- 1Department of Biology, Emory University, Atlanta, GA, United States
- 2Department of Physics, Emory University, Atlanta, GA, United States
Many insects selectively associate with specific microbes in long-term, symbiotic relationships. Maintaining these associations can be vital for the insect hosts’ development, but insects must also contend with potential coinfections from other microbes in the environment. Fending off microbial threats while maintaining mutualistic microbes has resulted in many insects developing specialized symbiotic organs to house beneficial microbes. Though locally concentrated in these organs, symbiont establishment can have global consequences for the insect, including influence over the success of coinfecting microbes in colonizing the insect host. We use a transcriptomic approach to examine how the mutualistic symbiosis between the agricultural pest Anasa tristis and bacteria in the genus Caballeronia affects insect gene expression locally within the symbiotic organs and in the insect host at large. We simultaneously determine whether Caballeronia colonization impacts insect host responses to infection with the plant pathogen Serratia marcescens, which it vectors to plants. We found that no significant differential gene expression was elicited by infection with S. marcescens. This was a surprising finding given previous work indicating that symbiotic A. tristis clear S. marcescens infection rapidly compared to aposymbiotic individuals. Our results indicate that symbiotic and nonsymbiotic tissues in A. tristis differ greatly in their gene expression, particularly following successful symbiont colonization. We found evidence for local downregulation of host immunity and upregulation of cell communication within the symbiotic organs, functions which can facilitate the success of the A. tristis-Caballeronia symbiosis.
Introduction
Herbivorous insects contend with continuous exposure to a variety of microorganisms as part of their daily lives. These interactions span the continuum of symbiosis and include beneficial microbes as well as potentially life-threatening pathogens. The threat of infection by potentially detrimental microorganisms has complicated the acquisition and accommodation of mutualistic microbes, many of which are necessary to supplement the nutritionally poor diets of their hosts (Gündüz and Douglas, 2008; Sudakaran et al., 2015). For insects that acquire mutualistic microbes from the environment, it is necessary to manage acquisition of mutualistic symbionts while excluding their detrimental counterparts (Douglas, 2014; Ohbayashi et al., 2020; Wang et al., 2023).
While some symbiotic microbes have adapted to the insect immune system in order to remain in symbiosis with their insect hosts (Russell and Castillo, 2020; Ganesan et al., 2022), many insects have resorted to confining their mutualists to specialized symbiotic organs (bacteriomes) or cells (bacteriocytes) for protection (Douglas, 2020). Conditions within these specialized organs are generally favorable for symbionts (Ferrarini et al., 2022), but hosts can still regulate the microbial populations within these tissues (Kim et al., 2013; Whittle et al., 2021). Because of their specialized function, these tissues can differ drastically from the rest of the insect host body (Heddi et al., 2005). Gene expression within bacteriomes and bacteriocytes often reflects the crosstalk between host and symbiont (Nakabachi et al., 2005; Price et al., 2011; Smith and Moran, 2020) as both partners balance their needs and concessions to maintain a beneficial relationship.
Inside their host, beneficial microbes can influence the outcomes of subsequent microbial infections. Some mutualists, known as defensive symbionts, protect their insect hosts from pathogens, parasitoids, and other natural enemies (Brownlie and Johnson, 2009; Oliver et al., 2012; Oliver and Perlman, 2020). Protective mechanisms vary from direct interactions with the microbial intruder to indirect effects modulated by the insect immune system (Gerardo and Parker, 2014). These microbial interactions within the insect host are of particular interest in herbivorous insects that vector pathogens. Work in various systems has shown that symbiotic microbes can alter the ability of insects to successfully vector pathogens (Moreira et al., 2009; Weiss and Aksoy, 2011; Gonella et al., 2018).
Here, we use transcriptomics to investigate how a mutualistic microbe alters gene expression locally within symbiotic tissues and globally within its insect host. We simultaneously investigate whether symbiotic and symbiont-free insects respond differently to infection with a vectored pathogen. For this study, we use the squash bug Anasa tristis, which has an established beneficial symbiosis with bacteria in the genus Caballeronia (Acevedo et al., 2021). A. tristis acquires Caballeronia from the environment de novo at each generation, usually early on in its development. Caballeronia symbionts then colonize a specialized region of A. tristis’ posterior midgut, known as the crypts. Here, they grow with little interference from other microbes. In addition to its mutualistic partner, A. tristis can also harbor the phytopathogen Serratia marcescens, which it vectors to commercially important cucurbit plants (i.e., squash, pumpkin, watermelon) causing Cucurbit Yellow Vine Disease (Bruton et al., 1998; Bextine et al., 2001). Previous work has established that A. tristis harboring Caballeronia symbionts have low titers of S. marcescens and clear infection rapidly (Mendiola et al., 2022). In Caballeronia-free, or aposymbiotic, individuals, S. marcescens titers can be up to 1000-fold higher and infections can last up to 10 times longer than in symbiotic individuals.
Given the stark differences between symbiotic and aposymbiotic insects (Acevedo et al., 2021; Mendiola et al., 2022), we expected to see differences in gene transcription in insects based on symbiont status as well as between their symbiotic and nonsymbiotic tissues. We further expected to see differential transcriptional responses that could explain why S. marcescens infection outcomes differ so drastically between symbiotic and aposymbiotic individuals. Though we found strong evidence of differential transcription between symbiotic and nonsymbiotic tissues as well as between symbiotic and aposymbiotic bugs, strikingly, we found minimal evidence for differential transcription between S. marcescens infected and uninfected insects. We conclude that differential regulation of symbiotic organs is essential for the successful establishment and persistence of the A. tristis-Caballeronia symbiosis. The successful establishment of symbiosis leads to further transcriptional changes in the symbiotic organs that have global consequences for the growth and development of A. tristis. Furthermore, our results support a neutral effect of the phytopathogen S. marcescens on its vector, which has implications for its long-term transmission potential.
Methods
Insect rearing
All insects were reared in an environmental chamber held at constant temperature (27°C) under a long day light cycle (16 hours light, 8 hours dark). We surface sterilized eggs collected from our existing A. tristis colony, founded by wild-caught adults from organic farms in Georgia and Florida, by alternately washing them in 70% ethanol and 10% bleach for one minute each and then rinsing with 70% ethanol for 10 seconds. We transferred newly hatched first instar insects to a sterile container and fed them slices of organic zucchini (Cucurbita pepo) fruit, surface sterilized with 70% ethanol and thinly wrapped in parafilm. We maintained insects in sterile containers with regular fruit changes until they molted to second instars.
Treatment administration
We collected one- to two-day-old second instar nymphs (n=180) hatched from eggs collected on the same day into sterile rearing boxes for administration of treatments. During the first feeding period, bugs to be maintained Caballeronia-free (‘Aposymbiotic bugs’) (n = 100), were fed on a liquid diet consisting of five mLs of sterile water mixed with 100 µL of blue dye, which facilitates confirmation of feeding. Symbiont-positive bugs (n = 80) were fed on five mL liquid diets with Caballeronia strain GAOX1 standardized to a concentration of 2x107 CFUs per mL using sterile water and 100 µL of blue dye. All groups were given access to liquid diets for 24 hours before being placed back on an organic zucchini diet. See Mendiola et al. (2022) for a more detailed feeding protocol.
At the third instar stage, insects were further separated into groups for the second feeding. Only individuals that molted within one day of each other were used. From these individuals, sixty aposymbiotic and forty symbiont-positive insects were allocated randomly into evenly sized groups for the second feeding (Serratia infection or no-pathogen control).
All individuals in the Serratia-infected groups were fed GFP-labeled Serratia marcescens Z01 (henceforth referred to as Z01) via vacuum-infused zucchini cubes (Bextine, 2001). This fluorescently labeled strain was used in previous work tracking S. marcescens persistence and titer in individual bugs (Mendiola et al., 2022). Controls were fed sterile water in the same manner. Insects had access to infused zucchini for 24 hours. Dissections and sample harvesting were done immediately following this feeding period. Experimental details are laid out schematically in Figure 1.
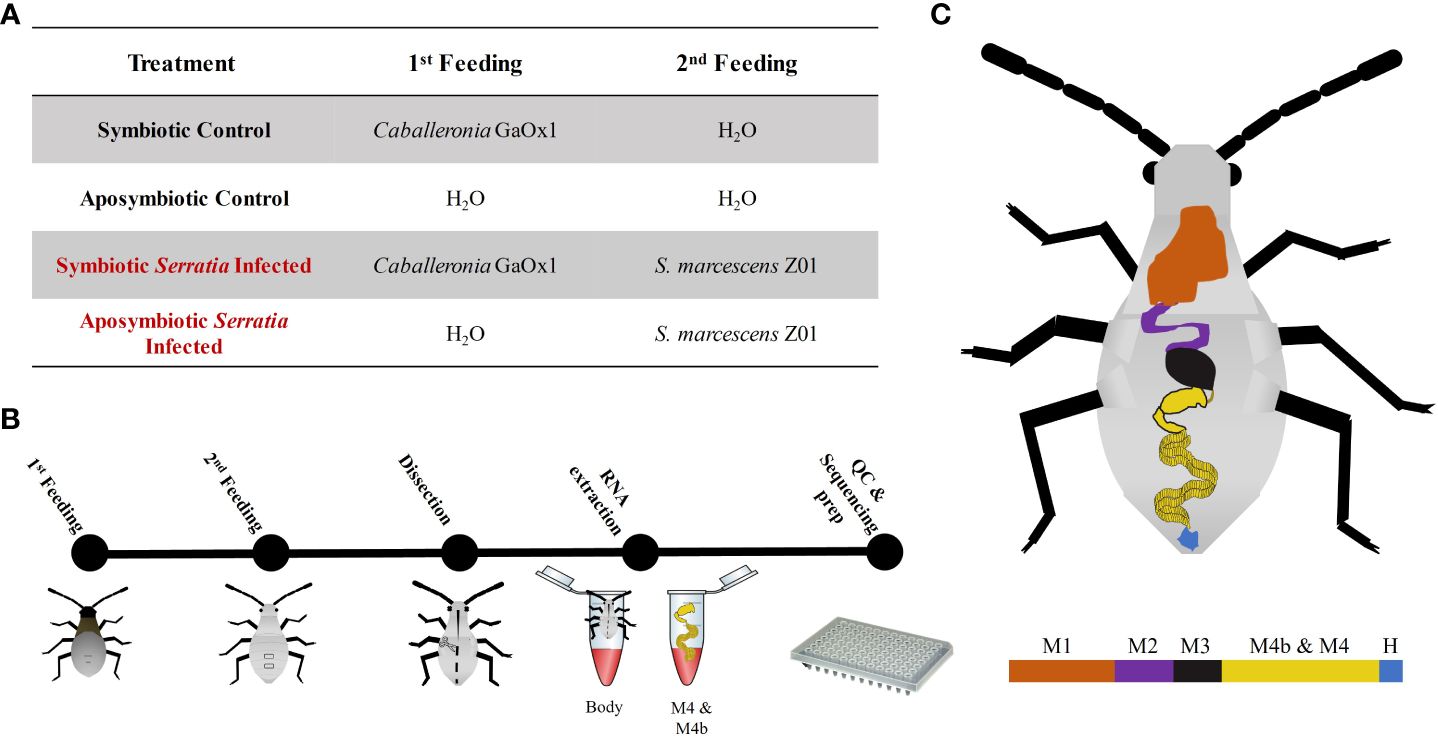
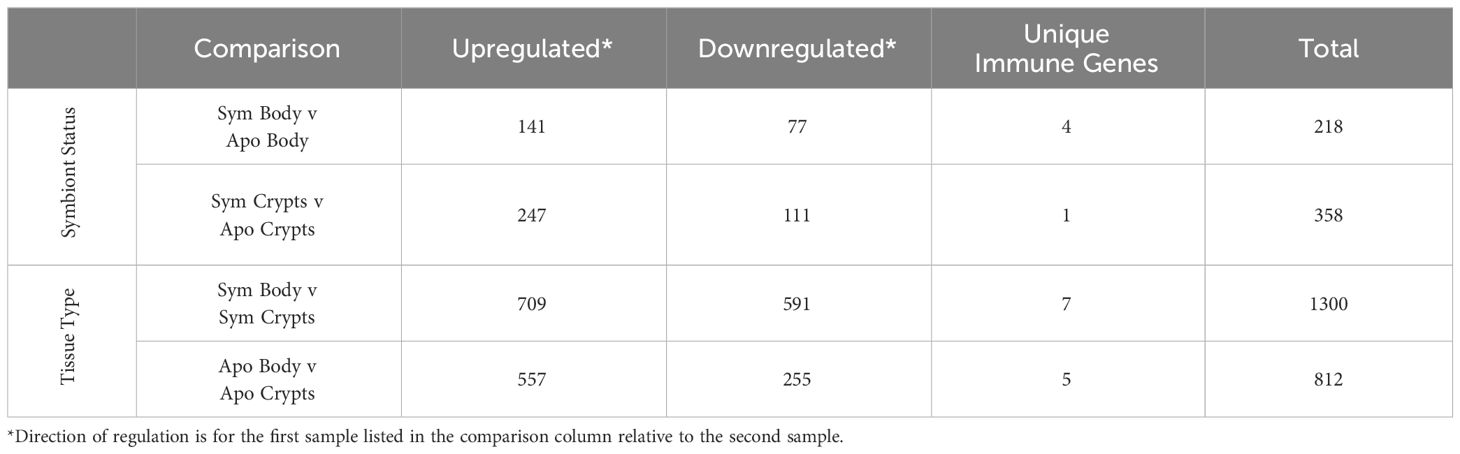
Figure 1 Schematic of experimental methods for host transcriptomics. (A) Table summarizing diets administered to each treatment group. (B) Relative timeline of experimental workflow. (C) Diagram of the A. tristis gut. Regions of the gut are as follows: M1 is midgut section one; M2 is midgut section two; M3 is midgut section three; M4b and M4, the symbiotic organs, are the pre-crypt bulb and the crypts, respectively; and, H is the hindgut. For our experiment, the M4b and M4 were dissected out and are collectively referred to as the crypts. The remainder of the body, including other sections of the gut are referred to simply as body.
