- 1 Cancer Research UK, Li Ka Shing Centre, Robinson Way, Cambridge, UK
- 2 Department of Oncology, University of Cambridge, Cambridge, UK
Hormonally regulated breast and prostate cancers are the most common cause of cancer in females and males respectively. FoxA1 acts as a pioneer factor for both androgen receptor (AR) and estrogen receptor-α (ER), dictating the binding location, and therefore function of these transcription factors. It is an essential protein for the transcriptional activity of both ER and AR, yet it has distinct roles with the two different nuclear receptors. In both malignancies, FoxA1 plays a pivotal role from early stage cancer through to drug resistant and metastatic disease. Due to this key role in mediating ER and AR function, FoxA1 is not only an attractive therapeutic target but could potentially function as a novel biomarker.
Breast and prostate cancer are hormonally fueled cancers driven by the sex steroid hormones estrogen and androgen, that activate their respective nuclear receptors estrogen receptor (ER) or androgen receptor (AR). These nuclear receptors do not function alone but with a plethora of other transcription factors and co-factors, which aid ER and AR to elicit a pro-tumorigenic gene expression program. FoxA1 is an interacting partner of both AR and ER where it plays a crucial role in the development and progression of breast and prostate cancer.
FoxA1 is one of three members of the FoxA family, a subset of the forkhead family of transcription factors which play vital roles in development (reviewed in Hannenhalli and Kaestner, 2009). Initially FoxA1 was discovered for its role in liver development (Costa et al., 1989) but has also been implicated in the development of a number of other organs (Bernardo and Keri, 2012) including the androgen and estrogen regulated tissues of the breast and prostate. From observations in the FoxA1−/− mice, normal mammary duct or prostate rudiment develops during embryogenesis but pubertal ductal morphogenesis does not occur in either organ (Gao et al., 2005; Bernardo et al., 2010). As is often the case, cancer mirrors development and FoxA1 plays a pivotal part in the development of breast and prostate cancer. This perspective aims to highlight the recent discoveries in the interplay of FoxA1 with AR and ER in cancer and the potential clinical implications of these interactions.
FoxA1 Facilitates Chromatin Accessibility for ER and AR at Cell Type Specific Locations
The first evidence of an interaction between ER and FoxA1 was shown at the estrogen regulated Vitellogenin B1 gene promoter after microinjection of the two proteins into Xenopus laevis oocytes (Robyr et al., 2000). However it was not until the advent of high-throughput technologies, that the elucidation of a wide spread association of FoxA1 with hormone nuclear receptors occurred. The first large scale assessment of ER–chromatin interactions involved chromatin immunoprecipitation (ChIP) enrichment of DNA bound by ER, coupled to microarrays containing all the non-repetitive regions of chromosome 21 and 22 (Carroll et al., 2005). From this initial experiment two key finding were made (a) ER mainly binds significant distances from the transcriptional start sites of the coding genes it regulates (regions termed distal enhancers) and (b) motif analysis of the DNA under the ER binding sites revealed EREs (estrogen responsive elements) and motifs for forkhead factors. With improvements in technology, from genome wide microarrays through to ChIP coupled to high-throughput sequencing (ChIP-seq), these two facts have held up in numerous cell line studies (Laganiere et al., 2005; Carroll et al., 2006; Ross-Innes et al., 2010; Hurtado et al., 2011), demonstrating the importance of the initial observation (Carroll et al., 2005). The first comprehensive ER ChIP-seq in primary breast cancer tissue has just been completed and forkhead motifs were again enriched in the ER binding events, particularly in binding events that occurred in breast cancer patients with a poor clinical outcome (Ross-Innes et al., 2012). An interaction between AR and FoxA1 was first shown at the promoter of the androgen regulated genes probasin and PSA (Gao et al., 2003). ChIP-chip and ChIP-seq of AR in LNCaP (Wang et al., 2007, 2009; Robinson et al., 2011; Tan et al., 2012) and C4-2B (Jia et al., 2008) cell lines confirmed the presence of forkhead motifs in AR binding events. The link between AR and FoxA1 has been reinforced by FoxA1 ChIP-seq experiments, which have shown that ∼50% ER binding sites and ∼70% AR binding events occur at regions also co-occupied by FoxA1 (Hurtado et al., 2011; Robinson et al., 2011; Sahu et al., 2011).
