- Department of Infection Biology, Faculty of Infectious and Tropical Diseases, London School of Hygiene & Tropical Medicine, London, United Kingdom
Membrane-located NAD(P) transhydrogenase (NTH) catalyses reversible hydride ion transfer between NAD(H) and NADP(H), simultaneously translocating a proton across the membrane. The enzyme is structurally conserved across prokaryotes and eukaryotes. In heterotrophic bacteria NTH proteins reside in the cytoplasmic membrane, whereas in animals they localise in the mitochondrial inner membrane. Eukaryotic NTH proteins exists in two distinct configurations (isoforms) and have non-mitochondrial functions in unicellular eukaryotes like Plasmodium, the causative agent of malaria. In this study, we carried out a systematic analysis of nth genes across eukaryotic life to determine its prevalence and distribution of isoforms. The results reveal that NTH is found across all major lineages, but that some organisms, notably plants, lack nth genes altogether. Isoform distribution and phylogenetic analysis reveals different nth gene loss scenarios in apicomplexan lineages, which sheds new light on the evolution of the Piroplasmida and Eimeriidae.
1 Introduction
Proton-translocating NAD(P) transhydrogenase (NTH), also known as nicotinamide nucleotide transhydrogenase (NNT) or pyridine nucleotide transhydrogenase (Pnt), is an integral membrane protein that catalyses the reversible hydride ion transfer between NAD(H) and NADP(H), whilst simultaneously translocating a proton across the membrane in which it is embedded (Figure 1A) (Anderson and Fisher, 1981; Hou et al., 1990; Hatefi and Yamaguchi, 1996). The enzyme is found in both prokaryotes and eukaryotes and is structurally conserved, possessing three functional domains: domain I that binds NAD(H); domain II that is made up of the transmembrane (TM) helices and has proton translocating activity; and domain III that binds NADP(H) (Figure 1A). In animals, NTH proteins reside in the mitochondrial inner membrane, whereas in heterotrophic bacteria they localise in the cytoplasmic membrane (Hatefi and Yamaguchi, 1996; Jackson, 2003), possibly reflecting the evolutionary origin of the mitochondrion from a proteobacterial endosymbiont (Andersson et al., 1998; Gray et al., 1999). The physiological role of NTH is widely considered to be generation of NADPH (the reduced form of NADP), an important cofactor used for redox homeostasis and for NADPH-dependent enzymatic activity.
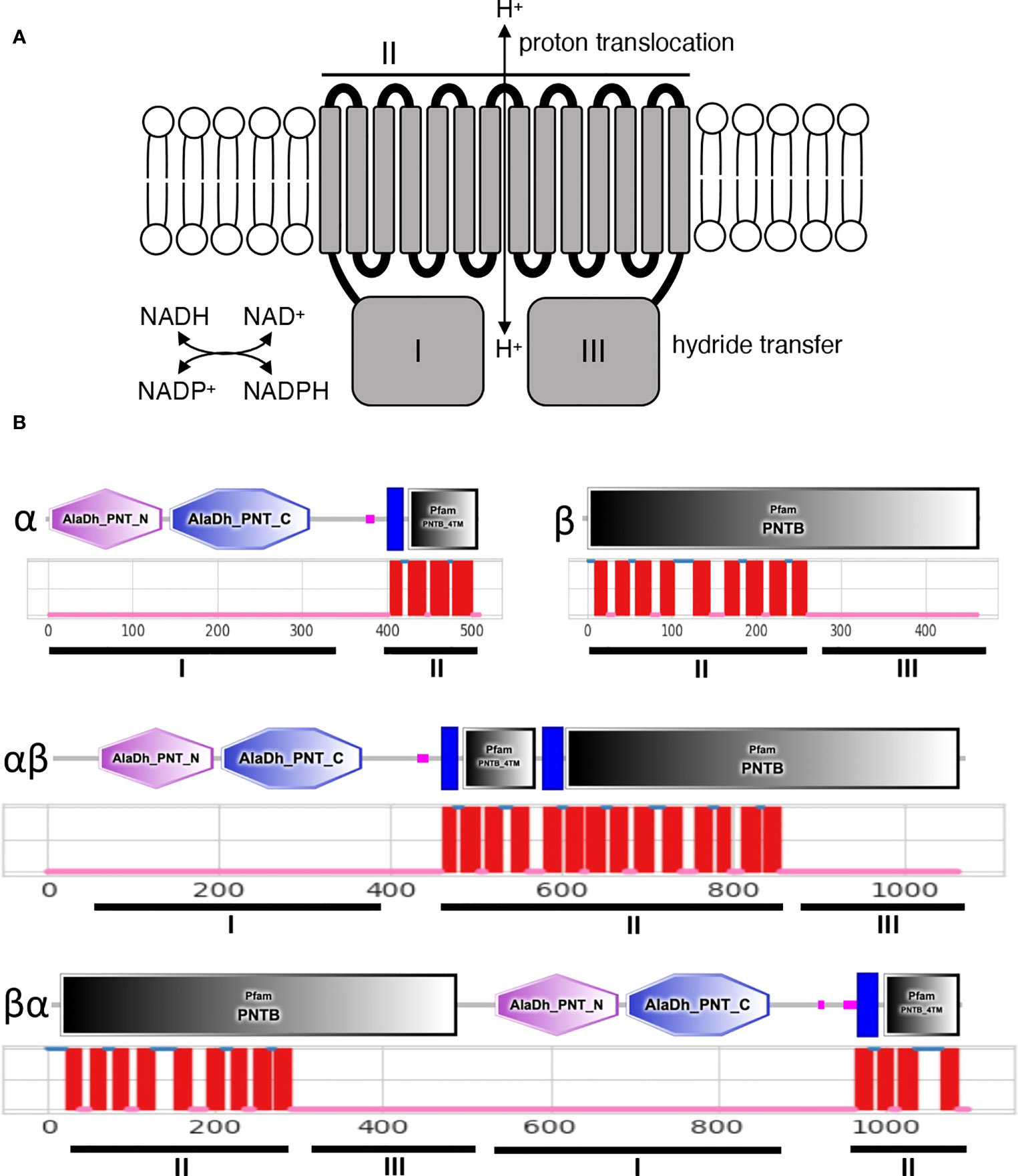
Figure 1 Structure of membrane bound NTH proteins. (A) Predicted functions of NTH in the lipid bilayer showing functional domains I-III and corresponding functional activities. (B) Organisation of functional NTH domains I-III in different NTH isoforms. Depicted are SMART and Pfam domains (https://smart.embl-heidelberg.dem) (top) and predicted transmembrane helices (https://dtu.biolib.com/DeepTMHMM) (middle).
