
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Ecol. Evol., 15 December 2023
Sec. Biogeography and Macroecology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1285115
This article is part of the Research TopicLong-Term Monitoring in Ecology and Evolution: Establishing a Sound Baseline to Help Inform our FutureView all 9 articles
 Pedro Manuel Carrasco De La Cruz1,2,3*
Pedro Manuel Carrasco De La Cruz1,2,3* Josie Antonucci Di Carvalho1,2
Josie Antonucci Di Carvalho1,2 Jana C. Massing1,2,3
Jana C. Massing1,2,3 Thilo Gross1,2,3
Thilo Gross1,2,3Long-term monitoring data is central for the analysis of biodiversity change and its drivers. Time series allow a more accurate evaluation of diversity indices, trait identification and community turnover. However, evaluating data collected across different monitoring programs remains complicated because of data discrepancies and inconsistencies. Here we propose a method for aggregating datasets using diffusion maps. The method is illustrated by aggregating long-term phytoplankton abundance data from the Wadden Sea and Southern North Sea gathered by two institutions located in Germany and The Netherlands. The aggregated data allowed us to infer species traits, to reconstruct the main trait axis which drives community functionality, ultimately quantifying functional diversity of the individual samples, having used only the co-occurrence of species in samples. Although functional diversity varies greatly among sampling stations, we detect a slight positive trend in German stations, which contrasts with the clear decreasing trend observed in most of the Dutch Wadden Sea stations. At the Terschelling transect, in Southern North Sea, the stations also showed contrasting estimations of functional diversity between off-shore and in-shore stations. Our research provides further evidence that traits and functional diversity can be robustly reconstructed from monitoring data alone, showing that data aggregation can increase the accuracy of this reconstruction, being able to aggregate heterogeneous datasets.
The climate crisis is increasingly impacting species distributions, changing macro-ecological patterns and reshuffling natural communities, which highlights biodiversity quantification as an essential task (Cardinale et al., 2012; Jonkers et al., 2019). However, the quantification of biodiversity variation remains challenging (Loreau et al., 2021). Most biodiversity indexes are based on taxonomic variation (Hill, 1973; Malavasi et al., 2004; Morin, 2009), which provides little information about species functionality or the effects on biological community structure (Bellwood et al., 2006; Tilman et al., 2006).
Several studies show that the importance of functional composition and functional richness tend to be larger than the importance of taxonomic richness in influencing ecosystem functions (Naeem and Wright, 2003; Petchey et al., 2004; Córdova-Tapia and Zambrano, 2015). Consequently, many indices were developed to measure functional diversity in an ecological community, using species traits (Petchey and Gaston, 2006). Rao’s quadratic entropy (Rao, 1982) is an important metric of functional diversity due to its mathematical simplicity and ability to analyze multiple traits. It is defined as:
where dij is the pair-wise distance between species i and j, and are the relative abundance of species i and j in sample k and the summation indices i, j run over all n species in the system (Botta-Dukát, 2005; Pavoine and Dolédec, 2005; Ricotta and Moretti, 2011).
The applicability of Rao’s index for functional diversity is presently limited by the availability of trait data. The term trait may refer to a number of closely related but subtly different concepts. In observational studies traits are morphological characteristics of taxa (McGill et al., 2006; Violle et al., 2007), whereas in modeling traits mostly refer to functional characteristics of modeled species (Huppert et al., 2002; Brännström et al., 2011). Bridging between these is the usage in data-analysis where traits are variables that are inferred from observational data, and thought to be informative of species functionality (Ryabov et al., 2022). Being able to infer species traits from observational data opens up the possibility to use existing long-term datasets to robustly quantify species traits (Mutshinda et al., 2017), obtaining a better species pairwise distances reconstruction, hence a more accurate computation of functional diversity (Botta-Dukát, 2005; Ricotta and Moretti, 2011).
An approach to infer species traits directly from monitoring datasets was proposed by Ryabov et al. (2022). Their approach adapts a manifold learning method known as diffusion maps (Coifman et al., 2005; Coifman and Lafon, 2006), which uses the observed multi-species distribution and species abundances to infer the functional traits that explain such distribution, turning around the traditional assessment of functionality of traits (Thomas et al., 2012; Kléparski et al., 2021). Once the trait space is reconstructed, Rao’s index is used to calculate the functional diversity of the community. Ultimately, this methodology provides a single-parameter algorithmic solution to identify important traits, being able to handle the high dimensionality of ecological datasets.
The accuracy in the reconstruction of the trait space should increase with the number of observations included in the analysis (Barter and Gross, 2019; Fahimipour and Gross, 2020). Hence, an important step to further develop this method is the combined analysis of different monitoring datasets. However, aggregating times series from different regions poses a major challenge due to heterogeneous sampling frequencies and methodologies, discrepancies in species taxonomic identification, or data access limitations (Benway et al., 2019). Therefore, it becomes necessary to develop a procedure to adequately aggregate data sets, that will improve the diffusion maps’ results while avoiding the limitations of individual data sets analysis.
In this work we introduce an approach for aggregating phytoplankton monitoring datasets for the diffusion map method proposed by Ryabov et al. (2022). The method is illustrated by the aggregation of two phytoplankton datasets gathered in different countries, as part of two extensive monitoring programs: one conducted in the Southern North Sea by Rijkswaterstaat, in the Netherlands, and the other by the Lower Saxony Water Management, Coastal Defence and Nature Conservation Agency (NLWKN), in Germany. Detailed description of the stations and sampling methods is given in Hanslik et al. (1998) for the German stations and in Prins et al. (2012) for the Dutch stations. The proposed method increases the accuracy of trait and biodiversity estimation for both of the datasets. Furthermore, it establishes common scales of traits and biodiversity, making it transferable between areas and regions.
We start by illustrating the diffusion mapping procedure using a single dataset. The phytoplankton dataset analyzed here is part of the extensive monitoring program conducted by Rijkswaterstaat, in the Netherlands (Baretta-Bekker et al., 2009). We used harmonized data from 18 stations, including 3691 samples and 366 species. The data harmonization consisted of first removing all species identified as purely heterotrophic, and second, homogenizing and updating phytoplankton species nomenclature using the WORMS website taxonomic database (Ahyong et al., 2023).
Following Ryabov et al. (2022) we begin the diffusion map process by calculating the similarity score between species over the set of samples. As our primary proxy for similarity between two species, species i and species j, we use the Spearman correlation (Spearman, 1987), building on the ecological principle that species tend to co-occur under the adequate environmental conditions (Hutchinson, 1959; Colwell and Rangel, 2009). The resulting similarity scores are gathered in a matrix, in which high values now indicate close similarity between the respective species.
Second, we threshold the similarity matrix to a set of ‘trusted comparisons,’ with the purpose of discarding the small similarity scores of our matrix. When comparing the entries in a high-dimensional space, a small similarity score provides very little information on the nature of the discrepancy (de la Porte et al., 2008; Barter and Gross, 2019). We therefore only consider such similarities as trusted when they are in the top-10 similarities for at least one of the compared species. As a result, we create a network in which each species is linked to at least the ten most similar species, a set of ‘trusted links.’
The set of species and trusted links now forms a complex network. This leads us to a new notion of similarity: Species are similar if they are close in the network of ‘trusted links.’ We can then define a system of proxy traits that describes where the respective species is located in a network. A natural coordinate system for a network is provided by the so-called Laplacian eigenmodes. To find them we construct the normalized Laplacian matrix as in Equation 2.
where Lij is the normalized similarity value between species i and j, obtained by weighting the Spearman similarity cij with the summatory of similarities in position j. Lij is 1 when the species is compared to itself.