Sample preparation
All squash bug dissections took place on the same day, and all individuals were still in the third instar stage. Only five bugs from each treatment underwent full dissection. An additional five bugs were flash frozen on dry ice and stored in RNALater at -80°C as back-ups. We also sacrificed five random bugs from each treatment to ensure that control bugs were not Z01 positive and that Z01 fed bugs were Z01 positive. This was done simply by homogenizing surface-sterilized bugs and plating out the homogenate to screen for GFP fluorescence; more detailed methods are available in (Mendiola et al., 2022). For those bugs that underwent dissection, we flash froze bugs individually on dry ice before surface sterilizing them in 70% ethanol for five minutes. Bugs were then dissected in ~200 µL of RNALater. The symbiotic organs (the crypts and the M4b, henceforth referred to as the crypts) and the remaining carcass of the bug were harvested separately and placed directly into RNA lysis buffer from the Promega Total RNA Extraction kit. Once in lysis buffer, samples were crushed immediately using sterile, nuclease-free micropestles. Fully homogenized samples were immediately frozen on dry ice before being stored at -80°C until RNA extraction.
RNA extraction and sequencing
All samples were processed within two days of dissection. Total RNA was extracted from all samples using the Promega Total RNA Extraction kit following manufacturer’s instructions, including the three-minute incubation period at 70°C. RNA was eluted into 100 µL of nuclease-free water and split into three ~30 µL aliquots, which were all stored at -80°C. We tested the quality of all extracted RNA using the Agilent 4200 TapeStation with high sensitivity RNA ScreenTapes. All samples exceeded the RNA integrity score of seven recommended for proceeding with RNAseq.
Total RNA was submitted to the Georgia Genomics and Bioinformatics Core for library prep and sequencing. Two samples per individual (crypts, remaining body tissues) for each of five individuals in each treatment (total of 40 samples) were submitted for sequencing insect host RNA. Quality control using the Agilent BioAnalyzer eliminated a handful of samples with low RNA concentrations. If one of the two samples per bug failed QC, we omitted all samples from that bug from our study. After QC, all symbiotic insect samples remained (five replicates for both S. marcescens infected and uninfected treatments), one replicate was removed from the uninfected aposymbiotic treatment (four final replicates) and two replicates were removed from the infected aposymbiotic treatment (three final replicates). Only samples that passed QC were sequenced. Stranded libraries were prepared using Kapa Biosystems RNA Hyperprep kit with polyA enrichment to exclude nontarget RNA. All samples were sequenced on the Illumina NextSeq 2000 platform with paired-end 100 bp reads. All samples were sequenced in the same run using a high-output, P3 flow cell.
In addition to data obtained from the host, we attempted to capture symbiont transcriptional profiles in vivo and in vitro. For in vitro samples, GAOX1 was streaked out from glycerol stocks onto nutrient agar (3g yeast extract, 5g peptone per L in 1.5% agar) and grown at 30°C for two days. Two individual colonies were then used to initiate each of five liquid cultures. Liquid cultures were grown in M9 glucose minimal media (made following Cold Spring Harbor Protocol) for 24 hours at 25°C shaking at 200 rpm. One and a half mL from each culture was spun down at 4°C for two minutes at 10,000 g to pellet bacterial cells. OD600 values of the cultures at the time that they were pelleted were between 0.35 and 0.6. Supernatant was removed and sample pellets were resuspended in 100 µL fresh TE buffer with 0.4 mg/mL of lysozyme. Resuspended pellets were incubated at room temperature for 5 minutes before 75 µL of RNA lysis buffer and 350 µL of RNA dilution buffer were added. Samples were mixed well by inverting tubes. After mixing, we proceeded with the same Promega Total RNA extraction protocol used previously. For in vivo samples, we used total RNA extracted from insects. Overall, 30 in vivo and five in vitro samples were submitted for sequencing of prokaryotic RNA. This included RNA from five bacterial cultures as well as two tissue samples per bug for five symbiont positive controls and 10 symbiont positive individuals with Z01 exposure. These samples were ribo-depleted to enrich for mRNA and were sequenced on the Illumina NextSeq 2000 platform in a single run using a P2 flow cell with 100 paired-end bp.
Data processing and analysis
We used Trimmomatic v0.39 set to TruSeq3 universal Illumina adapters to trim adapters from all raw reads before further filtering out any eukaryotic and prokaryotic ribosomal RNA reads using existing rRNA databases in sortmeRNA v4.3.6. Cleaned reads for both host and bacteria can be found in NCBI’s sequence read archive under BioProject PRJNA1062536.
Host transcriptomics
We used reads obtained from all insect-specific samples across treatments to assemble one reference A. tristis transcriptome de novo with Trinity v2.8.5. We assessed the quality of the transcriptome assembly with BUSCO v.5.4.2 for completeness and further verified read alignment from all our samples to the assembled transcriptome using bowtie2. Once the quality of the assembly was verified, we proceeded to conduct differential gene expression analyses.
We first performed transcript quantification using Trinity’s align_and_estimate_abundance.pl script with the pseudoaligner Kallisto. Initial data exploration was conducted in R version 4.3.0 using the “DESeq2” package. We excluded lowly expressed genes from further analyses by filtering out those genes with counts of less than 10 across all samples. After filtering, Kallisto gene count matrices were standardized using a variance stabilizing transformation for generation of heatmaps and principal component analysis. After data exploration, the untransformed gene count matrix generated in Trinity was used to conduct differential gene expression analyses using the run_DE_analysis.pl Trinity script employing the “DESeq2” method with default parameters. Results were used to generate volcano plots of relevant contrasts. Genes were labeled as significantly differentially expressed if they had log2 fold changes greater than two and adjusted p-values less than 0.05.
Subsets of significantly differentially expressed genes (DEGs) were included in Gene Ontology (GO) enrichment analysis using the enrichGO function in the “clusterProfiler” package in R. Using the assembled A. tristis transcriptome generated with Trinity, we used Transdecoder v5.7.0 to identify candidate protein-coding regions within our transcriptome. Because an annotated genome is not currently publicly available for A. tristis, we BLAST searched our candidate protein-coding genes against the genome of a more well-annotated insect through the Kyoto Encyclopedia of Genes and Genomes (KEGG) Automatic Annotation Server (KAAS) using the best bi-directional hit (BBH) method and applied any matching annotations to our candidate genes. We initially attempted to run the GO analysis with the most closely related insect for which we could find publicly available annotations, the brown marmorated stinkbug Halyomorpha halys. Unfortunately, the annotations were insufficient to carry out our analyses. For our final analysis, we settled on a more extensively annotated insect genome, that of the pea aphid Acyrthosiphon pisum. We applied the annotations from A. pisum to our assembly to carry out the GO analysis using the organismal database obtained from the “AnnotationHub” package for A. pisum. We used the default parameters for the enrichGO function: p-value cutoff of 0.05 with the Benjamini-Hochberg “BH” correction for multiple comparisons, q-value cutoff of 0.2, minimum gene size of 10, maximum gene size of 500. Our analyses included all three ontology groups: biological processes (BP), cellular components (CC), and molecular function (MF).
In addition to GO enrichment analysis, we also looked for evidence of differential expression of canonical insect innate immune genes in our subset of DEGs. We used the Interactive Database for Insect Innate Immunity (http://bf2i300.insa-lyon.fr:443/home) to obtain a list of 434 known innate immune genes in A. pisum and 391 known innate immune genes in Drosophila melanogaster, for a total of 825 known insect innate immune genes. We matched the AphidBase and FlyBase identifiers in the database back to NCBI’s Gene IDs using GenBank’s index of gene data. We then took our candidate gene hits, obtained from BLAST searching against the A. pisum and D. melanogaster genomes, and looked for immune-specific matches. Hits that mapped to both the list of immune genes and our identified squash bug genes were then used to create a database of canonical immune genes in the squash bug. We then used this database to identify differentially expressed innate immune genes amongst our treatment groups.