Bacterial NTH (PntAB) is a composite enzyme composed of an α subunit (PntA) and β subunit (PntB) (Figure 1B) encoded by pntA and pntB genes, respectively, typically located within polycistronic operons. In some prokaryotes, the α subunit is further split into α1 and α2 subunits (Leung et al., 2015). Domain I is located on the α subunit; domain II corresponds to the combined TM helices of subunits α and β; and domain III is located on the remainder of subunit β (Figure 1B). In contrast to bacterial NTH, eukaryotic NTH is formed by a single protein chain corresponding to a fusion of the α subunit portion at its carboxy-terminus with the amino-terminus of the β subunit portion (αβ configuration) (Figure 1B). We recently showed that Plasmodium species encode a single NTH protein that possesses an inverted structure in which the β subunit portion sits amino-terminally to the α subunit portion (βα configuration) (Saeed et al., 2020) (Figure 1B). The same was found for NTH of the unicellular eukaryote Entamoeba histolytica (Weston et al., 2002).
We have shown that Plasmodium NTH does not reside in the mitochondrion, but is instead found localised in crystalloid and apicoplast organelles (Saeed et al., 2020). These findings demonstrate that the NTH enzyme is in fact not functionally confined to mitochondria and has a wider and possibly more diverse role in eukaryotic biology. For this study, we conducted a systematic analysis of nth genes across eukaryotic life to determine the prevalence of proton-translocating NTH and the distribution of its isoforms. The distinct underlying protein configurations of eukaryotic NTH (Figure 1) provide an additional tool for evolutionary biology as it can be combined with conventional phylogenetic data. We use this approach to consider the evolutionary trajectory of the Apicomplexa.
2 Materials and methods
NTH amino acid sequences were identified using the Plasmodium falciparum NTH protein sequence as query in BlastP searches of nonredundant protein databases (including GenPept, RefSeq, Swiss-Prot, PIR, PRF, and PDB), or TblastN searches of transcriptome shotgun assembly (TSA) or whole genome shotgun (WGS) databases at NCBI (https://ncbi.nlm.nih.gov) and VEuPathDB (https://veupathdb.org) web servers. Sequences were quality assessed by SMART/Pfam domain analysis (Letunic et al., 2021) (https://smart.embl-heidelberg.de) and DeepTMHMM transmembrane topology prediction (Hallgren et al., 2022) (https://dtu.biolib.com/DeepTMHMM) to conform with proton-translocating NTH characteristics (Figure 1B). Geneious Prime software was used to carry out βα-type NTH phylogenetic reconstructions using default settings. Specifically, amino acid multiple sequence alignment was conducted with Clustal Omega (Sievers et al., 2011) (full distance matrix for guide-tree calculations and cluster size 100, mBed algorithm); PhyML (Guindon et al., 2010) was used for maximum likelihood tree building (Le Gascuel substitution model); MrBayes (Huelsenbeck and Ronquist, 2001) was used for Bayesian inference tree building (rate matrix: Poisson; rate variation: gamma, 4 categories; chain length: 1,100,000; heated chains: 4; Heated chain temp: 0.2; subsampling frequency: 200; burn-in length: 100,000; unconstrained branch lengths). Vitrella brassicaformis NTH (Vbra_8937) was used as outgroup for apicomplexan phylogenies, being a close relative (Chromerida). Stygiella incarcerata NTH (ANM86865) was used as outgroup for TSAR phylogenies, as a βα-type NTH from a non-TSAR clade of unicellular eukaryotes (Discoba).