This specific matrix is closely related to many natural processes such as different types of diffusion processes, heat conduction, or the spreading of vibrations (Pires et al., 2021). While a deeper discussion of the exact relation is beyond the scope of the current paper, the basic idea is that if we built the network as a mechanical object and repeatedly struck or heat random parts of it, the nodes that would in average warm or vibrate in sync must be in similar places (Yeakel et al., 2014; Delmas et al., 2019; Gibert and Yeakel, 2019). The actual matrix used here is not in exact correspondence to either of these physical processes, but a compromise chosen for its advantageous mathematical properties (Barter and Gross, 2019).
To extract the inferred proxy trait values for the species we compute the eigenvectors of the Laplacian. The eigenvectors contain one element for each of the species. Hence we can interpret the elements of an eigenvector as trait values of the species. Thus each eigenvector defines a trait axis, while the individual eigenvector elements are the respective trait values assigned to the individual species. Mathematically an eigenvector can be scaled arbitrarily. Common algorithms scale eigenvectors such that the length of the eigenvector is one. However, in a diffusion map, we want to scale the eigenvectors to reflect their respective importance. This importance is inversely proportional to the corresponding eigenvalue of L.
Laplacian matrices are positive semi-definite matrices, thus the eigenvalues are either positive or zero. The number of zero eigenvalues is identical to the number of components in the network of data points. If more than one zero eigenvalue exist, the network has become disconnected in the thresholding step. In that case, the analysis must be repeated with an increased number of threshold links. As the importance of an eigenvector is inversely related to the eigenvalue we could think that the zero-eigenvector is of infinite importance. However, in this eigenvector all elements are identical, the information that it tells us is just that all nodes are part of the same network component. We can hence ignore it in our analysis. Each of the remaining eigenvectors gives us a new trait axis for which the trait values of the individual species are given by the eigenvector elements (Ryabov et al., 2022).
To get an understanding of the results we consider two-dimensional plots of the eigenvector entries (Figure 1). The plot shows the traits constructed from eigenvector 1 and 2 (EV1 and EV2 respectively) and each dot represents a phytoplankton species used in the analysis. Diffusion mapping does not provide a biological interpretation of the eigenvectors, however, we can uncover such an interpretation by analyzing additional data. We used environmental data which were gathered during sampling (e.g., day of year, sea surface temperature, total NO3− concentration, total PO43− concentration, salinity, Dissolved Inorganic Nitrogen (DIN), Dissolved Inorganic Phosphorus (DIP), suspended particles), to estimate the species-specific environmental condition. We compute a weighted average of the gathered environmental parameters (Equation 3), using the abundance of species i in sample k, or , as a statistical weight of the sample
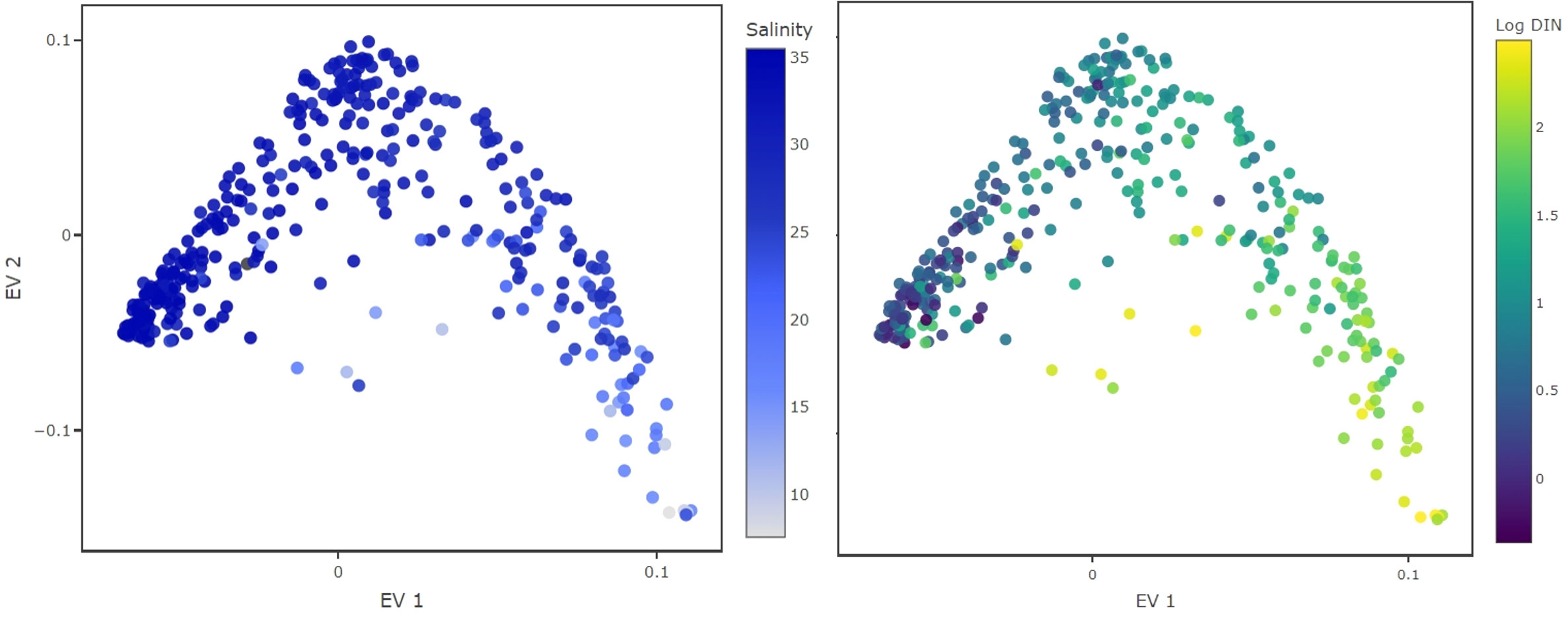
Figure 1 Inferred traits from the monitoring dataset. Color coded are environmental conditions under which the species were observed with high relative abundance. The EV1 aligns well with salinity (left) and DIN concentrations, displayed in logarithmic scale (right). This EV probably separates species by their adaptation to salinity levels or their nitrogen requirements.
where is the environmental factor in sample k, and m represent the number of samples. In this way we obtain the species-specific environmental value for each phytoplankton species.
Color coding the species in the reconstructed trait space (Figure 1) shows that the first i-trait aligns well with salinity and DIN concentrations, suggesting that this trait might represent adaptation to different levels of nutrient availability and water masses. This does not imply causality, but demonstrates the feasibility of our method to unveil the possible functional traits driving diversity in this phytoplankton community.
The analysis of individual datasets may limit our ability to construct a reliable network if the number of samples or the number of species is small. When this happens, we are forcing a comparison between dissimilar species, degrading the quality of trait space reconstruction (Barter and Gross, 2019; Fahimipour and Gross, 2020). Therefore, a recommended solution is to increase the data used in the analysis, which can be done by aggregating multiple long-term datasets.
