- Fishery College, Zhejiang Ocean University, Zhoushan, Zhejiang, China
Being the phylum with the second largest biodiversity, mollusks are widely distributed in marine, freshwater, and land, conquered almost all habitats on the earth. Throughout geological time, several molluscan lineages independently colonized freshwater and land, evolving independently in new habitats. Resolving the timing of their colonization for novel environments is the basis of understanding the complex evolution of Mollusca. Herein, we obtained an elaborate single–copy orthologs set including 107 genes with a total length of 3,447,675 amino acid sites across 134 mollusks to reconstruct the phylogeny of Mollusca. The phylogenetic analysis recovered the “Aculifera+ Conchifera” topology with well–supported. By the divergence times estimation with fossil calibration, we revealed Cambrian rapid diversification of all molluscan classes. What’s more, our results provided the times of three major independent colonization of novel environments and consistent with previous studies. The freshwater mussels (Bivalvia: Unionida) colonized freshwater about 233 Mya, ancient origin and late diversification make this order to become the main represent of the freshwater bivalves. The orders Stylommatophora and Basommatophora (Gastropoda: Pulmonata) respectively colonized land and freshwater about 201 Mya, the Triassic–Jurassic mass extinction may have provided the opportunities for their colonization. Ampullariidae (Gastropoda) colonized freshwater about 156 Mya, their lungs may have contributed to the adaption of tropical freshwater environment and make them survive. Our results probably offer the most comprehensive time–scale of molluscan evolution, could provide some insights into the habitat transition and evolution of Mollusca.
Introduction
The ocean is the cradle of life, but organisms have been expanding their habitat in the course of evolution. Evolutionary transitions to new habitats are a type of ecological specialization that can lead to speciation (Goulding et al., 2022). Although the evolutionary transitions to new habitats occurred only in a few animal groups, there is no denying that expansion into new habitats does promote biological diversity. Mollusca is a phyla including many species with successful evolutionary transitions, which containing more than one hundred thousand described species is the phylum with the second highest biodiversity (Haszprunar and Wanninger, 2012). Due to the high biodiversity, mollusks have always been an important group in biological studies, especially in their origin and phylogeny. Numerous fossil records, like small shelly fossils (SSF) (Parkhaev, 2008) have confirmed that mollusks originated in the Precambrian oceans. But currently, mollusks have expanded in almost all habitats all over the world, particularly being abundant in marine and freshwater (Ponder and Lindberg, 2008). Most classes of Mollusca remained in the ancestral marine niche, but at present, about 5,000 species in 34 gastropod families and 9 bivalve families invaded and colonized freshwater ecosystems at different times (Cummings and Lydeard, 2019), and become very crucial members of freshwater ecosystems for their contributions to the material cycle (Böhm et al., 2021). Moreover, some gastropods have truly become terrestrial, like a portion of Stylommatophora slugs, this order containing more than 112 families with a total of about 3,000 species, is a main group of Gastropoda (Saadi et al., 2021). In those several groups, their large different morphological characteristics and classifications suggest that they colonized novel environments independently (Yang et al., 2018; Hopper et al., 2021). However, the times of their colonization and the stimulative factors of colonization are quite uncertain. Solving these problems will be the basis for further revealing the complex evolutionary process of mollusks.
Throughout the course of biological evolution, many species colonized novel environment and survived. For example, several ecdysozoan lineages independently colonized land during the evolution, and have been important components of ecosystems (Kenrick et al., 2012), a comprehensive molecular study further revealed the relation to the ecdysozoan land colonization with the evolution of vascular plants and forests (Rota-Stabelli et al., 2013). In mammals, studies revealed that cetaceans are a typical group of secondary aquatic animals whose ancestors returned to the sea from land about 55 Mya (Thewissen et al., 2009). For mollusks, a part of molecular phylogeography studies of major freshwater groups have provided some insights into the evolution and habitats transitions, like the Gondwanan origin of Ampullariidae (Gastropoda) (Hayes et al., 2009) and the expansion of radicine pond snails (Gastropoda: Lymnaeidae) (Aksenova et al., 2018). Except for those entirely freshwater or terrestrial species, the study of intertidal onchidiid slugs (Gastropoda, Pulmonata) revealed the connection of habitat and microhabitat evolutionary transitions, suggesting that the freshwater and marginal environments may be the path of gastropod to land (Goulding et al., 2022). Those studies suggested that the evolutionary transitions for novel environment of mollusks may be more complex.
For the same selection pressure, the similar morphological convergence may occur among different taxa, this could lead the morphological approaches to the study the diversity processes of marine invertebrates may be limited (Vidal-García and Keogh, 2015). Thus, reconstruct a reliable molecular phylogenetic tree of Mollusca is the base of understanding the colonization of freshwater habitats by freshwater mollusks. The large differences in morphological and molecular results have been an important cause of controversy in the phylogenetic relationships of mollusks. In recent years, phylogenomic studies recovered much class–level phylogenetic relationships of mollusks with strong support, and made great progress. However, previous studies mainly focused on the internal phylogeny and the times estimation of mollusk’s major classes, even in some studies that included marine and freshwater species, the timing of colonization of new environments had not been discussed (Erwin et al., 2011; Zapata et al., 2014; Parkhaev, 2017; Kocot et al., 2020). But those studies proved that the phylogenomic is effective to resolve the deep phylogenetic relationships. Although phylogenetic genomics is very effective, however, it needs to rely on complex analytical methods and huge amounts of data. In any group of organisms, identifying the single–copy orthologs is a vital step in phylogenetic studies (Creevey et al., 2011). There are many databases of single–copy orthologs could be used to identify the homologous genes accurately and conveniently. Benchmarking Universal Single–Copy Orthologs (BUSCO) is a database covering the identified single–copy homologous genes of different species (Seppey et al., 2019). It is effective in deep and large–scale phylogenetic studies (Manni et al., 2021a), and it have been widely used in reconstructing the phylogeny (Manni et al., 2021b). Therefore, the phylogenomic approach is still the first choice to address the concerns about the mollusks’ novel environment transitions.
In this study, we collected all available genomic data of mollusks, which could represent all major classes of Mollusca. Our data set included the freshwater species like freshwater mussels (Unionida), apple snails (Ampullariidae) and so on, and terrestrial species from Pulmonata. BUSCO was used to reconstruct the phylogeny and we performed divergence time estimation with rigorous fossil calibration. Our comprehensive timescale of molluscan evolution could reveal the ages of mollusks’ colonization of novel environments. Moreover, we combined our results with historical ecological events to try to find the connection factors. This has significance in understanding the adaptive evolution of mollusks and the terrestrialization of animals.