3 Results
3.1 Distribution of NTH across eukaryotes
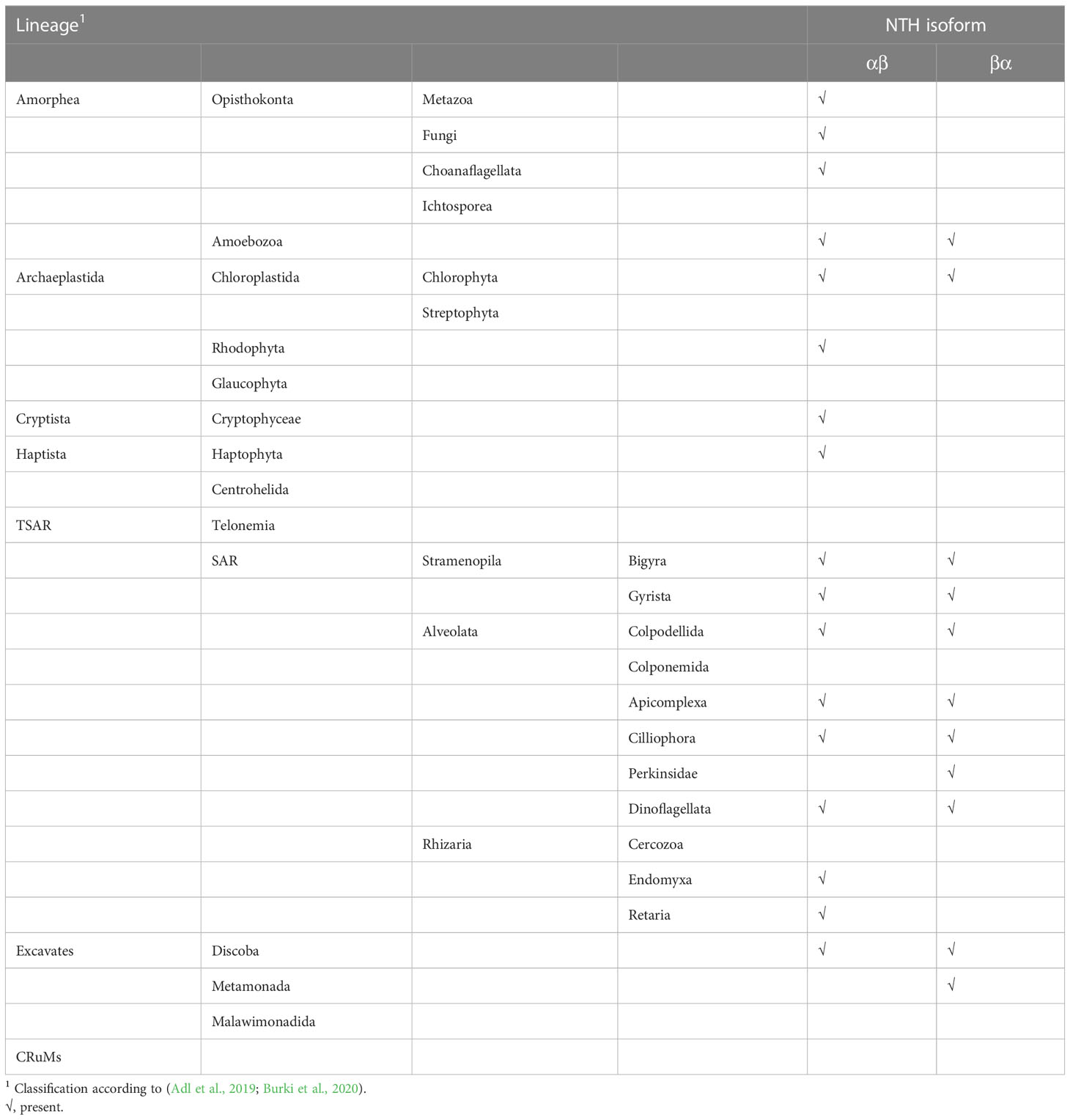
It is important to note that apparent absence of nth genes from taxa could be the result of incomplete genome/transcriptome coverage. Absence is therefore putative unless specified otherwise.
• Amorphea. This taxon is composed of opisthokonts (animals, fungi and their unicellular relatives) and Amoebozoa (classification according to (Adl et al., 2019; Burki et al., 2020)). We found nth genes exclusively of the αβ configuration in all animals (Metazoa) as diverse as stony corals to warm-blooded vertebrates (Table 1; Table S1). The fungi also encode NTH exclusively of the αβ configuration (Table 1; Table S1), but in contrast to animals not all fungal taxa encode NTH, including yeasts. In the Amoebozoa, a group of amoeboid protists sister to the opisthokonts, we found taxa encoding either αβ-type or βα-type NTH (Table 1; Table S1).
• Archaeplastida. This taxon includes plants (Streptophyta) and green, red and glaucophyte algae, which all contain photosynthetic plastids (chloroplasts) derived from primary endosymbiosis with a cyanobacterial ancestor (Raven and Allen, 2003). In sharp contrast to animals, we found no NTH proteins in plants (Table 1). Nonetheless, unsegmented nth genes of either a αβ or βα configuration are found in some primary algae belonging to Rhodophyta and Chlorophyta (Table 1; Table S1).
• TSAR. The constituent lineages of TSAR: Telonemia, Stramenopila, Alveolata and Rhizaria collectively are estimated to encompass up to half of all eukaryotic species. We found a mixed picture in this group of unicellular eukaryotes, with some genera lacking NTH, some having NTH of αβ configuration, and others possessing nth genes encoding βα-type NTH (Table 1; Table S1). Few genera possess genes encoding both αβ-type and βα-type NTH.
• Haptista. In this taxon of haptophyte algae we found organisms apparently without nth genes, as well as those encoding αβ-type NTH.
• Cryptista. This taxon of unicellular eukaryotes includes organisms that encode αβ-type NTH (Table 1; Table S1).
• Excavates. This is a newly proposed taxon that includes lineages Discoba, Metamonada and Malawimonadida (Burki et al., 2020). We identified several organisms in this clade encoding either αβ-type NTH or βα-type NTH (Table 1; Table S1).
• CRuMs. This represents a newly proposed small clade combining several lineages of unicellular eukaryote: Collodictyonidae, Rigifilida and Mantamonas. We found no nth-encoding genes in organisms belonging to this clade (Table 1; Table S1).
