development and breakthroughs in omics technologies have renewed our efforts to identify novel biomarkers for prostate cancer prognosis, prediction, and therapeutic response monitoring. In this perspective article, we overview the current status and highlight future prospects of biomarkers for prostate cancer, a disease that affects millions of men worldwide.
Introduction
The incidence of prostate cancer has increased dramatically in recent years, largely because of an aging population, the practice of prostate-specific antigen (PSA) testing and subsequent biopsy. In the United States, prostate cancer is the most commonly diagnosed cancer in men and second only to lung cancer in the number of cancer deaths, with a total of 241,740 new cases and 28,170 deaths from prostate cancer projected to occur in 2012 (Siegel et al., 2012). Worldwide, prostate cancer is the second most prevalent cancer diagnosis and the sixth leading cause of death from cancer in men, with a global incidence of 913,000 new cases and 258,000 deaths in 2008, according to the WHO GLOBOCAN database. Effective therapeutic options are available for early stage prostate cancer. However, despite the recent advent of anticancer agents, there is still no cure for the advanced stage of the disease. This article presents a multidisciplinary perspective on the role of biomarkers in prostate cancer drug development and patient care.
Types of Biomarkers
The National Cancer Institute defines a biomarker as “a biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process, or of a condition or disease.” Based on biological properties, biomarkers may also be specific cells, molecules, or genes, gene products, enzymes, or hormones etc (Biomarkers Definitions Working Group, 2001). For the purposes of developing drugs, three key types of biomarkers are used to guide clinical development – prognostic, predictive, and pharmacodynamic (PD) – each of which can influence decision making and aid in the rational development of anticancer agents. Prognostic biomarkers inform the natural course of the cancer in the absence of therapy or on standard chemotherapy regimens, distinguishing those patients with tumors that result in “good outcome” versus those with a “poor outcome.” They can be used to guide decisions of whom to treat, and how aggressively to treat. Predictive biomarkers differ in that they are used to identify (predict) those patients who are most likely to benefit from a particular treatment, enabling “personalized medicine.” PD biomarkers are used to assess the post-treatment effects of a drug on a tumor, confirm target or pathway modulation and mechanism of action, and can, at times, be used to guide or at least, increase confidence in the dosage that should be used in subsequent proof of concept and pivotal trials.
The Quest for Prostate Cancer Biomarkers
Active Surveillance Versus Chemoprevention
Prostate cancers display a variable range of clinical behaviors, from slow-growing tumors of no or little clinical significance, to aggressively metastatic and lethal diseases. In addition to improved cancer registries and transurethral prostatectomy, population-based PSA screening has been postulated to contribute to the increased incidence of prostate cancer, causing “over-diagnosis,” where a large number of detected prostate tumors are indolent. Aggressive treatment in patients with indolent diseases can result in side effects and reduce a patient’s quality of life for little to no benefit. One of the current clinical priorities is to develop prognostic biomarkers to identify those with indolent prostate diseases at low risk of progressing, so that these patients may better benefit from active surveillance or watchful waiting, thus avoiding unnecessary treatment and financial burden (Figure 1).
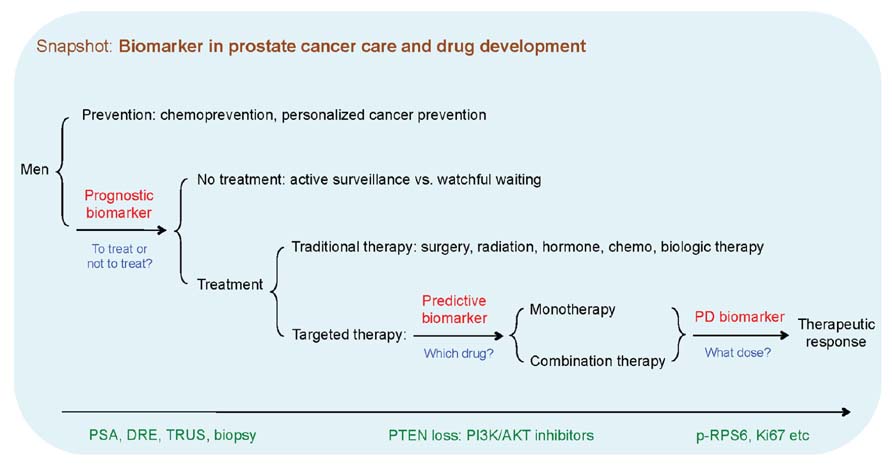
FIGURE 1. Types of biomarkers in prostate cancer prognosis, therapeutic response, and drug development. Biomarkers can be used to predict the natural course of a prostate disease (prognosis), help clinicians with decision making about the likelihood to respond to a given drug (predictive) and at what dose it might be most effective (pharmacodynamics). Red color denotes types of biomarker, blue color denotes the biomarker-based decision making, and green color indicates the exemplary actions or events. PSA, prostate-specific antigen; DRE, digital rectal examination; TRUS, transrectal ultrasound; PD biomarker, pharmacodynamic biomarker.