Our goal is now to demonstrate that datasets cannot be aggregated directly. For this purpose we use the previously introduced data set by Rijkswaterstaat, in the Netherlands (Baretta-Bekker et al., 2009), and the dataset collected from the monitoring program of the Lower Saxony Water Management, Coastal Defence and Nature Conservation Agency, in Germany (NLWKN, 2013), both gathered in the coastline of the Southern North Sea. Data was harmonized, according to the previous section, and phytoplankton abundance observations were added subsequently.
As a result, the EV1, which represents the primary pattern detected by the method in the data, clustered the species into two groups: those only observed in the Netherlands and those only observed in Germany (Figure 2). This is not the desired result but rather an artifact from the data gathering. Plankton monitoring is a difficult task, and attribution of different taxonomic identities, for similar observations, might happen due to the high number of taxa or their sometimes very high morphological similarity. Although a certain degree of local endemism is possible (de Jonge et al., 1993; Tillmann et al., 2000; Cadée and Hegeman, 2002; Loebl and van Beusekom, 2008; van Walraven et al., 2015), the geographical context makes this only a partial explanation. Consequently, what we see here is that the diffusion map picks up on an artefact that is rooted in the nature of the data collection and then exacerbated by the naive aggregation. This defines the need for an aggregation procedure that avoids such artefacts.
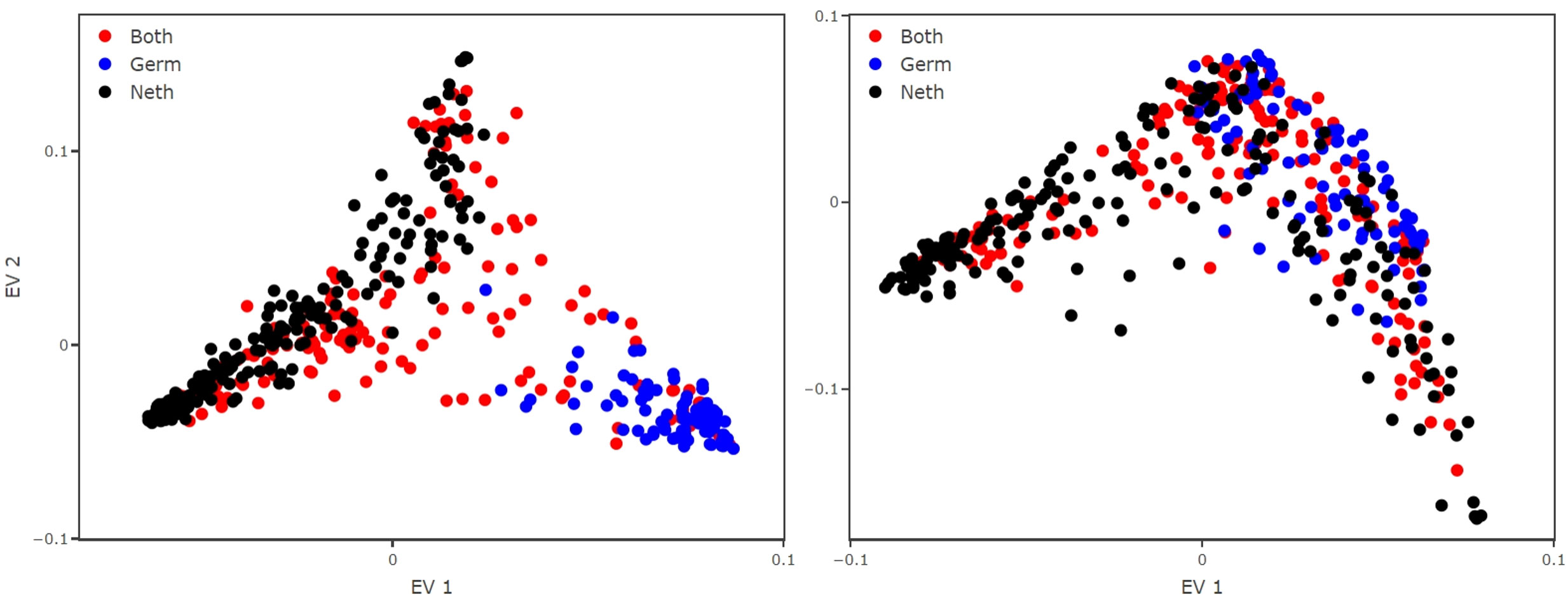
Figure 2 Reconstructed trait space from the aggregated monitoring dataset using the simple aggregation method (left panel) and our proposed aggregation method (right panel). Applying a naive aggregation makes the species (dots) cluster in species observed only in Germany (blue) and observed only in The Netherlands (black). The species (dots) that are common to both datasets are colored in red. Applying our aggregation method breaks the cluster, providing a better reconstructed trait space and avoiding data artefacts.
To find a better procedure for aggregation, let us analyze why the separation into Dutch and German species occurred in the naive attempt. When considering different monitoring datasets, the list of observed species in the respective areas may be different because some species are genuinely absent in one of the areas, however more likely the respective agencies have different equipment, procedures, and institutional cultures, which determine what can be observed and what taxonomic name is assigned to a given observation. It is easy to lament these differences between datasets, and call for more standardization. However, different cultures and capabilities may also open up different angles on a complex system that, when properly taken into account, reveal additional information.
We now recognize that if a species is not observed in a given sample this may indicate the actual absence of the species or it may signal that the species, while objectively present, was not able to be identified or was assigned a different name (Petchey and Gaston, 2002; Legras et al., 2020). In our naive merging procedure we interpreted the absence of an observation as evidence for the absence of the species from the respective sample. This assumption leads to an erroneous matrix of similarities which biases makes species that occur in only one of the regions appear different from the others.
We propose a more careful approach to dataset merging, which fixes the epistemological shortcomings of the naive procedure. We illustrate this approach using the datasets gathered by Rijkswaterstaat (Baretta-Bekker et al., 2009) and by NLWKN (NLWKN, 2013) (Figure 3). After basic data harmonization each of these datasets can be considered as internally consistent regarding its identification of taxa. Thus, we can safely construct and threshold the similarity matrices for the individual datasets as described above.