FoxA1 functions as a “pioneer factor” for transcription factors such as AR and ER, engaging with condensed chromatin and facilitating the association of other factors (recently reviewed in Zaret and Carroll, 2011). FoxA1 and the other FoxA family members have a so-called “winged helix” structure, which makes them perfectly adapted to this role. They perch in the major groove of DNA through an alpha helix domain with two loops, like wings, which make additional site specific contacts with the DNA (Clark et al., 1993) in a structure similar to that of the linker Histone H1. When FoxA1 is silenced in the ER+ breast cancer cell line MCF7, there is a significant loss in global chromatin accessibility coupled with a concurrent loss of ER binding at more than 90% of all ER binding events (Hurtado et al., 2011). This result emphasizes how essential FoxA1 is for ER to function in breast cancer. As will be discussed later, the interaction between AR and FoxA1 is more complicated and does not result in a simple loss of AR binding after silencing of FoxA1.
Androgen receptor and ER only bind to a very small subset of locations in the genome where their canonical motifs (ARE and EREs) occur, indicating that other factors must constrain their positioning. FoxA1 appears to provide cell type specificity for ER and AR binding and therefore, FoxA1 ultimately dictates the estrogen and androgen gene targets. Cell type specific FoxA1 sites in breast and prostate cancer cell lines often overlap with the driving nuclear receptor and are enriched proximal to genes that are expressed in that cancer type (Lupien et al., 2008; Eeckhoute et al., 2009). In fact, it is possible to predict a functional ER binding site by the presence of FoxA1 binding in conjunction with an ERE motif and the histone modification H3K4me1 (Joseph et al., 2010) which underscores the importance of FoxA1. That said there are many FoxA1 sites that do not recruit a nuclear receptor and equally a number of AR and ER sites where there is no FoxA1 binding. As such, although FoxA1 is a required parameter for ER binding, alone it is not sufficient and additional collaborating factors help contribute to functional enhancer elements.
Approximately 50% of FoxA1 sites are shared between different cell lines (Hurtado et al., 2011; Robinson et al., 2011); a specific binding pattern that appears to be influenced by the histone landscape. FoxA1 binds preferentially to the active histone marks H3K4me1 and H3K4me2 and less at regions possessing the H3K9me2 repressive mark (Lupien et al., 2008). The part that FoxA1 plays in the reading, maintenance, or creation of these chromatin modifications is still undetermined. However there is evidence to suggest that FoxA1 may be actively involved in the maintenance of these marks, since forced FoxA1 expression in FoxA1 negative MDA-MB-231 breast cancer cells results in binding at regions that subsequently acquire H3K4me2 marks (Sérandour et al., 2011). FoxA1 binding may also be influenced by DNA methylation, since it is enriched at regions of low methylation and FoxA1 binding occurs before differentiation induced demethylation in mouse pluripotent P19 cells (Sérandour et al., 2011).
FoxA1 has the Ability to Reprogram the Binding of AR and ER
As FoxA1 demarks the chromatin locations where AR binds and its removal results in loss of ER binding, one would presume that AR binding in the prostate would be lost in the absence of FoxA1. However, two groups showed that loss of FoxA1 results in redistribution of AR and initiation of a new transcriptional program (Sahu et al., 2011; Wang et al., 2011). Both studies used siRNA to deplete FoxA1 in LNCaP prostate cancer cells, then conducted AR ChIP-seq and compared the results to the parental cells. The results are markedly similar and show that loss of FoxA1 results in loss of ∼50% AR binding events (FoxA1 dependent), with the remaining 50% of AR sites still present, independently of FoxA1. Moreover, in the absence of FoxA1, there is a pronounced gain in novel AR binding events, resulting in threefold more AR binding in the absence of FoxA1. This unexpected finding was validated in another prostate cancer cell line, VCaP, and for another nuclear receptor, GR (Sahu et al., 2011). Gene expression changes and the production of eRNAs (small RNAs produced at active enhancers) echoed the same changes seen in genomic binding events and the new/lost binding events were enriched near these differentially expressed regions. It appears that FoxA1 normally represses AR binding at some locations, but the lack of FoxA1 binding at the regions where AR is reprogrammed suggests an indirect trans-repression mechanism (Wang et al., 2011).
