
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 16 February 2024
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 12 - 2024 | https://doi.org/10.3389/fevo.2024.1356316
 Jamael C. Abato1,2*
Jamael C. Abato1,2* Alexei V. Chernyshev3
Alexei V. Chernyshev3 Natsumi Hookabe4
Natsumi Hookabe4 Aoi Tsuyuki5
Aoi Tsuyuki5 Gauri Kaushik6
Gauri Kaushik6 Hiroshi Kajihara7*
Hiroshi Kajihara7*Three new species of the monostiliferous hoplonemertean genus Oerstedia Quatrefages, 1864, are herein described using morphological and molecular data—Oerstedia pseudoculata sp. nov., from Akkeshi Bay and Oshoro Bay, Hokkaido, Japan, and from Aniwa Bay, Sakhalin, Russia; Oerstedia rugosa sp. nov. from Sagami Bay, Misaki, Kanagawa, Japan, and Van Phong Bay, Vietnam; and Oerstedia viridifusca sp. nov. from Manazuru, Kanagawa, Japan. As to the external morphology, O. pseudoculata sp. nov. can be differentiated from O. oculata only by its bright-orange ocelli visible on both sides of the head, and a proboscis pore opening at the ventral tip of the head. These two sister species repeat each other’s color patterns, a phenomenon that can be explained by Vavilov’s law of homologous series. Oerstedia rugosa sp. nov. can be identified by its carmine or deep-red to brownish-red body with several longitudinal, intertwined white lines or wrinkles running from the head to the posterior body, and by 17–23 vaguely bordered white bands composed of variedly sized dots encircling the body, arranged at irregular intervals. Oerstedia viridifusca sp. nov. can be distinguished from other Oerstedia by (i) the entire body flecked with minute greenish-brown dots, especially densely on the anterior portion of the dorsal surface, but sparsely on the posterior half of the ventral surface; (ii) a collar-like portion encircling the body along the posterior cephalic furrow where the greenish-brown dots are absent; (iii) the anterolateral edges of the head lacking the greenish-brown dots; and (iv) the ocelli being brownish-orange in color. Oerstedia phoresiae (Kulikova, 1987) is reported for the first time from Japan, in addition to its previous distribution record in Russia and in South Korea. Phylogenetic analyses based on the 16S, 18S, 28S ribosomal RNA, cytochrome c oxidase subunit I, and histone H3 genes show that the new species are true congeners of the genus Oerstedia with O. pseudoculata sp. nov. and O. viridifusca sp. nov. nested within the clade Paroerstediella whereas O. rugosa sp. nov. in the clade Oerstedia. This taxonomic work emphasizes the importance of DNA barcode sequence in the taxonomy and systematics of the polymorphic congeners of the genus Oerstedia.
The monostiliferous hoplonemertean genus Oerstedia Quatrefages, 1864 (Quartrefages, 1846), is currently composed of 30 valid ribbon worm species (Norenburg et al., 2023; Abato et al., 2023). As to the external morphology, these worms are characterized by having (i) a small body, which is usually stout and cylindrical; (ii) four ocelli, of which the anterior pair are sometimes smaller than the posterior pair; and (iii) cephalic furrows that are either weakly developed or absent (Envall and Sundberg, 1993; Gibson, 1994). The internal characteristics include (i) the presence of accessory lateral nerves and (ii) the absence of a single mid-dorsal vascular plug, but both are not unique for the genus within Eumonostilifera (Chernyshev, 2004; Kajihara, 2017). Oerstedia species can be collected from the littoral, sublittoral, and bathyal zones (Iwata, 1954; Kulikova, 1987; Sundberg and Andersson, 1995; Chernyshev and Polyakova, 2022; Abato et al., 2023). Some congeners are known to exhibit intraspecific polymorphism in terms of internal and external features (Bürger, 1895; Berg, 1972; Sundberg, 1979; Sundberg, 1984; Envall and Sundberg, 1993; Sundberg et al., 2009; Akhmatova et al., 2012; Abato et al., 2023). In fact, the two genera Oerstediella Friedrich, 1935 (Friedrich, 1935), and Paroerstedia Friedrich, 1955 (Friedrich, 1955), were synonymized with Oerstedia because the internal characters supposed to define the two were found to vary within a single species (Envall and Sundberg, 1993), Oerstedia dorsalis (Abilgaard, 1806) (Abildgaard, 1806), the type species of the genus. The polymorphism within the genus (Bürger, 1895; Brunberg, 1964; Sundberg, 1984; Sundberg, 1988; Envall and Sundberg, 1993; Sundberg and Andersson, 1995; Zaslavskaya and Chernyshev, 2008; Sundberg et al., 2009; Akhmatova et al., 2012) makes the taxonomy of its members complicated and problematic.
The occurrence of banded and non-banded Oerstedia species in the Northwest Pacific has been reported by several authors. Of these, the banded forms include the nominal species Oerstedia fuscosparsa Abato et al., 2023 (Abato et al., 2023); Oerstedia polyorbis Iwata, 1954 (Iwata, 1954); Oerstediella valentinae Chernyshev, 1993 (Chernyshev, 1993); Oerstediella verae Chernyshev, 1993 (Chernyshev, 1993); Oerstediella zebra Chernyshev, 1993 (Chernyshev, 1993); and the specimen reported as Oerstedia zebra by Thollesson and Norenburg (2003). The non-banded forms include the nominal species Oerstedia venusta Iwata, 1954 (Iwata, 1954); Oerstediella oculata Kulikova, 1987 (Kulikova, 1987); Oerstediella phoresiae Kulikova, 1987 (Kulikova, 1987); and specimens reported as Oerstedia dorsalis by Iwata (1954) and Oerstedia venusta by Thollesson and Norenburg (2003). Because of the known polymorphism in the genus, a pioneering species-delimitation study on the Oerstedia occurring in the Far-Eastern seas of Russia was conducted based on the 28S ribosomal RNA (rRNA) gene, claiming that the nominal species O. oculata, O. zebra, and the specimens identified by Thollesson and Norenburg (2003) as O. venusta and O. zebra would be conspecific with O. polyorbis (Akhmatova et al., 2012). On the other hand, more recent and comprehensive phylogenetic studies of the genus involving the Pacific and Atlantic congeners—but excluding Thollesson and Norenburg’s (Thollesson and Norenburg, 2003) O. venusta and O. zebra—showed that O. oculata and O. polyorbis are different species, despite close similarities in their morphology (Chernyshev and Polyakova, 2022; Abato et al., 2023). The non-inclusion of Thollesson and Norenburg’s (Thollesson and Norenburg, 2003) Oerstedia in these recent phylogenetic studies makes the taxonomic identity and the phylogeny of these species within the genus still open for investigation.
Most of the Oerstedia species known in the Northwest Pacific were described by Iwata (1954), Kulikova (1987), and Chernyshev (1993), and their distribution is limited in the cold-temperate province (Briggs and Bowen, 2012). Only two have so far been reported from the warm-temperate Sino-Japanese province (Briggs and Bowen, 2012): O. fuscosparsa, established from Honshu, Japan (Abato et al., 2023), and O. phoresiae, reported from Jeju Island, South Korea (Akhmatova et al., 2012). Nemertean investigation efforts are therefore needed in the southern areas of the Northwest Pacific. Thus, in this study, members of the genus Oerstedia were surveyed in Kanagawa, Honshu, Japan, and Van Phong Bay, Vietnam.
In this paper, we employ integrative taxonomy to (i) ascertain the species identity of O. venusta and O. zebra sensu Thollesson and Norenburg (2003); (ii) describe new species from Japan and Vietnam; and (iii) confirm the occurrence of O. phoresiae in Japan. Furthermore, we also infer the phylogenetic positions of the new species within the genus using the newly determined 16S, 18S, 28S rRNA (16S, 18S, and 28S), cytochrome c oxidase subunit I (COI), and histone H3 (H3) gene sequences. This taxonomic work highlights the importance of using both traditional and molecular taxonomy in species identification and delimitation of the genus Oerstedia.
A total of 13 ribbon-worm individuals were collected (i) intertidally around Cape Crillon, Aniwa Bay, Sakhalin, Russia (one specimen); (ii) intertidally among brown algae in Akkeshi Bay, Japan (two specimens), along with the ones used in Thollesson and Norenburg (2003); (iii) intertidally among brown algae of the family Sargassaceae in Oshoro Bay, Japan (seven specimens); (iv) by dredging off Misaki, Japan (one specimen); (v) intertidally among calcareous algae in Manazuru, Japan (one specimen); and (vi) at a depth of 1 m among calcareous algae off Van Phong Bay, Vietnam (one specimen). The single specimen collected in Aniwa Bay was preserved in 99% ethanol. The two specimens collected in Akkeshi Bay were anesthetized in a MgCl2 solution and fixed in Bouin’s fluid, dehydrated in an ethanol series, cleared in xylene, embedded in paraffin wax, sectioned at 6 µm in thickness, and stained with the Mallory trichrome method. The remaining nine specimens collected in Japan were observed and photographed either alive or in anesthetized condition. For anesthetization, the examined live specimens were placed in a transparent Petri dish, to which a MgCl2 solution isotonic to seawater was slowly added. Photomicroscopy was mostly done under a Nikon SMZ1500 stereo microscope, with an attached Nikon D5600 digital camera and with external strobe lighting provided by a pair of Morris Hikaru Komachi Di flash units. When successfully extracted, the proboscis of the worms was observed under an Olympus BX51 compound light microscope for the documentation of the stylet apparatus and photographed using the D5600 digital camera. After morphological examination, the worms were preserved in 99% ethanol for genomic DNA extraction. The single specimen collected in Vietnam was cut into two after being photographed; the anterior half was processed in the same manner as for the specimens in Akkeshi Bay.
Nucleotide sequencing was done for 11 of the 13 specimens (the remaining two were fixed in Bouin’s fluid for histological sectioning). DNA extraction, PCR amplification, and sequencing followed Abato et al. (2023) except that this study used the primer pairs ar-L/br-H (Palumbi et al., 1991) for 16S amplification and sequencing. BLAST searches (Altschul et al., 1997) on the NCBI website (http://www.ncbi.nlm.nih.gov/) were performed to check for possible sequence contamination, to initially verify the genus affiliation of the specimens, and to confirm the identity of the already known Oerstedia species from the collection. Genetic variation between the 11 specimens was determined by calculating uncorrected pairwise genetic distances (p-distances) based on the 657-bp of COI sequences using MEGA ver. 11 (Tamura et al., 2021). A maximum-likelihood analysis based on the same gene from the specimens that resemble O. oculata that were collected from Oshoro Bay (six specimens), Aniwa Bay (one specimen) including the Oerstedia forms (two specimens) in Thollesson and Norenburg (2003), and O. oculata (two specimens) from Russia was performed in RAxML ver. 8.2.10 (Stamatakis, 2015) using the GTR + G model with branch support values calculated from 1,000 bootstrap pseudoreplicates. The same specimens were subjected to a species delimitation analysis using three methods: Assemble Species by Automatic Partitioning (ASAP) (Puillandre et al., 2021) Automatic Barcode Gap Discovery (ABGD) (Puillandre et al., 2012), and PTP (Poisson Tree Processes) (Zhang et al., 2013). ASAP was performed in webserver https://bioinfo.mnhn.fr/abi/public/asap/, ABGD in https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html, and PTP in the bPTP webserver https://species.h-its.org/ptp/—all in default settings. ASAP and ABGD analyses were based on the 657-bp COI sequences, whereas the bPTP analysis used an input tree generated by MrBayes ver. 3.2.6.x64 (parallel version) (Ronquist and Huelsenbeck, 2003; Altekar et al., 2004) based on concatenated 16S, 18S, 28S, COI, and H3 genes of selected Oerstedia species. A TCS haplotype network based on COI was created using PopART (Leigh and Bryant, 2015) for visualizing relationships between haplotypes.