Materials and methods
Data collection and transcriptomes de novo assembly
We collected all available published genomes and transcriptomes of Mollusca and one outgroup species Lingula anatina (Brachiopoda) from National Center for Biotechnology Information (NCBI) databases (https://www.ncbi.nlm.nih.gov) and MolluscDB (http://mgbase.qnlm.ac/) (before July 30, 2023). The transcriptomes were dismissed if the genome of this species has been already available. For the collected transcriptomes, after removing the reads with sequencing adapters, unknown nucleotides (N ratio > 10%) and low–quality reads (quality scores < 20) by Trimmomatic software (used Paired end mode [PE] and default parameters for others) (Bolger et al., 2014), we used Trinity v2.8.5 (Grabherr et al., 2011) to perform the transcriptome de novo assembly with 10,000 max introns (–seqType fq –max_memory 300G –SS_lib_type RF and used default parameters for others). Then, we used a custom Perl script (could be found in Supplementary File 2) to extract the unigenes whose length more than 200 bp for the next phylogenetic inference. In order to improve the accuracy of phylogeny inference, the transcriptomes whose contig N50 were below 1,000 were discarded. Moreover, we used SOAPdenovo 2.0 software with 147 k–mer (max_rd_len=100, avg_ins=400, reverse_seq=0, asm_flags=3, rank=1 and used default parameters for others) (Luo et al., 2012) to assembly a published raw data and got the draft genome of Laevipilina antarctica (Kocot et al., 2020) as a supplement in our study. In this way, a total of 138 genomes and transcriptomes across 134 mollusks and one outgroup species were retained. The information of genomes and transcriptomes used in this study can be found in Supplementary Table 1. The habitat information was retrieved from the primary literature and WoRMS database (https://www.marinespecies.org/).
Single–copy orthologous amino acid dataset processing
We used the metazoan_odb10 database BUSCO v5.0 (Manni et al., 2021a) to identify the single–copy orthologous genes of all collected genomes and transcriptomes. Due to the large number of samples and different assembly quality, we retained the orthologous genes that existed in at least 90% samples. Through this process, we obtained 107 orthologous genes. After alignment by MAFFT (–auto and used default parameters for others) (Katoh et al., 2009) and concatenation by SCaFos (Roure et al., 2007), these orthologous genes were complemented the deletion of amino acid sites by the gaps. Finally, we obtained a dataset including 107 orthologous genes with a total length of 3,447,675 amino acid sites across 135 species, covering one outgroup species and 134 mollusks (this dataset could be found in Supplementary File 1). This data set could represent all seven major Mollusca classes.
Phylogenetic analysis and molecular clock analysis
After the estimate of best–fit model for the amino acid data set by ProtTest 3 (Darriba et al., 2011), RAxML software v8.2.12 (Stamatakis, 2014) was used for maximum likelihood (ML) analysis, a total of 1,000 bootstrap replicates based on PROTGAMMAIJTT substitution model on all concatenated amino acid were performed. To evaluate the potential effects of substitution models of phylogeny inference, we performed the best–fit model test by ModelFinder (Kalyaanamoorthy et al., 2017) of each amino acid sequence and used IQ–TREE v2.2.0 (Nguyen et al., 2015) with the best–fit partition model (the best–fit model of each amino acid sequence can be founded in Supplementary Table 2) for ML analysis (–bb 100). Moreover, the Bayesian analysis (BA) was also carried out by MrBayes v3.2.7 (Huelsenbeck and Ronquist, 2001) with 200,000 metropolis–coupled Markov chain Monte Carlo (MCMCMC) generations with sampling every 100 generations and then the first 25% of the sampled generations were discarded as burn–in. After the running of 200,000 generations, the average standard deviation of split frequencies was less than 0.01. To estimate the diversification times of major molluscan classes and the mollusk colonization of novel environments, we employed PAML mcmctree v.4.9 (Yang, 2007) to perform molecular clock analysis. The best topology of RAxML analysis was used in mcmctree molecular clock analysis with the correlated–rates evolutionary clock model and HKY85 substitution model (this model is the most similar to the best substitution model PROTGAMMAIJTT and available in PAML). A total of 400,000 generations were performed, sampling every 100 generations, with a burn–in of 50,000 generations. The other parameters followed the PAML instruction. The software Tracer v1.7.1 (Rambaut et al., 2018) was used for the count of ESS value to estimate the convergence.
Divergence time calibration
To get the accurate divergence time of mollusks and the ages of colonization of novel environments, we used a set of calibrations from a recent study (Song et al., 2023), this set was obtained by the comprehensive reviews of all important molluscan fossils, which is well suited for the divergence time estimation of large–scale phylogenetic analysis. By referring to the minimum and maximum ages of the fossils given in the Supplementary Documents in the above study, we calibrated the divergence times of the major molluscan nodes. Moreover, we reviewed studies on the three groups that underwent habitat transitions in this study, in every major colonization event, we adopted one well–accepted fossils in the estimation. Similarly, we used the minimum– and maximum–age bounds of fossils as the prior on times to restrict the habitat transition nodes. The information of fossil constraints used in PAML mcmctree divergence time estimation could be found in Supplementary Table 3.
Results
The phylogeny of Mollusca
A data set including 107 orthologous genes with a total length of 3,447,675 amino acid sites, covering 135 mollusks, three species of Aplacophora, six species of Polyplacophora, one species of Monoplacophora, nine species of Cephalopoda, one species of Scaphopoda, 63 species of Bivalvia and 53 species of Gastropoda, which could represent seven major Mollusca classes, was used for phylogenetic analysis. Two ML trees that were reconstructed by RAxML based on PROTGAMMAIJTT model (Figure 1) and IQ–TREE based on best–fit partition model (Supplementary Figure 1) both revealed the same topology. In our study, the results of phylogenetic analysis using a single model for the concatenated alignmnet are consistent with the partition results of IQ–TREE, which has indicated that the different evolution rate of BUSCO genes has no effect on the topological results of the phylogenetic tree. Since the use of a single model and a partition model had no effect on the results of the phylogenetic analysis. Moreover, the BA tree also shown this same topology (Supplementary Figure 2) with ML trees. As a whole, our phylogenetic analysis recovered the Aculifera+Conchifera phylogenetic relationship. In the clade Conchifera, Bivalvia cluster with Gastropoda+Scaphopoda, and this clade cluster with the clade Monoplacophora+Cephalopoda.