The loss of FoxA1 in LNCaPs was accompanied by little change in the histone marks, H3K4me1 (Wang et al., 2011), H3K4me2 (Sahu et al., 2011), or H3K27Ac (Wang et al., 2011). In fact, 50% of the AR gained events did not have DNase I hypersensitivity (a marker for open chromatin; Sahu et al., 2011) and H2A.Z ChIP-seq suggested that there was no nucleosome remodeling at these new AR events (Wang et al., 2011). This leads to the question of how AR is able to bind to these regions of apparently less accessible chromatin yet still manage to elicit a change in gene expression. It hints toward a non-classical method of AR binding and activity. One explanation for this phenomenon is that FoxA1 may be required to maintain the structure of the chromatin in a certain manner, forcing AR to bind to specific sites and simultaneously preventing binding to numerous other regions, a hypothesis supported by the gain of the looping protein Med12 binding at the new sites (Wang et al., 2011). Alternatively another protein could be acting in place of FoxA1 to maintain the mark and recruit AR to its reprogrammed locations.
In contrast to AR in prostate cancer cells, it is the over-expression of FoxA1 that induces redistribution of ER. By forcing FoxA1 expression in U2OS-ER osteosarcoma cells, the ER binding profile changes from bone cancer-specific sites to locations normally observed in breast cancer (Hurtado et al., 2011). Similarly, the ectopic expression of ER, FoxA1, and an additional interacting transcription factor GATA3, can induce hormone responsive growth in the ER− breast cancer cell lines MDA-MB-231 and BT-549 (Kong et al., 2011). In this scenario, each factor had no effect in isolation, illustrating a clear co-operative interaction, which must occur for effective ER transcriptional response. This reprogramming ability highlights the complexity of FoxA1’s function as an initiation factor for ER yet it harbors some repressive properties in its interactions with AR as illustrated in Figure 1.
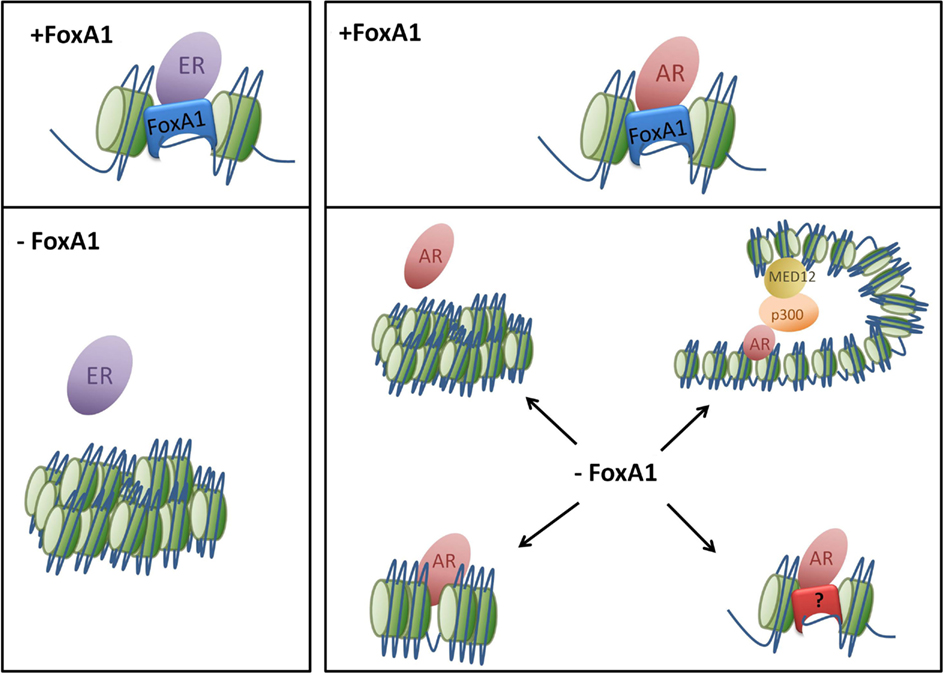
Figure 1. Loss of FoxA1 has disparate effects on nuclear receptor binding. FoxA1 acts as a pioneer factor for ER and AR allowing the nuclear receptors to make targeted contacts with specific genomic regions which induce a transcriptional response. The loss of FoxA1 results in condensation of the chromatin so ER and AR can no longer bind to DNA at specific locations. However AR still appears to be able to bind in an unorthodox manner, without nucleosome redistribution or active histone marks. This may be due to changes in chromatin architecture and the development of new loops or through an alternate transcription factor.