Our extensive knowledge of the etiology and progression of prostate cancer also makes it an ideal disease for cancer prevention. Androgen deprivation is the natural choice for prevention because of the established hormonally mediated pathogenesis for prostate diseases. However, chemoprevention with 5α-reductase inhibitors (5-ARI) has yielded encouraging yet ultimately confounding results in two landmark randomized, placebo-controlled prostate cancer prevention trials (Thompson et al., 2003; Andriole et al., 2010); it decreased the overall incidence of prostate cancer yet increasing the risk of developing high-grade prostate cancer in certain patients. The lingering uncertainty surrounding the risk of disease progression has thus far discouraged FDA approval (Walsh, 2010), while highlighting the necessity of identifying predictive biomarkers to discriminate between those who may or may not benefit from chemoprevention.
Cancer Treatment and Drug Resistance
Regardless of our current experience with PSA, early detection remains essential in treating clinically relevant prostate cancer. Localized prostate cancer, a tumor confined to the prostate gland, may be treated effectively with prostatectomy, radiation therapy, or other local treatments (Figure 1). For advanced prostate cancer that has recurred or spread beyond the prostate to lymph nodes, bones, or other sites, androgen deprivation therapy (ADT) remains the “gold standard” treatment, which yields considerable clinical benefits. However, similar to the development of acquired resistance in melanoma patients treated with Vemurafenib, most prostate cancer patients who initially respond to ADT subsequently become refractory. Amplification or mutations in androgen receptor (AR), AR-PI3K cross talk, reactivation of TMPRSS2:ERG fusion and the androgen-response pathway, and epithelial to mesenchymal transition (EMT) have been reported in castration-resistant prostate cancer (CRPC; Amler et al., 2000; Yap et al., 2011; Sun et al., 2012). Unlike KRAS mutations that are used to predict primary resistance to epidermal growth factor receptor (EGFR) targeted therapy in metastatic colorectal cancer, no biomarker has yet been identified to predict resistance to ADT in prostate cancer. Therefore, an improved understanding of molecular mechanisms of acquired resistance to ADT may provide valuable insights to enhance the efficacy of standard-of-care therapeutic intervention. The recent identification of AR splicing variants associated with acquired resistance to ADT in prostate cancer patients represents a landmark conceptual advancement in the prostate cancer field (Hu et al., 2010).
Very recently, there has been a rapid increase in the number of effective systemic agents for men with metastatic CRPC (mCRPC), including abiraterone acetate and MDV3100 (hormonal therapies), Sipuleucel-T (immunotherapy), Cabazitaxel (chemotherapy), Denosumab, and Radium 223 (bone microenvironment targeting agents; Armstrong et al., 2011). However, the high cost of these therapies, genetic heterogeneity of prostate cancer, and the increasing complexity of clinical decision making, have highlighted the need for effective biomarkers to identify the right therapy for the right patient and enable the monitoring of therapeutic response and disease progression.
Current Status of Prostate Cancer Biomarkers
Protein Biomarkers
Human prostatic acid phosphatase (PAP) was reportedly the first, though less sensitive, serum biomarker for prostate cancer (Gutman and Gutman, 1938). Recent advancements in proteomics, tissue microarray, and other biotechnologies have led to many protein biomarkers being reported, including, among others, chromogranin-A, interleukin-6, and prostate-specific cell antigen (Makarov et al., 2009; Madu and Lu, 2010). Here we will discuss, PSA, the current “gold standard” biomarker for prostate cancer.
Prostate-specific antigen, a 33 kDa serine protease produced by both normal and neoplastic prostate epithelial cells, belongs to the family of “Kallikrein genes.” The introduction of PSA has revolutionized prostate cancer screening, and PSA testing is currently a standard clinical application for staging and measuring disease progression and therapeutic response. Despite these advances, considerable debate still remains. The first and most important concern is the lack of cancer specificity as the rise in PSA level can reflect the presence of non-malignant disorders such as benign prostatic hyperplasia, infection, or chronic inflammation. Further, changes in the epidemiology of prostate cancer limit the correlation between PSA and the stage of prostate cancer where significant numbers of men with “normal” or very low PSA values may nonetheless harbor prostate cancer (Thompson et al., 2004). Moreover, the level of PSA expression appears to be inversely correlated with higher Gleason scores in certain studies (Aihara et al., 1994; Darson et al., 1997). As such, the United States Preventative Services Task Force has recently published draft recommendations against PSA-based prostate cancer screening. In the meantime, the concept of using PSA as a PD biomarker in therapeutic interventions is also changing, as evidenced by a lack of PSA response in CRPC patients receiving Sipuleucel-T, an immunomodulatory agent that prolongs life and is postulated to slow disease trajectory in part through effects on the prostate tumor microenvironment (Gulley and Drake, 2011). Taken together, serum PSA levels must be interpreted carefully and algorithms or nomograms that combine disease stage, grade, circulating tumor cells (CTCs), and PSA level for an individual patient rather than PSA alone should be used.
