- 1Science Unit, Lingnan University, Tuen Mun, Hong Kong SAR, China
- 2Museum für Naturkunde, Leibniz Institut für Evolutions- und Biodiversitätsforschung, Berlin, Germany
- 3Department of Zoology, Faculty of Science, Kasetsart University, Bangkok, Thailand
- 4Museum of Vertebrate Zoology, Department of Integrative Biology, University of California, Berkeley, Berkeley, CA, United States
- 5Animal Systematics Research Unit, Department of Biology, Faculty of Sciences, Chulalongkorn University, Bangkok, Thailand
- 6Florida Museum of Natural History, University of Florida, Gainesville, FL, United States
Editorial on the Research Topic
Recent advances in museomics: revolutionizing biodiversity research
Introduction
Museomics, a term coined by Drs. Stephan Schuster and Webb Miller in ~2009, refers to “the large-scale analysis of the DNA content of museum collections” (http://mammoth.psu.edu/museomics.html). Although such DNA studies existed before the term was first used, “museomics” highlighted the importance of specimens in biological studies.
Specimens in natural history collections (NHCs) have been collected for hundreds of years to document the spatial and temporal occurrences of species. It is estimated that NHCs worldwide house 3 billion specimens (Soberon, 1999). These specimens preserve a wealth of information, such as morphological and genetic data on the identity and phylogenetics of species, biogeographic and ecological data, and even biographical information of the collectors, and the contributions of NHCs extend well-beyond organismal biology research to fields such as public health (Suarez and Tsutsui, 2004; Cook et al., 2020) and education (Ellwood et al., 2020; Lendemer et al., 2020; National Academies of Sciences Engineering and Medicine, 2020). NHCs are valuable resources with unknown future potential, and there are countless examples of research made possible that was not the goal of the original collector (Heberling et al., 2019; Miller et al., 2020). We provide three examples. First, Moritz et al. (2008) compared modern specimens of small mammals to those collected ~100 years prior to document how climate change caused the distributions of some species to shift in elevation. Second, bird egg collections in museums were instrumental in showing the role of DDT in causing egg-shell thinning that adversely affected raptor and pelican populations (Ratcliffe, 1967; Hickey and Anderson, 1968). Lastly, Freelance et al. (2022) stress the importance of properly designing captive breeding programs, since the sensory organs of the endangered Lord Howe Island stick insect (Dryococelus australis) differed between wild specimens (>100 years old) and individuals bred in captivity. Given the accelerated rate of biodiversity loss, the role of NHCs will increase in prominence by being an archive of genetic and phenotypic diversity across space and time for many species that have gone extinct or where populations have vanished.
Similarly in terms of unexpected potential, the advent of DNA sequencing technology opened up new avenues for specimen-based research. Modern specimen preparation now includes special steps to preserve DNA/RNA in tissues (e.g., freezing or placing tissues in ethanol or other storage media) for genetic studies, while previously there were no special efforts to preserve the DNA. There are challenges working with these materials, such as DNA naturally degrading over time and the DNA of formalin-fixed specimens being cross-linked with proteins and other DNA (Raxworthy and Smith, 2021). Advances in laboratory methods and new sequencing technologies (e.g., high throughput short-read sequencing) have facilitated improvements in our ability to recover and sequence DNA from museum specimens.
There are four primary sources of DNA that we discuss here: ancient DNA (aDNA), historical DNA (hDNA), modern DNA, and archival DNA (Raxworthy and Smith, 2021). DNA extracted from samples that died under natural circumstances and were later recovered from the field are referred to as aDNA. Familiar examples of aDNA include samples obtained from species such as mammoths and cave bears, which can be quite old and are often >200 years in age. In contrast, DNA extracted from formalin-fixed or ethanol-fixed specimens that were preserved and stored in museum collections is referred to as hDNA (these specimens are usually <200 years old). DNA extracted from tissue samples specifically prepared with genetic analysis in mind is referred to as modern DNA and is usually <40 years old. Archival DNA refers to hDNA and modern DNA stored in museum specimens. The first studies from researchers using the word “museomics” sequenced mitochondrial genomes from the aDNA in hair of the extinct Siberian mammoth (Gilbert et al., 2008) and Tasmanian tiger (Miller et al., 2009).
