
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 15 February 2024
Sec. Conservation and Restoration Ecology
Volume 12 - 2024 | https://doi.org/10.3389/fevo.2024.1355018
Introduction: Non-perennial rivers and streams are increasingly present, in part because of climate change, even in the temperate climate. However, how the loss of connectivity and complete drying affect microphytobenthos in general and diatom communities in particular has gone mostly unstudied.
Methods: With this paper, we aim to close this gap, identifying diatom biodiversity through manual digital microscopy and rbcL amplicon sequencing, to observe a) which method is better suited to it and b) how the ecotone flow-pool-dry affects diatom diversity under duress. Three karstic, non-perennial rivers and streams with a gradient from natural to anthropogenically disturbed were sampled under flooding conditions and after a long and intense drought in 2022.
Results: Our results show that digital microscopy shows a higher diversity and species richness than amplicon sequencing. We posit that this might be due to a reduced pool of subaerophile taxa having been sequenced and being part of the reference database. Furthermore, the effect of drying only resulted in a reduction in diversity after this drought, although the biofilm was still alive under these conditions.
Discussion: To use amplicon sequencing for non-perennial river diatom diversity monitoring, the reference databases will have to be adapted to such systems, as most rivers may be subjected to drying regularly in the future.
Non-perennial rivers and streams make up over 60% of all riparian systems (Messager et al., 2021). There might even be a prospect of increase, leaving most rivers affected by it due to climate change (Blöschl et al., 2019; Bonaldo et al., 2023). As non-perennial streams have an important impact on ecosystems because of their changing hydrology, it is important to understand how they work (Datry et al., 2017). However, in the case of phytobenthic communities in general and diatom communities in particular, their study has been underrepresented and very localized (Delgado et al., 2012; Novais et al., 2014; B-Béres et al., 2019).
As the loss of longitudinal connectivity and subsequent loss of surface water creates hydrologically very distinct habitats, ecological communities are shaped by them. These habitats contain new ecological niches to fill: the riparian flow would favor phytobenthic communities, loss of connectivity creates pools, which further the addition of phytoplankton, and dry riverbeds should facilitate survival of sub-aerophile species (Burfeid Castellanos, 2018). Thus, non-perennial streams have the potential of being hotspots of diversity (Datry et al., 2014) and as such would be important to conserve and understand.
Furthermore, the European Water Framework Directive (European Commission, Directive 2000/60/EC, European Commission, 2000) ensures the ecological quality of flowing rivers. It has, nonetheless, neglected to cover drying and intermittent rivers outside the Mediterranean realm (Stubbington et al., 2018). Due to the previously described problematics and ever-increasing proportion of dry falling rivers at all latitudes, the biodiversity and ecological quality of the rivers are in danger. How these ecological parameters must be analyzed has not yet been assayed.
The traditional way of measuring this diatom biodiversity entails microscopy as a semi-quantitative tool (Szczepocka and Żelazna-Wieczorek, 2018). However, as an alternative to capture even more diversity, molecular tools such as amplicon sequencing can help to get a non-quantitative (presence–absence) overview of the taxa (Vasselon et al., 2017a; Bailet et al., 2019). For diatoms, two primer sets are most used, 18S [V4 or V9, mostly used in Germany because of availability of reference barcodes] and rbcL [mostly used outside of Germany] (Bailet et al., 2019). As rbcL is a more changing molecule, part of the RubisCo located in the chloroplast, it required the creation of five primers (three forward and two reverse) (Vasselon et al., 2017b; Bruce et al., 2021). Because of this, a higher variability and diversity might be caught, which is why we have selected rbcL for this study. Furthermore, as this marker has been most frequently used, a quantification coefficient has been created and could increase similarity to microscopy results (Vasselon et al., 2018; Tapolczai et al., 2019), and has been found to do so (Rimet et al., 2018).
However, similar the results of the methods might be, the combination of amplicon sequencing of benthic diatom communities in non-perennial streams and its functionality is yet unstudied. To fill this gap, we sampled 10 sites within three karstic, naturally drying rivers twice in the year 2022. We aimed to observe differences between high flow spring and a severely dry summer on the effectivity of both methods to capture biodiversity. Finally, we aimed to monitor the effects that the extreme changes had on the diatom community composition.
Thus, in this study, we have focused on how the diatom biodiversity of three non-perennial streams might be best monitored, using digital microscopy and one amplicon sequencing marker (rbcL) to identify the biodiversity along the changing hydrology. We hypothesize that the diatom community divergence will be determined by the hydrological state (moment of sampling) to a higher degree than because of the method that was used for the identification. Changes in connectivity have been found to significantly alter the biodiversity through the creation of new ecological niches (Datry et al., 2017). Regarding biodiversity, we expect that only connectivity will create the framework for correct and coherent identification using the different methods, as the amplicon sequencing method was developed for monitoring purposes and has been developed for flowing rivers. In non-flow and dry conditions, we expect a divergence of diatom biodiversity according to the method.
Diatoms were sampled in three streams of the Paderborn High Plateau (North-Rhine Westphalia, Germany) twice during the year 2022 (Figure 1). The streams (Menne, Talgosse, and Sauer) are karstic and characterized by a longitudinal and temporal drying pattern (Meyer and Meyer, 2000; Meyer et al., 2003). In the smaller and natural streams (Menne and Talgosse), three sampling sites were selected, namely, dry (located at the lower reach), pool, and flowing sites, based on the characterization of Meyer et al. In the Sauer river, a bigger catchment of approximately 110,000 km2 located under anthropogenic and agricultural influence, four sites were selected: a dry site at the lower reach; two pools, one of which was under direct influx of farmyard effluent; and a flowing site at the upper reach. The samples were taken in April, under flowing conditions with high flow (relative flooding), and in September, after an extreme drought affected Germany.
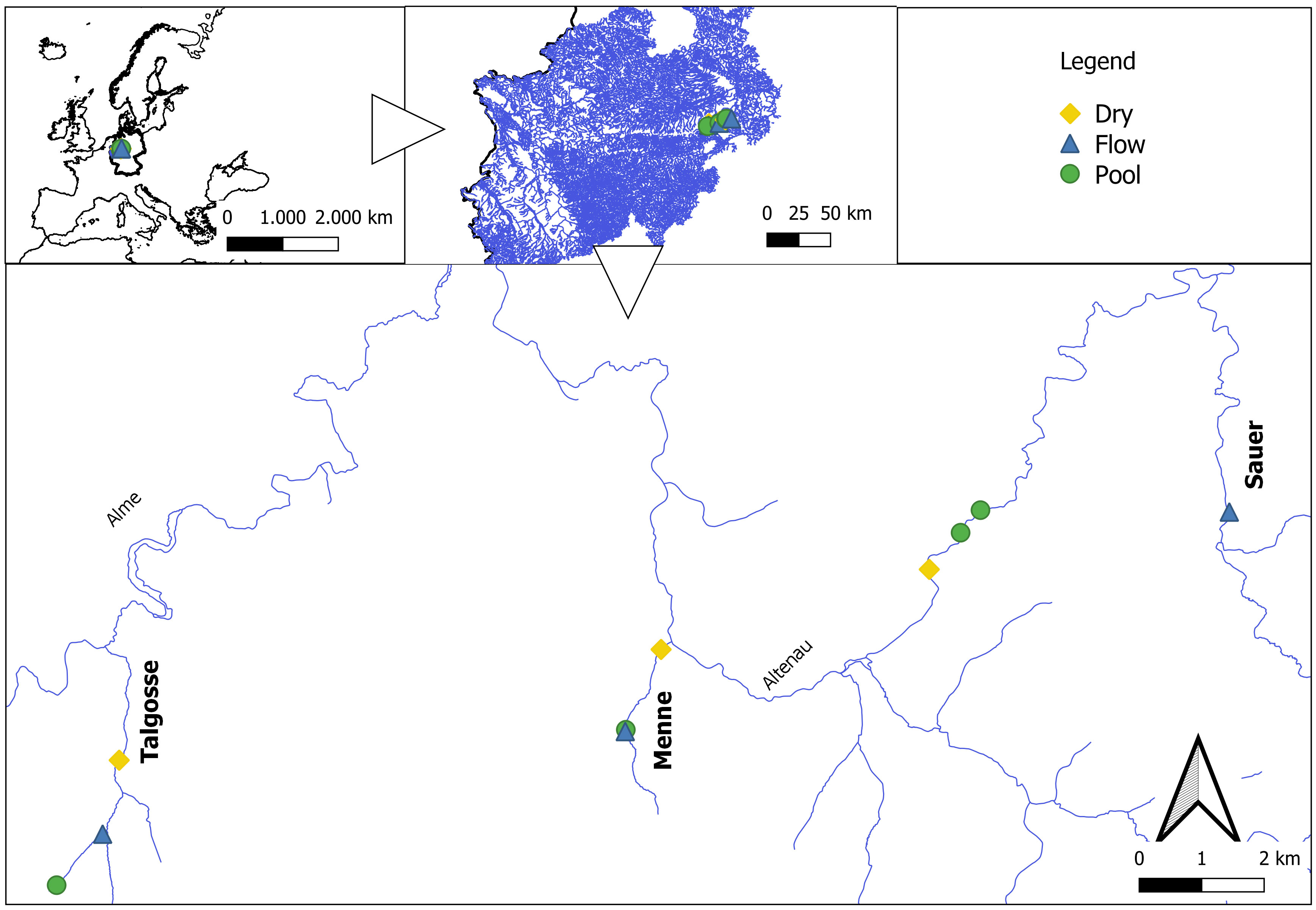
Figure 1 Sampling sites along the Alme catchment. The sites are described at the majoritarian aquatic state, not necessarily the state that the sites were found in.
