
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 18 June 2024
Sec. Biogeography and Macroecology
Volume 12 - 2024 | https://doi.org/10.3389/fevo.2024.1328569
Lyophyllum consists of rare edible and medicinal mushrooms. Considering this group’s economic and ecological significance, understanding its biodiversity could be strategically important. Our analysis involved an extensive examination of publicly available ITS sequences from NCBI-GenBank and fungal amplicon sequencing data obtained from NCBI-SRA. At a 98% sequence similarity level, we deliminated 88 Lyophyllum OTUs, 49 of which were not categorized as ‘known species’. The diversity of Lyophyllum is predominantly concentrated in the Northern Hemisphere and tends to display endemic distribution, and Europe is particularly notable for its high diversity of Lyophyllum. Given the escalating volume of data produced by amplicon sequencing, employing the amplicon dataset can facilitate an accurate survey of species diversity in Lyophyllum taxonomy. By using data from amplicon datasets, the fieldwork time and research funding for taxonomists can be saved, thereby significantly advancing the progress of the entire field of biodiversity research.
While fungi play diverse roles in ecosystems and are vital for biodiversity, ecological balance, and environmental health, researchers are just beginning to unravel the processes that shape the diversity and distribution of their global species (Dähncke et al., 2011; Tedersoo et al., 2014). Over the past decade, the discovery of several new genera is expected to complete the system of the Lyophyllaceae, including Australocybe T.J. Baroni, N. Fechner & van de Peppel, Calocybella Vizzini, Consiglio & Setti, Clitolyophyllum E. Sesli, Vizzini & Contu, Myochromella V. Hofst., Clémençon, Moncalvo & Redhead, Nigrocarnea P. Sparre & Læssøe, Phaeotephrocybe T.J. Baroni, T.W. Kuyper & van de Peppel, Praearthromyces T.J. Baroni, T.W. Kuyper & van de Peppel, Sagaranella V. Hofst., Clémençon, Moncalvo & Redhead, etc (Cooper, 2014; Hofstetter et al., 2014; Vizzini et al., 2015; Li et al., 2017; Xu et al., 2019; Ye et al., 2019; van de Peppel et al., 2022). The synapomorphy of Lyophyllum is the presence of siderophilous granulation in the basidia (Kühner, 1938; Joshi et al., 2021). Lyophyllum has a global distribution, encompassing more than 40 species (Kirk et al., 2008; He et al., 2019). It includes several rare edible fungi, such as L. decastes (Fr.) Singer, L. fumosum (Pers.) P.D. Orton, and L. shimeji (Kawam.) Hongo, etc. Lyophyllum species play crucial roles in various ecological functions. Traditionally, Lyophyllum has been regarded as saprophytic, playing important roles in ecosystems by promoting organic matter decomposition and nutrient cycling (Kobayashi et al., 2023). Furthermore, subsequent research has revealed that Lyophyllum shimeji exhibits ectomycorrhizal characteristics, which can enhance plant nutrient acquisition efficiency through a symbiotic relationship (Singer, 1986; Ohta, 1994; Kusuda et al., 2004; Tedersoo et al., 2010). Lyophyllum shimeji thrives predominantly in Pinus, Quercus, and alpine forests (Ohta, 1994), forming mycorrhizal symbiotic associations with plant roots (Cooper, 2014). This association enables L. shimeji to supply plants with water, inorganic salts, and nutrient absorption, thereby playing a vital role in the dynamics and evolution of forest ecosystems (Larsson and Sundberg, 2011).
Lyophyllum is known for its abundance of biologically active substances. Notably, three primary polysaccharides found in L. decastes have exhibited significant antitumor activities against Sarcoma 180 (Ukawa et al., 2000) and have also demonstrated lower blood sugar effects (Miura et al., 2002). In 2022, Ding et al. completed the structure identification and biological activities of the new polysaccharide of L. decastes (Ding et al., 2022). Several studies have identified various active components that can be isolated from Lyophyllum species (Zhang et al., 2010), making them a promising and valuable source for new natural medicines (Barros et al., 2008). Species within the L. decastes complex also exhibit numerous biological activities (Tel et al., 2015), including radiation protection, anti-fungal, and antioxidant effects. These effects are believed to be attributed to bioactive components in their fruiting bodies, such as ribonucleases and β-D-glucans. Wang et al. revealed that the polysaccharides of L.decastes could alter gut microbiota, contribute to the antiobesity effects, and provide insights into the potential therapeutic mechanism in modulating energy metabolism to enhance energy expenditure (Wang et al., 2022).
Lyophyllum is highly economically valued in Japan, with price ranking second only to Tricholoma matsutake (S. Ito & S. Imai) Singer (Larsson and Sundberg, 2011). However, recent factors such as habitat loss, pine wilt disease, and shifts in forest management have led to a decline in wild populations of L. shimeji in Japan (Li et al., 2019). Consequently, there has been a growing emphasis on the artificial cultivation of Lyophyllum species to meet commercial demand (Takaki et al., 2014). Furthermore, evidence indicates that Lyophyllum has gained commercial traction internationally (Estrada-Martínez et al., 2009; Abarenkov et al., 2010). Among them, Lyophyllum decastes Sing., also known as “Fried chicken mushroom” in Europe and “Laifumo” in China, has a rich and delicate texture, a fragrant aroma, and a delicious taste. It is abundant in trace elements and offers numerous medicinal benefits (Xu et al., 2023). In recent years, there has been a gradual increase in reports regarding cultivation techniques (Pokhrel and Ohga, 2007a; Pokhrel and Ohga, 2007b; Xu et al., 2023). Nevertheless, a notable dearth of studies focusing on species diversity and taxonomy remains. This could be attributed to the limited macroscopic morphological differences among Lyophyllum species, making them challenging to distinguish from one another. Additionally, the sampling challenges of Lyophyllum may pose difficulties for researchers in locating and documenting their presence.
Traditionally, species within the Lyophyllum have been delineated based on morphological characteristics and, more recently, through mating compatibility studies (Clémençon and Smith, 1983; Maeta et al., 2008; Vizzini and Contu, 2010). However, due to the challenges associated with identifying species solely based on macroscopic traits, there has been an increasing emphasis on understanding species boundaries within Lyophylloid clades through phylogenetic analyses. Studies on the Lyophylloid clade (Hofstetter et al., 2014; Wei et al., 2023), like many assessments of fungal diversity, heavily relied on data from the nuclear ribosomal operon, including the internal transcribed spacer region (ITS), the small subunits of mitochondrial ribosomes (mtSSU), and the nuclear ribosomal large subunit gene (nLSU) (Hofstetter et al., 2002). However, recent investigations have demonstrated that the internal transcribed spacer (ITS) and specific regions of the RNA polymerase II second largest subunit gene (RPB2) yield the most reliable combination for resolving the systematics of Lyophyllaceae (Bellanger et al., 2015). Although the mycological research community has increasingly employed the RPB2 marker in studies on Lyophylloid fungi, this locus has not yet been utilized in environmental amplicon sequencing studies. While the mycological research community continues to gather molecular data from multiple molecular markers on a global scale, current efforts have predominantly focused on characterizing fungal species richness using the ITS region, as agreed upon by the fungal research community (Schoch et al., 2012; Herr et al., 2015; Hibbett et al., 2016).
Considering the economic value and ecological significance of Lyophyllum, it is imperative to understand its global diversity and distribution comprehensively. We utilized a widely recognized environmental barcode marker, the internal transcribed spacer region (ITS), which has proven to be a valuable tool in fungal research (Tedersoo et al., 2014). Our analysis encompassed an extensive examination of publicly available ITS sequences sourced from NCBI-GenBank and fungal amplicon sequencing data acquired from NCBI-SRA. Through these efforts, we aimed to enhance our knowledge of the species richness and global distribution of Lyophyllum. Consequently, this study was undertaken with the following objectives: 1) Explore the potential species diversity of Lyophyllum on the global scale; 2) Provide a theoretical basis for taxonomy research; 3) Investigate the patterns and regularity of global distribution within Lyophyllum.
We created the dataset containing all publicly available Lyophyllum ITS sequences, queried NCBI-GenBank for “Lyophyllum” with “ITS”, and downloaded all nucleotide sequences. Then, the correct classification name is verified by homology 98% similarity comparison in NCBI-BLAST (https://blast.ncbi.nlm.nih.gov) (Tedersoo et al., 2014). We removed sequences deposited in NCBI under an incorrect genus-level designation (Schoch et al., 2020). Finally, all sequences with the identifier “Lyophyllum” in the UNITE database are also included in the dataset (Abarenkov et al., 2010).
All public fungal amplicon sequencing data datasets of the ITS region were downloaded from the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra, accessed on 22 March 2023). Keywords of “fungi”, “fungal”, “fungus”, and “ITS” were used to search for the ITS amplicon data. Amplicon data generated by various sequencing platforms, such as 454 pyrosequencing, Illumina, ABI SOLiD, BGISEQ, Ion Torrent, and PacBio SMRT, were all included in our analysis. This provided us with 269 412 samples, and they were downloaded using Amazon Simple Storage Service (Amazon S3) by each sample’s Run accessions number (https://www.ncbi.nlm.nih.gov/sra/docs/sra-aws-download/). Pair end reads were merged using vsearch v2.8.1 (https://github.com/torognes/vsearch) (Rognes et al., 2016). The resulting sequences were quality-checked using fastp software with default parameters to filter out low-quality reads (Chen et al., 2018). Chimera detection was performed using vsearch v2.8.1 and with denovo mode.