FoxA1 has a Vital Role in Molecular Apocrine (ER−AR+) Breast Cancer
Estrogen receptor negative (ER−) breast cancers constitute approximately one-third of all cases and were historically thought to be hormonally unresponsive. However a number of studies have identified a subset of ER− breast cancers which are driven by AR. These tumors have a more luminal like expression profile, typical to ER+ tumors, and all express AR which seems to act as a proxy for the lack of ER (Farmer et al., 2005; Doane et al., 2006; Teschendorff et al., 2007). This ER− breast cancer subtype has been validated in additional studies, each group coining their own name but for this review we will refer to them as molecular apocrine. The ER−AR+ breast cancer cell line MDA-MB-453 has a comparable gene expression profile to that of molecular apocrine tumors and is often used as a representative model. AR has been shown to be necessary for growth of MDA-MB-453 cells in vitro (Doane et al., 2006), in three-dimensional culture (Robinson et al., 2011), and in mouse xenografts (Ni et al., 2011). To understand the function of AR, the Brown lab and our lab conducted AR ChIP-seq in MDA-MB-453 cells (Ni et al., 2011; Robinson et al., 2011). Due to differences in experimental design, methodology, and sequencing platform, there was a 10-fold difference in the absolute number of AR binding events between the two datasets, but despite this discrepancy, motif analysis in both cases showed a strong association with forkhead motifs at the summit of the AR peaks.
FoxA1 is highly expressed in molecular apocrine tumors (Doane et al., 2006; Ni et al., 2011) and a comparison of genome wide FoxA1 occupancy in our study found an unprecedented level of concordance between AR and FoxA1 binding: 98% overlap (Robinson et al., 2011). Even though Ni et al. saw a lower level of overlap between AR and FoxA1 binding (37%), these co-bound sites were much more likely to be near testosterone regulated genes (p < 5.3 × 10−20) than AR unique sites (p < 0.011) implying that these are the more functional AR sites. We found that AR’s genomic locations were more similar to ER binding events in MCF7 cells than AR binding in the prostate cancer cell line LNCaP (Robinson et al., 2011). FoxA1 binding was shown to have a cell type specific pattern in MDA-MB-453 cells and therefore could be directing AR to these regions normally occupied by ER. Additionally, FoxA1 was required for expression of the growth promoting gene expression profile commonly seen in molecular apocrine breast cancer and growth of the cells. Since AR has a growth inhibitory function in ER+ breast cancer cells, but a growth promoting effect in ER− molecular apocrine breast cancer cells (Birrell et al., 1995), it would be of interest to map binding of AR in a ER+AR+ context to assess how the presence of ER influences AR binding on a global scale and how FoxA1 is involved in this cross-talk.
FoxA1 has Opposing Predictive Power in Breast and Prostate Cancer
FoxA1 has been repeatedly found to be a marker for good prognosis in ER+ breast cancer and it is an independent marker for recurrence free survival and overall survival (Badve et al., 2007). Initial studies suggest that it may have better prognostic significance than progesterone receptor, a classic marker for breast cancer outcome (Badve et al., 2007) or the proliferation marker Ki67 (Hisamatsu et al., 2011). Breast cancer is a very heterogeneous disease and FoxA1 may be useful as a biomarker for the stratification of patients with ER+ HER2− disease, a subset which currently suffers from over-treatment. Tumors within this cohort that have high FoxA1 levels respond equally well with or without chemotherapy (Hisamatsu et al., 2011) and a study with a large cohort of 4444 patients treated with tamoxifen showed that FoxA1 was predictive of response and therefore has the potential to identify a subgroup who could be treated less aggressively (Mehta et al., 2012).