To characterize the streams, the physicochemical conditions were measured in the field with a Multimeter sonde (PCE-PHD1, PCE Instruments, Meschede, Germany). Fresh water samples were taken at each site with flowing or standing surface water and analyzed spectrophotometrically in the laboratory after being filtered. The results are listed in Supplementary Table 1.
Diatoms were sampled from the periphyton following the CEN/TR 17244:288 (CEN, 2018). The stones to be sampled were first measured with a Benthotorch (BBE+ Moldaenke GmbH, Schwentinental, Germany) to confirm fluorescence and thus functional chlorophyll activity within the biofilm (Supplementary Table 2). A total of 100 cm2 of biofilm extracted from five stones was scraped with a new toothbrush for each site, pooled in a Falcon tube and directly fixated using molecular grade ethanol to a final concentration of 75%. In the dry sites, stones were wetted with deionized water to scrape the sample. The fixated sample was then kept cool during the sampling and put into the −20°C freezer upon arrival.
Diatoms were pre-washed after sampling using deionized water and centrifuged for 4 min at 1,300 rpm. This process was repeated three times. After this, the diatom samples were digested using the hot hydrogen peroxide (H2O2)– hydrochloric acid (HCl) method (Taylor et al., 2007). After the diatom samples were cleaned of organic and carbonic debris, the washing step was repeated equivalently to the pre-wash seven times. After a quantification step, the diluted samples were then dripped onto coverslips with an additional ammonium chloride (NH4Cl) and dried overnight in the lateral flow chamber. The coverslip was later attached to the slide using the artificial mounting resin, Naphrax ® (Thorns Biologie Bedarf) and let to harden for 2 weeks.
The prepared slides were then scanned into the Olympus Slideview VS200 (EVIDENT, Tokyo, Japan) using the pipeline described in the paper of Burfeid-Castellanos et al. (2022) on a surface of 16–25 mm² depending on density. The stitched and stacked virtual slides were then uploaded onto the browser-based annotation platform BIIGLE 2.0 (Langenkämper et al., 2017). An average of 420 diatom valves (half-shells) were identified manually in each slide for the counting using general bibliography (Bey and Ector, 2013; Cantonati et al., 2017; Peeters and Ector, 2017). As the dry locations presented sub-aerial diatom genera such as Luticola, Humidophila, and Gomphonema, we used genus-specific bibliography for their identification (Levkov et al., 2013; Kopalová et al., 2015; Levkov et al., 2016). The identified samples are available on here: https://biigle.de/project-invitations/b087efa0-37fc-445e-bfcb-2ccc4ea33d7d.
Diatom DNA was extracted with a NucleoSPIN soil Mini Kit (Macherey & Nagle). The primers used were rbcL primers (Vasselon et al., 2017a). The first PCR was performed using the protocol from Vautier et al. (2020). After the quality and quantity of DNA were confirmed electrophoretically, a second PCR was made to attach the MiSeq indices (Illumina) following a modified protocol for 16S Sequencing Library preparation (Illumina) for the amplicon attachment and cleaning of rbcL DNA. The PCR was adapted to the rbcL primer (see Supplementary Table). The cleaned and labeled DNA was then sent to CeGAT (https://cegat.com/) for sequencing. The data are available on Zenodo: https://doi.org/10.5281/zenodo.10351139.
The taxonomic assignation and statistics were made using RStudio (v. 2023.06.1 + 524) with R version 4.2.2. We followed the bioinformatic pipeline from Canino et al., modified from Keck (2020). Based on DADA2, it utilizes the diat.barcode database for taxonomic classification of the amplicon sequence variants (ASVs). Harmonization of the taxonomic names was made with the taxonomy tool and DNA barcodes reference library named Phytool (Canino et al., 2021). The unassigned diatom sequences were blasted on NCBI (https://www.ncbi.nlm.nih.gov/) and the assignations that surpassed 95% of similarity with over 98% if length were taken as species. The ASVs were clustered into taxa, and further unassigned to species or genus were taken out of the dataset.
The microscopy data were standardized with a logarithm for comparability; the amplicon reads were first calculated to proportion before standardization and analysis. Biodiversity measures were calculated following the Rimet and collaborator study setup (Rimet et al., 2018), in which alpha and gamma diversity equals the Shannon index using natural logarithms (Shannon, 1948). The diatom biodiversity measures (species richness and Shannon entropy), Jaccard dissimilarities, Bray–Curtis distances, Mantel test, and PCA were calculated using the vegan package (Oksanen et al., 2015). The bar- and boxplots were created using the ggplot2 package (Wickham, 2009). The heatmap distance matrices were calculated with FactoMineR (Husson et al., 2015).
Using microscopy, a total of 187 diatom taxa were identified, 99.99% to species (Supplementary Table 3). The division of taxa proportions according to sampling can be seen in Table 1. Through microscopy identification, the main diatom species were Achnanthidium minutissimum (Kützing) Czarnecki, Amphora pediculus (Kützing) Grunow, and Gomphonema micropus Kützing, which coincided with the predominant results of the rbcL dataset (Figure 2A). Microscopy had a predominance of subaerial taxa pertaining to Luticola and Humidophila, which were not found when amplicon sequencing.