Oerstedia sequences based on the five gene markers used in Abato et al. (2023) and Chernyshev and Polyakova (2022) and those sequences from specimens previously identified as O. venusta and O. zebra by Thollesson and Norenburg (2003) were downloaded from GenBank (Table 1) and used in the phylogenetic analyses together with the newly generated sequences in the present study. MAFFT ver. 7 online version (Katoh et al., 2019) was used for aligning 16S, 18S, and 28S sequences, applying the E-INS-i iterative refinement method in the advanced settings. The COI and H3 sequence datasets were aligned using the MUSCLE algorithm (Edgar, 2004) implemented in MEGA ver. 11 (Tamura et al., 2021) in default settings. Uncertainly aligned sites were trimmed using ClipKIT ver. 1.3.0 (Steenwyk et al., 2020) resulting in 475-bp 16S, 1,759-bp 18S, 2,880-bp 28S, 657-bp COI, and 330-bp H3 sequences, concatenated in MEGA ver. 11 (Tamura et al., 2021). The best-fit partitions and substitution models for maximum likelihood (ML) analyses and Bayesian inference (BI) were determined using PartitionFinder ver. 2.1.1 (Lanfear et al., 2016) employing the greedy algorithm (Lanfear et al., 2012) resulting in five subsets and corresponding models: subset 1, 28S, COI (first codon), and H3 (first codon), TRN + I + G; subset 2, COI (second codon), 18S, and H3 (second codon), TRNEF + I; subset 3, COI (second codon), TIM + G; subset 4, H3 (second codon), TVMEF + G; and subset 5, 16S, GTR + G. ML analyses were executed using IQ-Tree ver. 2.1.3 (Minh et al., 2020) applying the suggested model to each subset with branch support values derived from 1,000 ultrafast bootstrap (UFBoot) (Minh et al., 2013) and RAxML ver. 8.2.10 (Stamatakis, 2015) using the GTR + G model to all subsets with branch support values calculated from 1,000 bootstrap pseudoreplicates. BI was performed using MrBayes ver. 3.2.6 x64 (parallel version) (Ronquist and Huelsenbeck, 2003; Altekar et al., 2004) employing models GTR + I + G for subset 1; GTR + I for subset 2; and GTR + G for subsets 3, 4, and 5, launching two parallel runs with four Markov chains in each run (three cold and one hot) for 5 × 107 generations sampling every 100 generations from the chain. Run convergence was assessed by standard deviation of split frequencies (0.000307), estimated sample sizes for all parameters (>5671), and potential scale reduction factors for all parameters (1.000). FigTree ver. 1.4.4 (Rambaut, 2014) was used to visualize the trees obtained from the ML analyses and BI.
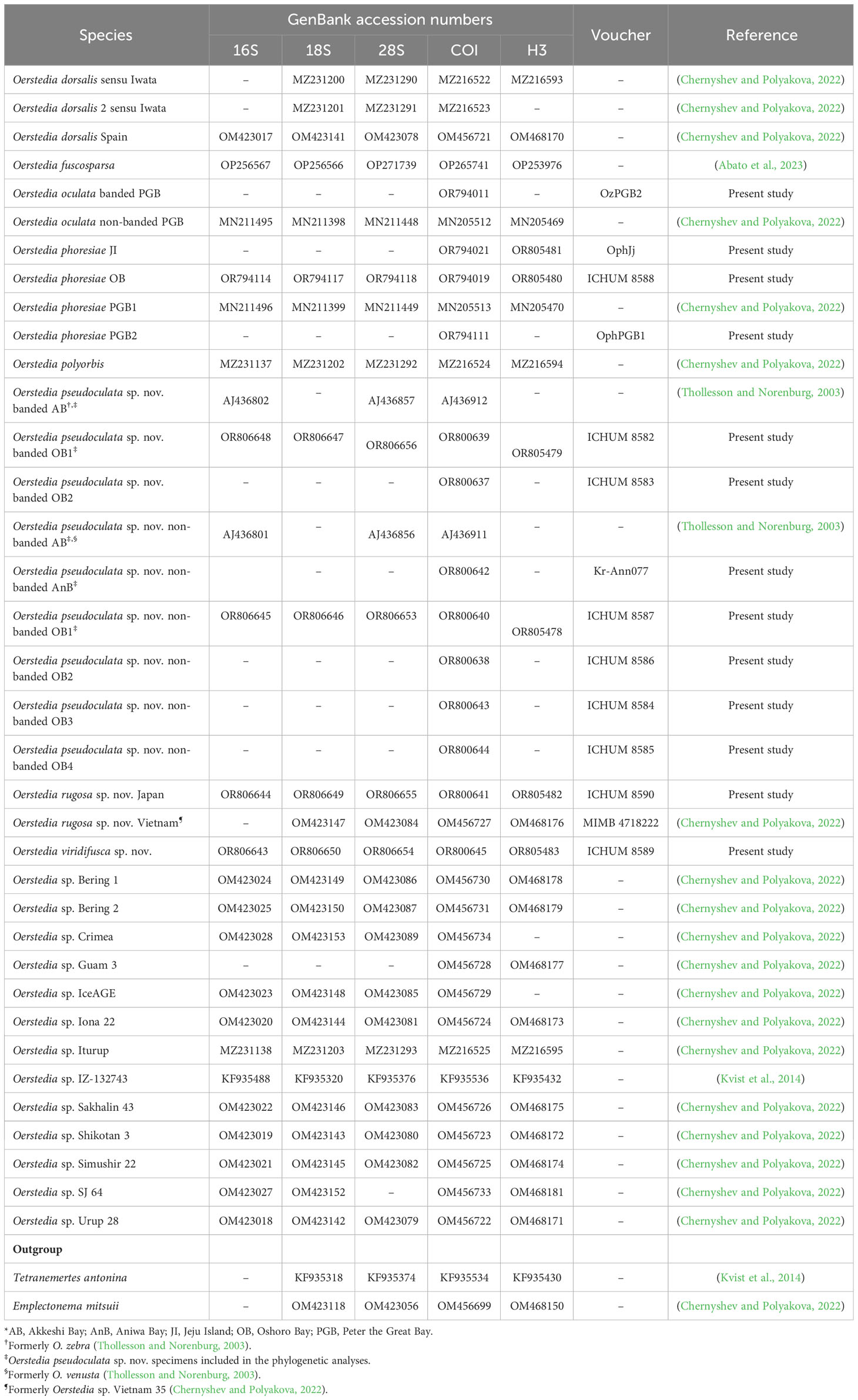
Table 1 List of Oerstedia species sequenced in the present study and/or included in the phylogenetic analyses, GenBank accession numbers for 16S, 18S, 28S rRNA (16S, 18S, and 28S), cytochrome c oxidase subunit I (COI), and histone H3 (H3) gene markers, catalogue numbers, and their references.
Voucher materials for the Oshoro and Akkeshi Bay Oerstedia have been deposited in the Invertebrate Collection of the Hokkaido University Museum (ICHUM), Sapporo, Hokkaido, Japan, whereas the specimen collected from Van Phong Bay has been deposited in the Museum of the Institute of Marine Biology (MIMB), National Scientific Center of Marine Biology, Far Eastern Branch, Russian Academy of Sciences, Vladivostok, Russia. The Aniwa Bay specimen has been deposited in Lomonosov Moscow State University, Moscow, Russia. Nucleotide sequences that were newly generated in this study and used in the phylogenetic analyses have been registered in GenBank. This work has been registered in ZooBank with the Life Science Identifier (LSID) http://zoobank.org/0705E574-771D-43A8-9C89-59397D2BD6C5.
The 13 Oerstedia specimens reported in this paper are composed of four species, of which three are new to science and the remaining one is already known and herein recorded for the first time in Japan. The new species are described below using an integrative approach, whereas their phylogenetic positions within the genus Oerstedia are inferred. This study updates the number and name of valid Oerstedia species reported from Japanese waters by establishing new species, fixing the identity of the species previously reported with uncertain taxonomic identification, and by reporting a new locality of an already known Oerstedia species.
Class Hoplonemertea Coe, 1905 (Coe, 1905).
Family Oerstediidae Chernyshev, 1993 (Chernyshev, 1993).
Genus Oerstedia Quatrefages, 1864 (Quartrefages, 1846).
Oerstedia polyorbis – Kajihara (2017), figure 16.5n; Kajihara (2021), figures 2h, i.
Oerstedia venusta – Thollesson and Norenburg (2003); Kajihara (2017), figure 16.5o.
Oerstedia zebra – Thollesson and Norenburg (2003).
ZooBank LSID. http://zoobank.org/1C729CE2-82C2-4D16-A915-FEDBBA8EA190.
Material examined. Holotype, ICHUM 8587, non-banded form (Figures 1A–H), extracted total DNA, collected by Jamael Abato and Hiroshi Kajihara among the brown algae in the family Sargassaceae, putatively Sargassum horneri (Turner) C. Agardh and/or Sargassum miyabei Yendo in Oshoro Bay (between 43.2093°N, 140.8585°E and 43.2095°N, 140.8582°E), Hokkaido, Japan, on 30/08/2022. Paratypes, extracted total DNA from six individuals and serial transverse sections representing two specimens: ICHUM 8586, non-banded form, DNA, collection data same as the holotype; ICHUM 8582 (Figures 1J–N), 8583, banded forms, DNA, collected by Gauri Kaushik and Hiroshi Kajihara from the topotype on 07/07/2023; ICHUM 8584 (Figure 1I), 8585, non-banded forms, DNA, collected by Gauri Kaushik and Hiroshi Kajihara from the topotype on 27/04/2023; ICHUM 3287, serial transverse sections, 6 µm, eight slides, non-banded form [originally labelled ‘Oerstedia venusta’], intertidal, among brown algae, Aininkappu (43.0038°N, 144.8575°E), Akkeshi, Hokkaido, Japan, collected by Svetlana A. Maslakova, Megan Schwartz, Jon L. Norenburg, and Hiroshi Kajihara on 25/07/1999; ICHUM 3288, serial transverse sections, 6 µm, seven slides, banded form [originally labelled ‘Oerstedia zebra’], collection data same as ICHUM 3287; Kr-Ann077, non-banded form, DNA, collected intertidally by Irina Ekimova and Darya Grishina around Cape Crillon, Aniwa Bay, Sakhalin, Russia, on 07/08/2023.