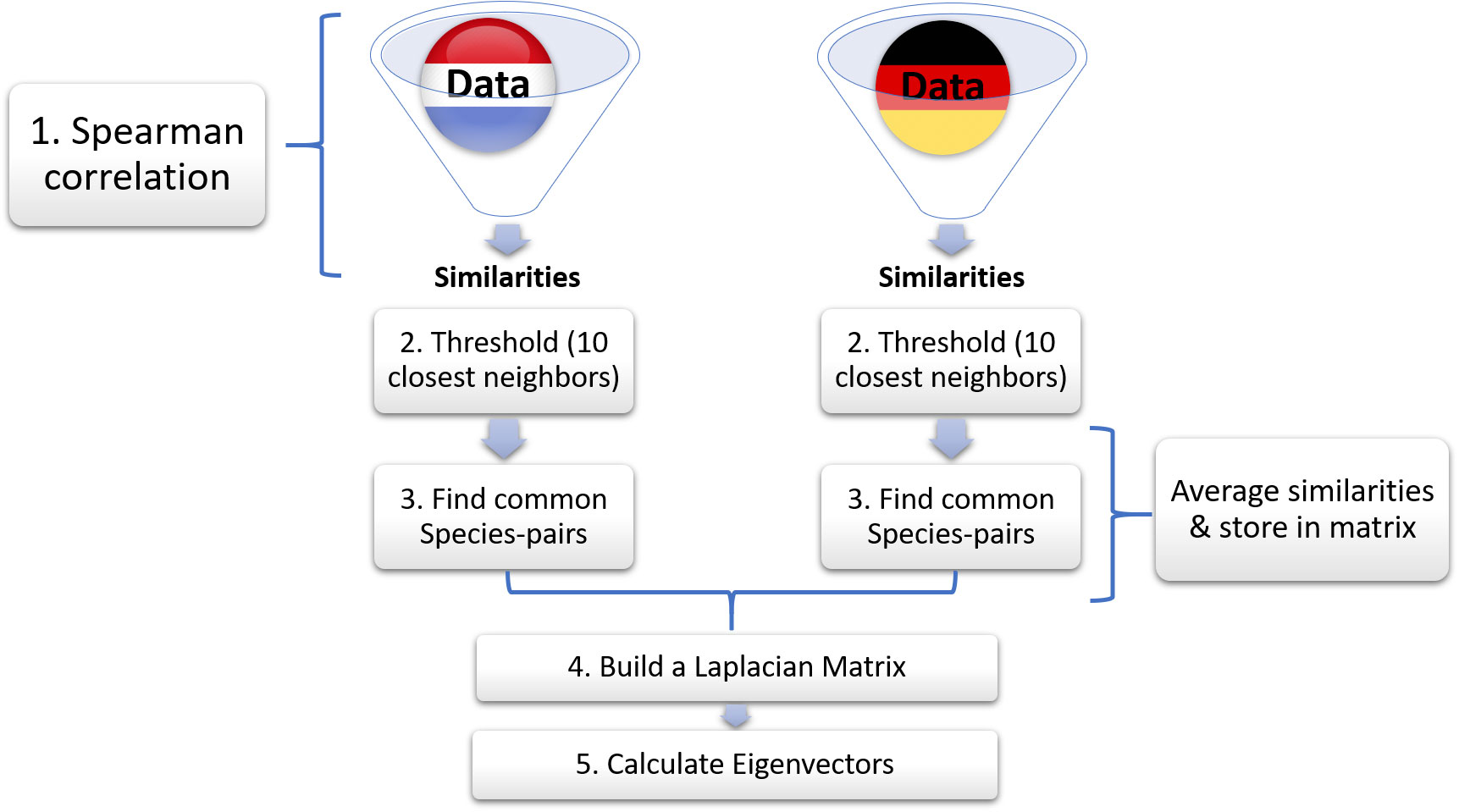
Figure 3 Schematic of proposed method for aggregating monitoring datasets. In step 1, we calculate similarities of German and Dutch phytoplankton abundance data separately. In step 2 we choose the 10 highest similarities (known as threshold). In step 3, after identifying the common species-pairs, we average their similarities and store them in a new matrix. The rest of the species-pairs are stored with their original similarity values. In step 4 we construct a Laplacian matrix, which is finally used to calculate the eigenvectors in step 5.
We then merge the processed similarity matrices as follows: We consider all possible pairs of species. For some of these pairs both species exist in both matrices. We interpret that as a sign that the corresponding species are reliably identified by both agencies and hence average the value of the respective similarities. For some pairs one or both of the species exist only in one of the matrices. We interpret this as an indication that only one of the agencies can make this comparison reliably and hence accept the value from the matrix where the comparison is possible. Finally, some comparisons cannot be made in either of the matrices because one species exists only in one of the matrices while the other species exists only in the other. In this case we set the similarity of the species to zero as no reliable comparison is possible.
The final choice means that we may assign some zeros to comparisons between similar species (or even between the same species which were identified by different taxonomic IDs). However, setting some comparisons wrongly to zero does not degrade the quality of the diffusion map result (Ryabov et al., 2022). The reconstructed trait space shows that the EV1 does no longer cluster the species into country of observation, rather we observe that they spread indistinctly over the manifold (Figure 2).
The first i-trait aligns well with DIN as well as with the water salinity (Figure 4). We conclude that this i-trait could represent adaptation to different water basin conditions (nutrient availability and salinity), which are different for the Wadden Sea and the Southern part of the North Sea (van Beusekom et al., 1999; van Beusekom and de Jonge, 2002). Such interpretation is likely, as it is being considered in the scientific literature (Carstensen et al., 2015; Jung et al., 2017).
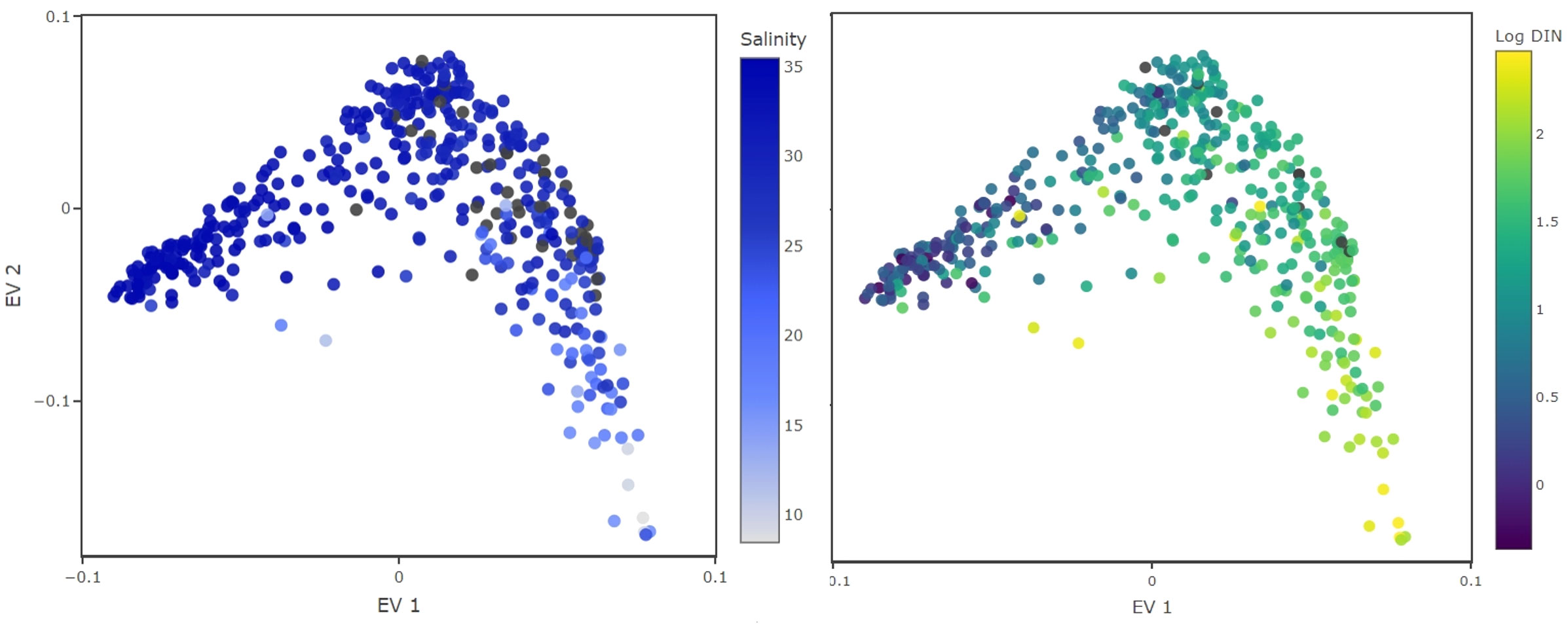
Figure 4 Inferred traits from the monitoring datasets. Color coded are environmental conditions under which the species were observed with high relative abundance. The EV1 aligns well with salinity (left) and DIN concentrations, displayed in logarithmic scale (right). This EV probably separates species by their adaptation to salinity levels or their nitrogen requirements.