Using the most closely related Gerhardtia as the outgroup, the sequence similarity between Gerhardtia and Lyophyllum was identified. Moreover, in order to obtain all Lyophyllum sequences as completely as possible, we used an even lower threshold of 70% similarity and 40% coverage as the local BLAST parameters (Johnson et al., 2008; Davis et al., 2015; Stover and Cavalcanti, 2017). At the same time, this method may also have potential problems, including noise and mismatch or a large amount of complex data that will impact the accuracy of downstream analysis, etc (Gold et al., 2023). The BLAST-2.2.29 software package provided by the National Center for Biotechnology Information (NCBI) of the United States was installed on the workstations in our institution to construct a local database (Cock et al., 2015). Both the ITS1 and ITS2 regions were blasted and extracted, respectively. The geographic location of each sequence was also extracted from studies containing Lyophyllum sequences. The extracted sequences were then aligned and trimmed to a proper length. Then the resulting sequences were clustered into OTUs with 98% similarity using USEARCH 11 (Edgar, 2010; Tedersoo et al., 2014).
The sequences of ITS1 and ITS2 were assembled and aligned with MAFFT using the -L-INS-i option (Katoh and Standley, 2013). Before performing phylogenetic analyses, start and end ambiguous sites were removed, and gaps were manually adjusted to optimize the alignment by BioEdit v7.1.3 (Hall, 1999). The data set was subjected to a maximum likelihood (ML) search using IQ-TREE (Bolyen et al., 2019), with a parametric bootstrap using 1000 replicates under the SYM+G4 model. The species of Gerhardtia, sister to Lyophyllum, were used to root this phylogenetic tree and to exclude the sequences that did not belong to Lyophyllum. We define a well-supported branch as having 70% or higher bootstrap support. The location associated with each species was mapped onto each phylogenetic tree using the ‘ggtree’ package (Yu et al., 2017) and the ‘UpSetR’ package (Conway et al., 2017) within the R 4.2.3 environment (R Core Team, 2021).
ITS1 region showed low discriminatory power for identifying Lyophyllum, and the ITS2 region with higher variability was used (Borman and Johnson, 2021). At a 98% sequence similarity level, 88 Lyophyllum OTUs were recovered, and the known species discovered so far are all in Clade A,of which Clade a (Lyophyllum decastes complex s.l.) contained 30 Lyophyllum OTUs, of which the species of L. fumosum, L. shimeji, L. decastes, L. subdecastes, Tephrocybe impexa, L. paelochroum, L. caerulescens, L. semitale can be annotated. Clade b contained 17 Lyophyllum OTUs, of which the sequence of T. cf. anthracophila can be annotated; Clade c contained 36 Lyophyllum OTUs, of which the species of L. cf. helvella, L. caerulescens, L. cf. pallidum, L. infumatum, L. rhombisporum, L. macrosporum, L. transforme, L. aemiliae, L. cf. aemiliae, L. pulvis-horrei, L. semitale, L. subalphinarum, L. maleolens can be annotated; Clade B contained 5 Lyophyllum OTUs, there are no species of the genus that can be annotated (Figure 1). By constructing a local BLAST using the sequences from published articles (Johnson et al., 2008), 49 Lyophyllum OTUs were discovered. Among them, there are 12 Lyophyllum OTUs in Clade a, accounting for 40% of the total species in Clade a. In Clade b, there are 16 Lyophyllum OTUs, representing 94.12% of the total number of species in Clade b. Clade c contains 14 Lyophyllum OTUs, making up 38.89% of the total number of species in Clade c. Finally, Clade B consists of 5 Lyophyllum OTUs, representing 100% of the total number of species in Clade B.
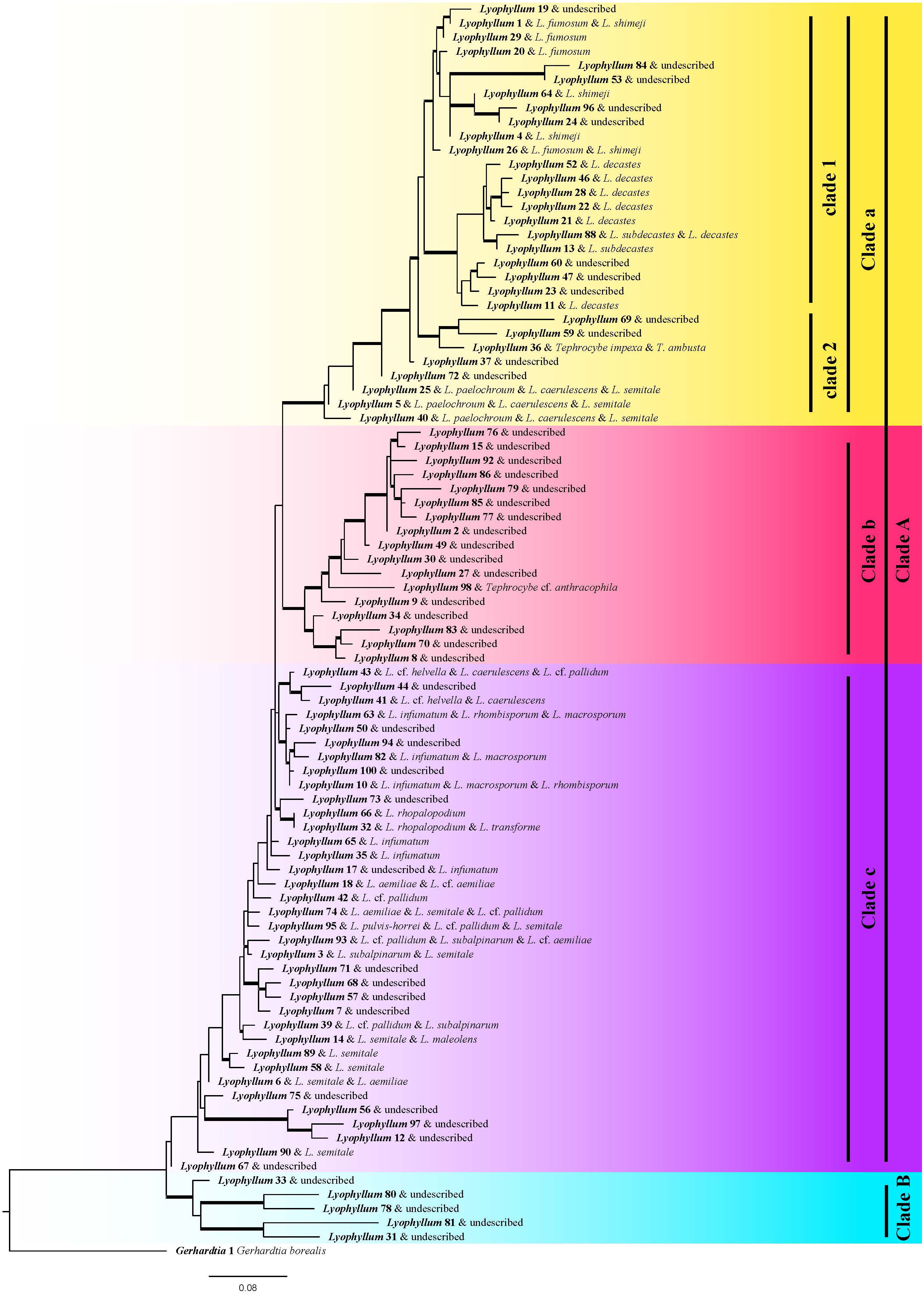
Figure 1 Phylogenetic trees illustrate the relationship of each species. Maximum likelihood phylogeny constructed from the ITS2 region using one exemplar sequence per species. One species of Gerhardtia, sister to all other Lyophyllum species, was used to root this phylogeny. Well-supported branches (greater than 70% bootstrap support) are thickened. The name of the annotation has a homology of over 98%, so there may be one or more names corresponding to OTUs. Different colors represent different groups, where Clade a represents Lyophyllum sect. Difforma, Clade b represents Lyophyllum sect. Tephrophana, Clade c represents Lyophyllum sect. Lyophyllum, Clade B represents previously undiscovered subgenus taxonomic groups.
Europe is the most species-rich geographical region, boasting 48 species. North America and Asia exhibit similar numbers of species, with 29 and 26, respectively. Conversely, the southern hemisphere demonstrates significantly lower species diversity, with six species. Substantial discrepancies in unique species were found across different regions (Supplementary Figure S3). Europe harbors 29 distinctive species, whereas Asia and North America possess 11 and 13, respectively. Oceania, in contrast, has only two exclusive species, Lyophyllum_66 and Lyophyllum_69. Within the northern hemisphere, there is a frequent exchange of Lyophyllum species. Among them, Asia, North America, and Europe share seven species; North America and Europe share five species; Asia and Oceania share one species; North America and Oceania share one species, namely Lyophyllum_27, which was not annotated by any known species. Europe and Oceania share one species, Lyophyllum_5, which was annotated by L. caerulescens, L. paelochroum, and L. semitale. Moreover, two species, namely Lyophyllum_1, and Lyophyllum_3, distributed across all four continents, Lyophyllum_1 was annotated L. shimeji and L. fumosum, Lyophyllum_3 was annotated L. subaloinarum and L. semitale (Supplementary Table S1).