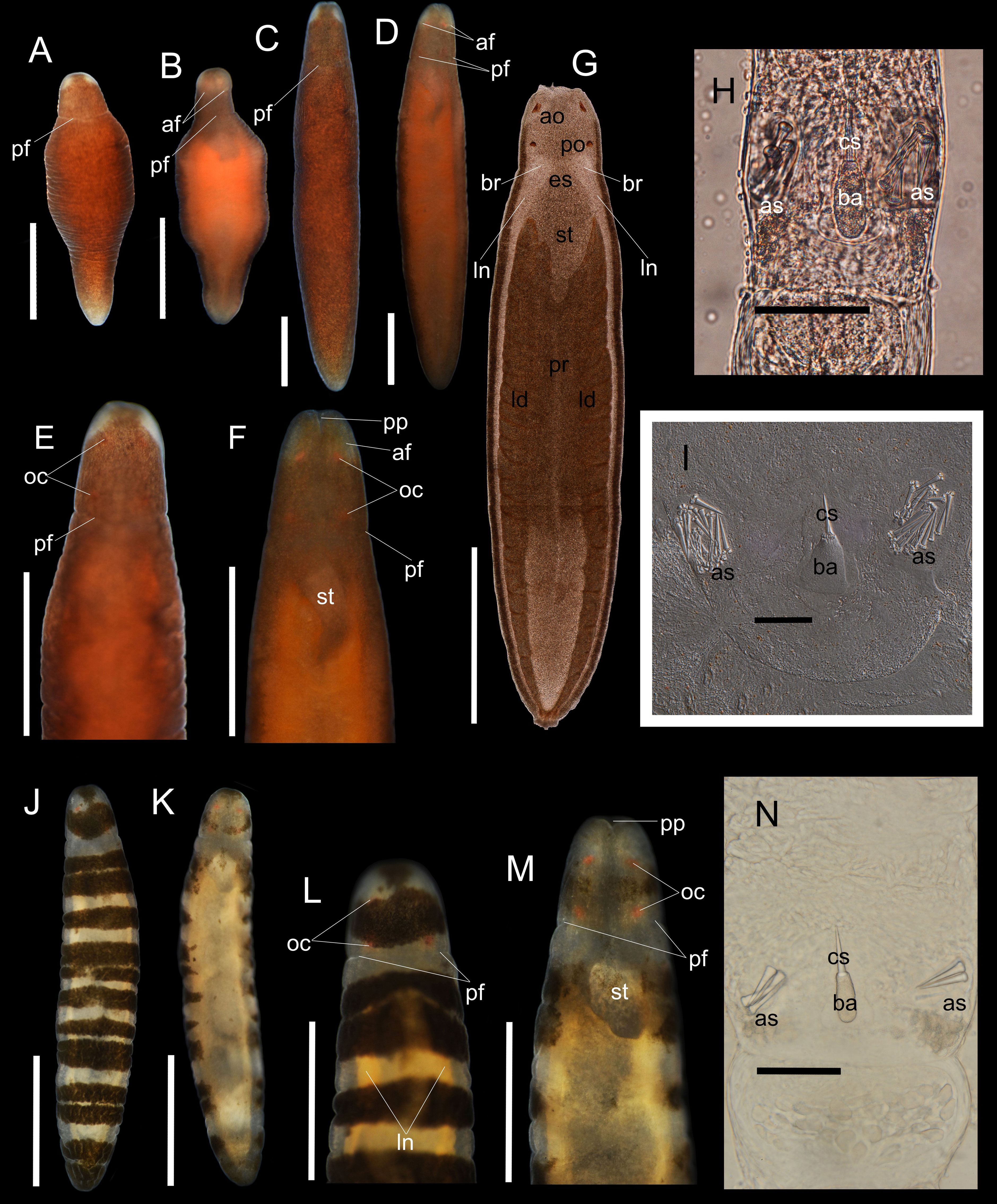
Figure 1 Oerstedia pseudoculata sp. nov., photographs and photomicrographs of the non-banded form, holotype, ICHUM 8587 (A–H), paratype, ICHUM 8584 (I) and the banded form, paratype, ICHUM 8582 (J–N), taken in living and anesthetized states. Entire body at dorsal view (A, J) and ventral view (B, K) in non-anesthetized condition; entire body at dorsal view (C) and ventral view (D) in anesthetized condition; magnification of the head at dorsal view (E, L) and ventral view (F, M) in anesthetized state; squeezed specimen at dorsal view (G); stylet apparatus (H, I, N). af, anterior cephalic furrow; ao, anterior ocelli; as, accessory stylet; ba, stylet basis; br, brain; cs, central stylet; es, esophagus; id; intestinal diverticula; ln, lateral nerves; oc, ocelli; pf, posterior cephalic furrow; po, posterior ocelli; pp, proboscis pore; pr, proboscis; st, stomach. Scale bars: (A, B, E, F, L, M) = 0.5 mm; (C, D, G, J, K) = 1 mm; (H, I, N) = 50 µm.
Etymology. The new specific name is an adjective (-us, -a, -um), a combination of the Latin words pseudo (meaning “falsely,” “resembling,” or “deceived”) and oculatus (meaning “adorned with eyes,” “having eyes,” or “eye-shaped”). In combination, pseudoculata pertains to the resemblance of the new species with another already established congener of the genus, O. oculata. The specific name directly translates as the new species “not being the true O. oculata”.
Diagnosis. An Oerstedia species with a brownish dorsal body pigmentation made up of minute brownish dots or with dorsolateral black transverse bands on a translucent body; anterolateral edges of the head non-pigmented; with bright-orange ocelli visible at the dorsal and ventral sides of the head; and a proboscis pore that opens ventrally near the tip of the head.
Description. External morphology. Body cylindrical, widest at middle portion; living worm highly contractile; holotype (ICHUM 8587) significantly shortened when contracted, 1.3 mm in length, 0.45 mm in width (Figures 1A, B); when anesthetized, holotype 5.6 mm in length, 0.97 mm in width (Figures 1C, D); paratypes 3.0–4.7 mm in length, 0.5–0.73 mm in width (n = 5, ICHUM 8282, 8283, 8584–8586). Dorsal body either banded or non-banded (Figures 1A, C, E, J, L). In non-banded form (ICHUM 8584–8587), dorsal surface uniformly dotted with numerous, fine, brownish dots (Figures 1A, C, E); posterior end opaque white; head similarly dotted with fine, brownish granules except for two, non-pigmented white spots at anterolateral sides, showing an impression of big, whitish, false eyes (Figures 1A, C, E); ventrally, body less pigmented, lighter than the dorsal, reflects brown-orange hue. In banded form (ICHUM 8582, 8583), background body color translucent; 12–14 dorsolateral, vaguely bordered, dark-brown, transverse bands of varying thickness present; ventral body translucent with a pale-yellow hue (Figures 1K, M); head dorsally with single, thick, well bordered, dark-brown transverse band, ventrally less pigmented. Ocelli four in number, bright-orange in color, arranged in square. Anterior cephalic furrows short, situated ventrolaterally, slightly notable in the non-banded form (Figures 1D, F) but not visible in the banded form. Posterior cephalic furrow encircling head (Figures 1A–F) (furrow positioned on the translucent collar of the banded form) (Figures 1J–M). Rhynchocoel extending for entire body length (Figure 1G). Proboscis pore opening at tip of head (Figures 1B, D, F, K, M).
Stylet apparatus. Central stylet 26.0 µm in length in holotype (21.3 µm and 27.2 µm in paratypes ICHUM 8586 and 8283, respectively; same in the following); basis cylindrical and narrowing at both ends, uniformly light colored, 34.6 µm in length (31.3 µm and 29.8 µm), 13.5 µm in width (11.3 µm and 11.0 µm); stylet to basis length ratio 0.75 (0.68 and 0.91); two accessory pouches each with four to five accessory stylets (5–7 and 4) (Figures 1H, N). One paratype female specimen (ICHUM 8584) had a strikingly different stylet apparatus morphology: central stylet 35.1 µm in length; basis triangular, uniformly light colored, 51.1 µm in length, 43.9 µm in maximum width at its posterior portion; stylet to basis length ratio 0.69; two accessory stylet pouches present with 15–18 accessory stylets, base of the stylets remarkably cleaved (Figure 1I).
Histological features. In paratypes (ICHUM 3287, 3288), proboscis with 10 nerves. Cephalic glands consisting of basophilic lobules, extending posteriorly to reach cerebral ring. Foregut with esophagus, stomach, and pylorus; stomach posteriorly turns dorsally to lead to pylorus; as pylorus slightly extends anteriorly beyond its ventral junction to stomach, there is a short pyloric cecum above stomach (Figure 2A); intestinal cecum (Figure 2C) anteriorly extends to reach posterior end of stomach, where it forks into two short dorsolateral pouches (Figure 2B), not reaching brain. Lateral nerve cords with accessory nerves extending backward to intestinal region (Figures 2A–C).

Figure 2 Oerstedia pseudoculata sp. nov., banded form, paratype, ICHUM 3288, photomicrographs of transverse histological sections through the foregut region showing the pyloric cecum above the stomach (A), the posterior foregut region at the stomach–pylorus junction (B), and the pyloric region (C). Abbreviations: an, accessory nerve; ic, intestinal cecum; ip, intestinal-cecum anterior dorsolateral pouch; pr, proboscis; py, pylorus; rh, rhynchocoel, st, stomach. Scale bars: (A–C) = 100 µm.
Type locality and distribution. The type locality of the species is in Oshoro Bay, Hokkaido, Japan. The species is also distributed in Akkeshi Bay, Hokkaido, and around Cape Crillon, Aniwa Bay, Sakhalin, Russia.
Oerstedia sp. Vietnam 35 – Chernyshev and Polyakova (2022).
ZooBank LSID. http://zoobank.org/36760C35-3A53-433D-BF09-1C0A3E5520E9.
Material examined. Holotype. ICHUM 8590 (Figures 3A–D), total DNA extracted from the 99% ethanol-preserved specimen; dredged at a depth of 38.1 m off Sagami Bay (35.1514°N, 139.6050°E), Misaki, Kanagawa, Japan, on 10/6/2022; collected by Natsumi Hookabe. Paratype. MIMB 4718222 (Figures 3E–H), total DNA extracted from the posterior body preserved in 99% ethanol; anterior body preserved in Bouin’s fluid then in 70% ethanol, sectioned for histology, four slides with transverse sections; collected at a depth of 1 m by Alexei V. Chernyshev, among red calcareous algae, off Van Phong Bay (2.6525°N, 109.3188°E), South China Sea, Vietnam, on 22/05/2010.
Etymology. The new specific name is an adjective (-us, -a, -um), meaning “wrinkled” in Latin. The specific name rugosa pertains to the longitudinal lines or wrinkles on the body of the species giving it a rough surface.
Diagnosis. An Oerstedia with carmine or deep-red to brownish-red body, decorated with vaguely bordered, transverse bands composed of white dots, and with prominent longitudinal and intertwined, white lines or wrinkles running from the head to the posterior end of the body; internally, the intestinal cecum has no anterior pouches and with short dorsolateral pouches.
Description. Holotype. Living specimen contractile (Figures 3A–D), 14.4–46.9 mm in length, 2.1 mm in width, tapering at both ends when stretched (tapering sometimes lost when worm contracts) (Figures 3C, D), firmly cylindrical in cross section, carmine or deep-red in color except white anterior end, with longitudinal, intertwined, white lines appearing as wrinkles, running from head to posterior end on all sides; entire body sparsely and irregularly decorated with white dots; fuzzily bordered, white, transverse bands (composed of unevenly sized and dispersed white dots) arranged at irregular interval along body; anterior-most band prominent, thicker than the rest, mid-dorsally and mid-ventrally bisected by a thin deep-red pigmentation (Figures 3A–D); succeeding bands vague, less thicker, more obvious on lateral sides (Figures 3A–D) and sometimes discontinuous on dorsal surface; number of recognizable transverse bands 17–23, depending on the degree of body contraction (Figures 3A–D). Head not demarcated from succeeding body; no ocelli observed; cephalic furrow not visible (Figures 3A–D); stylet apparatus not observed.