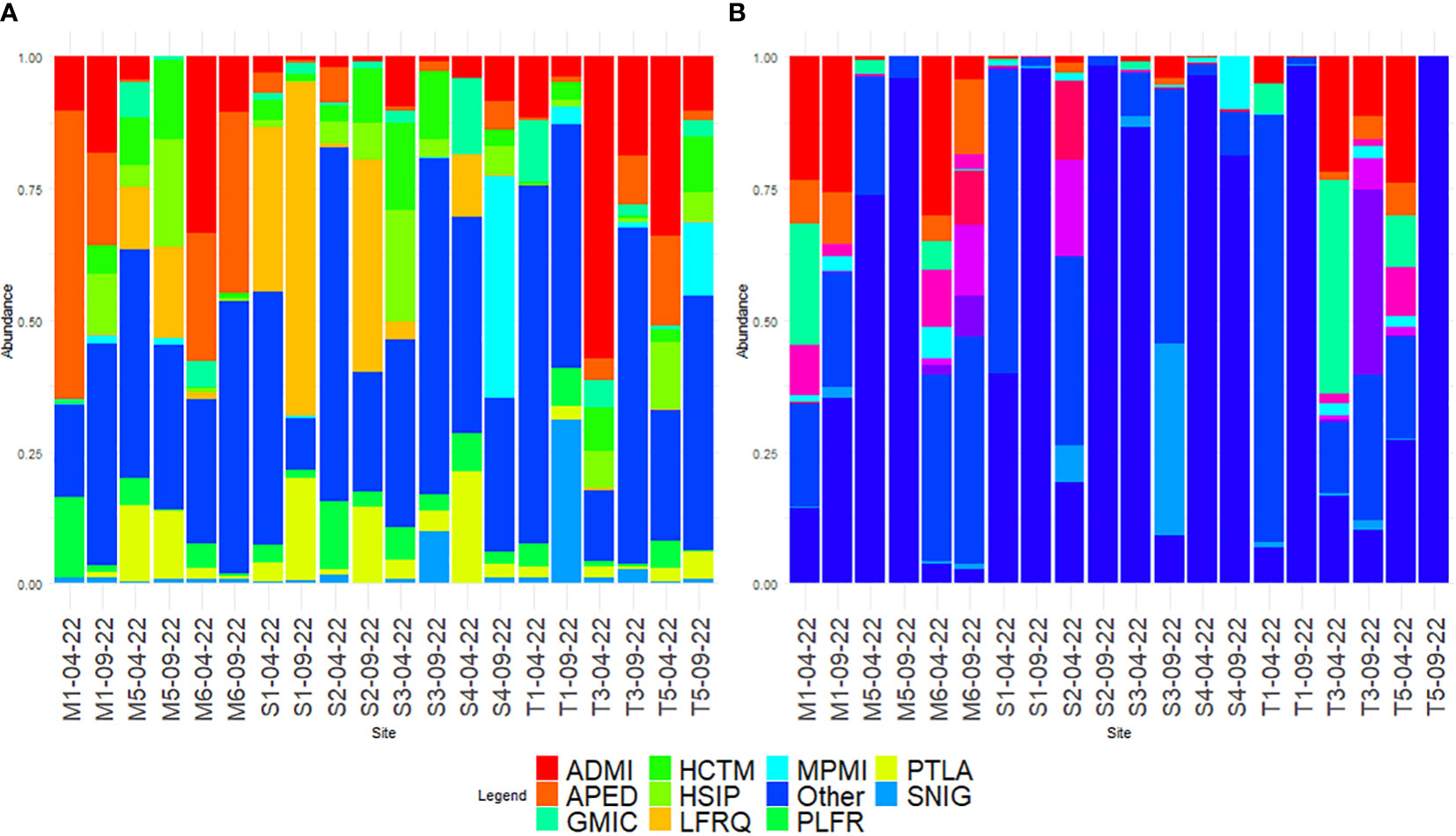
Figure 2 Comparison of the 10 predominant diatoms as identified with (A) microscopy and (B) rbcL amplicon sequencing, the rest is assigned to the “Other” category. Species codes based on OMNIDIA coding: ADMI, Achnanthidium minutissimum; APED, Amphora pediculus; GMIC, Gomphonema micropus; GPUM, Gomphonema pumilum; HCTM, Humidophila contemnata; HSIP, Humidophila simplex; LFQR, Luticola frequentissima; MPMI, Mayamaea permitis; MVAR, Melosira varians; NTPT, Navicula tripunctata; PLFR, Planothidium frequentissimum; PTLA, Planothidium lanceolatum; SNIG, Sellaphora nigri; XXXX, not assigned to taxon.
Based on amplicon sequencing, a total of 124 taxa were identified [43.36% species, 172 after blasting from a total of 286 molecular species (amplicon sequence variants, ASVs)] using a distance threshold of 95% for assignation through the diat.barcode reference library (v. 10.1). Further blasting only added 10 reads of low proportions to the before-mentioned subaerial taxa Luticola and five counts of Humidophila. A high proportion of the diatoms, including 100% of the T5 September sample, were not assigned to a taxon at dry locations (Figure 2B). However, the taxa assigned had some overlap with the microscopy results, as the three predominant taxa were the same. Some big taxa (Navicula tripunctata (O.F. Müller) Bory and Melosira varians C. Agardh) were found to be part of the predominant taxa with the amplicon assignation, although they were present in low numbers when using microscopy. A comparison of the proportions of the predominant diatoms according to the method of identification is listed in Table 2.
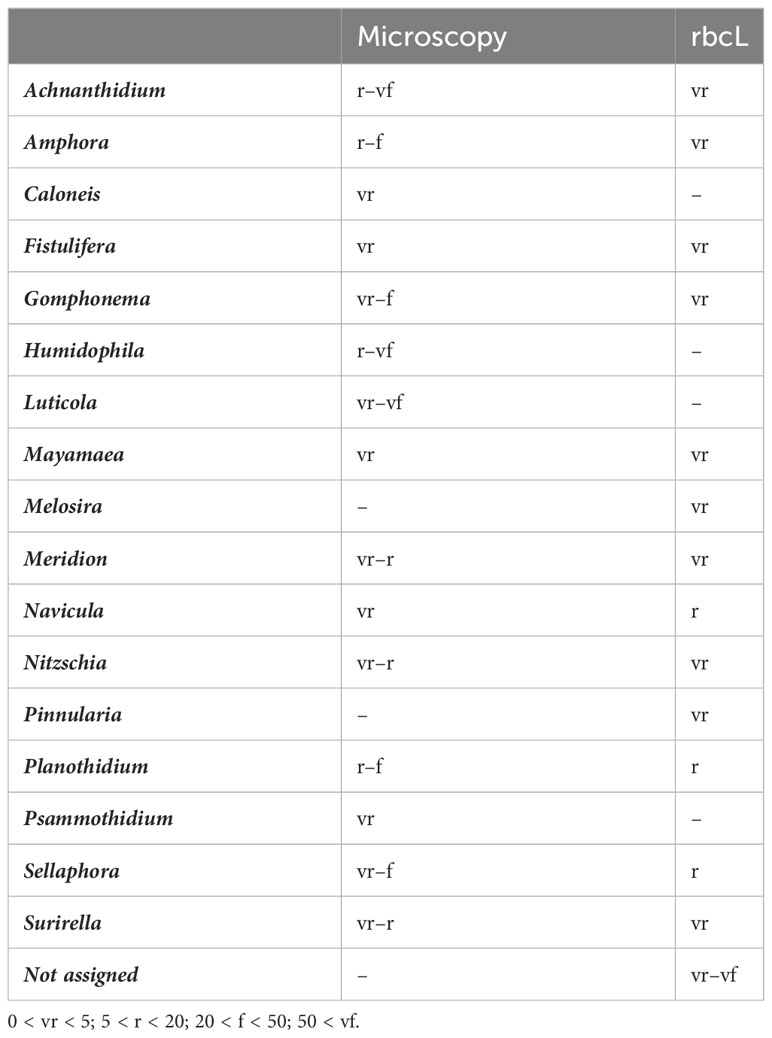
Table 2 Genus distribution according to method used for the biodiversity identification and median proportion.
To compare the distances between the diatom communities identified with the different methods, a mantel test was made on the Bray–Curtis distances from each site as calculated from DNA or microscopy method, which showed significant differences (r = 0.39, p-value = 0.00007). To confirm the similarities between methods, we compared the species and richness and Shannon diversity found using each method. The result showed that rbcL was usually not significantly different to the microscopy result (paired Student’s t-test: p-value > 0.0001). With further analysis, the differentiation of the spring sample and the summer samples showed that only summer had significant differences between the communities ascertained through microscopy or barcoding (Wilcoxon = 53, p-value = 0.002).
We calculated two principal component analyses (PCA) of the diatom composition as calculated from microscopy and amplicon sequencing (Supplementary Figure 1). The first two axes of the microscopy PCA represented 53.37% of the cumulative proportion, while the amplicon sequencing represented 73.64%. However, the correlation of both first axes of each method was not significant (ρ=0.19, p-value >0.05). An NMDS calculation (Figure 3) resulted in a clear separation of the methods used and, to some extent, the state of the sampled river site (Stress = 0.15). To better follow the site distribution, see Supplementary Figure 3.