Compared to other biogeographic regions, the prevalence of species sharing is higher among European biogeographic regions. The most notable species sharing was between Southern Europe and Western Europe. No significant differences were observed in the species sharing among other biogeographic regions within Europe (Figure 2A). In Asia, seven species exist between South Asia and East Asia; Southeast Asia, South Asia, and East Asia share two species, Lyophyllum_2 and Lyophyllum_77, which were not annotated by any known species (Figure 2B). North America, Canada, and the United States shared relatively more species, with eight shared species (Figure 2C). Only one species (Lyophyllum_27, which was not annotated by any known species) was shared in Oceania between Australia, New Zealand, and Papua New Guinea (Figure 2D). Four species were specifically distributed in Southern, Western, Northern, and Eastern Europe. In Asia, 14 regionally specific species were mainly distributed in East Asia. Eleven regionally specific species in North America are primarily distributed in the United States. Papua New Guinea is the primary location in the southern hemisphere, with four regionally specific species.
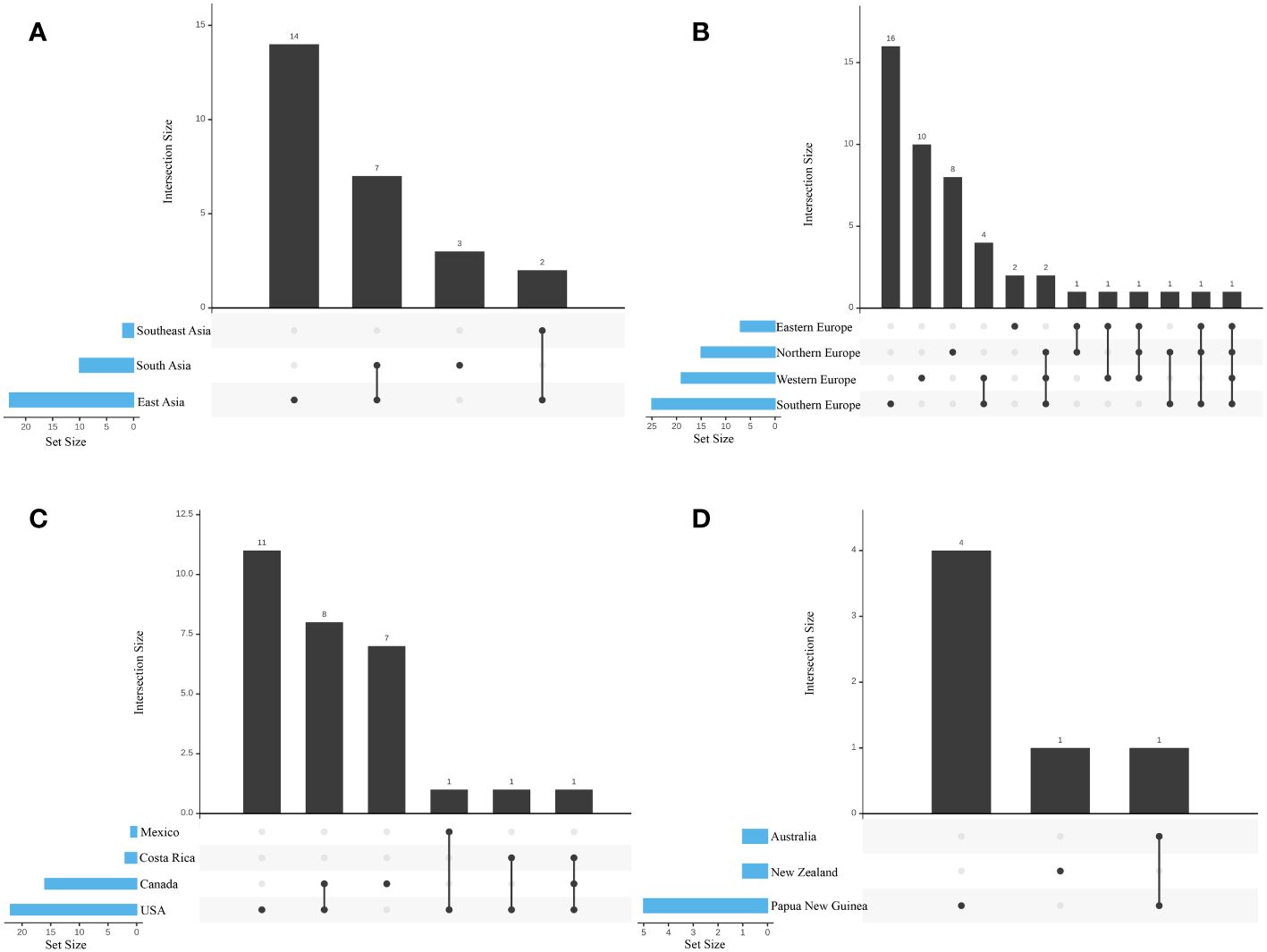
Figure 2 Species richness in each biogeographic region. Bar illustrated the total species richness within each biogeographic region and the combined richness across multiple regions. (A) Different regions of Europe; (B) Different regions of Asia; (C) Different countries of North America; (D) Different countries of Oceania. The species diversity represented by each vertical bar corresponds to the number of species exclusively found in the biogeographic regions matching the colored circles below. The species diversity depicted by the horizontal insets corresponds to the overall species number within each biogeographic region.
Eleven out of the 88 annotated species did not have geographic locations (Figure 3), and most of these Lyophyllum species were found to be distributed in the Northern temperate regions. The most abundant species were Lyophyllum_10, Lyophyllum_2, Lyophyllum_1, and Lyophyllum_11, each comprising over 50 geographic records, with 95, 94, 61, and 54 records, respectively. Furthermore, Lyophyllum species were also observed in tropical regions, such as South China and India in Asia, Costa Rica in North America, and Papua New Guinea in Oceania. Among these tropical regions, Papua New Guinea exhibited the highest number of species, with six identified species. Remarkably, our database even discovered Lyophyllum species in the Arctic region. These species include Lyophyllum_26 from Norway, Lyophyllum_27 from the United States, and Lyophyllum_43 from Finland. Lyophyllum_4 and Lyophyllum_26 fall within clade 1 in the phylogenetic analyses, representing the Lyophyllum decastes complex. Moreover, Lyophyllum_27, as a widely distributed species, spans all climatic zones outside of Antarctica. Such occurrences are relatively rare and have not been reported within the known taxonomic studies of the Lyophyllum (Figure 3). At the base of the phylogenetic tree, Lyophyllum_33, Lyophyllum_56, Lyophyllum_67, Lyophyllum_78, Lyophyllum_80, and Lyophyllum_90 were only found in Europe (Figure 4).
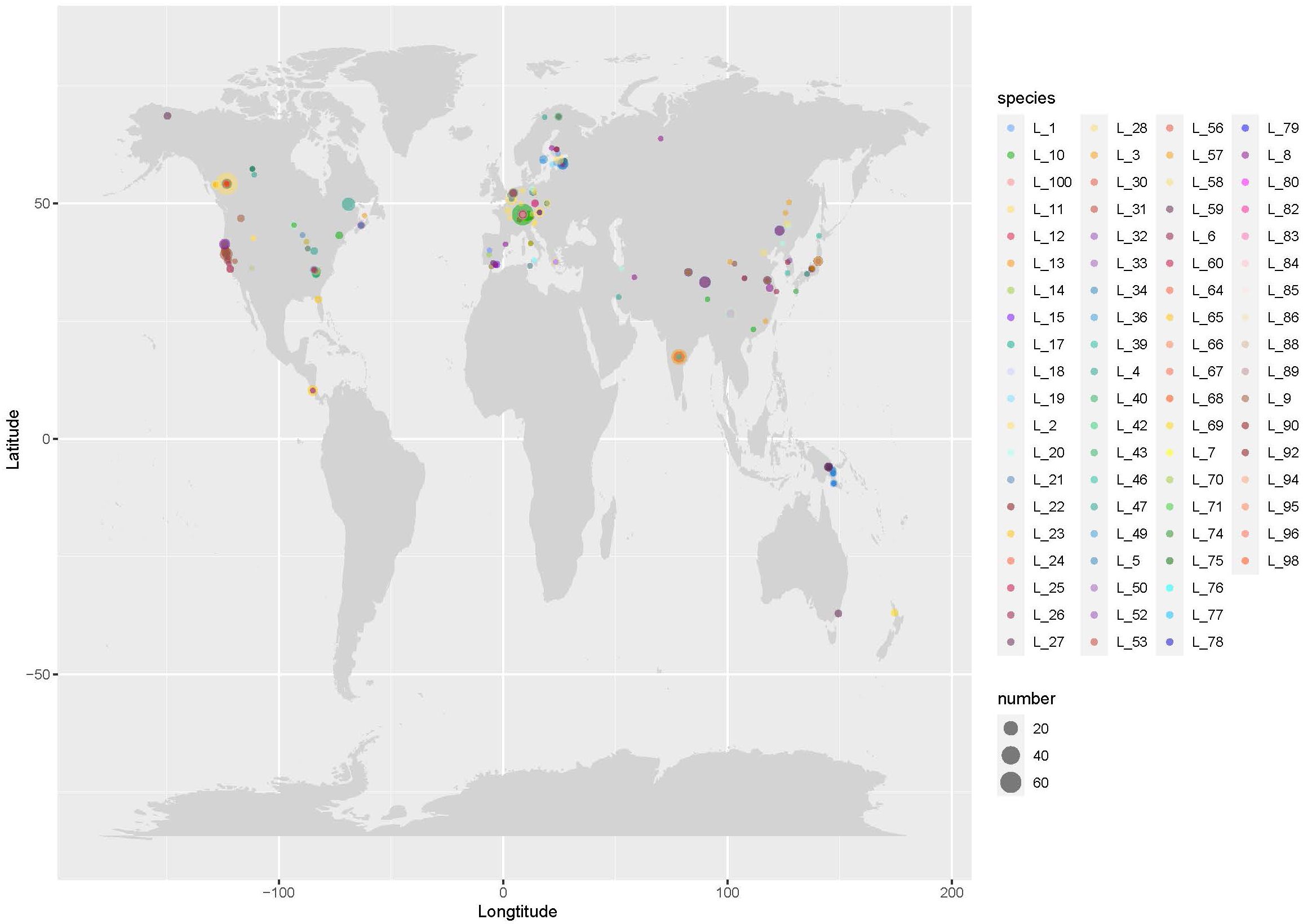
Figure 3 The worldwide distribution of the Lyophyllum species. Records of the species of Lyophyllum, see Supplementary Table S1. The size of the circles is scaled by the number of sequences grouped into a particular species from a particular region. ‘L’ stands for ‘Lyophyllum’ as an abbreviation.