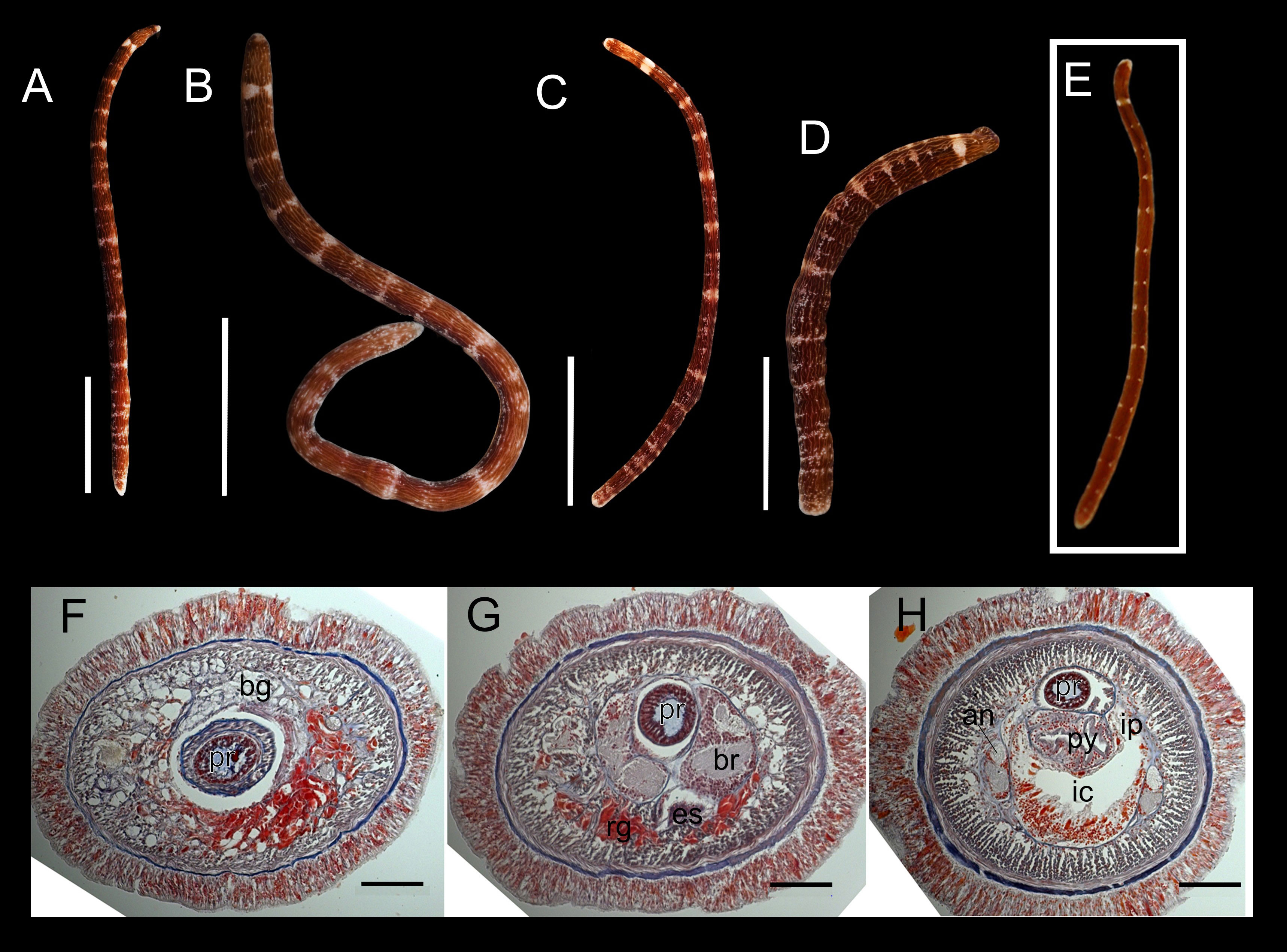
Figure 3 Oerstedia rugosa sp. nov., photographs of the entire worm at living state, holotype, ICHUM 8590 (A–D), paratype, MIMB 4718222 (E), and photomicrographs of the transverse histological sections, paratype, MIMB 4718222 (F–H). Entire body at dorsal (A, B, E), ventral (C), and right lateral view (D). Transverse histological section at precerebral region (F), brain region (G), and anterior pylorus region (H). an, accessory nerve; bg, basophilic glands; br, brain; es, esophagus; ic, intestinal cecum; ip, intestinal caecum pouches; py, pylorus; rg, red-stained glands. Scale bars: (A) = 10 mm; (B) = 5 mm; (C, D) = 2.5 mm; (F–H) = 100 µm.
Paratype. Body cylindrical, 14 mm long, with narrow head; body color reddish-brown; with 15 transverse white rings or bands discontinued dorsally and laterally; very small light longitudinal wrinkles and white dots rarely present; anterior tip of head white (Figure 3E).
Histological features (paratype). Proboscis with 10 nerves and claviform papillae; central stylet around 48 µm in length, basis oval, 50 µm in length and 20 µm in width. Cephalic glands not extending behind brain, including basophilic (colorless) lobules, and red-stained glands located ventrally (Figures 3F, G). Foregut with esophagus, stomach, and narrow pylorus; intestinal cecum without anterior pouches and with short dorsolateral pouches (Figure 3H); intestine with narrow and unbranched dorsolateral pouches. Lateral nerve cords with large (one and a half to two times less than the main neuropil) (Figure 3H) and long accessory nerve (at least 1/2 of the body length). The posterior half of the body was not sectioned.
Type locality and distribution. The type locality of the species is in Sagami Bay, Misaki, Kanagawa, Japan. It is also distributed in Van Phong Bay, Vietnam.
ZooBank LSID. http://zoobank.org/35279167-5BA1-42DD-A756-5469F4306947.
Material examined. Holotype. ICHUM 8589 (Figures 4A–F), female, total DNA extracted from a single specimen collected intertidally by Aoi Tsuyuki, among calcareous algae in a large tide pool on a rocky shore in Manazuru (35.1418°N, 139.1621°E), Kanagawa, Japan, on 22/04/2023.
Etymology. The new specific name is an adjective (-us, -a, -um), a combination of the Latin words viridis (meaning “green”) and fuscus (meaning “brown”). In combination, viridifusca pertains to the greenish-brown body pigmentation/freckles of the species.
Diagnosis. An Oerstedia with greenish-brown freckles on the entire body except for anterolateral edges of the head and a collar-like portion along the posterior cephalic furrow; freckles are denser on the anterior dorsal surface and sparser on the posterior three-fourths of the ventral surface; ocelli brownish-orange.
Description. Holotype. Non-anesthetized worm highly contractile, 4.3 mm in length (Figure 4A); anesthetized body 4.9 mm in length, 0.86 mm in width, cross-sectionally cylindrical in shape, entirely covered with greenish-brown freckles except for two anterolateral edges of head and the collar along posterior cephalic furrow; pigmentation more intense dorsally on anterior body near head and on lateral sides (Figures 4A, B, D); ventral body less freckled, greenish-brown freckles concentrated only laterally and on anterior one-fourths of ventral body, leaving entire ventral surface lighter than dorsal (Figures 4C, E); intestinal diverticula and gonads visible except at anterior body before head, (Figures 4C, E); rhynchocoel running for entire body length (Figure 4F); head narrower than, but not demarcated from, succeeding body; ocelli prominent at dorsal view, barely visible at ventral view, five in number, brownish-orange in color, and nearly equally sized, with two anterior ones situated more medially than two posterior ones, and additional one (probably representing an abnormal ocellus) situated between anterior pair (Figures 4A, B, D, F); pair of very short, laterally located, anterior cephalic furrows slightly visible; posterior cephalic furrow encircles the body, visible at the collar; proboscis pore evident at ventral side on tip of head (Figures 4C, E); stylet apparatus not observed.
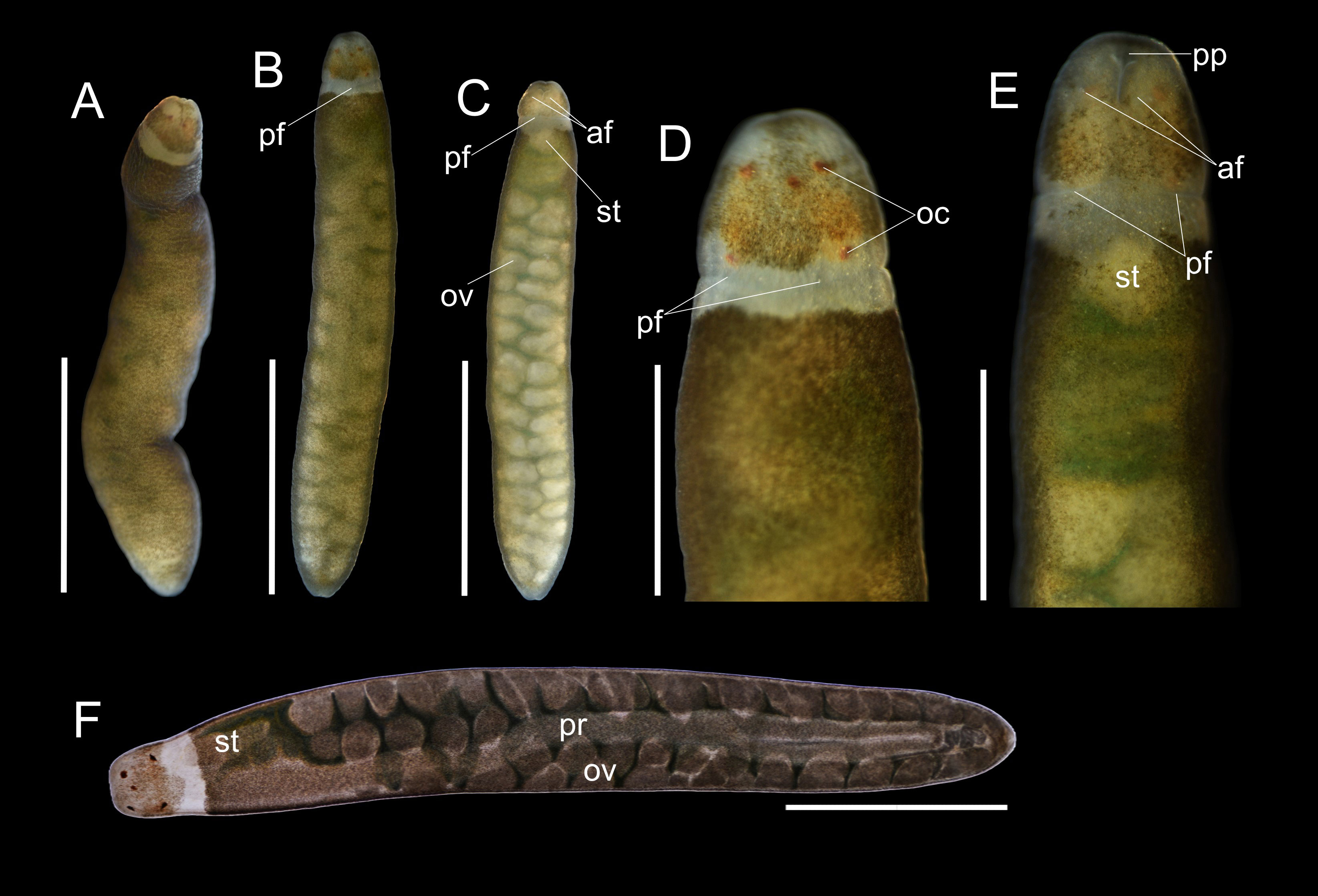
Figure 4 Oerstedia viridifusca sp. nov., holotype, ICHUM 8589, photographs of the entire worm in non-anesthetized state at dorsal view (A); anesthetized state (B–E), entire animal at dorsal (B) and ventral (C) views, and magnification of the head at dorsal (D) and ventral (E) views; and photomicrograph of the specimen compressed under a coverslip (F). af, anterior cephalic furrow; oc, ocelli; ov, ovary; pf, posterior cephalic furrow; pp, proboscis pore; pr, proboscis; st, stomach. Scale bars: (A–C) = 2 mm; (D–F) = 1 mm.
Type locality and distribution. The species is currently known only from its type locality—Manazuru, Kanagawa, Japan.
Oerstediella phoresiae Kulikova, 1987 (Kulikova, 1987).
Oerstedia phoresiae – Akhmatova et al. (2012).
Material examined. ICHUM 8588 (Figures 5A–C), total DNA extracted from a single specimen collected by Gauri Kaushik and Hiroshi Kajihara from brown algae in the family Sargassaceae, putatively Sargassum horneri (Turner) C. Agardh and/or Sargassum miyabei Yendo in Oshoro Bay (between 43.2093°N, 140.8585°E and 43.2095°N, 140.8582°E), Hokkaido, Japan, on 27/04/2023.
Description. Body in anesthetized state 4.0 mm in length, widest at middle portion, 0.37 mm in width, cross-sectionally cylindrical in shape, dorsally entirely covered with fine, brownish dots; head nearly square at dorsal view, wider than anterior body, 0.28 mm in width (Figure 5A); ocelli four in number, brownish in color, squarely arranged, poorly visible at dorsal view but prominent in a squeezed specimen (Figures 5A, B); posterior cephalic furrow dorsally continuous; ventral surface of head not examined in detail; rhynchocoel not apparent from dorsal body (Figure 5A) but barely visible in a squeezed specimen (Figure 5B). Central stylet 0.39 µm in length; basis cylindrical, narrows at both ends, lightly colored, 0.42 µm in length, 0.20 µm in width; stylet to basis length ratio 0.93; two accessory stylet pouches, each with four and six accessory stylets; stylet base cleaved (Figure 5C).