3.2 Distribution of NTH among apicomplexa
The Apicomplexa constitute a large eukaryotic lineage made up of microbial endosymbionts of animals, that includes medically important parasites such as Plasmodium and Toxoplasma species (causative agents of malaria and toxoplasmosis, respectively), as well as important pathogens of livestock such as Eimeria, Theileria and Babesia species. Though they constitute a diverse group of organisms with thousands of species across hundreds of genera, they share distinctive secretory organelles and cytoskeletal structures enabling motility, host cell interaction, invasion and egress (Frenal et al., 2017). Apicomplexans also share, with few exceptions, a relic four-membrane plastid called the apicoplast, which is of algal origin but no longer has photosynthetic activity (McFadden et al., 1996; Janouskovec et al., 2010; Lim and McFadden, 2010). Apicomplexans can be placed into different categories:
(1) ‘Core’ Apicomplexa. Organisms in this category are known for causing important human and animal diseases and are therefore best studied. They include two major lineages:
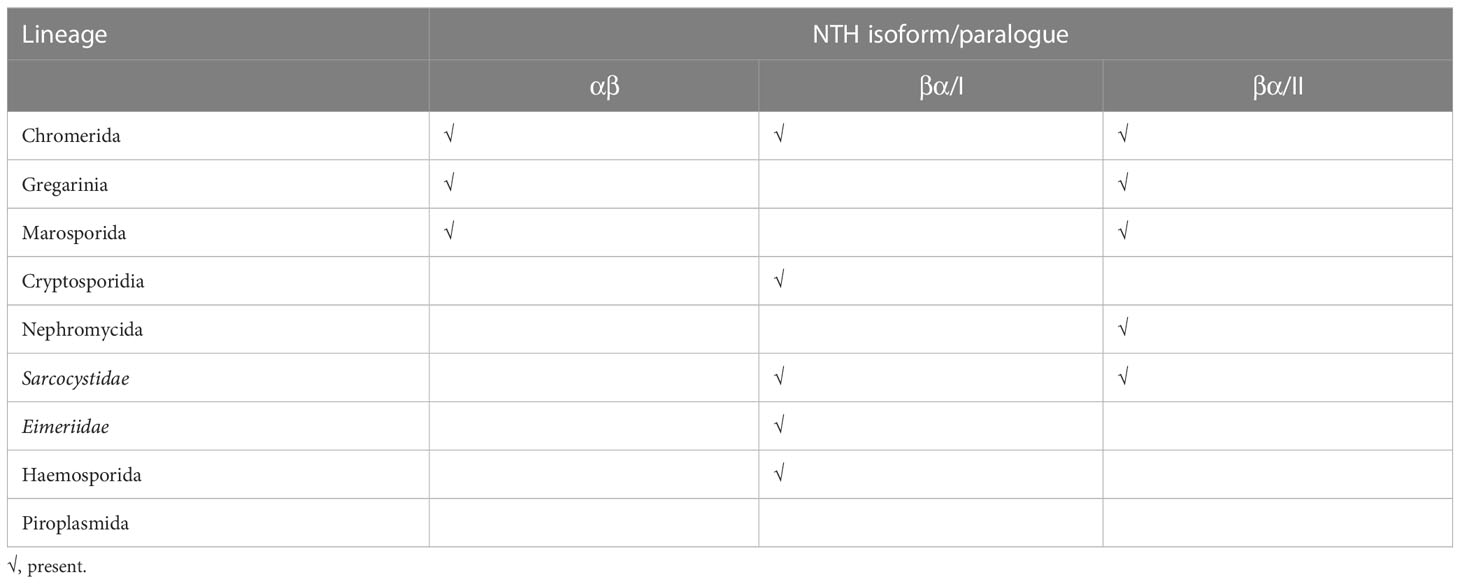
• Hematozoa: These are arthropod-transmitted intracellular blood parasites of vertebrates. Insect-transmitted Haemosporida (syn. Heamospororida) possess a single NTH protein of the βα-type that is both orthologous and syntenic between the three haemosporidan genera investigated (Table S2). By contrast, we did not find any NTH-encoding genes in organisms belonging to the tick-transmitted Piroplasmida (syn. Piroplasmorida) (Table S2). The robust sequence coverage of Babesia and Theileria genomes indicates that this reflects a genuine absence of NTH proteins in piroplasmids.
• Coccidia: The coccidians constitute non-vector-transmitted intracellular gut parasites of vertebrates and are grouped into families Sarcocystidae (e.g. genera Toxoplasma, Sarcocystis, Neospora, Hammondia, Besnoitia, Cystoisospora) and Eimeriidae (e.g. genera Eimeria and Cyclospora). This grouping is supported by the distinct representation of nth genes in these organisms, with the Sarcocystidae encoding two distinct βα-type proteins, while the Eimeriidae encode only a single NTH of the βα conformation (Table S2). The latter copy is in fact both orthologous and syntenic with one of the nth genes of the Sarcocystidae, confirming the close evolutionary relationship between these two coccidian lineages.
(2) ‘Basal’ Apicomplexa. Organisms in this category constitute deep-rooted lineages, including:
• Cryptosporidia: This lineage contains organisms of the genus Cryptosporidium that were once regarded coccidians based on their similar biology, but were later removed from the coccidian lineage based on molecular phylogenetic evidence. Cryptosporidium species possess two distinct and conserved nth genes, both of the βα configuration (Table S2).
• Gregarinia: Species belonging to this apicomplexan clade constitute a large and diverse group of extracellular, non-parasitic endosymbionts found in a wide range of terrestrial, marine and freshwater invertebrates (Rueckert et al., 2019). We identified both αβ- and βα-type NTH proteins in this lineage (Table S2).
• Marosporida: The Marosporida form a newly proposed deep-rooted apicomplexan monophyletic lineage (Mathur et al., 2021). Its taxa include species of the genera Rhytidocystis, Margolisiella, Aggregata and Merocystis, intracellular endosymbionts found in a variety of marine invertebrates including molluscs, annelids, whelks and crustaceans (Miroliubova et al., 2020). Analysis of transcriptomic sequence from Rhytidocystis species isolated from Ophelia limacina and Travisia forbesii (Janouskovec et al., 2019) identifies at least four distinct nth genes encoding both αβ- and βα-type NTH proteins (Table S2).
(3) ‘Other’ Apicomplexa. This category of apicomplexans includes the Nephromycida, species which are found in marine ascidian tunicates with which they have suspected mutualistic (e.g. genus Nephromyces) or parasitic (e.g. genus Cardiosporidium) relationships. Species of Nephromycida are unusual in that they accommodate and rely on bacterial endosymbionts (Hunter et al., 2020; Paight et al., 2022). At least two distinct βα-type NTH-encoding genes are present in the assembled genome of Cardiosporidium cionae species isolated from the hemolymph of the tunicate Ciona intestinalis (Hunter et al., 2020) (Table S2). In addition, we identified at least three distinct βα-type NTH-encoding genes from the metagenome of Nephromyces species isolated from the renal sac of the tunicate Molgula occidentalis (Munoz-Gomez et al., 2019) (Table S2).