Symbiont transcriptomics
During rRNA filtering, it became clear that we had insufficient bacterial mRNA reads for full transcriptomics analysis of all our in vivo samples, particularly those collected from insect body tissues. Because they had the best coverage and no contamination with host RNA, we used the filtered reads from the in vitro samples to assemble a reference transcriptome de novo with Trinity, which we assessed for completeness with BUSCO and bowtie2 as before. We were only interested in the transcriptional profile of Caballeronia symbionts, so we took further steps to ensure no exogenous bacteria were included in our in vivo samples. Specifically, all in vivo samples were subjected to additional cleaning using bbsplit from bbmap v38.18. This allowed us to screen our reads and only retain those that mapped to a reference symbiont genome Caballeronia zhejiangensis A33M4c (NCBI Accession GCF_022879815.1) for subsequent analysis. Our analysis of differential expression was conducted as above using “edgeR” in the Trinity script. GO enrichment analysis was conducted as before. This time, we applied annotations from the organismal database for Escherichia coli to our candidate protein-coding genes. Though our data on symbiont transcription is lacking and by no means definitive, we include it here as a preliminary step into understanding Caballeronia-A. tristis interactions.
Results
Sequencing data
We obtained a total of ~949 million sequences across all sequenced insect samples. Following adapter trimming and rRNA filtering, we had ~904 million sequences across these samples, with an average of ~26.5 million sequences per sample. Individual treatments averaged between 20.5 to 30.5 million sequences per sample and, with the exception of the aposymbiotic treatment infected with S. marcescens, we found that more reads were produced from body tissues compared to crypt tissues (Supplementary Table 1). Our transcriptome assembly was missing only two out of 255 BUSCO groups with a completeness score of 99.2%. Furthermore, over 90% of reads for all insect samples mapped to the assembled reference transcriptome.
Our symbiont sequencing produced ~921 million reads which were reduced to ~160 million after adapter trimming and filtering of both eukaryotic and prokaryotic rRNA. Following cleaning with bbsplit, we were left with ~25 million bacterial reads across all our treatments. These reads were primarily concentrated in the in vitro samples, which averaged 4.9 million reads per sample. Crypt tissues produced an average of 32,000 reads per sample. Body tissues showed the fewest symbiont transcripts, producing only 1,000 reads per sample on average. Our symbiont transcriptome assembly was missing 280 out of the 688 BUSCO groups searched in the Burkholderiales lineage orthologs (59% complete). Samples fared better when aligning to the symbiont transcriptome, with all in vitro samples and all in vivo crypt samples producing 99% alignment rates. In vivo body tissue samples were less predictable, with alignment rates ranging from six to 97%. This prompted us to exclude body tissue samples from further analyses of symbiont gene expression and focus only on comparisons between in vitro and in vivo crypt samples.
No effect of Serratia marcescens infection status on host sample clustering
Host transcript samples primarily clustered by tissue type in principal component analysis (PCA), with crypts and bodies tending to cluster on opposite sides of the PC1 axis (Figure 2). Samples were further stratified by symbiont status along PC2; separation of symbiont vs. non-symbiont samples was larger for crypts than for body tissues. Most notably, we observed no differential clustering by Serratia infection status; Serratia infected and uninfected insects clustered by their other treatment conditions (symbiont status and tissue type). A few of our samples did not cluster with their respective groups, possibly due to imperfect dissection of tissues or imperfect symbiont colonization.
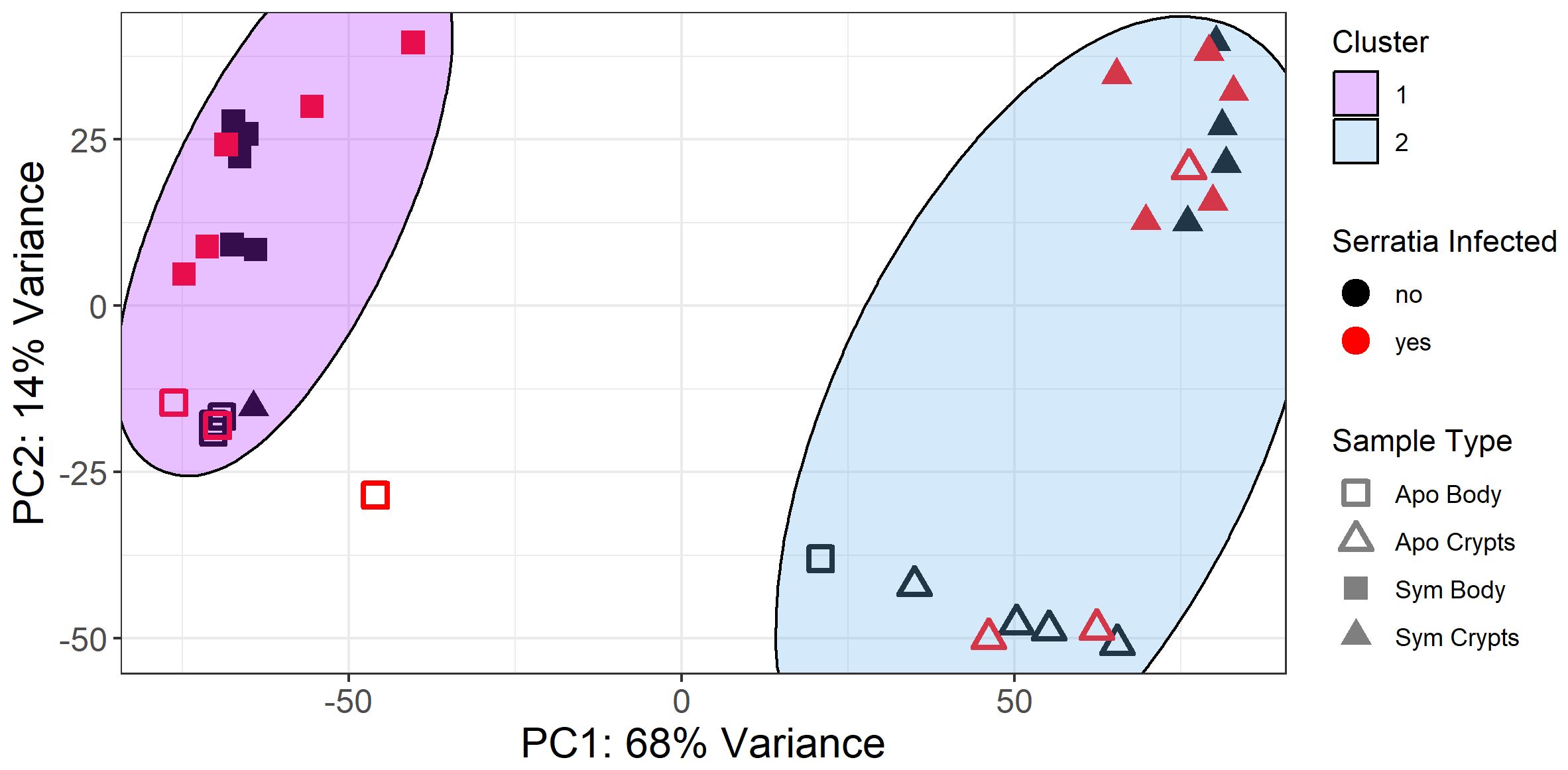
Figure 2 PCA plot of variance stabilized gene counts for all samples. Samples clustered primarily by tissue type (PC1, body vs crypts) and symbiont status (PC2, aposymbiotic = apo and sym = symbiotic). Kmeans clustering of gene counts revealed two primary sample clusters: Cluster 1 corresponds to body tissues, and Cluster 2 corresponds to crypt tissues. Notably, a few samples clustered with different groups, possibly due to imperfect symbiont colonization or imperfect dissection of tissue types. Serratia infection status had little influence on sample clustering. Specifically, Serratia infected and uninfected samples clustered together based on tissue type and symbiont status, with no distinct cluster correlated with infection status.
A heatmap of the 100 genes with the most transcripts across all our samples in the variance stabilized gene count matrix further corroborated patterns observed in the principal component analysis. Samples primarily clustered by tissue type with secondary clustering by symbiont status within tissue types. However, no obvious pattern emerged based on infection status (Figure 3). Furthermore, differential expression analysis of Serratia-infected and uninfected host samples, controlled for symbiont status and tissue type, revealed few DEGs with most comparisons having less than 10 (Supplementary Figure 1). The uninfected and infected aposymbiotic body tissue comparison was the exception with 25 DEGs. Unfortunately, we were unable to conduct any further analyses on these genes as the vast majority did not map back to genes of known function. Based on this and the lack of differential clustering by Serratia infection status, we decided to pool Serratia infected and uninfected samples into treatments based on their corresponding tissue type and symbiont status for further analysis.
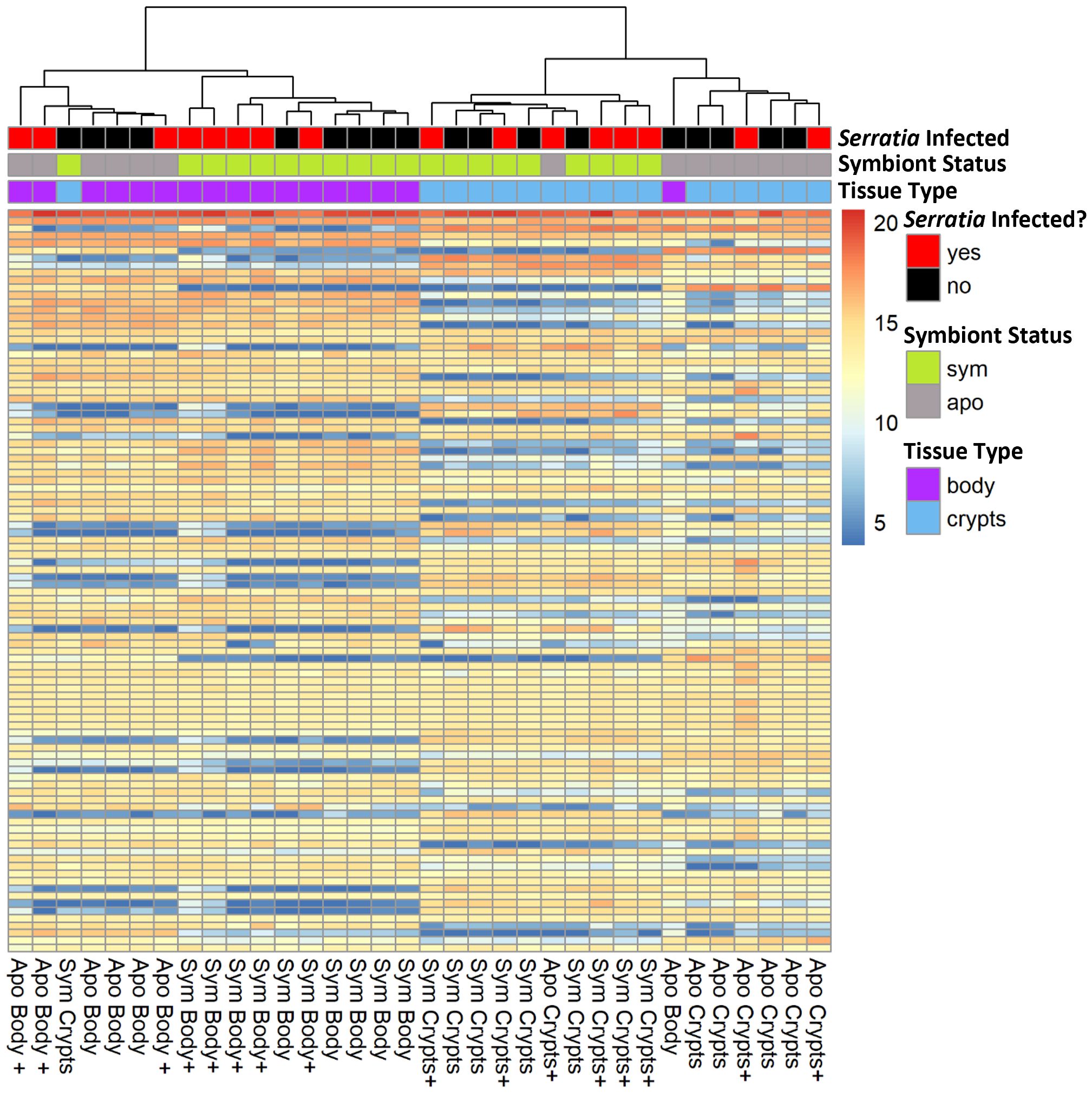
Figure 3 Expression heatmap of 100 genes with the most transcripts across all samples in the variance stabilized gene count matrix. Here, we show only the 100 genes with the most transcript counts across all samples. Samples clustered by tissue type and symbiont status, but the distribution of infected samples was haphazard. Column names denote the treatment groups of each sample. The “+” indicates positive Serratia infection.
