
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
DATA REPORT article
Front. Ecol. Evol., 10 August 2023
Sec. Phylogenetics, Phylogenomics, and Systematics
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1205557
Cyanobacteria are highly diverse photosynthetic prokaryotes that can widely adapt and survive in different environmental conditions. These blue-green algae are vastly proliferating in extreme environments whose mats usually develop on submerged stones or rocks. Their ability to survive in stressed environments make them good candidates for screening and isolation of bioactive metabolites that can be used for the development of therapeutics and/or those that can promote good health and well-being (UN SDG 3). Cyanobacteria are said to be the most promising producers of structurally novel and biologically active secondary metabolites of low molecular weight compounds, which are usually produced during their maximum stationary growth phase (Drobac-Čik et al., 2007). This makes them an interesting group of organisms that can be explored and harnessed for biotechnological applications (Nandagopal et al., 2021; Favas et al., 2022).
Nodosilinea is a filamentous cyanobacterium found in Europe, North America, and Asia. They grow in various habitats such as soil, freshwater bodies, sea water, and even in geothermal springs. Morphologically, they are capable of forming nodules within the sheath upon exposure to low light conditions (Perkerson Iii et al., 2011).
Available publications that identified this genus employed the polyphasic approach, such as morphological characterization and molecular identification using 16S rRNA gene phylogeny and 16S-23S ITS rRNA secondary structures (Perkerson Iii et al., 2011; Vázquez-Martínez et al., 2018; Radzi et al., 2019; Cai et al., 2022). Currently, only a few species of the filamentous cyanobacterium Nodosilinea have been documented and described.
A variety of next generation sequencing (NGS) tools make genome analysis feasible to come up with the assembly and analysis of sequences from diverse cyanobacteria, establish taxonomic classification, and phylogenetic relationships. The great advantage of whole genome sequencing is the discovery of secondary metabolites as sources of natural products. The bioinformatics tools for genome mining utilized by Micallef et al. (2015) revealed the diversity, abundance and complex nature of the biotherapeutic potential of the true-branching Subsection V filamentous cyanobacteria. Most of them synthesize non-ribosomal peptide synthetases (NRPS), polyketide synthases (PKS) (Shih et al., 2013), post-ribosomal peptide synthases (PRPS), mycosporine-like amino acids (MAAs) (Li et al., 2020), hydrocarbons, alkaloids and terpenes.
In this study, we report a newly sequenced filamentous cyanobacterium Nodosilinea sp. PGN35 isolated from a tailing storage facility of a mining site in Tuba, Benguet, Philippines. Using whole genome sequencing technology and bioinformatics tools, its genomic features and biosynthetic gene clusters were elucidated.
This study is the first to report the assembly, genome and functional annotation, as well as biosynthetic gene cluster prediction of Nodosilinea sp. isolated from a copper mine tailing storage facility in Tuba, Benguet, Philippines. The data and information generated would enable further understanding of this species’ phylogeny and biology.
The cyanobacterium Nodosilinea sp. used in this study was from the existing cyanobacterial cultures of the Plant Molecular Biology and Genetics Laboratory (PMBGL) of the Institute of Biology, University of the Philippines Diliman, Quezon City, Philippines. Sediment samples were previously collected from an active Tailing Storage Facility 3 (TSF3) of a large-scale mining site in Tuba, Benguet, Philippines. Upon collection, a portion of the sediment samples was subjected to serial dilution. The sediment solutions were filtered, and the filter used was inoculated in BG-11 (HiMedia, India) liquid medium to allow enrichment of cyanobacteria. The cultures were then placed under white fluorescent lamps for a 12-hour light-dark cycle. This served as the mother culture for cyanobacterial isolation. To obtain unialgal cultures, classical microbiological techniques were employed using both solid and liquid BG-11 media. Isolated cyanobacteria were transferred into sterile 250mL Erlenmeyer flasks with BG-11 medium for cultivation and maintenance.