Circulating Tumor Cells
Circulating tumor cells, which are rare in healthy adults, originate from the primary tumor or metastatic deposits after invading and intravasating through the tumor vasculature. It has been shown that CTCs isolated from patients with mCRPC exhibit features of prostate cancer, expressing PSA and alpha-methylacyl-CoA racemase (AMACR), and exhibiting genomic abnormalities such as AR amplification, PTEN loss, and TMPRSS:ETV fusion (Attard et al., 2009). Studies have shown that the enumeration of CTCs from whole blood is prognostic for overall survival in many tumor types including mCRPC (de Bono et al., 2008; Danila et al., 2011). In addition to providing prognostic information and a potential indicator of efficacy, CTCs also have the potential to provide a real-time snapshot of the molecular makeup of an individual patient’s tumor, to profile for determinants that predict for sensitivity or resistance to therapy (Punnoose et al., 2010). Enumeration of CTCs – as measured by the CellSearch assay – has been cleared by the FDA for use as an aid to monitor men with mCRPC in conjunction with other clinical assessments of response/benefit. Efforts are ongoing to qualify the measurement of CTCs for determining patient response and drug efficacy in mCRPC.
Genomic Biomarkers
Genomic biomarkers measure DNA or RNA characteristics that are indicative of biologic processes, disease progression, and/or response to therapeutic interventions. At the DNA level, genetic and epigenetic alterations are common events in prostate cancer. Recent genomics studies have shown promise in being able to subtype prostate tumors for diagnostic purposes by using genomic classification. Taylor et al. (2010) used an integrative genomic analysis to identify copy number variation as a biomarker to predict prostate cancer outcome. Ding et al. (2011) used comparative oncogenomics to derive a four-gene signature and an additional pathway-representative fourteen-gene panel that proved superior as a prognostic biomarker compared to PSA (Ding et al., 2012). Similarly, a population-based study demonstrated the use of a five-SNP panel to predict aggressive prostate cancer (Lin et al., 2011). At the epigenetic level, methylated genes including GSTP1, DNMT3A2, and EZH2 are emerging as attractive biomarkers for prostate cancer (Henrique and Jeronimo, 2004; Kobayashi et al., 2011).
At the level of gene transcription, genomic biomarkers measure RNA characteristics including, but not limited to, protein-coding gene expression, RNA processing (splicing and editing), and non-coding RNAs including small (microRNAs, snoRNAs) and long RNAs (antisense RNA, dsRNA, and long RNA species). Recent attempts to stratify or cluster prostate cancers on the basis of RNA gene signature and/or histology have yielded disappointing results, despite its proven success in breast cancer (Hegde et al., 2007). More recently, Tomlins et al. (2011) demonstrated that urine TMPRSS2:ERG fusion transcript, in combination with urine PCA3, a prostate cancer-specific non-coding mRNA, enhances the utility of serum PSA for predicting prostate cancer risk and clinically relevant cancer on biopsy.
Among the non-coding RNAs, microRNAs are small (an average of 22 nucleotides), single-stranded, highly conserved RNA molecules that repress gene expression in a sequence-dependent manner. In prostate cancer, deregulated microRNA expression correlates with clinically aggressive or metastatic cancer phenotypes (Tavazoie et al., 2008; Bryant et al., 2012), where circulating microRNAs such as miR-141 distinguish patients with prostate cancer from healthy controls (Mitchell et al., 2008) and miR-21 predicts the resistance to docetaxel-based chemotherapy in patients with mCRPC (Zhang et al., 2011). Currently, microRNA gene signatures are being evaluated in prostate cancer clinical trails (e.g., NCT01050504). The fact that microRNAs remain stable and detectable in archival, especially formalin-fixed paraffin embedded (FFPE) tissues, and in body fluids has made them attractive candidates as cancer biomarkers.
mRNAs and microRNAs are also detected in exosomes, the 30–100 nm membrane vesicles that are secreted by multiple cell types and detectable in blood, urine, amniotic fluid, and malignant ascites (Valadi et al., 2007; Mitchell et al., 2008). Recent studies demonstrate that the circulating exosomal microRNA signatures parallel the developmental origins of tumor cells (Taylor and Gercel-Taylor, 2008), and increased levels of exosomes found in blood of patients with late-stage prostate, ovarian, and lung cancer suggests a role for these membrane vesicles in cancer diagnosis and biomarker analysis (Duijvesz et al., 2011).