In prostate cancer however, it appears that the opposite is true and that high FoxA1 is a predictor of poor prognosis. Two large scale tissue microarray (TMA) studies have been carried out in the last year assessing FoxA1 expression across 288 and 350 cases respectively (Sahu et al., 2011; Gerhardt et al., 2012). Sahu et al. (2011) noted a strong correlation of FoxA1 staining with AR levels (r = 0.72, p < 0.0001) and high FoxA1 staining was a marker of shorter time to prostate cancer-specific death (hazard ratio = 2.89, p = 0.04). These discoveries were reproduced by a separate study (Gerhardt et al., 2012), which also found that patients with high FoxA1 levels have shorter time to biochemical recurrence, a common indicator of relapse, however, FoxA1 was not an independent prognostic marker. Interestingly high FoxA1 staining was observed in patients who have become resistant to hormonal therapy (castrate resistant prostate cancer – CRPC) and FoxA1 positivity was seen in 32 out of 39 lymph node and distant metastases. An independent study also found FoxA1 to be highly expressed in 90% of prostate cancer metastases (Jain et al., 2011), an observation that has also been seen in breast cancer metastases (Ross-Innes et al., 2012).
Clinical observations again highlight that FoxA1 interacts differently with ER and AR in primary cancer and are summarized in our schematics in Figure 2. With regard to ER in breast cancer, FoxA1 is a marker for good outcome, possibly because it enables a functional ER complex, resulting in increased sensitivity to ER antagonists, such as tamoxifen. Concordantly, it has been shown that FoxA1 is essential for tamoxifen to functional (Hurtado et al., 2011). However, in the case of prostate cancer, higher levels of FoxA1 are a marker for poor outcome, which may in part be due to the fact that FoxA1 correlates with AR levels. Increased AR is a poor prognosis marker in prostate cancer (Gerhardt et al., 2012) and amplification of the AR gene is one of the many ways that prostate cancer circumvents hormonal therapies (Brown et al., 2002; Edwards et al., 2003; Ford III et al., 2003). Since high FoxA1 is observed in both breast and prostate metastases, we hypothesize that there is a shared dependence on this transcription factor at secondary sites of breast and prostate metastasis. Furthermore, the increase of FoxA1 in CRPC patients (Gerhardt et al., 2012) is particularly interesting, since it may hint toward a novel mechanism for the reactivation of AR in these hormone deprived tumors. An alternate hypothesis, independent of AR, has been postulated in a LNCaP model of CRPC, where FoxA1 can drive G1–S phase transition through recruitment of MYBL2 or CREB1 to the Cyclin E2 and E2F1 genes (Zhang et al., 2011).

Figure 2. The clinical paradox of FoxA1 in relation to breast and prostate cancer. FoxA1 is a good prognostic marker in ER+ breast cancer yet in prostate cancer is a marker of poor outcome as indicated by the hypothetical survival curves. This may be due to differences in the hormonally responsive pathways activated by FoxA1. However for end-stage disease for both breast and prostate cancer, there is increased expression of FoxA1 suggesting dependence on the nuclear receptor complex.
FoxA1 is a Key Target in Breast and Prostate Cancer
Given the fact that FoxA1 is required for the growth of ER+ breast cancer cell lines (Laganiere et al., 2005), ER−AR+ breast cancer cell lines (Robinson et al., 2011), and prostate cancer cell lines (Zhang et al., 2011), FoxA1 constitutes an attractive therapeutic target in a range of different hormone dependent cancers. In particular, it may be a good second line therapy as FoxA1 is required for growth of drug resistant breast and prostate cancer cell lines (Hurtado et al., 2011; Zhang et al., 2011). Transcription factors are notoriously difficult to target, in part due to their ubiquitous nature, a problem which is partially relieved by the observation that FoxA1 is only expressed in a few adult tissues. Furthermore, the binding and transcriptional activity of another forkhead family member, FOXM1, has been inhibited by the natural product thiostrepton (Hegde et al., 2011) which raises the possibility of finding compounds that selectively target FoxA1.