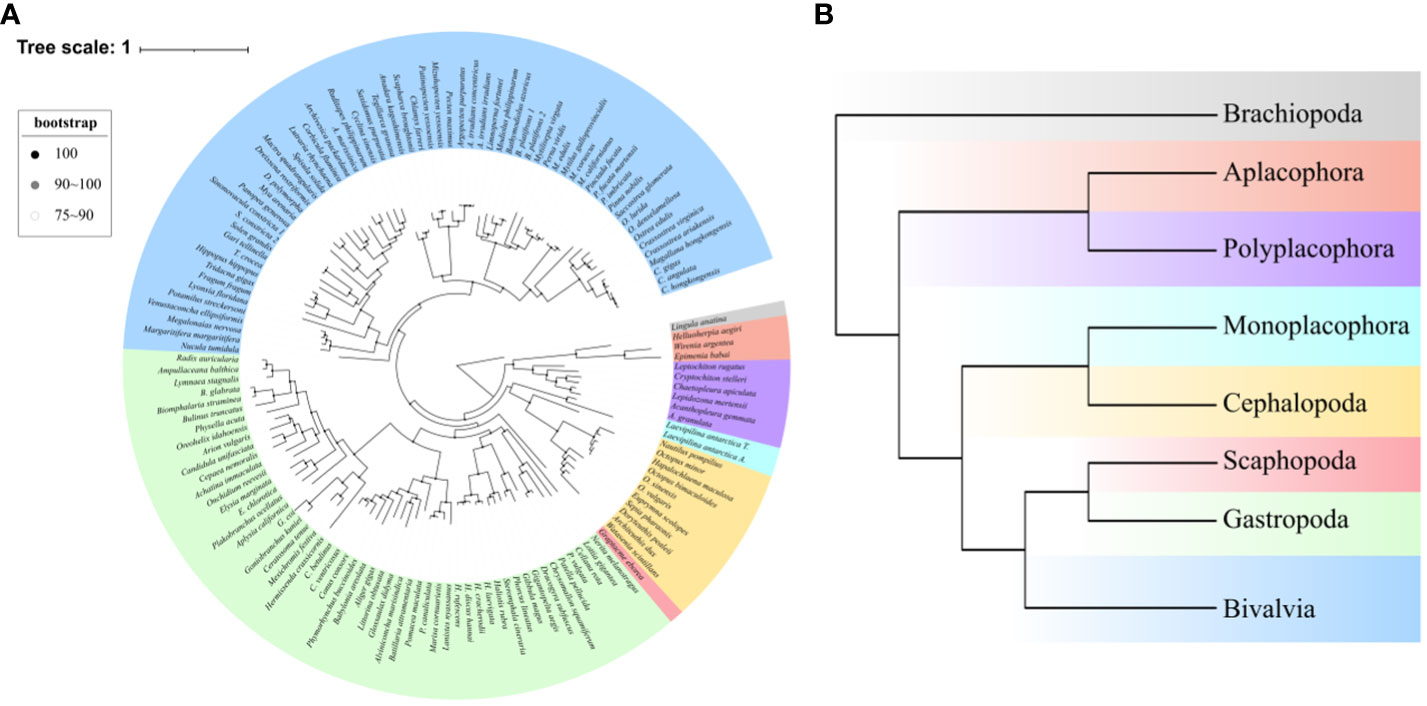
Figure 1 The ML phylognetic tree reconstructed by RAxML based on concatenated 107 orthologous genes with a total length of 3,447,675 amino acid sites. (A) The circle tree of all species in this study, bootstrap value at every node are shown through different colored dots. (B) The phylognetic tree of seven major molluscan classes, different colors represent different classes and correspond to Panel A.
The divergence time estimation and ages of mollusks’ transitions
Through rigorous fossils calibration, we obtained a comprehensive molluscan evolution time–scale (Figure 2). Aculifera and Conchifera split 526 Mya, and the diversification of tow subphyla occurred 475 and 516 Mya. The split times of other classes are shown in Figure 2. What’s more, we have listed three major groups that have undergone habitat transitions separately (see Roman numerals in Figure 2). In the Bivalvia, the freshwater mussels (Unionida) colonized freshwater about 233.21 Mya (95% HPD: 223.81, 239.61). The other species of freshwater bivalves, Corbicula fluminea and Limnoperna fortunei colonized freshwater about 99.84 Mya and 130.50 Mya. In the class Gastropoda, there are two main groups expanding to novel environments, one is the family Ampullariidae, this colonization occurred 156.21 Mya (95% HPD: 145.41, 164.10). The second group contains freshwater and terrestrial snails from Pulmonata, the evolutionary transition of the Pulmonata ancestors occurred 201.62 Mya (95% HPD: 175.03, 230.16).