Morphological examination of the isolated cyanobacteria was performed using a light compound microscope (Labomed LB-221, USA). Photomicrographs were taken with the aid of ScopeImage 9.0. One particular isolate was morphologically characterized by examining the filament and trichome width, cell color and sheath, apical cells, and the presence of nodules from 10 randomly selected replicate specimens. The examined features were compared to the descriptions from the literature (Anagnostidis and Komarek, 2005; Heidari et al., 2018; Vázquez-Martínez et al., 2018).
After two months of growth, planktonic colonies of the cultured cyanobacterium were collected for the extraction of the total genomic DNA using the Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, USA). Agarose gel electrophoresis was performed to detect the presence of the extracted DNA, while a spectrophotometer (Epoch™) was used to determine its purity. Furthermore, the 16S rRNA gene region of the extracted genomic DNA was amplified in a T100 Thermal Cycler (Bio-Rad, Singapore) using forward and reverse primers specific to cyanobacteria: CYA106F (5′ CGG ACG GGT GAG TAA CGC GTG A 3′) and CYA781R (mixed in equimolar quantities: (a) (5′- GAC TAC TGG GGT ATC TAA TCC CAT T 3′) and (b) (5′- GAC TAC TGG GGT ATC TAA TCC CAT T 3′) (Nübel et al., 1997). The PCR cycle conditions were set with an initial denaturation step at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 1 minute, an annealing step at 60°C for 1 minute, an extension step at 72°C for 1 minute, and a final elongation step at 72°C for 5 minutes. The amplicons were then visualized in 1% agarose gel at 100V for 30 minutes using 1kb universal DNA ladder (HyperLadder™). The gel was stained with gel red, and the image was recorded from a Bio-Rad Gel Doc EZ Gel Documentation System (Bio-Rad, USA). In addition, the quality of the extracted DNA was assessed using a microplate spectrophotometer (Epoch™). The PCR products were sent to Macrogen, Inc. (South Korea) for bidirectional capillary sequencing. The resulting sequences were assembled using Sequencher v5.4.6 (Gene Codes Corporation, 2017) and a nucleotide BLAST search was conducted on the NCBI website using the generated consensus sequences to identify the cyanobacterium. Upon confirmation, the genomic DNA of the cyanobacterial isolate was also sent to Macrogen, Inc. (South Korea) for whole genome shotgun sequencing (2x150 PE) on the Illumina NovaSeq platform.
Paired-end reads were filtered and trimmed with BBtools v.38.92 (Bushnell, 2014) and PRINSEQ (Preprocessing and Information and SEQuence 0.20.4) (Schmieder and Edwards, 2011). De novo assembly was performed using SPAdes 3.15.4 with k-mer sizes 21, 33, 55, 77, 99, and 127 (Bankevich et al., 2012). CheckM v1.1.6 (Parks et al., 2015) was used to assess the quality of assembled contigs. Since the culture was not axenic but unialgal, taxonomic binning was performed to remove contaminants (Teikari et al., 2022) using MetaBAT2 v1.7 (Kang et al., 2019), MaxBIN2 v2.2.4 (Wu et al., 2015), and CONCOCT v1.1 (Alneberg et al., 2014). The resulting bins were compared using DAS Tool v1.1.2 (Sieber et al., 2018). The highest quality bins were selected and subjected to GTDB-Tk v2.2.1 (Chaumeil et al., 2022) for taxonomic classification. The genome assembly was submitted to the Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al., 2016) at the National Center for Biotechnology Information (NCBI) for rapid annotation. Then, the cyanobacterial genome was functionally annotated using RASTtk server v2.0 (Brettin et al., 2015). Meanwhile, AntiSMASH 6.0 (Blin et al., 2019), DeepBGC 0.1.23 (Hannigan et al., 2019) and NaPDoS2 (Klau et al., 2022) were used to predict gene clusters related to secondary metabolite production. Default parameters were used unless specified.