3.3 NTH phylogeny
To shine more light on the underlying evolutionary relationships of the Apicomplexa, we conducted a phylogenetic examination of their βα-type NTH sequences, which are shared between all apicomplexan lineages except piroplasmids (Table S2). Phylogenetic reconstruction using maximum likelihood or Bayesian methods produced very similar trees, clustering the NTH sequences according to established apicomplexan groupings (Figures 2A, B). The only exception to this were the two cryptosporidian NTH sequences, indicating that the event that gave rise to these two NTH copies occurred early in apicomplexan evolution. Notably, βα-type NTH sequences split into two groups, I and II (Figures 2A, B), pointing to their descendance from ancestral nth paralogues. The Sarcocystidae are the only apicomplexan lineage to have retained nth copies from both paralogous groups (Figures 2A, B). By contrast, the Cryptosporidia, Haemosporida and Eimeriidae lineages have lost the group II nth copy, whereas the Gregarinia, Marosporida and Nephromycida lineages have lost the group I nth copy (Figures 2A, B).
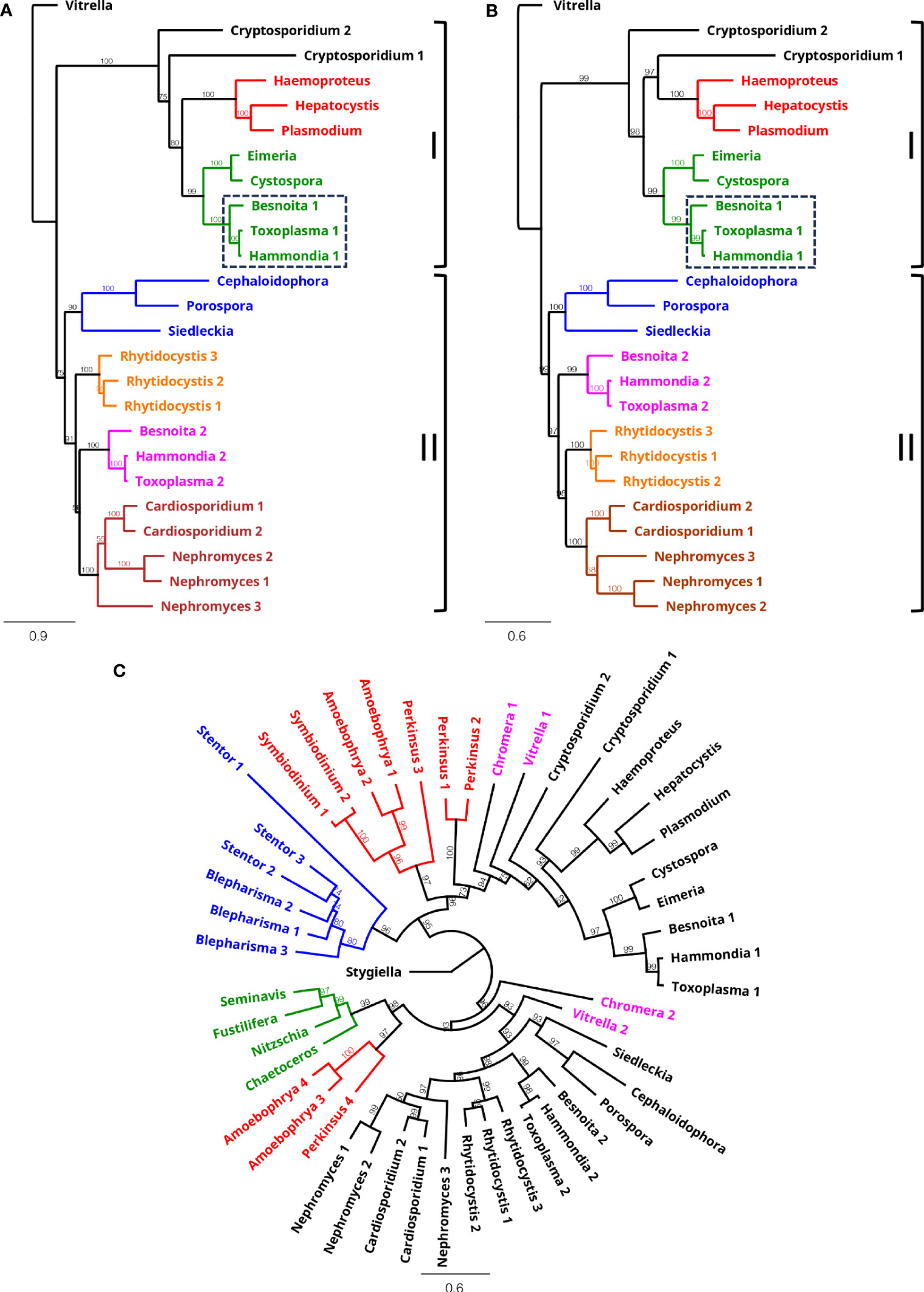
Figure 2 Phylogeny of βα-type NTH proteins. Phylogenetic reconstruction of apicomplexan NTH sequences using maximum likelihood (A) and Bayesian inference (B) methods. Colours in (A) and (B) represent Haemosporida (red), Coccidia (green, Sarcoscystidae indicated with dashed box), Gregarinia (blue), Marosporida (orange), Nephromycida (maroon) and Sarcocystidae (pink). Outgroup: Vitrella (Chromerida). Numbers behind genus names indicate distinct NTH homologues found. Paralogous groups I and II are indicated on right hand side. (C) Circular phylogram generated by Bayesian inference of apicomplexan NTH sequences (black) with inclusion of sequences from chromerid (pink), stramenopile (green), ciliate (blue) and dinoflagellate/perkinsid (red) lineages. Outgroup: Stygiella (Discoba). Bootstrap support (n=100) (A) and consensus support percentages (B, C) are shown to indicate branch support.