Metabolomics and Imaging Biomarkers
Metabolomics has recently emerged as a novel approach to early and non-invasive prostate cancer detection based on changes in the metabolic signatures, including citrate, polyamines, lactate, choline, and creatine (Spratlin et al., 2009; Roberts et al., 2011), in addition to sarcosine and alanine (Tessem et al., 2008; Sreekumar et al., 2009).
The field of imaging in prostate cancer care is continuously evolving. FDG-PET imaging, which measures the extent of change in glucose utilization in many cancer types, appears to be associated with increasing Gleason grade, clinical stage, and serum PSA level (Jadvar, 2009). ImmunoPET imaging for antibody drug conjugates offers exciting potential diagnostic applications (Nakajima et al., 2011). The routine clinical use of transrectal ultrasound, magnetic resonance imaging (MRI), computed tomography (CT), radionuclide bone scanning, and positron emission tomography (PET) in prostate cancer has been described in excellent reviews (Hricak et al., 2007; Pinto et al., 2012).
However, the biomarkers mentioned above are based on relatively small cohorts of patients and the results merit further prospective validation.
Archival Tissues in Predictive Diagnostics
Clinical decision for prostate cancer often relies on the initial diagnostic tumor biopsy. However, such archival tissues may not be representative of the mutational landscape of the entire tumor bulk due to intratumor heterogeneity. They often do not carry the genomic alterations accumulated in the metastatic deposits, which may arise years later. Fresh biopsies would be preferable but obviously are difficult to obtain in mCRPC, given its particular propensity to metastasize to bone and induce blastic bone lesions. Surrogate biomarkers, such as CTCs, circulating tumor DNA, and microRNA, may serve as a non-invasive “liquid biopsy” to complement the use of archival tissues and PSA for predictive diagnostics.
Personalized Cancer Therapy
The landmark discoveries of Trastuzumab in breast cancer and Imatinib in chronic myeloid leukemia (CML) have opened the door to personalized medicine, treating patients individually by targeting the pathways that are present or activated in their tumors. In this regard, biomarker-based patient stratification is a key factor toward the development of targeted therapeutics for prostate cancer (Figure 1).
The PI3K-PTEN signaling pathway is a noteworthy example. PTEN loss of function has been observed in 40–70% of advanced prostate cancer, accompanied by frequent alterations in the pathway network such as INPP4B, PHLPP, and PIK3R1 (Taylor et al., 2010). The fact that PTEN-null prostate tumors respond to inhibition of PI3K has demonstrated the possible role of PTEN loss as a predictive biomarker in clinical trials testing PI3K pathway inhibitors (Jia et al., 2008, 2009; Lackner, 2010). Like other targeted therapies, resistance eventually occurs, leading to treatment failure. For instance, c-Myc elevation confers resistance to PI3K inhibitors (Ilic et al., 2011; Liu et al., 2011) and overrides the mTOR dependence of prostate lesions arising from constitutive AKT activation (Clegg et al., 2011). More importantly, allelic loss of PTEN and gain of c-Myc co-exist in 3% (4/126) human prostate cancer (Zafarana et al., 2012), suggesting innate resistance to PI3K inhibitors in this type of tumor. Thus, deciphering the genetic make-up of prostate tumors may facilitate patient stratification for PI3K targeted therapies. In deed, biomarker-driven drug development has been encouraged by the regulatory authorities as exemplified by the fast-track approval of Trastuzumab for the treatment of Her2-positive breast cancer.
When Prostate Meets Omics: The Next-Generation Biomarkers
The breakthroughs of omics-based technology development have revolutionized drug development and cancer care for prostate cancer. The enormous deluge of omics data generated over the past 5 years has easily surpassed the sum of wet bench data from the past several decades, providing unprecedented opportunities to re-visit the long-standing problem of biomarker development in prostate and other common human cancers. The use of proteomic technologies in micro-dissected clinical specimens would help to reveal the intricate tumor-stroma signaling networks that cross-talk in prostate cancer. Massively parallel sequencing technologies, in particular, the application of next-generation sequencing (NGS) of matched localized prostate tumors and mCRPC, coupled with genetic and epigenetic profiling, have vastly improved our capacity to more comprehensively uncover the multistage, stochastic and highly heterogeneous nature of prostate cancer. The recent identification of AR splicing variants in CRPC is an excellent example of NGS-driven prostate cancer biomarker discovery (Hu et al., 2010).
The considerable challenge that remains is the functional interpretation of the genetic and epigenetic changes derived from the vast amounts of omics data. The sophisticated computational algorithms and systems biology approach that are currently being explored will aid in the identification of the driving oncogenic events from passenger events, deriving novel biomarkers to detect and guide the treatment of prostate cancer.