The phylogenetic tree construction was performed using the 16S rRNA partial sequence of Nodosilinea sp. PGN35, including the representative sequences from the genera of filamentous cyanobacteria: Leptolyngbya (16), Nodosilinea (13), Arthronema (3), Lyngbya (5), and Phormidesmis (5). Gloeobacter violaceus was used as the outgroup. Gene alignment was performed using ClustalW. The nucleotide substitution model utilized was Kimura two parameter with gamma (K2+G) distribution, as determined by MEGA11 (Tamura et al., 2021). The phylogenetic tree was also generated using MEGA11; running bootstrap method with 1,000 replications. For further analysis, a phylogenomic tree of Nodosilinea sp. PGN35 was constructed using the Species Tree App present in KBase (Arkin et al., 2018). All parameters were set at default.
The cyanobacterial isolate was unialgal and non-axenized. It has uniseriate, unbranched filaments, which during nodule formation becomes multiseriate. It also has a blue-green trichome enclosed in a thin, colorless sheath. Each cell is longer than wide. The cell length measured 4.01 μm and the cell width was 3.45 μm. Their apical cells are rounded and the filaments form nodules (Figures 1A–C).
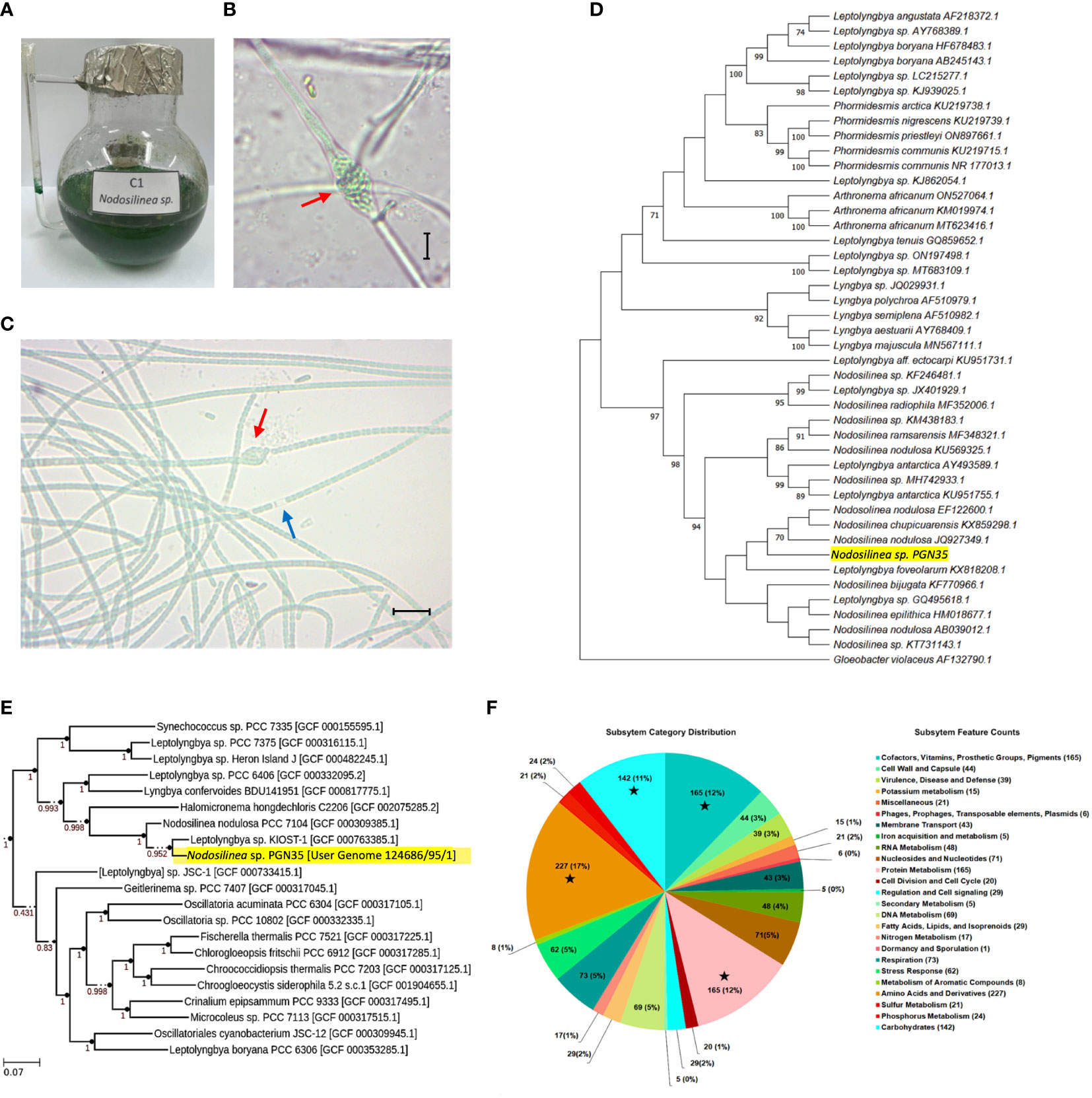
Figure 1 Morphological and molecular characterization of Nodosilinea sp. PGN35. (A) Blue-green aggregates of Nodosllinea culture in BG-11 medium. (B) Characteristic nodule (red arrow) measuring 17.53 μm long and 10.63 μm wide. Microscopic figures scale -10μm. (C) Unbranched filaments enclosed in a sheath (blue arrow) with nodule formation (red arrow) and Individual cells measuring 4.01 μm in length and 3.45 μm in width. (D) Maximum Likelihood (ML) phylogenetic tree of Nodosilinea sp. PGN35 (highlighted in yellow) using 16S rRNA gene sequencing based on K2+G substitution