- Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Bunkyo-ku, Tokyo, Japan
In our previous report, we demonstrated the possibility that various regulatory neuropeptides influence feeding behavior in the silkworm, Bombyx mori. Among these feeding-related neuropeptides, short neuropeptide F (sNPF) exhibited feeding-accelerating activity when injected into B. mori larvae. Like other insect sNPFs, the deduced amino acid sequence of the cDNA encoding the sNPF precursor appears to produce multiple sNPF and sNPF-related peptides in B. mori. The presence of three sNPFs, sNPF-1, sNPF-2, and sNPF-3, in the brain of B. mori larvae was confirmed by direct MALDI-TOF mass spectrometric profiling. In addition, all three sNPFs are present in other larval ganglia. The presence of sNPF mRNA in the central nervous system (CNS) was also confirmed by Reverse transcription-polymerase chain reaction. Semi-quantitative analyses of sNPFs in the larval brain using matrix-assisted laser desorption ionization time-of-flight mass spectrometry further revealed that brain sNPF levels decrease in response to starvation, and that they recover with the resumption of feeding. These data suggest that sNPFs were depleted by the starvation process. Furthermore, food deprivation decreased the transcriptional levels of the sNPF receptor (BNGR-A10) in the brain and CNS, suggesting that the sNPF system is dependent on the feeding state of the insect and that the sNPF system may be linked to locomotor activity associated with foraging behavior. Since the injection of sNPFs accelerated the onset of feeding in B. mori larvae, we concluded that sNPFs are strongly related to feeding behavior. In addition, semi-quantitative MS analyses revealed that allatostatin, which is present in the larval brain, is also reduced in response to starvation, whereas the peptide level of Bommyosuppressin was not affected by different feeding states.
Introduction
Although phytophagous insects feed on their preferred host plants, feeding is not continuous. A number of biochemical and behavioral studies have found that chemical components extracted from host plants serve as attractants or deterrents for plant feeders (Harborne, 1993; Hanson, 2003), and thus contribute to host selection. Therefore, it is believed that feeding behavior in phytophagous insects may be initiated by the chemical components present in their preferred host plants.
The biology and physiology of the mechanisms that regulate insect feeding behavior have been extensively investigated (Simpson, 1995). To date, sequential behaviors in regularly occurring feeding patterns have been monitored using a number of phytophagous insects, including orthopteran (Blaney et al., 1973; Simpson, 1982; Simpson and Ludlow, 1986) and lepidopteran species (Reynolds et al., 1986; Bowdan, 1988a,b; Timmins et al., 1988; Bernays and Woods, 2000; Nagata and Nagasawa, 2006). In the case of the silkworm, Bombyx mori, larvae exhibit characteristic feeding behavioral patterns that are generated independent of circadian rhythms at regular intervals of approximately every 2 h (Nagata and Nagasawa, 2006). Since these characteristic behaviors are primarily timed by the mode-changing process from quiescent status to feeding status, we assumed that some process might be involved in the decision to initiate feeding activity.
Based on previous physiological reports, the feeding state of an insect can be defined according to the following two indicators: (1) duration elapsed from the end of the previous meal and (2) head-swaying behavior immediately before the onset of eating or under feeding-motivated states (Nagata et al., 2011a). These two physiologically defined feeding states, hunger and satiety, support the evaluation of feeding behavior in B. mori larvae by measuring the latency period prior to the initiation of feeding (Nagata et al., 2011b). We were previously able to identify several biologically active peptides as feeding modulators by measuring the length of the latency period prior to the initiation of feeding in B. mori larvae (Nagata et al., 2009, 2011a,b). Among these peptides, two classes of peptides with different biological activities on feeding behavior were characterized: (1) inhibitory peptides, which prolong the latency to the initiation of feeding, and (2) acceleratory peptides, which shorten the latency to feeding initiation. Consistent with a previous proposal regarding control of invertebrate feeding behavior (Simpson, 1995), we found that FMRFamide-related peptides (FaRPs) and myosuppressin have inhibitory effects on the initiation of feeding behavior by peptide administration in B. mori larvae (Nagata et al., 2011b). In contrast, the administration of short neuropeptide F (sNPF) was shown to shorten the latency period, possibly resulting from the activation of locomotor activity. Quantitative analysis of sNPF levels during different feeding states, however, has not been fully examined.
The compiled genomic information from a number of insect species has allowed us to identify in silico various conserved factors, including the identification of sNPFs from a number of insect species. sNPFs belong to a very diverse neuropeptide group sharing a C-terminal RF-amide or RY-amide. In vertebrates, a representative RY-amide peptide, neuropeptide Y (NPY), has been characterized as a neuromodulator controlling feeding behaviors (Tatemoto et al., 1982). In invertebrates, a NPY homologous peptide has been identified in the tapeworm, Moniezia expansa, as a factor that regulates myo-stimulatory effects (Maule et al., 1991). By using an antibody against NPF [antiserum against Arg-X-Arg-Phe/Tyr-NH2: serum code 792(3)], shorter peptides that share the C-terminal RF-amide sequence, subsequently designated as short NPF, have been identified in the Colorado potato beetle (Spittaels et al., 1996). However, the detailed function of sNPF in vivo remains to be elucidated.
A cDNA encoding sNPF was first identified in the fruit fly, Drosophila melanogaster (Vanden Broeck, 2001). The deduced amino acid sequence contained four sNPFs, which were predicted to be produced after the post-translational modification of a dibasic amino acid cleavage site and C-terminal amidation (Vanden Broeck, 2001). In D. melanogaster, sNPF has been shown to be pleiotropic including regulation of feeding behavior (Lee et al., 2004, 2008; Nassel and Wegener, 2011). sNPFs are conserved across arthropod species with the number of sNPF isoforms encoded by a single cDNA varying depending on the insect species (Nassel and Wegener, 2011). In B. mori, sNPF has been identified as a juvenile hormone (JH) regulating peptide that functions in tandem with allatotropin to regulate the secretion of JH from the corpora allata (Yamanaka et al., 2008). Identification of a cDNA encoding sNPFs revealed the presence of three sNPFs in B. mori (Mita et al., 2003; Roller et al., 2008; Yamanaka et al., 2008), which is consistent with reports in other insects of multiple sNPFs.
