
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 04 July 2024
Sec. Ecophysiology
Volume 12 - 2024 | https://doi.org/10.3389/fevo.2024.1412124
 Mercedes Berlanga1*†
Mercedes Berlanga1*† Pere Picart1
Pere Picart1 Arnau Blasco2
Arnau Blasco2 Robert Benaiges-Fernandez3
Robert Benaiges-Fernandez3 Ricardo Guerrero4
Ricardo Guerrero4 Andrea Butturini2
Andrea Butturini2 Jordi Urmeneta3
Jordi Urmeneta3La Muerte lagoon is an ephemeral endorheic water body located in the Monegros desert, Zaragoza, Spain. Amplicon sequencing of the 16S rRNA gene was performed to analyze the bacterial and archaeal communities in biofilm-sediment samples over three years, to understand the dynamic changes in the microbial community. PICRUSt and shotgun metagenomics were used to examine energy production and carbohydrate metabolism pathways. The dominant bacterial phyla were Actinobacteriota, Bacteroidota, Cyanobacteriota, and Pseudomonadota, while Halobacteriota was the predominant archaeal phylum. Despite seasonal environmental fluctuations, the biofilm community remained stable over time, suggesting resilience. The Calvin-Benson cycle was the main carbon fixation pathway, carried out by Cyanobacteria and purple non-sulfur bacteria. Nitrogen fixation by diazotrophs supplied an important nitrogen source. Organic carbon was derived primarily from autotrophs, with little use of allochthonous plant material. The comparison of biofilm-sediment and water column biotopes showed distinct but related prokaryote communities. Biofilm-sediments showed higher taxonomic diversity and different proportions of microbial phyla compared to the water column. This study provides initial insights into the complex microbial life in endorheic lagoons and underscores the importance of protecting these globally threatened habitats. The limited sample size in this study warrants further investigation with a more comprehensive sampling strategy to fully characterize the microbial communities and their functional roles in the different biotopes of La Muerte lagoon.
Endorheic lakes are inland water bodies found in arid and semiarid regions around the world. Unlike exorheic lakes, endorheic lakes lack natural outflows and are characterized by the accumulation of water and dissolved salts (Yechieli and Wood, 2002). With high evaporation rates, these systems often become hypersaline environments with salt concentrations exceeding those of seawater. The ionic composition of endorheic athalassohaline lakes depends largely on the geology of the surrounding region, having no connection with an ocean (Yapiyev et al., 2017). Small saline water bodies tend to be relatively shallow, with an average depth of less than 3 m. Evaporation leads to the precipitation of dissolved salts and the formation of salt crusts and flats along the lake margins (Briere, 2000). The mineral composition of these deposits depends on the hydrochemistry of the lake. Salt flats create distinctive landscapes, with polygonal cracks, salt ridges, and colorful salt crystallization patterns (Lasser et al., 2020).
The endorheic athalassohaline wetlands of the Monegros desert are located in the Central Ebro Basin (NE Spain), which is formed of materials deposited in the upper Oligocene and lower Miocene. The wetlands comprise almost 100 scattered depressions ranging in size from <2 Ha to 239 Ha, excavated on gypsum-rich bedrock with a low‐permeability matrix, and they are closely connected with the two main aquifers in the area. The upper aquifer consists of lutitic limestone interleaved with layers of gypsum, whereas the lower aquifer contains gypsum and limestone with intercalations of lutite (Castañeda and Herrero, 2005). The hottest period of the year coincides with the dry season, from June to September, whereas the wet season extends from October to May (Menéndez-Serra et al., 2021). The water regime of the salt flats depends on direct precipitation, the limited surface runoff, and above all, on the contribution of groundwater, which varies according to the location and the duration of the wet cycle. Thus, some salt flats flood every year, whereas others do so rarely. Nevertheless, despite the marked water deficit, all seem to have a humid floor, even in summer, probably due to the formation of a salt crust, as observed in “La Salineta” or “La Playa” (playa lakes) in the Monegros zone (Castañeda and Herrero, 2005; Mees et al., 2011).
Planktonic microbial communities (water column biotope) have been studied in several lakes in this desert region, including the ephemeral La Muerte lagoon, which encompass a wide gradient of salinities and temperatures (Casamayor et al., 2013; Menéndez-Serra et al., 2021). Peak eukaryotic diversity was recorded at intermediate salinities, whereas bacterial and archaeal diversity was related to salt concentration. It was concluded that the extreme dynamism of these shallow ephemeral lakes, which undergo wet-dry cycles, might select for more versatile microbes capable of tolerating fluctuating osmotic conditions. However, the diversity and functionality of benthic microbial communities in the La Muerte lagoon has remained unexplored. Benthic habitats are known to harbor significant microbial biomass and diversity, which contributes to ecosystem processes (Bolhuis et al., 2014; Prieto-Barajas et al., 2018; Berlanga et al., 2022). Sediment biofilms represent active zones of organic matter mineralization and nutrient regeneration (Guerrero et al., 2002; Berlanga and Guerrero, 2016; Prieto-Barajas et al., 2018).
Nine DNA extraction samples were categorized into three groups, MS5 (January 2020), MS4 (September 2021), and MS6 (July 2022), and each sample was collected from a distinct zone within the lagoon to analyze the dynamic changes in population and potential function in biofilm–sediment samples biotope. To ascertain function, we studied the general pathways associated with energy production and carbohydrate metabolism. Additionally, comparison of the microbiota of biofilm–sediment with water samples could reflect whether there is a differentiation in the composition in the two biotopes of the lagoon taking into account that resuspension of microbes from sediments likely was one of the major contributions to the microbial composition of surface waters (Brandt et al., 2021). For study of water column microbiota, previously published data were used (bioproject PRJNA429605; Menéndez-Serra et al., 2021). It is important to note that the water column data were collected several years prior to the biofilm–sediment sampling conducted in this study. This temporal separation introduces potential limitations in directly comparing the two datasets. However, it is acknowledged that such comparisons can still offer valuable preliminary insights into the broader differences between these two biotopes.
Previous research suggests that the dynamic nature of Monegros lagoons, experiencing wet–dry cycles, likely favors adaptable microbes capable of enduring fluctuating osmotic conditions (Menéndez-Serra et al., 2021). The substrate supporting biofilm–sediments in the lagoon is homogeneous, primarily composed of gypsum (71.1%), hexahydrite (9.8%), calcite (6.9%), silicates (3.1%), and dolomite (2.7%) (Personal communication Esther Sanz, Universidad Complutense Madrid). Observations by the Spanish Meteorological Agency (AEMET) over the past 8 years indicate minimal impact from storms on the Monegros region, suggesting a stable environmental trend (source: www.aemet.es/es/conocermas/borrascas). With these observations, it can be hypothesized that biofilm assemblages may exhibit a dynamic yet stable composition over the years and locations analyzed, despite “normal” variations due to seasonal changes in environmental conditions.
Sediment samples were collected over 3 years at noon during different months from various locations in the endorheic La Muerte lagoon (area 0.3 km2) (Figure 1), which is situated at latitude 41.4013 and longitude −0.2614. A total of nine samples’ biofilm-sediment cores of 5-cm diameter were collected to a depth of 1 cm. Nine samples of DNA extractions were clustered into three groups: MS5 (January 2020, wet biofilm–sediment); MS4 (September 2021, dry biofilm–sediment); and MS6 (July 2022, dry biofilm–sediment). Each one was picked up in a different zone inside the lagoon (approximately 30 m, 90 m, and 135 m) to determine potential higher-scale heterogeneity (Osman et al., 2019) (Figure 1). Upon collection, the samples were immediately frozen in liquid nitrogen, transported to the laboratory, and stored at –80°C until further processing. The original intent of the study was to conduct sampling in January and July at three different points over the years 2020, 2021, and 2022. These months were chosen to represent extreme conditions: January with potential wet, cold, and low irradiance conditions, and July with dry, warm, and high irradiance conditions. Unfortunately, due to COVID-19 lockdowns, it was unable to collect samples in July 2020. Furthermore, between 2021 and 2022, mobility restrictions due to the pandemic limited also our ability to collect samples.