The closest living relatives of Apicomplexa are the Chromerida (now grouped within Colpodellida lineage): coral-associated photosynthetic algae whose plastids share many similarities with the apicoplast including four limiting membranes (Moore et al., 2008; Obornik et al., 2012; Janouskovec et al., 2015). Given these features and the notion that the ancestral apicomplexan was itself capable of photosynthesis, the chromerids are considered to share a most recent common ancestor with the Apicomplexa, a view that is robustly supported by phylogenomics (Janouskovec et al., 2019; Mathur et al., 2019; Munoz-Gomez et al., 2019; Salomaki et al., 2021). This is indeed consistent with the NTH isoform distribution among chromerids, encoding both αβ-type and βα-type NTH (Table S1; Table S3). Inclusion of βα-type NTH sequences from the chromerid lineages (Chromera and Vitrella) in the phylogenetic reconstruction revealed similar paralogous origins shared with the Apicomplexa (Figure 2C), indicating that the most recent common ancestor of apicomplexans and chromerids possessed at least two βα-type NTH paralogues (Table 2). Based on this NTH isoform distribution, different lineage-specific scenarios of nth gene loss/retention in extant apicomplexans can be inferred (Table 2). In addition to chromerids, other βα-type NTH sequences from the TSAR clade (Table S3) clustered broadly according to lineage and positioned basal to the Apicomplexa (Figure 2C) in agreement with established phylogenies (Janouskovec et al., 2019; Mathur et al., 2019; Salomaki et al., 2021). NTH sequences from dinoflagellates and the closely related Perkinsidae (Zhang et al., 2011) were also found distributed across both paralogous βα-type NTH groups.
4 Discussion
Our systematic analysis of nth genes across eukaryotic life reveals that NTH is found across all major lineages, but equally that many organisms, notably plants, lack nth genes, indicating that the selective pressures on maintaining NTH activity vary widely between different organisms. Our analysis provides evidence for recent loss and recent gain of NTH-encoding genes. One example of nth gene loss is provided by the Eimeriidae lineage, which possesses only one nth gene that is orthologous and syntenic with an nth gene in the closely related Sarcocystidae. The second NTH copy present in the Sarcocystidae, which only shares distant homology with the first (Figure 2A), is absent from the Eimeriidae (Table S2), providing evidence that the Eimeriidae evolved from the Sarcocystidae lineage within the coccidian clade. Another example of recent nth gene loss comes from the Piroplasmida (Table 2) that are lacking NTH altogether, whereas the closely related Haemosporida lineage possess NTH encoded by a single gene (Table 2). This observation strongly indicates that the Piroplasmida evolved from the haemosporidan lineage within the Hematozoa, and not the other way around. An example of recent gain of NTH-encoding genes via gene duplication comes from Porospora sp. that possess two virtually identical βα-type nth genes that are tandemly located, as well as three tandem and virtually identical αβ-type nth genes (Table 2; Table S2). We found no evidence to suggest that αβ-type nth genes readily convert into βα-type genes or vice versa, for example via rearrangement after gene duplication. Only older (unicellular) lineages possess αβ and βα-type nth genes, and among those very few organisms possess both αβ and βα-type NTH simultaneously (Table 1; Table S1). This suggests that events that led to the acquisition of the αβ- and βα-type nth genes happened early in eukaryotic evolution and that these isoforms have subsequently been maintained through vertical gene transfer/inheritance.
How did the unsegmented nth gene end up in eukaryotes? One possibility is that the eukaryotic unsegmented nth gene was obtained by lateral gene transfer from prokaryotic endosymbionts that gave rise to the mitochondrion (Andersson et al., 1998; Gray et al., 1999), followed by gene fusion. Increasing prevalence of gene fusions in eukaryotes over prokaryotes has also been observed for genes involved in purine biosynthesis (Chua and Fraser, 2020). There is evidence that mitochondrial evolution involved multiple bacterial endosymbionts (Baughn and Malamy, 2002) so lateral gene transfer could have occurred more than once. This scenario would offer an explanation for the apparent conservation of both location and function between prokaryotic and mitochondrial NTH, as well as for the creation of the two different NTH configurations.
Recent phylogenomic studies strongly support the grouping of coccidians, hematozoans, cryptosporidians and gregarines as four distinct monophyletic apicomplexan lineages (Janouskovec et al., 2019; Mathur et al., 2019; Salomaki et al., 2021). However, the evolutionary relationship between these four main apicomplexan clades remains controversial, Mathur et al. supporting monophyly of Cryptosporidia with Coccidia and Hematozoa (Mathur et al., 2019); Janouskovec et al. favouring monophyly of Gregarinia with Cryptosporidia (Janouskovec et al., 2019); and Salomaki and colleagues recovering monophyly of Gregarinia with Coccidia and Hematozoa (Salomaki et al., 2021). Among the Apicomplexa, only the gregarines and Marosporida possess NTH proteins of the αβ configuration, a feature shared with the chromerids (Table 2). It is tempting to speculate that a single event of αβ-type nth gene loss from the apicomplexan lineage, after the divergence of Marosporida and Gregarinia, led to the absence of αβ-type NTH from the other apicomplexan lineages (Table 2). This scenario would place the Marosporida and Gregarinia most basal in the apicomplexan phylogeny and thus favour the tree topology proposed by Mathur and colleagues (Mathur et al., 2019). However, it is also possible that αβ-type nth gene loss occurred multiple times during apicomplexan evolution, as appears to have been the case for the βα-type nth genes (Table 2), and therefore the question which apicomplexan lineage was first to diverge remains unresolved. Nonetheless, the NTH isoform distribution identified here (Table 2) argues against the proposed groupings of Nephromycida with Haemosporida and Piroplasmida, or of Cryptosporidia with the gregarines (Adl et al., 2019).