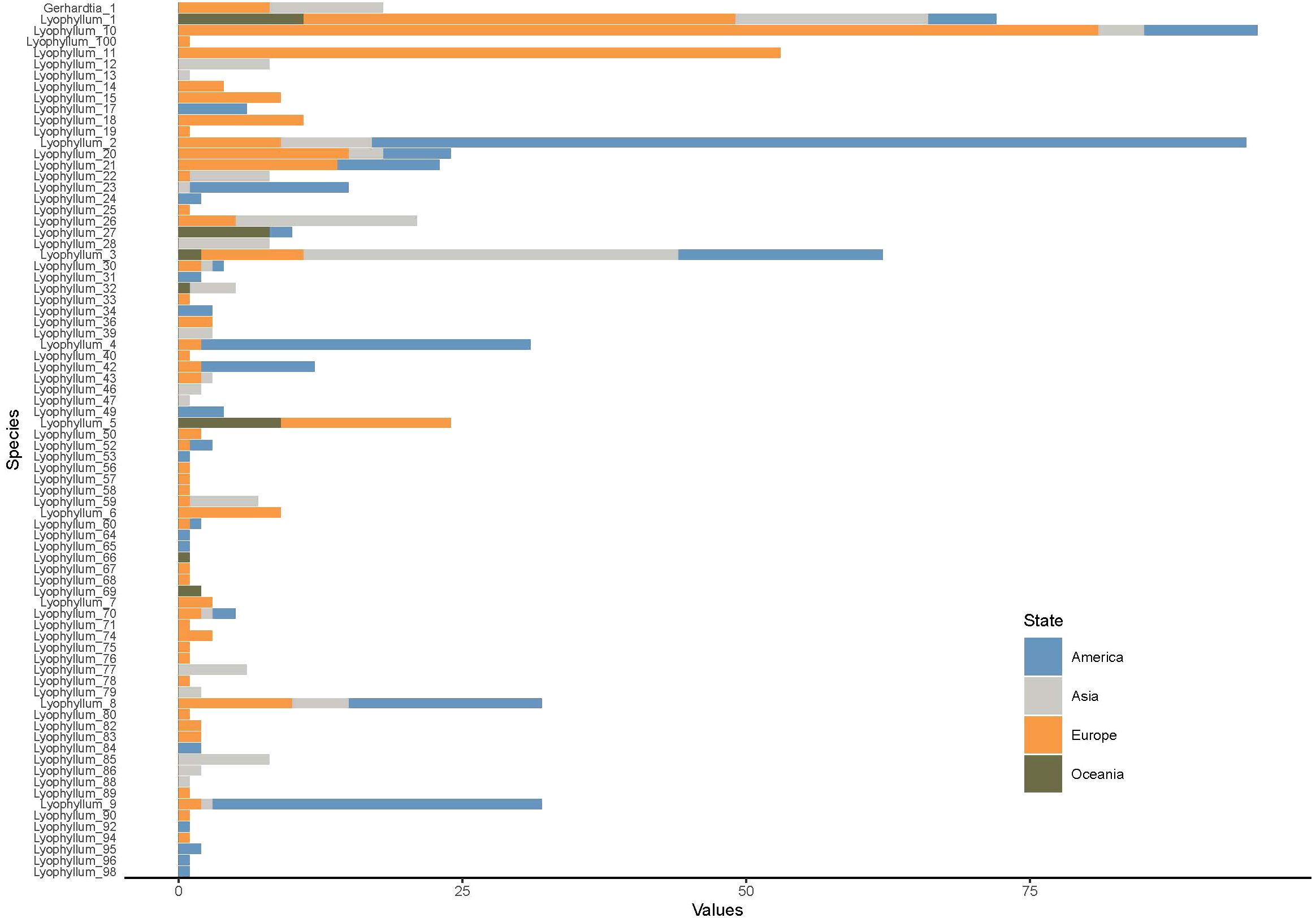
Figure 4 Stacking bar chart of potential species in the Lyophyllum and distribution linked to each sequence. The color of the corresponding columnar corresponds to the biogeographic region. The size of the columnar is scaled by the number of sequences that were grouped into a particular potential species from a particular region.
Previously, morphological, isoenzyme testing, and phylogenetic analyses were primarily employed to assess diversity and global abundance in Lyophyllum (van de Peppel et al., 2022; Wei et al., 2023). In this study, we utilized the ITS marker region of amplicon sequencing studies to explore global potential species richness and distribution (Supplementary Figure S2). The ITS region is widely applied in fungal molecular ecology and is the most widely used in public databases (Baldrian et al., 2022). Moreover, there is a growing recognition of the importance of using ITS for identifying and building comprehensive databases for barcoding macrofungal collections (Li and Wang, 2019). Therefore, the advantage of this approach lies in its ability to capitalize on an increasing number of publicly available datasets, particularly those utilizing the ITS region, derived from fungal ribosomal DNA barcoding and metabolic code encoding. Despite potential errors in fungal community assessments, ITS amplicon sequencing remains widely utilized in current research (Yang et al., 2018). Consequently, future studies should strive to promote the use of diverse targets in sequencing-based fungal ecology research. The increasing number of investigations will resolve fungal diversity through high-throughput amplicon sequencing (Taylor et al., 2016; De Filippis et al., 2017; Johnston et al., 2017). Thus, accurate and reliable community sampling is essential for deriving meaningful insights and exploring ecological and biological studies (Koch and Herr, 2021).
Using ITS as a marker for Lyophyllum phylogenetic studies has limitations due to its relative conservatism, which renders it unable to distinguish similar species (Bellanger et al., 2015). Our findings confirm this point, especially when distinguishing L. infumatum, L. macrosporum, L. semitale, and L. aemiliae, etc. (Figure 1). However, an alternative explanation for this observation may not be poor marker resolution but rather the misutilization of names deposited in GenBank (Bidartondo, 2008). Although our results indicate that ITS can provide similar delineation results to morphologically based classifications in Lyophyllum, ITS is overestimated in many other species delimitations (Bachy et al., 2013). Species delineation is best achieved using large datasets with multiple gene markers featuring diverse characteristics–a strategy that has shown success in Lyophyllum. Previous studies emphasized the necessity for additional fungal ecological markers and sequence analyses developed from fungal genomics and transcriptome research (Pérez-Izquierdo et al., 2017; Kauserud, 2023). We must admit the limitations of determined potential species by ITS marker regions, as well as limitations of single-gene phylogenetic analyses in previous studies and questions about the reliability of GenBank data submission information. Thus, we propose that further taxonomic efforts are best achieved through an integrative approach that combines morphology, mitochondrial genomics, and systematic genomic data (Zhang et al., 2020; Li et al., 2021).
OTUs to differentiate potential species boundaries is a challenging problem in fungal ecology, taxonomy, and phylogeny (Mysara et al., 2017). The potential species were similar to the reference sequences for the ITS region in the UNITE database and were generated by clustering homologous sequences with a similarity of 98.0% (Eisfeldt et al., 2020). Similar to the UNITE database and other databases of ribosomal operons, the potential species are very close to the real species, but the assignment is arbitrary (Koch and Herr, 2021). Here, we manually match species names for potential species or OTUs. Although we assign them in Figure 1, these should only be considered accurate when incorporating multiple factors, such as multilocus or genome sequence data, as well as morphology and mating studies (Hibbett et al., 2016).