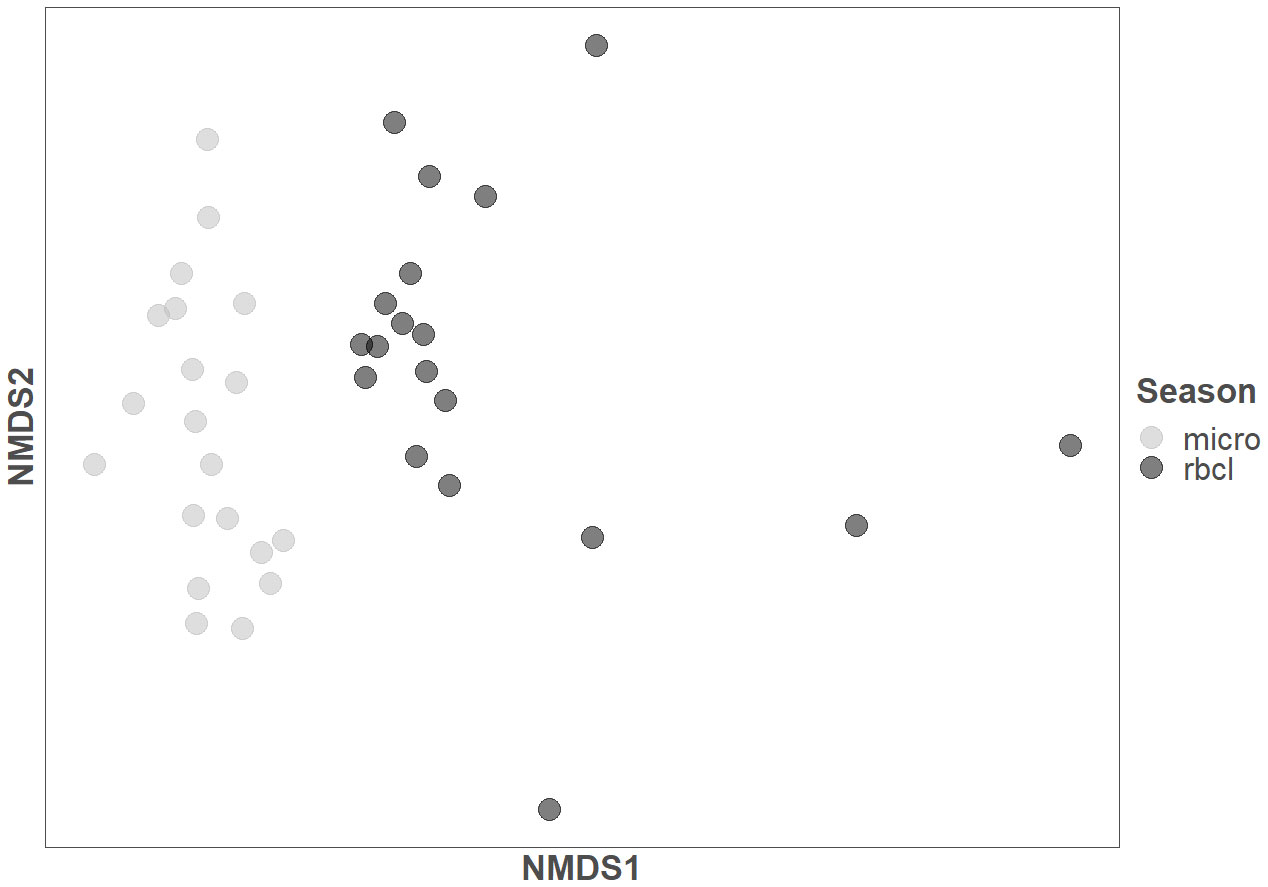
Figure 3 Non-metric multidimensional scaling (NMDS) of the samples identified with microscopy (micro) and amplicon sequencing (rbcL).
When comparing the local alpha-biodiversity measured in September 2022 with the values measured in April, there is a clear tendency of reduction (Figure 4). This had also been seen through the reduction in functional biomass measured in situ (Supplementary Table 2) and the increase in broken frustule and valve fragments (not shown). Both alpha and gamma diversities were significantly reduced using the amplicon sequencing method of diatom identification.
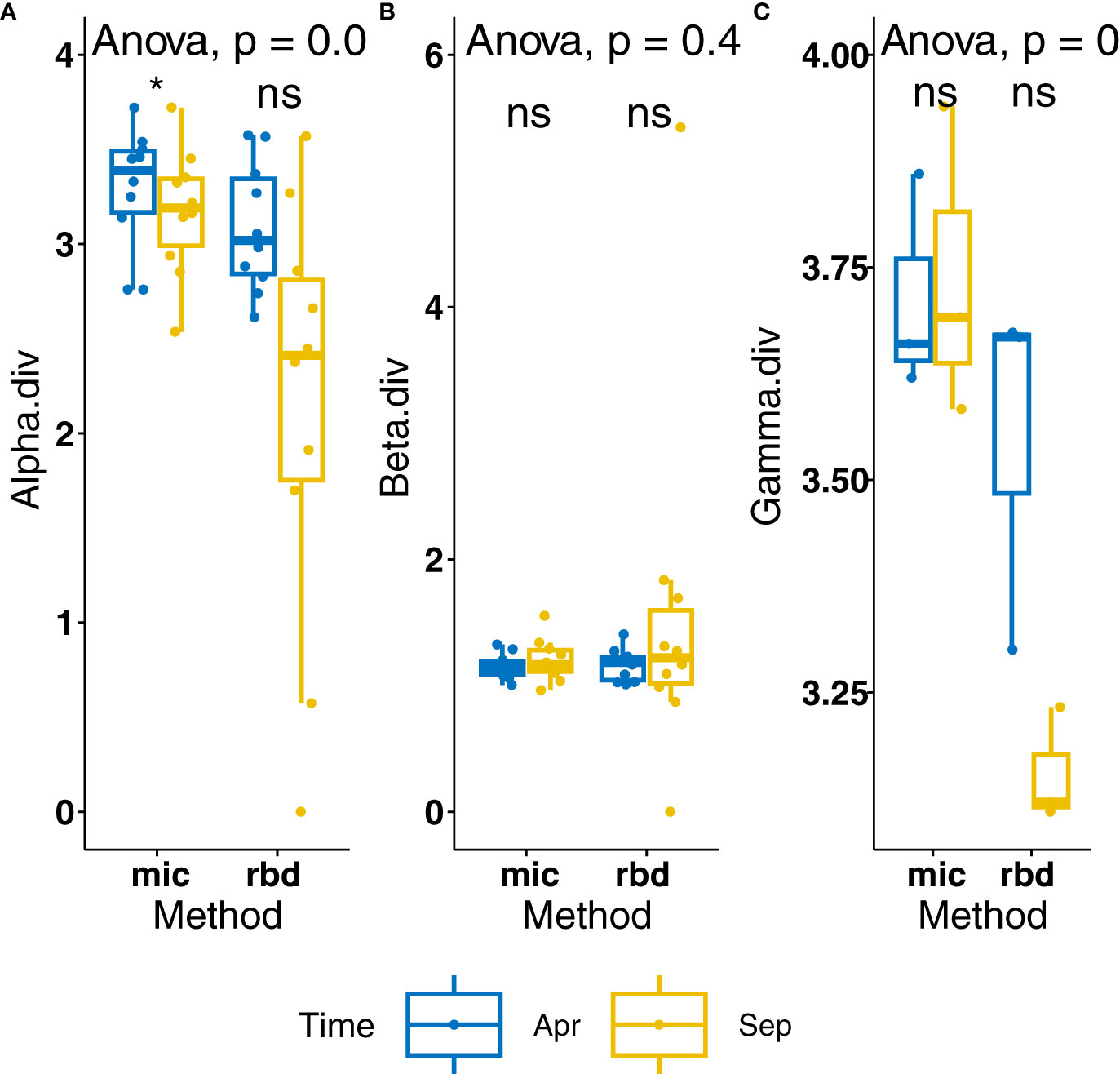
Figure 4 Comparison of diatom biodiversity depending on method of obtention. (A) Alpha diversity per method and time, (B) beta diversity per method and time, (C) gamma diversity per method and time. * = significant difference, p-value < 0.05. ns = not significant (p-values >0.05).