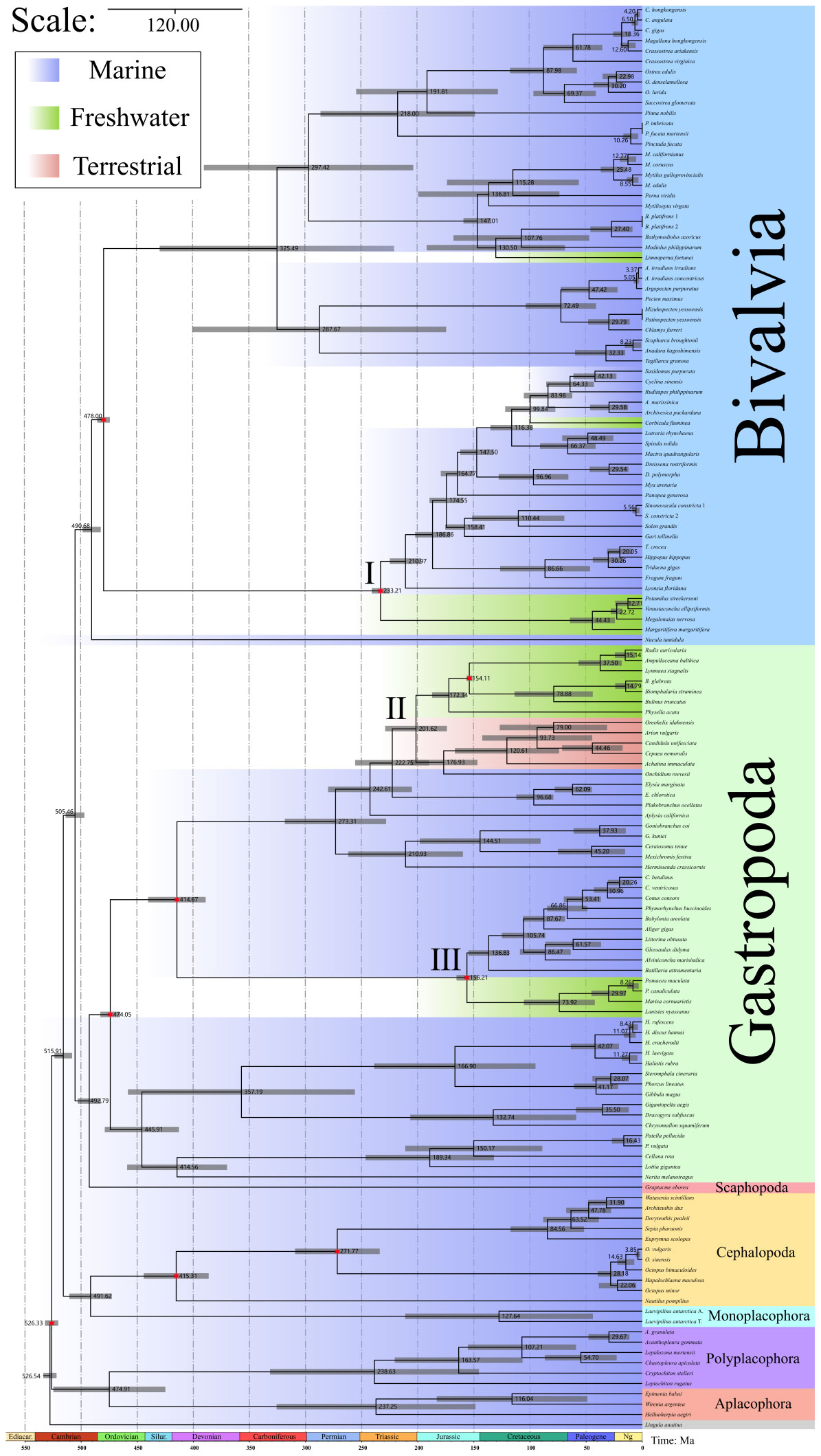
Figure 2 The divergence time estimation of 135 mollusks by mcmctree function in PAML v.4.9. Gray bars show the 95% highest probability density (HPD). The number marked at the node represents the average divergence times (Mya). Red six–pointed stars represent nodes calibrated using fossils. The different genetic data from the same species are indicated in different Roman numerals.
Discussion
When more species were included, our phylogenetic analysis still recovered the same phylogeny of Aculifera+Conchifera with well support, conforming to the most of phylogenetic studies of mollusks based on molecular data (Smith et al., 2013; Kocot et al., 2020; Song et al., 2023), thereby illustrating this phylogenetic relationship is reliable. The clade Aculifera contains the sister lineages Aplacophora and Polyplacophora. In the clade Conchifera, our results support the view that Monoplacophora was the sister to Cephalopoda (Smith et al., 2013), but some phylogenetic studies recovered Monoplacophora as the basal clade of all other shelled mollusks (Smith et al., 2013; Zapata et al., 2014; Kocot et al., 2017). In the other three classes Bivalvia, Scaphopoda and Gastropoda, the phylogenetic relationships were long–term controversy for various hypothesis (Kocot et al., 2017; Kocot et al., 2020). In our results, we recovered the clade Scaphoda+Gastropoda being sister group to Bivalvia with well supported, which was supported by some molecular studies (Smith et al., 2013; Kocot et al., 2020). The morphological studies revealed the clade Diasoma (Scaphoda+Bivalvia) (Runnegar and Pojeta, 1974), and a recent molecular study recovered this clade with well supported genomics data (Song et al., 2023), however, this result was not supported by our phylogenetic trees. The same results based on IQ–tree analysis revealed that model potential influence was avoided in our analysis, and BA tree suggested that the different methods have no influence on the results. In the past molecular studies of molluscan phylogeny, recovering well–accepted phylogenetic relationship is difficult, this could be due to a number of reasons (Kocot et al., 2011). Fortunately, dividing mollusks into two major clades, Aculifera and Conchifera had become the consensus (Kocot et al., 2020). However, the major controversial remained in the internal phylogenetic relationships of Conchifera, especially in Gastropoda, Bivalvia and Scaphopoda (Smith et al., 2013; Sumner-Rooney et al., 2015; Kocot et al., 2017; Kocot et al., 2020). Our result recovered the clade Aculifera+Conchifera and consistent with some past studies (Smith et al., 2013; Kocot et al., 2020), therefore, although the controversial still remained in the phylogeny of Mollusca, our ML topology could fit the current molluscan phylogenetic consensus.
Our divergence time estimation based on PAML mcmctree of ML tree obtained the basically consistent results of the major molluscan diversification nodes with previous molecular studies (Kocot et al., 2020; Song et al., 2023). The molecular clock analysis indicated that mollusks split from Brachiopoda about Precambrian 526.54 Mya (95% HPD: 519.61, 531.44), this time is a little bit earlier than some molecular studies (Kocot et al., 2020; Song et al., 2023). Our analysis suggests that this may be due to the data set we used, the orthologous of both outgroup species and mollusks are all derived from metazoan_odb10 database. But our results still conformed to the Precambrian origin of Mollusca (Parkhaev, 2008). The split of Aplacophora and Polyplacophora time was consistent with the fossil studies (Nielsen et al., 2007; Parkhaev and Demidenko, 2010). Cephalopods split from Monoplacophora 491.62 Mya (95% HPD: 472.58, 510.49), this roughly coincides with the emergence times of the earliest fossil cephalopods (Nishiguchi and Mapes, 2008). Our molecular analysis revealed Bivalves originated about 505.46 Mya (95% HPD: 497.01, 513.72) which was confirmed with the first Bivalves fossil record (Parkhaev, 2008). In addition, there was the diversification of Bivalves occurred 490.68 Mya (95% HPD: 482.77, 499.07), the fossil study also suggested Bivalves had a noticeable diversification in the Ordovician (Babin, 2000). The origin of gastropods is still a matter of debate (Parkhaev, 2017), our results reveal that gastropods is sister to Scaphopoda, and originate in the Cambrian period, which coincides with the date of the oldest gastropods fossil (Parkhaev, 2002; Parkhaev, 2017).