In phototrophic cyanobacteria, NTH is located in the thylakoid membranes (Kamarainen et al., 2017). However, the absence of NTH from all multicellular plants and most unicellular algae (Table 1; Table S1) indicates that NTH was not maintained in the cyanobacterium-derived chloroplasts. This contrasts with the apparent reliance on NTH in animals (Table 1). What could be the reason for this? One explanation is that plants, which are fully autotrophic, use a portion of the ATP generated during photosynthesis to reduce NADP+ to NADPH for the synthesis of organic compounds. Arguably therefore, there is less need for plants to generate NADPH via NTH activity. In support of this hypothesis, transhydrogenase activity in the cyanobacterium Synechocystis is dispensable for its growth under autotrophic (normal light) conditions, while knockout of its transhydrogenase (pntA) gene leads to growth defects under low light conditions (Kamarainen et al., 2017). Thus, autotrophic organisms (including chemotrophs) may be less reliant on NTH than their heterotrophic counterparts. It should be noted that the presence of αβ- and βα-type nth genes in some species of primary algae (Table 1; Table S1) indicates that the ancestral plant cell possessed both types. Accordingly, these genes could have ended up in organisms such as Plasmodium (derived from secondary endosymbiosis) by horizontal gene transfer from the primary algal endosymbiont, perhaps reflecting the residence of Plasmodium NTH in the apicoplast (Saeed et al., 2020).
For this study we used a new algorithm for transmembrane helix (TM) prediction, DeepTMHMM (Hallgren et al., 2022). This software consistently predicted, across all eukaryotic taxa, a single stretch of 14 TM helices in αβ-type NTH proteins, or two stretches of typically 9 and 4 TM helices, respectively, in βα-type NTH proteins (Figure 1B). This differed from TMHMM predictions (Krogh et al., 2001) that were more variable (e.g. see Plasmodium NTH (Saeed et al., 2020)) and that often calculated an odd number of TM helices in αβ-type NTH proteins (data not shown). The latter cannot be correct as it would position functional domains I and III on opposite sides of the membrane (Figure 1A), rendering the enzyme non-functional. Thus, DeepTMHMM appears a more reliable TM prediction tool than TMHMM.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
All authors contributed to the study conception and design. Data collection and analysis were performed by AT, SS and JD. The first draft of the manuscript was written by JD and all authors commented on previous versions of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Biotechnology and Biological Sciences research Council (grant BB/V006428) and the UK Medical Research Council (grant MR/P021611).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1216385/full#supplementary-material
References
Adl S. M., Bass D., Lane C. E., Lukes J., Schoch C. L., Smirnov A., et al. (2019). Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66 (1), 4–119. doi: 10.1111/jeu.12691
Anderson W. M., Fisher R. R. (1981). The subunit structure of bovine heart mitochondrial transhydrogenase. Biochim. Biophys. Acta 635 (1), 194–199. doi: 10.1016/0005-2728(81)90018-9
Andersson S. G., Zomorodipour A., Andersson J. O., Sicheritz-Ponten T., Alsmark U. C., Podowski R. M., et al. (1998). The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396 (6707), 133–140. doi: 10.1038/24094
Baughn A. D., Malamy M. H. (2002). A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. USA 99 (7), 4662–4667. doi: 10.1073/pnas.052710199
Burki F., Roger A. J., Brown M. W., Simpson A. G. B. (2020). The new tree of eukaryotes. Trends Ecol. Evol. 35 (1), 43–55. doi: 10.1016/j.tree.2019.08.008
Chua S. M., Fraser J. A. (2020). Surveying purine biosynthesis across the domains of life unveils promising drug targets in pathogens. Immunol. Cell Biol. 98 (10), 819–831. doi: 10.1111/imcb.12389
Frenal K., Dubremetz J. F., Lebrun M., Soldati-Favre D. (2017). Gliding motility powers invasion and egress in apicomplexa. Nat. Rev. Microbiol. 15 (11), 645–660. doi: 10.1038/nrmicro.2017.86
Gray M. W., Burger G., Lang B. F. (1999). Mitochondrial evolution. Science 283 (5407), 1476–1481. doi: 10.1126/science.283.5407.1476
Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59 (3), 307–321. doi: 10.1093/sysbio/syq010
Hallgren J., Tsirigos K. D., Pedersen M. D., Armenteros J. J. A., Marcatili P., Nielsen H., et al. (2022). Deep TMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv. doi: 10.1101/2022.04.08.487609
Hatefi Y., Yamaguchi M. (1996). Nicotinamide nucleotide transhydrogenase: a model for utilization of substrate binding energy for proton translocation. FASEB J. 10 (4), 444–452. doi: 10.1096/fasebj.10.4.8647343
Hou C., Potier M., Bragg P. D. (1990). Crosslinking and radiation inactivation analysis of the subunit structure of the pyridine nucleotide transhydrogenase of Escherichia coli. Biochim. Biophys. Acta 1018 (1), 61–66. doi: 10.1016/0005-2728(90)90110-p
Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 (8), 754–755. doi: 10.1093/bioinformatics/17.8.754
Hunter E. S., Paight C., Lane C. E. (2020). Metabolic contributions of an alphaproteobacterial endosymbiont in the apicomplexan Cardiosporidium cionae. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.580719
Jackson J. B. (2003). Proton translocation by transhydrogenase. FEBS Lett. 555 (1), 176–177. doi: 10.1016/s0014-5793(03)01123-2