Mbareche et al. employed two eukaryotic genomic ribosomal DNA regions, ITS1 and ITS2, to describe fungal diversity in aerosol samples using amplicon datasets (Mbareche et al., 2020). They indicated that the barcodes evaluated were not flawless in distinguishing all species, but both barcodes provided a more comprehensive understanding of fungal aerosol populations. In contrast, we aim to ensure the rationality of potential species diversity predictions and the adequacy of using ITS as a marker to characterize the diversity of Lyophyllum. The high-throughput sequencing of PCR amplicons derived from environmental DNA has become an indispensable aspect of fungal ecology, aiding in the comprehensive characterization of fungal communities (Itskovich et al., 2013; Tedersoo et al., 2014; Mikryukov et al., 2023). In most cases, the ITS region serves as a DNA barcode marker with a longstanding history in fungal systematics (Schoch et al., 2012). Despite advancements in sequencing technology and bioinformatics methods, the utilization of single-gene locus DNA markers still presents inherent limitations frequently overlooked, as a single molecular marker has certain constraints in species delineation within the field of taxonomy (Liu et al., 2021; Zhao et al., 2023). Therefore, when employing ITS amplicons, consideration must be given to the fundamental characteristics of the ITS region, as well as the errors and biases introduced during amplification (Koch and Herr, 2021). Another possible scenario that could lead to changes is the grouping of ASVs within OTUs. However, Zhang et al. have found that ASV grouping has a relatively minor impact on studies at higher taxonomic levels (phylum) and can also enhance the explanatory power of environmental factors on microbial communities (Zhong et al., 2022). Ongoing research involves the extensive survey of ITS sequences in public databases, leading to the design of new ITS primers. These primers, compared to existing ones, exhibit improved coverage across different fungal taxa, enabling selective investigation of fungal communities in mycorrhizal, soil, and other environmental samples (Mourgela et al., 2023). This contributes to laying the groundwork for ecological studies on fungal diversity and community structure in the era of large-scale DNA sequencing.
Research on species within the Lyophyllum has predominantly focused on morphology, biological characteristics, and phylogenetic analyses (Ohta, 1994; Bellanger et al., 2015; Wei et al., 2023). However, no global-scale study has been undertaken to comprehensively explore the diversity of Lyophyllum. In this study, we adopted an integrated approach utilizing the global amplicon sequencing database to delve into the worldwide diversity and distribution of Lyophyllum species. This encompasses the data obtained through amplicon sequencing, which will facilitate species identification and contribute to creating a more comprehensive database (Kõljalg et al., 2019). An advantage of this approach lies in the capacity to harness an expanding array of publicly available datasets. However, compared to other genera within the Agaricales, Lyophyllum exhibits a relatively limited number of species (Clericuzio et al., 2013; Wu et al., 2019). Furthermore, current community sequencing efforts may not fully represent all the basidiocarps of Lyophyllum species, implying that with increased sampling efforts, additional species are likely yet to be uncovered (Figure 5) (Hawksworth and Lücking, 2017).

Figure 5 Field photographs of selected species of Lyophyllum decastes complex, showing the indistinguishable morphological characteristics.
Furthermore, the massive number of amplicon sequencing samples means a vast number of geographic locations sampled. Our method is not limited to the study of Lyophyllum, and other fungal potential diversity; in general, research on the diversity of bacteria, fungi, and plant pathogens can also benefit from this approach (Koch and Herr, 2021; Li P. et al., 2023; Mourgela et al., 2023). Given the increasing amount of data generated by amplicon sequencing, although resources can be saved based on existing sequence databases, it should be noted that amplicon sequencing itself is costly and has clear disadvantages compared to field research, such as difficulty in accurately identifying species, and the greater difficulty of distinguishing closely related species (Lam et al., 2020). Therefore, to overcome the limitations of amplicon data in species identification and ecological surveys, future research could combine amplicon data with field investigation as much as possible. Field investigation can provide valuable sample, ecological, and climate information, while high-throughput sequencing technology of amplicon data can help better understand the distribution of potential species. This can fully leverage the advantages of both methods, improve the accuracy and reliability of species identification and ecological surveys, and provide a more effective scientific basis for biodiversity conservation and ecological environment monitoring (Vierheilig et al., 2015; Banchi et al., 2022).
The data derived from the analysis of the ITS2 region showed the presence of 88 species within the Lyophyllum, of which 49 were unannotated to the ‘known species’. These findings complemented the results obtained from previous taxonomic and phylogenetic studies. In previous studies, researchers found more than 40 species of Lyophyllum globally. However, due to significant morphological differences, there are cases of synonymous names, misidentification, etc. (e.g. L. decastes was once thought to be multiple species). In the NCBI database, there are 44 species with uploaded sequence information, including some species that have been separated from Lyophyllum, such as L. connatum (Leucocybe connata), L. favrei (Calocybe favrei), L. leucophaeatum (C. gangraenosa) (Ma et al., 2022). Considering our rigorous filtering scheme for short sequences as well as the exclusion of species containing fewer than three sequences, it is likely that our results still underestimate the species richness of Lyophyllum. Thus, our findings suggested a high potential for species diversity within the Lyophyllum, indicating the existence of numerous undiscovered species. Moreover, as the increase in environmental amplicon sequencing studies, particularly in under-sampled regions, it is conceivable that we will uncover even more incredible species richness. Future environmental amplicon sequencing data, which has expanded geographical sampling, will enable us to unveil the true extent of species richness within the Lyophyllum, surpassing what is reported in this study.
In taxonomy, phylogenetic analyses, and ecological research, collecting a substantial number of sequences and differentiating species boundaries present significant challenges (Baldrian et al., 2022). The species generated in this study were achieved through clustering homologous sequences with a 98% similarity threshold (Tedersoo et al., 2021a). The ITS region, serving as a marker for the fungi, may exhibit deviations due to its conservative nature as a widely used marker, leading to challenges in differentiating certain species (Moncalvo et al., 1993). Our result confirmed this observation, particularly regarding the distinction between species such as L. fumosum and L. shimeji in section Difformia (Clade a), as well as L. infumosum, L. macrosporum, and L. rhombiosporum in the section Lyophyllum (Clade c) (Figure 1). Neither the ITS2 nor the full-length ITS marker region enhanced these species’ resolution. However, it is important to note that confusion may also stem from the incorrect names being uploaded to the public database (Meiklejohn et al., 2019). Accurate species delimitations should only be considered when multiple factors, such as multigene markers, genomic sequence data, and morphological studies, are considered (Borman and Johnson, 2021). However, our primary objective was to acquire insights into diversity and distribution patterns rather than specifically address taxonomic inquiries within Lyophyllum.
The 49 ‘unannotated’ species may be new species that have not been reported, or may be reported the species of through macroscopic or microscopic morphological methods (eg. L. fuscbrunneum, L. impudicum, etc) (Dähncke et al., 2011; Sesli et al., 2015). Insufficient reference sequences can lead to numerous unidentifiable species within a clade, as evidenced by the annotation of only one known species: Lyophyllum_98, among the 17 species in Clade b (Figure 1). Another exciting finding is Clade B, an undiscovered branch (Hofstetter et al., 2014); this clade may be a currently undiscovered new group with distinct morphological characteristics. As a L.decastes complex, Clade a has 12 species most similar to Lyophyllum decastes, namely Lyophyllum (11, 13, 21, 22, 23, 28, 46, 47, 52, 60, 88). This result is broadly consistent with and surpasses those based on the phylogenetic analyses and morphology studies, which found 5 and 8 species, respectively (Moncalvo et al., 1993; Ma et al., 2022, 2023), also proving the reliability of our results.
Through studying the distribution of potential species of Lyophyllum in different temperature zones, it is found that the unique species in the northern temperate zone are the most abundant, with 62 species, while there are 2 unique species in the tropical region, 1 unique species in the southern temperate zone, and no unique species in the northern frigid zone. However, there are 3 species shared between the northern temperate zone and the northern frigid zone, and the number of species shared between the northern temperate zone and the tropical zone is relatively abundant, with 8 species (Supplementary Figure S4), this study demonstrates that the species richness of the Lyophyllum is mainly concentrated in the Northern Hemisphere, particularly in Europe, while the tropical and cold regions display lower species richness. This phenomenon could be attributed to extensive soil sampling efforts in these regions (Figure 2), or it may be a characteristic feature of the global distribution pattern of this genus. Africa and South America have rarely been found to have a distribution of species belonging to the Lyophyllum. This could be due to the barrier posed by the Sahara Desert and the Central American land bridge, hindering the dispersal of Lyophyllum species or fewer soil sampling efforts in these regions. Overall, although variant encoding based on ribosomal genes is still a good first approach to document the eukaryotic clades, there are still some other unknown factors involved, so future studies need to address the problem of primers, and provide an unbiased understanding of species diversity in these regions (Useros et al., 2024). At the base of the phylogenetic tree, Lyophyllum_33, Lyophyllum_56, Lyophyllum_67, Lyophyllum_78, Lyophyllum_80, and Lyophyllum_90 are exclusively found in Europe (Figure 4), indicating that the part of species within this genus may be geographically restricted to Europe. Considering that Europe exhibits the highest diversity of the Lyophyllum in this study, it is reasonable to speculate that the Eurasian continent might have served as the ancestral range for this group (Varga et al., 2019). These findings align with that Eurasia serves as a center of species diversity. There is an unusually high species richness in Spain (W-3.36, N 36.95); this observation confirmed the research conducted by Dähncke et al. on this region (Dähncke et al., 2011). China is vast and resource-rich, with extensive land and diverse climate conditions (Liu et al., 2023). The geographical environment and climate diversity provide favorable conditions for the survival and reproduction of fungi. Among the large fungal resources, Lyophyllum is an important fungal group, but it is difficult to obtain due to its strict growth conditions and the similarity of its pileus color to fallen leaves, which provides them with good camouflage, and this group includes many important edible and medicinal fungi, and the species diversity of Lyophyllum in China is not yet fully understood. Therefore, exploring and protecting China’s Lyophyllum resources is significant for maintaining ecological balance and biodiversity (Li S. et al., 2023). In our research, we found that there may be more than 15 potential species of Lyophyllum in China, and more than half of them may be found in Qinghai. It is reasonable to believe that the Qinghai-Tibet Plateau and the southwestern region may be hotspots for the diversity of Lyophyllum species. Through in-depth research on the potential species diversity of Lyophyllum, we can further understand this important group’s ecological functions, species distribution, and evolutionary history.