Both symbiont status and host tissue type significantly affect host gene expression
We looked for DEGs among four treatment groups stratified by symbiont status and tissue type: symbiotic body tissue, symbiotic crypts, aposymbiotic body tissue, and aposymbiotic crypts. Table 1 provides a summary of DEGs that met our cutoff criteria (adjusted p value ¾ 0.05 and log2 fold change > 2). These results are visualized in volcano plots in Figure 4. We compared the same host tissue types across symbiont status (Figures 4A, B) and different host tissue types within symbiont status (Figures 4C, D). All four comparisons yielded DEGs, though generally, we noted that contrasting by host tissue type while controlling for symbiont status produced more DEGs than contrasting by host symbiont status controlling for tissue type.
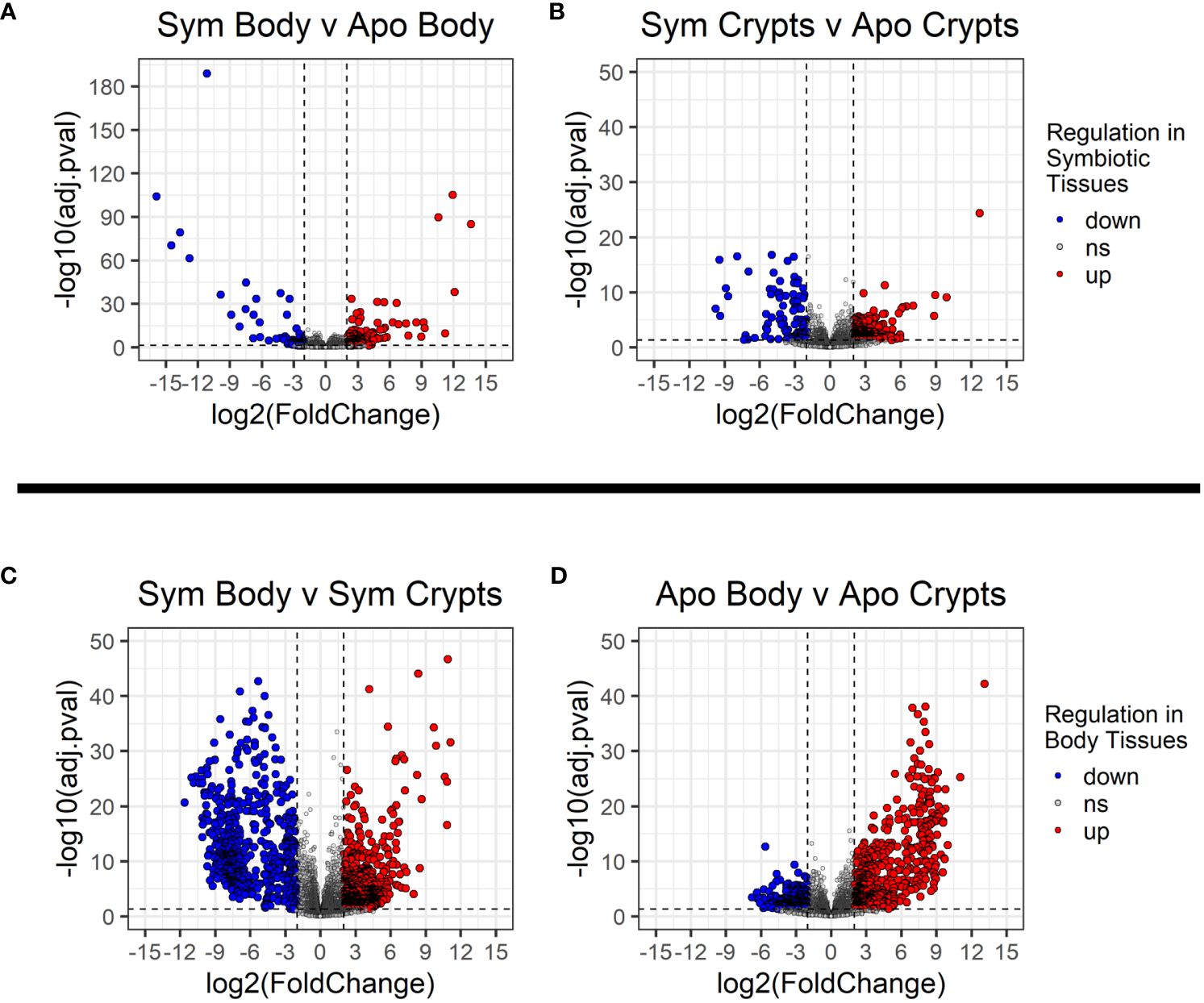
Figure 4 Volcano plots showing differentially expressed genes across treatment groups. (A, B) Contrasts for the same tissue type across symbiont status. (C, D) contrasts across tissue types within the same symbiont status. Dashed vertical lines indicate the cut off for log2 fold changes greater than 2, while the horizontal dashed line corresponds to an adjusted p-value of 0.05. All significantly differentially expressed genes are colored either red for upregulated or blue for downregulated. The direction of regulation is always for the first sample listed in the contrast relative to the second sample. Thus, in (A, B), blue indicates genes significantly downregulated in individuals with symbionts, and in (C, D), blue indicates genes significantly downregulated in the rest of the body relative to the crypt tissues.
Differential regulation of host metabolic and biosynthetic pathways across symbiont status and host tissue types
We performed GO enrichment analysis on DEGs from each contrast to gain insight into their functions. First, we examined contrasts between symbiotic and aposymbiotic samples within a tissue type. We found upregulation of metabolic and catabolic pathways in symbiotic body tissues and crypts as compared to aposymbiotic samples. In crypts specifically, we observed up-regulation of organic acid biosynthesis, monocarboxylic acid biosynthesis, and fatty acid biosynthesis (Figure 5). In crypts and body tissues, symbiotic samples showed lower expression of some specific enzymatic activities (lipase, ester hydrolase) and increased expression of others (e.g. glycosyl hydrolase). Membrane-associated ion channel activity was also increased in symbiotic vs aposymbiotic crypts.
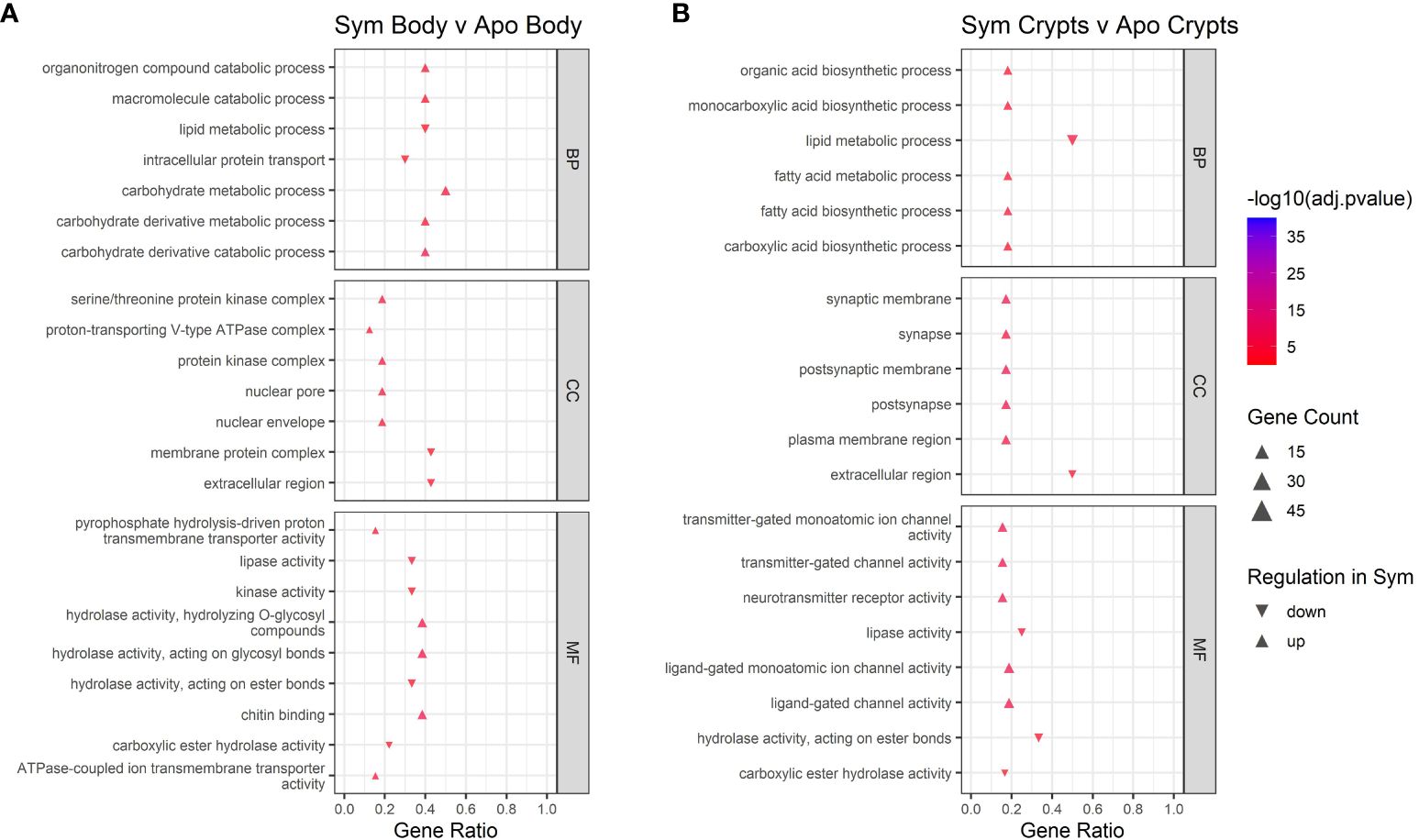
Figure 5 GO enrichment analysis for contrasts across symbiont status. (A) contrast of body tissue across symbiont status. (B) contrast of crypt tissue across symbiont status. Only the top five up and downregulated categories in each ontology group are plotted. Ontology groups include Biological Process (BP), Cellular Component (CC), and Molecular Function. Not all categories had unidirectional gene regulation, resulting in categories with both up and down regulation. Note that adjusted p-values have been log10 transformed for better visualization of results. Gene ratio was calculated as the percentage of total DEGs identified in the given GO term (restricted to only genes assigned to a GO term).
