
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 04 August 2023
Sec. Interdisciplinary Climate Studies
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1224849
This article is part of the Research Topic Agricultural Production in a Warmer World: Challenges and Sustainable Development Strategies View all 8 articles
 Xia Li1,2
Xia Li1,2 Guang He3
Guang He3 Dandan Li2
Dandan Li2 Shuikuan Bei2,4
Shuikuan Bei2,4 Dongdong Luan2
Dongdong Luan2 Xinzhan Sun2
Xinzhan Sun2 Gaiqiang Yang5
Gaiqiang Yang5 Lijuan Huo5
Lijuan Huo5 Lina Zhen1
Lina Zhen1 Ruotong Zhao2*
Ruotong Zhao2*Nitrous oxide (N2O) is a potent greenhouse gas, and agricultural soils represent a major anthropogenic source. Crop residues provide nutrients for plants but also act as hotspots of N2O production. The hyphae of arbuscular mycorrhizal fungi (AMF) could proliferate in organic patches, utilize released N from the organic patches, and potentially mitigate N2O emissions. However, the effect of AMF on N2O emissions in degraded residue patches and the possible microbial mechanism remain uncertain. Here, a mesocosm experiment was conducted to investigate the impact of AMF (Funneliformis mosseae) inoculation on N2O emissions, availabilities of carbon and nitrogen, extracellular enzyme activities, and the abundance of key N-cycling genes in degraded residue patches. Our results showed that AMF hyphae significantly reduced N2O emissions from degraded residue patches. Quantitative PCR analysis of key functional genes involved in N2O production (amoA, nirK, nirS) and consumption (nosZ) showed that AMF significantly reduced the abundance of the bacterial amoA and nirS genes. NH4+, NO3−, total dissolved nitrogen (TDN), total nitrogen (TN), and dissolved organic carbon (DOC) contents decreased drastically in the presence of AMF. In addition, the activities of all tested extracellular enzymes were significantly decreased by AMF and positively correlated with DOC content. Multiple stepwise regression analysis demonstrated that the abundance of the nirS gene primarily influenced N2O emissions and was positively correlated with DOC content in degraded residue patches. Our findings indicate that AMF suppressed N2O producers, particularly nirS-type denitrifiers, by slowing down the release of C and N from degraded residues, thereby leading to a cascade effect on the decrease of N2O emissions. This study provides a promising approach to mitigate N2O emissions by enhancing AMF in the agroecosystems.
Nitrous oxide (N2O) is a potent, long-lived greenhouse gas, with 273 times more warming potential than CO2 per molecule, and accelerates ozone depletion (Ravishankara et al., 2009; Arias et al., 2021). Agricultural soils are the major source of N2O, accounting for 50% of anthropogenic N2O emissions (Tian et al., 2020). Microbial processes, such as nitrification and denitrification, are the major pathways for N2O production in agricultural soil (Butterbach-Bahl et al., 2013). The oxidation of ammonia into nitrite via hydroxylamine was the first step of nitrification which was primarily carried out by two types of microbes: ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA). N2O is produced as a byproduct during the decomposition of hydroxylamine (NH2OH) (Prosser and Nicol, 2012). Nitrification predominates under aerobic conditions (Dobbie et al., 1999). Denitrification is the stepwise reduction of NO3− to N2 via NO2−, NO, and N2O and is performed by a diverse range of facultatively or obligate anaerobic organisms when O2 becomes scarce. N2O is an obligate intermediate of denitrification, and its net emissions depend on the balance between N2O-producing and N2O-consuming rates. The nirK/nirS genes encoding nitrite reductases are key genes involved in the reduction of NO2− to NO, and their abundance is often used to predict N2O-producing rates. The nosZ gene, encoding N2O reductase, is the only known biotic sink for N2O, and its abundance is an indicator of N2O-consuming rates (Chapuis-Lardy et al., 2007; Hallin et al., 2018). In reality, N2O emissions from agricultural soils exhibit fluctuations and spatiotemporal variability, and both nitrification and denitrification processes likely occur simultaneously (Kravchenko et al., 2017).
The majority of N2O production in agricultural soils occurs within small soil volumes (<1 cm3) associated with crop residues (Kravchenko et al., 2017). For example, more than 80% of total N2O emissions were traced to an 80-mg plant remnant inside the soil core (Parkin, 1987). A large amount of crop residues is generated during agricultural production and annual production reaching approximately 990 million tons in China and 5 billion tons globally (Dai et al., 2018; Roberto et al., 2018). Returning crop residues to agricultural soils can alleviate soil erosion, promote nutrient cycling, and increase soil carbon sequestration. However, the decomposition of crop residues can also result in higher N2O emissions. During residue decomposition, released NH4+, NO2−, and NO3− become available to nitrifiers and denitrifiers, and dissolved organic carbon (DOC) serves as a carbon source and electron donor for denitrifiers (Korom, 1992). Moreover, water absorption from surrounding soils by crop residues can create an anaerobic microenvironment favoring denitrification (Kravchenko et al., 2017). The annual N2O emissions derived from crop residues amount to 0.4 Tg N, partially offsetting the benefits of increased soil carbon sequestration from residues (Mosier et al., 1998).
Arbuscular mycorrhizal fungi (AMF) are ubiquitous in soils and form symbiotic associations with more than two-thirds of terrestrial plants (Smith and Read, 2008), playing a crucial role in N cycling (Veresoglou et al., 2012). Recent studies have found that AMF can reduce N2O emissions by altering nitrogen substrate availability, water content, or the community of nitrifying and denitrifying microorganisms (Bender et al., 2014; Lazcano et al., 2014; Bender et al., 2015; Zhang et al., 2015; Storer et al., 2017; Gui et al., 2021; Shen and Zhu, 2021). The extraradical hyphae of AMF can extend into organic patches away from the roots to acquire nutrients (Leigh et al., 2009; Hodge and Fitter, 2010). Although AMF lack the lytic exo-enzymatic repertoire to exploit organic patches alone (Joner and Johansen, 2000; Tisserant et al., 2012; Tisserant et al., 2013), they can influence the decomposition of plant residues by interacting with saprophytic microorganisms (Nuccio et al., 2013) or impacting the soil structure (Rillig and Mummey, 2006). Accumulating evidence suggests that AMF promote the decomposition and N release of residues and acquire the released N for their own growth or transfer some to the host plant (Hodge et al., 2001; Hodge and Fitter, 2010). However, results regarding the release of carbon in crop residues affected by AMF are inconsistent (Hodge et al., 2001; Hodge, 2014; Verbruggen et al., 2016), mainly due to the differences in the C/N and/or the labile-to-recalcitrant fraction of carbon (Cornwell et al., 2008; Hattenschwiler et al., 2011). Generally, it is acknowledged that AMF stimulate the decomposition of fresh residues, whereas for older or degraded residues, AMF tend to inhibit decomposition and slow down the release of DOC by promoting soil aggregates (Hodge et al., 2001; Leifheit et al., 2015; Verbruggen et al., 2016). Taken together, the changes in C and N substrate availability during residue decomposition affected by AMF can trigger cascading effects on soil nitrification and denitrification. However, it remains unclear 1) whether AMF-mediated N2O emission is related to the decomposition of plant residues and 2) the microbial mechanisms of AMF regulating N2O emission during the residue decomposition. Here, we conducted a mesocosm experiment to assess the impact of AMF on N2O emissions from the organic patches after approximately 30 days of crop residue degradation. Soil mineral nitrogen and the abundance of key genes involved in nitrification and denitrification were also analyzed. Given that AMF promote N absorption and slow down the release of DOC from older or degraded residues, we hypothesized that i) AMF reduce the availability of nitrogen substrates in organic patches and slow down the release of DOC from degraded residues and ii) AMF inhibit nitrification and denitrification due to the low availability of C and N, thereby reducing N2O emissions in the organic patches.
The soil was collected from a long-term experimental field at the Quzhou Experimental Station (36.87°N, 115.02°E) in Quzhou County, Hebei Province, North China. The soil is classified as a Cambisol with a silt loam texture. The background soil properties were as follows: pH 7.24 (H2O), 7.95 g kg−1 organic carbon (SOC), 0.90 g kg−1 total N (TN), 12.01 mg kg−1 Olsen-P (AP), and 176.2 mg kg−1 available K (AK) (Zhang et al., 2016). The collected soil was sieved through a 2-mm sieve, air-dried, and mixed with sand to a soil-to-sand ratio of 1:1 (m/m) as the cultivated medium. The medium was gamma-irradiated with a maximum dose of 32 kGy to eliminate indigenous AMF. To ensure nutrient supply, 100 mg kg−1 of N (calcium nitrate tetrahydrate), 20 mg kg−1 of P (monopotassium phosphate), and 100 mg kg−1 of K (monopotassium phosphate) were added to the medium prior to the experiment.