Conclusion
FoxA1 is a critical mediator of the function of both AR and ER in the context of breast and prostate cancer, from initial tumorigenesis through to hormone resistant, metastatic disease. Although FoxA1 appears to be a critical determinant of ER and AR binding and function, it does not work alone, but in concert with a host of other AR and ER interacting partners, that are required to make a fully functional transcriptional complex capable of responding to hormonal activation. That said, FoxA1 is a factor that appears to play a critical and central role in mediating ER and AR interactions with chromatin. In the future, further exploration into the potential of FoxA1 in the clinic as either a drug target or a new biomarker may prove to be clinically beneficial.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Professor David Neal for critical reading. We would like to acknowledge the support of University of Cambridge and Cancer Research UK.
References
Badve, S., Turbin, D., Thorat, M. A., Morimiya, A., Nielsen, T. O., Perou, C. M., Dunn, S., Huntsman, D. G., and Nakshatri, H. (2007). FOXA1 expression in breast cancer – correlation with luminal subtype A and survival. Clin. Cancer Res. 13, 4415–4421.
Bernardo, G. M., and Keri, R. A. (2012). FOXA1: a transcription factor with parallel functions in development and cancer. Biosci. Rep. 32, 113–130.
Bernardo, G. M., Lozada, K. L., Miedler, J. D., Harburg, G., Hewitt, S. C., Mosley, J. D., Godwin, A. K., Korach, K. S., Visvader, J. E., Kaestner, K. H., Abdul-Karim, F. W., Montano, M. M., and Keri, R. A. (2010). FOXA1 is an essential determinant of ERα expression and mammary ductal morphogenesis. Development 137, 2045–2054.
Birrell, S. N., Bentel, J. M., Hickey, T. E., Ricciardelli, C., Weger, M. A., Horsfall, D. J., and Tilley, W. D. (1995). Androgens induce divergent proliferative responses in human breast cancer cell lines. J. Steroid Biochem. Mol. Biol. 52, 459–467.
Brown, R. S. D., Edwards, J., Dogan, A., Payne, H., Harland, S. J., Bartlett, J. M. S., and Masters, J. R. W. (2002). Amplification of the androgen receptor gene in bone metastases from hormone-refractory prostate cancer. J. Pathol. 198, 237–244.
Carroll, J. S., Liu, X. S., Brodsky, A. S., Li, W., Meyer, C. A., Szary, A. J., Eeckhoute, J., Shao, W., Hestermann, E. V., Geistlinger, T. R., Fox, E. A., Silver, P. A., and Brown, M. (2005). Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43.
Carroll, J. S., Meyer, C. A., Song, J., Li, W., Geistlinger, T. R., Eeckhoute, J., Brodsky, A. S., Keeton, E. K., Fertuck, K. C., Hall, G. F., Wang, Q., Bekiranov, S., Sementchenko, V., Fox, E. A., Silver, P. A., Gingeras, T. R., Liu, X. S., and Brown, M. (2006). Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38, 1289–1297.
Clark, K. L., Halay, E. D., Lai, E., and Burley, S. K. (1993). Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420.
Costa, R. H., Grayson, D. R., and Darnell, J. E. (1989). Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol. Cell. Biol. 9, 1415–1425.
Doane, A. S., Danso, M., Lal, P., Donaton, M., Zhang, L., Hudis, C., and Gerald, W. L. (2006). An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 25, 3994–4008.
Edwards, J., Krishna, N. S., Grigor, K. M., and Bartlett, J. M. S. (2003). Androgen receptor gene amplification and protein expression in hormone refractory prostate cancer. Br. J. Cancer 89, 552–556.
Eeckhoute, J., Lupien, M., Meyer, C. A., Verzi, M. P., Shivdasani, R. A., Liu, X. S., and Brown, M. (2009). Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 19, 372–380.
Farmer, P., Bonnefoi, H., Becette, V., Tubiana-Hulin, M., Fumoleau, P., Larsimont, D., Macgrogan, G., Bergh, J., Cameron, D., Goldstein, D., Duss, S., Nicoulaz, A.-L., Brisken, C., Fiche, M., Delorenzi, M., and Iggo, R. (2005). Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 24, 4660–4671.
Ford, O. H. III, Gregory, C. W., Kim, D., Smitherman, A. B., and Mohler, J. L. (2003). Androgen receptor gene amplification and protein expression in recurrent prostate cancer. J. Urol. 170, 1817–1821.