The development of next-generation sequencing can enable high-throughput diversity analysis of species groups with potential value or unexplored environments, mainly through targeted sequencing of ITS gene amplicons. Relatively low cost, high efficiency, and simplified methods have led to a growing body of research covering a variety of settings around the world (Ptaszyńska et al., 2021; Tedersoo et al., 2021b). However, integrating a large amount of information and reusing it as a unified resource for new issues is a powerful, energy-saving, and cost-effective method. Over the years, the available datasets have continued to grow, and novel databases and tools have been introduced, making querying global sequencing repositories easier (Delavaux et al., 2020; Giusti et al., 2021; Delavaux et al., 2022). In this study, we demonstrate that a dataset with more than 10 TB amplicon sequencing data allows us to easily explore the global potential species diversity of Lyophyllum. Therefore, future work should concentrate on targeted field investigations to unearth additional members within this group, uncover their diverse and valuable applications, and thoroughly investigate the ecological factors contributing to their high diversity. Furthermore, increased automation of these methods will enhance our understanding of important biological communities, although these methods’ isolation and classification limitations pose challenges.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
SW: Formal analysis, Investigation, Software, Validation, Visualization, Writing – original draft. BQ: Data curation, Formal analysis, Methodology, Resources, Validation, Writing – original draft. XZ: Investigation, Software, Validation, Visualization, Writing – review & editing. ZP: Data curation, Formal analysis, Resources, Writing – review & editing. YL: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing. QW: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Key R&D Program of China (2021YFD1600401), Jilin Provincial Science and Technology Development Project (20220402051GH), the Science and Technology Development Project of Jilin Province (20190201197JC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1328569/full#supplementary-material
Supplementary Figure 1 | Schematic delineating the protocols and methodologies for retrieving sequences from public repositories.
Supplementary Figure 2 | Distribution of the top ten potential species of Lyophyllum worldwide in the amplicon database.
Supplementary Figure 3 | Venn diagram of shared and unique species between four Continents.
Supplementary Figure 4 | Venn diagram of shared and unique species between four different temperature zones.
Abarenkov K., Nilsson R. H., Larsson K. H., Alexander I. J., Eberhardt U., Erland S., et al. (2010). The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Bachy C., Dolan J. R., López-García P., Deschamps P., Moreira D. (2013). Accuracy of protist diversity assessments: morphology compared with cloning and direct pyrosequencing of 18S rRNA genes and ITS regions using the conspicuous tintinnid ciliates as a case study. ISME J. 7, 244–255. doi: 10.1038/ismej.2012.106
Baldrian P., Větrovský T., Lepinay C., Kohout P. (2022). High-throughput sequencing view on the magnitude of global fungal diversity. Fungal Divers. 114, 539–547. doi: 10.1007/s13225-021-00472-y
Banchi E., Manna V., Fonti V., Fabbro C., Celussi M. (2022). Improving environmental monitoring of Vibrionaceae in coastal ecosystems through 16S rRNA gene amplicon sequencing. Environ. Sci. pollut. R. 29, 67466–67482. doi: 10.1007/s11356-022-22752-z
Barros L., Cruz T., Baptista P., Estevinho L. M., Ferreira I. C. (2008). Wild and commercial mushrooms as source of nutrients and nutraceuticals. Food Chem. Toxicol. 46, 2742–2747. doi: 10.1016/j.fct.2008.04.030
Bellanger J. M., Moreau P. A., Corriol G., Bidaud A., Chalange R., Dudova Z., et al. (2015). Plunging hands into the mushroom jar: a phylogenetic framework for Lyophyllaceae (Agaricales, Basidiomycota). Genetica 143, 169–194. doi: 10.1007/s10709-015-9823-8
Bidartondo M. I. (2008). Preserving accuracy in genBank. Science 319, 1616. doi: 10.1126/science.319.5870.1616a
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Borman A. M., Johnson E. M. (2021). Sequence-based identification and classification of fungi. Trends systematics bacteria fungi 108, 198–216. doi: 10.1079/9781789244984.019
Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Clémençon H., Smith A. H. (1983). New species of Lyophyllum (Agaricales) from North America and a key to the known staining species. Mycotaxon 18, 379–437.
Clericuzio M., Negri R., Cossi M., Gilardoni G., Gozzini D., Vidari G. (2013). Cadinane sesquiterpenes from the mushroom Lyophyllum transforme. Phytochemistry 93, 192–198. doi: 10.1016/j.phytochem.2013.03.019
Cock P. J., Chilton J. M., Grüning B., Johnson J. E., Soranzo N. (2015). NCBI BLAST+ integrated into Galaxy. Gigascience 4, 39. doi: 10.1186/s13742-015-0080-7
Conway J. R., Lex A., Gehlenborg N. (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940. doi: 10.1093/bioinformatics/btx364
Cooper J. (2014). New species and combinations of some New Zealand agarics belonging to Clitopilus, Lyophyllum, Gerhardtia, Clitocybe, Hydnangium, Mycena, Rhodocollybia and Gerronema. Mycosphere 5, 263–288. doi: 10.5943/mycosphere/5/2/2
Dähncke R. M., Contu M., Vizzini A. (2011). Two new species of Lyophyllum sl (Basidiomycota, Agaricomycetes) from La Palma (Canary Islands, Spain). Mycotaxon 115, 65–71. doi: 10.5248/115.65
Davis C., Kota K., Baldhandapani V., Gong W., Abubucker S., Becker E., et al. (2015). mBLAST: keeping up with the sequencing explosion for (meta) genome analysis. J. Data Min. Genomics Proteomics 4, 135. doi: 10.4172/2153–0602.1000135
De Filippis F., Laiola M., Blaiotta G., Ercolini D. (2017). Different amplicon targets for sequencing-based studies of fungal diversity. Appl. Environ. Microb. 83, e00905–e00917. doi: 10.1128/AEM.00905-17
Delavaux C. S., Ramos R. J., Sturmer S. L., Bever J. D. (2022). Environmental identification of arbuscular mycorrhizal fungi using the LSU rDNA gene region: an expanded database and improved pipeline. Mycorrhiza 32, 145–153. doi: 10.1007/s00572-022-01068-3
Delavaux C. S., Sturmer S. L., Wagner M. R., Schütte U., Morton J. B., Bever J. D. (2020). Utility of large subunit for environmental sequencing of arbuscular mycorrhizal fungi: a new reference database and pipeline. New Phytol. 229, 3048–3052. doi: 10.1111/nph.17080
Ding X., Liu Y., Hou Y. (2022). Structure identification and biological activities of a new polysaccharide isolated from Lyophyllum decastes (Fr.) Sing. Pharmacogn. Mag. 18, 112–120. doi: 10.4103/pm.pm_185_21
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eisfeldt J., Mårtensson G., Ameur A., Nilsson D., Lindstrand A. (2020). Discovery of novel sequences in 1,000 Swedish genomes. Mol. Biol. Evol. 37, 18–30. doi: 10.1093/molbev/msz176
Estrada-Martínez E., Guzmán G., Cibrián Tovar D., Ortega Paczka R. (2009). Contribución al conocimiento etnomicológico de los hongos comestibles silvestres de mercados regionales y comunidades de la Sierra Nevada (México). Interciencia 34, 25–33.
Giusti A., Ricci E., Gasperetti L., Galgani M., Polidori L., Verdigi F., et al. (2021). Building of an Internal Transcribed Spacer (ITS) gene dataset to support the Italian Health Service in mushroom identification. Foods 10, 1193. doi: 10.3390/foods10061193
Gold Z., Shelton A. O., Casendino H. R., Duprey J., Gallego R., Van Cise A., et al. (2023). Signal and noise in metabarcoding data. PloS One 18, e0285674. doi: 10.1371/journal.pone.0285674
Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Ser. 41, 95–98.