The experiment followed a full factorial design with two factors: 1) patch types: organic patch and control patch and 2) inoculation treatments: inoculated with AMF (+AMF) and without AMF (−AMF).
Mesocosm units were constructed by two joining compartments (one host compartment, 3 × 10 × 15 cm3; one hyphal compartment, 10 × 10 × 15 cm3) via a 30-μm mesh that allowed hyphae, but not plant roots, access to the hyphal compartment (Supplementary Figure 1). The host compartment contained 450 g of the sterilized medium and 50 g of inoculum of Funneliformis mosseae in the +AMF treatment or 50 g of the sterilized inoculum with microbial filtrate from the unsterilized inoculum in the −AMF treatment. Funneliformis mosseae is a common AMF in Chinese agricultural soils. The inoculum consisted of spores, mycelium, and fine root segments and was propagated using the same cultivated medium and planted with Zea mays L. for 4 months in a greenhouse. The hyphal compartment contained 1,500 g of the sterilized medium. A sterile PVC pipe (3.3 cm wide, 11 cm high) was inserted into the hyphal compartment 3 cm from the host compartment and 4 cm from the base of the hyphal compartment. Two opposing aperture windows (2.5 cm wide, 4 cm high) were opened 1 cm from the base of the PVC pipe. These windows and the base of the pipe were covered with a 30-μm mesh (Supplementary Figure 1).
Two pregerminated maize seeds were grown in each host compartment and thinned to one plant after emergence. The patch was added to the PVC pipe 29 days after maize planting. The patch materials contained 14 g of the sterilized medium mixed with 7 g of milled dried roots of maize plants (total carbon 40.62%, total N 1.43%, C:N ratio 28.41) as the organic patch or 50 g of the sterilized medium as the control patch (to ensure equal volume with the organic patch). Then, 5 mL of microbial filtrate was pipetted into each patch to equalize the initial microbial communities. The microbial filtrate was produced from the same fresh field soil according to the previously described method (Koide and Li, 1989; van der Heijden et al., 2006). The mesocosm experiment was conducted in a greenhouse at China Agricultural University, Beijing. Soil moisture was maintained at approximately 50%–60% of water-filled pore space (WFPS) with deionized water.
Thirty days after patch addition, the PVC pipe was sealed at the top with a rubber plug that included a syringe needle and a three-way valve to form a gas sampling port (5 cm high). The total internal volume of the gas port was approximately 35 cm3 (Supplementary Figure 1). Two days after sealing the pipe, the gas within the pipe was sampled with a syringe through the three-way valve. A 10-cm3 gas sample was slowly removed from the pipe, waiting for an additional 5 s to allow the sample to mix inside the syringes before removing the syringes. The N2O concentrations of the gas samples were determined using a model 7890A gas chromatograph (Agilent, Santa Clara, CA, USA).
Mesocosms were destructively sampled after gas sampling. Samples of patches in the hyphal compartment were collected and divided into three portions: one for the determination of soil gravimetric water content, mineral nitrogen, and DOC and total dissolved nitrogen (TDN) contents was stored at 4°C; one for the analysis of total carbon (TC) and TN contents and mycorrhizal hyphal length density (HLD) was stored at room temperature; and the remaining soil for molecular analysis was immediately frozen with liquid nitrogen and stored at −80°C.
In the host compartment, the shoots of the maize plants were removed from the soil surface. The roots of the plants were extracted from the medium, washed, and divided into three portions: a fraction for the estimation of AMF root colonization was soaked in 50% ethanol, and the rest of the roots and shoots were dried (65°C; 48 h) for the determination of N contents and dry weights.
The gravimetric water content of the patches was measured using the oven-dried method. The ammonium (NH4+-N) and nitrate (NO3−-N) contents of the patches were measured using a continuous flow analysis (TRAACS 2000, Bran and Luebbe, Norderstedt, Germany) after the fresh patch samples were extracted with 0.01 mol L−1 of CaCl2 solution (Peng et al., 2013). DOC and TDN in the extracts were measured by infrared spectrometry after combustion at 850°C. The total N and C contents of the plant shoots, roots, and patch samples were determined using a FLASH Elemental Analyzer 1112 (Thermo Finnigan, Waltham, MA, USA) after the samples were ground in a ball mill.
The subsamples of roots were cleared with 10% KOH, acidified with 1% HCl, and stained with 0.05% trypan blue (Phillips and Hayman, 1970). Thirty pieces of roots from each sample were randomly selected, arranged on slides, and then mounted under a light microscope (Axioplan, Zeiss, Germany). The AMF root colonization rate was determined using the magnified intersection method (Mcgonigle et al., 1990).
The HLD of AMF in patches was determined by a modified aqueous extraction and membrane filter technique using air-dried samples of patches in triplicate (Jakobsen et al., 1992). Hyphal length density was calculated according to the modified Newman formula for calculating root length (Tennant, 1975).
The potential extracellular enzyme activities of the patch samples were measured using fluorogenically labeled substrates (Bell et al., 2013). Five fluorogenic enzyme substrates based on 4-methylumbelliferone (MUF) were used to assess enzyme activities: MUB-α-D-glucopyranoside for α-1,4-glucosidase, MUF-β-D-glucopyranoside for β-1,4-glucosidase, MUF-β-D-cellobioside for β-cellobiosidase, MUB-β-D-xylopyranoside for β-1,4-xylosidase, and MUB-N-acetyl-β-D-glucosaminide dehydrate for β-1,4-N-acetylglucosaminidase. L-Leucine-7-amido-4-methylcoumarin was used to analyze the activity of leucine amino peptidase. Briefly, fresh patch samples (equivalent to 1 g dry weight) were suspended in 100 mL of 50 mM sodium acetate buffer (pH 6.0) by shaking for 30 min, and then 200 μL of the suspensions and 50 μL of a 200-μM substrate (labeled with the MUB) were pipetted into a 96-well microplate and incubated in the dark at 25°C for 4 h. All solutions and suspensions were prepared with sterile water. A negative control was arranged with blanks (without soil suspension). After a 4-h incubation period, the microplates were measured fluorometrically with an automated luminescence spectrophotometer (FLx800 microplate, Bio-Tek Instruments, Winooski, VT, USA; emission at 450 nm, extinction at 345 nm). After correcting for the negative control and quenching effects, enzyme activities were expressed in units of nmol h−1 g−1 dry sample as MUB release.
The genomic DNA was extracted from fresh patch samples using the Fast DNA® SPIN Kit for soil (MP Biomedicals, Cleveland, OH, USA) according to the manufacturer’s instructions. The quantity and quality of DNA were checked using a Nanodrop spectrophotometer (Thermo Fisher, Waltham, MA, USA). Real-time quantitative polymerase chain reactions (qPCRs) of the bacterial and archaeal amoA genes involved in nitrification and nirK, nirS, and nosZ genes involved in denitrification were conducted using a CFX96 Optical Real-Time detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each reaction system was performed in a 20-µL solution containing 10 µL of SYBR Premix Ex Taq (Tli RNaseH Plus, 2×, Takara Bio, Shiga, Japan), 1 µL of DNA template, 0.5 µL of each primer (10 mM), and 8 µL of ddH2O. The qPCR primer pairs and thermal conditions are shown in Supplementary Table 1. Standard curves were obtained using a 10-fold dilution series (102–108 copies) of plasmid DNA containing target genes with known copy numbers. Each sample was performed in technical triplicate. The amplification efficiencies were 90%–105%, and the R2 value of the standard curves was 0.99–1.00 for all genes.
All data were first tested for normality and homogeneity of variances. To fulfill normality and homogeneity of variances, the copy numbers of N-cycling genes were log10(x)-transformed, and AM fungal variables (AMF root colonization rate and HLD) were transformed by square roots. The effects of patch types, inoculation treatments and their interactions on plant and AMF variables, patch N2O concentration, and gene copies were examined by two-way analysis of variance (ANOVA). The effects of patch types, inoculation treatments, and their interactions on soil physicochemical properties and activities of extracellular enzymes in patches which clearly did not fit the homogeneity of variances were examined by the Scheirer–Ray–Hare test. The effects of inoculation treatments on the variables within each patch type were determined by the two-tailed unpaired t-test. Multiple stepwise regression was employed to identify the most influential variables affecting N2O concentration. Independent variables include NH4+ and NO3– contents, as well as the copy numbers of N-cycling genes. To assess the overall extracellular enzyme activities, we calculate the standardized scores (Z scores) of six extracellular enzyme activities and sum them up to obtain a single index (Z value). Pearson correlation analysis was conducted to test the relationships between the copy numbers of N-cycling genes, HLD, physiochemical properties, and the Z value of extracellular enzyme activities. The above Scheirer–Ray–Hare test and Pearson correlation analysis were conducted using R version 4.0.4. The rest of the statistical analysis was performed using the SPSS statistical software package version 22 (SPSS, Inc., Chicago, IL, USA).