Janouskovec J., Horak A., Obornik M., Lukes J., Keeling P. J. (2010). A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl. Acad. Sci. USA 107 (24), 10949–10954. doi: 10.1073/pnas.1003335107
Janouskovec J., Paskerova G. G., Miroliubova T. S., Mikhailov K. V., Birley T., Aleoshin V. V., et al. (2019). Apicomplexan-like parasites are polyphyletic and widely but selectively dependent on cryptic plastid organelles. Elife 8, e49662. doi: 10.7554/eLife.49662
Janouskovec J., Tikhonenkov D. V., Burki F., Howe A. T., Kolisko M., Mylnikov A. P., et al. (2015). Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc. Natl. Acad. Sci. USA 112 (33), 10200–10207. doi: 10.1073/pnas.1423790112
Kamarainen J., Huokko T., Kreula S., Jones P. R., Aro E. M., Kallio P. (2017). Pyridine nucleotide transhydrogenase PntAB is essential for optimal growth and photosynthetic integrity under low-light mixotrophic conditions in synechocystis sp. PCC 6803. New Phytol. 214 (1), 194–204. doi: 10.1111/nph.14353
Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305 (3), 567–580. doi: 10.1006/jmbi.2000.4315
Letunic I., Khedkar S., Bork P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49 (D1), D458–D460. doi: 10.1093/nar/gkaa937
Leung J. H., Schurig-Briccio L. A., Yamaguchi M., Moeller A., Speir J. A., Gennis R. B., et al. (2015). Structural biology. division of labor in transhydrogenase by alternating proton translocation and hydride transfer. Science 347 (6218), 178–181. doi: 10.1126/science.1260451
Lim L., McFadden G. I. (2010). The evolution, metabolism and functions of the apicoplast. Philos. Trans. R Soc. Lond B Biol. Sci. 365 (1541), 749–763. doi: 10.1098/rstb.2009.0273
Mathur V., Kolisko M., Hehenberger E., Irwin N. A. T., Leander B. S., Kristmundsson A., et al. (2019). Multiple independent origins of apicomplexan-like parasites. Curr. Biol. 29 (17), 2936–2941.e2935. doi: 10.1016/j.cub.2019.07.019
Mathur V., Kwong W. K., Husnik F., Irwin N. A. T., Kristmundsson A., Gestal C., et al. (2021). Phylogenomics identifies a new major subgroup of apicomplexans, marosporida class nov., with extreme apicoplast genome reduction. Genome Biol. Evol. 13 (2), evaa244. doi: 10.1093/gbe/evaa244
McFadden G. I., Reith M. E., Munholland J., Lang-Unnasch N. (1996). Plastid in human parasites. Nature 381 (6582), 482. doi: 10.1038/381482a0
Miroliubova T. S., Simdyanov T. G., Mikhailov K. V., Aleoshin V. V., Janouskovec J., Belova P. A., et al. (2020). Polyphyletic origin, intracellular invasion, and meiotic genes in the putatively asexual agamococcidians (Apicomplexa incertae sedis). Sci. Rep. 10 (1), 15847. doi: 10.1038/s41598-020-72287-x
Moore R. B., Obornik M., Janouskovec J., Chrudimsky T., Vancova M., Green D. H., et al. (2008). A photosynthetic alveolate closely related to apicomplexan parasites. Nature 451 (7181), 959–963. doi: 10.1038/nature06635
Munoz-Gomez S. A., Durnin K., Eme L., Paight C., Lane C. E., Saffo M. B., et al. (2019). Nephromyces represents a diverse and novel lineage of the apicomplexa that has retained apicoplasts. Genome Biol. Evol. 11 (10), 2727–2740. doi: 10.1093/gbe/evz155
Obornik M., Modry D., Lukes M., Cernotikova-Stribrna E., Cihlar J., Tesarova M., et al. (2012). Morphology, ultrastructure and life cycle of Vitrella brassicaformis n. sp., n. gen., a novel chromerid from the great barrier reef. Protist 163 (2), 306–323. doi: 10.1016/j.protis.2011.09.001
Paight C., Hunter E. S., Lane C. E. (2022). Codependence of individuals in the Nephromyces species swarm requires heterospecific bacterial endosymbionts. Curr. Biol. 32 (13), 2948–2955.e2944. doi: 10.1016/j.cub.2022.05.007
Raven J. A., Allen J. F. (2003). Genomics and chloroplast evolution: what did cyanobacteria do for plants? Genome Biol. 4 (3), 209. doi: 10.1186/gb-2003-4-3-209
Rueckert S., Betts E. L., Tsaousis A. D. (2019). The symbiotic spectrum: where do the gregarines fit? Trends Parasitol. 35 (9), 687–694. doi: 10.1016/j.pt.2019.06.013
Saeed S., Tremp A. Z., Sharma V., Lasonder E., Dessens J. T. (2020). NAD(P) transhydrogenase has vital non-mitochondrial functions in malaria parasite transmission. EMBO Rep. 21, e47832. doi: 10.15252/embr.201947832
Salomaki E. D., Terpis K. X., Rueckert S., Kotyk M., Varadinova Z. K., Cepicka I., et al. (2021). Gregarine single-cell transcriptomics reveals differential mitochondrial remodeling and adaptation in apicomplexans. BMC Biol. 19 (1), 77. doi: 10.1186/s12915-021-01007-2
Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Weston C. J., Venning J. D., Jackson J. B. (2002). The membrane-peripheral subunits of transhydrogenase from entamoeba histolytica are functional only when dimerized. J. Biol. Chem. 277 (29), 26163–26170. doi: 10.1074/jbc.M203514200
Keywords: transhydrogenase, isoform, TSAR, alveolates, apicomplexa, Plasmodium
Citation: Tremp AZ, Saeed S and Dessens JT (2023) NAD(P) transhydrogenase isoform distribution provides insight into apicomplexan evolution. Front. Ecol. Evol. 11:1216385. doi: 10.3389/fevo.2023.1216385
Received: 03 May 2023; Accepted: 14 June 2023;
Published: 28 June 2023.
Edited by:
Michael Tessler, St. Francis College, United StatesCopyright © 2023 Tremp, Saeed and Dessens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johannes T. Dessens, Sm9oYW5uZXMuZGVzc2Vuc0Bsc2h0bS5hYy51aw==
†These authors have contributed equally to this work and share first authorship
 Annie Z. Tremp
Annie Z. Tremp Sadia Saeed
Sadia Saeed Johannes T. Dessens
Johannes T. Dessens