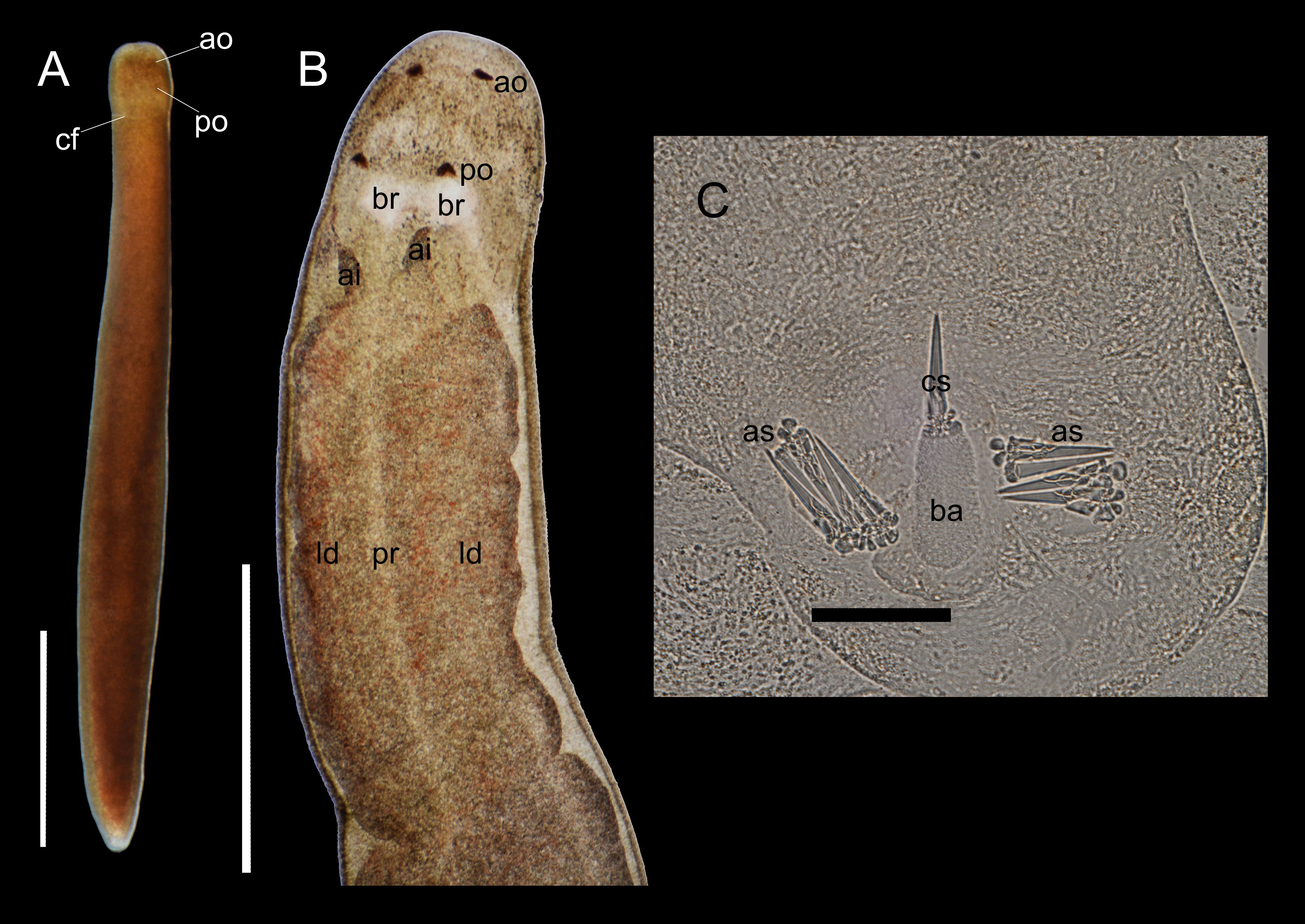
Figure 5 Oerstedia phoresiae (Kulikova, 1987), non-type specimen, ICHUM 8588, photograph of the entire specimen at dorsal view in an anesthetized state (A); photomicrographs of the compressed specimen showing the anterior half of the dorsal body (B) and its stylet apparatus (C). ai, anterior pouch of the intestinal cecum; ao, anterior ocellus; as, accessory stylets; ba, stylet basis; br, brain; cf, cephalic furrow; cs, central stylet; id, intestinal diverticulum; po, posterior ocellus; pr, proboscis. Scale bars: (A, B) = 1 mm; (C) = 50 µm.
Type locality and distribution. The type locality of this species is in Avangard Bight in Vostok Bay, Russia. It is also distributed in Peter the Great Bay, Russia, and Jeju Island, South Korea. A new locality, Oshoro Bay, Hokkaido, Japan, is herein reported.
The result of the uncorrected pairwise COI genetic distance analysis between the species collected in the present study is presented in Table 2. Oerstedia oculata (banded and non-banded forms) from Peter the Great Bay and O. phoresiae both from Peter the Great Bay and Jeju Island, O. venusta and O. zebra sensu Thollesson and Norenburg (2003) from Akkeshi Bay, and O. polyorbis from Kuril Island are also included in the genetic distance analysis. The 11 Oerstedia specimens collected and sequenced in the present study are suggested to comprise four species based on the p-distance analysis (≥ 3%): (i) Oerstedia phoresiae, (ii) Oerstedia pseudoculata sp. nov., (iii) Oerstedia rugosa sp. nov., and (iv) Oerstedia viridifusca sp. nov. The specimen identified as O. phoresiae reported from Oshoro Bay and that of specimens from Peter the Great Bay and Jeju Island are suggested to be conspecific (0.49%–0.76% p-distance).

Table 2 Uncorrected pairwise genetic distances (p-distances) between the Oerstedia species collected in the present study and with some previously described species using the 657-bp cytochrome c oxidase subunit I (COI) barcode sequence. Individuals with p-distance ≥3% are treated as different species.
The remaining six (i.e., two banded and four non-banded) specimens from Oshoro Bay, the two Oerstedia forms from Akkeshi Bay in Thollesson and Norenburg (2003), and a single specimen obtained from Sakhalin Island are all suggested to be conspecific (0%–0.76% p-distance) and different from the two O. oculata specimens from Peter the Great Bay (p-distance of 4.6%–4.8%). The maximum-likelihood analysis based on the partial COI sequences from these specimens (with O. phoresiae as outgroup) shows that the former nine specimens forms a highly supported clade that is sister to another clade composed of O. oculata. That O. pseudoculata sp. nov. is different from and sister to O. oculata is corroborated by the ABGD (prior maximal distance P = 0.035938; barcode distance gap = 0.27; distance JC69 Jukes-Cantor MinSlope = 1.500,000); ASAP (ASAP score = 1.00; P-val(rank) = 0.0453(1); W(rank) = 0.378(1); and bPTP (support value = 0.93 for both O. pseudoculata sp. nov. and O. oculata) species delimitation analyses (Figure 6B). A TCS haplotype network further shows the relationships among O. pseudoculata sp. nov. and O. oculata specimens considered in the present study (Figure 6A). Based on the three species-delimitation methods, we confirm that (i) O. venusta and O. zebra sensu Thollesson and Norenburg (2003), (ii) the two banded and four non-banded Oerstedia from Oshoro Bay, and (iii) the non-banded specimen from Aniwa Bay belong to a single species, established herein as O. pseudoculata sp. nov. The new species is genetically distinct from O. oculata despite a very close resemblance in morphology. Furthermore, the phylogenetic analyses reveal that the two species exhibit a sister–taxon relationship in the clade Paroerstediella with high support and are together sister to another dorsally banded species from Kuril Island in the same clade, i.e., O. polyorbis, with full support. The holotype Japanese specimen of Oerstedia rugosa sp. nov. is found to be conspecific with what was formerly an unidentified specimen, Oerstedia sp. Vietnam 35 (2.3% p-distance between the Japanese and Vietnamese individuals) (Table 2), nested within the clade Oerstedia of the genus and equally closely related to the undescribed Oerstedia species from Guam and Crimea. Oerstedia viridifusca sp. nov. is within the clade Paroerstediella and is sister to O. phoresiae with full support (Figure 7).

Figure 6 Delimiting Oerstedia oculata, O. pseudoculata sp. nov., O. venusta and O. zebra sensu Thollesson and Norenburg. A TCS haplotype network generated in PopART using partial sequences (657-bp) of the COI (A). The number of black bars corresponds to the number of nucleotide substitution between haplotypes; numbers inside each circle show the number of specimen(s) representing each haplotype; p-distance value represents the uncorrected pairwise genetic distance calculated within and between species; dashed boxes represent the species. A COI-based ML tree (B). The number near each node represents the RAxML maximum-likelihood bootstrap value (BS) with closed circles indicated by 100%BS. Vertical bars show the results of the ABGD, ASAP, and PTP species delimitation analyses.
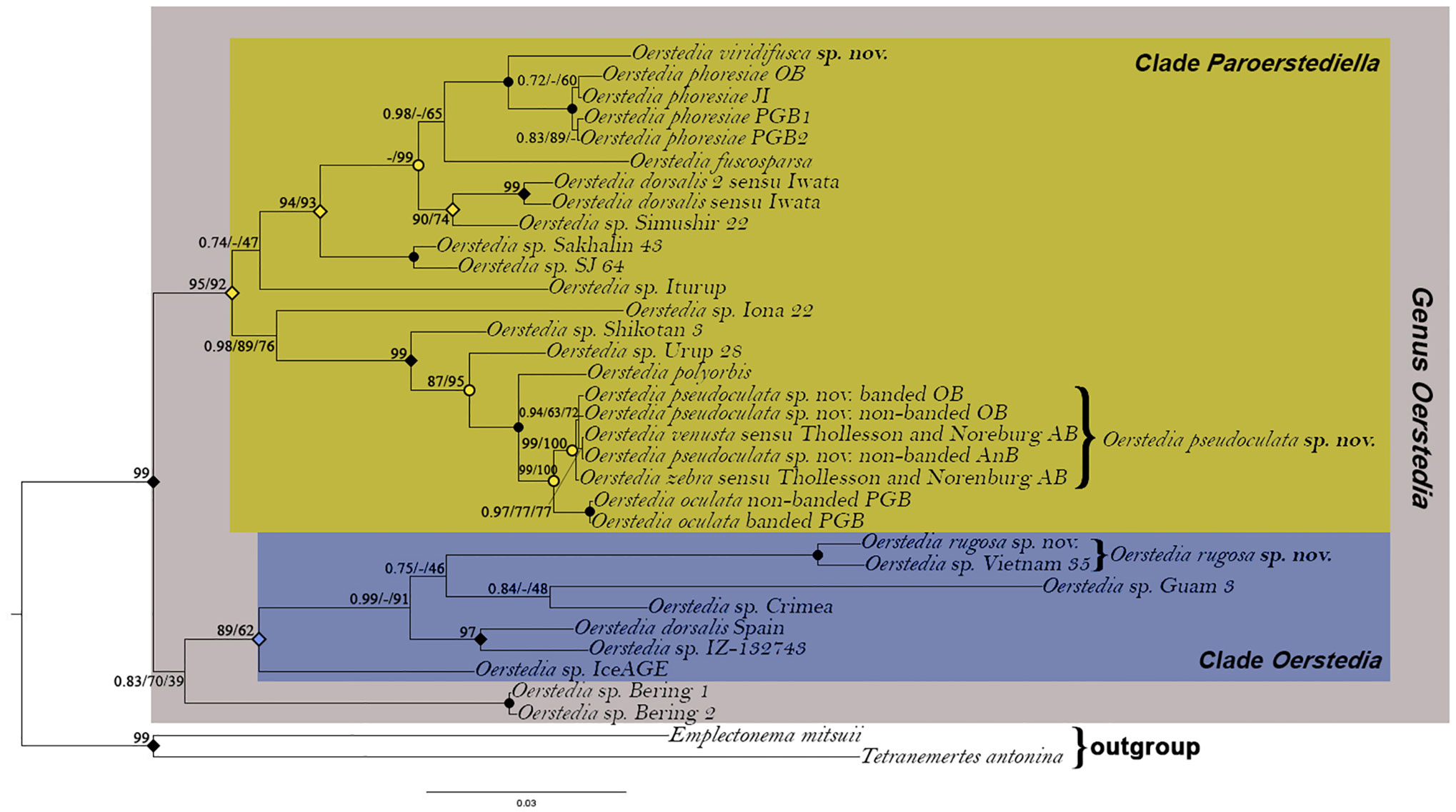
Figure 7 Rooted Bayesian phylogenetic tree showing the position of Oerstedia pseudoculata sp. nov., Oerstedia rugosa sp. nov., and Oerstedia viridifusca sp. nov. within the genus based on concatenated sequences of five gene markers (16S, 18S, 28S, COI, and H3). Number at each node represents Bayesian posterior probability (PP)/IQ-Tree maximum-likelihood ultrafast bootstrap value (UFBoot)/RAxML maximum-likelihood bootstrap value (BS). Solid circles indicate full support (1.00 PP/100% UFBoot/100% BS); open circles indicate 1.00 PP; solid diamonds indicate 1.00 PP/100% UFBoot; open diamonds indicate 0.99 PP.