In D. melanogaster, the presence of sNPFs isoforms has been characterized by direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) profiling (Predel et al., 2004; Baggerman et al., 2005; Yew et al., 2009; Carlsson et al., 2010; Wegener et al., 2011). Although sNPF-2 and a partial fragment of sNPF-3 were purified from B. mori brain extracts (Yamanaka et al., 2008), there is currently no experimental evidence for the presence of all three sNPF isoforms at the peptide level in B. mori. We consequently performed direct mass spectrometric profiling of larval brains to reassess the presence of sNPFs. In addition, to further address the mechanisms underlying sNPF-mediated feeding behaviors, we semi-quantitatively measured the levels sNPFs during different feeding states in B. mori.
Materials and Methods
Chemicals and Reagents
Chemicals and reagents, α-cyano-4-hydroxycinnamic acid (CCA), ethidium bromide, agarose for electrophoresis and Trizma-base, and EDTA used in the present study were purchased from Nacalai-tesk (Osaka, Japan). Methanol, diethylether, formic acid, trifluoro acetic acid (TFA), and acetonitrile for reversed-phase HPLC (RP-HPLC) were purchased from Kanto Chemicals (Tokyo, Japan). Fmoc derivatives of amino acids were purchased from Watanabe Chemical Industries (Hiroshima, Japan).
Insects
Silkworm eggs from the hybrid B. mori strain (Kinshu × Showa) were purchased from UEDA SANSHU Ltd. (Ueda, Japan). Larvae were reared in plastic containers at 26 ± 1°C with 70 ± 10% relative humidity under long-day lighting conditions (16L: 8D) using SILKMATE 2S artificial diet purchased from NIPPON NOSAN Co. Ltd. (Yokohama, Japan). Larvae were provided with fresh diet on a daily basis. Only larvae whose growth was synchronized were utilized for the present experiments.
Preparation of Starved and Re-Fed B. mori Larvae
Bombyx mori larvae were starved by food deprivation with feces frequently removed from the containers throughout starvation. Food deprivation was initiated in synchronously growing fifth instar, day-2 larvae (2.7 ± 0.3 g). Resumption of feeding was initiated with addition of an artificial diet block. One hour after resumption of feeding, the re-fed-larvae were anesthetized on ice. For mass spectrometric analyses, brains were dissected out from the prepared larvae.
Preparation of Synthetic Peptides
Short neuropeptide Fs were chemically synthesized based on the Fmoc method using an automated peptide synthesizer (APEX-SC, apex396, Advanced ChemTech, Louisville, KT, USA) and deprotection as previously reported (Nagata et al., 2011b). In brief, after deprotection and cleavage from resins, the synthetic peptides in diethylether were collected by centrifugation. The resulting crude synthetic peptides were partially purified using Sep-Pak C18. The adsorbed sample was eluted with 60% acetonitrile/0.1% TFA. The eluate was subjected to RP-HPLC (Jasco SC 802, PU-880, UV-875, Jasco; Tokyo, Japan) on a Senshu Pak Pegasil-300 ODS column (4.6 mm i.d. × 250 mm, Shenshu Kagaku, Tokyo, Japan) with a 25-min linear gradient of 10–60% acetonitrile containing 0.05% TFA at a flow rate of 1.0 ml/min. The purity of the synthetic peptide was confirmed by MALDI-TOF MS using a Voyager-DE™ STR (Applied Biosystems, CA, USA) in the positive ion mode and CCA as the matrix. A highly purified byproduct (m/z 983) of sNPF synthesis was used for semi-quantitative analyses in the present study.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from fat body, silk glands, foregut, midgut, hindgut, Malpighian tubules, hemocytes, central nervous ganglia, brain, ovary, and testis using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. After treatment with DNase I (TaKaRaBio, Shiga, Japan), cDNA was synthesized using a reverse transcriptase, Superscript III (Invitrogen), and an oligo-dT30 primer. Gene specific primers were sNPF-Fw (5′-GATGCTCCTAAAGAAGACAAT-3′) and sNPF-Rv (5′-TCAGGCTTTCGGTGCTTCCG-3′). Primers for sNPFR (BNGR-A10) were sNPFR-Fw (5′-GCTACAATCCATTCCTCTATGCTT-3′) and sNPFR-Rv (5′-AGGTTCCTTGTCTGAAGAAATCAC-3′). Ribosomal protein 49 (rp49) was used as an experimental control with the following gene specific primer: Rp-Fw (5′-GACCTGTTTACAGGCCGACAATCG-3′) and Rp-Rv (5′-TTATATTTATTCATTCTCCTGGGAGCGGAG-3′). Amplified fragments were electrophoresed on a 1.2% agarose gel and detected by staining with ethidium bromide. The saturation of PCR products were observed after 35 cycles of amplification. We consequently performed our analyses at 30 cycles. To quantitate the sNPF expression levels (data not shown), gel images were analyzed using the public domain NIH Image program (the U.S. National Institutes of Health at http://rsb.info.nih.gov/nih-image/). The analyzed PCR product bands were normalized to bands derived from the PCR experimental control, rp49.