Hawksworth D. L., Lücking R. (2017). Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 5, 1–17. doi: 10.1128/microbiolspec.FUNK-0052-2016
He M. Q., Zhao R. L., Hyde K. D., Begerow D., Kemler M., Yurkov A., et al. (2019). Notes, outline and divergence times of Basidiomycota. Fungal Divers. 99, 105–367. doi: 10.1007/s13225-019-00435-4
Herr J. R., Öpik M., Hibbett D. S. (2015). Towards the unification of sequence-based classification and sequence-based identification of host-associated microorganisms. New Phytol. 205, 27–31. doi: 10.1111/nph.13180
Hibbett D., Abarenkov K., Kõljalg U., Öpik M., Chai B., Cole J., et al. (2016). Sequence-based classification and identification of Fungi. Mycologia 108, 1049–1068. doi: 10.3852/16–130
Hofstetter V., Clémençon H., Vilgalys R., Moncalvo J. M. (2002). Phylogenetic analyses of the Lyophylleae (Agaricales, Basidiomycota) based on nuclear and mitochondrial rDNA sequences. Mycol. Res. 106, 1043–1059. doi: 10.1017/S095375620200641X
Hofstetter V., Redhead S. A., Kauff F., Moncalvo J. M., Matheny P. B., Vilgalys R. (2014). Taxonomic revision and examination of ecological transitions of the Lyophyllaceae (Basidiomycota, Agaricales) based on a multigene phylogeny. Cryptogamie Mycol. 35, 399–425. doi: 10.7872/crym.v35.iss4.2014.399
Itskovich V., Kaluzhnaya O., Ostrovsky I., McCormack G. (2013). The number of endemic species of freshwater sponges (Malawispongiidae; Spongillina; Porifera) from Lake Kinneret is overestimated. J. Zool. Syst. Evol. Res. 51, 252–257. doi: 10.1111/jzs.12022
Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Johnston P. R., Park D., Smissen R. D. (2017). Comparing diversity of fungi from living leaves using culturing and high-throughput environmental sequencing. Mycologia 109, 643–654. doi: 10.1080/00275514.2017.1384712
Joshi M., Bhargava P., Bhatt M., Kadri S., Shri M., Joshi C. G. (2021). “Lyophyllaceae,” in Mushrooms of gujarat (Springer, Singapore), 31–40. doi: 10.1007/978-981-16-4999-8
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kauserud H. (2023). ITS alchemy: on the use of ITS as a DNA marker in fungal ecology. Fungal Ecol. 65, 101274. doi: 10.1016/j.funeco.2023.101274
Kirk P. M., Cannon P. F., Minter D. W., Stalpers J. A. (2008). Dictionary of the fungi. 10th (Wallingford: CABI).
Kobayashi Y., Shibata T. F., Hirakawa H., Nishiyama T., Yamada A., Hasebe M., et al. (2023). The genome of Lyophyllum shimeji provides insight into the initial evolution of ectomycorrhizal fungal genomes. DNA Res. 30, dsac053. doi: 10.1093/dnares/dsac053
Koch R. A., Herr J. R. (2021). Global distribution and richness of armillaria and related species inferred from public databases and amplicon sequencing datasets. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.733159
Kõljalg U., Abarenkov K., Nilsson R. H., Larsson K. H., Taylor A. F. (2019). The UNITE database for molecular identification and for communicating fungal species. Biodiversity Inf. Sci. Standards 5, 791–797. doi: 10.3897/biss.3.37402
Kühner R. (1938). Utilisation du carmin acétique dans la classification des agarics leucosporés. Publications la Société Linnéenne Lyon 7, 204–211.
Kusuda M., Ueda M., Nakazawa M., Miyatake K., Konishi Y., Shirasaka N., et al. (2004). Characterization of extracellular glucoamylase from the ectomycorrhizal mushroom Lyophyllum shimeji. Mycoscience 45, 383–389. doi: 10.1007/S10267-004-0196-6
Lam T. Y., Mei R., Wu Z., Lee P. K., Liu W. T., Lee P. H. (2020). Superior resolution characterisation of microbial diversity in anaerobic digesters using full-length 16S rRNA gene amplicon sequencing. Water Res. 178, 115815. doi: 10.1016/j.watres.2020.115815
Larsson E., Sundberg H. (2011). Lyophyllum shimeji, a species associated with lichen pine forest in northern Fennoscandia. Mycoscience 52, 289–295. doi: 10.1007/S10267-010-0104-1
Li J. J., Wu S. Y., Yu X. D., Zhang S. B., Cao D. X. (2017). Three new species of Calocybe (Agaricales, Basidiomycota) from northeastern China are supported by morphological and molecular data. Mycologia 109, 55–63. doi: 10.1080/00275514.2017.1286570
Li P., Tedersoo L., Crowther T. W., Wang B., Shi Y., Kuang L., et al. (2023). Global diversity and biogeography of potential phytopathogenic fungi in a changing world. Nat. Commun. 14, 6482. doi: 10.1038/s41467-023-42142-4
Li Q., Wang Q., Jin X., Chen Z., Xiong C., Li P., et al. (2019). Characterization and comparison of the mitochondrial genomes from two Lyophyllum fungal species and insights into phylogeny of Agaricomycetes. Int. J. Biol. Macromol. 121, 364–372. doi: 10.1016/j.ijbiomac.2018.10.037
Li S., Tang S., He J., Zhou D. (2023). Two new edible lyophyllum species from tibetan areas, China. Diversity 15, 1027. doi: 10.3390/d15091027
Li Y., Steenwyk J. L., Chang Y., Wang Y., James T. Y., Stajich J. E., et al. (2021). A genome-scale phylogeny of the kingdom Fungi. Curr. Biol. 31, 1653–1665. doi: 10.1016/j.cub.2021.01.074
Li Y., Wang Q. (2019). The development of the Fungal Resources Conservation System One District, One Herbarium and Five Banks and its application on Qinghai-Tibet Plateau. Agric. Sci. Euro-North-East 20, 29–35. doi: 10.30766/2072-9081.2019.20.1.29-35
Liu M., Braun U., Takamatsu S., Hambleton S., Shoukouhi P., Bisson K. R., et al. (2021). Taxonomic revision of Blumeria based on multi-gene DNA sequences, host preferences and morphology. Mycoscience 62, 143–165. doi: 10.47371/mycosci.2020.12.003
Liu S., Dong Y., Liu H., Wang F., Yu L. (2023). Review of valuation of forest ecosystem services and realization approaches in China. Land 12, 1102. doi: 10.3390/land12051102
Ma Y., Liu P., Zhao Z., Chen W., Zhao Y. (2022). Lyophyllum pallidofumosum sp. nov.(Lyophyllaceae, Agaricales), from southwestern China. Phytotaxa 576, 173–183. doi: 10.11646/phytotaxa.576.2.3
Ma Y., Liu P., Zhao Z., Chen W., Zhao Y. (2023). Lyophyllum deqinense (Lyophyllaceae, Agaricales), a new species from southwestern China. Phytotaxa 598, 219–228. doi: 10.11646/phytotaxa.598.3.3
Maeta K., Koike A., Mukaiyama H., Ochi T., Terashita T., Kitamoto Y., et al. (2008). Preparation and crossing of mating-capable monokaryons via protoplasting of the dikaryotic mycelia of a mycorrhizal mushroom, Lyophyllum shimeji. J. Wood Sci. 54, 337–340. doi: 10.1007/s10086-008-0949-5
Mbareche H., Veillette M., Bilodeau G., Duchaine C. (2020). Comparison of the performance of ITS1 and ITS2 as barcodes in amplicon-based sequencing of bioaerosols. PeerJ 8, e8523. doi: 10.7717/peerj.8523
Meiklejohn K. A., Damaso N., Robertson J. M. (2019). Assessment of BOLD and GenBank - Their accuracy and reliability for the identification of biological materials. PloS One 14, e0217084. doi: 10.1371/journal.pone.0217084
Mikryukov V., Dulya O., Zizka A., Bahram M., Hagh-Doust N., Anslan S., et al. (2023). Connecting the multiple dimensions of global soil fungal diversity. Sci. Adv. 9, eadj8016. doi: 10.1126/sciadv.adj8016
Miura T., Kubo M., Itoh Y., Iwamoto N., Kato M., Park S. R., et al. (2002). Antidiabetic activity of Lyophyllum decastes in genetically type 2 diabetic mice. Biol. Pharm. Bull. 25, 1234–1237. doi: 10.1248/bpb.25.1234
Moncalvo J. M., Rehner S. A., Vilgalys R. (1993). Systematics of Lyophyllum section Difformia based on evidence from culture studies and ribosomal DNA sequences. Mycologia 85, 788–794. doi: 10.1080/00275514.1993.12026333
Mourgela R. N., Kioukis A., Pourjam M., Lagkouvardos I. (2023). Large-scale integration of amplicon data reveals massive diversity within saprospirales, mostly originating from saline environments. Microorganisms 11, 1767. doi: 10.3390/microorganisms11071767
Mysara M., Vandamme P., Props R., Kerckhof F. M., Leys N., Boon N., et al. (2017). Reconciliation between operational taxonomic units and species boundaries. FEMS Microbiol. Ecol. 93, fix029. doi: 10.1093/femsec/fix029
Ohta A. (1994). Some cultural characteristics of mycelia of a mycorrhizal fungus, Lyophyllum shimeji. Mycoscience 35, 83–87. doi: 10.1007/BF02268533
Pérez-Izquierdo L., Morin E., Maurice J. P., Martin F., Rincón A., Buée M. (2017). A new promising phylogenetic marker to study the diversity of fungal communities: The Glycoside Hydrolase 63 gene. Mol. Ecol. Resour. 17, e1–e11. doi: 10.1111/1755-0998.12678
Pokhrel C. P., Ohga S. (2007a). Submerged culture conditions for mycelial yield and polysaccharides production by Lyophyllum decastes. Food Chem. 105, 641–646. doi: 10.1016/j.foodchem.2007.04.033
Pokhrel C. P., Ohga S. (2007b). Synthetic cultivation of Lyophyllum decastes on a combination of cattle livestock compost and corn cobs. Mushroom Sci. Biotechnol. 15, 123–128. doi: 10.1016/j.foodchem.2007.04.033
Ptaszyńska A. A., Latoch P., Hurd P. J., Polaszek A., Michalska-Madej J., Grochowalski Ł., et al. (2021). Amplicon sequencing of variable 16S rRNA from bacteria and ITS2 regions from fungi and plants, reveals honeybee susceptibility to diseases results from their forage availability under anthropogenic landscapes. Pathogens 10, 381. doi: 10.3390/pathogens10030381
R Core Team (2021) R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available online at: https://www.R-project.org/ (Accessed 26th July 2023).