Next we examined contrasts between tissue types. GO analysis of different host tissue types differed for symbiotic and aposymbiotic insects, particularly when looking at Biological Processes and Molecular Function (Figure 6). For symbiotic individuals, we found downregulation of signaling, cell communication, and signaling receptor activity in the body tissues relative to the crypts. The monooxygenase and iron ion binding activity categories did not show unidirectional regulation. This could be due to certain genes within those categories being upregulated while others are downregulated. The same analysis for aposymbiotic tissues showed little in common with regulation occurring in symbiotic tissues. Transporter and transmembrane transporter activity was upregulated in aposymbiotic body tissues relative to crypts, and lipid metabolic processes were downregulated in body tissues relative to the crypts.
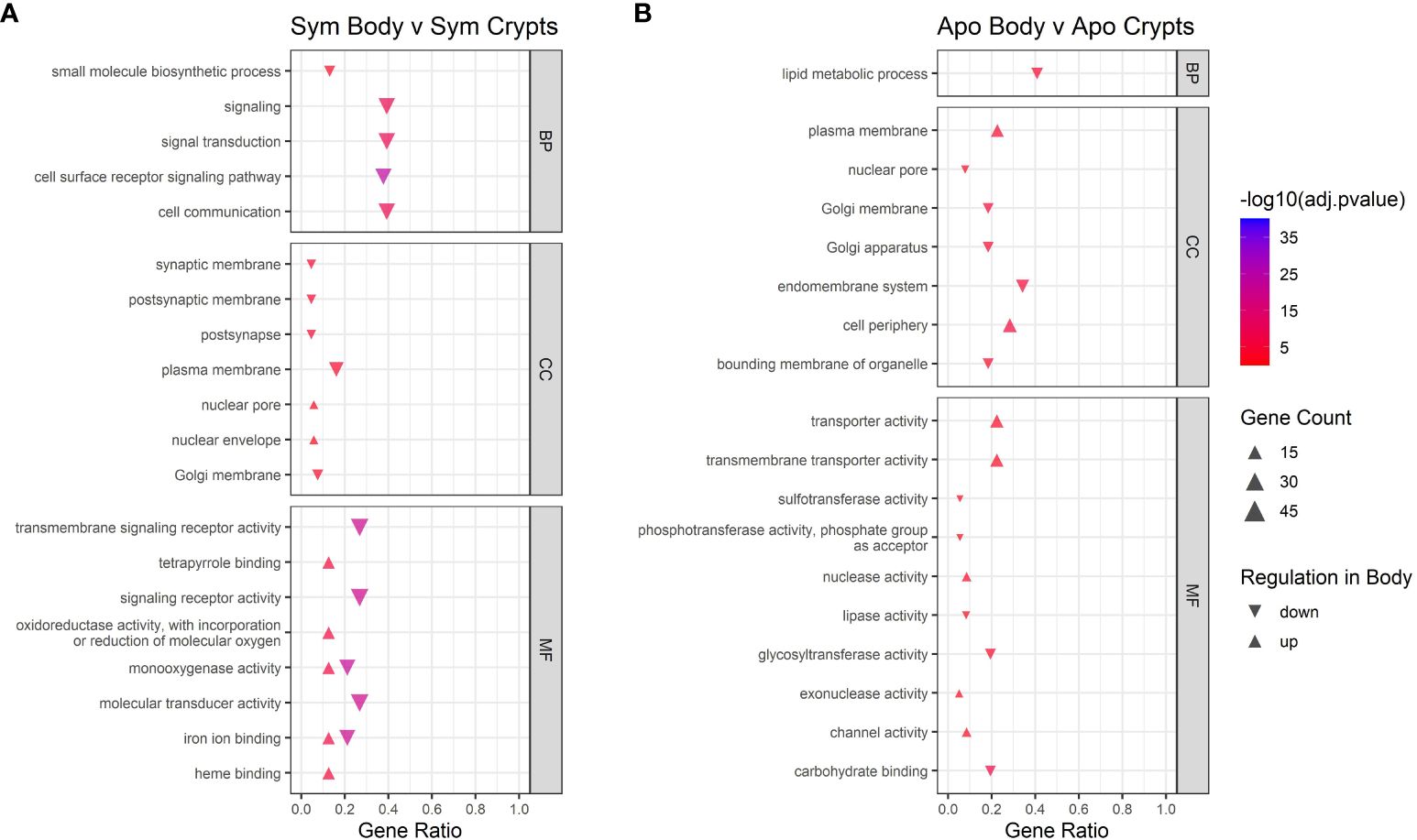
Figure 6 GO enrichment analysis for contrasts across tissue types. Contrast of tissue types in (A) symbiotic insects and (B) aposymbiotic insects. Only the top five up and downregulated categories in each ontology group are plotted. Ontology groups include Biological Process (BP), Cellular Component (CC), and Molecular Function. Not all categories had unidirectional gene regulation, resulting in categories with both up and down regulation. Adjusted p-values have been log10 transformed for better visualization of results. Gene ratio was calculated as the percentage of total DEGs identified in the given GO term (restricted to only gene assigned to a GO term).
Known insect immune genes are upregulated in body tissues relative to crypts, regardless of symbiont status
We further examined subsets of DEGs for evidence of immune gene activity. Using our database of canonical insect immune genes, we were able to detect immune genes among the significant DEGs for all comparisons (Table 1). Immune gene hits for each comparison are summarized in Tables 2, 3. Some A. tristis genes mapped to multiple, similar known immune genes in D. melanogaster and A. pisum. We filtered out immune genes that duplicated Gene ID, role, pathway, and name in order to present only unique immune gene hits here.
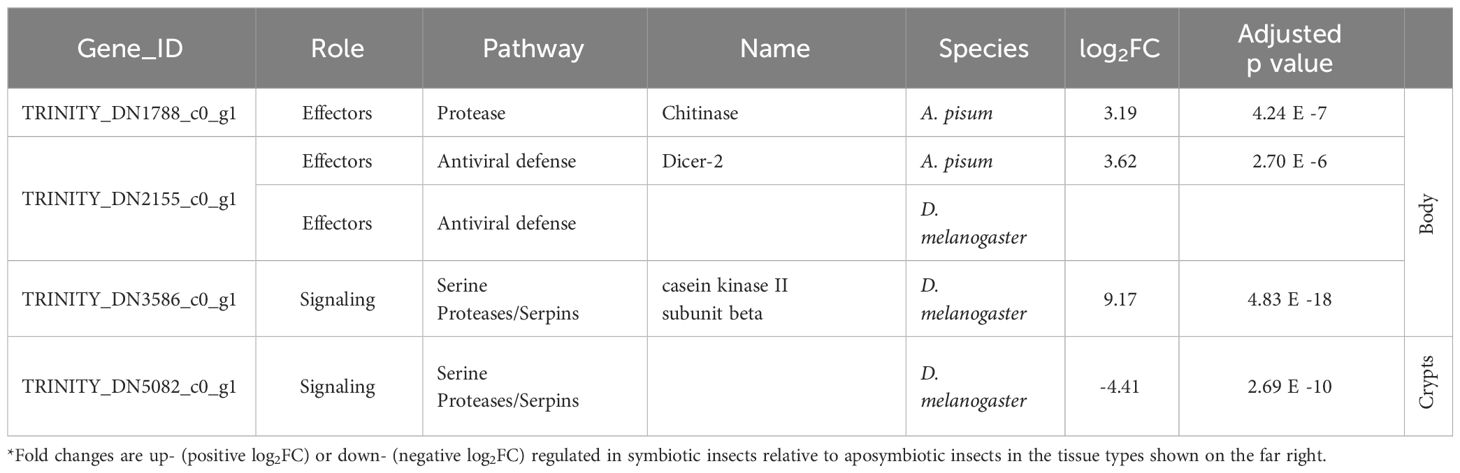
Table 2 Summary of host immune genes differentially expressed in symbiotic insects vs. aposymbiotic insects.
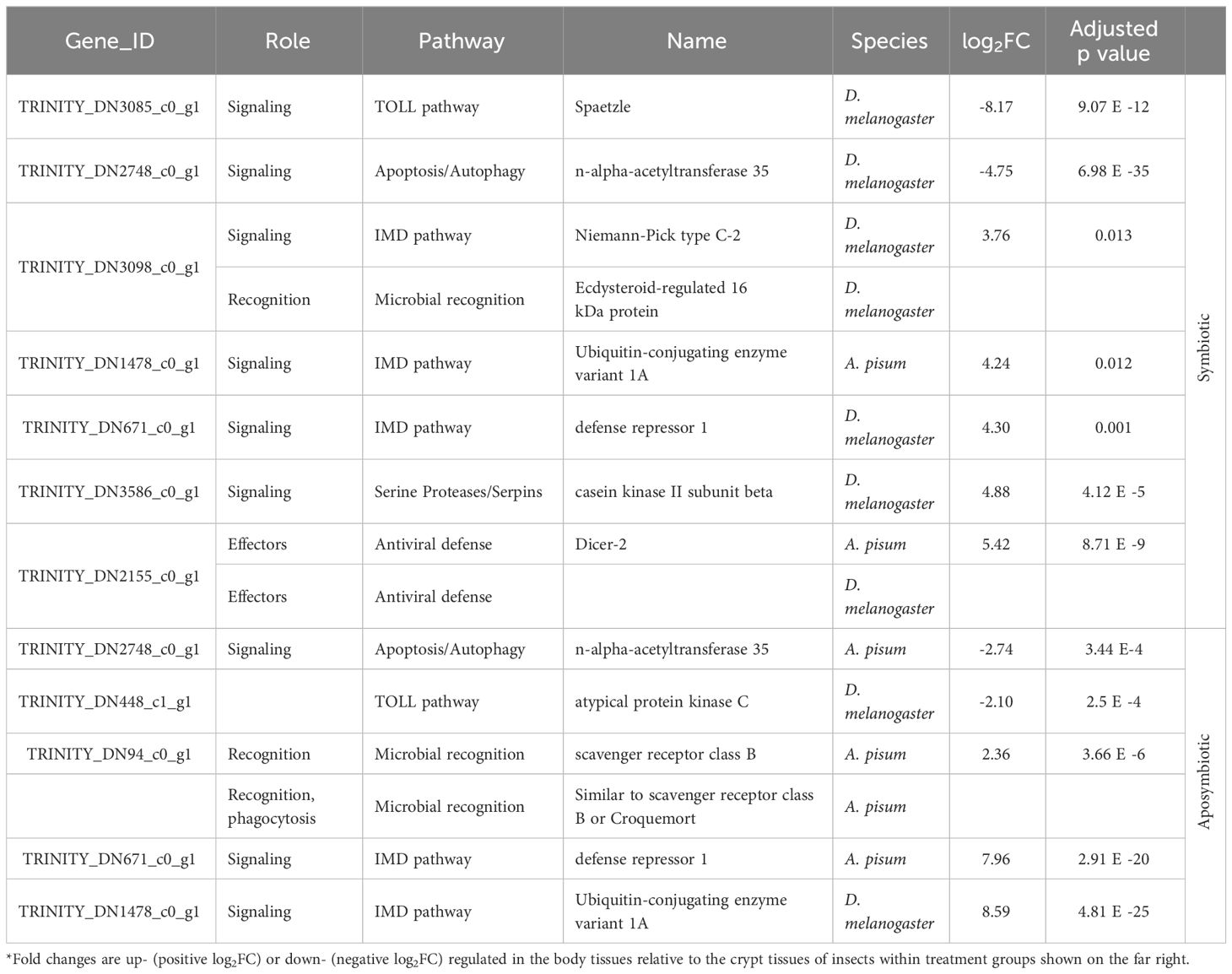
Table 3 Summary of host immune genes differentially expressed in body vs. crypt tissues in insects of different symbiont status.