Quantitative RT-PCR
Extraction of total RNA and RT reaction were performed as described above. The primers for Quantitative RT-PCR (qRT-PCR) for sNPFR were: forward 5′-TGCTTGGCTCAACGAAAACT-3′; reverse 5′-GAAAGCACCCAAACATGGAA-3′. As a control, the following primers for rp49 were used: forward 5′-TGGGAGGTTTCCCCCATT-3′; reverse 5′-CCATGCAGCAACCCTTGAT-3′. PCR was performed using a 7300 Real-Time PCR system (Applied Biosystems). Thermal cycling conditions were as follows: after initial denaturation at 95°C for 10 min, 40 cycles of 95°C, 15 s; 60°C, 1 min. The singly targeted qRT-PCR product was confirmed by constructing a melt curve to all reactions. The reaction was performed using Power SYBR® Green PCR Master Mix (Applied Biosystems) and 50 nM of each primer. Data were analyzed using the 7300 Real-Time PCR system software v1.3 (Applied Biosystems). Results are represented as a ratio of sNPFR and RpL3 by Ct method.
Direct MALDI-TOF MS Analysis
Brain and other ganglia were isolated from B. mori larvae and placed in a drop of distilled water. After removal of excess water, the matrix solution was covered with the dried brain or ganglia prior to mass spectrometry analysis. Mass spectra were measured on a MALDI-TOF mass spectrometer with CCA as matrix in the positive ion mode. The matrix solution was prepared by saturating CCA in 60% acetonitrile containing 0.1% TFA.
Semi-Quantitative Analyses Using MALDI-TOF MS
For semi-quantitative analyses, the intensities of the mass spectra ion peaks were evaluated by comparing with an exogenously added internal standard peptide as previously reported (Jimenez et al., 1997). The internal standard peptide, which is not observed in the spectrum of brain extracts, has a m/z 983 and is the byproduct of Fmoc chemical synthesis of sNPF-2 with Fmoc at the N-terminus of sNPF-2. The internal standard was added in extraction solution (90% methanol in 0.1% formic acid). The relative intensities of each ion peak were compared with the intensity of the ion peak corresponding to the internal standard peptide. Five samples were measured for analyses of ion peak intensities. At least three measurements were carried out per sample. Single measurement of MALDI-TOF MS was carried out by accumulation of more than 50 shots using the same laser desorption settings.
Statistical Analysis
The intensities of the semi-quantitative analyses of sNPF were statistically compared with control samples derived from brains of larvae fed ad libitum using one-way ANOVA. Significances were determined with Dunnett’s post hoc test (data from larvae fed ad libitum were used as experimental controls for comparison).
Results
sNPF in the Silkworm, Bombyx mori
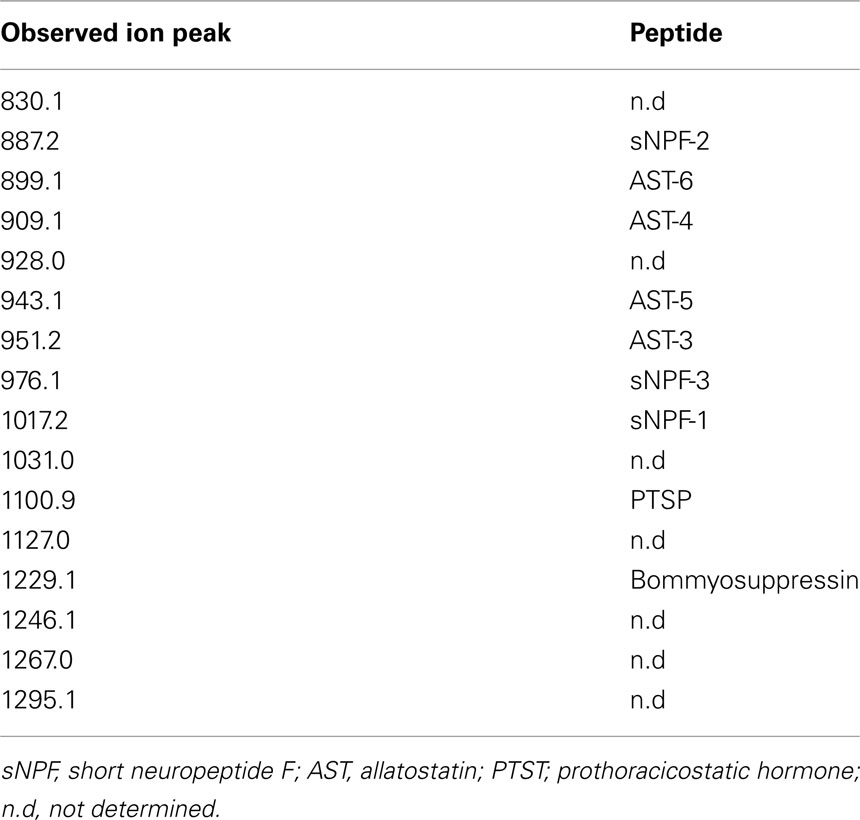
In B. mori, three sNPFs, sNPF-1, sNPF-2, and sNPF-3, are encoded by a single cDNA sequence (Yamanaka et al., 2008). Of these three sNPFs, sNPF-2 and a partial fragment of sNPF-3 were purified from brain extracts (Yamanaka et al., 2008). To confirm the presence of sNPFs in the central nervous system (CNS), we performed direct MALDI-TOF mass spectrometric analyses using the isolated brain of a last instar larva (Figure 1). In total, more than 15 detectable ion peaks were observed. As deduced from the cDNA sequence encoding the sNPF precursor, ion peaks at m/z 1017.2, 887.2, and 976.1 corresponding to sNPF-1, sNPF-2, and sNPF-3, respectively, were observed. These data suggest the presence of all three forms of sNPF in the brain. A database search revealed that these ion peaks coincided with those of known neuropeptides, including sNPFs (Figure 1).