The roots of the maize plants were successfully colonized by AMF. The AMF root colonization rates were greatly increased in the +AMF treatment but not significantly affected by patch types (Figure 1A and Supplementary Table 2).
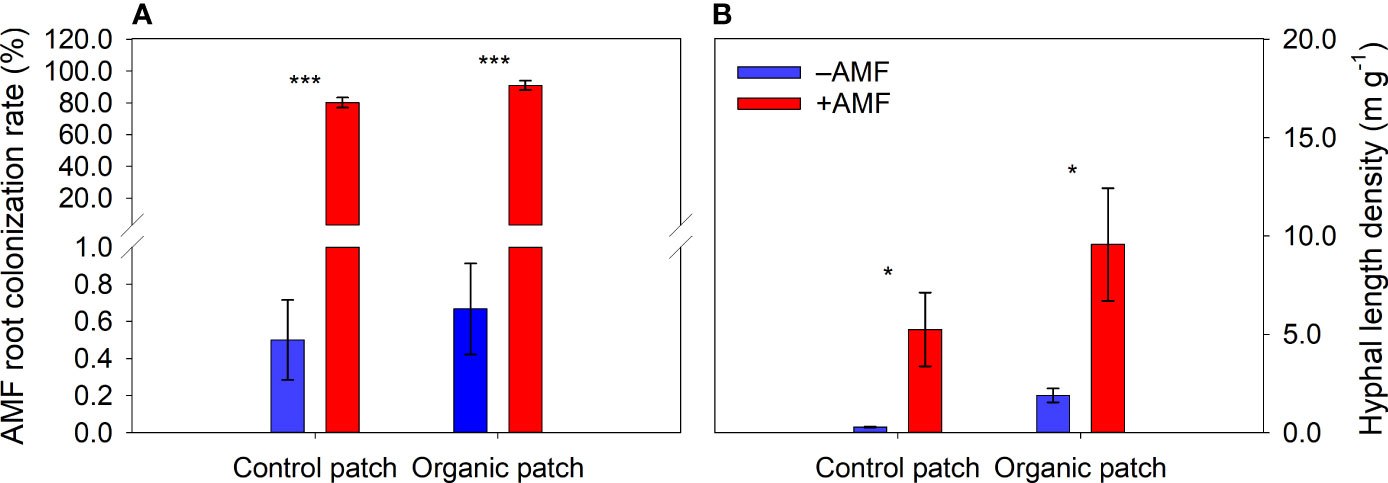
Figure 1 Arbuscular mycorrhizal fungi (AMF) root colonization rate of maize plants (A) and hyphal length density in patches (B) between +AMF and −AMF treatments under different patch types. Values represent means ± standard error, n = 4. +AMF and −AMF indicate the inoculation with and without Funneliformis mosseae in the host compartment, respectively. Asterisks indicate significant differences between +AMF and −AMF treatments within each patch type according to the two-tailed unpaired t-test (*, P < 0.05; ***, P < 0.001).
The HLD in the +AMF treatment was 7.41 m g−1 on average, which was significantly higher than that in the –AMF treatment, where a small number of extraradical hyphae were detected. The HLD values in the organic patches were higher than those in the control patches (Figure 1B and Supplementary Table 2).
The water, NH4+, DOC, TDN, TC, and TN contents in the organic patches were much higher than those in the control patches (Figures 2, 3 and Supplementary Table 3). AMF significantly reduced the NH4+, NO3−, DOC, TDN, and TN contents in the organic patches. In the control patches, only the NO3− and TDN contents were reduced by AMF. The patch water and TC contents were not affected by AMF, regardless of patch type (Figures 2, 3 and Supplementary Table 3).
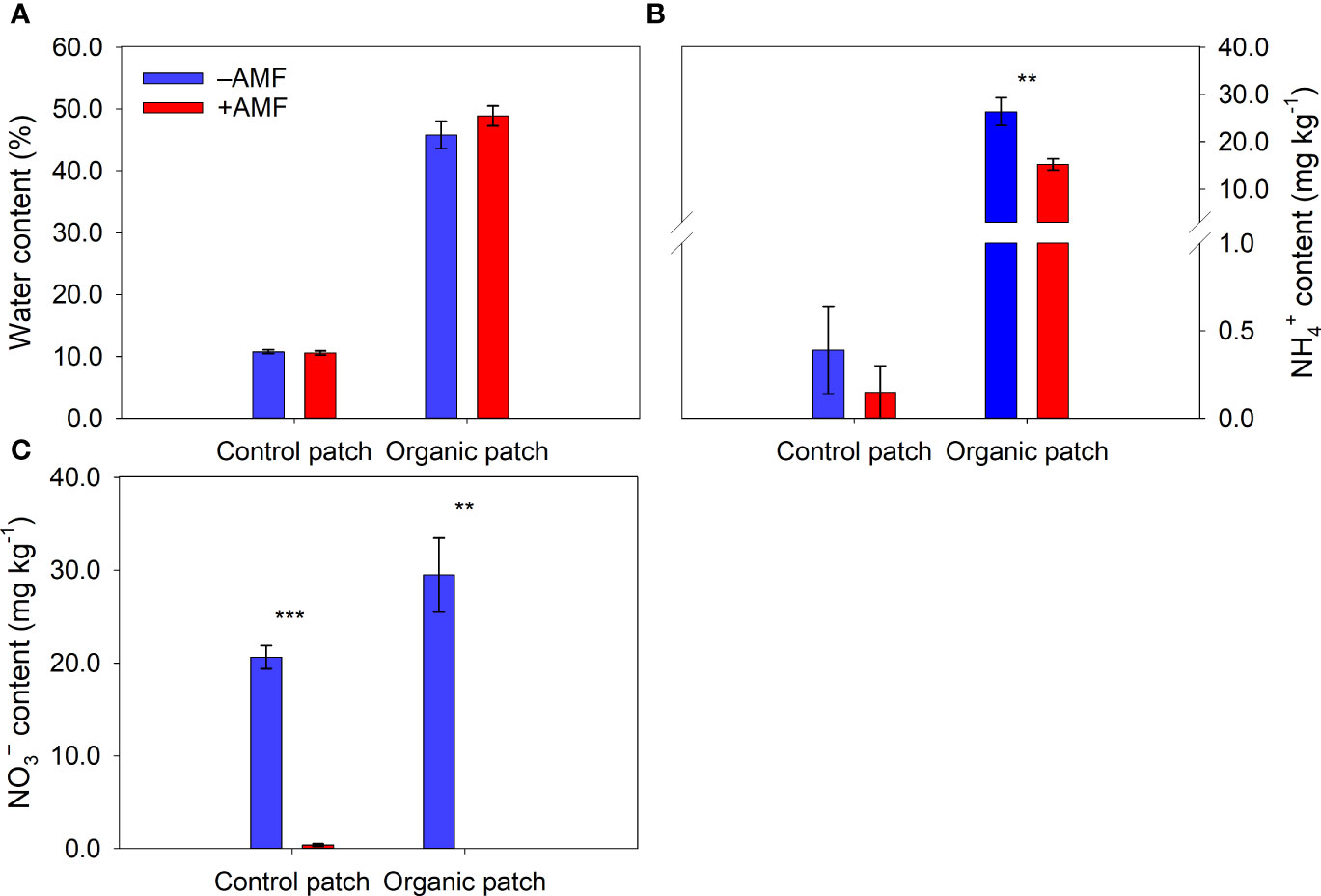
Figure 2 Water (A), NH4+-N (B), and NO3−-N (C) contents in patches of different types between +AMF and −AMF treatments. Values represent means ± standard error, n = 4. +AMF and −AMF indicate the inoculation with and without Funneliformis mosseae in the host compartment, respectively. Asterisks indicate significant differences between +AMF and −AMF treatments within each patch type according to the two-tailed unpaired t-test (**, P < 0.01; ***, P < 0.001).