model. The bootstrap support values for the nodes were obtained from 1,000 replicates. The scale bar represents the number of nucleotide substitutions per site. (E) ML phylogenomic tree of Nodosilinea sp. PGN35 (highlighted in yellow) generated by Species Tree App along with closely related genomes selected from the public KBase genomes Imported from NCBI-RefSeq. (F) RASTtk subsystem category distribution. The pie chart represents the cellular subsystem category and the number of protein-coding genes (in parentheses) that are predicted to be involved in the cellular process. ★ indicates the top four subsystem groups with the greatest number of protein-coding genes.
The whole genome sequencing of Nodosilinea sp. PGN35 generated a total of 22,265,312 reads. After quality control and assembly, 25 contigs were generated. The cyanobacterial genome has an estimated size of 6,390,537 bp and a GC content of 59.3%. Furthermore, the assembled genome showed a 98.37% completeness and 1.54% contamination.
The annotated genome using the PGAP revealed a total of 5,688 genes, 5617 protein coding genes (CDS), 45 rRNAs, 41 tRNAs, 4 ncRNAs, 26 pseudo genes, and 1 CRISPR arrays (Table 1A).
A circular graphical display from the RASTtk annotation is shown in Figure 1F. There were 17% proteins annotated by the RASTtk server. Annotations were grouped according to their likelihood to be involved in a cellular process or subsystem. Following the pie chart clockwise, subsystem categories are listed in the legend from top to bottom. Among all the cellular subsystem components, (1st) the Amino Acids and Derivatives, (2nd) Protein Metabolism, (3rd) Cofactors, Vitamins, Prosthetics Groups, Pigments and (4th) the Carbohydrates were the biggest groups, with 227, 165, 158, and 142 annotated protein-coding genes, respectively.