Looking at Jaccard similarities of the molecular and microscopic workflow (Figure 5), differences become clear. A clearer subdivision of diatom communities according to time sampled is found in the microscopy-identified samples, while the rbcL-amplicon-identified samples have more random dissimilarities, rarely following the drying pattern. Furthermore, when comparing the whole dataset (molecular and microscopic), the sites were clustered by methodology, with only the rbcL T1-09-22 sample clustering with the microscopy (Supplementary Figure 2).
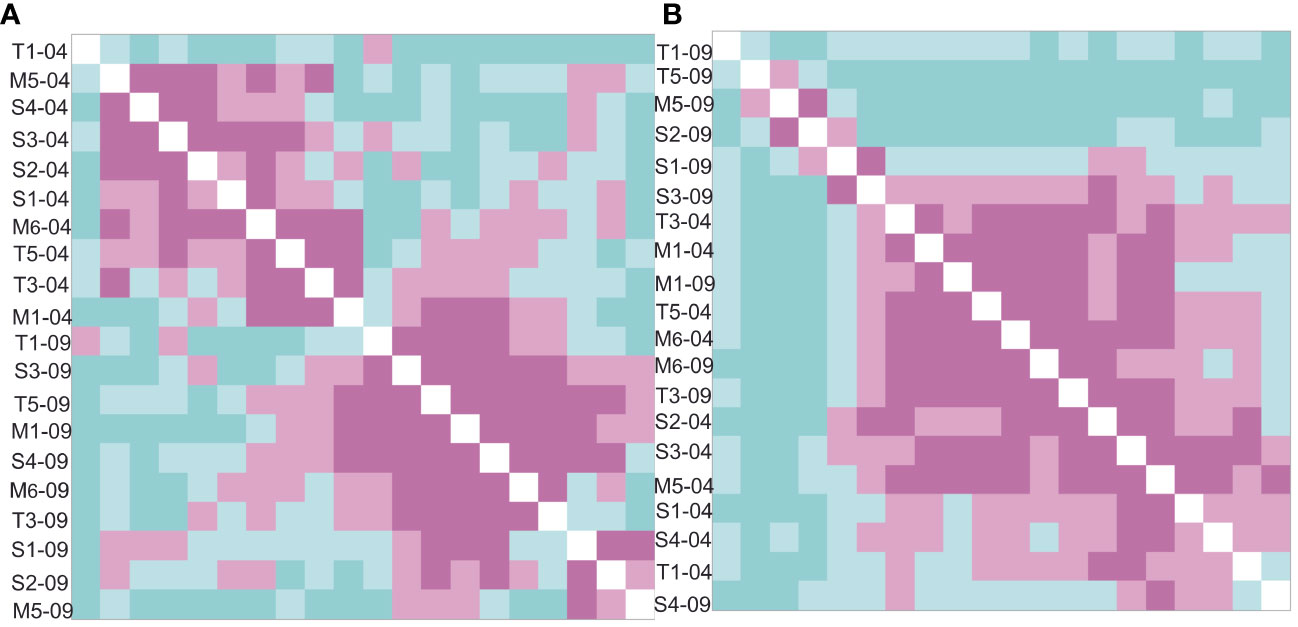
Figure 5 Ordered Jaccard dissimilarity matrix of the (A) microscopic and (B) rbcL-amplicon-detected diatom communities. The microscopy shows a clearer time separation (April = black, September, color) than the rbcl-amplicon-identified community. Microscopy also shows a better similarity of similar flow state. Menne = hexagons and circles, Sauer = triangles and reptiles, Talgosse = fish scales (black and color).
The biodiversity was highest in the pool locations, followed by the flowing river state. The dry river had the lowest biodiversity in general (Figure 6). Further comparing the methods of identification, microscopy usually showed a higher value of diversity and a narrower range, which was more spread for amplicon sequencing. In the dry state, the proportion of alpha diversity was even significantly higher. Table 3 shows the alpha, beta, and gamma diversity according to the method and time of sampling.
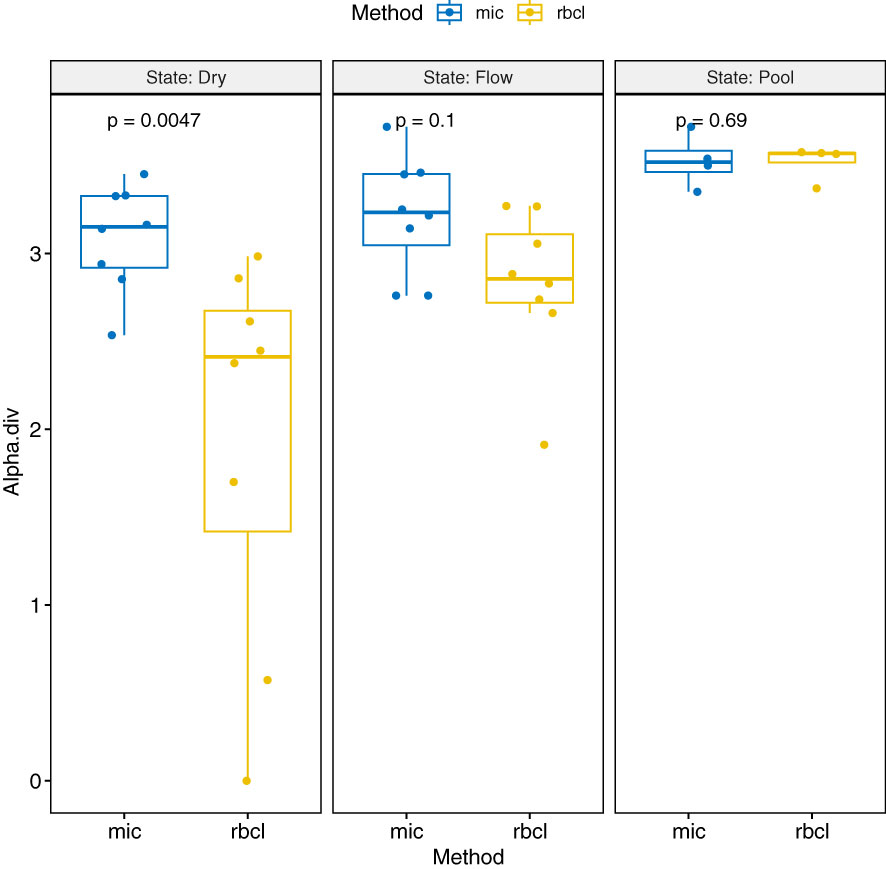
Figure 6 Boxplot with ANOVA values of the alpha diversity values depending on the method of obtaining (mic, microscopy; rbcl, rbcL amplicon) and the state of the river when sampled.