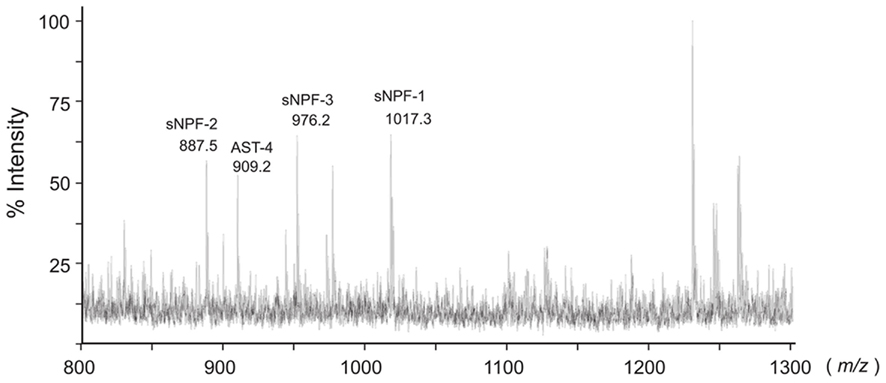
Figure 1. Bombyx mori sNPF precursor and MS spectrum of B. mori larval brain. Representative MS spectrum of the B. mori larval brain. A fifth instar B. mori larval brain was isolated and directly analyzed by MALDI-TOF MS. The B. mori genomic database was used to facilitate identification of ion peaks in the spectrum.
RT-PCR Analyses of B. mori sNPFs
We subsequently confirmed the expression sites of the sNPF precursor transcript in B. mori larvae. Reverse transcription-polymerase chain reaction (RT-PCR) using specific primers for the sNPF precursor cDNA sequence revealed that the expression of sNPFs was observed in the brain and CNS (Figure 2A). To analyze the detailed location of sNPF expression, we examined the expression profile in each ganglion. We found that all ganglia expressed the sNPF precursor mRNA, even though biased cDNA amplification was observed (Figure 2B). These RT-PCR data are consistent with our MS data.

Figure 2. Reverse transcription-polymerase chain reaction-based tissue expression profile of the sNPF precursor. (A) Tissues were isolated from 2-day-old fifth instar larvae: brain (lane 1), central nervous system (CNS) (lane 2), prothoracic glands (lane 3), silk gland (lane 4), foregut (lane 5), midgut (lane 6), hindgut (lane 7), Malpighian tubules (lane 8), fat body (lane 9), muscle (lane 10), hemocytes (lane 11), testis (lane 12), and ovary (lane 13). PCR control consisted of rp49 amplification. (B) RT-PCR of the sNPF precursor in each ganglion from a fifth instar B. mori larvae. Brain (lane 1), subesophageal ganglion (lane 2), thoracic ganglia (lanes 3–6), abdominal ganglia (lanes 7–12).
sNPF Expression during Different Feeding States
While determining the sNPF precursor expression profile, we observed clear differences in expression levels. The resulting RT-PCR products were compared between the tissues of ad libitum fed larvae and starved larvae. These results showed similar levels for the sNPF precursor transcript in the brain and CNS in the different feeding states (Figure 3A). Furthermore, the transcriptional expression level of the sNPF receptor (sNPFR, BNGR-A10) was also analyzed by RT-PCR. The widely distributed expression sites of the sNPFR were reconfirmed, supporting previous reports (Yamanaka et al., 2008). Comparison of the transcriptional levels of the sNPFR between RT-PCR products derived from tissues of ad libitum fed larvae and starved larvae revealed that starvation reduces sNPFR transcript levels (Figure 3B). The reduced levels of sNPFR transcript by starvation were confirmed using several sNPFR expressing tissues, brain, CNS, midgut, and hindgut by qRT-PCR (Figure 3C). We observed a significant reduction in sNPFR expression levels following starvation in all tissues examined except for the brain, which likewise showed a slight albeit not significant reduction in sNPFR.
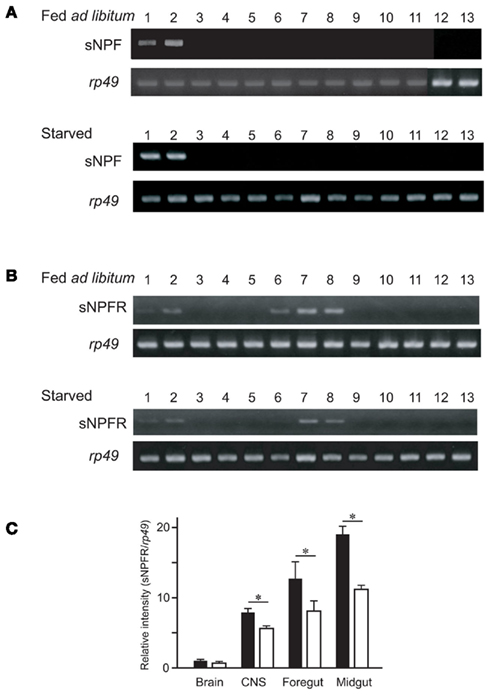
Figure 3. Reverse transcription-polymerase chain reaction analyses of the sNPF precursor and sNPFR (BNGR-A10) in B. mori in ad libitum fed larvae and starved larvae. Tissues were isolated from 2-day-old fifth instar larvae: brain (lane 1), central nervous system (CNS) (lane 2), foregut (lane 3), midgut (anterior, medial, posterior, lanes 4–6), hindgut (lane 7), Malpighian tubules (lane 8), fat body (lane 9), silk glands (lane 10), testis (lane 11), ovary (lane 12), and hemocytes (lane 13). RT-PCR analyses were performed for the sNPF precursor (A) and sNPFR (BNGR-A10) (B). PCR control consisted of rp49 amplification. (C) qRT-PCR analyses for sNPFR of brain, CNS, foregut, and midgut. Black and white bars represent relative expression levels of sNPFR compared with rp49 in the larvae fed ad libitum and starved, respectively. N = 5, Mean ± SEM, *P < 0.05, t-test.