Once the i-trait space has been successfully reconstructed for the aggregated data sets, we can use it to first calculate the distance in trait space for each species pair, i and j (Equation 4). Such distance, defined as dij, is calculated by using the euclidean distance in the reconstructed trait space, where the species traits are now given by the eigenvector elements corresponding to the species, re-scaled by the respective eigenvalue, as in:
where vk,i and vk,j are the species corresponding eigenvectors, λk is their corresponding eigenvalue and k is the respective trait.
For each sample, we then use the distances between the species in the i-trait space to compute the Rao index (F), introduced previously in Equation 1.
Multiple fluctuations can be observed in functional diversity estimations of samples, having dramatic inter-annual, as well as inter-station variations. However, when considered over the entire period, clearer patterns emerge. On the one hand, significant functional diversity losses occur at most Dutch Wadden Sea stations, with fastest decrease observed at the Marsdiep basin (MARSDND and DOOVBWT stations) and off the coast of Groningen, Lauwers basin (ZUIDOLWOT station). On the other hand, there is a mild increase of functional diversity in the German Wadden Sea stations, with the fastest increase at the Weser estuary, WeMu_W_1 station (Figure 5).
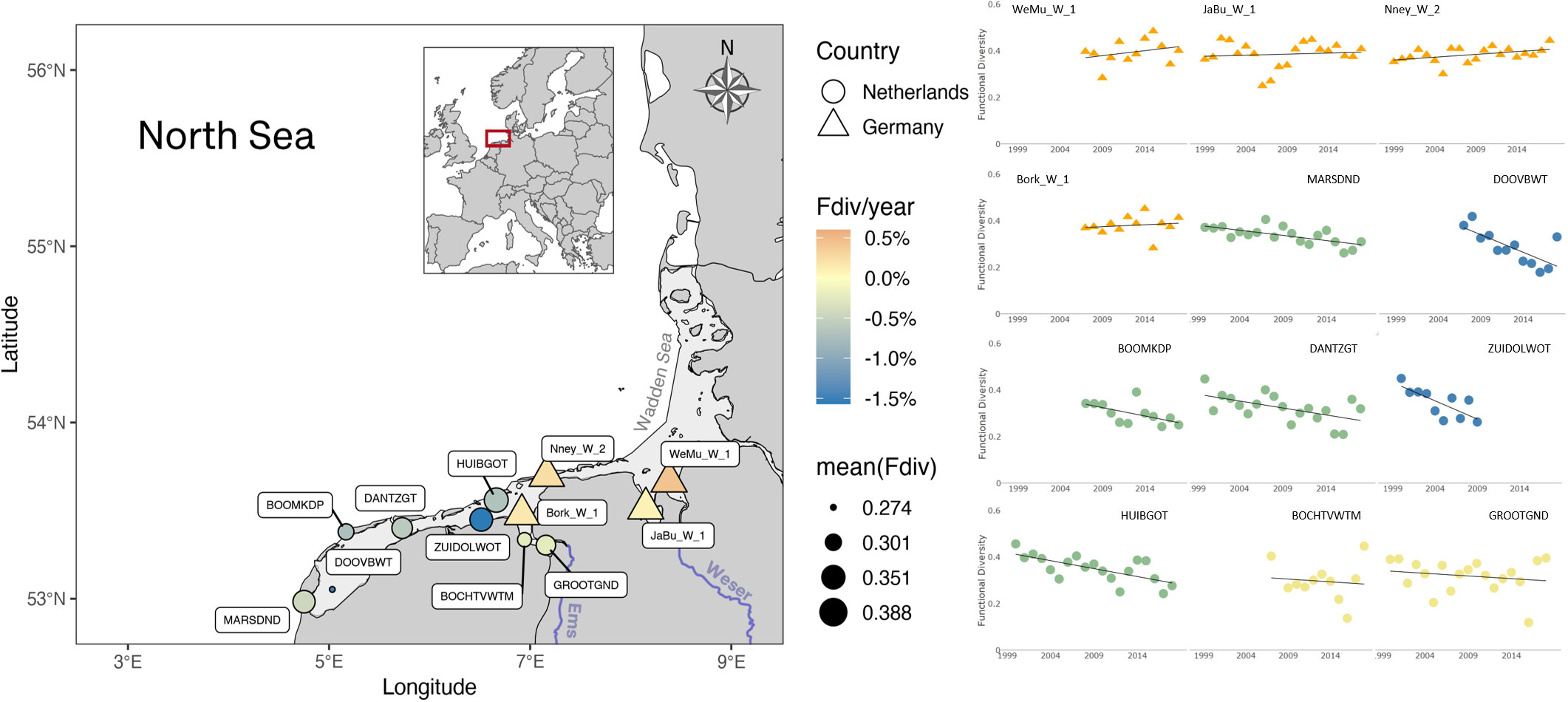
Figure 5 Phytoplankton functional diversity in the Wadden Sea. A decrease in functional diversity (% Fdiv per year) is observed over the measurement period at all Dutch stations (circles), whereas a mild increase (warmer colors) can be observed at the German stations (triangles). The fastest decrease rate (colder colors) is found at coastal stations on the Marsdiep and off Groningen. German Wadden Sea stations are in average the most functionally diverse (larger diameter).
Once catalogued as a ‘Changed Ecosystem’ (de Jonge et al., 1993), the Wadden Sea experienced a consistent decreasing trend in eutrophication starting in the 1990s (Cadée and Hegeman, 2002). However, contrasting recent reports have found significant signs of increasing eutrophication, persistent algal blooms, and phytoplankton diversity alteration in the Western Wadden Sea (Wolff et al., 2010; Carstensen et al., 2015; van Beusekom et al., 2019; Jacobs et al., 2020; Dajka et al., 2022). The declining diversity in the Marsdiep basin is likely explained by the dominance of Phaeocystis globosa spring and summer blooms (Cadée and Hegeman, 2002; Niu et al., 2015). The inter-annual variability among stations also suggests a blooming limitation by nutrients or light, which triggered the prevalence of fast-growing nutrient opportunist, C-strategist or R-strategist phytoplankton species such as Micromonas pusila, Thalassiosira sp., Chaetoceros sp., particularly in the second half of last decade (Smayda and Reynolds, 2001; Reynolds, 2006; Zhang et al., 2022).
Stations at the Terschelling transect, in the Southern North Sea, also showed contrasting estimations of functional diversity between off-shore stations (TERSLG235 to TERSLG100) and in-shore stations (TERSLG50, TERSLG10 and TERSLG4). Whereas off-shore stations had no significant trend variation, the in-shore stations had a clear negative trend (Figure 6). A possible explanation for this is the existence of a ‘line-of-no-return’ off the sand barrier islands of the Wadden Sea (Postma, 1984), which decreases the exchange between water masses and increases the accumulation of suspended matter in the coastal zone (de Jong and de Jong, 2002). Jung et al. (2017) recently estimated this line somewhere between 10 and 100 km at the Terschelling transect, thus having stations inside the ‘line-of-no-return’ highly influenced by the Wadden Sea dynamics and its environmental conditions. Therefore, the negative trend in functional diversity observed in the in-shore stations, as well as in the ROTTMP transect stations, might be due to seasonal exchange with the Wadden Sea phytoplanktonic community.