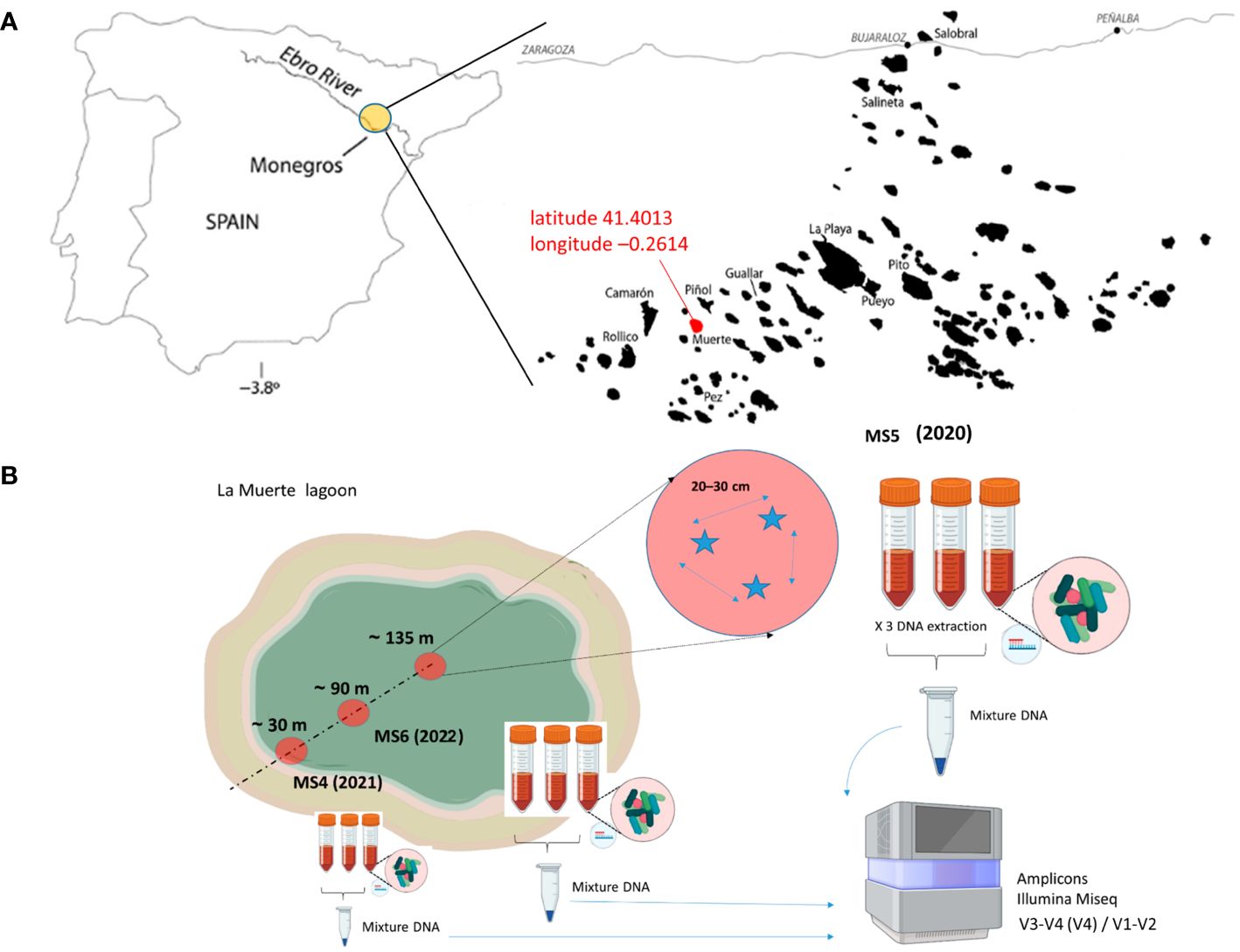
Figure 1 (A) Geographical location of the Monegros desert endorheic area within the semiarid Central Ebro Basin, NE Spain. Sampling campaign location at the La Muerte lagoon is shown in red. This map image is based on Casamayor et al. (2013). (B) Sampling strategy used.
To characterize biodiversity patterns and ecosystem functions in the water column biotope, sequences previously deposited in the GenBank database BioProject PRJNA429605 were utilized (Menéndez-Serra et al., 2021). To perform a robust comparative and integrative study, it is generally preferred to collect water and sediment samples in parallel under similar environmental conditions. Nevertheless, comparisons of non-parallel water and sediment samples can still provide meaningful insights, particularly when analyzing relative trends or patterns rather than absolute values, especially those related to a specific taxon Table 1 shows environmental characteristics of biofilm-sediment and water column samples.
DNA extraction was based on the protocol described by Biver and Vandenbol (2013) with modifications. One gram of biofilm-sediment sample was ground using liquid nitrogen, suspended in 8 mL of DNA extraction buffer (100 mM Tris–HCl pH 8, 100 mM sodium phosphate pH 8, 100 mM sodium EDTA pH 8, 1.5 M NaCl, and lysozyme 100 mg/mL) (Thermo Fisher Scientific, Madrid, Spain), and incubated at 37°C for 1 h. Incubation was continued, first at 65°C for 75 min after adding 50 µL proteinase K (10 mg/mL) (Thermo Fisher Scientific) and 180 µL SDS 20% (wt/vol) (Thermo Fisher Scientific), then for a further 75 min after adding 580 µL SDS 20% (wt/vol), and finally for 15 min after adding 750 µL CTAB 10% (wt/vol) (cetyltrimethylammonium bromide) (Thermo Fisher Scientific). The supernatant was collected after centrifugation at 9,800×g for 10 min and mixed with an equal volume of phenol–chloroform–isoamyl alcohol (25:24:1) (Thermo Fisher Scientific). After centrifugation at 9,800×g for 10 min, the upper phase was collected and mixed with an equal volume of chloroform–isoamyl alcohol (24:1) (Thermo Fisher Scientific). The aqueous phase was recovered by centrifugation (10 min, 9,800×g), mixed with one volume of 20% (wt/vol) polyethylene glycol 6000 and 1.6 M NaCl, and incubated for 2 h at room temperature in the dark. To pellet the DNA, samples were centrifuged at 9,800×g for 10 min. The supernatant was discarded, and the pellet was dissolved in TE (10 mM Tris–HCl, 1 mM sodium EDTA, pH 8) (Thermo Fisher Scientific).
The DNA concentration was estimated using BioDrop µLite (Biotech, Madrid, Spain). To determine the microbial community composition in biofilm-sediment samples, 16S rRNA gene sequencing (V3–V4 region for Bacteria and V1–V2 region for Archaea) was performed on the Illumina MiSeq platform [MiSeq Nanorun 500 (2 × 250)]. The primer pairs for region V3–V4 for Bacteria were 341F-CCTACGGGNGGCWGCAG and 805R-GACTACHVGGGTATCTAATCC (Varliero et al., 2023). The primer pairs for regions V1–V2 for Archaea were 1ArF-TCCGGTTGATCCYGCBRG and 280ArR-TCAGWNYCCNWCTCSRGG (Bahram et al., 2019). Library preparation for amplicon sequencing was performed following Illumina protocols. Briefly, 16S rRNA amplicon libraries for bacteria were generated by analyzing the V3–V4 variable regions of the prokaryotic 16S ribosomal RNA (rRNA) gene. In the first PCR step, specific primers with overhang adapters flanking the V3–V4 regions were used to amplify templates from DNA samples in a thermal cycler under the following conditions: 95°C for 3 min, 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and 72°C for 5 min. In the second PCR step, sequencing adapters and dual-index barcodes were added to the amplicons using the Nextera XT DNA Index Kit (Illumina, Paris, France) in a limited-cycle PCR. PCR conditions were 95°C for 3 min, 8 cycles of (95°C for 30 s, 55°C for 30 s, and 72°C for 30 s), and 72°C for 5 min. Subsequently, paired reads were generated on the MiSeq platform using MiSeq v2 reagents (Illumina), with the ends of each read overlapped to produce high-quality reads of the V3–V4 regions. Library preparation and sequencing were performed using the Illumina MiSeq platform by the Genomic and Bioinformatic Service of the Autonomous University of Barcelona.
Shotgun metagenomics analysis was performed on the biofilm-sediment sample collected at location MS4 (corresponding to three pooled samples) using the Illumina MiSeq_RunMicro300 cycles (2 × 150) platform by the Genomic and Bioinformatic Service of the Autonomous University of Barcelona.
Sequence reads were processed by the amplicon analysis pipeline of the SILVA project (SILVAngs 1.4) (Quast et al., 2013). Reads shorter than 50 aligned nucleotides and reads with more than 2% of ambiguities or 2% of homopolymers were excluded from further processing. After these initial steps of quality control, identical reads were identified (dereplication), the unique reads were clustered (OTUs), and the reference read of each OTU was classified at 0.97% of similarity. Dereplication and clustering were done using VSEARCH (version 2.17.0) (Rognes et al., 2016). A rarefaction curve was generated to determine the relationship between the number of OTUs and the number of sequences (Supplementary Figure S1). The classification was performed by BLASTn (2.11.0) with standard settings using the non-redundant version of the SILVA SSU Ref dataset reference 138.1 (http://www.arb-silva.de).
Before using the PICRUSt tool, it was necessary to construct an OTU table that contained all the reads for each sample. To obtain the OTU table, the QIIME program was used (Caporaso et al., 2010). Singletons were removed, and the data were rarified based on the number of reads from each sample. The resulting OTU table was used to calculate the alpha diversity and the core OTUs (Caporaso et al., 2010) and to predict functionality based on the database of KEGG Orthologs (KOs) using the PICRUSt script predict_metagenomes.py (Langille et al., 2013; Douglas et al., 2020). Functional profiles of microbial communities based on their taxonomic composition PICRUSt do not adequately represent extreme environments due to abundant unknown microorganisms, but this method can aid in understanding aids in elucidating potential metabolic pathways and ecosystem functions, albeit with caution in extrapolating to extreme environments (Sun et al., 2020). We used PICRUSt to compare potential functionality in biofilm-sediment and water column biotopes.
Shotgun metagenomics was performed for the MS4 sample. Paired-end fastq files containing sequences generated by Illumina MiSeq_RunMicro300 cycles were assembled using the MEGAHIT tool (Li et al., 2015) in Galaxy pipeline Version 1.2.9 [https://usegalaxy.eu/] (Hiltemann et al., 2023). The assembled sequences were submitted to the DOE-JGI Metagenome Annotation Pipeline (MAP v.4) (https://gold.jgi.doe.gov/) (Huntemann et al., 2016), using the Velvet algorithm package for genomic assembly and short read sequencing alignments. Protein-coding genes were identified using a consensus of four different ab initio gene prediction tools: prokaryotic GeneMark.hmm (v.2.8), MetaGeneAnnotator (v. Aug 2008), Prodigal (v. 2.6.2), and FragGeneScan. Protein-coding genes with translations shorter than 32 amino acids were deleted. Assignment was done if 90% of the KO gene sequence was covered by the alignment (Huntemann et al., 2016).
OTUs were defined as groups of sequences with at least 97% similarity. Reference OTU fasta sequences of approximately 250-bp length were obtained using the Silva database (https://ngs.arb-silva.de/silvangs/) (Quast et al., 2013) to reveal ecotypes by cluster analysis. OTUs were aligned using MEGA-X (Molecular Evolutionary Genetics Analysis) (Kumar et al., 2018). A Neighbor-Joining tree using the Jukes–Cantor nucleotide substitution model and 1,000 bootstrap replications was generated with the MEGA-X program. Each OTU within a clade was considered an ecotype of the same species (Cardoso et al., 2019).