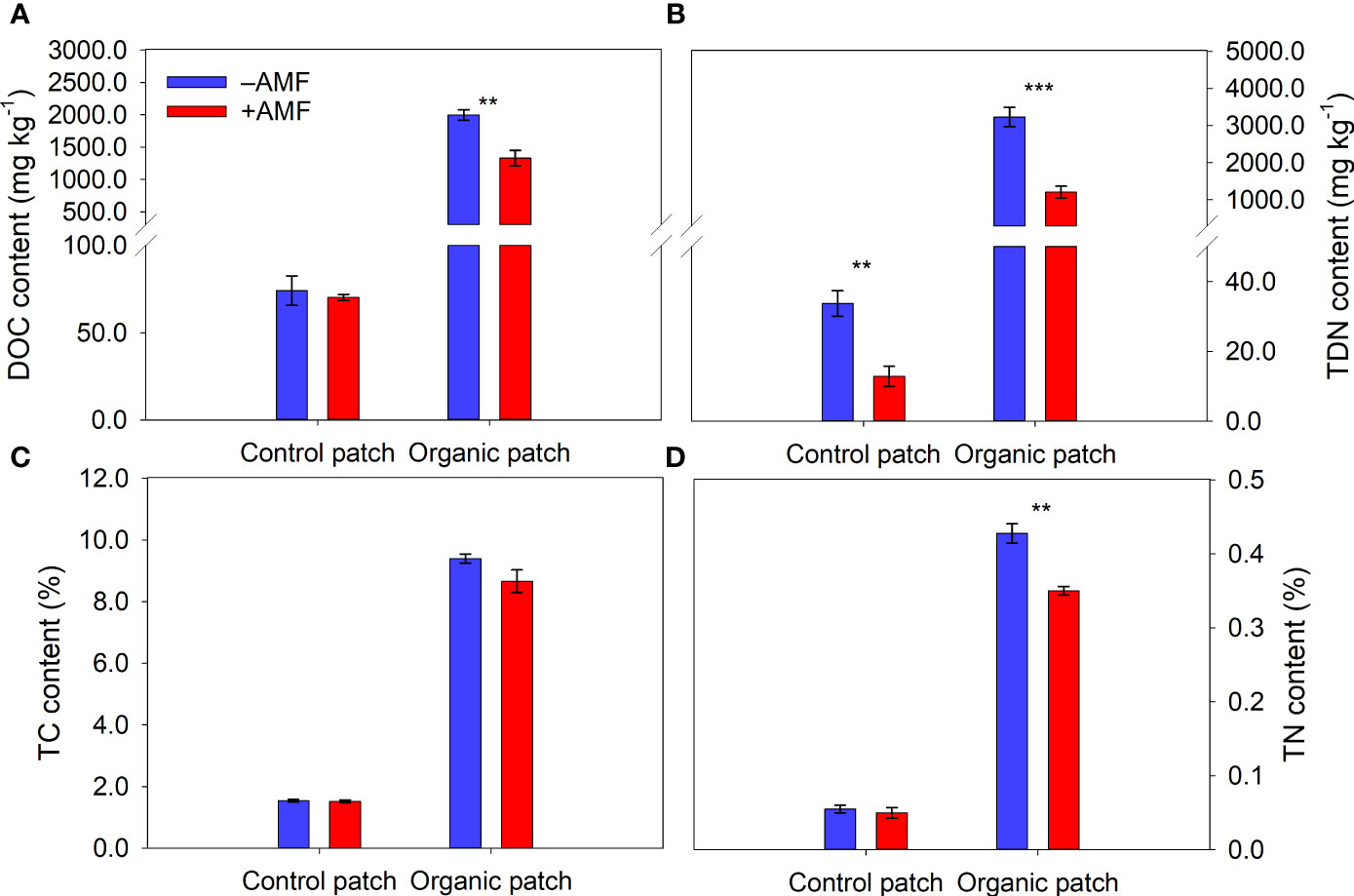
Figure 3 Dissolved organic carbon (DOC) (A), total dissolved nitrogen (TDN) (B), total carbon (TC) (C), and total nitrogen (TN) (D) in patches of different types between +AMF and −AMF treatments. Values represent means ± standard error, n = 4. +AMF and −AMF indicate the inoculation with and without Funneliformis mosseae in the host compartment, respectively. Asterisks indicate significant difference between +AMF and −AMF treatments within each patch type according to the two-tailed unpaired t-test (**, P < 0.01; ***, P < 0.001).
The activities of the six extracellular enzymes in the organic patches (α-1,4-glucosidase, β-1,4-glucosidase, β-cellobiosidase, β-1,4-xylosidase, β-1,4-N-acetyl-glucosamidase, and leucine amino peptidase) were approximately 30–230 times higher than those in the control patches. AMF significantly reduced the activities of the six extracellular enzymes in the organic patches by 38% to 70%. In the control patches, the activities of α-1,4-glucosidase, β-1,4-glucosidase, and leucine amino peptidase were significantly reduced, while the activity of β-1,4-N-acetylglucosaminidase was increased by AMF. The activities of the other two extracellular enzymes (β-cellobiosidase and β-1,4-xylosidase) were not significantly affected by AMF in the control patches (Figure 4 and Supplementary Table 3).
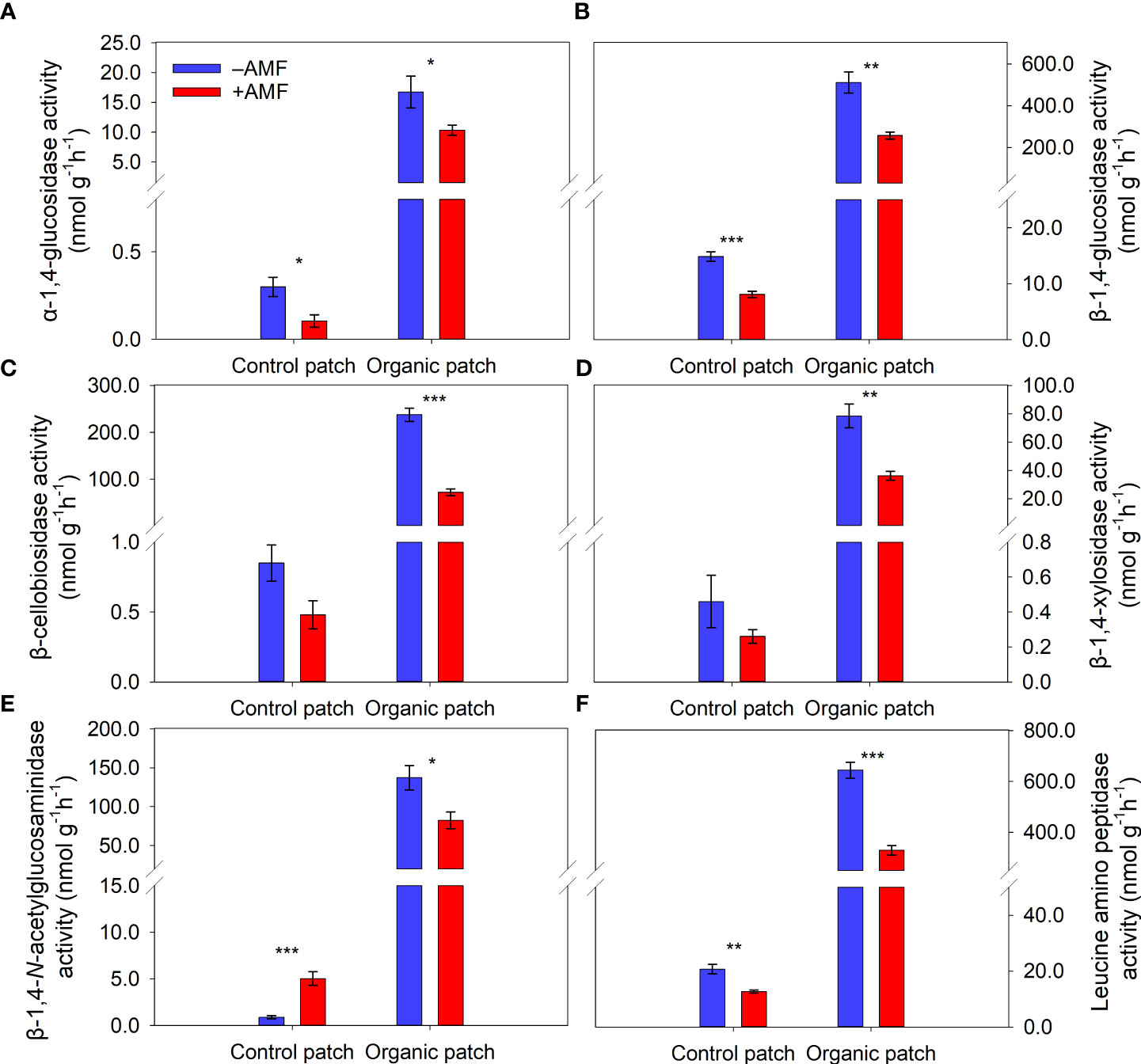
Figure 4 Activities of extracellular enzymes α-1,4-glucosidase (A), β-1,4-glucosidase (B), β-cellobiosidase (C), β-1,4-xylosidase (D), β-1,4-N-acetylglucosaminidase (E), and leucine amino peptidase (F) in patches of different types between +AMF and −AMF treatments. Values represent means ± standard error, n = 4. +AMF and −AMF indicate the inoculation with and without Funneliformis mosseae in the host compartment, respectively. Asterisks indicate significant differences between +AMF and −AMF treatments within each patch type according to the two-tailed unpaired t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
AMF significantly reduced the N2O concentration from the organic patches and did not significantly affect the N2O concentration from the control patches (Figure 5A and Supplementary Table 2). The abundance of the bacterial amoA gene ranged from 8.60 × 105 to 3.56 × 106 copies g−1 dry soil. The values were several times lower than those of the archaeal amoA gene (2.56 × 106 to 1.35 × 107 copies g−1 dry soil). In the organic patches, AMF significantly reduced the abundance of the bacterial amoA gene and did not affect the abundance of the archaeal amoA gene, while in the control patches, AMF significantly increased the abundance of the bacterial and archaeal amoA genes (Figures 5B, C and Supplementary Table 2).