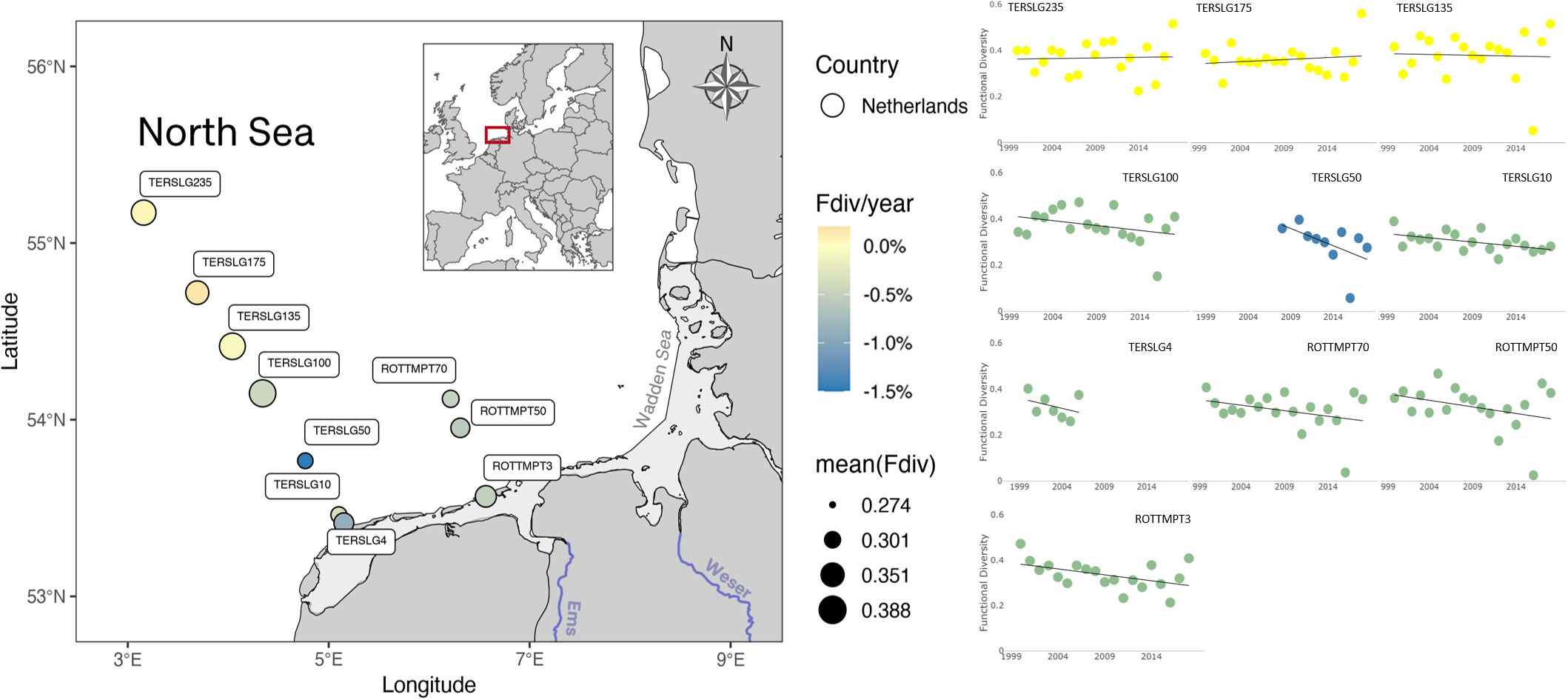
Figure 6 Phytoplankton functional diversity in Southern North Sea off the Dutch sand barrier islands. Offshore stations (pale-yellow color) show no significant functional diversity trend (% Fdiv per year), contrary to those stations located closer to barrier islands, which show a mild decrease rate (colder color). Offshore stations are on average the most functionally diverse (larger diameter).
Lastly, the estimations of functional diversity were consistent with expectations based on species composition. The low functional diversity in samples of 2006 and 2015 coincides with the dominance of the flagellate Micromonas pusila, with numbers over 90% of the total phytoplankton abundance (Figure 5). Similarly, low values of functional diversity in Dutch off-shore waters is due to a major dominance of Phaeocystis sp., whose numbers got to represent up to 99% of the total phytoplankton abundance in 2016 (Figure 6). On the contrary, the period of increased functional diversity in German samples are due to the community being dominated by two to three species constituting together more than 50% of the total abundance. Among this species were Lithodesmium undulatum, Paralia sulcata, Leptocylindrus minimus, Skeletonema costatum and other diatoms. The number of non-dominant species with relative abundances less than 10% also increased.
In this paper, we proposed a method to aggregate phytoplankton abundance datasets from different origins to reconstruct i-traits using diffusion maps. This aggregated data improved the reconstruction of trait axes and the subsequent estimation of functional diversity from monitoring data. Our approach enables a robust estimation of functional diversity within the system based solely on species abundances.
We demonstrated that failure of naive aggregation is rooted in the nature of the data collection and then exacerbated to the point of clustering those species unique to individual datasets, hence conflicting the trait reconstruction. If some species are not reported in a dataset, it can be assumed that these species were never present there or could not be identified, but total certainty for any alternative is unlikely. Our approach to data aggregation avoids assuming a total absence of those no-reported species by averaging similarity values of only those species common to both data sets, obtaining a better reconstructed trait space.
The final result is a better estimation of functional diversity for both data sets and for the entire analyzed geographical area. Significant declining estimations of functional diversity in the West Wadden Sea are in line with recent reports (Wolff et al., 2010; van Beusekom et al., 2019; Jacobs et al., 2020) and showed the ever prevalence of fast-growing nutrient opportunist phytoplankton species in this ecosystem. Additionally, the difference in the functional diversity trends of the Southern North Sea stations might be explained by the existence of a ‘line-of-no-return’ off the sand barrier islands of the Wadden Sea (Postma, 1984; Jung et al., 2017), which might isolate off-shore stations and their phytoplanktonic community.
We envision the possibility of large-scale aggregation of many different monitoring datasets, moving from local to regional, and even to global scales. Successful application of diffusion maps to large-scale aggregated data could ultimately provide a unified standard of functional diversity that can be used to map the functional diversity of samples on a fixed scale.
The datasets analyzed and generated, as well as the Julia Program used for this study, can be found in the ZENODO public repository via this link: https://zenodo.org/records/10209871, DOI 10.5281/zenodo.10209870.
The manuscript presents research on animals that do not require ethical approval for their study.