We identified three immune-related DEGs when comparing symbiotic to aposymbiotic body tissues. All three of these genes were upregulated in the symbiotic samples. We identified only one immune-related DEG when comparing symbiotic to aposymbiotic crypts, and it was downregulated in the symbiotic crypts (Table 2). Differences in immune gene expression were more apparent between insect host tissue types. We identified the most immune-related DEGs when comparing body tissues to crypt tissues in symbiotic bugs (Table 3). The majority of these genes were upregulated in body tissues relative to the crypts. When comparing aposymbiotic host tissues, we found those immune genes upregulated in body tissues to have higher log fold changes than downregulated genes (Table 3). In both symbiotic and aposymbiotic comparisons, we found genes associated with apoptosis and autophagy to be downregulated in body tissues relative to the crypts.
Differential gene expression of Caballeronia symbionts grown in vivo and in vitro
We compared transcripts from Caballeronia symbionts found within the crypts of symbiotic insects that were either infected or uninfected with S. marcescens as well as symbionts grown in liquid culture (in the absence of S. marcescens). Principal component analysis of transcripts revealed clustering of bacterial samples based on whether Caballeronia was grown in the insect crypts or in vitro (Figure 7A), but we noted no pattern of clustering based on whether Caballeronia was in hosts exposed to S. marcescens. Within the in vivo crypt populations that we were able to examine, we detected no differential gene expression when comparing crypt symbionts in insects exposed to S. marcescens to crypt symbionts in unexposed insects (Supplementary Figure 3). Growth condition (i.e., growth in vivo or in vitro) was the most pertinent factor in determining sample clustering, explaining 56% of the variance in our data. Because we noted no effect of S. marcescens exposure status on crypt symbionts, subsequent analyses compare all crypt symbiont samples, regardless of S. marcescens exposure, to in vitro samples.
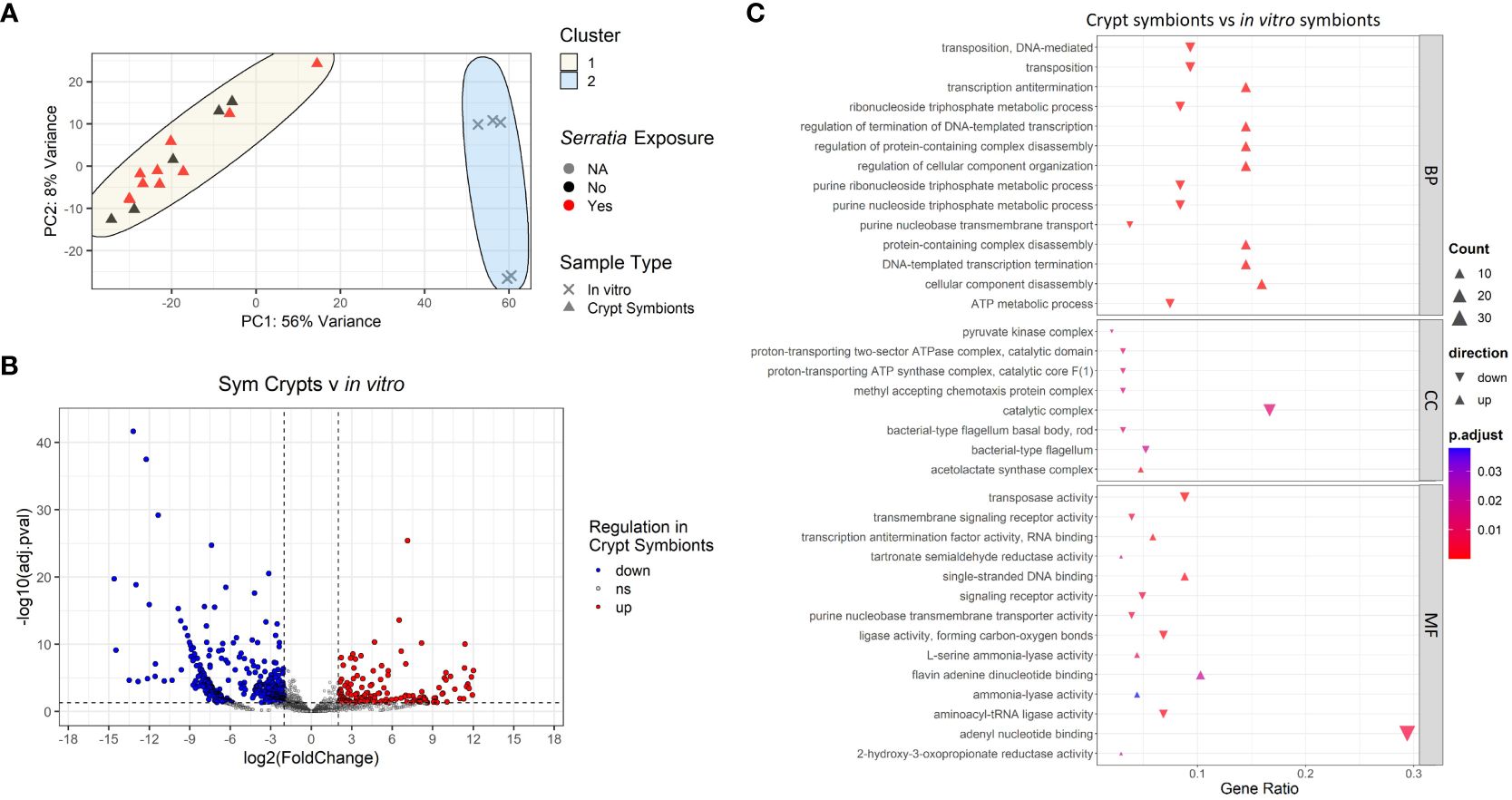
Figure 7 Transcriptomic analyses of Caballeronia symbionts growing in symbiotic insect crypts and in vitro. (A) Principal component analysis of gene transcript counts of Caballeronia growing in symbiotic insect crypts with and without exposure to S. marcescens and in vitro in liquid culture. Sample clustering was apparent by sample type, but differential S. marcescens exposure of the insect host did not result in differential clustering of symbionts. (B) Volcano plot of differentially expressed genes across samples in different growth conditions (crypts vs in vitro culture). Dashed vertical lines indicate cut off for log2 fold changes greater than 2, while the horizontal dashed line corresponds to an adjusted p-value of 0.05. All significantly differentially expressed genes are colored either red for upregulated in crypts samples or blue for downregulated in crypt samples. (C) GO enrichment analysis of differentially expressed genes of Caballeronia symbionts from the crypts vs. in vitro. Only the top five up and downregulated categories in each ontology group are plotted. Ontology groups are Biological Process (BP), Cellular Component (CC), and Molecular Function. Gene ratio was calculated as the percentage of total DEGs identified in the given GO term (restricted to only genes we could annotate).
Despite our depleted samples, we were able to detect evidence of significant differential gene expression corresponding to symbiont growth condition (Figure 7B). GO enrichment analysis of these DEGs showed downregulation of cellular components related to bacterial motility (i.e., bacterial-type flagellum) and processes associated with rapid growth (nucleotide and ATP metabolism) in Caballeronia residing in the crypts relative to those grown in vitro in liquid culture (Figure 7C). We also noted upregulation of transcription antitermination and regulation of DNA-templated transcription in crypt symbionts relative to in vitro symbionts. Furthermore, we observed upregulation of the acetolactate synthase complex—an enzyme that catalyzes the synthesis of three essential amino acids: valine, leucine, and isoleucine—in crypt symbionts relative to symbionts grown in vitro.
Discussion
Our work explores the role of bacterial Caballeronia symbionts in altering the gene expression of their insect host (A. tristis). Despite stark differences in pathogen-clearing abilities in symbiotic and aposymbiotic A. tristis (Mendiola et al., 2022), we found minimal evidence that this phenomenon is due to transcriptional changes elicited by symbiont colonization of the insect host. Comparisons of aposymbiotic and symbiotic insects revealed differences in metabolism and biosynthesis, but comparisons across insect tissue types, not symbiont status, exhibited the greatest differentiation. Lastly, we attempted to gain insight into the in vivo Caballeronia transcriptome. In this regard, our findings are limited, though we can say that there are differences between symbionts growing in vivo and in vitro that are worth exploring further.
Differential clustering by S. marcescens infection status was not apparent in our analyses. Gene expression in S. marcescens infected and uninfected samples showed few differentially expressed genes (DEGs) in either symbiotic or aposymbiotic insects (Supplementary Figure 1), indicating a lackluster response to infection. Even in aposymbiotic insects, where S. marcescens infections can reach high titers (Mendiola et al., 2022), we observed a minimal differential immune response to mitigate S. marcescens infection and no gene expression response that would suggesting that S. marcescens is influencing other host processes. We also found only a handful of known immune genes to be significantly differentially expressed in symbiotic and aposymbiotic individuals, suggesting that Caballeronia infection is not priming the host immune system through upregulation of immune genes in such a way as to mitigate S. marcescens clearance upon infection. This was surprising given the stark differences in S. marcescens clearance between aposymbiotic and symbiotic insects that have been previously documented (Mendiola et al., 2022). While some S. marcescens strains are recognized as virulent insect pathogens (Omoya and Kelly, 2014; Wang and Rozen, 2018), phytopathogenic S. marcescens strains are able to persistently colonize A. tristis (Pair et al., 2004; Wayadande et al., 2005; Mendiola et al., 2022) without ill effects (Mendiola et al., 2022). Mounting an immune response can be energetically costly for the insect host (Ardia et al., 2012), and in the case of vectored pathogens, evolution toward reduced antagonism would favor longer host infection times and could contribute to higher transmission success. As a whole, there is broad evidence for a net neutral effect of vectored pathogens on their vectors (Santiago et al., 2023). The lack of transcriptional differences between S. marcescens infected and uninfected insects that we observed suggests that this may be the case for A. tristis as well.