Bacterial communities and their potential correlations with environmental factors were determined using non-metric multidimensional scaling (NMDS) analysis and canonical correspondence analysis (CCA) using R with the package vegan v2.6–6 (https://github.com/vegandevs/vegan).
Ecological diversity is considered a function of both the number of different types (richness or variety) and the relative importance of individual elements among these types (evenness or equitability). Estimated by the Shannon index, diversity was 5.25 for the MS4 sample, 5.35 for the MS5 sample, and 5.05 for the MS6 sample. Analysis of the 16S rRNA gene and shotgun metagenomic sequencing revealed 10 bacterial phyla with a proportion higher than 1%. The most abundant phyla were Actinobacteria, Bacteroidota, Cyanobacteria, and Pseudomonadota (Figure 2). The classes Pseudomonadota Alpha-, Gamma-, and Deltaproteobacteria displayed ascending relative abundance. Differences in phylum-level proportions were observed between amplicon and shotgun metagenomics sequencing, particularly within Actinobacteria, Bacteroidota, Cyanobacteriota, and Pseudomonadota–Alphaproteobacteria. Variations in taxonomic profiling associated with methodology have been previously reported (Fierer et al., 2012; Berlangaet al., 2017).
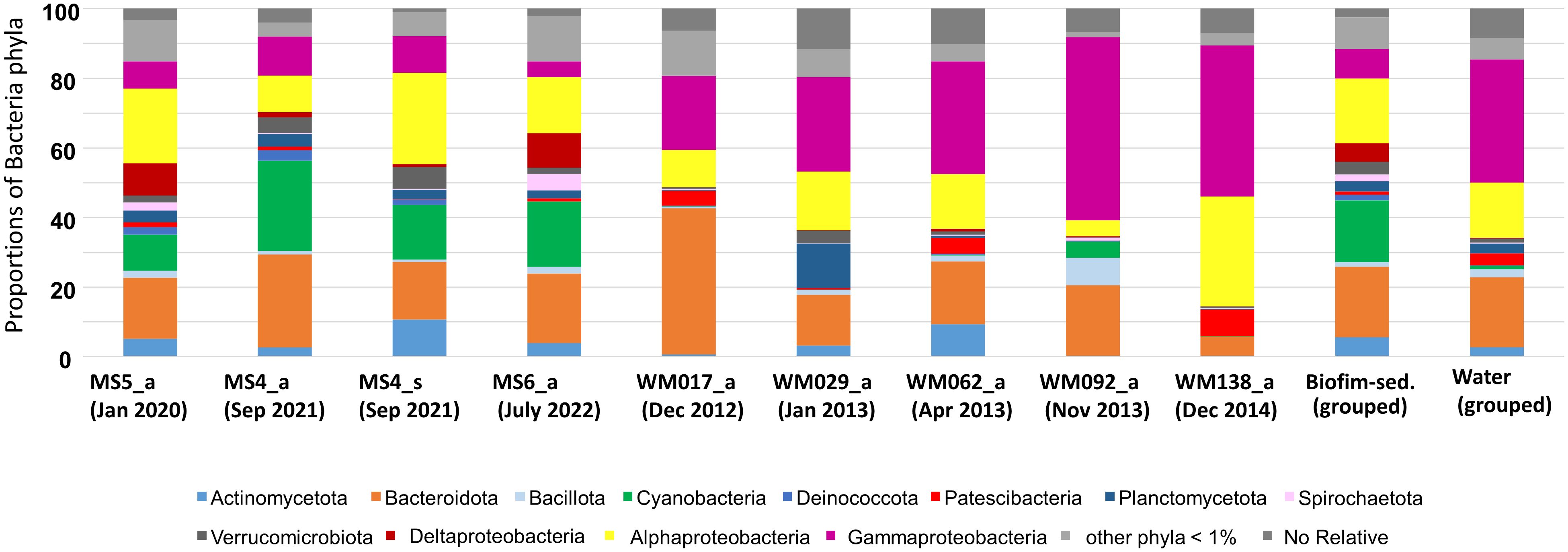
Figure 2 Proportions of bacterial phyla in biofilm-sediment samples, in water column, grouped biofilm-sediment and grouped water column samples. Data were obtained by amplicon and shotgun metagenomics sequencing. MS represents sampling sites for biofilm-sediment in the La Muerte lagoon (each sampling site corresponds to three pooled subsamples). The biofilm-sediment group corresponds to samples MS4 (Sep. 2021), MS5 (Jan 2020), and MS6 (July 2022). The water column group corresponds to samples MW017 (Dec 2012), MW029 (Jan 2013), MW062 (Apr 2013), MW092 (Nov 2013), and MW138 (Dec 2014). The water column samples were analyzed using amplicon sequencing data from BioProject PRJNA429605.
In the family-level analysis, over 40 families with more than 1% relative abundance were found in biofilm–sediment samples. Key families include Ilumatobacteraceae and Nitriliruptoraceae for Actinobacteria, Microcystaceae for Cyanobacteriota, Beijerinckiaceae and Rhodobacteraceae for Alphaproteobacteria, and Marinobacteraceae for Gammaproteobacteria across all samples (Supplementary Figure S2). Non-metric multidimensional scaling (NMDS) showed close similarities among biofilm–sediment samples (Figure 3), regardless of dry–wet state or temperature variations (ranging from 10°C for MS5 to nearly 30°C in MS4 and MS6). Despite the limited sample size, the microbial composition in biofilm–sediment showed little variation across diverse environmental conditions of temperature, light exposure, and potentially salinity. Sample MS4 showed slight divergence from other sediment samples, likely due to its proximity to the lagoon’s edge, although this deviation did not reach statistical significance.
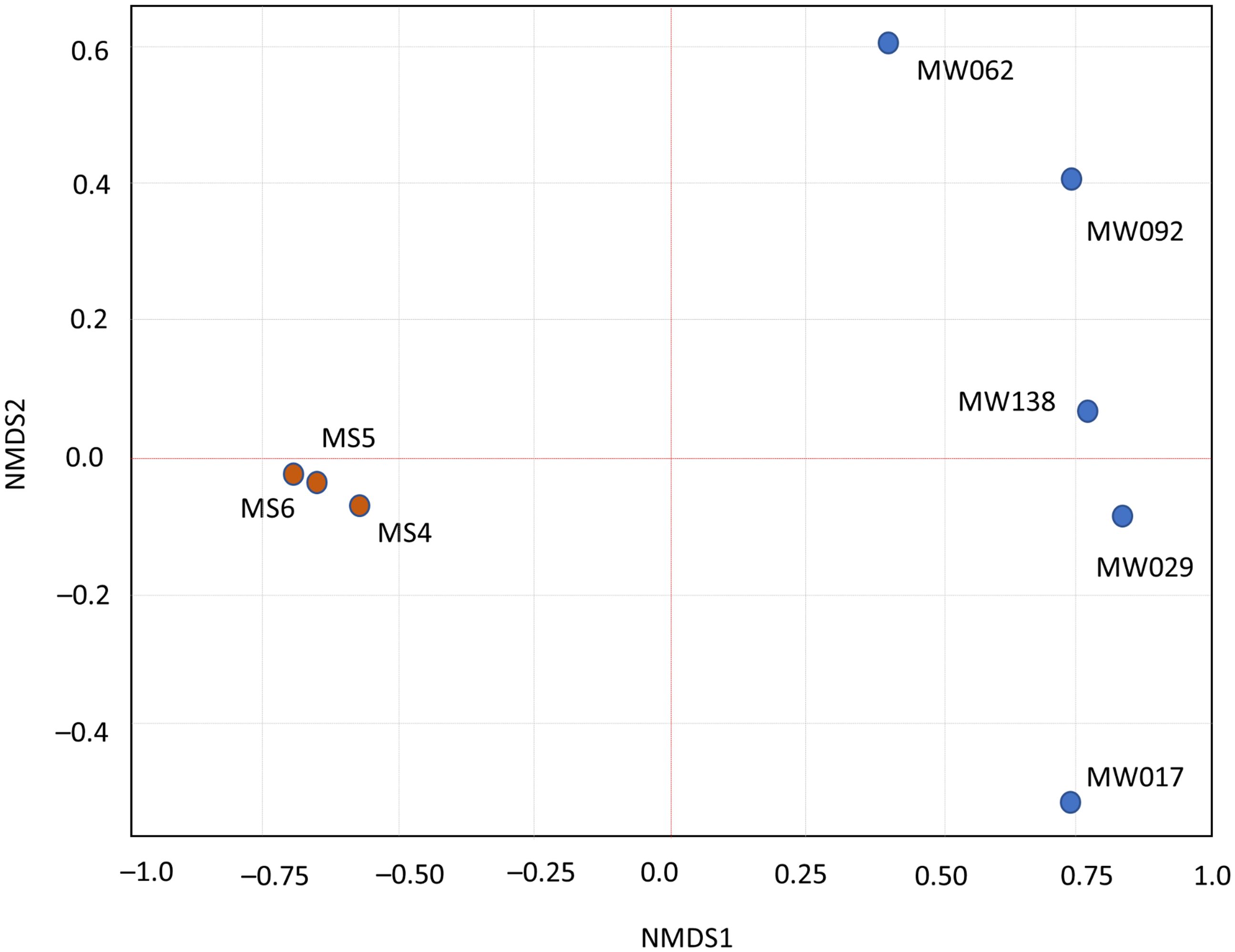
Figure 3 Non-metric multidimensional scaling (NMDS) analysis of microbial community data. This figure presents an NMDS analysis of the microbial communities across biofilm–sediment and column water samples (MS4, MS5, MS6, MW17, MW29, MW62, MW92, and MW138), based on operational taxonomic units (OTUs). Bray–Curtis dissimilarity was employed to measure the differences in community composition. Each point represents a distinct sample, with the spatial arrangement reflecting the dissimilarity in microbial composition—closer points indicate more similar communities. Stress factor for NMDS was 0.1046.