PC: Conceptualization, Formal Analysis, Investigation, Writing – original draft, Project administration, Data curation. JAC: Data curation, Validation, Visualization, Writing – review & editing. JM: Software, Validation, Visualization, Writing – review & editing, Methodology. TG: Conceptualization, Project administration, Supervision, Writing – review & editing, Methodology, Validation.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We are grateful to all those who have contributed to this study by sampling, analyzing and providing data. We would like to thank the Niedersächsischer Landesbetrieb für Wasserwirtschaft, Küstenund Naturschutz (NLWKN) and Rijkswaterstaat (Netherlands) for the data provision. HIFMB is a collaboration between the Alfred-Wegener-Institute, Helmholtz-Center for Polar and Marine Research, and the Carl-von-Ossietzky University Oldenburg, initially funded by the Ministry for Science and Culture of Lower Saxony and the Volkswagen Foundation through the ‘Niedersächsisches Vorab’ grant program (grant number ZN3285).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahyong S., Boyko C., Bailly N., Bernot J., Bieler R., Brandao S., et al. (2023). World register of marine species, Dataset. (Ostend, Belgium: VLIZ). doi: 10.14284/170
Baretta-Bekker J., Baretta J., Latuhihin M., Desmit X., Prins T. (2009). Description of the long-term (1991–2005) temporal and spatial distribution of phytoplankton carbon biomass in the Dutch North Sea. J. Sea Res. 61, 50–59. doi: 10.1016/j.seares.2008.10.007
Barter E., Gross T. (2019). Manifold cities: social variables of urban areas in the UK. Proc. R. Soc. A: Mathematical Phys. Eng. Sci. 475, 20180615. doi: 10.1126/science.1224836
Bellwood D., Wainwright P., Fulton C., Hoey A. (2006). Functional versatility supports coral reef biodiversity. Proc. R. Soc. B: Biol. Sci. 273, 101–107. doi: 10.1098/rspb.2005.3276
Benway H., Lorenzoni L., White A., Fiedler B., Levine N., Nicholson D., et al. (2019). Ocean time series observations of changing marine ecosystems: an era of integration, synthesis, and societal applications. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00393
Botta-Dukát Z. (2005). Rao’s quadratic entropy as a measure of functional diversity based on multiple traits. J. Vegetation Sci. 16, 533–540. doi: 10.1111/j.1654-1103.2005.tb02393.x
Brännström A., Gross T., Blasius B., Dieckmann U. (2011). Consequences of fluctuating group size for the evolution of cooperation. J. Math. Biol. 63, 263–281. doi: 10.1007/s00285-010-0367-3
Cadée G., Hegeman J. (2002). Phytoplankton in the marsdiep at the end of the 20th century; 30 years monitoring biomass, primary production, and phaeocystis blooms. J. Sea Res. 48, 97–110. doi: 10.1016/S1385-1101(02)00161-2
Cardinale B., Duffy E., Gonzalez A., Hooper D., Perrings C., Venail P., et al. (2012). Biodiversity loss and its impact on humanity. Nature 486, 59–67. doi: 10.1038/nature11148
Carstensen J., Klais R., Cloern J. (2015). Phytoplankton blooms in estuarine and coastal waters: Seasonal patterns and key species. Estuarine Coast. Shelf Sci. 162, 98–109. doi: 10.1016/j.ecss.2015.05.005
Coifman R., Lafon S. (2006). Diffusion maps. Appl. Comput. Harmonic Anal. 21, 5–30. doi: 10.1016/j.acha.2006.04.006
Coifman R., Lafon S., Lee A., Maggioni M., Nadler B., Warner F., et al. (2005). Geometric diffusions as a tool for harmonic analysis and structure definition of data: Diffusion maps. Proc. Natl. Acad. Sci. 102, 7426–7431. doi: 10.1073/pnas.0500334102
Colwell R., Rangel T. (2009). Hutchinson’s duality: the once and future niche. Proc. Natl. Acad. Sci. 106, 19651–19658. doi: 10.1073/pnas.0901650106
Córdova-Tapia F., Zambrano L. (2015). La diversidad funcional en la ecologia de comunidades. Ecosistemas 24, 78–87. doi: 10.7818/ECOS.2015.24-3.10
Dajka J.-C., di Carvalho J. A., Ryabov A., Scheiffarth G., Rönn L., Dekker R., et al. (2022). Modeling drivers of biodiversity change emphasizes the need for multivariate assessments and rescaled targeting for management. Conserv. Sci. Pract. 4, e12794. doi: 10.1111/csp2.12794
de Jong V., de Jong D. (2002). ‘Global change’ impact of inter-annual variation in water discharge as a driving factor to dredging and spoil disposal in the River Rhine system and of turbidity in the Sadden Sea. Estuarine Coast. Shelf Sci. 55, 969–991. doi: 10.1006/ecss.2002.1039
de Jonge V., Essink K., Boddeke R. (1993). The Dutch Wadden Sea: A changed ecosystem. Hydrobiologia 265, 45–71. doi: 10.1007/BF00007262
de la Porte J., Herbst B., Hereman W., Walt S. V. D. (2008). “An introduction to diffusion maps,” in Proceedings of the 19th symposium of the pattern recognition association of South Africa, 15–25 (South Africa: Member of the International Association of Pattern Recognition (IAPR).
Delmas E., Besson M., Brice M.-H., Burkle L., Riva G. D., Fortin M.-J., et al. (2019). Analysing ecological networks of species interactions. Biol. Rev. 94, 16–36. doi: 10.1111/brv.12433
Fahimipour A., Gross T. (2020). Mapping the bacterial metabolic niche space. Nat. Commun. 11, 1–8. doi: 10.1038/s41467-020-18695-z
Gibert J., Yeakel J. (2019). Laplacian matrices and turing bifurcations: revisiting levin 1974 and the consequences of spatial structure and movement for ecological dynamics. Theor. Ecol. 12, 265–281. doi: 10.1007/s12080-018-0403-2
Hanslik M., Rahmel J., Bätje M., Knieriemen S., Schneider G., Dick S. (1998). Der jahresgang blütenbildender und toxischer algen an der niedersächsischen küste seit 1982. Umweltbundesamt Texte Bonn 47, 221–236.
Hill M. (1973). Diversity and evenness: a unifying notation and its consequences. Ecology 54, 427–432. doi: 10.2307/1934352
Huppert A., Blasius B., Stone L. (2002). A model of phytoplankton blooms. Am. Nat. 159, 156–171. doi: 10.1086/324789
Hutchinson G. (1959). Homage to santa rosalia or why are there so many kinds of animals? Am. Nat. 93, 145–159. doi: 10.1086/282070
Jacobs P., Kromkamp J., van Leeuwen S., Philippart C. (2020). Planktonic primary production in the western Dutch Wadden Sea. Mar. Ecol. Prog. Ser. 639, 53–71. doi: 10.3354/meps13267
Jonkers L., Hillebrand H., Kucera M. (2019). Global change drives modern plankton communities away from the pre-industrial state. Nature 570, 372–375. doi: 10.1038/s41586-019-1230-3
Jung A., Bijkerk R., van der Veer H., Philippart C. (2017). Spatial and temporal trends in order richness of marine phytoplankton as a tracer for the exchange zone between coastal and open waters. J. Mar. Biol. Assoc. United Kingdom 97, 477–489. doi: 10.1017/S0025315416001326
Kléparski L., Beaugrand G., Edwards M. (2021). Plankton biogeography in the north atlantic ocean and its adjacent seas: Species assemblages and environmental signatures. Ecol. Evol. 11, 5135–5149. doi: 10.1002/ece3.7406
Legras G., Loiseau N., Gaertner J., Poggiale J.-C., Gaertner-Mazouni N. (2020). Assessing functional diversity: the influence of the number of the functional traits. Theor. Ecol. 13, 117–126. doi: 10.1007/s12080-019-00433-x
Loebl M., van Beusekom J. (2008). Seasonality of microzooplankton grazing in the northern Wadden Sea. J. Sea Res. 59, 203–216. doi: 10.1016/j.seares.2008.01.001
Loreau M., Barbier M., Filotas E., Gravel D., Isbell F., Miller S., et al. (2021). Biodiversity as insurance: from concept to measurement and application. Biol. Rev. 96, 2333–2354. doi: 10.1111/brv.12756
Malavasi S., Fiorin R., Franco A., Franzoi P., Granzotto A., Riccato F., et al. (2004). Fish assemblages of venice lagoon shallow waters: an analysis based on species, families and functional guilds. J. Mar. Syst. 51, 19–31. doi: 10.1016/j.jmarsys.2004.05.006
McGill B., Enquist B., Weiher E., Westoby M. (2006). Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21, 178–185. doi: 10.1016/j.tree.2006.02.002
Mutshinda C., Finkel Z., Widdicombe C., Irwin A. (2017). Phytoplankton traits from long-term oceanographic time-series. Mar. Ecol. Prog. Ser. 576, 11–25. doi: 10.3354/meps12220
Naeem S., Wright J. (2003). Disentangling biodiversity effects on ecosystem functioning: deriving solutions to a seemingly insurmountable problem. Ecol. Lett. 6, 567–579. doi: 10.1046/j.1461-0248.2003.00471.x
Niu L., van Gelder P., Zhang C., Guan Y., Vrijling J. (2015). Statistical analysis of phytoplankton biomass in coastal waters: Case study of the Wadden Sea near Lauwersoog (the Netherlands) from 2000 to 2009. Ecol. Inf. 30, 12–19. doi: 10.1016/j.ecoinf.2015.08.003
NLWKN (2013). Gewässerüberwachungssystem niedersachsen, gütemessnetz Übergangs-und küstengewässer Vol. 6 (Location, Germany: Küstengewässer und Ästuare), 1–50.