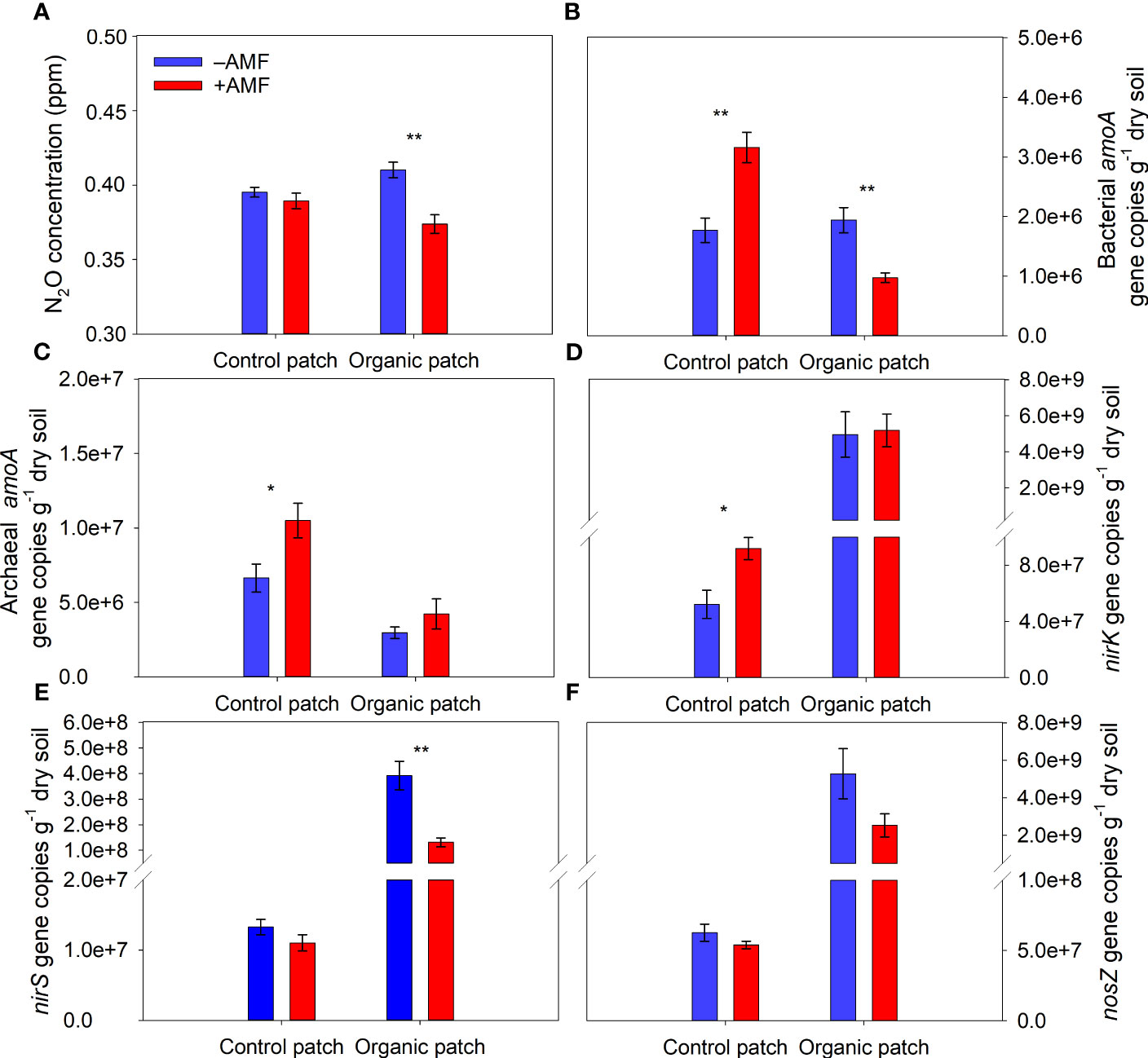
Figure 5 N2O concentration (A) and copies of the bacterial amoA (B), archaeal amoA (C), nirK (D), nirS (E), and nosZ (F) genes in patches of different types between +AMF and −AMF treatments. Values represent means ± standard error, n = 4. +AMF and −AMF indicate the inoculation with and without Funneliformis mosseae in the host compartment, respectively. Asterisks indicate significant differences between +AMF and −AMF treatments within each patch type according to the two-tailed unpaired t-test (*, P < 0.05; **, P < 0.01).
The abundance of the nirK gene ranged from 2.82 × 107 to 7.88 × 109 copies g−1 soil, which was approximately an order of magnitude higher than that of the nirS gene in patches (ranged from 7.58 × 106 to 5.50 × 108 copies g−1 soil). The abundance of the nirK, nirS, and nosZ genes in the organic patches was higher than that in the control patches. In the organic patches, AMF significantly reduced the nirS gene abundance by 67% on average and did not significantly affect the abundance of the nirK and nosZ genes. In the control patches, AMF significantly increased the nirK gene abundance and did not significantly affect the abundance of the nirS and nosZ genes (Figures 5D–F and Supplementary Table 2).
Multiple stepwise regression analysis showed that the abundance of the nirS gene alone was the main factor controlling the N2O concentration in the organic patches, explaining 68.2% of the variation in N2O concentration. In the control patches, no relevant independent variables were entered into the model (Supplementary Table 4). The abundance of the bacterial amoA and nirS genes was negatively correlated with HLD in the organic patches. In the control patches, HLD was positively correlated with the abundance of the bacterial amoA gene and showed no significant linear correlation with the nirS gene abundance (Figures 6A, C). The abundance of the bacterial amoA gene was positively correlated with NH4+ content (r = 0.72, P = 0.043), and the abundance of the nirS gene was positively correlated with DOC content (r = 0.76, P = 0.028) in the organic patches but not in the control patches (Figures 6B, D). The Z value of extracellular enzyme activities was positively correlated with DOC content in the organic patches (r = 0.75, P = 0.031; Figure 6E).
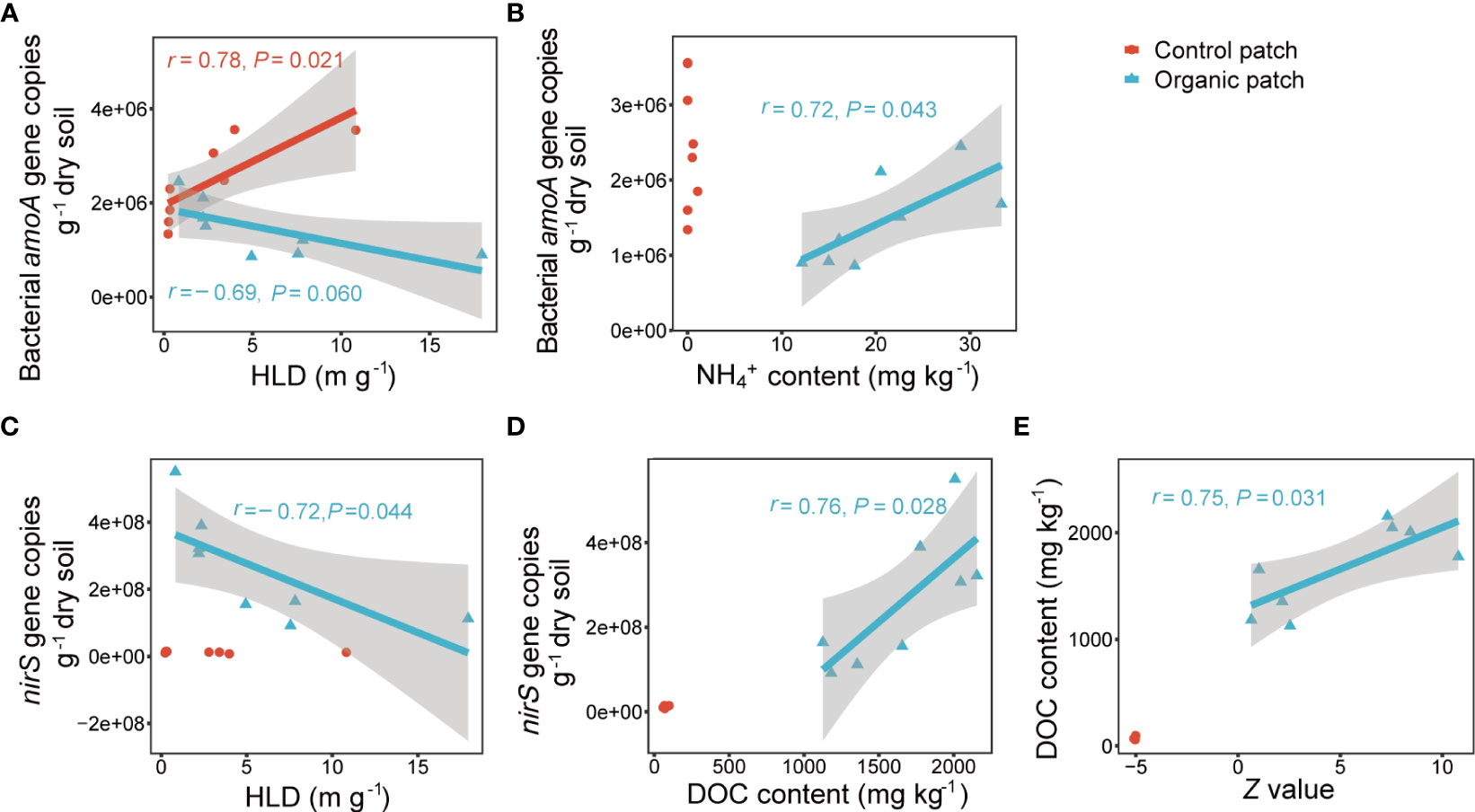
Figure 6 Correlations between the bacterial amoA gene copies with HLD (A) and NH4+ content (B), between nirS gene copies with HLD (C) and DOC content (D), and between DOC content with the Z value of extracellular enzyme activities (E). HLD indicates hyphal length density, DOC indicates dissolved organic carbon. Correlation analysis based on Pearson correlation coefficient. Gray shades visualize the 95% confidence interval and correlations with P < 0.1 are listed.