Microorganisms must adapt to the seasonal fluctuations of their environment in order to survive. Their physiological plasticity or tolerance to changing conditions is reflected by the presence of new and permanent OTUs. Co-occurring Cyanobacteria ecotypes have been described previously (Sohm et al., 2016; Wörmer et al., 2020; Thompson et al., 2021; Berlanga et al., 2022) also observed in the phylum Spirochaeota (Berlanga et al., 2008). Using reference OTUs from the Silva database, the majority of OTUs in each sample were not found in other samples. In the case of Cyanobacteria, especially the genera Coleofasciculus, no clear clusters were observed based on environmental conditions such as the cold–wet and hot–dry periods (Supplementary Figure S3). In the case of Spirochaeota we also did not observe clusters of “ecotypes” based on dry-hot and wet-cold period (Supplementary Figure S4).
Analysis of archaeal prokaryotes using the 16S rRNA gene revealed three major phyla in the biofilm-sediments: Halobacteriota (previously reported as members of Euryarchaeota), Nanorarchaeota, and Nanohaloarchaeota (members of the proposed superphyla DPANN) (Rinke et al., 2021). The proportion of the phyla Aenigmarchaeota, Thermoplasmatota, and Euryarchaeota–Methanobacteriaceae was less than 0.5%. Shotgun analysis of biofilm-sediment samples revealed Halobacteriota to be the Archaea predominant phylum, representing 99% of the detected relative sequences of the Archaea domain (Figure 4).
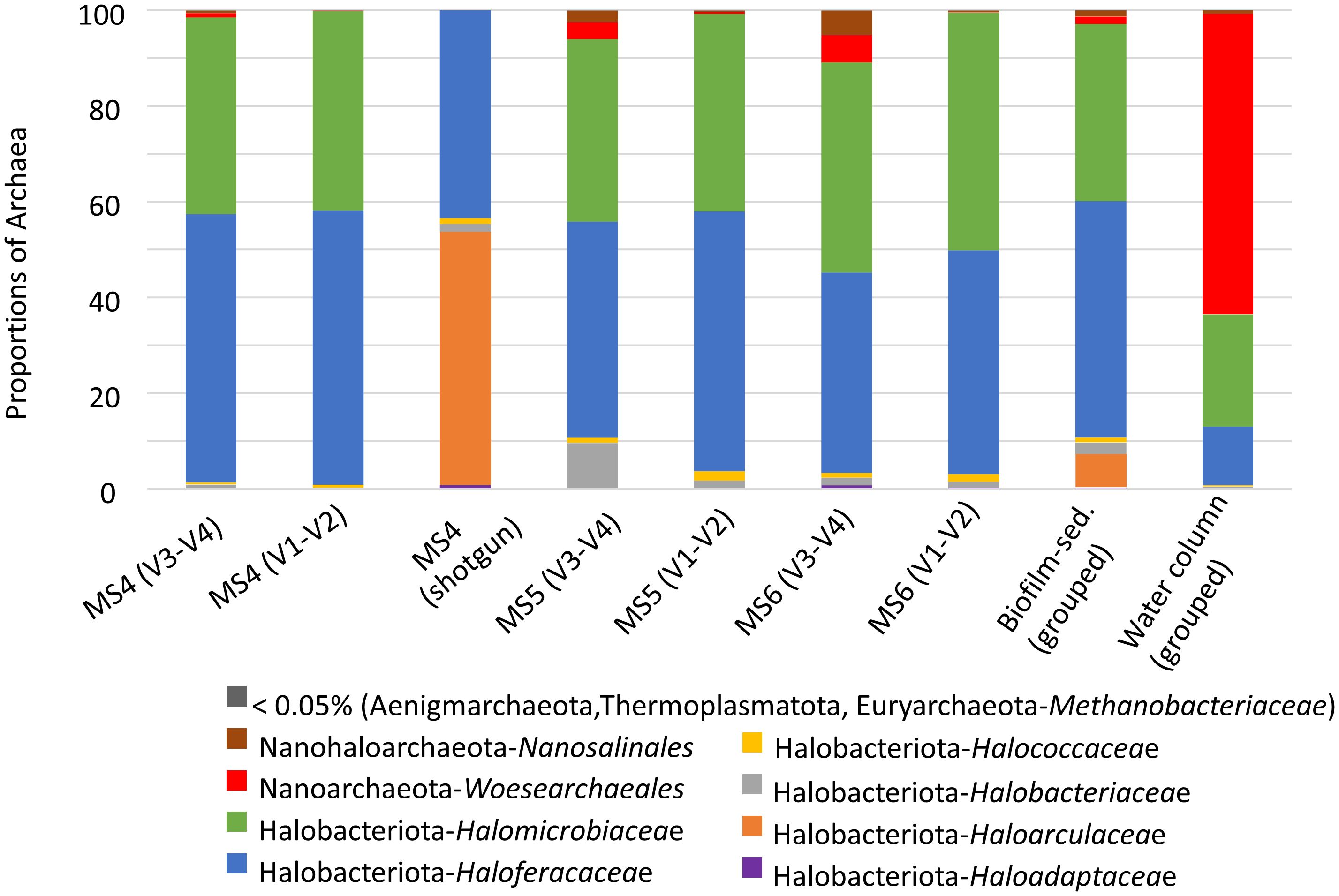
Figure 4 Proportions of archaeal phyla/families in biofilm-sediment and water column samples. MS represents sampling sites for biofilm-sediments of the La Muerte lagoon (each sampling site corresponds to three pooled subsamples). For Archaea, two 16S rRNA regions were studied (V1–V2 and V3–V4) and shotgun metagenomics in biofilm-sediment samples. The biofilm-sediment group corresponds to samples MS4 (Sep. 2021), MS5 (Jan 2020), and MS6 (July 2022). The water column group corresponds to samples MW017, MW029, MW062, MW092, and MW138. The water column samples were analyzed using amplicon sequencing data from BioProject PRJNA429605.
The core phyla in the biofilm-sediments were Halobacteria (11 OTUs), Actinobacteriota (11 OTUs), Rhodothermaeota (a new phylum previously included within the Bacteroidota) (11 OTUs), Bacteroidota (27 OTUs), Cyanobacteriota (11), Alphaproteobacteria (Rhodobacteriaceae, Rhodospirillaceae, and others) (55 OTUs), Gammaproteobacteria (20 OTUs), Deltaproteobacteria (12 OTUs), Verrucomicrobiota (11 OTUs), Planctomycetota (7 OTUs), the super group candidate phyla radiation [CRP], (5 OTUs), Spirochaetota (5 OTUs), Clostridia (7 OTUs), and Chloroflexi (1 OTU).
The functional properties of the microbial communities were analyzed using PICRUSt and shotgun metagenomics (Supplementary Figure S5). The biological processes essential to sustain prokaryotic life in the environment, such as replication, repair, transcription, and translation, were of similar abundance in both biotopes (both methodologies gave similar results). Shotgun metagenomics showed greater proportion of genes related to studied pathways with respect to PICRUSt analysis. Some metabolic pathways in the microbial community (PICRUSt results) were lightly more active in the hot–dry period than in the cold–wet period but, using the t-Wilcoxon statistics, showed no significant differences (p > 0.05) (Supplementary Figure S5). In microbial assemblages, the temperature can influence selection and adaptation and can determine the generation time of microorganisms, which is indirectly related to metabolic rates (Weiser et al., 2019). An increase in the relative abundance of KOs associated with glycolysis and the tricarboxylic acid cycle in the hot-dry period could be connected with enhanced oxidative phosphorylation in this period.
Regarding carbon sources in the biofilm-sediment biotope, several predicted KOs related to the Calvin–Benson cycle (e.g. K01601, K01602), Wood–Ljungdahl pathway (e.g., K14138, K00198, K00192), and 3-hydroxypropionate bicycle (e.g. K14468, K08691, K01961) were detected, but none involved in the reverse tricarboxylic acid cycle (K15234, K15230, K15231). The Calvin–Benson cycle was the predominant pathway for carbon fixation in La Muerte lagoon (Figure 5).