Semi-Quantitative Analyses of sNPFs in the Brain
We next examined the peptide levels of sNPFs in the brain during different feeding states. The amount of brain sNPFs were semi-quantitatively analyzed by comparing the relative levels of sNPFs in crude larval brain extracts with an exogenous internal standard peptide. A byproduct of sNPF-2 chemical synthesis was used as an exogenous standard peptide after separation from sNPF-2 by RP-HPLC. In the resulting spectra, we observed ion peaks at m/z 1017.2, 887.2, and 976.1 corresponded to sNPF-1, sNPF-2, and sNPF-3, respectively. Reduced levels of sNPF-1 and sNPF-2 were observed in starved larval brains. The levels of sNPF-1 and sNPF-2 further decreased with an increase in the starvation period (Figure 4). A resumption of feeding returned the levels of brain sNPF-1 and sNPF-2 back to basal levels. It is noteworthy that sNPF-1 and sNPF-2 were significantly reduced by starvation, whereas sNPF-3 was not as clearly affected. However, there did appear to be a similar trend, albeit not statistically significant, in the reduction sNPF-3 levels, most noticeably after 2 days of starvation. The less pronounced reduction in sNPF-3 levels could be due to a lower ionization efficiency.
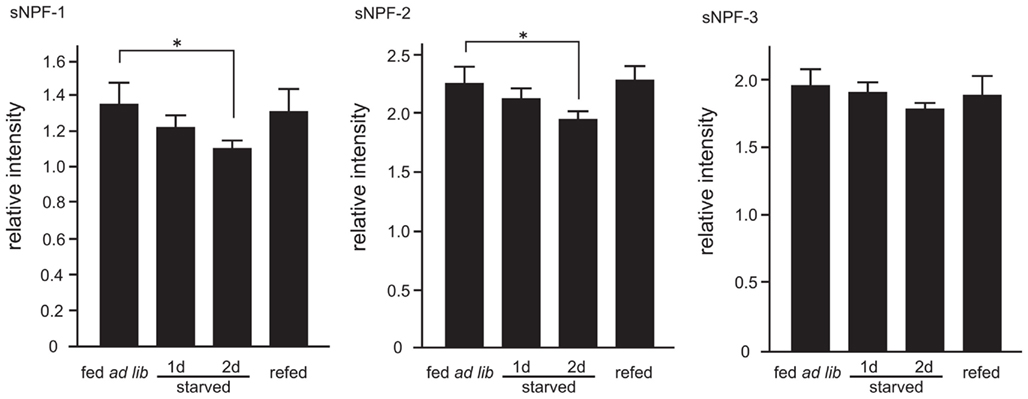
Figure 4. Semi-quantitative mass spectrometry analyses of sNPFs. The sNPFs levels were measured by comparing with an exogenous nonsense peptide. Samples were prepared from 2-day-old fifth instar larvae. Larvae were starved for 1 and 2 days. Re-fed-larvae were prepared as described in the Materials and Methods. N = 5, Mean ± SE. Data were statistically analyzed using a one-way ANOVA with significant differences determined with a post hoc Dunnett’s test, *P < 0.05.
The mass spectral profiles (Table 1) obtained while performing the semi-quantitative analyses of sNPFs allowed us to examine the effects of starvation two other significant ion peaks. These ion peaks correspond to Bommyosuppressin (BMS) at m/z 1229.1 and allatostatin-4 (AST-4) at m/z 909.1, which have been suggested to be feeding-related peptides (Audsley and Weaver, 2009; Nagata et al., 2011b). We found that, similar to sNPF-1 and sNPF-2, AST-4 levels were reduced by starvation and were recovered following a resumption of feeding (Figure 5). In contrast, the levels of brain of BMS did not change significantly.
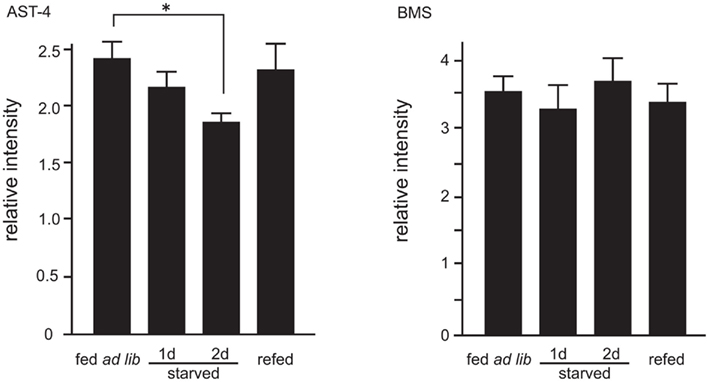
Figure 5. Semi-quantitative analyses of allatostatin (AST)-4 and Bommyosuppressin (BMS). Ion peaks corresponding to AST-4 at m/z 909.1 and BMS at m/z 1229.1 were analyzed. Analytical conditions were the same as those for sNPFs analyses. N = 5, Mean ± SE. Data was statistically analyzed by one-way ANOVA. Significant differences were determined by the post hoc Dunnett’s test, *P < 0.05.
Discussion
In B. mori, post-translational cleavage of the sNPF precursor yields three sNPF forms. In the current study, we examined the presence and expression profile of these three cleavage products and have shown that sNPF mRNA is expressed throughout the CNS with all three sNPF forms in each B. mori larval ganglion. Since sNPFs have been linked to the regulation of feeding behavior (Nagata et al., 2011b; Nassel and Wegener, 2011), the presence of sNPFs in all ganglia is consistent with a putative role in regulating feeding behavior in B. mori.