The shoot and root biomass and nitrogen uptake of maize plants were significantly affected by the inoculation treatment but not by patch type (Supplementary Table 5). AMF significantly increased the shoot and root biomass of host plants by 1.79 and 1.65 times, respectively. The shoot and root N contents of maize plants were not significantly affected by AMF except for a slight but significant decrease in shoot N content in the control patches. AMF also significantly enhanced the shoot and root N uptake of maize plants by 1.58 and 1.73 times, respectively (Supplementary Table 6).
AMF hyphae can penetrate into crop residue patches to search for patchily distributed nutrients and alter carbon and nitrogen cycling during the decomposition of crop residues (Hodge et al., 2001). Growing evidence shows that AMF reduce soil N2O emissions in the mycorrhizosphere or hyphosphere (Bender et al., 2014; Zhang et al., 2015; Storer et al., 2017; Gui et al., 2021; Zhao et al., 2021). However, the reasons for the decrease were variable and environmentally dependent. The successful colonization of extraradical hyphae in patches is a prerequisite for the effect of AMF on patch N2O emissions. The hyphal growth rate of Funneliformis mosseae after initial infection with host plants is only about 3 mm d–1 (Smith and Read, 2008), and time is needed for substantial hyphae to proliferate in patches. In the present study, the average HLD in the +AMF treatment after approximately 30 days of patch addition was 7.41 m g–1, much higher compared to the –AMF treatment (Figure 1B), which is consistent with previous reports (Hodge and Fitter, 2010; Gui et al., 2021). In the present study, AMF reduced N2O concentration from the organic patches after approximately 30 days of patch addition (Figure 5A), when crop residues in the organic patches had undergone a period of rapid degradation (Wu et al., 2019). This reduction in N2O emissions from degraded residue patches by AMF could potentially be attributed to the decreased abundance of nirS-type denitrifiers and ammonia-oxidizing bacteria, as well as the decrease in N-substrate availability, which will be discussed in detail below.
AMF significantly reduced the abundance of the nirS gene in organic patches which negatively correlated with HLD (Figures 5E, 6C). Multiple stepwise regression analysis suggested that the abundance of the nirS gene best explained the decrease of N2O concentration from organic patches (Supplementary Table 4). Denitrification is carried out by microbes as an alternate respiration to conserve energy under O2 constraints (Zumft, 1997), with either organic carbon (for heterotrophic denitrification) or reduced inorganic compounds (for autotrophic denitrification) as electron donors (Korom, 1992). In the present study, the DOC content in the organic patches was significantly decreased by AMF and positively correlated with the abundance of the nirS gene (Figures 3A, 6D). This result indicates that the growth of the nirS-type denitrifiers may be limited by the availability of organic carbon, which is a result of AMF interference in residue degradation. This finding is inconsistent with a previous study that found that AMF increased the content of DOC in patches, possibly due to hyphal exudates (Li et al., 2023). The possible explanation for this inconsistency is that the ratio of crop residues in the present study was 33%, which is much higher than in the previous study (1%), and the contribution of carbon released from degraded residues outweighs the carbon exuded by AMF hyphae in the patch DOC pool.
In the present study, inoculation with AMF significantly reduced all tested extracellular enzyme activities in the organic patches (Figure 4). This demonstrates that the presence of AMF inhibits the activity of decomposers in degraded residue after approximately 30 days of incubation. This finding is consistent with previous studies in which AMF suppressed the decomposition of woody plant litters and wheat stubbles after incubation for 3 months in the field (Leifheit et al., 2015; Verbruggen et al., 2016). In contrast, AMF have also been reported to accelerate litter decomposition (Hodge et al., 2001). The discrepancy may be attributed to the quality, quantity, composition, and decomposing time of the residues. AMF could initially stimulate decomposers by providing hyphal exudates in the early stages of residue decomposition (Herman et al., 2012; Kaiser et al., 2015). Over time, AMF hyphae efficiently scavenge mineral nutrients, such as N released from the organic patches, and potentially suppress these decomposers by competing substrates (Leifheit et al., 2015). This is consistent with lower N availability including NH4+, NO3−, TDN, and TN in the organic patches due to AMF in the present study (Figures 2, 3). The low decomposition rate in the presence of AMF may mitigate the release of DOC in plant residues and offer great potential to occlude C within soil aggregates (Leifheit et al., 2015; Verbruggen et al., 2016). The decrease in DOC content by AMF in the organic patches, which is positively correlated with the Z value of extracellular enzyme activities, supports this speculation (Figures 3A, 6E). However, AMF only significantly decreased the abundance of the nirS gene, but not the nirK gene, in the organic patches (Figures 5D, E). Compared with nirK-type denitrifiers, nirS-type denitrifiers are more sensitive to DOC, and their populations increase with soil DOC content within a certain range. Meanwhile, nirK-type denitrifiers have a broader profile of carbon substrate utilization and do not significantly respond to alterations in soil DOC content (Hou et al., 2018; Langarica-Fuentes et al., 2018). In addition, one limitation of the study is that it did not adequately reflect the impact of AMF on the early residue degradation phase. Future studies should focus on improving the experimental design and the dynamic monitoring throughout the entire cultivation period to obtain a more comprehensive understanding of the impact of AMF on residue degradation.
In the present study, AMF significantly decreased NH4+ and NO3− contents in the organic patches resulting in low N-substrate availability to nitrifying and denitrifying microbes (Figure 2). AMF can detect, proliferate, and acquire N from decomposing organic matter such as clover, cellulose, albumin, DNA, and chitin (Bukovska et al., 2018) and, consequently, utilize this N primarily for their own growth and transfer some to the host plant. This is supported by the significantly improved N uptake of maize plants in the presence of AMF (Supplementary Table 6). It is estimated that nearly a third of the total N detected in the AMF hyphae is derived from the organic patches (Hodge and Fitter, 2010).
In the present study, AMF hyphae significantly reduced the abundance of the bacterial amoA gene in the organic patches (Figure 5B), which is consistent with previous studies that showed AMF suppressed AOB and soil potential nitrification rates (Veresoglou et al., 2011; Bukovska et al., 2018). AMF can absorb NH4+, NO3−, and some simple organic N from the soil, with a preference for NH4+ (Govindarajulu et al., 2005; Tanaka and Yano, 2005). AOB are a group of slow-growing microbes and generally thought to be weak competitors for NH4+ compared with AMF (Bollmann et al., 2002; Storer et al., 2017). In the present study, AMF hyphae proliferated extensively in the organic patches and increased the N uptake of maize plants (Figure 1B and Supplementary Table 6). The NH4+ content in the organic patches decreased nearly by 50% in the presence of AMF, which was also positively correlated with the abundance of the bacterial amoA gene (Figures 2B, 6B). This demonstrates that AMF outclassed AOB in acquiring NH4+ in degraded residue patches and subsequently suppressed the growth of AOB. In contrast to the organic patches, AOB and AOA abundance was promoted by AMF in the control patches (Figures 5B, C), which is consistent with increased AOB abundance after urea application by native AMF in a pot experiment (Teutscherova et al., 2019). A possible explanation is that AMF promoted soil aeration by improving soil aggregation (Rillig and Mummey, 2006; Morris et al., 2019).