Gao, N., Ishii, K., Mirosevich, J., Kuwajima, S., Oppenheimer, S. R., Roberts, R. L., Jiang, M., Yu, X., Shappell, S. B., Caprioli, R. M., Stoffel, M., Hayward, S. W., and Matusik, R. J. (2005). Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development 132, 3431–3443.
Gao, N., Zhang, J., Rao, M. A., Case, T. C., Mirosevich, J., Wang, Y., Jin, R., Gupta, A., Rennie, P. S., and Matusik, R. J. (2003). The role of hepatocyte nuclear factor-3 alpha (forkhead box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol. Endocrinol. 17, 1484–1507.
Gerhardt, J., Montani, M., Wild, P., Beer, M., Huber, F., Hermanns, T., Müntener, M., and Kristiansen, G. (2012). FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castrate-resistant prostate cancer. Am. J. Pathol. 180, 848–861.
Hannenhalli, S., and Kaestner, K. H. (2009). The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 10, 233–240.
Hegde, N. S., Sanders, D. A., Rodriguez, R., and Balasubramanian, S. (2011). The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat. Chem. 3, 829–829.
Hisamatsu, Y., Tokunaga, E., Yamashita, N., Akiyoshi, S., Okada, S., Nakashima, Y., Aishima, S., Morita, M., Kakeji, Y., and Maehara, Y. (2011). Impact of FoxA1 expression on the prognosis of patients with hormone receptor-positive breast cancer. Ann. Surg. Oncol. 19, 1145–1152.
Hurtado, A., Holmes, K. A., Ross-Innes, C. S., Schmidt, D., and Carroll, J. S. (2011). FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33.
Jain, R. K., Mehta, R. J., Nakshatri, H., Idrees, M. T., and Badve, S. S. (2011). High-level expression of forkhead-box protein A1 in metastatic prostate cancer. Histopathology 58, 766–772.
Jia, L., Berman, B. P., Jariwala, U., Yan, X., Cogan, J. P., Walters, A., Chen, T., Buchanan, G., Frenkel, B., and Coetzee, G. A. (2008). Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS ONE 3, e3645. doi:10.1371/journal.pone.0003645
Joseph, R., Orlov, Y. L., Huss, M., Sun, W., Li Kong, S., Ukil, L., Fu Pan, Y., Li, G., Lim, M., Thomsen, J. S., Ruan, Y., Clarke, N. D., Prabhakar, S., Cheung, E., and Liu, E. T. (2010). Integrative model of genomic factors for determining binding site selection by estrogen receptor-[alpha]. Mol. Syst. Biol. 6, 456.
Kong, S. L., Li, G., Loh, S. L., Sung, W.-K., and Liu, E. T. (2011). Cellular reprogramming by the conjoint action of ER[alpha], FOXA1, and GATA3 to a ligand-inducible growth state. Mol. Syst. Biol. 7, 526.
Laganiere, J. E., Deblois, G. V., Lefebvre, C. L., Bataille, A. R., Robert, F. O., and Giguere, V. (2005). Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. U.S.A. 102, 11651–11656.
Lupien, M., Eeckhoute, J., Meyer, C. A., Wang, Q., Zhang, Y., Li, W., Carroll, J. S., Liu, X. S., and Brown, M. (2008). FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970.
Mehta, R., Jain, R., Leung, S., Choo, J., Nielsen, T., Huntsman, D., Nakshatri, H., and Badve, S. (2012). FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Cancer Res. Treat. 131, 881–890.
Ni, M., Chen, Y., Lim, E., Wimberly, H., Bailey, S. T., Imai, Y., Rimm, D. L., Liu, S. X., and Brown, M. (2011). Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell 20, 119–131.
Robinson, J. L. L., Macarthur, S., Ross-Innes, C. S., Tilley, W. D., Neal, D. E., Mills, I. G., and Carroll, J. S. (2011). Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 30, 3019–3027.
Robyr, D., Gegonne, A., Wolffe, A. P., and Wahli, W. (2000). Determinants of vitellogenin B1 promoter architecture. J. Biol. Chem. 275, 28291–28300.