At present, the species composition of the Japanese Oerstedia include O. dorsalis sensu Iwata (1954), O. fuscosparsa (Abato et al., 2023), O. phoresiae [present study], O. polyorbis (Iwata, 1954), O. pseudoculata sp. nov. [present study], O. rugosa sp. nov. [present study], O. venusta (Iwata, 1954), and O. viridifusca sp. nov. [present study]. These species together with O. oculata (Kulikova, 1987) (with O. valentinae (Chernyshev, 1993) and O. zebra (Chernyshev, 1993) being its junior synonyms) and O. verae (Chernyshev, 1993) constitute the banded and non-banded Oerstedia in the Northwest Pacific region.
We performed species-delimitation analyses using COI (for two non-tree-based methods) and a combined 16S, 18S, 28S, COI, and H3 genes (for a tree-based method) for unknown banded and non-banded forms of Oerstedia from Oshoro Bay and Aniwa Bay that morphologically resemble O. oculata. For this purpose, we included in the analyses sequences of O. oculata from the Peter the Great Bay, as well as O. venusta and O. zebra in Thollesson and Norenburg (2003). Our analyses revealed that (i) the O. venusta and O. zebra sensu Thollesson and Norenburg (2003) and the present specimens from Oshoro Bay and Aniwa Bay are all conspecific (p-distance 0–0.76%), and (ii) these are different from O. oculata found in Peter the Great Bay (p-distance 4.6%–4.8%), despite very similar morphological features. As we discuss below, O. venusta s.str. has anterior cephalic furrows extending dorsally, a character state that is quite unusual among Oerstedia congeners, suggesting that it may not even affiliate with the genus. Oerstedia valentinae and O. zebra are now considered synonymous with O. oculata (Zaslavskaya and Chernyshev, 2008; Akhmatova et al., 2012). Oerstedia polyorbis is distinct from O. oculata and O. pseudoculata sp. nov. (Chernyshev and Polyakova, 2022; Abato et al., 2023; present study). As we found it difficult to apply any existing Linnaean binomen to the species in question, we established O. pseudoculata sp. nov. for this species.
The taxonomic history of O. pseudoculata sp. nov. has been complicated and its identity has remained uncertain for 21 years (Figure 8). Iwata (1954) established the earliest Oerstedia in the Northwest Pacific—a dorsally banded O. polyorbis (with 30 transverse bands composed of minute black dots) and a non-banded, dorsally brownish O. venusta, both from Hokkaido Island. Later, Kulikova (1987) established a non-banded, dorsally brownish to dark-brown O. oculata from Vostok Bay. This species has two symmetrical white spots notable on the anterolateral edges of the dorsal head. Chernyshev (1993) described two species of dorsally banded Oerstedia solely based on morphology as O. valentinae (with 11–15 dark-brown stripes) and O. zebra (with 10–18 bands) from Peter the Great Bay. From their Akkeshi Bay nemertean collections, Thollesson and Norenburg (2003) identified two Oerstedia species. The authors applied the name O. zebra to the dorsally banded forms (with 9–16 stripes) and used the name O. venusta to the non-banded forms (Figure 8). Both forms have the same morphological features, except that one is banded and the other is not (Kajihara, 2007; Kajihara, 2021).
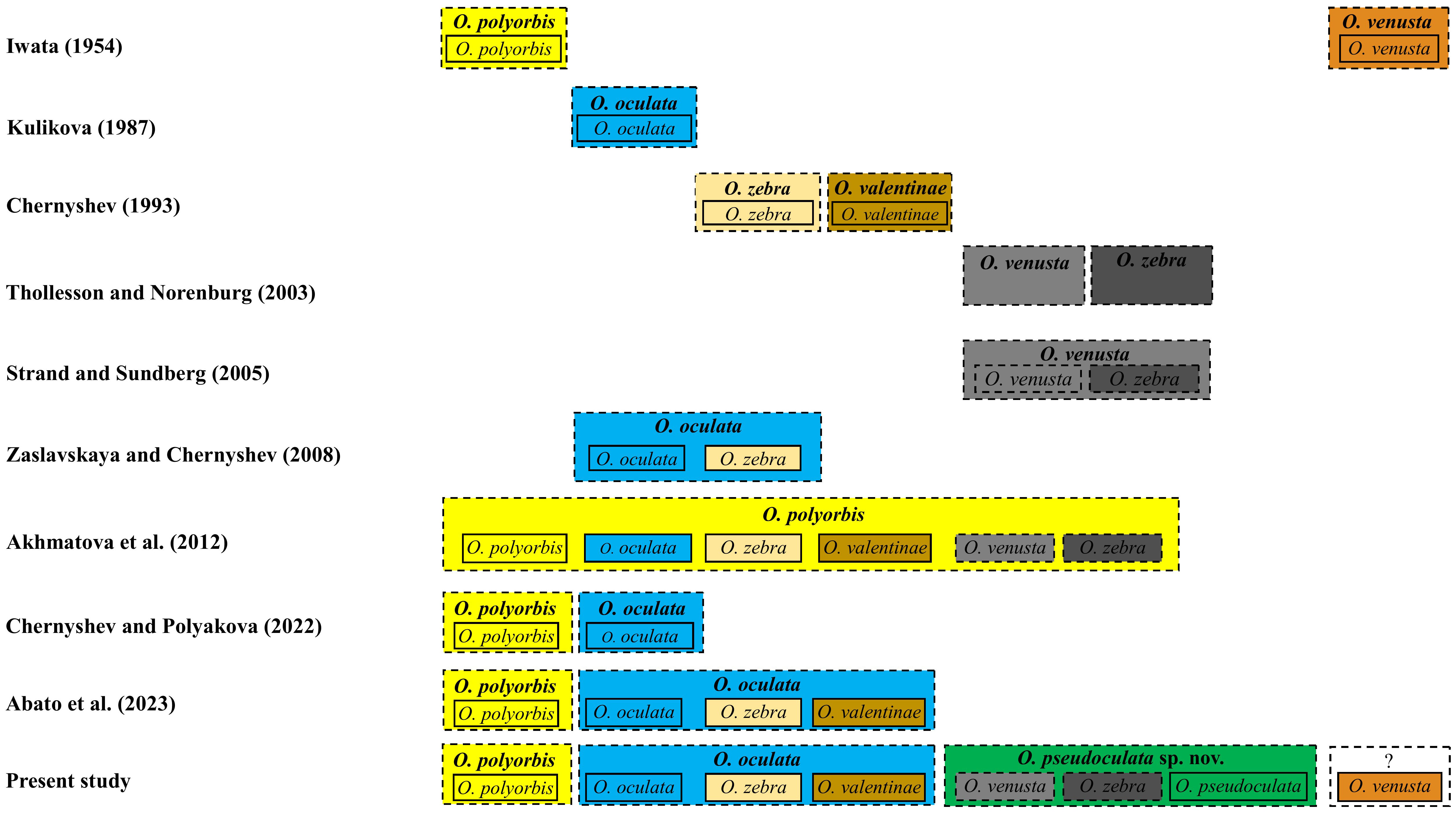
Figure 8 Taxonomic names applied to some banded and non-banded Oerstedia from the Northwest Pacific according to several taxonomic works. Solid box represents nominal species (each denoted by a name bearing type specimen and an available name), and dashed box represents the species concept proposed by the author(s).
The first molecular species delimitation including Thollesson and Norenburg’s (Thollesson and Norenburg, 2003) Oerstedia was conducted by Strand and Sundberg (2005) based on a haplotype network analysis of Oerstedia from the Atlantic Ocean (O. striata Sundberg, 1988 (Sundberg, 1988) and O. dorsalis) and from the Pacific Ocean, including O. venusta and O. zebra sensu Thollesson and Norenburg (2003), using partial 539-bp COI sequences. Strand and Sundberg (2005) showed that the two Oerstedia forms in Thollesson and Norenburg (2003) differed only by one substitution in their COI sequence (i.e., they belong to the same species); therefore, the former authors regarded these Akkeshi Bay Oerstedia as O. venusta; they regarded the name O. zebra in Thollesson and Norenburg (2003) as a junior synonym of O. venusta (Figure 8), despite not being consistent with Iwata’s external description of O. venusta. This synonymization was doubted by Kajihara (2007) since the identity of O. venusta sensu Thollesson and Norenburg (2003) was not resolved.
In the continued efforts to ascertain the Oerstedia species composition of the waters in the Northwest Pacific, Zaslavskaya and Chernyshev (2008) proposed that O. zebra is just a color variation of O. oculata based on the similarities of their allozyme markers. Using 28S sequences of the banded and non-banded Oerstedia from Peter the Great Bay and Thollesson and Norenburg’s (Thollesson and Norenburg, 2003) Oerstedia from Akkeshi Bay, Akhmatova et al. (2012) concluded that the forms identified as O. venusta and O. zebra sensu Thollesson and Norenburg (2003), the species O. oculata, O. valentinae, O. zebra from Russia and the one identified as O. polyorbis from Kuril Island are all conspecific—with the former names considered as junior synonyms of O. polyorbis. However, this synonymization was not conclusive since molecular species delimitation in metazoans, including ribbon worms, is accurately and reliably achieved using fast-evolving COI barcode sequences (Hebert et al., 2003a; Hebert et al., 2003b; Sundberg et al., 2016). Chernyshev and Polyakova (2022) showed that O. oculata and O. polyorbis from Kuril Island are two different species based on a phylogenetic analysis using a concatenated 16S, 18S, 28S, COI, and H3 sequences. Having the same phylogenetic evidence, Abato et al. (2023) stressed that it is more correct and valid to only synonymize O. oculata, O. valentinae, and O. zebra, with precedence given to O. oculata (Figure 8). Although the two recent phylogenetic studies of the genus were conclusive (Chernyshev and Polyakova, 2022; Abato et al., 2023), both failed to include Thollesson and Norenburg’s (Thollesson and Norenburg, 2003) Oerstedia in their phylogenetic analyses, making the species’ phylogenetic relationship with O. oculata, its potential closest relative based on morphological resemblance, not established.
This study settled and ascertained the taxonomic identity of a cryptic species hiding within banded and non-banded Oerstedia in the cold-temperate province of the Northwest Pacific. The use of barcode sequence data and its subsequent correct interpretation and application in the taxonomy of the genus Oerstedia (in conjunction with traditional taxonomy) is necessary to prevent taxonomic problems in the genus. Oerstedia pseudoculata sp. nov. in Hokkaido/Sakhalin Islands and O. oculata in Primorsky Krai are very similar in their external features—both have banded and non-banded forms. However, a few observed morphological differences between the two species are pointed out. In O. pseudoculata sp. nov., the ocelli are brilliant-orange and are notable on the dorsal and ventral sides of the head; the proboscis pore opens at the ventral tip of the head; the dorsally located transverse bands are 9–16 in number; the body is shorter and thinner; the stylet basis is shorter, varies in shaped from cylindrical to triangular; the accessory stylets are usually 4–7 (sometimes as many as 15–18) in number. On the other hand, in O. oculata, the black ocelli are not clearly visible on both sides of the head; the proboscis pore is located further below the tip of the head; the dorsally located transverse bands are 10–18 in number; the body is longer and wider; the stylet basis is longer and cylindrical; and the accessory stylets are 1–4 (sometimes up to 9) in number.