This Research Topic is a collection of studies highlighting advances in museomics, both in demonstrating applications and refining methodologies. Some applications demonstrated in this Research Topic include using DNA barcoding of a degraded whale sample to identify it to subspecies (Ren et al.), obtaining data from a holotype to verify the existence of an undescribed rodent genus (Castañeda-Rico et al.), obtaining DNA from hundreds of herbarium specimens to elucidate the phylogeography of the genus Dalbergia (Sotuyo et al.), and using target capture to understand the phylogenetic placement of two rare shark species (Agne, Naylor et al.). These studies are diverse in the DNA type used (hDNA and modern DNA), taxa studied, objectives, and approaches. A variety of factors have been identified that affect the performance of sequencing DNA from specimens, and a major goal of museomics is to develop a set of best practices to maximize success (Raxworthy and Smith, 2021). Efforts are being made to document and understand these factors (e.g., Irestedt et al., 2022), and this Research Topic was initiated to further this cause. As an overview of this Research Topic, we identify several factors being addressed across the articles (Figure 1). Following the terminology of Roycroft et al., we organize these factors temporally in the research process as pre-sequencing and post-sequencing (Figure 1). This list of factors is not exhaustive, but rather highlights those that are addressed in this Research Topic. We note that findings in different studies may contradict each other, highlighting the dynamic state of the field and the need for more exhaustive research on this topic.
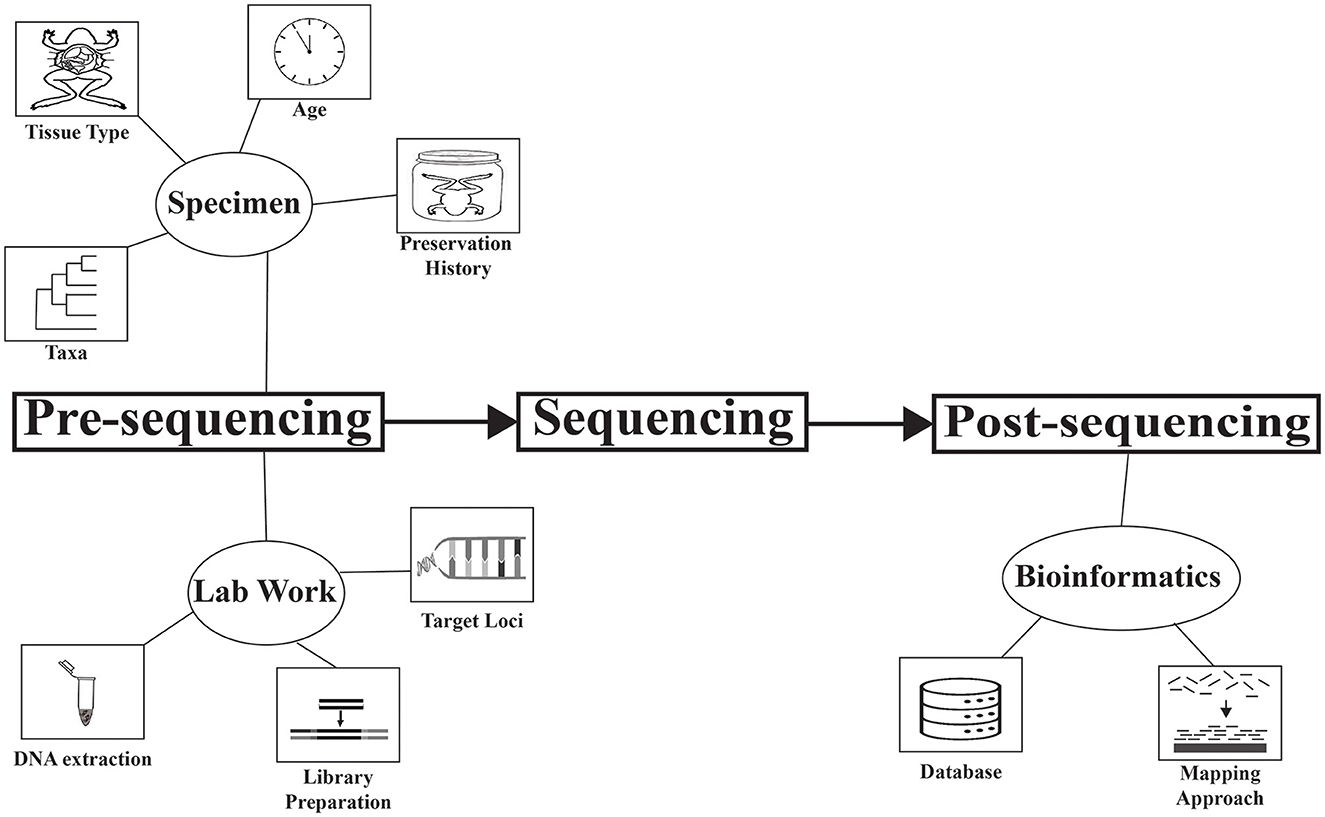
Figure 1. Factors that influence the data-quality and success of museomic studies, addressed in this Research Topic. Factors are organized temporally in the research process: pre-sequencing and post-sequencing.