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Schoch C. L., Ciufo S., Domrachev M., Hotton C. L., Kannan S., Khovanskaya R., et al. (2020). NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database 2020, baaa062. doi: 10.1093/database/baaa062
Schoch C. L., Seifert K. A., Huhndorf S., Robert V., Spouge J. L., Levesque C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. P. Natl. Acad. Sci. U.S.A. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Sesli E., Vizzini A., Contu M. (2015). Lyophyllum turcicum (Agaricomycetes: Lyophyllaceae), a new species from Turkey. Turk. J. Bot. 39, 512–519. doi: 10.3906/bot-1407-16
Stover N. A., Cavalcanti A. R. (2017). Using NCBI BLAST. Curr. Protoc. Essential Lab. Techniques 14, 11.1.1–1.1.34. doi: 10.1002/cpet.8
Takaki K., Yoshida K., Saito T., Kusaka T., Yamaguchi R., Takahashi K., et al. (2014). Effect of electrical stimulation on fruit body formation in cultivating mushrooms. Microorganisms 2, 58–72. doi: 10.3390/microorganisms2010058
Taylor D. L., Walters W. A., Lennon N. J., Bochicchio J., Krohn A., Caporaso J. G., et al. (2016). Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for Illumina amplicon sequencing. Appl. Environ.Microb. 82, 7217–7226. doi: 10.1128/AEM.02576-16
Tedersoo L., Albertsen M., Anslan S., Callahan B. (2021a). Perspectives and benefits of high-throughput long-read sequencing in microbial ecology. Appl. Environ.Microb. 87, e00626–e00621. doi: 10.1128/AEM.00626-21
Tedersoo L., Bahram M., Põlme S., Kõljalg U., Yorou N. S., Wijesundera R., et al. (2014). Global diversity and geography of soil fungi. Science 346, 1256688. doi: 10.1126/science.1256688
Tedersoo L., May T. W., Smith M. E. (2010). Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 20, 217–263. doi: 10.1007/s00572-009-0274-x
Tedersoo L., Mikryukov V., Anslan S., Bahram M., Khalid A. N., Corrales A., et al. (2021b). The Global Soil Mycobiome consortium dataset for boosting fungal diversity research. Fungal Divers. 111, 573–588. doi: 10.1007/s13225–021-00493–7
Tel G., Ozturk M., Duru M. E., Turkoglu A. (2015). Antioxidant and anticholinesterase activities of five wild mushroom species with total bioactive contents. Pharm. Biol. 53, 824–830. doi: 10.3109/13880209.2014.943245
Ukawa Y., Ito H., Hisamatsu M. (2000). Antitumor effects of (1→ 3)-β-D-glucan and (1→ 6)-β-D-glucan purified from newly cultivated mushroom, Hatakeshimeji (Lyophyllum decastes Sing.). J. Biosci. Bioeng. 90, 98–104. doi: 10.1016/S1389-1723(00)80041-9
Useros F., García-Cunchillos I., Henry N., Berney C., Lara E. (2024). How good are global DNA-based environmental surveys for detecting all protist diversity? Arcellinida as an example of biased representation. Environ. Microbiol. 26, e16606. doi: 10.1111/1462-2920.16606
van de Peppel L. J., Aime M. C., Læssøe T., Pedersen O. S., Coimbra V. R., Kuyper T. W., et al. (2022). Four new genera and six new species of lyophylloid agarics (Agaricales, Basidiomycota) from three different continents. Mycol. Prog. 21, 85. doi: 10.1007/s11557-022-01836-7
Varga T., Krizsán K., Földi C., Dima B., Sánchez-García M., Sánchez-Ramírez S., et al. (2019). Megaphylogeny resolves global patterns of mushroom evolution. Nat. Ecol. Evol. 3, 668–678. doi: 10.1038/s41559-019-0834-1
Vierheilig J., Savio D., Ley R. E., Mach R. L., Farnleitner A. H., Reischer G. (2015). Potential applications of next generation DNA sequencing of 16S rRNA gene amplicons in microbial water quality monitoring. Water Sci. Technol. 72, 1962–1972. doi: 10.2166/wst.2015.407
Vizzini A., Consiglio G., Setti L., Ercole E. (2015). Calocybella, a new genus for Rugosomyces pudicus (Agaricales, Lyophyllaceae) and emendation of the genus Gerhardtia. IMA fungus 6, 1–11. doi: 10.5598/imafungus.2015.06.01.01
Vizzini A., Contu M. (2010). Lyophyllum rosae-mariae sp. nov.(Basidiomycota, Agaricomycetes) from La Palma (Canary Islands, Spain). Mycosphere 1, 83–86.
Wang T., Han J., Dai H., Sun J., Ren J., Wang W., et al. (2022). Polysaccharides from Lyophyllum decastes reduce obesity by altering gut microbiota and increasing energy expenditure. Carbohyd. Polym. 295, 119862. doi: 10.1016/j.carbpol.2022.119862
Wei S. W., Lu B. Y., Wang Y., Dou W. J., Wang Q., Li Y. (2023). Morphology and phylogeny of lyophylloid mushrooms in China with description of four new species. J. Fungi 9, 77. doi: 10.3390/jof9010077
Wu F., Zhou L. W., Yang Z. L., Bau T., Li T. H., Dai Y. C. (2019). Resource diversity of Chinese macrofungi: edible, medicinal and poisonous species. Fungal Divers. 98, 1–76. doi: 10.1007/s13225-019-00432-7
Xu J., Yu X., Zhang C., Li Y. (2019). Two new species of Calocybe (Lyophyllaceae) from northeast China. Phytotaxa 425, 219–232. doi: 10.11646/phytotaxa.425.4.3
Xu L., Yang W., Qiu T., Gao X., Zhang H., Zhang S., et al. (2023). Complete genome sequences and comparative secretomic analysis for the industrially cultivated edible mushroom Lyophyllum decastes reveals insights on evolution and lignocellulose degradation potential. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1137162
Yang R. H., Su J. H., Shang J. J., Wu Y. Y., Li Y., Bao D. P., et al. (2018). Evaluation of the ribosomal DNA internal transcribed spacer (ITS), specifically ITS1 and ITS2, for the analysis of fungal diversity by deep sequencing. PloS One 13, e0206428. doi: 10.1371/journal.pone.0206428
Ye L., Karunarathna S. C., Li H., Xu J., Hyde K. D., Mortimer P. E. (2019). A survey of Termitomyces (Lyophyllaceae, Agaricales), including a new species, from a subtropical forest in Xishuangbanna, China. Mycobiology 47, 391–400. doi: 10.1080/12298093.2019.1682449
Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T. Y. (2017). ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36. doi: 10.1111/2041-210X.12628
Zhang R., Zhang G., Hu D., Wang H., Ng T. (2010). A novel ribonuclease with antiproliferative activity from fresh fruiting bodies of the edible mushroom Lyophyllum shimeiji. Biochem. Genet. 48, 658–668. doi: 10.1007/s10528-010-9347-y
Zhang Y., Yang G., Fang M., Deng C., Zhang K. Q., Yu Z., et al. (2020). Comparative analyses of mitochondrial genomes provide evolutionary insights into nematode-trapping fungi. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00617
Zhao C., Qu M., Huang R., Karunarathna S. (2023). Multi-gene phylogeny and taxonomy of the wood-rotting fungal genus Phlebia sensu lato (Polyporales, Basidiomycota). J. Fungi 9, 320. doi: 10.3390/jof9030320
Keywords: amplicon sequencing, geographic, global diversity, internal transcribed spacer, Lyophyllum
Citation: Wei S, Qi B, Zhang X, Peng Z, Li Y and Wang Q (2024) Global diversity and biogeography of Lyophyllum inferred from amplicon datasets. Front. Ecol. Evol. 12:1328569. doi: 10.3389/fevo.2024.1328569
Received: 17 November 2023; Accepted: 13 May 2024;
Published: 18 June 2024.
Edited by:
Qiang Li, Chengdu University, ChinaReviewed by:
Shanshan Yang, Wageningen University and Research, NetherlandsCopyright © 2024 Wei, Qi, Zhang, Peng, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhanwu Peng, cGVuZ3poYW53dUBqbGF1LmVkdS5jbg==; Yu Li, eXVsaTk2NkAxMjYuY29t; Qi Wang, cWl3YW5nQGpsYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.