Personalized Cancer Prevention
Despite the advent of new cancer therapies, the future of prostate cancer care will depend on early detection and effective cancer prevention. Targeted or personalized therapy is increasingly becoming part of clinical practice for many types of advanced cancers, however, this approach has not been adopted in the setting of prostate cancer prevention, where certain oncogenic events already occur at the very early stage of prostate cancer. The recent breakthroughs in decoding the “$1,000 genome” have renewed our efforts to identify novel biomarkers to sub-classify prostate cancer patients and prevent prostate cancer with a more targeted or “personalized” approach. Here we propose the concept of “biomarker-driven, personalized prevention of prostate cancer,” where biomarkers will guide the clinical decisions of whom to treat and which molecularly targeted intervention to be used (Figure 2).
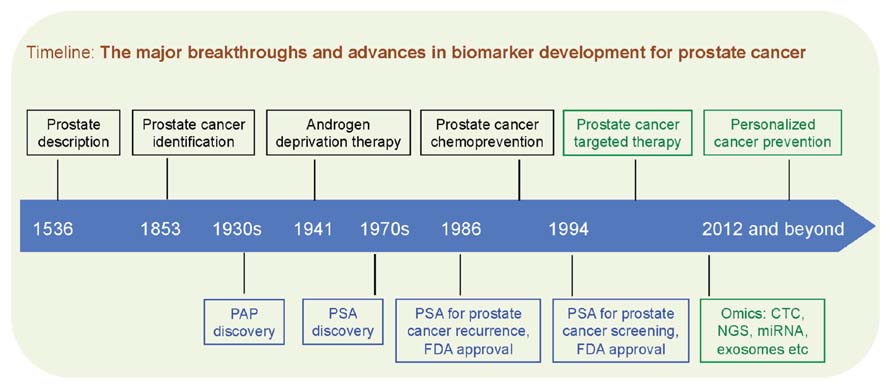
FIGURE 2. Black boxes denote conceptual breakthroughs, blue boxes denote biomarker advances, and green boxes denote therapeutic and technological outlook. PAP, prostatic acid phosphatase; PSA, prostate-specific antigen; FDA, US Food and Drug Administration; CTC, circulating tumor cells; NGS, next-generation sequencing; miRNA, microRNA.
Closing Remarks
Prostate cancer is poised to enter a new era of personalized health care, where biomarkers act as a central hub in cancer detection, treatment, and prevention. The next breakthroughs in therapeutics and biomarkers will only occur with an in-depth understanding of fundamental disease mechanisms, such as prostate cancer initiation and progression, response to therapy, and mechanisms of action of anticancer agents. Such “disease orientation” will require a greater collaboration between industry, academia, regulatory agencies, and patients. The integrative, interdisciplinary systems biology approach, armed with omics technologies, is offering promise in the quest of next-generation biomarkers. In the coming decades, the identification, qualification, and application of novel biomarkers will remain as the aspiration of patient care and the drug discovery paradigm for prostate cancer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Lukas Amler, Mark Lackner, Yibing Yan, and Zhenhai Gao for in-depth discussions and valuable inputs into the manuscript. We apologize to colleagues whose primary papers were not cited due to space constraints.
The views or opinions presented in this article are solely those of the authors and do not necessarily represent the official views of their employers.
No writing assistance was utilized in the production of this manuscript.
References
Aihara, M., Lebovitz, R. M., Wheeler, T. M., Kinner, B. M., Ohori, M., and Scardino, P. T. (1994). Prostate specific antigen and gleason grade: an immunohistochemical study of prostate cancer. J. Urol. 151, 1558–1564.
Amler, L. C., Agus, D. B., Leduc, C., Sapinoso, M. L., Fox, W. D., Kern, S., Lee, D., Wang, V., Leysens, M., Higgins, B., Martin, J., Gerald, W., Dracopoli, N., Cordon-Cardo, C., Scher, H. I., and Hampton, G. M. (2000). Dysregulated expression of androgen-responsive and nonresponsive genes in the androgen-independent prostate cancer xenograft model CWR22-R1. Cancer Res. 60, 6134–6141.
Andriole, G. L., Bostwick, D. G., Brawley, O. W., Gomella, L. G., Marberger, M., Montorsi, F., Pettaway, C. A., Tammela, T. L., Teloken, C., Tindall, D. J., Somerville, M. C., Wilson, T. H., Fowler, I. L., and Rittmaster, R. S. (2010). Effect of dutasteride on the risk of prostate cancer. N. Engl. J. Med. 362, 1192–1202.
Armstrong, A. J., Eisenberger, M. A., Halabi, S., Oudard, S., Nanus, D. M., Petrylak, D. P., Sartor, A. O., and Scher, H. I. (2011). Biomarkers in the management and treatment of men with metastatic castration-resistant prostate cancer. Eur. Urol. 61, 549–559.