In other insect species, multiple sNPF forms (from one to five) are present (Nassel and Wegener, 2011). This is consistent with evidence showing the presence of three sNPFs in B. mori. For example, based on the deduced amino acid sequence of the sNPF precursor, the D. melanogaster sNPF gene appears to encode four separate sNPF forms (Vanden Broeck, 2001). The presence of these four sNPFs at the peptide level has been confirmed by direct mass spectrometry analyses (Predel et al., 2004; Baggerman et al., 2005; Yew et al., 2009; Carlsson et al., 2010). In the mass spectrum profiling, trypsin-like enzymatic processing of sNPFs appeared to generate shorter sNPF-like products (Baggerman et al., 2005; Wegener et al., 2011), which would suggest that at least six sNPF isoforms are present in the D. melanogaster larval CNS. In contrast, as demonstrated in Figure 1, no ion peaks corresponding to sNPF precursor-derived peptides were observed in the crude B. mori larval brain extract.
Both our direct mass spectrometry measurements and RT-PCR analyses of the B. mori CNS indicated that sNPF expression is localized, if not restricted to, the nervous tissues. However, it has been reported that, in addition to the CNS, D. melanogaster sNPFs are also expressed in the midgut (Reiher et al., 2011). In the case of B. mori, we were unable to detect any ion signals for sNPFs in the crude extract of the larval midgut nor could we detect sNPF precursor mRNA in the midgut via RT-PCR. As sNPFs exert a number of biological events, including regulating feeding behavior, sNPFs in the midgut might function as enterogastric peptides in D. melanogaster. Although sNPF expression was not observed in the B. mori larval midgut, sNPFs secreted from the individual ganglia could potentially function in all tissues because receptor expression has been reported to be widespread in B. mori larvae (Yamanaka et al., 2008). Additionally, antiserum against RF-amide, which can recognize sNPFs in addition to other RF-amide peptides, indicated that RF-amide peptides are preset in many neurons (Walker et al., 2009; Nassel and Wegener, 2011). Consequently, even though sNPFs are not expressed in the midgut of B. mori, it is possible that they function in gastric control.
Even though sNPFs were initially identified based on cross-reactivity with an anti-neuropeptide F (NPF) antibody, our knowledge of their functionality remains limited. A number of studies, however, have proposed possible functions for sNPFs (Nassel and Wegener, 2011). For example, in D. melanogaster, sNPFs are produced in numerous small neurons in the brain that are thought to be associated with locomotor modulation and regulation as well as other various functions (Hanesch et al., 1989; Strauss, 2002; Kahsai and Winther, 2011). This possible function supports our previous data, which showed that the injection of sNPFs into B. mori larvae influenced the initiation of feeding behavior (Nagata et al., 2011b). sNPFs sometimes co-express with other peptide hormones in D. melanogaster, such as tachykinin and ion transport peptides, resulting in a proposal that sNPFs may regulate hormone release (De Lange et al., 1997; de Jong-Brink et al., 2001; Kahsai et al., 2010). In B. mori, sNPFs also appear to negatively act on the allatotropin producing organ, the corpora allata, such that JH secretion and biosynthesis are decreased (Yamanaka et al., 2008).
To have a better understanding of how sNPF may function in B. mori, we estimated the amount of sNPFs at both the transcriptional and peptide levels. The present data indicate that starvation has no effect on the transcriptional level of sNPFs (data not shown) but did reduce the amount of mature sNPF peptides in the brain. Although the sNPF content of the remaining ganglia was not measured in this study, we found that the brain sNPF levels changed significantly in response to starvation (Figure 4). We speculate that a reduction in brain sNPFs, which likely results from functional consumption of the peptides, would trigger several biologically important events in B. mori larvae such as the activation of locomotor activity accompanied with other functions. This would be consistent with the observation that sNPFs drive locomotor activity in D. melanogaster (Kahsai et al., 2010).
Intriguingly, the sNPF receptor transcript levels were reduced in response to starvation (Figures 3B,C). Furthermore, different behavioral states were observed after short versus long starvation, with the latter showing much lower levels of locomotor activity (Wolfgang and Riddiford, 1987; Nagata and Nagasawa, 2006). As sNPFs are possible locomotor modulators (Kahsai et al., 2010), the reduction in sNPFR levels is consistent with the fact that larvae starved for longer periods would exhibit decreased locomotor activity as a means of conserving crucial energy stores.
Database-assisted transcriptomic analyses revealed that several GPCRs might be allocated to receptors for RF-amide peptides in most insect species, including D. melanogaster (Mertens et al., 2002) and Anopheles gambiae (Hill et al., 2002). In B. mori, there are three sNPF receptors, BNGR-A10, BNGR-A11, and BNGR-A7 (data not shown, manuscript in preparation). Interestingly, these three receptors are expressed in almost all tissues of the larval body, but exhibit biases in distribution (Yamanaka et al., 2008; unpublished data by our group). This observation suggests that each receptor has different ligands or different functions in vivo. Therefore, careful analyses of sNPF expression levels, in addition to that of the associated receptors, are required to further develop our understanding of the mechanisms underlying sNPF-modulated feeding behavior.
Finally, as observed in sNPFs, there are a number of similar cases where several homologous peptides are produced from a single cDNA, such as AST (Secher et al., 2001) and tachykinin (Roller et al., 2008). Interestingly, such multiple peptides have several different receptors with high similarity (Roller et al., 2008; Yamanaka et al., 2008). Hence, further information about how various ligands may be recognized by various receptors requires further elucidation.