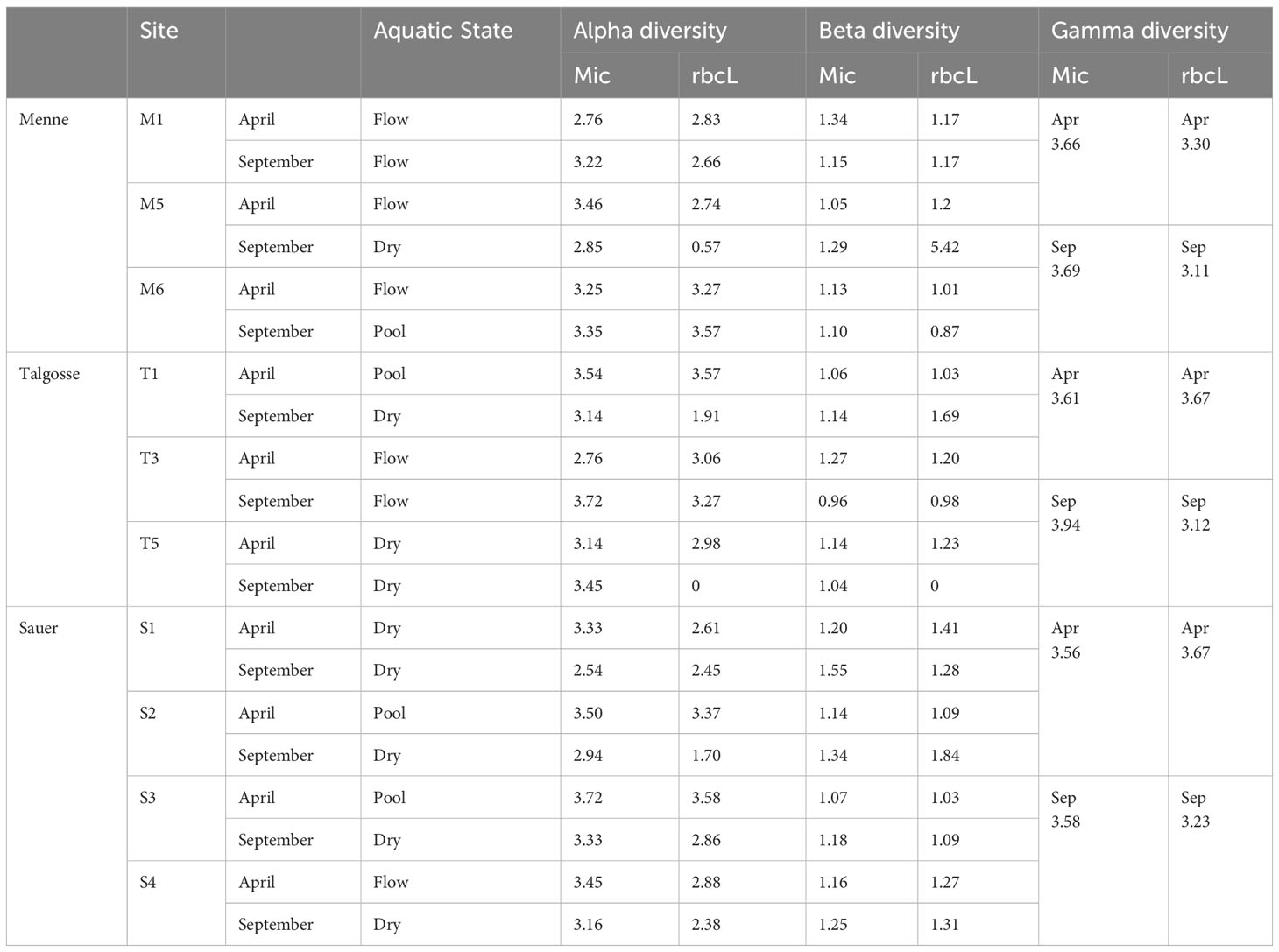
Table 3 Comparison of the calculated alpha, beta, and gamma diversity according to sample and identification method.
This study aimed to confirm and to compare the effectiveness of both (digital) microscopy and amplicon sequencing in determining the biodiversity in diatom communities of intermittent rivers. We have found that river intermittency has a big impact on the taxon assignation accuracy of the molecular method, confirming the stated hypotheses. This will mainly be due to the reference library being mostly based on fully humidophilous taxa and lacking the subaerial and terrestrial diatoms that are characteristic of these types of streams. Until now, only a handful of studies have investigated intermittent streams using metabarcoding (Pissaridou et al., 2021). Although the assignation rate was not ascertained, the level of assignation was mostly at taxonomic class level.
Even though amplicon sequencing resulted in a higher quantity of amplicon sequence variants (ASVs), the total of assigned taxa was smaller than the number of diatom taxa identified using digital microscopy. Furthermore, even after blasting, the number of assigned diatom taxa was still smaller. In addition, some differences were found between the datasets; for instance, a higher proportion of centric diatoms was found in the amplicon sequences (Supplementary Material). This can be due to the differential quantity of chloroplasts between centric (high quantity and small) and pennate diatoms (low quantity and big, Nonoyama et al., 2019). Although the DADA2 pipeline contains the quantification coefficient (Vasselon et al., 2018) to avoid quantitative errors, the quantity of chloroplasts might still be affecting the read count.
Nevertheless, the chloroplast and biovolume difference might also be the reason why the species composition was partially differential. The higher proportion of big taxa, Navicula tripunctata and Melosira varians, although present, were found in a lower proportion in the microscopy dataset. However, the predominant taxa, Achnanthidium minutissimum, Amphora pediculus, and Gomphonema micropus were found in all sites, although the variability of percentage found according to the method of identification soared (Supplementary Table 3). The change in proportion and even inversion of proportion of two taxa are not uncommon in taxa not pertaining to the most abundant ones (Vidakovic et al., in prep.). This might already point to the unsuitability of the amplicon-sequencing-based identification for biodiversity studies in non-perennial rivers as long as these biomass divergences persist.
Because of these differences between the identification method results, it is not surprising that the similarities between the site and method were quite small (Supplementary Figure 2). Only the site T1 sampled in September, the original pool site found in a dry state, was too different to anything else and clustered only with the microscopy data. Meanwhile, T5, original flowing state sampled dry in September, did not even feature because of the amplicon sequences only amplifying other algae sequences (mostly of the order Vaucheriales). As these sites were dry at the time of sampling, after a heavy drought, the quality of the DNA sampled could have deteriorated, showing the death of the diatoms within the biofilm (Smith et al., 2013; Smith et al., 2019). The dissimilarity method used did not change the pattern of dissimilarity, as no variability was observed when re-calculating the distances of the sites with other formulas (not shown).
This leads to the question, how many of the diatoms were still alive at the time of sampling? Both methods have some degree of bias to identify the living diatoms from the dead ones. As diatom microscopy looks at diatoms only after the oxidation of the cells, the identification of diatom state can, to some extent, be ascertained through the brokenness of the frustules (shells). To circumvent this, before preparation, we observed an aliquot of each sample under the microscope to confirm the proportion of living cells and used the Benthotorch for biomass estimation in the field (Supplementary Table 2). Through this, we can confirm that although the biomass and general quantity of diatoms were low at these sites, a significant proportion of diatoms was still alive at the time of sampling and should have been caught by amplicon sequencing. However, as explained above, the subaerial taxa are mostly missing from the reference databases and such can be the explanation of the differences (Kulaš et al., 2022).
Due to the low proportion of assigned taxa, alpha and gamma diversity were significantly reduced when using amplicon sequencing (Figure 3). This means that at the moment of this analysis, the use of amplicon sequencing to ascertain biodiversity would have dramatically undershot. This may be a reason for the increase in use of taxonomy-free amplicon sequencing (Apothéloz-Perret-Gentil et al., 2017; Tapolczai et al., 2019; Gregersen et al., 2023). Taxonomy assignments work best where they have been created, and a geographical bias is already visible in permanent rivers. In the case of the rbcL marker, the proportion of assigned taxa names gets gradually smaller the higher the distance is to France, where the marker was initially created (Mugnai et al., 2023). This phenomenon is enhanced when the riparian lotic ecosystem changes to lentic, confirming our first and second hypotheses. Therefore, at this stage, amplicon sequencing would not be encouraged to sole identification of diatom biodiversity.
In order to be able to use amplicon sequencing meaningfully in diatom biodiversity monitoring in non-perennial rivers, the reference databases will need to be updated with the subaerial and aerial taxa. Furthermore, to equate it to “traditional” monitoring based on proportional species distribution, the biomass correction factors should be expanded and updated accordingly.
When looking at the effects of the hydrological conditions within the catchments, clear differences between them appear. The point in time in sampling had very different effects on the sampled rivers, as in April, the Menne was mainly flooded, homogenizing the flow type of the whole catchment. In the meantime, in September, the drought mostly affected the Sauer and the Talgosse, leaving all visited sites with either extremely reduced flow (T3), hyporheic (S3 and S2), or completely dry (S1, T1, and T5). However, although reduced, the amount of biomass measured was still high, including a predominance of diatoms (Supplementary Table 2).
Both methods show a significant clustering of the dry sites. The sampling date also creates a clustering of the spring samples for both methods, while the summer samples were clustering better in the microscopy samples. As the summer samples were taken during an intense drought, the taxa found in excess were mostly subaerial. As already mentioned, they were thus underrepresented in the diat.barcode datasets due to it being adapted to flowing river biomonitoring (Keck et al., 2019; Rimet et al., 2019).