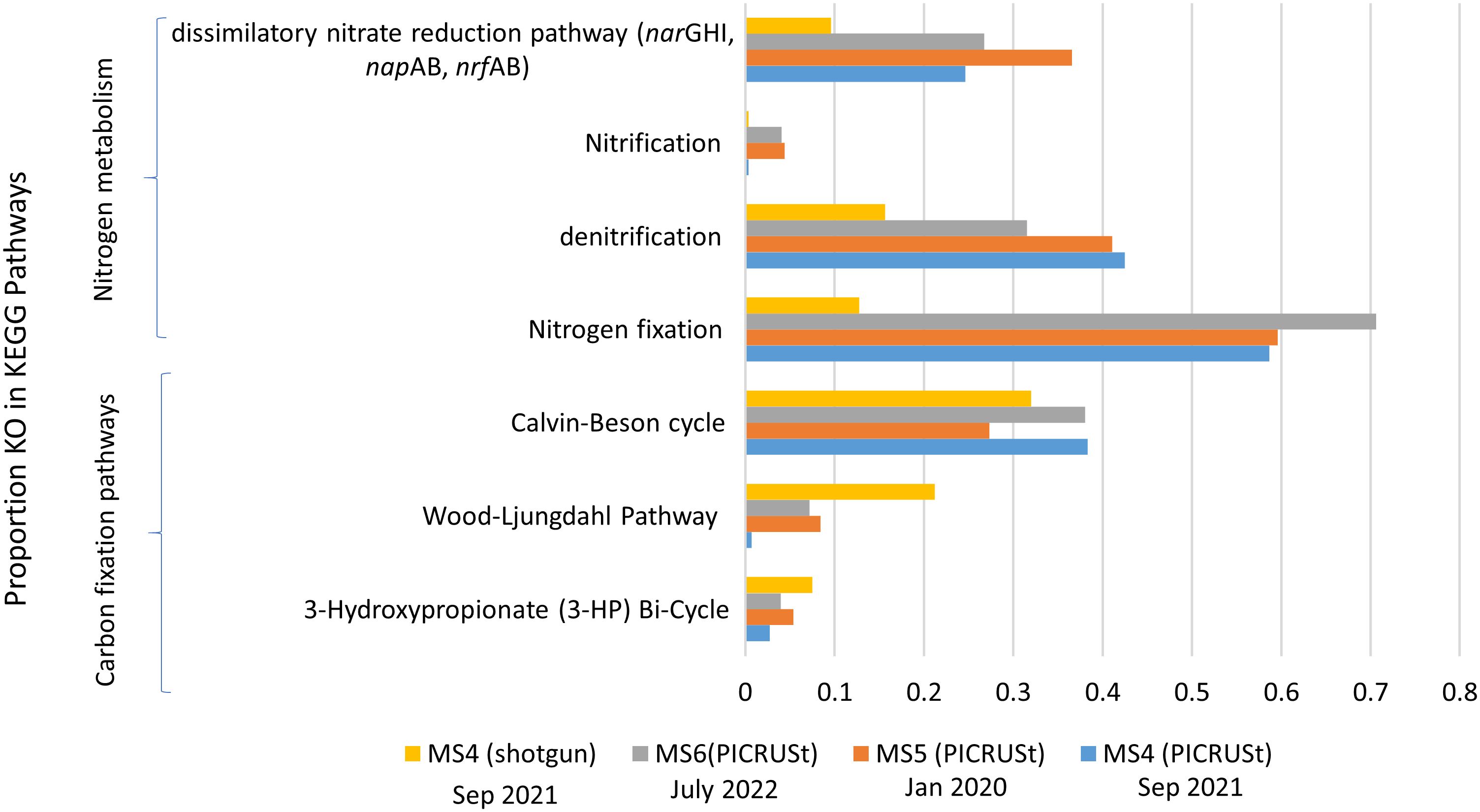
Figure 5 Proportion of KEGG pathways related to “energy metabolism,” such as nitrogen metabolism and carbon fixation pathways, in biofilm-sediment samples, MS4 (Sep. 2021), MS5 (Jan 2020), and MS6 (July 2022). Data were obtained by PICRUSt and shotgun metagenomics.
Relative abundance of KOs for β-glucosidase vs. phosphatase (GLU: PHOS), β-xylosidase vs. β-glucosidase (XYL: GLU), and β-glucosidase vs. phenol oxidase (GLU: PHEN) was calculated. Ratios of 1:1 were obtained for GLU: PHOS (by PICRUSt and shotgun metagenomics), suggesting an equilibrium between available organic matter, microbial biomass, and nutrient assimilation (Pastor et al., 2019). The XYL: GLU ratio was 0.158 (PICRUSt) and 0.16 (shotgun), suggesting that allochthonous plant material was not used in these communities (Pastor et al., 2019). Finally, the ratio of GLU: PHEN was 5.8 (PICRUSt) or 4.33 (shotgun), potentially indicating a greater use of recalcitrant organic matter in the former (Pastor et al., 2019).
The major pathway involving nitrogen was nitrogen fixation (e.g., K02586, K02591, K02588, K00531) providing an important input of nitrogen into the ecosystem. Nitrification (e.g., K10535, K10944, K10945, and K10946), which is the oxidation of ammonium to nitrite, was the lower represented pathway on biofilm–sediment samples. Denitrification (e.g., K00374, K02567, K02568, K00368, K02305, K00376) seemed to exhibit higher activity in dry-hot biofilm samples than wet-cold samples (Figure 5).
It was decided to group the samples from both biofilm-sediment and water column to facilitate their comparison. A comparative analysis revealed a higher proportion of the following phyla in the biofilm-sediment (grouped samples) biotope compared with the water column (grouped samples) biotope: Bacillota, Cyanobacteriota, Pseudomonadota–Deltaproteobacteria, Spirochaetota, and Verrucomicrobiota. Conversely, the water column had a higher proportion of Patescibacteria, Pseudomonadota–Alphaproteobacteria, and especially Pseudomonadota–Gammaproteobacteria (Figure 2).
In biofilm-sediment samples, the Cyanobacteriota genera observed were Coleofasciculus (10.7% relative abundance), Cyanothece (43%), and Phormidium (12.1%) whereas Leptolyngbya (5.6%) and Nodosilinea (8.3%) were less abundant. In water samples, Cyanobacteriota represented 0.5% of the total Bacteria, the major Cyanobacteriota genera detected being Nodosilinea (18.6%) and Spirulina (1.4%). The most abundant genera from Alphaproteobacteria in the biofilm-sediment samples were Salinarimonas, Tropicimonas, Rhodovibrio, Roseobacter, and Roseovarius, whereas in the water column, they were Marivita, Porphyrobacter, Pseudoruegeria, and Sphingomonas. Nevertheless, the high proportion of Rhodobacterales in the water column, not classified at the genus level, should be emphasized. Regarding Gammaproteobacteria, Marinobacter was the major genus when considering both biotopes and Halochromatium was well represented in the biofilm-sediment samples, whereas Methylophaga was the predominant genus observed in the water samples.
Haloferacaceae and Halomicrobiaceae were the most representative families from Halobacteriota (Archaea prokaryotes) in both biofilm-sediment and water column biotopes. Nanoarchaeota–Woesearchaeales was also well represented in water column samples (Figure 4).
Shannon and Simpson indices revealed that biofilm-sediments were taxonomically more diverse than the water column. The Shannon index of bacterial diversity was found to be 5.23 in biofilm-sediments (pooled samples) compared with 4.27 in the water column (pooled samples). Similarly, the Simpson index for bacterial diversity was 0.02 in biofilm-sediments and 0.08 in water column samples. For Archaea, the Shannon index was 4.31 versus 3.99 and the Simpson index was 0.062 versus 0.078 for the biofilm-sediments and water column, respectively.
Analysis of the core community of both biotopes revealed the following composition: Halobacteriota (2 OTUs) for Archaea, Actinobacteriota (3 OTUs), Deinococcales (1 OTUs), Rhodothermales (3 OTUs), Bacteroidota (4 OTUs), Cyanobacteriota (2 OTUs), Alphaproteobacteria (Rhodobacteriaceae, Rhodospirillaceae, others) (12 OTUs), Gammaproteobacteria (2 OTUs), and Verrucomicrobiota (2 OTUs) for Bacteria.
The two biotopes contained distinct but related prokaryote communities (Figure 3). The environmental factors could have important influence on the community composition and diversity (Shen et al., 2016). To find the potential correlations between the distribution of bacterial assemblages from the two biotopes and the environmental factors of the examined habitat, CCA test was conducted based on the recovered bacterial 16S rRNA gene sequences and the measured physicochemical parameters (figure not showed). None of the tested environmental factors (temperature, salinity, irradiance) showed a statistically significant effect on the dependent variable at typical significance levels (ANOVA p < 0.05). The high p-values in all cases suggest that variations in these factors did not significantly explain the variations in the dependent variable measured in this study. This indicates that these factors truly had no significant effects.
The analysis of KEGG pathways using PICRUSt revealed some differences in energy and carbohydrate metabolism between biofilm-sediment and water samples, although without statistical significance (Figure 6A). However, when looking at the overall metabolism, the aggregate of the small differences in individual functions resulted in a significant difference between the two biotopes (t-Wilcoxon p < 0.05) (Figure 6A), reflecting higher functionality in sediments versus the water column biotope. Noticeably, pathways related to DNA repair, two-component regulatory systems, ABC transporters and bacterial chemotaxis could reflect higher capability of population in the biofilm-sediment biotope to adapt to different environmental or stress conditions. Taxon contributions to carbohydrate and energy metabolisms differed between the two biotopes. Energy metabolism in biofilm-sediments was performed primarily by Gammaproteobacteria, Alphaproteobacteria, and Cyanobacteriota, whereas in the water column it was dominated by Alphaproteobacteria. Carbohydrate metabolism in biofilm-sediment was mainly driven by Gammaproteobacteria, Alphaproteobacteria, and Bacteroidota, and in water samples, by Alphaproteobacteria, Gammaproteobacteria, and Bacteroidota (Figure 6B). Similar functions were performed by different taxa.