While there may be immune processes that we cannot capture through gene expression analyses (i.e., changes in the number of immune cells), we hypothesize that the rapid clearance of S. marcescens observed in symbiotic individuals is a byproduct of other changes in the host elicited by the symbiosis, or of indirect bacterial interactions within the host, rather than a result of A. tristis mounting a direct immune response upon S. marcescens infection. Symbiosis with Caballeronia has been shown to result in diverse benefits across many insect species (Kim et al., 2015; Kaltenpoth and Flórez, 2020; Hunter et al., 2022; Lee and Lee, 2023). Such changes could influence the outcome of phytopathogen infection, even when not directly tied to host immunity. Though the exact localization of S. marcescens within A. tristis tissues is unknown, we know it is excluded from the crypts, where Caballeronia is the most abundant microbe (Acevedo et al., 2021; Stoy et al., 2023). The apparent lack of opportunity for significant direct interaction suggests instead that S. marcescens and Caballeronia may engage in indirect competition for host resources (Gerardo and Parker, 2014). In this case Caballeronia may have the advantage of procuring host resources, interfering with the long-term establishment of S. marcescens. Alternatively, S. marcescens may not be able to proliferate in the face of a suite of developmental changes that occur upon Caballeronia infection, which include changes in molting rate (Acevedo et al., 2021) and gut development. In the absence of Caballeronia, however, S. marcescens can grow and establish without interference. Future research should determine where S. marcescens proliferates in its vectors and how tissue development and metabolites vary in these areas in Caballeronia-infected and Caballeronia-uninfected individuals. Unfortunately, our own attempt to characterize the transcriptome of Caballeronia symbionts in vivo was unsuccessful and could shed little light into the specific mechanisms at play.
Transcriptional differences between symbiotic and aposymbiotic insects can give us clues as to how hosts respond to symbiosis. We found upregulation of metabolic processes in the bodies of symbiotic insects relative to the bodies of aposymbiotic insects. This relative increase in metabolism may reflect the fact that symbiotic A. tristis dedicate more time to feeding than aposymbiotic insects (Villa et al., 2023) and grow more rapidly (Acevedo et al., 2021), requiring more energy to sustain their rapid development. We further found several biosynthetic processes upregulated in the symbiotic crypts relative to the aposymbiotic crypts. As a symbiotic organ, the crypts in A. tristis are at the interface of host-symbiont interactions. Within the crypts, the host must balance the need to provision its symbionts with necessary nutrients to sustain healthy symbiont populations as well as to glean benefits from the conversion of those nutrients into necessary byproducts that the host itself cannot produce (Smith and Moran, 2020).
In addition to differences in tissues across symbiont status, we observed differential gene expression between crypt and body tissues within both aposymbiotic and symbiotic individuals. This suggests that the symbiotic organs may be primed for microbial occupation prior to host colonization by Caballeronia in a way that the remainder of the insect body is not. When comparing tissues within symbiotic individuals, we found downregulation of cell-cell communication and signaling in the body relative to the crypts. This is consistent with the crypts’ function as a symbiotic organ where coordination between A. tristis and Caballeronia is paramount for successful symbiosis. Enhanced cell communication and signaling in the crypts thus reflects the need of the host to dynamically manage its symbiotic population (Chomicki et al., 2020). We failed to see similar regulation of signaling when comparing body and crypt tissues in aposymbiotic insects suggesting that the presence of Caballeronia is necessary to drive these transcriptional changes. This is consistent with work in the R. pedestris-Caballeronia symbiosis, which shows that symbiont colonization triggers morphological and transcriptional changes in crypt tissues (Kikuchi et al., 2020; Jang et al., 2023), a phenomenon also seen in other animals (Montgomery and McFall-Ngai, 1994; Couret et al., 2019). In A. tristis, morphological differences in the crypts of symbiotic and aposymbiotic individuals are apparent even at early developmental stages with symbiotic individuals showing robust, invaginated crypts while the crypts of aposymbiotic bugs appear atrophied (Supplementary Figure 2).
Our investigation of immune gene expression supports the local downregulation of insect immunity within the symbiotic organs, particularly in symbiotic individuals. We identified three immune-related DEGs when comparing symbiotic to aposymbiotic bodies, all three of which were upregulated in the symbiotic samples. While these genes could be central to more rapid clearance of S. marcescens in symbiotic than aposymbiotic insects, this seems unlikely given their putative activity (chitinase and antiviral defense). We further identified one differentially expressed immune gene between symbiotic and aposymbiotic crypts which was downregulated in the symbiotic crypts. When we compared body and crypt tissues in symbiotic insects, we found most immune genes to be upregulated in the insect body. Particularly, genes in the Immune Deficiency (IMD) pathway, which responds primarily to Gram negative bacteria like Caballeronia, were downregulated in the crypts. A similar trend was also apparent when comparing body and crypt tissues in aposymbiotic insects.
Unfortunately, our experimental design was unable to capture a complete picture of Caballeronia gene expression patterns. We were unable to glean much insight into Caballeronia symbionts residing in body tissues outside of the crypts, most likely due to low abundance of Caballeronia in other insect body tissues and high abundance of RNA from other microbes and the insect host. There were substantial DEGs when comparing Caballeronia symbionts growing in vivo in the insect crypts and in vitro. Though our study lacks the power to examine these differences in depth, we did find downregulation of genes related to bacterial motility in crypt symbionts relative to those in culture, as well as upregulation of genes necessary for the production of essential amino acids that are lacking in the insect plant sap diet (Douglas, 2016; Michalik et al., 2023). Nutritional symbioses are common in herbivorous insects (Gündüz and Douglas, 2008; Shan et al., 2021; Stever et al., 2021), often providing otherwise limiting nutrients that enable host development. Though preliminary, these findings provide a first insight into the specific role of Caballeronia symbionts in A. tristis’ development.
Our work adds to the growing body of evidence that shows differentiation among symbiotic and nonsymbiotic tissues across eukaryotic organisms (Nakabachi et al., 2005; Moriano-Gutierrez et al., 2019; Tang et al., 2021). In horizontally transmitted mutualisms, these differences are further exacerbated upon successful symbiont colonization (Jang et al., 2023) and most likely enable the persistence of the microbial population within the insect host. Furthermore, we found that Caballeronia alters its gene expression when in symbiosis with A. tristis versus its free-living state. Such transcriptional changes have been documented in other symbionts with free-living stages (Becker et al., 2004; Bellantuono et al., 2019; Maor-Landaw et al., 2020) and are hypothesized to facilitate the cooperation of host and symbiont. A more in-depth examination of these pathways could identify the genes underlying the successful symbiotic interaction between Caballeronia and A. tristis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA, SRR27463935–SRR27463968; NCBI BioProject, PRJNA1062536.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
SM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. JC: Data curation, Investigation, Writing – review & editing. BL: Writing – review & editing, Data curation, Formal analysis, Investigation. DC: Conceptualization, Supervision, Visualization, Writing – review & editing. NV: Conceptualization, Supervision, Writing – review & editing, Methodology, Visualization. NG: Conceptualization, Funding acquisition, Resources, Supervision, Visualization, Writing – review & editing, Methodology.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding for this work was provided by the United States Department of Agriculture in a grant to NG (USDA NIFA 2019-67013-29371). SM was supported by a USDA NIFA Predoctoral Fellowship (project 2021-67034-35147).
Acknowledgments
We thank the Georgia Genomics and Bioinformatics Core (GGBC) at the University of Georgia in Athens for carrying out sample library prep and sequencing (RRID: SCR_010994). We also thank Gabe DuBose for advice on data analysis and coding genius Virgille Arquisola for assistance with code.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1390625/full#supplementary-material
References
Acevedo T. S., Fricker G. P., Garcia J. R., Alcaide T., Berasategui A., Stoy K. S., et al. (2021). The importance of environmentally acquired bacterial symbionts for the squash bug (Anasa tristis), a significant agricultural pest. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.719112
Ardia D. R., Gantz J. E., Brent C., Schneider, Strebel S. (2012). Costs of immunity in insects: an induced immune response increases metabolic rate and decreases antimicrobial activity. Funct. Ecol. 26, 732–739. doi: 10.1111/j.1365-2435.2012.01989.x
Becker A., Bergès H., Krol E., Bruand C., Rüberg S., Capela D., et al. (2004). Global Changes in Gene Expression in Sinorhizobium meliloti 1021 under Microoxic and Symbiotic Conditions. MPMI 17, 292–303. doi: 10.1094/MPMI.2004.17.3.292
Bellantuono A. J., Dougan K. E., Granados-Cifuentes C., Rodriguez-Lanetty M. (2019). Free-living and symbiotic lifestyles of a thermotolerant coral endosymbiont display profoundly distinct transcriptomes under both stable and heat stress conditions. Mol. Ecol. 28, 5265–5281. doi: 10.1111/mec.15300
Bextine B. R. (2001) Insect transmission of Serratia marcescens, the causal agent of cucurbit yellow vine disease (United States – Oklahoma: Oklahoma State University). Available online at: https://www.proquest.com/docview/275865498/abstract/6267CD27D0AD4BB0PQ/1 (Accessed 26, 2023).