General patterns emerged, as the combination of drought and sampling a dry location reduced alpha diversity of the molecular results significantly. In contrast, in wet environments (both pools and flowing river), the alpha diversity was more similar between the used methods. As this includes both sampling efforts, the time since drying did not seem to have a deleterious effect on the diversity. Furthermore, we had expected the flooding to affect the maturity of the biofilm, as increased flow and turbulence could move the substrate, which would be repeatedly colonized as a result, which we did not observe much, other than the (small) increase in colonizing taxa in those sites (Figure 2). We believe that this confirms our hypothesis that drying would create a divergence between the methods.
Comparing hydrology, we found that through extreme drought (e.g., the dry locations found in the second sampling in September), the diversity suffered. The alpha diversity was found to be significantly lower in the dry locations, and the gamma diversity was somewhat lower, confirming our third hypothesis.
The similarity of amplicon sequencing samples was increased in dry locations and mostly in the September sampling. We posit that we are observing the effect of taxon homogenization due to drying. On the other hand, the recurring impact of missing subaerial taxa, which varied according to the microscopic identification, may also be responsible for this (artificial) similarity. Up to this time few comparative studies of microscopic and amplicon sequencing identifications have been made in intermittent rivers and streams, but those that exist have found similar effects (Kulaš et al., 2022).
In conclusion, drying does affect diatom communities and reduce their diversity after prolonged drought. Using different methods, we have found that digital microscopy showed a higher resolution and diversity in intermittent streams due to the incomplete reference databases for dry(ing) habitats. In the future, the use of both combined methods to calculate bioindex values might be of interest, as regrettably, the trend of intermittency is reaching ever further in the European continent, and as it currently stands, a lot of rivers might fall out of the regulation of the Water Framework Directive because of it. However, for the time being, the unique use of amplicon sequencing for non-perennial rivers would not be advisable, as information on subaerial taxa and proportion of the taxa within the community would be missing.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
AB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. PM: Data curation, Formal analysis, Writing – review & editing. MD: Data curation, Formal analysis, Writing – review & editing. BB: Funding acquisition, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. SSC Postdoctoral seed funding - supported the sampling and sequencing financially. This funding is part of the internal funding of the University of Duisburg-Essen (Postdoc Seed Funding/ Programm zur Förderung des Exzellenten wissenschaftlichen Nachwuchses) and does not have a grant number.
We want to thank Marzena Spyra, Danijela Vidakovic and Carolina van Gelder for their help in the lab and with the molecular methods. We also want to acknowledge the support of Kreis Paderborn, as this institution gave us the permits for sampling in the nature reserves. The funding of this project was due to the Science Support Centre of the University of Duisburg-Essen, through a now called “Postdoc Seed Funding” (at the time called “Funding for the support of excellent Early Career Researchers”). We thank two reviewers for their help in improving this manuscript. We acknowledge support by the Open Access Publication Fund of the University of Duisburg-Essen.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1355018/full#supplementary-material
Apothéloz-Perret-Gentil L., Cordonier A., Straub F., Iseli J., Esling P., Pawlowski J. (2017). Taxonomy-free molecular diatom index for high-throughput eDNA biomonitoring. Mol. Ecol. Resour. 17, 1231–1242. doi: 10.1111/1755-0998.12668
Bailet B., Bouchez A., Franc A., Frigerio J.-M., Keck F., Karjalainen S.-M., et al. (2019). Molecular versus morphological data for benthic diatoms biomonitoring in Northern Europe freshwater and consequences for ecological status. Metabarcoding and Metagenomics (MBMG) 3, 21–35. doi: 10.3897/mbmg.3.34002
B-Béres V., Tóthmérész B., Bácsi I., Borics G., Abonyi A., Tapolczai K., et al. (2019). Autumn drought drives functional diversity of benthic diatom assemblages of continental intermittent streams. Adv. Water Resour. 126, 129–136. doi: 10.1016/j.advwatres.2019.02.010
Bey M.-Y., Ector L. (2013). Atlas des diatomées des cours d’eau de la région Rhône-Alpes (Caluire, France: Tomes 1-6. DREAL Rhône-Alpes).
Blöschl G., Hall J., Viglione A., Perdigão R. A. P., Parajka J., Merz B., et al. (2019). Changing climate both increases and decreases European river floods. Nature 573, 108–111. doi: 10.1038/s41586-019-1495-6
Bonaldo D., Bellafiore D., Ferrarin C., Ferretti R., Ricchi A., Sangelantoni L., et al. (2023). The summer 2022 drought: a taste of future climate for the Po valley (Italy)? Reg. Environ. Change 23, 1–6. doi: 10.1007/s10113-022-02004-z
Bruce K., Blackman R., Bourlat S. J., Hellström A. M., Bakker J., Bista I., et al. (2021). A practical guide to DNA-based methods for biodiversity assessment. Pensoft Publishers 1, e68634. doi: 10.3897/ab.e68634
Burfeid Castellanos A. M. (2018). Ecological factors and diatom diversity at rivers of the Iberian Mediterranean river basins: macro-scale, meso-scale and micro-scale = Factores ecológicos y diversidad de diatomeas en los ríos de las cuencas hidrográficas del mediterráneo ibérico: maro-escala, meso-escala y micro-escala (Ph.D. Thesis) (Barcelona, Spain: Universitat de Barcelona). Available at: https://tdx.cat/handle/10803/663475.
Burfeid-Castellanos A. M., Kloster M., Beszteri S., Postel U., Spyra M., Zurowietz M., et al. (2022). A digital light microscopic method for diatom surveys using embedded acid-cleaned samples. Water 14, 3332. doi: 10.3390/w14203332
Canino A., Bouchez A., Laplace-Treyture C., Domaizon I., Rimet F., Bouchez A., et al. (2021). Phytool, a ShinyApp to homogenise taxonomy of freshwater microalgae from DNA barcodes and microscopic observations. Metabarcoding and Metagenomics (MBMG) Vol. 5, e74096. doi: 10.3897/mbmg.5.74096
Cantonati M., Kelly M. G., Lange-Bertalot H. (Eds.) (2017). Freshwater benthic diatoms of Central Europe: over 800 common species used in ecological assessment, 1st ed (Oberreifenberg, Germany: Koeltz Scientific Books).
CEN (2018). Water quality - Technical report for the management of diatom barcodes. Available at: https://standards.iteh.ai/catalog/standards/cen/b4f19219-1ab9-4ec7-9457-47b02fd12649/cen-tr-17244-2018 (Accessed 9 Feb 2023).
Datry T., Bonada N., Boulton A. J. (Eds.) (2017). Intermittent Rivers and Ephemeral Streams -. Ecology and Management, 1st Edition (London, UK: Academic Press).
Datry T., Larned S. T., Fritz K., Bogan M. T., Wood P. J., Meyer E. I., et al. (2014). Broad-scale patterns of invertebrate richness and community composition in temporary rivers: effects of flow intermittence. Ecography 37, 94–104. doi: 10.1111/j.1600-0587.2013.00287.x
Delgado C., Pardo I., García L. (2012). Diatom communities as indicators of ecological status in Mediterranean temporary streams (Balearic Islands, Spain). Ecol. Indic. 15, 131–139. doi: 10.1016/j.ecolind.2011.09.037
European Commission (2000). Directive 2000/60/EC of the European Parliament and of the Council of 23 October 2000 establishing a framework for community action in the field of water policy. Official Journal of the European Union L 327, 22.12.2000. (Luxembourg, Luxembourg: WFD).
Gregersen R., Pearman J. K., Atalah J., Waters S., Vandergoes M. J., Howarth J. D., et al. (2023). A taxonomy-free diatom eDNA-based technique for assessing lake trophic level using lake sediments. J. Environ. Manage. 345, 118885. doi: 10.1016/j.jenvman.2023.118885
Husson F., Josse J., Le S., Mazet J. (2015). Package ‘ FactoMineR ‘. Available at: https://cran.r-project.org/web/packages/FactoMineR/FactoMineR.pdf.
Illumina. 16S Sample Preparation Guide. Available at: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf.
Keck F. (2020). DADA2_diatoms_pipeline. for R. github: Keck, F. Available at: https://github.com/fkeck/DADA2_diatoms_pipeline.