Pavoine S., Dolédec S. (2005). The apportionment of quadratic entropy: a useful alternative for partitioning diversity in ecological data. Environ. Ecol. Stat 12, 125–138. doi: 10.1007/s10651-005-1037-2
Petchey O., Gaston K. (2002). Functional diversity (fd), species richness and community composition. Ecol. Lett. 5, 402–411. doi: 10.1046/j.1461-0248.2002.00339.x
Petchey O., Gaston K. (2006). Functional diversity: back to basics and looking forward. Ecol. Lett. 9, 741–758. doi: 10.1111/j.1461-0248.2006.00924.x
Petchey O., Hector A., Gaston K. (2004). How do different measures of functional diversity perform? Ecology 85, 847–857. doi: 10.1890/03-0226
Pires M., Crokidakis N., Queirós S. (2021). Diffusion plays an unusual role in ecological quasi-neutral competition in metapopulations. Nonlinear Dynamics 103, 1219–1228. doi: 10.1007/s11071-020-06105-4
Postma H. (1984). Introduction to the symposium on organic matter in the Wadden Sea. Netherlands Institute Sea Res. Publ. Ser. 10, 15–22.
Prins T., Desmit X., Baretta-Bekker J. (2012). Phytoplankton composition in dutch coastal waters responds to changes in riverine nutrient loads. J. Sea Res. 73, 49–62. doi: 10.1016/j.seares.2012.06.009
Rao R. (1982). Diversity and dissimilarity coefficients: a unified approach. Theor. Population Biol. 21, 24–43. doi: 10.1016/0040-5809(82)90004-1
Reynolds C. (2006). The ecology of phytoplankton (United States of America: Cambridge University Press).
Ricotta C., Moretti M. (2011). Cwm and rao’s quadratic diversity: a unified framework for functional ecology. Oecologia 167, 181–188. doi: 10.1007/s00442-011-1965-5
Ryabov A., Blasius B., Hillebrand H., Olenina I., Gross T. (2022). Estimation of functional diversity and species traits from ecological monitoring data. Proc. Natl. Acad. Sci. 119, e2118156119. doi: 10.1073/pnas.2118156119
Smayda T., Reynolds C. (2001). Community assembly in marine phytoplankton: Application of recent models to harmful dinoflagellate blooms. J. Plankton Res. 23, 447–461. doi: 10.1093/plankt/23.5.447
Spearman C. (1987). The proof and measurement of association between two things. Am. J. Psychol. 100, 441–471. doi: 10.2307/1422689
Thomas M., Kremer C., Klausmeier C., Litchman E. (2012). A global pattern of thermal adaptation in marine phytoplankton. Science 338, 1085–1088. doi: 10.1126/science.1224836
Tillmann U., Hesse K.-J., Colijn F. (2000). Planktonic primary production in the German Wadden Sea. J. Plankton Res. 22, 1253–1276. doi: 10.1093/plankt/22.7.1253
Tilman D., Reich P., Knops J. (2006). Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 441, 629–632. doi: 10.1038/nature04742
van Beusekom J., Brockmann U., Hesse K.-J., Poremba K., Tillmann U. (1999). The importance of sediments in the transformation and turnover of nutrients and organic matter in the Wadden Sea and German Bight. Deutsche Hydrographische Z. 51, 245–266. doi: 10.1007/BF02764176
van Beusekom J., Carstensen J., Dolch T., Grage A., Hofmeister R., Lenhart H., et al. (2019). Wadden Sea eutrophication: long-term trends and regional differences. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00370
van Beusekom J., de Jonge V. (2002). “Long-term changes in Wadden Sea nutrient cycles: importance of organic matter import from the North Sea,” in Nutrients and eutrophication in estuaries and coastal waters. Eds. Orive E., Eliott M., Jonge V. (Bilbao, Spain: Springer), 185–194.
van Walraven L., Langenberg V., Dapper R., Witte J., Zuur A., van der Veer H. (2015). Long-term patterns in 50 years of Scyphomedusae catches in the western Dutch Wadden Sea in relation to climate change and eutrophication. J. Plankton Res. 37, 151–167. doi: 10.1093/plankt/fbu088
Violle C., Navas M.-L., Vile D., Kazakou E., Fortunel C., Hummel I., et al. (2007). Let the concept of trait be functional! Oikos 116, 882–892. doi: 10.1111/j.0030-1299.2007.15559.x
Wolff W., Bakker J., Laursen K., Reise K. (2010). “The Wadden Sea quality status report – synthesis report 2010,” in The wadden sea 2010 (Wilhelmshaven, Germany: Common Wadden Sea Secretariat (CWSS), 25–74.
Yeakel J., Moore J. P. G., de Aguiar M. (2014). Synchronisation and stability in river metapopulation networks. Ecol. Lett. 17, 273–283. doi: 10.1111/ele.12228
Keywords: functional diversity, long-term phytoplankton monitoring, diffusion map, Wadden Sea, North Sea
Citation: Carrasco De La Cruz PM, Antonucci Di Carvalho J, Massing JC and Gross T (2023) Aggregation of monitoring datasets for functional diversity estimation. Front. Ecol. Evol. 11:1285115. doi: 10.3389/fevo.2023.1285115
Received: 29 August 2023; Accepted: 28 November 2023;
Published: 15 December 2023.
Edited by:
Mauro Fois, University of Cagliari, ItalyReviewed by:
Loïck Kléparski, Marine Biological Association, United KingdomCopyright © 2023 Carrasco De La Cruz, Antonucci Di Carvalho, Massing and Gross. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pedro Manuel Carrasco De La Cruz, cGVkcm8uY2FycmFzY29AaGlmbWIuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.