Crop residues serve as hotspots of N2O emissions in agriculture. Here, we showed that AMF inhibited all the tested extracellular enzyme activities in degraded residue patches. AMF hyphae significantly reduced N2O emissions, the abundance of the bacterial amoA and nirS genes (functional genes involved in N2O production), and N-substrate availability in degraded residue patches after approximately 30 days of incubation. Additionally, the decrease of the nirS gene abundance by AMF had substantial consequences for reduced N2O emissions in degraded residue patches and was positively correlated with DOC contents. However, the reduction of AMF abundance and diversity in intensive agricultural practices might have cascading effects on residue decomposition and N cycling. Agricultural management strategies aimed at strengthening hyphal networks for sustainable agriculture will reinforce the interaction between AMF and the N-cycling microbiome, leading to increased N uptake, carbon sequestration, and mitigation of N2O emissions.
XL and RZ conceived the research and drafted the initial version of the manuscript. XL and GH conducted the experiments. SB and GY performed the data analysis. All authors contributed to the discussion and writing of the manuscript. All author(s) read and approved the final manuscript.
This study was funded by the National Natural Science Foundation of China (Grant nos. 31872182 and 42007032); the Scientific and Technological Cooperation and Exchange Project of Shanxi Province (202204041101018); the Key Research and Development Project of Pingcheng District, Datong City (202003); and the Doctoral Research Project of Shanxi Datong University (2022-B-02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1224849/full#supplementary-material
Arias P., Bellouin N., Coppola E., Jones R., Krinner G., Marotzke J., et al. (2021). Climate change 2021: The physical science basis. Contribution of working group I to the sixth assessment report of the intergovernmental panel on climate change. Cambridge University Press. Available at: https://www.ipcc.ch/report/ar6/wg1/.
Bell C. W., Fricks B. E., Rocca J. D., Steinweg J. M., McMahon S. K., Wallenstein M. D. (2013). High-throughput fluorometric measurement of potential soil extracellular enzyme activities. Jove-Journal Visualized Experiments, 81, e50961. doi: 10.3791/50961
Bender S. F., Conen F., van der Heijden M. G. A. (2015). Mycorrhizal effects on nutrient cycling, nutrient leaching and N2O production in experimental grassland. Soil Biol. Biochem. 80, 283–292. doi: 10.1016/j.soilbio.2014.10.016
Bender S. F., Plantenga F., Neftel A., Jocher M., Oberholzer H. R., Kohl L., et al. (2014). Symbiotic relationships between soil fungi and plants reduce N2O emissions from soil. ISME J. 8, 1336–1345. doi: 10.1038/ismej.2013.224
Bollmann A., Bar-Gilissen M. J., Laanbroek H. J. (2002). Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68, 4751–4757. doi: 10.1128/Aem.68.10.4751-4757.2002
Bukovska P., Bonkowski M., Konvalinkova T., Beskid O., Hujslova M., Puschel D., et al. (2018). Utilization of organic nitrogen by arbuscular mycorrhizal fungi-is there a specific role for protists and ammonia oxidizers? Mycorrhiza 28, 269–283. doi: 10.1007/s00572-018-0825-0
Butterbach-Bahl K., Baggs E. M., Dannenmann M., Kiese R., Zechmeister-Boltenstern S. (2013). Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Philos. Trans. R. Soc. B-Biological Sci. 368, 20130122. doi: 10.1098/rstb.2013.0122
Cavagnaro T. R., Barrios-Masias F. H., Jackson L. E. (2012). Arbuscular mycorrhizas and their role in plant growth, nitrogen interception and soil gas efflux in an organic production system. Plant Soil 353, 181–194. doi: 10.1007/s11104-011-1021-6
Chapuis-Lardy L., Wrage N., Metay A., Chotte J.-L., Bernoux M. (2007). Soils, a sink for N2O? A review. Global Change Biol. 13, 1–17. doi: 10.1111/j.1365-2486.2006.01280.x
Cornwell W. K., Cornelissen J. H. C., Amatangelo K., Dorrepaal E., Eviner V. T., Godoy O., et al. (2008). Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecol. Lett. 11, 1065–1071. doi: 10.1111/j.1461-0248.2008.01219.x
Dai Y. J., Sun Q. Y., Wang W. S., Lu L., Liu M., Li J. J., et al. (2018). Utilizations of agricultural waste as adsorbent for the removal of contaminants: A review. Chemosphere 211, 235–253. doi: 10.1016/j.chemosphere.2018.06.179
Dobbie K. E., McTaggart I. P., Smith K. A. (1999). Nitrous oxide emissions from intensive agricultural systems: Variations between crops and seasons, key driving variables, and mean emission factors. J. Geophys. Research-Atmospheres 104, 26891–26899. doi: 10.1029/1999jd900378
Govindarajulu M., Pfeffer P. E., Jin H. R., Abubaker J., Douds D. D., Allen J. W., et al. (2005). Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435, 819–823. doi: 10.1038/nature03610
Gui H., Gao Y., Wang Z., Shi L., Yan K., Xu J. (2021). Arbuscular mycorrhizal fungi potentially regulate N2O emissions from agricultural soils via altered expression of denitrification genes. Sci. Total Environ. 774, 145133. doi: 10.1016/j.scitotenv.2021.145133
Hallin S., Philippot L., Löffler F. E., Sanford R. A., Jones C. M. (2018). Genomics and ecology of novel N2O-reducing microorganisms. Trends Microbiol. 26, 43–55. doi: 10.1016/j.tim.2017.07.003
Hattenschwiler S., Coq S., Barantal S., Handa I. T. (2011). Leaf traits and decomposition in tropical rainforests: revisiting some commonly held views and towards a new hypothesis. New Phytol. 189, 950–965. doi: 10.1111/j.1469-8137.2010.03483.x
Herman D. J., Firestone M. K., Nuccio E., Hodge A. (2012). Interactions between an arbuscular mycorrhizal fungus and a soil microbial community mediating litter decomposition. FEMS Microbiol. Ecol. 80, 236–247. doi: 10.1111/j.1574-6941.2011.01292.x
Hodge A. (2014). Interactions between arbuscular mycorrhizal fungi and organic material substrates. Adv. Appl. Microbiol. 89, 47–99. doi: 10.1016/B978-0-12-800259-9.00002-0
Hodge A., Campbell C. D., Fitter A. H. (2001). An arbuscular mycorrhizal fungus accelerates decomposition and acquires nitrogen directly from organic material. Nature 413, 297–299. doi: 10.1038/35095041
Hodge A., Fitter A. H. (2010). Substantial nitrogen acquisition by arbuscular mycorrhizal fungi from organic material has implications for N cycling. Proc. Natl. Acad. Sci. United States America 107 (31), 13754–13759. doi: 10.1016/B978-0-12-800259-9.00002-0
Hou S., Ai C., Zhou W., Liang G., He P. (2018). Structure and assembly cues for rhizospheric nirK- and nirS-type denitrifier communities in long-term fertilized soils. Soil Biol. Biochem. 119, 32–40. doi: 10.1016/j.soilbio.2018.01.007
Jakobsen I., Abbott L. K., Robson A. D. (1992). External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L.: 1. Spread of hyphae and phosphorus inflow into roots. New Phytol. 120, 371–380. doi: 10.1111/j.1469-8137.1992.tb01077.x
Joner E. J., Johansen A. (2000). Phosphatase activity of external hyphae of two arbuscular mycorrhizal fungi. Mycological Res. 104, 81–86. doi: 10.1017/s0953756299001240
Kaiser C., Kilburn M. R., Clode P. L., Fuchslueger L., Koranda M., Cliff J. B., et al. (2015). Exploring the transfer of recent plant photosynthates to soil microbes: mycorrhizal pathway vs direct root exudation. New Phytol. 205, 1537–1551. doi: 10.1111/nph.13138
Koide R. T., Li M. G. (1989). Appropriate controls for vesicular arbuscular mycorrhiza research. New Phytol. 111, 35–44. doi: 10.1111/j.1469-8137.1989.tb04215.x
Korom S. F. (1992). Natural denitrification in the saturated zone - a review. Water Resour. Res. 28, 1657–1668. doi: 10.1029/92wr00252