Bextine B., Wayadande A., Bruton B. D., Pair S. D., Mitchell F., Fletcher J. (2001). Effect of insect exclusion on the incidence of yellow vine disease and of the associated bacterium in squash. Plant Dis. 85, 875–878. doi: 10.1094/PDIS.2001.85.8.875
Brownlie J. C., Johnson K. N. (2009). Symbiont-mediated protection in insect hosts. Trends Microbiol. 17, 348–354. doi: 10.1016/j.tim.2009.05.005
Bruton B. D., Fletcher J., Pair S. D., Shaw M., Sittertz-Bhatkar H. (1998). Association of a phloem-limited bacterium with yellow vine disease in cucurbits. Plant Dis. 82, 512–520. doi: 10.1094/PDIS.1998.82.5.512
Couret J., Huynh-Griffin L., Antolic-Soban I., Acevedo-Gonzalez T. S., Gerardo N. M. (2019). Even obligate symbioses show signs of ecological contingency: Impacts of symbiosis for an invasive stinkbug are mediated by host plant context. Ecol. Evol. 9, 9087–9099. doi: 10.1002/ece3.5454
Chomicki G., Werner G. D. A., West S. A., Kiers E. T. (2020). Compartmentalization drives the evolution of symbiotic cooperation. Philosophical Transactions of the Royal Society B: Biological Sciences 375, 20190602. doi: 10.1098/rstb.2019.0602
Douglas A. E. (2014). The molecular basis of bacterial–insect symbiosis. J. Mol. Biol. 426, 3830–3837. doi: 10.1016/j.jmb.2014.04.005
Douglas A. E. (2016). How multi-partner endosymbioses function. Nat. Rev. Microbiol. 14, 731–743. doi: 10.1038/nrmicro.2016.151
Douglas A. E. (2020). Housing microbial symbionts: evolutionary origins and diversification of symbiotic organs in animals. Philos. Trans. R. Soc. B: Biol. Sci. 375, 20190603. doi: 10.1098/rstb.2019.0603
Ferrarini M. G., Dell’Aglio E., Vallier A., Balmand S., Vincent-Monégat C., Hughes S., et al. (2022). Efficient compartmentalization in insect bacteriomes protects symbiotic bacteria from host immune system. Microbiome 10, 156. doi: 10.1186/s40168-022-01334-8
Ganesan R., Wierz J. C., Kaltenpoth M., Flórez L. V. (2022). How it all begins: bacterial factors mediating the colonization of invertebrate hosts by beneficial symbionts. Microbiol. Mol. Biol. Rev. 86, e00126–e00121. doi: 10.1128/mmbr.00126-21
Gerardo N. M., Parker B. J. (2014). Mechanisms of symbiont-conferred protection against natural enemies: an ecological and evolutionary framework. Curr. Opin. Insect Sci. 4, 8–14. doi: 10.1016/j.cois.2014.08.002
Gonella E., Crotti E., Mandrioli M., Daffonchio D., Alma A. (2018). Asaia symbionts interfere with infection by Flavescence dorée phytoplasma in leafhoppers. J. Pest Sci. 91, 1033–1046. doi: 10.1007/s10340-018-0973-1
Gündüz E. A., Douglas A. E. (2008). Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc. R. Soc. B: Biol. Sci. 276, 987–991. doi: 10.1098/rspb.2008.1476
Heddi A., Vallier A., Anselme C., Xin H., Rahbe Y., Wäckers F. (2005). Molecular and cellular profiles of insect bacteriocytes: mutualism and harm at the initial evolutionary step of symbiogenesis. Cell. Microbiol. 7, 293–305. doi: 10.1111/cmi.2005.7.issue-2
Hunter M. S., Umanzor E. F., Kelly S. E., Whitaker S. M., Ravenscraft A. (2022). Development of common leaf-footed bug pests depends on the presence and identity of their environmentally acquired symbionts. Appl. Environ. Microbiol. 88, e01778–e01721. doi: 10.1128/aem.01778-21
Jang S., Matsuura Y., Ishigami K., Mergaert P., Kikuchi Y. (2023). Symbiont coordinates stem cell proliferation, apoptosis, and morphogenesis of gut symbiotic organ in the stinkbug-Caballeronia symbiosis. Front. Physiol. 13. doi: 10.3389/fphys.2022.1071987
Kaltenpoth M., Flórez L. V. (2020). Versatile and dynamic symbioses between insects and burkholderia bacteria. Annu. Rev. Entomology 65, 145–170. doi: 10.1146/annurev-ento-011019-025025
Kikuchi Y., Ohbayashi T., Jang S., Mergaert P. (2020). Burkholderia insecticola triggers midgut closure in the bean bug Riptortus pedestris to prevent secondary bacterial infections of midgut crypts. ISME J. 14, 1627–1638. doi: 10.1038/s41396-020-0633-3
Kim J. K., Kim N. H., Jang H. A., Kikuchi Y., Kim C.-H., Fukatsu T., et al. (2013). Specific midgut region controlling the symbiont population in an insect-microbe gut symbiotic association. Appl. Environ. Microbiol. 79, 7229–7233. doi: 10.1128/AEM.02152-13
Kim J. K., Lee J. B., Huh Y. R., Jang H. A., Kim C.-H., Yoo J. W., et al. (2015). Burkholderia gut symbionts enhance the innate immunity of host Riptortus pedestris. Dev. Comp. Immunol. 53, 265–269. doi: 10.1016/j.dci.2015.07.006
Lee J., Lee D.-W. (2023). Burkholderia gut symbiont induces insect host fecundity by modulating Kr-h1 gene expression. Arch. Insect Biochem. Physiol. 112, e21987. doi: 10.1002/arch.21987
Maor-Landaw K., van Oppen M. J. H., McFadden G. I. (2020). Symbiotic lifestyle triggers drastic changes in the gene expression of the algal endosymbiont Breviolum minutum (Symbiodiniaceae). Ecol. Evol. 10, 451–466. doi: 10.1002/ece3.5910
Mendiola S. Y., Stoy K. S., DiSalvo S., Wynn C. L., Civitello D. J., Gerardo N. M. (2022). Competitive exclusion of phytopathogenic serratia marcescens from squash bug vectors by the gut endosymbiont caballeronia. Appl. Environ. Microbiol. 88, e01550–e01521. doi: 10.1128/AEM.01550-21
Michalik A., Bauer E., Szklarzewicz T., Kaltenpoth M. (2023). Nutrient supplementation by genome-eroded Burkholderia symbionts of scale insects. ISME J. 17, 2221–2231. doi: 10.1038/s41396-023-01528-4
Montgomery M. K., McFall-Ngai M. (1994). Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120, 1719–1729. doi: 10.1242/dev.120.7.1719
Moreira L. A., Iturbe-Ormaetxe I., Jeffery J. A., Lu G. J., Pyke A. T., Hedges L. M., et al. (2009). A Wolbachia symbiont in Aedes aEgypti limits infection with dengue, chikungunya, and Plasmodium. Cell 139, 1268–1278. doi: 10.1016/j.cell.2009.11.042
Moriano-Gutierrez S., Koch E. J., Bussan H., Romano K., Belcaid M., Rey F. E., et al. (2019). Critical symbiont signals drive both local and systemic changes in diel and developmental host gene expression. Proc. Natl. Acad. Sci. 116, 7990–7999. doi: 10.1073/pnas.1819897116
Nakabachi A., Shigenobu S., Sakazume N., Shiraki T., Hayashizaki Y., Carninci P., et al. (2005). Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc. Natl. Acad. Sci. 102, 5477–5482. doi: 10.1073/pnas.0409034102
Ohbayashi T., Mergaert P., Kikuchi Y. (2020). “Chapter 2 - Host-symbiont specificity in insects: Underpinning mechanisms and evolution,” in Advances in Insect Physiology. Eds. Oliver K. M., Russell J. A. (Elsevier, 125 London Wall, London, EC2Y 5AS, United Kingdom: Academic Press), 27–62. doi: 10.1016/bs.aiip.2020.03.002
Oliver K. M., Noge K., Huang E. M., Campos J. M., Becerra J. X., Hunter M. S. (2012). Parasitic wasp responses to symbiont-based defense in aphids. BMC Biol. 10, 11. doi: 10.1186/1741-7007-10-11
Oliver K. M., Perlman S. J. (2020). “Chapter Eight - Toxin-mediated protection against natural enemies by insect defensive symbionts,” in Advances in Insect Physiology. Eds. Oliver K. M., Russell J. A. (Academic Press), 277–316. doi: 10.1016/bs.aiip.2020.03.005
Omoya F. O., Kelly B. A. (2014). Variability of the potency of some selected entomopathogenic bacteria (Bacillus spp. and Serratia spp.) on termites, Macrotermes bellicosus (Isoptera: Termitidae) after exposure to magnetic fields. Int. J. Trop. Insect Sci. 34, 98–105. doi: 10.1017/S1742758414000253
Pair S. D., Bruton B. D., Mitchell F., Fletcher J., Wayadande A., Melcher U. (2004). Overwintering squash bugs harbor and transmit the causal agent of cucurbit yellow vine disease. J. Economic Entomology 97, 74–78. doi: 10.1093/jee/97.1.74
Price D. R. G., Duncan R. P., Shigenobu S., Wilson A. C. C. (2011). Genome expansion and differential expression of amino acid transporters at the aphid/buchnera symbiotic interface. Mol. Biol. Evol. 28, 3113–3126. doi: 10.1093/molbev/msr140
Russell S. L., Castillo J. R. (2020). “Trends in Symbiont-Induced Host Cellular Differentiation,” in Symbiosis: Cellular, Molecular, Medical and Evolutionary Aspects. Ed. Kloc M. (Springer International Publishing, Cham), 137–176. doi: 10.1007/978–3-030–51849-3_5
Santiago Ma. F.M., King K. C., Drew G. C. (2023). Interactions between insect vectors and plant pathogens span the parasitism–mutualism continuum. Biol. Lett. 19, 20220453. doi: 10.1098/rsbl.2022.0453
Shan H., Wu W., Sun Z., Chen J., Li H. (2021). The gut microbiota of the insect infraorder pentatomomorpha (Hemiptera: heteroptera) for the light of ecology and evolution. Microorganisms 9, 464. doi: 10.3390/microorganisms9020464
Smith T. E., Moran N. A. (2020). Coordination of host and symbiont gene expression reveals a metabolic tug-of-war between aphids and Buchnera. Proc. Natl. Acad. Sci. 117, 2113–2121. doi: 10.1073/pnas.1916748117
Stever H., Eiben J., Bennett G. M. (2021). Hawaiian nysius insects rely on an obligate symbiont with a reduced genome that retains a discrete nutritional profile to match their plant seed diet. Genome Biol. Evol. 13, evab189. doi: 10.1093/gbe/evab189
Stoy K. S., Chavez J., De Las Casas V., Talla V., Berasategui A., Morran L. T., et al. (2023). Evaluating coevolution in a horizontally transmitted mutualism. Evolution 77, 166–185. doi: 10.1093/evolut/qpac009
Sudakaran S., Retz F., Kikuchi Y., Kost C., Kaltenpoth M. (2015). Evolutionary transition in symbiotic syndromes enabled diversification of phytophagous insects on an imbalanced diet. ISME J. 9, 2587–2604. doi: 10.1038/ismej.2015.75
Tang N., Lebreton A., Xu W., Dai Y., Yu F., Martin F. M. (2021). Transcriptome profiling reveals differential gene expression of secreted proteases and highly specific gene repertoires involved in lactarius–pinus symbioses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.714393
Villa S. M., Chen J. Z., Kwong Z., Acosta A., Vega N. M., Gerardo N. M. (2023). Specialized acquisition behaviors maintain reliable environmental transmission in an insect-microbial mutualism. Curr. Biol. 33, 2830–2838.e4. doi: 10.1016/j.cub.2023.05.062
Wang Y., Rozen D. E. (2018). Gut microbiota in the burying beetle, Nicrophorus vespilloides, provide colonization resistance against larval bacterial pathogens. Ecol. Evol. 8, 1646–1654. doi: 10.1002/ece3.3589
Wang Z., Yong H., Zhang S., Liu Z., Zhao Y. (2023). Colonization resistance of symbionts in their insect hosts. Insects 14, 594. doi: 10.3390/insects14070594
Wayadande A., Bruton B., Fletcher J., Pair S., Mitchell F. (2005). Retention of Cucurbit Yellow Vine Disease Bacterium Serratia marcescens Through Transstadial Molt of Vector Anasa tristis (Hemiptera: Coreidae). Ann. Entomological Soc. America 98, 770–774. doi: 10.1603/0013-8746(2005)098[0770:ROCYVD]2.0.CO;2
Weiss B., Aksoy S. (2011). Microbiome influences on insect host vector competence. Trends Parasitol. 27, 514. doi: 10.1016/j.pt.2011.05.001
Keywords: symbiosis, Caballeronia, Serratia marcescens, coinfection, transcriptomics
Citation: Mendiola SY, Chen JZ, Lukubye B, Civitello DJ, Vega NM and Gerardo NM (2024) Differential gene expression in the insect vector Anasa tristis in response to symbiont colonization but not infection with a vectored phytopathogen. Front. Ecol. Evol. 12:1390625. doi: 10.3389/fevo.2024.1390625
Received: 23 February 2024; Accepted: 07 May 2024;
Published: 22 May 2024.
Edited by:
Jing Chen, Institute of Zoology (CAS), ChinaReviewed by:
Morten Schiøtt, Technical University of Denmark, DenmarkAb. Matteen Rafiqi, Bezmialem Vakıf University, Türkiye
Copyright © 2024 Mendiola, Chen, Lukubye, Civitello, Vega and Gerardo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandra Y. Mendiola, c2FuZHJhLnkubWVuZGlvbGFAZ21haWwuY29t
†Present address: Sandra Y. Mendiola, Department of Entomology, University of Georgia, Athens, GA, United States
 Sandra Y. Mendiola
Sandra Y. Mendiola Jason Z. Chen1
Jason Z. Chen1 Nicole M. Gerardo
Nicole M. Gerardo