Pre-sequencing
Pre-sequencing factors dealt with in these studies are either related to the specimen or methodological advances to improve our ability to obtain DNA from historical collections.
Specimen-related factors
Four specimen-related factors are addressed: taxa, tissue type, age, and preservation history. A diversity of taxa was targeted across studies (mammals, insects, gastropods, bony fish, cartilaginous fish, reptiles, sponges, polychaetes, crustaceans, amphibians, plants, arachnids, birds), with mammals being the most frequent focal group (six studies). Agne, Preick et al. included samples from nine classes of animals and found lower success with crustaceans, insects, and cartilaginous fish, and higher success with sponges, gastropods, polychaetes, and amphibians. Another study on gastropods (Clewing et al.) noted that mollusks can be difficult to work with because their tissues are high in mucopolysaccharides, which can hinder DNA extraction.
Several studies compared the performance of different tissue types. In a study of wolf specimens comparing tissue types (jaw bone, nasal bone, skin), skin had the best performance and should be preferred because it is less destructive to the specimen (Pacheco et al.). In contrast, Roycroft et al. found in their mammal study that DNA extraction from toe pad and bone tissue performed better than with skin.
The importance of the age of specimens was commonly explored in these studies, with both types of archival DNA (hDNA and modern DNA) investigated across studies. The oldest specimen included was 192 years old (Agne, Preick et al.). Some studies found a negative correlation between age and DNA yield (Bernstein and Ruane; Hawkins et al.; Roycroft et al.), while others found no relationship (Nunes et al.; Pacheco et al.; Pavlek et al.).
Preservation history is an important factor that can be difficult to evaluate because the entire preservation process is usually not fully documented. Frozen tissue, as expected, preserves DNA better than other methods (Speer et al.). Agne, Preick et al. found that dry specimens performed better than wet across a variety of taxa, while Nunes et al. found the opposite for insects where ethanol-preserved specimens performed better than dry papered and pinned specimens. Variation within preservation types, obscuring trends, is potentially confounded by the time between euthanization and preservation (Speer et al.).
Lab work-related factors
Three lab work-related factors are target loci, DNA extraction protocol, and method of library preparation.
For target loci, four major approaches were used—target capture, barcoding, shotgun sequencing, and cDNA sequencing. The approach used was largely determined by the objective of the study. One common theme is that the loci targeted are short in length, due to the tendency of DNA to fragment over time in historical and ancient tissues.
For DNA extraction, Hawkins et al. compared four methods (spin column, spin column with aDNA modifications, magnetic beads, and phenol chloroform) and found that the spin column and phenol chloroform methods outperformed magnetic beads. The spin column with aDNA modifications retained smaller fragments but took more time and was more expensive. Taking into consideration performance, cost, time, and toxicity, they recommended the spin column method.
For library preparation, Roycroft et al. compared the performance of single and dual barcoded library indexing strategies. They found that sequencing performance was better with dual barcoded libraries, having more reads and lower heterozygosity (=less cross contamination) compared to single barcoded libraries.
Post-sequencing
Post-sequencing factors addressed in these studies are related to the bioinformatic approaches.