Kravchenko A. N., Toosi E. R., Guber A. K., Ostrom N. E., Yu J., Azeem K., et al. (2017). Hotspots of soil N2O emission enhanced through water absorption by plant residue. Nat. Geosci. 10, 496–500. doi: 10.1038/ngeo2963
Langarica-Fuentes A., Manrubia M., Giles M. E., Mitchell S., Daniell T. J. (2018). Effect of model root exudate on denitrifier community dynamics and activity at different water-filled pore space levels in a fertilised soil. Soil Biol. Biochem. 120, 70–79. doi: 10.1016/j.soilbio.2018.01.034
Lazcano C., Barrios-Masias F. H., Jackson L. E. (2014). Arbuscular mycorrhizal effects on plant water relations and soil greenhouse gas emissions under changing moisture regimes. Soil Biol. Biochem. 74, 184–192. doi: 10.1016/j.soilbio.2014.03.010
Leifheit E. F., Verbruggen E., Rillig M. C. (2015). Arbuscular mycorrhizal fungi reduce decomposition of woody plant litter while increasing soil aggregation. Soil Biol. Biochem. 81, 323–328. doi: 10.1016/j.soilbio.2014.12.003
Leigh J., Hodge A., Fitter A. H. (2009). Arbuscular mycorrhizal fungi can transfer substantial amounts of nitrogen to their host plant from organic material. New Phytol. 181, 199–207. doi: 10.1111/j.1469-8137.2008.02630.x
Li X., Zhao R., Li D., Wang G., Bei S., Ju X., et al. (2023). Mycorrhiza-mediated recruitment of complete denitrifying Pseudomonas reduces N2O emissions from soil. Microbiome 11, 1–18. doi: 10.1186/s40168-023-01466-5
Mcgonigle T. P., Miller M. H., Evans D. G., Fairchild G. L., Swan J. A. (1990). A new method which gives an objective-measure of colonization of roots by vesicular arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x
Morris E. K., Morris D. J. P., Vogt S., Gleber S. C., Bigalke M., Wilcke W., et al. (2019). Visualizing the dynamics of soil aggregation as affected by arbuscular mycorrhiz. ISME J. 13, 1639–1646. doi: 10.1038/s41396-019-0369-0
Mosier A., Kroeze C., Nevison C., Oenema O., Seitzinger S., van Cleemput O. (1998). Closing the global N2O budget: nitrous oxide emissions through the agricultural nitrogen cycle. Nutrient Cycling Agroecosystems 52, 225–248. doi: 10.1023/a:1009740530221
Nuccio E. E., Hodge A., Pett-Ridge J., Herman D. J., Weber P. K., Firestone M. K. (2013). An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Environ. Microbiol. 15, 1870–1881. doi: 10.1111/1462-2920.12081
Parkin T. B. (1987). Soil microsites as a source of denitrification variability. Soil Sci. Soc. America J. 51, 1194–1199. doi: 10.2136/sssaj1987.03615995005100050019x
Peng Y., Yu P., Li X., Li C. (2013). Determination of the critical soil mineral nitrogen concentration for maximizing maize grain yield. Plant Soil 372, 41–51. doi: 10.1007/s11104-013-1678-0
Phillips J. M., Hayman D. S. (1970). Improved procedures for clearing roots and staining parasitic and vesicular-arbuscularmycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycological Soc. 55, 158–160. doi: 10.1016/S0007-1536(70)80110-3
Prosser J. I., Nicol G. W. (2012). Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 20, 523–531. doi: 10.1016/j.tim.2012.08.001
Ravishankara A. R., Daniel J. S., Portmann R. W. (2009). Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125. doi: 10.1126/science.1176985
Rillig M. C., Mummey D. L. (2006). Mycorrhizas and soil structure. New Phytol. 171, 41–53. doi: 10.1111/j.1469-8137.2006.01750.x
Roberto C. M., Silva O., Josefine F. B., Gouveia P. L., Pinheiro L. I., Regina G. M., et al. (2018). Crop residue harvest for bioenergy production and its implications on soil functioning and plant growth: a review. Agriculture 75, 255–272. doi: 10.1590/1678-992x-2016-0459
Shen Y., Zhu B. (2021). Arbuscular mycorrhizal fungi reduce soil nitrous oxide emission. Geoderma 402, 115179. doi: 10.1016/j.geoderma.2021.115179
Storer K., Coggan A., Ineson P., Hodge A. (2017). Arbuscular mycorrhizal fungi reduce nitrous oxide emissions from N2O hotspots. New Phytol. 220, 1285–1295. doi: 10.1111/nph.14931
Tanaka Y., Yano K. (2005). Nitrogen delivery to maize via mycorrhizal hyphae depends on the form of N supplied. Plant Cell Environ. 28, 1247–1254. doi: 10.1111/j.1365-3040.2005.01360.x
Tennant D. (1975). A test of a modified line intersect method of estimating root length. J. Ecol. 6, 995–1001. doi: 10.2307/2258617
Teutscherova N., Vazquez E., Arango J., Arevalo A., Benito M., Pulleman M. (2019). Native arbuscular mycorrhizal fungi increase the abundance of ammonia-oxidizing bacteria, but suppress nitrous oxide emissions shortly after urea application. Geoderma 338, 493–501. doi: 10.1016/j.geoderma.2018.09.023
Tian H., Xu R., Canadell J. G., Thompson R. L., Winiwarter W., Suntharalingam P., et al. (2020). A comprehensive quantification of global nitrous oxide sources and sinks. Nature 586, 248–256. doi: 10.1038/s41586-020-2780-0
Tisserant E., Kohler A., Dozolme-Seddas P., Balestrini R., Benabdellah K., Colard A., et al. (2012). The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol. 193, 755–769. doi: 10.1111/j.1469-8137.2011.03948.x
Tisserant E., Malbreil M., Kuo A., Kohler A., Symeonidi A., Balestrini R., et al. (2013). Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. United States America 110, 20117–20122. doi: 10.1073/pnas.1313452110
van der Heijden M. G. A., Streitwolf-Engel R., Riedl R., Siegrist S., Neudecker A., Ineichen K., et al. (2006). The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland. New Phytol. 172, 739–752. doi: 10.1111/j.1469-8137.2006.01862.x
Verbruggen E., Jansa J., Hammer E. C., Rillig M. C., de Vries F. (2016). Do arbuscular mycorrhizal fungi stabilize litter-derived carbon in soil? J. Ecol. 104, 261–269. doi: 10.1111/1365-2745.12496
Veresoglou S. D., Chen B. D., Rillig M. C. (2012). Arbuscular mycorrhiza and soil nitrogen cycling. Soil Biol. Biochem. 46, 53–62. doi: 10.1016/j.soilbio.2011.11.018
Veresoglou S. D., Sen R., Mamolos A. P., Veresoglou D. S. (2011). Plant species identity and arbuscular mycorrhizal status modulate potential nitrification rates in nitrogen-limited grassland soils. J. Ecol. 99, 1339–1349. doi: 10.1111/j.1365-2745.2011.01863.x
Wu L., Zhang W. J., Wei W. J., He Z. L., Kuzyakov Y., Bol R., et al. (2019). Soil organic matter priming and carbon balance after straw addition is regulated by long-term fertilization. Soil Biol. Biochem. 135, 383–391. doi: 10.1016/j.soilbio.2019.06.003
Zhang Y., Li C., Wang Y., Hu Y., Christie P., Zhang J., et al. (2016). Maize yield and soil fertility with combined use of compost and inorganic fertilizers on a calcareous soil on the North China Plain. Soil Tillage Res. 155, 85–94. doi: 10.1016/j.still.2015.08.006
Zhang X., Wang L., Ma F., Shan D. (2015). Effects of arbuscular mycorrhizal fungi on N2O emissions from rice paddies. Water Air Soil pollut. 226, 1–10. doi: 10.1007/s11270-015-2493-4
Zhao R., Li X., Bei S., Li D., Li H., Christie P., et al. (2021). Enrichment of nosZ-type denitrifiers by arbuscular mycorrhizal fungi mitigates N2O emissions from soybean stubbles. Environ. Microbiol. 23, 6587–6602. doi: 10.1111/1462-2920.15815
Keywords: nitrous oxide, mycorrhizal fungi, crop residue, denitrification, extracellular enzyme activities
Citation: Li X, He G, Li D, Bei S, Luan D, Sun X, Yang G, Huo L, Zhen L and Zhao R (2023) Arbuscular mycorrhizal fungi reduce N2O emissions from degraded residue patches. Front. Ecol. Evol. 11:1224849. doi: 10.3389/fevo.2023.1224849
Received: 18 May 2023; Accepted: 07 July 2023;
Published: 04 August 2023.
Edited by:
Yuchuan Fan, University of Florida, United StatesReviewed by:
Xiaoliang Li, Chinese Academy of Tropical Agricultural Sciences, ChinaCopyright © 2023 Li, He, Li, Bei, Luan, Sun, Yang, Huo, Zhen and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruotong Zhao, YjIwMjAzMDMwMjU5QGNhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.