With credible fossils to calibrate our time–tree of our molecular clock analysis, we reveal the time–scale of the major mollusks’ habitat transitions events (Figure 2). I. Unionida (Bivalvia): this habitat transition event occurred Triassic about 233.21 Mya. Unionida are widely distributed in freshwater lakes, rivers and ponds on all continents except Antarctica (Huang et al., 2019), containing over 900 species (Lopes-Lima et al., 2017). This order could represent the major radiation of Bivalvia in freshwater ecosystems (Froufe et al., 2020). It is generally accepted that the family Silesunionidae (Early Triassic) should be considered the MRCA of this order (Skawina and Dzik, 2011), and recent phylogenetic reconstructions also supported the Triassic origin of the order Unionida (Crouch et al., 2020; Froufe et al., 2020). According to the geological and paleontological literature, the earliest freshwater ecosystems on Earth may have formed during the Ordovician period for the warm climate (Robin and Cocks, 2020). The Ordovician oldest freshwater jawless fish ostracoderms (Gregory, 1935) also illustrated that the stable freshwater ecosystem had emerged before the expansion of Unionida. The fossil record of the earliest freshwater plants (before 480 Mya) proved the presence of the producers in contemporaneous freshwater ecosystems (Strother and Foster, 2021). All this suggests that Earth had a freshwater environment capable of supporting these mollusks before Triassic, allowing this order to colonize and survive in freshwater environment, but the reason for its later appearance relative to other freshwater organisms remains unexplained. For the distribution of Unionida, our results revealed the same late diversification of a super family Unionidae+Margaritiferidae (Froufe et al., 2020), and Huang et al., (Huang et al., 2019) also revealed the close relationship of the Unionidae distributed in different regions, like in China and North America, based on ancestral range estimation. Although the genomic data and ancestral habitat tests for some Unionida are lacking in our study, our molecular clock analysis revealed that the expansion to freshwater of the Unionida occurred about Triassic, which is same as some previous molecular studies (Bolotov et al., 2017; Froufe et al., 2020).
II: Pulmonata (Gastropoda), in this group, we used the earliest records of lymnaeids Lymnaea morrisonensis (Gray, 1988; Aksenova et al., 2018) to calibrate the divergence times. From our results, the second habitat transition event occurred the late Triassic about 201.62 Mya. This period is after the Triassic–Jurassic mass extinction event (about 199 Mya) (Cohen et al., 2013). Volcanic eruptions in the Central Atlantic Magmatic Province, along with global warming, ocean acidification and oxygen deprivation, are widely believed to have caused the mass extinction (Preto et al., 2010; Whiteside et al., 2010; Bond and Grasby, 2017). The mass extinction event caused great damage to both marine and terrestrial ecosystems and a sharp decline in biodiversity (Lucas and Tanner, 2015). Therefore, we speculate that the emergence of a large number of niche vacancies in terrestrial and freshwater environments after this extinction event provided the opportunity to expand new environments, the same conclusion was also revealed in a study of the expansion of bivalve families (Crouch et al., 2020). Moreover, in the order Stylommatophora, one molecular study have revealed that intertidal Onchidiidae are a transitional species of snails from the sea to land (Goulding et al., 2022), and our phylogenetic analysis also suggested this family is the basal family of Stylommatophora, which support above conclusion. Three families freshwater snails are the intermediate hosts of Schistosoma, showing a high degree of relatedness, the family Physidae is native to the North America, Planorbidae is from the South America and Lymnaeidae comes from the Africa (Bakry et al., 2015; Aksenova et al., 2017; Pila et al., 2017; Zeng et al., 2017; Review et al., 2019; Kivistik et al., 2022). According to our molecular clock analysis, they diversified 173.34 Mya. This period is in the breakup of the first stage of Gondwana (about 180 Mya) (Burrett et al., 2014). And late, these freshwater snails split in the next about 20 million years, in this time, Africa has split off from America. Therefore, we speculate that the ancestors of these freshwater snails colonized the freshwater ecosystem before the breakup of Gondwana. Later, with the breakup of Gondwana, they each stayed on a different continent.
III: Ampullariidae (Gastropoda), this habitat transition event occurred about 156.21 Mya, containing African Lanistes nyassanus, South American Marisa cornuarietis, Pomacea maculata and P. canaliculata (Hayes et al., 2015). From our revealed divergence times, we support the Gondwanan origin for Ampullariidae (Berthold, 1991), and three South American apple snails diversified about 30 Mya, which conforms with the hypothesis that the diversification of New World ampullariids probably occurred after the separation of America and Africa (Jokat et al., 2003). The anatomic studies revealed the fact that Ampullariidae have both gills and lungs, this allows them to live temporarily out of water (Mueck et al., 2020), which we think this may have been an important condition for the colonization of freshwater during this period. Because since the Carboniferous, the Earth’s climate was warm and wet, and forests and swamps began to appear on a large scale (Thomas and Cleal, 2015). During this period, amphibians began to diversify rapidly, like Labyrinthodontia (Witzmann et al., 2017) and Eryops (Konietzko-Meier et al., 2015). Research shows that the Carboniferous amphibian explosion had a lot to do with their ability to breathe with their lungs and adapt to the heat and oxygen deprivation of the time (Long and Gordon, 2004). So do Ampullariidae, the emergence of lungs would allow them to adapt to this high temperature and low oxygen environment. Their native habitat was mainly in the rainforests of South America, suggesting that they had similar environmental adaptations to their ancestors. This also explains why they have established populations in modern times as invasive species only in other tropical regions besides South America (Hayes et al., 2008; Lv et al., 2009).