Bioinformatic approaches
Two bioinformatic approaches were addressed in these studies: database and mapping approach.
Databases are important in genetic studies, especially when identifying an unknown sample or determining its evolutionary relationship with other taxa. Existing data in a database may affect the resolution of genetic analyses. Nakazato and Jinbo compared two commonly used DNA databases (GenBank and BOLD) and found that data for barcode loci are not the same in each database, despite each database importing from each other. This finding highlights the need of researchers to cross reference databases for relevant data.
To identify the genetic location of sequence reads and compare homologous loci, a mapping approach can be used. Erroneous read mapping can impact the results of a population genetics study, such as estimation of selection or genetic parameters. Roycroft et al. compared the effect of two different mapping approaches (sample-specific historical de novo assembly vs. high-quality “closest sister” de novo assembly) and found that data quality was better when mapping to a high-quality “closest sister” de novo assembly.
Other specimen-based research
Lastly, we note one study that in the strict sense may not qualify as “museomics”, since it is not a genetic study. Balmaki et al. studied plant-pollinator relationships by preparing pollen slides, taking photographs, and using an artificial neural network to help in identification. This approach, compared to metabarcoding, had greater resolution when identifying plant species. We include this study in the Research Topic because it exemplifies the spirit of developing novel research uses of specimens.
Conclusion
In the early 1900s, natural history museums were recognized as an “indispensable feature of modern civilization” due to the growing public interest in nature, their recognition of evolutionary trends in nature, and concerns regarding disappearing biodiversity (Farrington, 1915). Despite their popularity and importance (Allmon, 1994; Suarez and Tsutsui, 2004), NHCs are currently facing a survival crisis of their own due to shrinking budgets (Dalton, 2003; Gropp, 2004; Pennisi, 2020). To survive, NHCs need to find creative ways to publicize and acknowledge the usefulness of specimens and their data (Schindel and Cook, 2018; Miller et al., 2020; National Academies of Sciences Engineering and Medicine, 2020). Some ideas proposed are to develop an “extended specimen network” digitizing and linking all associated data to a specimen (Lendemer et al., 2020) and to recognize NHCs as coauthors on research articles (Rouhan et al., 2017). We are heartened to see museomics helping to expand interest in specimen-based research while showcasing the importance of natural history collections, and we look forward to seeing how newly developed technologies are used to study existing specimens.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
We would like to thank all the contributors to this Research Topic.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allmon, W. D. (1994). The value of natural history collections. Curator 37, 83–89. doi: 10.1111/j.2151-6952.1994.tb01011.x
Cook, J. A., Arai, S., Armien, B., Bates, J., Bonilla, C. A. C., Cortez, M. B. D. S., et al. (2020). Integrating biodiversity infrastructure into pathogen discovery and mitigation of emerging infectious diseases. Bioscience 70, 531–534. doi: 10.1093/biosci/biaa064
Dalton, R. (2003). Natural history collections in crisis as funding is slashed. Nature 423, 575. doi: 10.1038/423575a
Ellwood, E. R., Sessa, J. A., Abraham, J. K., Budden, A. E., Douglas, N., Guralnick, R., et al. (2020). Biodiversity science and the twenty-first century workforce. Bioscience 70, 119–121. doi: 10.1093/biosci/biz147
Farrington, O. C. (1915). The rise of natural history museums. Science 42, 197–208. doi: 10.1126/science.42.1076.197
Freelance, C. B., Magrath, M. J. L., Elgar, M. A., and Wong, B. B. M. (2022). Long-term captivity is associated with changes to sensory organ morphology in a critically endangered insect. J. Appl. Ecol. 59, 504–513. doi: 10.1111/1365-2664.14069