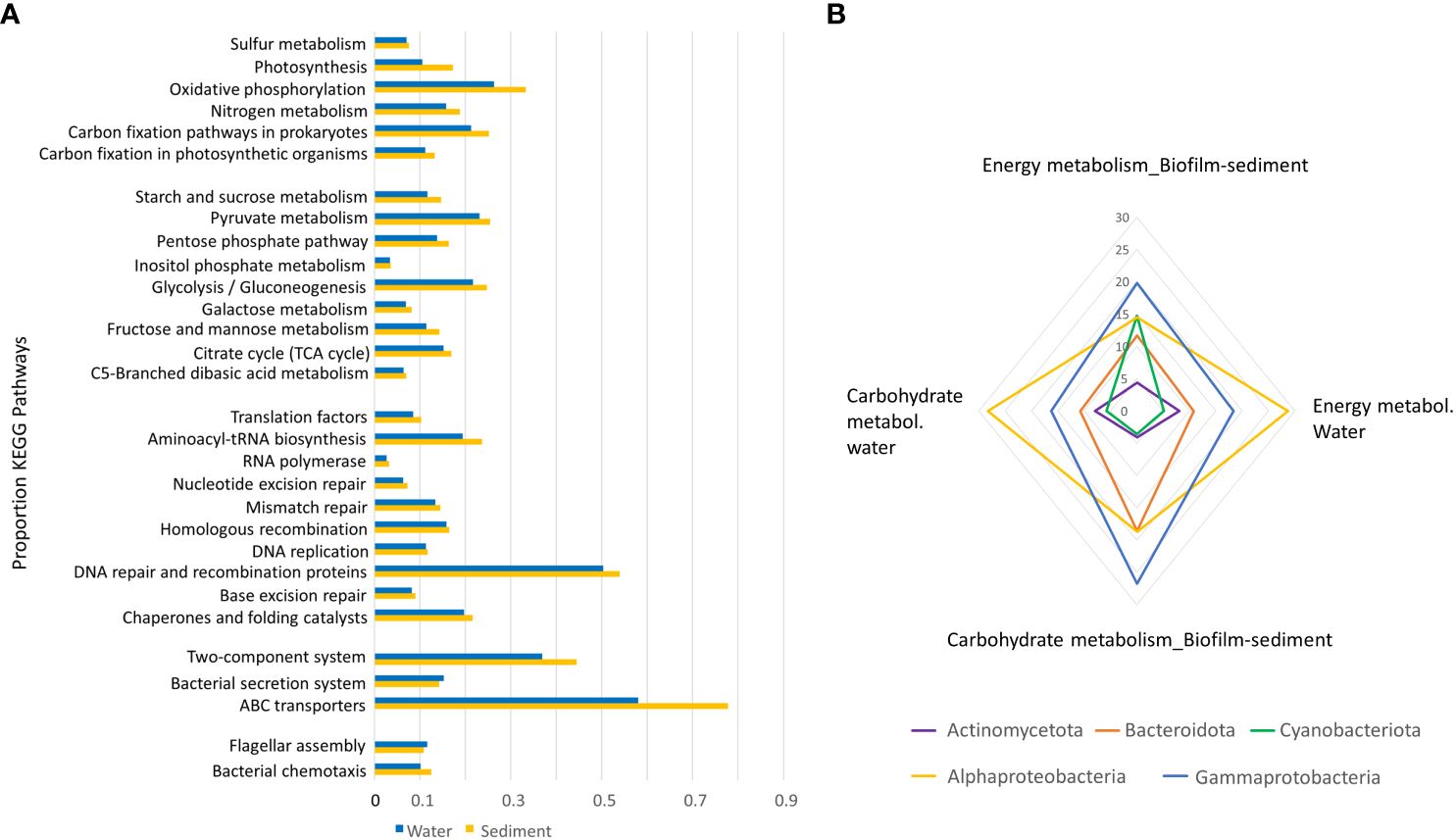
Figure 6 (A) Relative abundances of KEGG Orthologs (KOs) related to pathways of “energy metabolism,” “carbohydrate metabolism,” “Genetic Information Processing,” “Environmental Information Processing,” and “Cellular Processes” in grouped biofilm–sediment and grouped water column samples from the La Muerte lagoon. Data obtained by PICRUSt. (B) The radial graphic displays the relative contributions of major phyla to carbohydrate and energy metabolisms from grouped biofilm–sediment samples and grouped water column samples.
The unique characteristics of hypersaline environments have significant implications for the biodiversity of their microbial communities. The Monegros Desert wetlands in Spain are an example of an athalassohaline hypersaline environment where the salinity of groundwater is highly variable, depending on the direct precipitation and limited surface runoff. Microbial organisms in such endorheic lakes are also exposed to other extreme conditions, such as low oxygen concentration, alkalinity, high UV irradiation, and extreme temperatures.
Based on shotgun metagenomics, the microbial profile of the biofilm-sediment in sample MS4 was dominated by Bacteria (82%), followed by Archaea (17%) and Eukarya (0.1%) (Figures 2, 4). The proportion of Bacteria and Archaea may depend on the degree of salinity (Menéndez-Serra et al., 2020, 2021; Guan et al., 2023).
Bacterial communities showed significant differences (t-Wilcoxon p < 0.05) between sediment and water column samples, whereas no such differences were seen in Archaea. Despite this, the prokaryotic communities found in the two biotopes were distinct but related (Figures 2, 3). This temporal separation introduces potential limitations in directly comparing the two datasets, as environmental conditions and community compositions may have shifted over time. However, such comparisons can still offer valuable preliminary insights into the broader differences between these two biotopes within the La Muerte lagoon ecosystem.
Microorganisms present in surface waters are likely derived from the resuspension of sediment-associated microorganisms, as evidenced by the shared core community between these biotopes (Brandt et al., 2021). However, only certain populations can adapt to planktonic conditions. Oxygen concentration may play a significant role in differentiating sediment and water column diversity, community structure, and metabolic functions (Chen et al., 2022). The deeper layers of the biofilm–sediment biotope may be anaerobic. Studies in the Salada de la Playa (Monegros desert) revealed notable day/night oxygen fluctuations, with daytime photosynthesis slightly oxygenating interstitial waters (Boadella et al., 2023). Similar patterns were observed in microbial mats (Wieland and Kühl, 2006). Temperature influences physicochemical processes such as solute diffusion and solubility across the biofilm–sediment-water interface. For instance, between 10°C and 25°C, the molecular O2 diffusion coefficient increases by 54%, whereas O2 solubility decreases by 24% (Wieland and Kühl, 2006). Salinity also affects O2 diffusion (Meier et al., 2021), and a lower proportion of Cyanobacteriota observed on water column may contribute to “semiaerobic” conditions in the water column. Stand out Rhodobacterales, a prominent group in the water column, includes purple non-sulfur bacteria capable of photoautotrophic growth under anaerobic conditions and photoheterotrophy under semiaerobic conditions, thus contributing to primary production (Pohlner et al., 2019). The candidate phyla radiation (CPR) group was more abundant in the water column (5% relative abundance) than in the biofilm–sediment biotope (1%). This group includes Saccharibacteria (TM7), Parcubacteria (OD1), and Ojkabacteria (WS6), which form part of the core microbiota in both compartments. CPR members are characterized by small cell sizes, streamlined genomes, and potentially anaerobic lifestyles, often surviving in episymbiotic relationships with bacteria and archaea (Ji et al., 2022). Despite sharing a fermentative lifestyle, CPR bacteria are metabolically diverse and play crucial roles in biogeochemical cycles, including denitrification in the nitrogen cycle (Ji et al., 2022). Their interactions with other organisms are essential in shaping the natural microbiome function. The physical and chemical conditions of sediment versus water biotopes may create distinct environments that can favor different cyanobacteria adapted to those specific conditions. Coleofasciculus, Phormidium, and Leptolyngbya, etc., are all adapted to thrive in sediment environments that can attach to surfaces and form biofilms. The mechanism behind the spatial ordering of cyanobacterial filaments is unclear but may involve photosensory motile behavior or geometric constraints related to filament reproduction (Fenchel and Kühl, 2000; Wierzchos et al., 2006). Filamentous cyanobacteria could have difficulty maintaining buoyancy and would have a tendency to settle.
The biodiversity of hypersaline lakes is shaped by combination of extremes. Microbial communities drive biogeochemical cycling in endorheic lakes (Bryanskaya et al., 2022; Guan et al., 2023). Core populations in the biofilm-sediment biotope were maintained through years in this biotope. It seemed that the biofilm-sediment community was stable and the extreme seasonal fluctuations (wet-cold or dry-warm periods) in the La Muerte lagoon do not substantially disrupt the well-adapted microbial community.
Biofilm-sediments contribute to primary production through phototroph and autotroph populations, thus participating in nutrient cycling and providing diverse microbial habitats. Labor division in these communities is primarily achieved by interactions between taxonomic groups occupying specific niches based on their physiology and functions. Thus, the presence of one group facilitates the survival of others (Van Gestel et al., 2015; Berlanga et al., 2022).
Carbon fixation enables inorganic carbon to be converted to organic matter such as carbohydrates, which is subsequently metabolized by heterotrophic microorganisms. Several autotrophic carbon fixation pathways are described in prokaryotes (Berg, 2011; Fang et al., 2022). The La Muerte lagoon may not depend on external organic matter. Primary production involving the fixation of inorganic carbon (CO2) was found to be performed mainly by Cyanobacteriota and purple non-sulfur Alphaproteobacteria through the Calvin–Benson cycle (Figure 5). Although detected, purple sulfur Gammaproteobacteria (also fixing CO2 via the Calvin–Benson cycle) were scarce in biofilm-sediments. Chloroflexi may use the 3-hydroxypropionate bicycle, whereas the chemotrophic Spirochaetota can employ the Wood–Ljungdahl pathway. In the water column, phototrophs, purple bacteria, and Cyanobacteriota also fixed carbon but were far less abundant than in biofilm-sediment biotope. The Calvin–Benson cycle is the predominant mechanism for carbon fixation in both sediment and water biotope, whereas the Wood–Ljungdahl pathway is typical of microbial mats (Berlanga et al., 2017; Gutiérrez-Preciado et al., 2018; Berlanga et al., 2022). Debris from bacterial predation by the Deltaproteobacteria families Bdellovibrionaceae and Bradymonadaceae, and viruses can constitute another carbon source for heterotrophs (Mu et al., 2020).
According to the relative abundance of KOs for several enzymes related to organic carbon processing, such as β-glucosidase, β-xylosidase, and phenol oxidase, allochthonous plant material was not used in these communities and “simple” polysaccharides released by photosynthetic microorganisms may be the principal source of organic carbon used by chemoheterotrophs. The phenol oxidase values obtained seemed to be related to the low presence of recalcitrant organic matter (or organic matter with low aromaticity) in the La Muerte lagoon. Similar findings have been described in La Playa lagoon, another endorheic water body of the Monegros Desert (Boadella et al., 2023).