Conclusion
In this study, we present a phylogenetic tree of Mollusca based on amount of genomic data and, through rigorous fossil calibration, present what may be the most comprehensive time–scale of molluscan evolution. It provides phylogenomic support for recovering the phylogeny of mollusks with “Aculifera+ Conchifera” topology. Our study elucidates the timing of several independent habitat transition events during the evolution of mollusks and explains possible inducers through historical ecological events. Revealed three major habitat transition events could represent the main expansion of Mollusca into freshwater and land ecosystems. It is of great significance to study the complex evolutionary process of mollusks and the terrestrialization of animals.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
XH: Formal analysis, Writing – original draft, Writing – review & editing. SS: Formal analysis, Writing – original draft, Writing – review & editing. YW: Formal analysis, Writing – original draft, Writing – review & editing. ML: Data curation, Writing – review & editing. BS: Conceptualization, Investigation, Writing – review & editing. ZH: Conceptualization, Investigation, Software, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LR21D060003).
Acknowledgments
We thank Qi Liu (Wuhan Onemore–tech Co., Ltd.) for the help of the sequencing efforts.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1327007/full#supplementary-material
References
Aksenova O. V., Bolotov I. N., Gofarov M. Y., Kondakov A. V., Vinarski M. V., Bespalaya Y. V., et al. (2018). Species richness, molecular taxonomy and biogeography of the Radicine Pond snails (Gastropoda: Lymnaeidae) in the Old World. Sci. Rep. 8, 11199. doi: 10.1038/s41598-018-29451-1
Aksenova O., Vinarski M., Bolotov I., Kondakov A., Bespalaya Y., Tomilova A., et al. (2017). Two Radix spp. (Gastropoda: Lymnaeidae) endemic to thermal springs around Lake Baikal represent ecotypes of the widespread Radix auricularia. J. Zool. Syst. Evol. Res. 55, 298–309. doi: 10.1111/jzs.12174
Babin C. (2000). Ordovician to Devonian diversification of the Bivalvia. Am. Malacol. Bull. 15, 167–178. doi: 10.1016/S0016-6995(99)80029-1
Bakry F., Ismail S., Abd El-Atti M. (2015). Glyphosate herbicide induces genotoxic effect and physiological disturbances in Bulinus truncatus snails. Pestic. Biochem. Phys. 123, 23–40. doi: 10.1016/j.pestbp.2015.01.015
Berthold T. (1991). “Vergleichende Anatomie, Phylogenie und historische Biogeographie der Ampullariidae (Mollusca, Gastropoda),” in Abhandlungen des Naturwissenschaftlichen Vereins in Hamburg NF, Hamburg, Germany 311–338.
Böhm M., Dewhurst-Richman N., Seddon M., Ledger S., Albrecht C., Allen D., et al. (2021). The conservation status of the world’s freshwater molluscs. Hydrobiologia 848, 3231–3254. doi: 10.1007/s10750-020-04385-w
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolotov I. N., Kondakov A. V., Vikhrev I. V., Aksenova O. V., Bespalaya Y. V., Gofarov M. Y., et al. (2017). Ancient river inference explains exceptional oriental freshwater mussel Radiations. Sci. Rep. 7, 2135. doi: 10.1038/s41598-017-02312-z
Bond D. P. G., Grasby S. E. (2017). On the causes of mass extinctions. Palaeogeogr. Palaeoclimatol. Palaeoecol. 478, 3–29. doi: 10.1016/j.palaeo.2016.11.005
Burrett C. F., Zaw K., Meffre S., Lai C., Khositanont S., Chaodumrong P., et al. (2014). The configuration of Greater Gondwana—Evidence from LA ICPMS, U–Pb geochronology of detrital zircons from the Palaeozoic and Mesozoic of Southeast Asia and China. Gondwana Res. 26, 31–51. doi: 10.1016/j.gr.2013.05.020
Cohen K., Finney S., Gibbard P., Fan J. (2013). The ICS International Chronostratigraphic Chart. Episodes 36, 199–204. Episodes 36. doi: 10.18814/epiiugs/2013/v36i3/002
Creevey C., Muller J., Doerks T., Thompson J., Arendt D., Bork P. (2011). Identifying single copy orthologs in Metazoa. PloS Comput. Biol. 7, e1002269. doi: 10.1371/journal.pcbi.1002269
Crouch N., Edie S., Collins K., Jablonski D. (2020). Calibrating phylogenies assuming bifurcation or budding alters inferred macroevolutionary dynamics in a densely sampled phylogeny of bivalve families. Proc. R. Soc B 288. doi: 10.1098/rspb.2021.2178
Cummings K., Lydeard C. (2019). Freshwater mollusks of the World. A distribution atlas. Baltimore, Maryland, USA: Johns Hopkins University Press
Darriba D., Taboada G., Doallo R., Posada D. (2011). ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Erwin D., Laflamme M., Tweedt S., Sperling E., Pisani D., Peterson K. (2011). The Cambrian conundrum: Early divergence and later ecological success in the early history of animals. Science 334, 1091–1097. doi: 10.1126/science.1206375
Froufe E., Bolotov I., Aldridge D. C., Bogan A. E., Breton S., Gan H. M., et al. (2020). Mesozoic mitogenome rearrangements and freshwater mussel (Bivalvia: Unionoidea) macroevolution. Hered. (Edinb) 124, 182–196. doi: 10.1038/s41437-019-0242-y
Goulding T. C., Khalil M., Tan S. H., Cumming R. A., Dayrat B. (2022). Global diversification and evolutionary history of onchidiid slugs (Gastropoda, Pulmonata). Mol. Phylogenet. Evol. 168, 107360. doi: 10.1016/j.ympev.2021.107360
Grabherr M., Haas B., Yassour M., Levin J., Thompson D., Amit I., et al. (2011). Full-Length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gray J. (1988). Evolution of the freshwater ecosystem: The fossil record. Palaeogeogr. Palaeoclimatol. Palaeoecol. 62, 1–214. doi: 10.1016/0031-0182(88)90054-5
Gregory W. K. (1935). On the evolution of the skulls of vertebrates with special reference to heritable changes in proportional diameters (Anisomerism). PNAS 21 1, 1–8. doi: 10.1073/pnas.21.1.1
Haszprunar G., Wanninger A. (2012). Molluscs. Curr. Biol. 22, 510–514. doi: 10.1016/j.cub.2012.05.039
Hayes K., Burks R., Castro-Vazquez A., Darby P., Heras H., Martín P., et al. (2015). Insights from an integrated view of the biology of Apple snails (Caenogastropoda: Ampullariidae). Malacologia 58, 245–302. doi: 10.4002/040.058.0209
Hayes K. A., Cowie R. H., Thiengo S. C. (2009). A global phylogeny of apple snails: Gondwanan origin, generic relationships, and the influence of outgroup choice (Caenogastropoda: Ampullariidae). Biol. J. Linn. Soc 98, 61–76. doi: 10.1111/j.1095-8312.2009.01246.x
Hayes K., Joshi R., Thiengo S., Cowie R. (2008). Out of South America: Multiple origins of non-native apple snails in Asia. Divers. Distrib. 14, 701–712. doi: 10.1111/j.1472-4642.2008.00483.x
Hopper G., Dickinson G., Atkinson C. (2021). Associations among elements in freshwater mussel shells (Unionidae) and their relation to morphology and life history. Freshw. Biol. 66, 1980–1991. doi: 10.1111/fwb.13807
Huang X. C., Su J. H., Ouyang J. X., Ouyang S., Zhou C. H., Wu X. P. (2019). Towards a global phylogeny of freshwater mussels (Bivalvia: Unionida): Species delimitation of Chinese taxa, mitochondrial phylogenomics, and diversification patterns. Mol. Phylogenet. Evol. 130, 45–59. doi: 10.1016/j.ympev.2018.09.019
Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jokat W., Boebel T., König M., Meyer U. (2003). Timing and geometry of early Gondwana breakup. JGR 108, 2428. doi: 10.1029/2002JB001802
Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh K., Asimenos G., Toh H. (2009). “Multiple alignment of DNA sequences with MAFFT,” in Bioinformatics for DNA Sequence Analysis. Ed. Posada D. (Totowa, NJ: Humana Press), 39–64.