Keck F., Rimet F., Vasselon V., Bouchez A. (2019). A ready-to-use database for DADA2: Diat.barcode_rbcL_312bp_DADA2 based on Diat.barcode v7. doi: 10.15454/HNI1EK
Kopalová K., Kociolek J. P., Lowe R. L., Zidarova R., van de Vijver B. (2015). Five new species of the genus Humidophila (Bacillariophyta) from the Maritime Antarctic Region. Diatom. Res. 30, 117–131. doi: 10.1080/0269249X.2014.998714
Kulaš A., Gligora Udovič M., Tapolczai K., Žutinić P., Orlić S., Levkov Z. (2022). Diatom eDNA metabarcoding and morphological methods for bioassessment of karstic river. Sci. Total Environ. 829, 154536. doi: 10.1016/j.scitotenv.2022.154536
Langenkämper D., Zurowietz M., Schoening T., Nattkemper T. W. (2017). BIIGLE 2.0 - browsing and annotating large marine image collections. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00083
Levkov Z., Metzeltin D., Pavlov A. (2013). Luticola and Luticolopsis, Diatoms of Diatoms of Europe. Ed. Lange-Bertalot H. (Oberreifenberg, Germany: Koeltz botanical books).
Levkov Z., Mitic-Kopanja D., Reichardt E. (2016). Diatoms of Europe, Vol 8: The Diatom Genus Gomphonema from the Republic of Macedonia. Ed. Lange-Bertalot H. (Oberreifenberg, Germany: Koeltz botanical books).
Messager M. L., Lehner B., Cockburn C., Lamouroux N., Pella H., Snelder T., et al. (2021). Global prevalence of non-perennial rivers and streams. Nature 594, 391–397. doi: 10.1038/s41586-021-03565-5
Meyer A., Meyer E. I. (2000). Discharge regime and the effect of drying on macroinvertebrate communities in a temporary karst stream in East Westphalia (Germany). Aquat. Sci. 62, 216–231. doi: 10.1007/PL00001333
Meyer A., Meyer E. I., Meyer C. (2003). Lotic communities of two small temporary karstic stream systems (East Westphalia, Germany) along a longitudinal gradient of hydrological intermittency. Limnologica 33, 271–279. doi: 10.1016/S0075-9511(03)80022-1
Mugnai F., Costantini F., Chenuil A., Leduc M., Gutiérrez Ortega J. M., Meglécz E. (2023). Be positive: customized reference databases and new, local barcodes balance false taxonomic assignments in metabarcoding studies. PeerJ 11, e14616. doi: 10.7717/peerj.14616
Nonoyama T., Kazamia E., Nawaly H., Gao X., Tsuji Y., Matsuda Y., et al. (2019). Metabolic innovations underpinning the origin and diversification of the diatom chloroplast. Biomolecules 9. doi: 10.3390/biom9080322
Novais M. H., Morais M. M., Rosado J., Dias L. S., Hoffmann L., Ector L. (2014). Diatoms of temporary and permanent watercourses in southern Europe (Portugal). River Res. Appl. 30, 1216–1232. doi: 10.1002/rra.2818
Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., Hara R. B. O., et al. (2015). Ordination methods, diversity analysis and other functiuons for community and vegetation ecologists, Community ecology package version 1.
Peeters V., Ector L. (2017). Atlas des Diatomées des cours d’eau du territoire Bourguignon, DREAL Bourgogne-Franche-Comté.
Pissaridou P., Vasselon V., Christou A., Chonova T., Papatheodoulou A., Drakou K., et al. (2021). Cyprus’ diatom diversity and the association of environmental and anthropogenic influences for ecological assessment of rivers using DNA metabarcoding. Chemosphere 272, 129814. doi: 10.1016/j.chemosphere.2021.129814
Rimet F., Gusev E., Kahlert M., Kelly M. G., Kulikovskiy M., Maltsev Y., et al. (2019). Diat.barcode, an open-access curated barcode library for diatoms. Sci. Rep. 9, 15116. doi: 10.1038/s41598-019-51500-6
Rimet F., Vasselon V., A.-Keszte B., Bouchez A. (2018). Do we similarly assess diversity with microscopy and high-throughput sequencing? Case of microalgae in lakes. Org Divers. Evol. 18, 51–62. doi: 10.1007/s13127-018-0359-5
Shannon C. E. (1948). A Mathematical Theory of Communication. The Bell System Technical Journal 27, 623–656.
Smith M. W., Herfort L., Rivers A. R., Simon H. M. (2019). Genomic signatures for sedimentary microbial utilization of phytoplankton detritus in a fast-flowing estuary. Front. Microbiol. 10, 2475. doi: 10.3389/fmicb.2019.02475
Smith M. W., Zeigler Allen L., Allen A. E., Herfort L., Simon H. M. (2013). Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front. Microbiol. 4, 120. doi: 10.3389/fmicb.2013.00120
Stubbington R., Chadd R., Cid N., Csabai Z., Miliša M., Morais M., et al. (2018). Biomonitoring of intermittent rivers and ephemeral streams in Europe: Current practice and priorities to enhance ecological status assessments. Sci. Total Environ. 618, 1096–1113. doi: 10.1016/j.scitotenv.2017.09.137
Szczepocka E., Żelazna-Wieczorek J. (2018). Diatom biomonitoring – scientific foundations, commonly discussed issues and frequently made errors. Oceanol. Hydrobiol. Stud. 47, 313–325. doi: 10.1515/ohs-2018-0030
Tapolczai K., Keck F., Bouchez A., Rimet F., Kahlert M., Vasselon V. (2019). Diatom DNA metabarcoding for biomonitoring: strategies to avoid major taxonomical and bioinformatical biases limiting molecular indices capacities. Front. Ecol. Evol. 7. doi: 10.3389/fevo.2019.00409
Taylor J. C., Harding W. R., Archibald C. G. M. (2007). A methods manual for the collection, preparation and analysis of diatom samples (Pretoria, South Africa: WRC Report TT282/07).
Vasselon V., Bouchez A., Rimet F., Jacquet S., Trobajo R., Corniquel M., et al. (2018). Avoiding quantification bias in metabarcoding: Application of a cell biovolume correction factor in diatom molecular biomonitoring. Methods Ecol. Evol. 9, 1060–1069. doi: 10.1111/2041-210X.12960
Vasselon V., Domaizon I., Rimet F., Kahlert M., Bouchez A. (2017a). Application of high-throughput sequencing (HTS) metabarcoding to diatom biomonitoring: Do DNA extraction methods matter? Freshw. Sci. 36, 162–177. doi: 10.1086/690649
Vasselon V., Rimet F., Tapolczai K., Bouchez A. (2017b). Assessing ecological status with diatoms DNA metabarcoding: Scaling-up on a WFD monitoring network (Mayotte island, France). Ecol. Indic. 82, 1–12. doi: 10.1016/j.ecolind.2017.06.024
Vautier M., Vasselon V., Chardon C., Rimet F., Bouchez A., Domaizon I. (2020). DNA extraction from environmental biofilm using the NucleoSpin® Soil kit (MACHEREY-NAGEL) v1. doi: 10.17504/protocols.io.bd52i88e
Keywords: IRES, intermittent rivers and ephemeral streams, temporary rivers, benthic diatoms, freshwater diatoms, metabarcoding, molecular method, ASV
Citation: Burfeid-Castellanos AM, Mones P, Dani M and Beszteri B (2024) Non-perennial rivers and streams in extreme hydrological conditions—comparing the effectiveness of amplicon sequencing and digital microscopy for diatom biodiversity appraisal. Front. Ecol. Evol. 12:1355018. doi: 10.3389/fevo.2024.1355018
Received: 13 December 2023; Accepted: 22 January 2024;
Published: 15 February 2024.
Edited by:
Evangelia Smeti, Hellenic Centre for Marine Research (HCMR), GreeceReviewed by:
John Patrick Kociolek, University of Colorado Boulder, United StatesCopyright © 2024 Burfeid-Castellanos, Mones, Dani and Beszteri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea M. Burfeid-Castellanos, YW5kcmVhLmJ1cmZlaWQtY2FzdGVsbGFub3NAdW5pLWR1ZS5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.