Attard, G., Swennenhuis, J. F., Olmos, D., Reid, A. H., Vickers, E., A’hern, R., Levink, R., Coumans, F., Moreira, J., Riisnaes, R., Oommen, N. B., Hawche, G., Jameson, C., Thompson, E., Sipkema, R., Carden, C. P., Parker, C., Dearnaley, D., Kaye, S. B., Cooper, C. S., Molina, A., Cox, M. E., Terstappen, L. W., and de Bono, J. S. (2009). Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 69, 2912–2918.
Biomarkers Definitions Working Group. (2001). Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 69, 89–95.
Bryant, R. J., Pawlowski, T., Catto, J. W., Marsden, G., Vessella, R. L., Rhees, B., Kuslich, C., Visakorpi, T., and Hamdy, F. C. (2012). Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 106, 768–774.
Clegg, N. J., Couto, S. S., Wongvipat, J., Hieronymus, H., Carver, B. S., Taylor, B. S., Ellwood-Yen, K., Gerald, W. L., Sander, C., and Sawyers, C. L. (2011). MYC cooperates with AKT in prostate tumorigenesis and alters sensitivity to mTOR inhibitors. PLoS ONE 6, e17449. doi: 10.1371/journal.pone.0017449
Danila, D. C., Pantel, K., Fleisher, M., and Scher, H. I. (2011). Circulating tumors cells as biomarkers: progress toward biomarker qualification. Cancer J. 17, 438–450.
Darson, M. F., Pacelli, A., Roche, P., Rittenhouse, H. G., Wolfert, R. L., Young, C. Y., Klee, G. G., Tindall, D. J., and Bostwick, D. G. (1997). Human glandular kallikrein 2 (hK2) expression in prostatic intraepithelial neoplasia and adenocarcinoma: a novel prostate cancer marker. Urology 49, 857–862.
de Bono, J. S., Scher, H. I., Montgomery, R. B., Parker, C., Miller, M. C., Tissing, H., Doyle, G. V., Terstappen, L. W., Pienta, K. J., and Raghavan, D. (2008). Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 14, 6302–6309.
Ding, Z., Wu, C. J., Chu, G. C., Xiao, Y., Ho, D., Zhang, J., Perry, S. R., Labrot, E. S., Wu, X., Lis, R., Hoshida, Y., Hiller, D., Hu, B., Jiang, S., Zheng, H., Stegh, A. H., Scott, K. L., Signoretti, S., Bardeesy, N., Wang, Y. A., Hill, D. E., Golub, T. R., Stampfer, M. J., Wong, W. H., Loda, M., Mucci, L., Chin, L., and Depinho, R. A. (2011). SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470, 269–273.
Ding, Z., Wu, C. J., Jaskelioff, M., Ivanova, E., Kost-Alimova, M., Protopopov, A., Chu, G. C., Wang, G., Lu, X., Labrot, E. S., Hu, J., Wang, W., Xiao, Y., Zhang, H., Zhang, J., Gan, B., Perry, S. R., Jiang, S., Li, L., Horner, J. W., Wang, Y. A., Chin, L., and Depinho, R. A. (2012). Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 148, 896–907.
Duijvesz, D., Luider, T., Bangma, C. H., and Jenster, G. (2011). Exosomes as biomarker treasure chests for prostate cancer. Eur. Urol. 59, 823–831.
Gulley, J. L., and Drake, C. G. (2011). Immunotherapy for prostate cancer: recent advances, lessons learned, and areas for further research. Clin. Cancer Res. 17, 3884–3891.
Gutman, A. B., and Gutman, E. B. (1938). An “Acid” phosphatase occurring in the serum of patients with metastasizing carcinoma of the prostate gland. J. Clin. Invest. 17, 473–478.
Hegde, P. S., Rusnak, D., Bertiaux, M., Alligood, K., Strum, J., Gagnon, R., and Gilmer, T. M. (2007). Delineation of molecular mechanisms of sensitivity to lapatinib in breast cancer cell lines using global gene expression profiles. Mol. Cancer Ther. 6, 1629–1640.
Henrique, R., and Jeronimo, C. (2004). Molecular detection of prostate cancer: a role for GSTP1 hypermethylation. Eur. Urol. 46, 660–669; discussion 669.
Hricak, H., Choyke, P. L., Eberhardt, S. C., Leibel, S. A., and Scardino, P. T. (2007). Imaging prostate cancer: a multidisciplinary perspective. Radiology 243, 28–53.
Hu, R., Denmeade, S. R., and Luo, J. (2010). Molecular processes leading to aberrant androgen receptor signaling and castration resistance in prostate cancer. Expert Rev. Endocrinol. Metab. 5, 753–764.
Ilic, N., Utermark, T., Widlund, H. R., and Roberts, T. M. (2011). PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc. Natl. Acad. Sci. U.S.A. 108, E699–E708.