Conclusion
In the present study, we monitored sNPF levels in the CNS and brain of B. mori larvae. The level of sNPFs in the B. mori brain was influenced by the feeding state of the specimens examined. As previously proposed, sNPFs may function as possible neuromodulators of locomotion in B. mori larvae. We suggest that one of the pleiotropic functions of sNPFs is neuromodulatory in nature in that they potentially drive feeding behavior associated locomotor activity in response to the animal’s feeding state.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported partly by Grants-in-Aid for Scientific Research [#18780083 and 22780099 (Shinji Nagata); #22-4193 (Nobukatsu Morooka); #22-1424 (Chiaki Nagai)] from the Ministry of Education, Science, Sports, and Culture of Japan.
References
Audsley, N., and Weaver, R. J. (2009). Neuropeptides associated with the regulation of feeding in insects. Gen. Comp. Endocrinol. 162, 93–104.
Baggerman, G., Boonen, K., Verleyen, P., De Loof, A., and Schoofs, L. (2005). Peptidomic analysis of the larval Drosophila melanogaster central nervous system by two-dimensional capillary liquid chromatography quadrupole time-of-flight mass spectrometry. J. Mass Spectrom. 40, 250–260.
Bernays, E. A., and Woods, H. A. (2000). Foraging in nature by larvae of Manduca sexta influenced by an endogenous oscillation. J. Insect Physiol. 46, 825–836.
Blaney, W. M., Chapman, R. F., and Wilson, A. (1973). The pattern of feeding of Locusta migratoria (L.) (Orthoptera, Acrididae). Acrida 2, 119–137.
Bowdan, E. (1988a). The effect of deprivation on the microstructure of feeding by the tobacco hornworm caterpillar. J. Insect Behav. 1, 31–50.
Bowdan, E. (1988b). Microstructure of feeding by tobacco hornworm caterpillar, Manduca sexta. Entomol. Exper. Appl. 47, 127–136.
Carlsson, M. A., Diesner, M., Schachtner, J., and Nassel, D. R. (2010). Multiple neuropeptides in the Drosophila antennal lobe suggest complex modulatory circuits. J. Comp. Neurol. 518, 3359–3380.
De Lange, R., van Golen, F. A., and can Minnen, J. (1997). Diversity in cell specific co-expression of four neuropeptide genes involved in control of male copulation behaviour in Lymnaea stagnalis. Neuroscience 78, 289–299.
de Jong-Brink, M., ter Maat, A., and Tensen, C. P. (2001). NPY in invertebrates: molecular answers to altered functions during evolution. Peptides 22, 309–315.
Hanesch, U., Fischbach, K. F., and Heisenberg, M. (1989). Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res. 257. 343–366.
Hanson, J. B. (2003). “Phenolics,” in Natural Products, Their Chemistry and Biological Significance, eds J. Mann, J. B. Davidson, J. B. Hobbs, D. V. Banthorpe, and J. B. Harborne (Upper Saddle River, NJ: Prentice Hall), 362–388.
Hill, C. A., Fox, A. N., Pitts, R. J., Kent, L. B., Tan, P. L., Chrystal, M. A., Cravchik, A., Collins, F. H., Robertson, H. M., and Zwiebel, L. J. (2002). G protein-coupled receptors in Anopheles gambiae. Science 298, 176–178.
Jimenez, C. R., Li, K. W., Dreisewerd, K., Mansvelder, H. D., Brussaard, A. B., Reinhold, B. B., Van der Schors, R. C., Karas, M., Hillenkamp, F., Burbach, J. P. H., Costello, C. E., and Geraerts, W. P. M. (1997). Pattern changes of pituitary peptides in rat after salt-loading as detected by means of direct, semiquantitative mass spectrometric profiling. Proc. Natl. Acad. Sci. U.S.A. 94, 9481–9486.
Kahsai, L., Martin, J. R., and Winther, A. M. (2010). Neuropeptides in the Drosophila central complex in modulation of locomotor behavior. J. Exp. Biol. 213, 2256–2265.
Kahsai, L., and Winther, A. M. (2011). Chemical neuroanatomy of the Drosophila central complex: distribution of multiple neuropeptides in relation to neurotransmitters. J. Comp. Neurol. 519, 290–315.
Lee, K. S., Kwon, O. Y., Lee, J. H., Kwon, K., Min, K. J., Jung, S. A., Kim, A. K., You, K. H., Tatar, M., and Yu, K. (2008). Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat. Cell Biol. 10, 468–475.
Lee, K. S., You, K. H., Choo, J. K., Han, Y. M., and Yu, K. (2004). Drosophila short neuropeptide F regulates food intake and body size. J. Biol. Chem. 279, 50781–50789.
Maule, A. G., Shaw, C., Halton, D. W., Thim, L., Johnston, C. F., Fairweather, I., and Buchanana, K. D. (1991). Neuropeptide F: a novel parasitic flatworm regulatory peptide from Moniezia expansa (Cestoda: Cyclophyllidae). Parasitology 102, 309–316.
Mertens, I., Meeusen, T., Huybrechts, R., De Loof, A., and Schoofs, L. (2002). Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 297, 1140–1148.
Mita, K., Morimyo, M., Okano, K., Koike, Y., Nohata, J., Kawasaki, H., Kadono-Okuda, K., Yamamoto, K., Suzuki, M. G., Shimada, T., Goldsmith, M. R., and Maeda, S. (2003). The construction of an EST database for Bombyx mori and its application. Proc. Natl. Acad. Sci. U.S.A. 100, 14121–14126.
Nagata, S., Morooka, N., Asaoka, K., and Nagasawa, H. (2011a). Identification of a novel hemolymph peptide that modulates silkworm feeding motivation. J. Biol. Chem. 286, 7161–7170.