Aside from these differences, the geographic distribution and perhaps habitat preference of these two very closely related species are different. Oerstedia pseudoculata sp. nov. seems to be common in Hokkaido Island. The recent collection of a single specimen of the same species from Aniwa Bay, Sakhalin Island, supports our hypothesis that the new species is an island species. This hypothesis on habitat preference is supported by its absence in the Primorsky Krai, despite the intensive ribbon-worm studies in this region. Likewise, O. oculata has never been documented in several ribbon-worm surveys conducted in Hokkaido Island, Kuril Islands, and Sakhalin Island, perhaps indicating that this species is not an island species.
Oerstedia rugosa sp. nov. is defined by its unique body pigmentation, banding patterns, and the presence of wrinkles on its body surface. The COI genetic distance analysis revealed that this new species is conspecific with Oerstedia sp. Vietnam 35. In this regard, this Vietnam specimen is also referred to in this paper as O. rugosa sp. nov. Although the Japan and Vietnam specimens are conspecific, the two slightly differ in external features—the Japan specimen is longer, with body surface and texture appearing rougher or intensely wrinkled, whereas the Vietnam specimen is less wrinkled, showing a smoother body surface; the most anterior white transverse band in the Japan specimen is bisected at mid by a thinner, deep-red pigmentation whereas it is thicker in the Vietnam specimen; the white bands in the Japan specimen appears completely transverse (i.e., encircling the body) whereas these bands are not completely transverse in the Vietnam specimen (except for the most anterior band); when viewed dorsally, the bands appear to be situated on the lateral sides only, with mid-dorsal body completely deep red in color in the Vietnam material. The absence of anterior pouches in the intestinal cecum and having short dorsolateral intestinal pouches brings O. rugosa sp. nov. closer to the species of the genus Friedrichia Kirsteuer, 1965 (Kirsteuer, 1965; Chernyshev, 1993), which have intestinal cecum with no anterior and lateral pouches. That is, Friedrichia is likely a junior synonym of Oerstedia.
Oerstedia viridifusca sp. nov. is identifiable by its stout, brownish-green-pigmented body; thick, opaque white collar; and the presence of transverse cephalic band lighter than the body—these make it unique from the currently identified Northwest Pacific Oerstedia and the rest of the congeners. Although closely related to O. phoresiae, the two species can be easily discriminated against one another based on body pigmentation, body shape, and head shape (Figures 4, 5). However, a “viridifusca-like” form of O. phoresiae, collected from Jeju Island, South Korea (Chernyshev pers.obs.), expresses external features that are quite similar with O. viridifusca sp. nov. This specimen is less slender; the body pigmentation pattern in the dorsal and ventral sides appears to be slightly similar to the new species except that the dorsal color is brownish and the ventral body darker; the head shape is similar to the new species but narrower; the tip of the head is opaque white dorsoventrally; and it has an opaque white collar. Other Oerstedia species with white collar include O. polyorbis, the banded form of O. oculata (Akhmatova et al., 2012), and the banded form of O. pseudoculata sp. nov. Oerstedia viridifusca sp. nov. can be distinguished from these collared Oerstedia by its body pigmentation pattern and color. Having three anterior ocelli and two posterior ones could not be a taxonomic character state unique to the new species; instead, this could be a result of a developmental defect that occurred in the collected specimen. Oerstedia species generally only have four ocelli. Whether or not having five ocelli is a trait attributed to O. viridifusca sp. nov. can be verified only if multiple specimens of this species are collected in the future. Not until this character state is consistently observed in other O. viridifusca sp. nov. specimens, in this paper, we consider it an aberration.
The two monophyletic clades within the genus Oerstedia are defined based on a close relationship with O. phoresiae (i.e., clade Paroerstediella) or with O. dorsalis s.str. (i.e., clade Oerstedia) (Chernyshev and Polyakova, 2022). Two members of the clade Paroerstediella do not have a free-swimming larval stage—O. phoresiae and O. dorsalis sensu Iwata (Iwata, 1960; Chernyshev, 2014). Oerstedia pseudoculata sp. nov. and O. viridifusca sp. nov. having nested in the clade Paroerstediella, could have the same larval development as to the two former species. This could lead to their limited dispersal and distribution, and perhaps, faster speciation. Coe (1943) reported that O. dorsalis s.str. has a free-swimming larval stage. Having nested in the clade Oerstedia, the developmental pattern of O. rugosa sp. nov. could be like that of O. dorsalis s.str. With a free-swimming larval stage, the species can be easily dispersed and, thus, widely distributed. This could probably explain its distribution in Japanese and Vietnamese waters.
Since its establishment in 1954, O. venusta s.str. has never been recollected to date. Based on external morphology, it can be identified by the presence of “[t]wo pairs of the cephalic grooves running obliquely from the ventral side toward the dorsal side of the body” (Iwata, 1954: pp. 15, 16, figure 3). Envall and Sunberg (Envall and Sundberg, 1993) pointed out that it is not possible from Iwata’s (Iwata, 1954) brief description of O. venusta to identify it to the genus Oerstedia. The generic diagnosis for Oerstedia includes furrows that are absent or weakly developed (Envall and Sundberg, 1993). Having a pair of well-developed anterior cephalic furrows is unusual for a true Oerstedia. The description of Iwata’s (Iwata, 1954) O. venusta is vague; hence, its genus affiliation is uncertain. His species description appears ambiguous—color of the ocelli not specified, presence of accessory lateral nerve (a trait common to Oerstedia) not mentioned. Iwata’s (Iwata, 1954) taxonomic concept of O. venusta cannot be challenged; however, its genus affiliation can be further studied or verified, only possible when this species is recollected from its type locality and subjected to a more comprehensive internal morphology examination, sequenced, and most importantly, put through a phylogenetic analysis. A phylogenetics study involving Iwata’s (Iwata, 1954) O. venusta could result to two possible outcomes that can either resolve its taxonomy or redefine the diagnoses of the genus Oerstedia: (i) if nested within the genus, it is undoubtedly a genuine member of Oerstedia, indicating that the presence of a well-developed anterior cephalic furrows should also define the genus, or (ii) if not nested within the genus Oerstedia, it is definitely not a valid congener and the presence of a well-developed anterior furrows is absolutely not a trait of the genus, and therefore, Iwata’s (Iwata, 1954) O. venusta needs to be transferred to its proper genus.
Intraspecific morphological variation or polymorphism is common among Oerstedia species. This phenomenon when not carefully considered in the taxonomic study of the genus could result in a taxonomic chaos—polymorphism can either lump two different biological species into a single taxonomic name (as in the case of O. dorsalis s.str. and the previously considered form A of the same species in Sundberg (1984), now known as O. striata; the taxonomic issue related to Thollesson and Norenburg’s Oerstedia) or split similar biological species into different taxonomic names, as in the case of O. oculata, O. valentinae, and O. zebra (now under the name O. oculata due to synonymy). In extreme cases, as reported in this paper, polymorphism in the genus could result to two different species with very similar morphology or feature (resemblance between O. oculata and O. pseudoculata sp. nov.; “viridifusca-like” O. phoresiae and O. viridifusca sp. nov.; O. phoresiae, non-banded O. oculata, and non-banded O. pseudoculata sp. nov.; and O. polyorbis and the banded forms of O. oculata and O. pseudoculata sp. nov.). It is noteworthy that the sister species, O. oculata and O. pseudoculata sp. nov., having exactly very similar pigmentation patterns (both being banded and non-banded in the same manner), likely inherited an ancestral polymorphism. This phenomenon can be explained by Vavilov’s law of homologous series (Vavilov, 1922; Rozhnov, 2006; Trapezov, 2007; Bulatova, 2020).
Ribbon-worm taxonomists should regard polymorphism when establishing new Oerstedia species and even identifying genus congeners. In Oerstedia, both the similarities and differences in external features between closely related members have two implications—they are either conspecific or different species. This confuses taxonomists in species delineation, identification, and the subsequent name application. To avoid such chaos in the genus and to make future species establishment and identification taxonomically correct and reliable, an integrative approach in their taxonomy is highly recommended, i.e., employing both traditional morphological and modern molecular taxonomy. In the aspect of traditional taxonomy, turbo taxonomy alone coupled with molecular taxonomy is usually sufficient. Molecular species delineation should also rely on the fast-evolving mitochondrial COI gene since the 16S gene of O. oculata and O. pseudoculata sp. nov. was found to differ only by two substitutions, suggesting that the latter mitochondrial gene is more conservative and does not differ among sibling species. Using our specimens, we have shown the necessity of DNA sequence data in the taxonomy of the genus and in resolving the taxonomic problems arising from the polymorphic species with uncertain identities.
The genus Oerstedia is composed of some congeners that are highly polymorphic. This polymorphism makes the taxonomy and systematics of the genus challenging. Some species in the genus can deceive taxonomists by their variable external morphology, resulting in problematic species demarcation, identification, and name application. Employing the so-called traditional taxonomy for Oerstedia species is undoubtedly inadequate and can possibly result in some taxonomic problems and inconsistencies. The role of DNA sequence data in the taxonomy and systematics of the genus Oerstedia is always indispensable. With these, it is important to revisit those already established Oerstedia and investigate them for potential polymorphism and generate barcode sequence data from a greater representative of the genus for more comprehensive future molecular taxonomy and phylogenetics studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The manuscript presents research on animals that do not require ethical approval for their study.
JA: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. AC: Data curation, Supervision, Validation, Writing – original draft, Writing – review & editing. NH: Writing – original draft, Writing – review & editing. AT: Writing – original draft, Writing – review & editing. GK: Writing – original draft, Writing – review & editing. HK: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the Society of Systematic Biologists Graduate Student Research Award (year 2022) for JCA and the Japan Society for Promotion of Science KAKENHI grant number 20K06780 for HK. The COI sequence of O. pseudoculata sp. nov. from Sakhalin Island was provided by Irina Ekimova (Lomonosov Moscow State University) and was obtained in the frame of the project #3/2023-gr granted by the non-profit Charitable Foundation “Support for Biological Research”.
JA would like to thank and acknowledge Mindanao State University–Marawi City for the APDP grant, Japan’s Ministry of Education, Culture, Sports, Science and Technology (MEXT) for the scholarship grant. AC sincerely thanks Alina Akhmatova, Irina Ekimova, and Neonila Polyakova for providing sequences for some samples used in the phylogenetic analyses. HK thanks Svetlana A. Maslakova, Megan Schwartz, and Jon L. Norenburg for help in field sampling; and staffs at Oshoro and Akkeshi Marine Stations for providing facilities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abato J., Yoshida R., Kajihara H. (2023). Species description and phylogenetics of Oerstedia fuscosparsa sp. nov. (Nemertea: Monostilifera: Oerstediidae) from Japan. Zootaxa 5249 (5), 589–597. doi: 10.11646/zootaxa.5249.5.6
Abildgaard P. C. (1806). “Planaria dorsalis,” in Zoologica Danica seu animalium Daniae et Norvegiae rariorum ac minus notorum descriptiones et historia. Christensen. Ed. Müller O. F. (Havnia: Hauniae, Sumtibus Weygandinis), 25.