Populations able to fix nitrogen (diazotrophs) in the biofilm-sediments consisted of Alphaproteobacteria, Cyanobacteriota, Deltaproteobacteria, and Spirochaetota. Denitrification was mainly associated with Rhodobacterales (Alphaproteobacteria), and, to a lesser extent, Deltaproteobacteria, Gammaproteobacteria (Alteromonadales, Pseudomonadales), and Bacteroidota (Flavobacteriales). All these taxa have been previously described as able to perform both anaerobic and aerobic denitrification (Ji et al., 2015; Coban et al., 2021; Hao et al., 2022) (Figure 5). Nitrification was the least represented function in the La Muerte lagoon samples. Although ammonia monooxygenase (amoA), a key enzyme in nitrification, is present in both ammonia-oxidizing Archaea and Bacteria, it was not observed in the studied samples. The hzoA/hzoB genes are ubiquitous and highly expressed in anammox bacteria (Planctomycetota), which perform anaerobic ammonium oxidation, but they were not detected by the PICRUSt tool and shotgun metagenomic, despite the presence of the phylum in the biofilm-sediment.
A comparison between the two biotypes shows differences in composition (Figure 3) and function (Figure 6). The biofilm–sediment exhibited greater diversity than the water column which could be related to a greater proportion of KEGG metabolic pathways in the case of the biofilm-sediment (Figure 6). The greater diversity of bacterial taxa in biofilm-sediments support the notion that biofilm provides a protective environment for bacteria to grow and survive, exchange nutrients, and adapt to harsh environments (Guerrero and Berlanga, 2016; Boadella et al., 2023).
The work provides an initial exploration of the microbial communities present in the sediment biofilms of La Muerte Lagoon. However, the conclusions drawn were based on limited samples collected and may not fully represent the spatial and temporal variations within these communities. Interestingly, the taxonomic composition of the biofilm-sediment samples did not appear to change significantly (t-Wilcoxon test) due to changes in location from the La Muerte lagoon, suggesting no spatial heterogeneity (at distances of 135 m, 90 m, and 50 m from the lagoon’s border). Additionally, the microbial communities did not notably fluctuate over different periods despite varying environmental conditions, such as changes in temperature and irradiance. Future research requires a more comprehensive sampling strategy to better characterize the microbial communities in La Muerte Lagoon, such as greater spatial and temporal coverage, including samples collected during both dry and wet seasons from various locations within the lagoon.
This study provides initial insights into the distinct yet interconnected microbial communities inhabiting the biofilm-sediments and water column of the extreme endorheic La Muerte lagoon. However, it is important to note that the limited sample size in this study imposes constraints on the generalizability of the findings. The observed differences between the water column and sediment–biofilm communities should be interpreted as preliminary observations that warrant further investigation with a more comprehensive sampling strategy. As hypersaline lakes and their surrounding environments are being negatively affected by various factors, including undesired freshening, sewage effluents, pollution due to agricultural runoff, urbanization, and global climate change (Varun and Mormile, 2017), it is of paramount importance to understand their unique characteristics and challenges. The results of this study contribute to a better understanding of the microbial ecology of extreme hypersaline environments and the roles and interactions of prokaryotes in these habitats while highlighting the need for further research to accurately characterize these globally threatened ecosystems.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA962491. Shotgun data: JGJ database Gold pj Ga0557956 (https://gold.jgi.doe.gov/analysis_project?id=Ga0557956).
MB: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. PP: Methodology, Writing – review & editing. ABl: Formal analysis, Methodology, Writing – review & editing. RB-F: Data curation, Formal analysis, Writing – review & editing. RG: Conceptualization, Supervision, Writing – review & editing. ABu: Funding acquisition, Supervision, Writing – review & editing. JU: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study is funded by MCIN/AEI/10.13039/501100011033 (project PID2021–123735OB-C).
JGI staff members who contributed to obtaining the analysis of metagenome data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1412124/full#supplementary-material
Supplementary Figure 1 | Rarefaction curves from 16S rRNA amplicons from 8 samples. Rarefaction was performed at 97% identity.
Supplementary Figure 2 | Proportions of bacterial family (represented those > 1%) from major phyla in biofilm-sediment samples. MS4 (Sep. 2021), MS5 (Jan 2020), and MS6 (July 2022).
Supplementary Figure 3 | Phylogenetic tree based on reference OTUs affiliated with the cyanobacterium Coleofasciculus using the Neighbor-Joining tree and the Jukes-Cantor nucleotide substitution model. In parentheses, the number of OTUs found repeatedly. One thousand bootstrap trees were generated; bootstrap confidence levels, as percentages (only values >50%), are shown at tree nodes.
Supplementary Figure 4 | Phylogenetic tree based on reference OTUs affiliated with the Spirochaetota using the Neighbor-Joining tree and the Jukes-Cantor nucleotide substitution model. In parentheses, the number of OTUs found repeatedly. One thousand bootstrap trees were generated; bootstrap confidence levels, as percentages (only values >50%), are shown at tree nodes.
Supplementary Figure 5 | Proportion of KEGG pathways related to “energy metabolism”, “carbohydrate metabolism”, in biofilm-sediment samples, MS4 (Sep 2021), MS5 (Jan 2020) and MS6 (July 2022). Data were obtained by PICRUSt and shotgun metagenomics.
Bahram M., Anslan S., Hildebrand F., Bork P., Tedersoo L. (2019). Newly designed 16S rRNA metabarcoding primers amplify diverse and novel archaeal taxa from the environment. Environ. Microbiol. Rep. 11, 487–494. doi: 10.1111/1758-2229.12684
Berg I. (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol. 77, 1925–1936. doi: 10.1128/AEM.02473-10
Berlanga M., Aas J. A., Paster B. J., Boumenna T., Dewhirst F. E., Guerrero R. (2008). Phylogenetic diversity and temporal variation in the Spirochaeta populations from two Mediterranean microbial mats. Int. Microbiol. 11, 267–274. doi: 10.2436/20.1501.71
Berlanga M., Guerrero R. (2016). Living together in biofilms: the microbial cell factory and its biotechnological implications. Microbial. Cell Factories 15, 165. doi: 10.1186/s12934-016-0569-5
Berlanga M., Palau M., Guerrero R. (2017). Functional stability and community dynamics during spring and autumn seasons over 3 years in Camargue microbial mats. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02619
Berlanga M., Palau M., Guerrero R. (2022). Community homeostasis of coastal microbial mats from the Camargue during winter (cold) and summer (hot) seasons. Ecosphere 13, e3922. doi: 10.1002/ecs2.3922
Biver S., Vandenbol M. (2013). Characterization of three new carboxylic ester hydrolases isolated by functional screening of a forest soil metagenomic library. J. Ind. Microbiol. Biotechnol. 40, 191–200. doi: 10.1007/s10295-012-1217-7
Boadella J., Rodríguez-Baliu C., Butturini A., Romaní A. M. (2023). Day-night variation of microbial organic matter use in sediments of saline shallow lake. Freshw. Biol. 68, 711–725. doi: 10.1111/fwb.14058
Bolhuis H., Cretoiu M., Stal L. J. (2014). Molecular ecology of microbial mats. FEMS Microbiol. Ecol. 90, 1–16. doi: 10.1111/1574-6941.12408
Brandt M. I., Pradillon F., Trouche B., Henry N., Liautard-Haag C., Cambon-Bonavita A., et al. (2021). Evaluating sediment and water sampling methods for the estimation of Deep-sea biodiversity using environmental DNA. Sci. Rep. 11, 7856. doi: 10.1038/s41598-021-86396-8
Briere P. R. (2000). Playa, playa lake, sabkha: proposed definitions for old terms. J. Arid Environ. 45, 1–7. doi: 10.1006/jare.2000.0633
Bryanskaya A. V., Shipova A. A., Rozanov A., Kolpakova O. A., Lazareva E. V., Uvarova Y. E., et al. (2022). Diversity and metabolism of microbial communities in a hypersaline lake along a geochemical gradient. Biol. (Basel) 11, 605. doi: 10.3390/biology11040605
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cardoso D. C., Cretoiu M. S., Stal L. J., Bolhuis H. (2019). Seasonal development of coastal microbial mat. Sci. Rep. 9, 9035. doi: 10.1038/s41598-019-45490-8
Casamayor E. O., Triadó-Margarit X., Castañeda C. (2013). Microbial biodiversity in saline shallow lakes of the Monegros Desert, Spain. FEMS Microbiol. Ecol. 85, 503–518. doi: 10.1111/1574-6941.12139
Castañeda C., Herrero J. (2005). The water regime of the Monegros playa-lakes as established from ground and satellite data. J. Hydrol. 310, 95–110. doi: 10.1016/j.jhydrol.2004.12.007
Chen Y.-J., Leung P. M., Cook P. L. M., Wong W. W., Hutchinson T., Eate V., et al. (2022). Hydrodynamic disturbance controls microbial community assembly and biogeochemical processes in coastal sediment. ISME J. 16, 750–763. doi: 10.1038/s41396-021-01111-9
Coban O., Rasigraf O., de Jong A. E. E., Spott O., Bebout B. M. (2021). Quantifying potential N turnover rates in hypersaline microbial mats by using 15N tracer techniques. Appli Environ. Microbiol. 87, e03118–e03120. doi: 10.1128/AEM.03118-20