Nagata, S., Morooka, N., Matsumoto, S., Kawai, T., and Nagasawa, H. (2011b). Effects of neuropeptides on feeding initiation in larvae of the silkworm, Bombyx mori. Gen. Comp. Endocrinol. 172, 90–95.
Nagata, S., Morooka, N., Matsumoto, S., and Nagasawa, H. (2009). Characterization of feeding-delaying factors from the silkworm Bombyx mori. Ann. N. Y. Acad. Sci. 1163, 481–483.
Nagata, S., and Nagasawa, H. (2006). Effects of diet-deprivation and physical stimulation on the feeding behaviour of the larvae of the silkworm, Bombyx mori. J. Insect Physiol. 52, 807–815.
Nassel, D. R., and Wegener, C. (2011). A comparative review of short and long neuropeptide F signaling in invertebrates: any similarities to vertebrate neuropeptide Y signaling? Peptides 32, 1335–1355.
Predel, R., Wegener, C., Russell, W. K., Tichy, S. E., Russell, D. H., and Nachman, R. J. (2004). Peptidedomics of CNS-associated neurohemal systems of adult Drosophila melanogaster: a mass spectrometric survey of peptides from individual flies. J. Comp. Neurol. 474, 379–392.
Reynolds, S. E., Yeomans, M. R., and Timmins, W. A. (1986). The feeding behaviour of caterpillars (Manduca sexta) on tobacco and on artificial diet. Physiol. Entomol. 11, 39–51.
Reiher, W., Shirras, C., Kahnt, J., Baumeister, S., Isaac, R. E., and Wegener, C. (2011) Peptidomics and peptide hormone processing in the Drosophila midgut. J. Proteo. Res. 10, 1881–1892.
Roller, L., Yamanaka, N., Watanabe, K., Daubnerova, I., Zitnan, D., Kataoka, H., and Tanaka, Y. (2008). The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38, 1147–1157.
Secher, T., Lenz, C., Cazzamali, G., Sorensen, G., Williamson, M., Hansen, G. N., Svane, P., and Grimmelikhuijzen, C. J. (2001). Molecular cloning of a functional allatostatin gut/brain receptor and an allatostatin preprohormone from the silkworm Bombyx mori. J. Biol. Chem. 276, 47052–47060.
Simpson, S. J. (1982). Patterns in feeding: a behavioural analysis using Locusta migratoria nymphs. Physiol. Entomol. 7. 325–336.
Simpson, S. J. (1995). “The control of meals in chewing insects,” in Regulatory Mechanisms of Insect Feeding, eds R. F. Chapman and J. de Boer (New York: Chapman & Hall), 137–156.
Simpson, S. J., and Ludlow, A. R. (1986). Why locusts start to feed? A comparison of causal factors. Anim. Behav. 34, 480–496.
Spittaels, K., Verhaert, P., Shaw, C., Johnston, R. N., Devreese, B., and Van Beeumen, J. (1996). Insect neuropeptide F (NPF)-related peptides: isolation from Colorado potato beetle (Leptinotarsa decemlineata) brain. Insect Biochem. Mol. Biol. 26, 375–382.
Strauss, R. (2002). The central complex and the genetic dissection of locomotor behaviour. Curr. Opin. Neurobiol. 12, 633–638.
Tatemoto, K., Carlquist, M., and Mutt, V. (1982). Neuropeptide Y-a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature 296, 659–660.
Timmins, W. A., Bellward, K., Stamp, A. J., and Reynolds, S. E. (1988). Food intake, conversion efficiency and feeding behaviour of tobacco hornworm caterpillar given artificial diet of carrying nutrient and water content. Physiol. Entomol. 13, 303–314.
Vanden Broeck, J. (2001). Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides 22, 241–254.
Walker, R. J., Papioannou, S., and Holden-Dye, L. (2009). A review of FMRFamide- and RFamide-like peptides in metazoan. Invert. Neurosci. 9, 111–153.
Wegener, C., Hervert, H., Kahnt, J., Bender, M., and Rhea, J. M. (2011). Deficiency of prohormone convertase dPC2 (AMONTILLADO) results in impaired production f bioactive neuropeptide hormones in Drosophila. J. Neurochem. 118. 581–595.
Wolfgang, W. J., and Riddiford, L. M. (1987). Cuticular mechanics during larval development of the tobacco hornworm, Manduca sexta. J. Exp. Biol. 128, 19–33.
Yamanaka, N., Yamamoto, S., Zitnan, D., Watanabe, K., Kawada, T., Satake, H., Kaneko, Y., Hiruma, K., Tanaka, Y., Shinoda, T., and Kataoka, H. (2008). Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS ONE 3, e3048. doi:10.1371/journal.pone.0003048
Keywords: Bombyx mori, feeding behavior, peptide, short neuropeptide F
Citation: Nagata S, Matsumoto S, Nakane T, Ohara A, Morooka N, Konuma T, Nagai C and Nagasawa H (2012) Effects of starvation on brain short neuropeptide F-1, -2, and -3 levels and short neuropeptide F receptor expression levels of the silkworm, Bombyx mori. Front. Endocrin. 3:3. doi: 10.3389/fendo.2012.00003
Received: 15 September 2011;
Accepted: 04 January 2012;
Published online: 18 January 2012.
Edited by:
Joe Hull, USDA Agricultural Research Service, USAReviewed by:
Hironori Ando, Niigata University, JapanJoe Hull, USDA Agricultural Research Service, USA
Åsa M. E. Winther, Karolinska Institutet, Sweden
Copyright: © 2012 Nagata, Matsumoto, Nakane, Ohara, Morooka, Konuma, Nagai and Nagasawa. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Shinji Nagata, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan. e-mail:YW5hZ2FzaGlAbWFpbC5lY2MudS10b2t5by5hYy5qcA==

 Sumihiro Matsumoto
Sumihiro Matsumoto