Kenrick P., Wellman C. H., Schneider H., Edgecombe G. D. (2012). A timeline for terrestrialization: consequences for the carbon cycle in the Palaeozoic. Philos. Trans. R. Soc Lond. B. Biol. Sci. 367, 519–536. doi: 10.1098/rstb.2011.0271
Kivistik C., Käiro K., Tammert H., Sokolova I. M., Kisand V., Herlemann D. P. R. (2022). Distinct stages of the intestinal bacterial community of Ampullaceana balthica after salinization. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.767334
Kocot K. M., Cannon J. T., Todt C., Citarella M. R., Kohn A. B., Meyer A., et al. (2011). Phylogenomics reveals deep molluscan relationships. Nature 477, 452–456. doi: 10.1038/nature10382
Kocot K. M., Poustka A. J., Stoger I., Halanych K. M., Schrodl M. (2020). New data from Monoplacophora and a carefully-curated dataset resolve molluscan relationships. Sci. Rep. 10, 101–107. doi: 10.1038/s41598-019-56728-w
Kocot K. M., Struck T. H., Merkel J., Waits D. S., Todt C., Brannock P. M., et al. (2017). Phylogenomics of Lophotrochozoa with consideration of systematic error. Syst. Biol. 66, 256–282. doi: 10.1093/sysbio/syw079
Konietzko-Meier D., Shelton C., Sander P. (2015). The discrepancy between morphological and microanatomical patterns of anamniotic stegocephalian postcrania from the Early Permian Briar Creek Bonebed (Texas). CR Palevol. 15, 103–114. doi: 10.1016/j.crpv.2015.06.005
Long J., Gordon M. (2004). The greatest step in vertebrate history: A oaleobiological review of the fish-tetrapod transition. Physiol. Biochem. Zool. 77, 700–719. doi: 10.1086/425183
Lopes-Lima M., Froufe E., Do V. T., Ghamizi M., Mock K. E., Kebapçı Ü., et al. (2017). Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): Defining modern subfamilies and tribes. Mol. Phylogenet. Evol. 106, 174–191. doi: 10.1016/j.ympev.2016.08.021
Lucas S., Tanner L. (2015). End-Triassic nonmarine biotic events. J. Palaeogeogr. 4, 331–348. doi: 10.1016/j.jop.2015.08.010
Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18–23. doi: 10.1186/2047-217x-1-18
Lv S., Zhang Y., Liu H. X., Hu L., Yang K., Steinmann P., et al. (2009). Invasive snails and an emerging infectious disease: Results from the first national survey on Angiostrongylus cantonensis in China. PloS Negl. Trop. Dis. 3, e368. doi: 10.1371/journal.pntd.0000368
Manni M., Berkeley M. R., Seppey M., Simao F. A., Zdobnov E. M. (2021b). BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654. doi: 10.1093/molbev/msab199
Manni M., Berkeley M., Seppey M., Zdobnov E. (2021a). BUSCO: Assessing genomic data quality and beyond. Curr. Protoc. Bioinf. 1, e323. doi: 10.1002/cpz1.323
Mueck K., Deaton L., Lee A. (2020). Microscopic anatomy of the gill and lung of the Apple snail Pomacea maculata, with Notes on the Volume of the Lung. J. Shellfish Res. 39, 125. doi: 10.2983/035.039.0112
Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nielsen C., Haszprunar G., Ruthensteiner B., Wanninger A. (2007). Early development of the aplacophoran mollusc Chaetoderma. Acta Zool. Stockholm 88, 231–247. doi: 10.1111/j.1463-6395.2007.00270.x
Nishiguchi M. K., Mapes R. H. (2008). “Cephalopoda,” in Phylogeny and evolution of the Mollusca. Ed. Ponder W. (Oakland, California, USA: University of California Press), 162–163.
Parkhaev P. (2002). Phylogenesis and the system of the Cambrian univalved mollusks. Paleontol. J+ 36, 25–36. doi: 10.1134/S003103011706003X
Parkhaev P. Y. (2008). “The early cambrian radiation of mollusca,” in Phylogeny and Evolution of the Mollusca. Ed. Ponder W. (Oakland, California, USA: University of California Press), 33.
Parkhaev P. Y. (2017). Origin and the early evolution of the Phylum mollusca. Paleontol. J+ 51, 663–686. doi: 10.1134/s003103011706003x
Parkhaev P. Y., Demidenko Y. E. (2010). Zooproblematica and mollusca from the Lower Cambrian Meishucun section (Yunnan, China) and taxonomy and systematics of the Cambrian small shelly fossils of China. Paleontol. J+ 44, 883–1161. doi: 10.1134/S0031030110080010
Pila E., Peck S., Hanington P. (2017). The protein pheromone temptin is an attractant of the gastropod Biomphalaria glabrata. J. Comp. Physiol. 203, 855–866. doi: 10.1007/s00359-017-1198-0
Ponder W. F., Lindberg D. R. (2008). “Molluscan evolution and phylogeny: an introduction,” in Phylogeny and Evolution of the Mollusca. Ed. Ponder W. (Oakland, California, USA: University of California Press), 1–17.