Moreover, three automated genome mining pipelines were used to annotate the major biosynthetic gene clusters (BGCs) of Nodosilinea sp. PGN35 (Table 1B). Analysis using AntiSMASH resulted in the identification of four BGCs which correspond to one cluster of arylpolyene, one cluster of resorcinol and two clusters of terpenes. It can be noted that these BGCs have not been characterized, and are currently referred to as orphan/cryptic, thus, warrants further exploration to determine whether these gene clusters are novel for this cyanobacterial strain. Aryl polyene is expected to be a prominent secondary metabolite for the prokaryotic Nodosilinea sp. since it is a widespread metabolite found in gram-negative bacteria (Lee et al., 2021) that functions as antioxidative carotenoids (Schöner et al., 2016). Meanwhile, resorcinol and its derivative is a class pf polyketide with antioxidant and antibacterial properties (Hiasa et al., 2013), hence commonly used as antiseptic and disinfectant. On the other hand, terpene synthases and its precursor pathways were expressed in cyanobacteria efficiently converting light and carbon dioxide to terpene HC and terpenoids (Lin and Pakrasi, 2019), which in turn, has vast applications in the pharmaceutical industry, as antiviral among others. Comparatively, DeepBGC was also able to predict two terpenes, the product class corresponding to saccharide and polyketide, as well as BGCs with predicted antibacterial and cytotoxic activity. The ribosomally synthesized and post-translationally modified peptides (RiPPs) are the dominant class of cyclic peptides reported to possess high antimicrobial activity (Carpine and Sieber, 2021). Meanwhile, NaPDoS2 was able to detect two ketosynthase domains identified as type II fatty acid synthase.
Phylogenetic tree construction of PGN35 using maximum likelihood of partial 16S rRNA gene sequences revealed that it belongs to the Nodosilinea genus. Likewise, it shows that it is related to the species of nodulosa and chupicuarensis with a similarity of 70% (Figure 1D). Meanwhile, phylogenomic analysis grouped PGN35 with one representative genome of the genus Leptolyngbya with 95.2% bootstrap support (Figure 1E). Although phylogenetic and phylogenomic trees of Nodosilinea sp. PGN35 showed high similarity to other Nodosilinea species, a close relationship with species of Leptolyngbya was also observed. This may suggest that additional genomic data is needed to further delineate the genus of Nodosilinea from Leptolyngbya.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, SAMN32710271.
DU, LS, and EC contributed equally to this work from enriching and maintenance of cultures, sequencing, and bioinformatics analysis, writing and revision of the manuscript. All authors contributed to the article and approved the submitted version.
We are grateful for the financial support for this study through a grant from the University of the Philippines Office of the Vice-President for Academic Affairs (UP OVPAA) through the Emerging Inter-Disciplinary Research (EIDR) Program (OVPAA-EIDR-C09-01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alneberg J., Bjarnason B. S., de Bruijn I., Schirmer M., Quick J., Ijaz U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11 (11), 1144–1146. doi: 10.1038/nmeth.3103
Anagnostidis K., Komarek J. (2005) Teil 2: Oscillatoriales Süßwasserflora von Mitteleuropa. Available at: https://link.springer.com/book/10.1007/978-3-662-56269-7.
Arkin A. P., Cottingham R. W., Henry C. S., Harris N. L., Stevens R. L., Maslov S., et al. (2018). KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 36 (7), 566–569. doi: 10.1038/nbt.4163
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 (5), Article 5. doi: 10.1089/cmb.2012.0021
Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., et al. (2019). antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47 (W1), Article W1. doi: 10.1093/nar/gkz310
Brettin T., Davis J. J., Disz T., Edwards R. A., Gerdes S., Olsen G. J., et al. (2015). RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5 (1), Article 1. doi: 10.1038/srep08365
Bushnell B. (2014). BBTools (United States: DOE Joint Genome Institute). Available at: https://jgi.doe.gov/data-and-tools/software-tools/bbtools/.