Akhmatova A. F., Chernyshev A. V., Zaslavskaya N. I. (2012). The species composition of the nemertean genus Oerstedia (Nemertea: Hoplonemertea) in the Far Eastern seas of Russia. Russ. J. Mar. Biol. 38 (6), 423–430. doi: 10.1134/S1063074012060028
Altekar G., Dwarkadas S., Huelsenbeck J. P., Ronquist F. (2004). Parallel Metropolis-coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 20, 407–415. doi: 10.1093/bioinformatics/btg427
Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 (17), 3389–3402. doi: 10.1093/nar/25.17.3389
Berg G. (1972). Studies on Nipponnemertes Friedrich, 1968 (Nemertini, Hoplonemertini). Redescription of Nipponnemertes pulcher (Johnston, 1837) with special reference to intraspecific variation of the taxonomical characters. Zool. Scr. 1, 211–225. doi: 10.1111/j.1463-6409.1972.tb00580.x
Briggs J. C., Bowen B. W. (2012). A realignment of marine biogeographic provinces with particular reference to fish distributions. J. Biogeogr. 39, 12–30. doi: 10.1111/j.1365-2699.2011.02613.x
Brunberg L. (1964). On the nemertean fauna of Danish waters. Ophelia. 1, 77–111. doi: 10.1080/00785326.1964.10416273
Bulatova N. (2020). Notable homologous variation in chromosomal races of the common shrew. Comp. Cytogenet. 14 (3), 313–318. doi: 10.3897/CompCytogen.v14i3.54526
Bürger O. (1895). Die Nemertinen des Golfes von Neapel und der angrenzenden Meeres-Abschnitte. Fauna und Flora des Golfes von Neapel und der Angrenzenden Meeresabschnitte 22, 1–714.
Chernyshev A. V. (1993). A review of the genera of nemerteans allied to Oerstedia (Monostilifera, Tetrastemmatidae) with descriptions of four new species. Zool. Zhur. 72 (3), 11–20.
Chernyshev A. V. (2004). Two new genera of nemertean worms of the family Tetrastemmatidae (Nemertea: Monostilifera). Zoosyst. Ross. 12 (2), 151–156. doi: 10.31610/zsr/2003.12.2.151
Chernyshev A. V. (2014). “Nemertean biodiversity in the Sea of Japan and adjacent areas,” in Marine biodiversity and ecosystem dynamics of the North-Western Pacific Ocean. Eds. Sun S., Adrianov A. V., Lutaenko K. A., Sun X. X. (Beijing, China: Science Press), 119–135.
Chernyshev A. V., Polyakova N. E. (2022). Nemerteans collected in the Bering Sea during the research cruises aboard the R/VAkademik MA Lavrentyev in 2016, 2018, and 2021 with an analysis of deep-sea heteronemertean and hoplonemertean species. Deep Sea Res. Part II Top. Stud. Oceanogr. 199, 105081. doi: 10.1016/j.dsr2.2022.105081
Coe W. R. (1905). Nemerteans of the west and northwest coasts of North America. Bull. Mus. Comp. Zool. 44, 1–318.
Coe W. R. (1943). Biology of the nemerteans of the Atlantic coast of North America. Trans. Conn. Ac. 35, 129–328.
Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi: 10.1093/nar/gkh340
Envall M., Sundberg P. (1993). Intraspecific variation in nemerteans (Nemertea): synonymization of genera Paroerstedia and Oerstediella with Oerstedia. J. Zool. 230, 2, 293–318. doi: 10.1111/j.1469-7998.1993.tb02687.x
Friedrich H. (1935). Studien zur Morphologie, Systematik und Ökologie der Nemertinen der Kieler Bucht. Archiv für Naturgeschichte. 4, 293–375.
Friedrich H. (1955). Beitrage zu einer Synopsis der Gattungen der Nemertini monastilifera nebst Bestimmungsschlussel. Z. wiss. Zool. 158, 733–745.
Gibson R. (1994). “Nemerteans. No. 24,” in Synopses of the british fauns (New series), 2nd ed. (Shrewsbury: Field Studies Council), vii + 224.
Hebert P. D. N., Cywinska A., Ball S. L., de Waard J. R. (2003a). Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B: Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hebert P. D. N., Ratnasingham S., de Ward J. R. (2003b). Barcoding animal life: cytochrome c oxidase subunit I divergences among closely related species. Proc. R. Soc. Lond. Ser. B: Biol. Sci. 270, 596–599. doi: 10.1098/rsbl.2003.0025
Iwata F. (1954). The fauna of Akkeshi Bay XX. Nemertini in Hokkaido (revised report). J. Fac. Sci. Hokkaido Univ. Zool. 12 (6), 11–39.
Iwata F. (1960). Studies on the comparative embryology of nemerteans with special reference to their interrelationships. Publ. Akkeshi Mar. Biol. Stat. 10, 2–51.
Kajihara H. (2007). A taxonomic catalogue of Japanese nemerteans (phylum Nemertea). Zool. Sci. 24 (4), 287–326. doi: 10.2108/zsj.24.287
Kajihara H. (2017). “Species diversity of Japanese ribbon worms (Nemertea),” in Species diversity of animals in Japan, vol. 2017 . Eds. Motokawa M., Kajihara H. (Tokyo, Japan: Springer), 419–444.
Kajihara H. (2021). Higher classification of the monostilifera (Nemertea: hoplonemertea). Zootaxa. 4920 (2), 151–199. doi: 10.11646/zootaxa.4920.2.1
Katoh K., Rozewicki J., Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20 (4), 1160–1166. doi: 10.1093/bib/bbx108
Kirsteuer E. (1965). Über das Vorkommen von Nemertinen in einem tropischen Korallenriff. 4. Hoplonemertini Monostilifera. Zool. Jb. Abt. Ayst. Ökol. Geogr. Tiere. 92, 289–326.
Kulikova V. I. (1987). New species of Oerstediella (Nemertini, Hoplonemertini) of the Vostok Bay, Sea of Japan. Izv. Akad. Nauk. SSSR. 6 (Ser Biol), 828–836.
Kvist S., Laumer C. E., Junoy J., Giribet G. (2014). New insights into the phylogeny, systematics and DNA barcoding of Nemertea. Invertebr. Syst. 28 (3), 287–308. doi: 10.1071/IS1306
Lanfear R., Calcott B., Ho S. Y. (2012). Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29 (6), 1695–1701. doi: 10.1093/molbev/mss020
Lanfear R., Frandsen P. B., Wright A. M., Senfeld T. (2016). Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34 (3), 772–773. doi: 10.1093/molbev/msw260
Leigh J. W., Bryant D. (2015). Popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Minh B. Q., Nguyen M. A. T., Haeseler A. V. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30 (5), 1188–1195. doi: 10.1093/molbev/mst024
Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., Haeseler A. V., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 (5), 1530–1534. doi: 10.1093/molbev/msaa015
Norenburg J., Gibson R., Herrera Bachiller A., Strand M. (2023). World nemertea database. Oerstedia quatrefages. Available at: https://www.marinespecies.org/aphia.php?p=taxdetails&id=122423.
Palumbi S. R., Martin A., Romano S., McMillan W. O., Stice L., Grabowski G. (1991). The simple fool’s guide to PCR, v. 2.0 (Honolulu: Department of Zoology, Kewalo Marine Laboratory, University of Hawaii), 1–45.
Puillandre N., Brouillet S., Achaz G. (2021). ASAP: assemble species by automatic partitioning. Mol. Ecol. Resour. 1 (2), 609–620. doi: 10.1111/1755-0998.13281
Puillandre N., Lambert A., Brouillet S., Achaz G. (2012). ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 21, 1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x
Quartrefages A. (1846). Études sur les types inférieurs de l’émbranchement des Annelés. Mémoire sur la famille des Némertiens (Nemertea). Ann. Sci. Nat. Zool. 3 (6), 173–303.
Rambaut A. (2014). FigTree v1.4.4, a graphical viewer of phylogenetic trees (Java). Available at: http://beast.bio.ed.ac.uk/figtree.
Ronquist F., Huelsenbeck J. P. (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rozhnov S. V. N. (2006). “Vavilov’s law of homologous series and archaic diversity accrording to the paleontological data,” in Evoliutsiia biosfery i bioraznobraziia. K 70-letiiu A.Yu. Rozanova. Ed. Rozhnov S. V. (Moscow: KMK), 134–146.
Stamatakis A. (2015). Using RAxML to infer phylogenies. Curr. Protoc. Bioinf. 51 (1), 6–14. doi: 10.1002/0471250953.bi0614s51
Steenwyk J. L., Buida I. I. I. J., Li Y., Shen X. X., Rokas A. (2020). ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PloS Biol. 18 (12), e3001007. doi: 10.1371/journal.pbio.3001007
Strand M., Sundberg P. (2005). Delimiting species in the hoplonemertean genus Tetrastemma (phylum Nemertea): morphology is not concordant with phylogeny as evidenced from mtDNA sequences. Biol. J. Linn. Soc. 86 (2), 201–212. doi: 10.1111/j.1095-8312.2005.00535.x
Sundberg P. (1979). Statistical analysis of variation in characters in Tetrastemma laminariae (Nemertini), with a redescription of the species. J. Zool. 189 (1), 39–56. doi: 10.1111/j.1469-7998.1979.tb03952.x
Sundberg P. (1984). Multivariate analysis of polymorphism in the hoplonemertean Oerstedia dorsalis (Abildgaard, 1806). J. Exp. Mar. Biol. Ecol. 78, 1–22. doi: 10.1016/0022-0981(84)90068-6
Sundberg P. (1988). A new monostiliferous hoplonemertean (Nemertea), Oerstedia striata sp. n., from the west coast of Sweden. Zool. Scr. 17 (2), 135–139. doi: 10.1111/j.1463-6409.1988.tb00090.x
Sundberg P., Andersson S. (1995). Random amplified polymorphic DNA (RAPD) and intraspecific variation in Oerstedia dorsalis (Hoplonemertea, Nemertea). J. Mar. Biol. Assoc. UK 75 (2), 483–490. doi: 10.1017/S0025315400018324
Sundberg P., Kvist S., Strand M. (2016). Evaluating the utility of single-locus DNA barcoding for the identification of ribbon worms (phylum Nemertea). PloS One 11 (5), e0155541. doi: 10.1371/journal.pone.0155541
Sundberg P., Vodoti E. T., Zhou H., Strand M. (2009). Polymorphism hides cryptic species in Oerstedia dorsalis (Nemertea, Hoplonemertea). Biol. J. Linn. Soc. 98 (3), 556–567. doi: 10.1111/j.1095-8312.2009.01310.x
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Thollesson M., Norenburg J. L. (2003). Ribbon worm relationships: a phylogeny of the phylum Nemertea. Proc. R. Soc. Lond. Ser. B: Biol. Sci. 270 (1513), 407–415. doi: 10.1098/rspb.2002.2254
Trapezov O. V. (2007). Homologous series of fur color variability in American mink (Mustela vison Schreber, 1777) detected under domestication. Vestnik VOGiS 11 (3/4), 547–560.
Zaslavskaya N. I., Chernyshev A. V. (2008). Allozyme comparison of three nemertean species of the genus Oerstedia (Nemertea: Monostilifera) from the Sea of Japan. Biochem. Syst. Ecol. 36 (7), 554–558. doi: 10.1016/j.bse.2008.03.013
Keywords: biodiversity, DNA barcoding, invertebrate, JAMBIO, monostilifera, ribbon worm
Citation: Abato JC, Chernyshev AV, Hookabe N, Tsuyuki A, Kaushik G and Kajihara H (2024) Delimiting the polymorphic congeners of the genus Oerstedia Quatrefages, 1864 (Nemertea, Hoplonemertea), and descriptions of three new species from the Northwest Pacific. Front. Ecol. Evol. 12:1356316. doi: 10.3389/fevo.2024.1356316
Received: 15 December 2023; Accepted: 22 January 2024;
Published: 16 February 2024.
Edited by:
Paulo Cesar Paiva, Federal University of Rio de Janeiro, BrazilReviewed by:
Sonia Andrade, University of São Paulo, BrazilCopyright © 2024 Abato, Chernyshev, Hookabe, Tsuyuki, Kaushik and Kajihara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jamael C. Abato, amFtYWVsYWJhdG9AZ21haWwuY29t; Hiroshi Kajihara, a2FqaWhhcmFAZWlzLmhva3VkYWkuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.