Jia, S., Liu, Z., Zhang, S., Liu, P., Zhang, L., Lee, S. H., Zhang, J., Signoretti, S., Loda, M., Roberts, T. M., and Zhao, J. J. (2008). Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature 454, 776–779.
Jia, S., Roberts, T. M., and Zhao, J. J. (2009). Should individual PI3 kinase isoforms be targeted in cancer? Curr. Opin. Cell Biol. 21, 199–208.
Kobayashi, Y., Absher, D. M., Gulzar, Z. G., Young, S. R., Mckenney, J. K., Peehl, D. M., Brooks, J. D., Myers, R. M., and Sherlock, G. (2011). DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 21, 1017–1027.
Lackner, M. R. (2010). Prospects for personalized medicine with inhibitors targeting the RAS and PI3K pathways. Expert Rev. Mol. Diagn. 10, 75–87.
Lin, D. W., Fitzgerald, L. M., Fu, R., Kwon, E. M., Zheng, S. L., Kolb, S., Wiklund, F., Stattin, P., Isaacs, W. B., Xu, J., Ostrander, E. A., Feng, Z., Gronberg, H., and Stanford, J. L. (2011). Genetic variants in the LEPR, CRY1, RNASEL, IL4, and ARVCF genes are prognostic markers of prostate cancer-specific mortality. Cancer Epidemiol. Biomarkers Prev. 20, 1928–1936.
Liu, P., Cheng, H., Santiago, S., Raeder, M., Zhang, F., Isabella, A., Yang, J., Semaan, D. J., Chen, C., Fox, E. A., Gray, N. S., Monahan, J., Schlegel, R., Beroukhim, R., Mills, G. B., and Zhao, J. J. (2011). Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat. Med. 17, 1116–1120.
Madu, C. O., and Lu, Y. (2010). Novel diagnostic biomarkers for prostate cancer. J. Cancer 1, 150–177.
Makarov, D. V., Loeb, S., Getzenberg, R. H., and Partin, A. W. (2009). Biomarkers for prostate cancer. Annu. Rev. Med. 60, 139–151.
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L., Peterson, A., Noteboom, J., O’briant, K. C., Allen, A., Lin, D. W., Urban, N., Drescher, C. W., Knudsen, B. S., Stirewalt, D. L., Gentleman, R., Vessella, R. L., Nelson, P. S., Martin, D. B., and Tewari, M. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518.
Nakajima, T., Mitsunaga, M., Bander, N. H., Heston, W. D., Choyke, P. L., and Kobayashi, H. (2011). Targeted, activatable, in vivo fluorescence imaging of prostate-specific membrane antigen (PSMA) positive tumors using the quenched humanized J591 antibody-indocyanine green (ICG) conjugate. Bioconjug. Chem. 22, 1700–1705.
Pinto, F., Totaro, A., Palermo, G., Calarco, A., Sacco, E., D’addessi, A., Racioppi, M., Valentini, A., Gui, B., and Bassi, P. (2012). Imaging in prostate cancer staging: present role and future perspectives. Urol. Int. 88, 125–136.
Punnoose, E. A., Atwal, S. K., Spoerke, J. M., Savage, H., Pandita, A., Yeh, R. F., Pirzkall, A., Fine, B. M., Amler, L. C., Chen, D. S., and Lackner, M. R. (2010). Molecular biomarker analyses using circulating tumor cells. PLoS ONE 5, e12517. doi: 10.1371/journal.pone.0012517
Roberts, M. J., Schirra, H. J., Lavin, M. F., and Gardiner, R. A. (2011). Metabolomics: a novel approach to early and noninvasive prostate cancer detection. Korean J. Urol. 52, 79–89.
Siegel, R., Naishadham, D., and Jemal, A. (2012). Cancer statistics, 2012. CA Cancer J. Clin. 62, 10–29.
Spratlin, J. L., Serkova, N. J., and Eckhardt, S. G. (2009). Clinical applications of metabolomics in oncology: a review. Clin. Cancer Res. 15, 431–440.
Sreekumar, A., Poisson, L. M., Rajendiran, T. M., Khan, A. P., Cao, Q., Yu, J., Laxman, B., Mehra, R., Lonigro, R. J., Li, Y., Nyati, M. K., Ahsan, A., Kalyana-Sundaram, S., Han, B., Cao, X., Byun, J., Omenn, G. S., Ghosh, D., Pennathur, S., Alexander, D. C., Berger, A., Shuster, J. R., Wei, J. T., Varambally, S., Beecher, C., and Chinnaiyan, A. M. (2009). Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914.
Sun, Y., Wang, B. E., Leong, K. G., Yue, P., Li, L., Jhunjhunwala, S., Chen, D., Seo, K., Modrusan, Z., Gao, W. Q., Settleman, J., and Johnson, L. (2012). Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 72, 527–536.