Cai F., Li S., Zhang H., Yu G., Li R. (2022). Nodosilinea hunanesis sp. Nov. (Prochlorotrichaceae, Synechococcales) from a freshwater pond in China based on a polyphasic approach. Diversity 14 (5), Article 5. doi: 10.3390/d14050364
Carpine R., Sieber S. (2021). Antibacterial and antiviral metabolites from cyanobacteria: their application and their impact on human health. Curr. Res. Biotechnol. 3, 65–81. doi: 10.1016/j.crbiot.2021.03.001
Chaumeil P. A., Mussig A. J., Hugenholtz P., Parks D. H. (2022). GTDB-Tk v2: Memory friendly classification with the genome taxonomy database. Bioinformatics 38 (23), 5315–5316. doi: 10.1093/bioinformatics/btac672
Drobac-Čik V., Dulic T., Stojanović D., Svircev B. (2007). The importance of extremophile cyanobacteria in the production of biologically active compounds. Zbornik Matice Srpske Za Prirodne Nauke (112), 57–66. doi: 10.2298/ZMSPN0712057D
Favas R., Morone J., Martins R., Vasconcelos V., Lopes G. (2022). Cyanobacteria secondary metabolites as biotechnological ingredients in natural anti-aging cosmetics: potential to overcome hyperpigmentation, loss of skin density and uv radiation-deleterious effects. Mar. Drugs 20 (3), Article 3. doi: 10.3390/md20030183
Hannigan G. D., Prihoda D., Palicka A., Soukup J., Klempir O., Rampula L., et al. (2019). A deep learning genome-mining strategy for biosynthetic gene cluster prediction. Nucleic Acids Res. 47 (18), e110–e110. doi: 10.1093/nar/gkz654
Heidari F., Zima J. Jr., Riahi H., Hauer T. (2018). New simple trichal cyanobacterial taxa isolated from radioactive thermal springs. J. Czech Phycological Soc. 18, 137–149. doi: 10.5507/fot.2017.024
Hiasa M., Kurokawa M., Ohta K., Esumi T., Akita H., Niki K., et al. (2013). Identification and purification of resorcinol, an antioxidant specific to Awa-ban (pickled and anaerobically fermented) tea. Food Res. Int. 54 (1), 72–80. doi: 10.1016/j.foodres.2013.05.036
Kang D. D., Li F., Kirton E., Thomas A., Egan R., An H., et al. (2019). MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7. doi: 10.7717/peerj.7359
Klau L. J., Podell S., Creamer K. E., Demko A. M., Singh H. W., Allen E. E., et al. (2022). The Natural Product Domain Seeker version 2 (NaPDoS2) webtool relates ketosynthase phylogeny to biosynthetic function. J. Biol. Chem. 298 (10). doi: 10.1016/j.jbc.2022.102480
Lee W. C., Choi S., Jang A., Yeon J., Hwang E., Kim Y. (2021). Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis. Sci. Rep. 11 (1), Article 1. doi: 10.1038/s41598-021-95890-y
Li Y., Naman C. B., Alexander K. L., Guan H., Gerwick W. H. (2020). The chemistry, biochemistry and pharmacology of marine natural products from Leptolyngbya, a chemically endowed genus of cyanobacteria. Mar. Drugs 18 (10). doi: 10.3390/md18100508
Lin P. C., Pakrasi H. B. (2019). Engineering cyanobacteria for production of terpenoids. Planta 249 (1), 145–154. doi: 10.1007/s00425-018-3047-y
Micallef M. L., D’Agostino P. M., Sharma D., Viswanathan R., Moffitt M. C. (2015). Genome mining for natural product biosynthetic gene clusters in the Subsection V cyanobacteria. BMC Genomics 16 (1), Article 1. doi: 10.1186/s12864-015-1855-z
Nandagopal P., Steven A. N., Chan L.-W., Rahmat Z., Jamaluddin H., Mohd Noh N. I. (2021). Bioactive metabolites produced by cyanobacteria for growth adaptation and their pharmacological properties. Biology 10 (10), 1061. doi: 10.3390/biology10101061