Preto N., Kustatscher E., Wignall P. (2010). Triassic climates — State of the art and perspectives. Palaeogeogr. Palaeoclimatol. Palaeoecol. 291, 1–10. doi: 10.1016/j.palaeo.2010.03.015
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Review I., Skála V., Walker A., Horák P. (2019). Snail defence responses to parasite infection: The Lymnaea stagnalis-Trichobilharzia szidati model. Dev. Comp. Immunol. 102, 103464. doi: 10.1016/j.dci.2019.103464
Robin L., Cocks M. (2020). Ordovician palaeogeography and climate change. Gondwana Res. 100, 53–72. doi: 10.1016/j.gr.2020.09.008
Rota-Stabelli O., Allison C., Pisani D. (2013). Molecular timetrees reveal a Cambrian colonization of land and a new scenario for Ecdysozoan evolution. Curr. Biol. 23, 392–398. doi: 10.1016/j.cub.2013.01.026
Roure B., Rodriguez-Ezpeleta N., Philippe H. (2007). SCaFoS: a tool for Selection, Concatenation and Fusion of Sequences for phylogenomics. BMC Evol. Biol. 7 Suppl 1, S2. doi: 10.1186/1471-2148-7-S1-S2
Runnegar B., Pojeta J. (1974). Molluscan phylogeny: The Paleontological viewpoint. Science 186, 311–317. doi: 10.1126/science.186.4161.311
Saadi A. J., Mordan P. B., Wade C. M. (2021). Molecular phylogeny of the Orthurethra (Panpulmonata: Stylommatophora). Zool. J. Linn. Soc 193, 1126–1140. doi: 10.1093/zoolinnean/zlaa168
Seppey M., Manni M., Zdobnov E. M. (2019). “BUSCO: Assessing genome assembly and annotation completeness,” in Gene prediction: Methods and protocols. Ed. Kollmar M. (New York, NY: Springer New York), 227–245.
Skawina A., Dzik J. (2011). Umbonal musculature and relationships of the Late Triassic filibranch unionoid bivalves. Zool. J. Linn. Soc 163, 863–883. doi: 10.1111/J.1096-3642.2011.00728.X
Smith S., Wilson N., Goetz F., Feehery C., Andrade S., Rouse G., et al. (2013). Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 493, 708–708. doi: 10.1038/nature11736
Song H., Wang Y., Shao H., Li Z., Hu P., Yap-Chiongco M., et al. (2023). Scaphopoda is the sister taxon to Bivalvia: Evidence of ancient incomplete lineage sorting. PNAS 120, e2302361120. doi: 10.1073/pnas.2302361120
Stamatakis A. (2014). RAxML Version 8: A Tool for Phylogenetic Analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Strother P., Foster C. (2021). A fossil record of land plant origins from charophyte algae. Science 373, 792–796. doi: 10.1126/science.abj2927
Sumner-Rooney L. H., Schrödl M., Lodde-Bensch E., Lindberg D. R., Heß M., Brennan G. P., et al. (2015). A neurophylogenetic approach provides new insight to the evolution of Scaphopoda. Evol. Dev. 17, 337–346. doi: 10.1111/ede.12164
Thewissen J. G. M., Cooper L. N., George J. C., Bajpai S. (2009). From land to water: The origin of Whales, Dolphins, and Porpoises. Evol.: Educ. Outreach 2, 272–288. doi: 10.1007/s12052-009-0135-2
Thomas B., Cleal C. (2015). Cyclones and the formation of plant beds in late Carboniferous tropical swamps. Palaeobiodivers. Palaeoenviron. 95, 531–536. doi: 10.1007/s12549-015-0191-2
Vidal-García M., Keogh J. S. (2015). Convergent evolution across the Australian continent: ecotype diversification drives morphological convergence in two distantly related clades of Australian frogs. J. Evol. Biol. 28, 2136–2151. doi: 10.1111/jeb.12746
Whiteside J., Olsen P., Eglinton T., Brookfield M., Sambrotto R. (2010). Compound-specific carbon isotopes from Earth’s largest flood basalt eruptions directly linked to the end-Triassic mass extinction. PNAS 107, 6721–6725. doi: 10.1073/pnas.1001706107
Witzmann F., Werneburg R., Milner A. (2017). A partial skull roof of an embolomere from Linton, Ohio (Middle Pennsylvanian) and its phylogenetic affinities. PalZ 91, 399–408. doi: 10.1007/s12542-017-0374-4
Yang Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang H., Zhang J.-E., Xia J., Yang J., Guo J., Deng Z., et al. (2018). Comparative characterization of the complete mitochondrial genomes of the three Apple snails (Gastropoda: Ampullariidae) and the phylogenetic analyses. Int. J. Mol. Sci. 19, 3646. doi: 10.3390/ijms19113646
Zapata F., Wilson N. G., Howison M., Andrade S. C. S., Jorger K. M., Schrodl M., et al. (2014). Phylogenomic analyses of deep gastropod relationships reject Orthogastropoda. Proc. R. Soc B 281, 20141739. doi: 10.1098/rspb.2014.1739
Keywords: Mollusca, phylogeny, divergence time, expansion, ecological event
Citation: Han X, Sun S, Wang Y, Liu M, Shui B and Han Z (2024) Out of the ocean: the timescale of molluscan evolution based on phylogenomics revealed the ages of mollusks’ evolutionary transitions into the novel environment. Front. Ecol. Evol. 12:1327007. doi: 10.3389/fevo.2024.1327007
Received: 24 October 2023; Accepted: 23 January 2024;
Published: 12 February 2024.
Edited by:
Diego Baldo, CONICET Institute of Subtropical Biology (IBS), ArgentinaReviewed by:
Federico Plazzi, University of Bologna, ItalyGabriella Trucco, University of Milan, Italy
Copyright © 2024 Han, Sun, Wang, Liu, Shui and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bonian Shui, c2h1aWJvbmlhbkAxNjMuY29t; Zhiqiang Han, aGFuenFAempvdS5lZHUuY24=
 Xiaolu Han
Xiaolu Han Shaolei Sun
Shaolei Sun Yiting Wang
Yiting Wang Mengyuan Liu
Mengyuan Liu Zhiqiang Han
Zhiqiang Han