Tavazoie, S. F., Alarcon, C., Oskarsson, T., Padua, D., Wang, Q., Bos, P. D., Gerald, W. L., and Massague, J. (2008). Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152.
Taylor, B. S., Schultz, N., Hieronymus, H., Gopalan, A., Xiao, Y., Carver, B. S., Arora, V. K., Kaushik, P., Cerami, E., Reva, B., Antipin, Y., Mitsiades, N., Landers, T., Dolgalev, I., Major, J. E., Wilson, M., Socci, N. D., Lash, A. E., Heguy, A., Eastham, J. A., Scher, H. I., Reuter, V. E., Scardino, P. T., Sander, C., Sawyers, C. L., and Gerald, W. L. (2010). Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22.
Taylor, D. D., and Gercel-Taylor, C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21.
Tessem, M. B., Swanson, M. G., Keshari, K. R., Albers, M. J., Joun, D., Tabatabai, Z. L., Simko, J. P., Shinohara, K., Nelson, S. J., Vigneron, D. B., Gribbestad, I. S., and Kurhanewicz, J. (2008). Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using 1H HR-MAS spectroscopy of biopsy tissues. Magn. Reson. Med. 60, 510–516.
Thompson, I. M., Goodman, P. J., Tangen, C. M., Lucia, M. S., Miller, G. J., Ford, L. G., Lieber, M. M., Cespedes, R. D., Atkins, J. N., Lippman, S. M., Carlin, S. M., Ryan, A., Szczepanek, C. M., Crowley, J. J., and Coltman, C. A. Jr. (2003). The influence of finasteride on the development of prostate cancer. N. Engl. J. Med. 349, 215–224.
Thompson, I. M., Pauler, D. K., Goodman, P. J., Tangen, C. M., Lucia, M. S., Parnes, H. L., Minasian, L. M., Ford, L. G., Lippman, S. M., Crawford, E. D., Crowley, J. J., and Coltman, C. A. Jr. (2004). Prevalence of prostate cancer among men with a prostate-specific antigen level <4.0 ng per milliliter. N. Engl. J. Med. 350, 2239–2246.
Tomlins, S. A., Aubin, S. M., Siddiqui, J., Lonigro, R. J., Sefton-Miller, L., Miick, S., Williamsen, S., Hodge, P., Meinke, J., Blase, A., Penabella, Y., Day, J. R., Varambally, R., Han, B., Wood, D., Wang, L., Sanda, M. G., Rubin, M. A., Rhodes, D. R., Hollenbeck, B., Sakamoto, K., Silberstein, J. L., Fradet, Y., Amberson, J. B., Meyers, S., Palanisamy, N., Rittenhouse, H., Wei, J. T., Groskopf, J., and Chinnaiyan, A. M. (2011). Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci. Transl. Med. 3, 94ra72.
Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J. J., and Lotvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659.
Yap, T. A., Zivi, A., Omlin, A., and de Bono, J. S. (2011). The changing therapeutic landscape of castration-resistant prostate cancer. Nat. Rev. Clin. Oncol. 8, 597–610.
Zafarana, G., Ishkanian, A. S., Malloff, C. A., Locke, J. A., Sykes, J., Thoms, J., Lam, W. L., Squire, J. A., Yoshimoto, M., Ramnarine, V. R., Meng, A., Ahmed, O., Jurisca, I., Milosevic, M., Pintilie, M., Van Der Kwast, T., and Bristow, R. G. (2012). Copy number alterations of c-MYC and PTEN are prognostic factors for relapse after prostate cancer radiotherapy. Cancer. doi: 10.1002/cncr.26729 [Epub ahead of print].
Zhang, H. L., Yang, L. F., Zhu, Y., Yao, X. D., Zhang, S. L., Dai, B., Zhu, Y. P., Shen, Y. J., Shi, G. H., and Ye, D. W. (2011). Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential pre- dictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 71, 326–331.
Keywords: biomarker, prostate cancer, personalized medicine
Citation: Liu Y, Hegde P, Zhang F, Hampton G and Jia S (2012) Prostate cancer – a biomarker perspective. Front. Endocrin. 3:72. doi: 10.3389/fendo.2012.00072
Received: 11 March 2012; Paper pending published: 10 April 2012;
Accepted: 08 May 2012; Published online: 29 May 2012.
Edited by:
Carmen Priolo, Brigham and Women’s Hospital, Harvard Medical School, USAReviewed by:
Alessandra Mosca, University Hospital “Maggiore della Carità”, ItalyPietro Ameri, Università degli Studi di Genova, Italy
Copyright: © 2012 Liu, Hegde, Zhang, Hampton and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Shidong Jia, Department of Oncology Biomarker Development, Genentech Inc., 1 DNA Way, South San Francisco, CA 94080, USA. e-mail:amlhLnNoaWRvbmdAZ2VuZS5jb20=
 Yanqiu Liu1
Yanqiu Liu1