Nübel U., Garcia-Pichel F., Muyzer G. (1997). PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63 (8), Article 8. doi: 10.1128/aem.63.8.3327-3332.1997
Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W. (2015). CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25 (7), 1043–1055. doi: 10.1101/gr.186072.114
Perkerson Iii R. B., Johansen J. R., Kovácik L., Brand J., Kaštovský J., Casamatta D. A. (2011). A unique pseudanabaenalean (cyanobacteria) genus Nodosilinea gen. Nov. based on morphological and molecular data. J. Phycology 47 (6), 1397–1412. doi: 10.1111/j.1529-8817.2011.01077.x
Radzi R., Muangmai N., Broady P., Wan Omar W. M., Lavoue S., Convey P., et al. (2019). Nodosilinea signiensis sp. nov. (Leptolyngbyaceae, Synechococcales), a new terrestrial cyanobacterium isolated from mats collected on Signy Island, South Orkney Islands, Antarctica. PloS One 14 (11), e0224395. doi: 10.1371/journal.pone.0224395
Schmieder R., Edwards R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinf. (Oxford England) 27 (6), Article 6. doi: 10.1093/bioinformatics/btr026
Schöner T. A., Gassel S., Osawa A., Tobias N. J., Okuno Y., Sakakibara Y., et al. (2016). Aryl polyenes, a highly abundant class of bacterial natural products, are functionally related to antioxidative carotenoids. Chembiochem: A Eur. J. Chem. Biol. 17 (3), 247–253. doi: 10.1002/cbic.201500474
Gene Codes Corporation (2017). Sequencher version 5.4.6 DNA sequence Analysis software. Ann Arbor, MI USA: Gene Codes Corporation. Available at: http://www.genecodes.com
Shih P. M., Wu D., Latifi A., Axen S. D., Fewer D. P., Talla E., et al. (2013). Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. United States America 110 (3), 1053–1058. doi: 10.1073/pnas.1217107110
Sieber C. M. K., Probst A. J., Sharrar A., Thomas B. C., Hess M., Tringe S. G., et al. (2018). Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3 (7), Article 7. doi: 10.1038/s41564-018-0171-1
Tamura K., Stecher G., Kumar S. (2021). Mega11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E. P., Zaslavsky L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44 (14), 6614–6624. doi: 10.1093/nar/gkw569
Teikari J., Baunach M., Dittmann E. (2022). Cyanobacterial genome sequencing, annotation, and bioinformatics. Methods Mol. Biol., 2489, 269–287. doi: 10.1007/978-1-0716-2273-5_14
Vázquez-Martínez J., Gutierrez-Villagomez J. M., Fonseca-García C., Ramírez-Chávez E., Mondragón-Sánchez M. L., Partida-Martínez L., et al. (2018). Nodosilinea chupicuarensis sp. nov. (Leptolyngbyaceae, Synechococcales) a subaerial cyanobacterium isolated from a stone monument in central Mexico. Phytotaxa 334 (2), Article 2. doi: 10.11646/phytotaxa.334.2.6
Keywords: Nodosilinea sp., cyanobacteria, genomics, secondary metabolites, biosynthetic gene clusters
Citation: Untiveros DPM, Sanchez LRS and Cao EP (2023) Whole genome sequence analysis of the filamentous Nodosilinea sp. PGN35 isolated from a mining site in Tuba, Benguet, Philippines. Front. Ecol. Evol. 11:1205557. doi: 10.3389/fevo.2023.1205557
Received: 14 April 2023; Accepted: 24 July 2023;
Published: 10 August 2023.
Edited by:
Zheng Wang, Yale University, United StatesReviewed by:
Yen-Wen Wang, Yale University, United StatesCopyright © 2023 Untiveros, Sanchez and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Libertine Rose S. Sanchez, bHNzYW5jaGV6QHVwLmVkdS5waA==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.