Ross-Innes, C. S., Stark, R., Holmes, K. A., Schmidt, D., Spyrou, C., Russell, R., Massie, C. E., Vowler, S. L., Eldridge, M., and Carroll, J. S. (2010). Cooperative interaction between retinoic acid receptor-α and estrogen receptor in breast cancer. Genes Dev. 24, 171–182.
Ross-Innes, C. S., Stark, R., Teschendorff, A. E., Holmes, K. A., Ali, H. R., Dunning, M. J., Brown, G. D., Gojis, O., Ellis, I. O., Green, A. R., Ali, S., Chin, S.-F., Palmieri, C., Caldas, C., and Carroll, J. S. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393.
Sahu, B., Laakso, M., Ovaska, K., Mirtti, T., Lundin, J., Rannikko, A., Sankila, A., Turunen, J.-P., Lundin, M., Konsti, J., Vesterinen, T., Nordling, S., Kallioniemi, O., Hautaniemi, S., and Janne, O. A. (2011). Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 30, 3962–3976.
Sérandour, A. L. A., Avner, S. P., Percevault, F. D. R., Demay, F., Bizot, M., Lucchetti-Miganeh, C. L., Barloy-Hubler, F. D. R., Brown, M., Lupien, M., Metivier, R. L., Salbert, G., and Eeckhoute, J. R. M. (2011). Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 21, 555–565.
Tan, P. Y., Chang, C. W., Chng, K. R., Wansa, K. D. S. A., Sung, W.-K., and Cheung, E. (2012). Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol. Cell. Biol. 32, 399–414.
Teschendorff, A., Miremadi, A., Pinder, S., Ellis, I., and Caldas, C. (2007). An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 8, R157.
Wang, D., Garcia-Bassets, I., Benner, C., Li, W., Su, X., Zhou, Y., Qiu, J., Liu, W., Kaikkonen, M. U., Ohgi, K. A., Glass, C. K., Rosenfeld, M. G., and Fu, X.-D. (2011). Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394.
Wang, Q., Li, W., Liu, X. S., Carroll, J. S., Janne, O. A., Keeton, E. K., Chinnaiyan, A. M., Pienta, K. J., and Brown, M. (2007). A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell 27, 380–392.
Wang, Q., Li, W., Zhang, Y., Yuan, X., Xu, K., Yu, J., Chen, Z., Beroukhim, R., Wang, H., Lupien, M., Wu, T., Regan, M. M., Meyer, C. A., Carroll, J. S., Manrai, A. K., Janne, O. A., Balk, S. P., Mehra, R., Han, B., Chinnaiyan, A. M., Rubin, M. A., True, L., Fiorentino, M., Fiore, C., Loda, M., Kantoff, P. W., Liu, X. S., and Brown, M. (2009). Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138, 245–256.
Zaret, K. S., and Carroll, J. S. (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241.
Zhang, C., Wang, L., Wu, D., Chen, H., Chen, Z., Thomas-Ahner, J. M., Zynger, D. L., Eeckhoute, J., Yu, J., Luo, J., Brown, M., Clinton, S. K., Nephew, K. P., Huang, T. H.-M., Li, W., and Wang, Q. (2011). Definition of a FoxA1 cistrome that is crucial for G1 to S-phase cell-cycle transit in castration-resistant prostate cancer. Cancer Res. 71, 6738–6748.
Keywords: FoxA1, breast cancer, prostate cancer, chromatin, ChIP-seq, transcription, drug response
Citation: Robinson JLL and Carroll JS (2012) FoxA1 is a key mediator of hormonal response in breast and prostate cancer. Front. Endocrin. 3:68. doi: 10.3389/fendo.2012.00068
Received: 31 January 2012; Paper pending published: 12 March 2012;
Accepted: 30 April 2012; Published online: 18 May 2012.
Edited by:
Carmen Priolo, Brigham and Women’s Hospital, USAReviewed by:
Silvia Migliaccio, Sapienza Università di Roma, ItalyErik Nelson, Dana Farber Cancer Institute, USA
Copyright: © 2012 Robinson and Carroll. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Jason S. Carroll, Cancer Research UK, Li Ka Shing Centre, Robinson Way, Cambridge CB2 0RE, UK e-mail:jason.carroll@cancer.org.uk