Douglas G. M., Maffei V. J., Zaneveld J. R., Yurgel S. N., Brown J. R., Taylor C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 36, 685–688. doi: 10.1038/s41587-020-0548-6
Fang Y., Liu J., Yang J., Wu G., Hua Z., Dong H., et al. (2022). Compositional and metabolic responses of autotrophic microbial community to salinity in lacustrine environments. mSystems 7, e0033522. doi: 10.1128/msystems.00335-22
Fenchel T., Kühl M. (2000). Artificial cyanobacterial mats: growth, structure, and vertical zonation patterns. Microb. Ecol. 40, 85–93. doi: 10.1007/s002480000062
Fierer N., Leff J. W., Adams B. J., Nielsen U. N., Bates S. T., Lauber C. L., et al. (2012). Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. PNAS 109, 21390–21395. doi: 10.1073/pnas.1215210110
Guan X., Zhao Z., Jiang J., Fu L., Pan Y., Gao S., et al. (2023). Succession and assembly mechanisms of seawater prokaryotic communities along an extremely wide salinity gradient. Environ. Microbiol. Rep. 15, 545–556. doi: 10.1111/1758-2229.13188
Guerrero R., Berlanga M. (2016). From the cell to the ecosystem: the physical evolution of symbiosis. Evol. Biol. 43, 543–552. doi: 10.1007/s11692-015-9360-5
Guerrero R., Piqueras M., Berlanga M. (2002). Microbial mats and the search for minimal ecosystems. Int. Microbiol. 5, 177–188. doi: 10.1007/s10123-002-0094-8
Gutiérrez-Preciado A., Saghaï A., Moreira D., Zivanovic Y., Deschamps P., Lopez-García P. (2018). Functional shifts in microbial mats recapitulate early earth metabolic transitions. Nat. Ecol. Evol. 2, 1700–1708. doi: 10.1038/s41559-018-0683-3
Hao Z.-L., Ali A., Ren Y., Su J.-F., Wang Z. (2022). A mechanistic review on aerobic denitrification for nitrogen removal in water treatment. Scie Total Environ. 847, 157452. doi: 10.1016/j.scitotenv.2022.157452
Hiltemann S., Rasche H., Gladman S., Hotz H.-R., Larivière D., Blankenberg D., et al. (2023). Galaxy Training: A powerful framework for teaching! PloS Comput. Biol. 19, e1010752. doi: 10.1371/journal.pcbi.1010752
Huntemann M., Ivanova N. N., Mavromatis K., Tripp H. J., Paez-Espino D., Tennenssen K., et al. (2016). The standard operation procedure of the DOE-JGI metagenome annotation pipeline (MAP v.4). Standards Gen. Sci. 11, 17. doi: 10.1186/s40793-016-0138-x
Ji B., Yang K., Zhu L., Jiang Y., Wang H., Zhou J., et al. (2015). Aerobic denitrification: A review of important advances of the last 30 years. Biotech. Bioprocess Eng. 20, 643–651. doi: 10.1007/s12257-015-0009-0
Ji Y., Zhang P., Zhou S., Gao P., Wang B., Jiang J. (2022). Widespread but poorly understood bacteria: Candidate Phyla Radiation. Microorganisms 10, 2232. doi: 10.3390/microorganisms10112232
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Langille M. G. I., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA maker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lasser J., Nield J. M., Goehring L. (2020). Surface and subsurface characterization of salt pans expressing polygonal patterns. Earth Sys. Sci. Data 12, 2881–2020. doi: 10.5194/essd-12-2881-2020
Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W. (2015). MEGAHIT: an ultra-fast single node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Mees F., Castañeda C., Van Ranst E. (2011). Sedimentary and diagenetic features in saline lake deposits of the Monegros region, northern Spain. Catena 85, 245–252. doi: 10.1016/j.catena.2011.01.010
Meier D. V., Greve A. J., Chennu A., van Erk M. R., Muthukrishnan T., Abed R. M. M. D., et al. (2021). Limitation of microbial processes at saturation-level salinities in a microbial mat covering a coastal salt flat. Appl. Environm. Microbiol. 87, e0069821. doi: 10.1128/AEM.00698-21
Menéndez-Serra M., Ontiveros V. J., Triadó-Margarit X., Alonso D., Casamayor E. O. (2020). Dynamics and ecological distributions of the Archaea microbiome from inland saline lakes (Monegros desert, Spain). FEMS Microbiol. Ecol. 96, fiaa019. doi: 10.1093/femsec/fiaa019
Menéndez-Serra M., Triadó-Margarit X., Casamayor E. O. (2021). Ecological and metabolic thresholds in the bacterial, protist, and fungal microbiome of ephemeral saline lakes (Monegros desert, Spain). Microb. Ecol. 82, 885–896. doi: 10.1007/s00248-021-01732-9
Mu D.-S., Wang S., Liang Q.-Y., Du Z.-Z., Tian R., Ouyang Y., et al. (2020). Bradymobacteria, a novel bacterial predator group with versatile survival strategies in saline environments. Microbiome 8, 126. doi: 10.1186/s40168-020-00902-0
Osman J. R., Regeard C., Badel C., Fernandes G., DuBow M. S. (2019). Variation of bacterial biodiversity from saline soils and estuary sediments present near the Mediterranean sea coast of Camargue (France). Anton van Leeuw. 112, 351–365. doi: 10.1007/s10482-018-1164-z
Pastor A., Freixa A., Skovsholt L. J., Wu N., Romaní A. M., Riis T. (2019). Microbial organic matter utilization in high-Artic streams: key enzymatic controls. Microb. Ecol. 78, 539–554. doi: 10.1007/s00248-019-01330-w
Pohlner M., Dlugosch L., Wemheuer B., Mills H., Engelen B., Reese B. K. (2019). The majority of active Rhodobacterraceae in marine sediments belong to uncultured genera: A molecular approach to link their distribution to environmental conditions. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00659
Prieto-Barajas C., Valencia-Cantero E., Santoyo G. (2018). Microbial mat ecosystems: Structure types, functional diversity, and biotechnological application. Electronic J. Biotechnol. 31, 48–56. doi: 10.1016/j.ejbt.2017.11.001
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rinke C., ChuvoChina M., Mussing A. J., Chaumeil P.-A., Davín A. A., Waite D. W., et al. (2021). A standardized archaeal taxonomy for the genome taxonomy database. Nat. Microbiol. 6, 946–959. doi: 10.1038/s41564-021-00918-8
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. Peer J. 4, e2584. doi: 10.7717/peerj.2584
Shen L.-D., Wu H.-S., Gao Z.-Q., Liu X., Li Ji. (2016). Comparison of community structures of Candidatus Methylomirabilis oxyfera-like bacteria of NC10 phylum in differnent freshwater habitats. Sci. Rep. 6, 25647. doi: 10.1038/srep25647
Sohm J. A., Ahlgren N. A., Thomson Z. J., Williams C., Moffett J. W., Saito M. A., et al. (2016). Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, Macronutrients and Iron. ISME J. 10, 333–345. doi: 10.1038/ismej.2015.115
Sun S., Jones R. B., Fodor A. A. (2020). Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome 8, 46. doi: 10.1186/s40168-020-00815-y
Thompson A. W., Kouba K., Ahlgren N. A. (2021). Niche partitioning of low-light adapted Prochlorococcus subecotypes across oceanographic gradients of the North Pacific subtropical front. Limnol. Oceanograph 66, 1548–1562. doi: 10.1002/lno.11703
Van Gestel J., Vlamakis H., Kolter R. (2015). Division of labor: The ecology of cell differentiation. Microbiol. Spectr. 3. doi: 10.1128/microbiolspec.mb-0002-2014
Varliero G., Lebre P. H., Stevens M. I., Czchowski C., Makhalanyane T., Cowan D. A. (2023). The use of different 16S rRNA gene variable regions in biogeographical studies. Environ. Microbiol. Rep. 15, 216–228. doi: 10.1111/1758-2229.13145
Varun G. P., Mormile M. R. (2017). A case for the protection of saline and hypersaline environments: a microbiological perspective. FEMS Microbiol. Ecol. 93, fix091. doi: 10.1093/femsec/fix091
Weiser M. D., Ning D., Buzzard V., Michaletz S. T., He Z., Enquist B. J., et al. (2019). Thermal disruption of soil bacterial assemblages decreases diversity and assemblage similarity. Ecosphere 10, e02598. doi: 10.1002/ecs2.2598
Wieland A., Kühl M. (2006). Regulation of photosynthesis and oxygen consumption in a hypersaline cyanobacterial mat (Camargue, France) by irradiance, temperature and salinity. FEMS Microb. Ecol. 55, 195–210. doi: 10.1111/fem.2006.55.issue-2
Wierzchos J., Berlanga M., Ascaso C., Guerrero R. (2006). Micromorphological characterization and lithification of microbial mats from the Ebro Delta (Spain). Int. Microbiol. 9, 289–295.
Wörmer L., Gajendra N., Schubotz F., Matys E. D., Evans T. W., Summons R. E., et al. (2020). A Micrometer-scale snapshot on phototroph spatial distributions: Mass spectrometry imaging of microbial mats in Octopus Spring, Yellowstone National Park. Geobiology 18, 742–759. doi: 10.1111/gbi.12411
Yapiyev V., Sagintayev Z., Inglezakis V. ,. J., Samarkhanov K., Verhoef A. (2017). Essentials of endorheic basins and lakes: A review in the context of current and future water resource management and mitigation activities in central Asia. Water 9, 798. doi: 10.3390/w9100798
Keywords: endorheic saline lagoons, bacterial community diversity, bacterial community functionality, biofilm-sediment biotope, water column biotope
Citation: Berlanga M, Picart P, Blasco A, Benaiges-Fernandez R, Guerrero R, Butturini A and Urmeneta J (2024) Biodiversity and potential functionality of biofilm-sediment biotope in La Muerte lagoon, Monegros Desert, Spain. Front. Ecol. Evol. 12:1412124. doi: 10.3389/fevo.2024.1412124
Received: 04 April 2024; Accepted: 10 June 2024;
Published: 04 July 2024.
Edited by:
Thanh Luu Pham, Vietnam Academy of Science and Technology, VietnamReviewed by:
Maxine Allayne Darlene Mowe, National University of Singapore, SingaporeCopyright © 2024 Berlanga, Picart, Blasco, Benaiges-Fernandez, Guerrero, Butturini and Urmeneta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mercedes Berlanga, bWJlcmxhbmdhQHViLmVkdQ==
†ORCID: Mercedes Berlanga, orcid.org/0000-0002-7305-7046
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.