Gilbert, M. T. P., Tomsho, L. P., Rendulic, S., Packard, M., Drautz, D. I., Sher, A., et al. (2008). Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science 317, 1927–1930. doi: 10.1126/science.1146971
Gropp, R. E. (2004). Budget cuts affecting natural history. Science 306, 811. doi: 10.1126/science.306.5697.811b
Heberling, J. M., Prather, L. A., and Tonsor, S. J. (2019). The changing uses of herbarium data in an era of global change: an overview using automated content analysis. Bioscience 69, 812–822. doi: 10.1093/biosci/biz094
Hickey, J. J., and Anderson, D. W. (1968). Chlorinated hydrocarbons and eggshell changes in raptorial and fish-eating birds. Science 162, 271–273. doi: 10.1126/science.162.3850.271
Irestedt, M., Thorn, F., Muller, I. A., Jonsson, K. A., Ericson, E. G. P., and Blom, M. P. K. (2022). A guide to avian museomics: insights gained from resequencing hundreds of avian study skins. Mol. Ecol. Resour. 22, 2672–2684. doi: 10.1111/1755-0998.13660
Lendemer, J., Thiers, B., Monfils, A. K., Zaspel, J., Ellwood, E. R., Bentley, A., et al. (2020). The extended specimen network: a strategy to enhance US biodiversity collections, promote research and education. Bioscience 70, 23–30. doi: 10.1093/biosci/biz140
Miller, S. E., Barrow, L. N., Ehlman, S. M., Goodheart, J. A., Greiman, S. E., Lutz, H. L., et al. (2020). Building natural history collections for the twenty-first century and beyond. Bioscience 70, 674–687. doi: 10.1093/biosci/biaa069
Miller, W., Drautz, D. I., Janecka, J. E., Lesk, A. M., Ratan, A., Tomsho, L. P., et al. (2009). The mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus). Genome Res. 19, 213–220. doi: 10.1101/gr.082628.108
Moritz, C., Patton, J. L., Conroy, C. J., Parra, J. L., White, G. C., and Beissenger, S. R. (2008). Impact of a century of climate change on small-mammal communities in Yosemite National Park, U. S. A. Science 322, 261–264. doi: 10.1126/science.1163428
National Academies of Sciences Engineering and Medicine (2020). Biological Collections: Ensuring Critical Research and Education for the 21st Century. Washington, DC: The National Academies Press.
Pennisi, E. (2020). Shuttered natural history museums fight for survival. Science 368, 1042–1043. doi: 10.1126/science.368.6495.1042
Ratcliffe, D. A. (1967). Decrease in eggshell weight in certain birds of prey. Nature 215, 208–210. doi: 10.1038/215208a0
Raxworthy, C. J., and Smith, B. T. (2021). Mining museums for historical DNA: advances and challenges in museomics. Trends Ecol. Evol. 36, 1049–1060. doi: 10.1016/j.tree.2021.07.009
Rouhan, G., Dorr, L. J., Gauthier, L., Clerc, P., Muller, S., and Gaudeul, M. (2017). The time has come for natural history collections to claim co-authorship of research articles. Taxon 66, 1014–1016. doi: 10.12705/665.2
Schindel, D. E., and Cook, J. A. (2018). The next generation of natural history collections. PLoS Biol. 16, e2006125. doi: 10.1371/journal.pbio.2006125
Soberon, J. (1999). Linking biodiversity information sources. TREE 14, 291. doi: 10.1016/S0169-5347(99)01617-1
Keywords: museomics, natural history collection (NHC), historical DNA, archival DNA, DNA barcoding, formalin extraction, target capture, herbaria
Citation: Fong JJ, Blom MPK, Aowphol A, McGuire JA, Sutcharit C and Soltis PS (2023) Editorial: Recent advances in museomics: revolutionizing biodiversity research. Front. Ecol. Evol. 11:1188172. doi: 10.3389/fevo.2023.1188172
Received: 17 March 2023; Accepted: 03 April 2023;
Published: 03 May 2023.
Edited and reviewed by: Mark A. Elgar, The University of Melbourne, Australia
Copyright © 2023 Fong, Blom, Aowphol, McGuire, Sutcharit and Soltis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan J. Fong, am9uZm9uZ0Bsbi5lZHUuaGs=
 Jonathan J. Fong
Jonathan J. Fong Mozes P. K. Blom
Mozes P. K. Blom Anchalee Aowphol
Anchalee Aowphol Jimmy A. McGuire
Jimmy A. McGuire Chirasak Sutcharit
Chirasak Sutcharit Pamela S. Soltis
Pamela S. Soltis