- 1College of Animal Science and Technology, Tarim University, Alar, China
- 2Key Laboratory of Tarim Animal Husbandry Science and Technology, Xinjiang Production and Construction Corps, Alar, China
Hetian sheep have a long history and a wide distribution. They are renowned for their carpet-grade wool, which makes them a valuable genetic resource in China. The survey revealed that Hetian sheep primarily inhabit three distinct ecological environments: mountains (MTS), mountain–grasslands (MGTS), and grasslands (GTS). To understand the evolutionary relationships and germplasm characteristics of Hetian sheep in these diverse environments, we randomly selected 84 healthy adult ewes from each of these ecological regions. We obtained the Illumina Ovine SNP50K BeadChip by extracting DNA from Hetian sheep ear tissue using the phenol-chloroform method. Afterward, we conducted a population genetic structure and genetic diversity analysis of Hetian sheep using the Illumina Ovine SNP50K Beadchip. Principal component analysis (PCA) and neighbor-joining (NJ) phylogenetic analysis indicated that Hetian sheep in three different ecological environments exhibit distinct genetic distances. Admixture analysis indicated that MGTS and GTS share a common ancestral origins. Additionally, the linkage disequilibrium (LD) analysis indicated that M had the highest decay rate, while MG had the lowest decay rate. Furthermore, we identified the overlapping homozygous genomic regions of Hetian sheep in the three ecological environments through runs of homozygosity (ROH) analysis. We subsequently performed gene annotation and enrichment analysis on these overlapping genomic regions. In the MTS environment, we identified 31 candidate genes associated with high-altitude environmental adaptation. These genes are involved in bone cell generation, differentiation, and the maintenance of bone homeostasis (WNT6, WNT10A, and CHSY1); tooth and tongue development (LEF1, TP63, and PRDM16); and hearing and visual functions (RBP4, ATF6, and JAG1). In the GTS environment, we identified 22 candidate genes related to economic traits, including those associated with reproduction (PLA2G4F, ACVR1, and ADCY2) and growth (CAPN3, YAP1, and FGF9). Research indicates that Hetian sheep can be divided at the genomic level into three subtypes: MTS, MGTS, and GTS. This enhances the genetic diversity of Hetian sheep germplasm resources and provides guidance for the conservation of Hetian sheep breeds. Additionally, we have identified genes related to multiparous traits in MGTS and GTS, offering insights for the selection and breeding of multiparous Hetian sheep.
1 Introduction
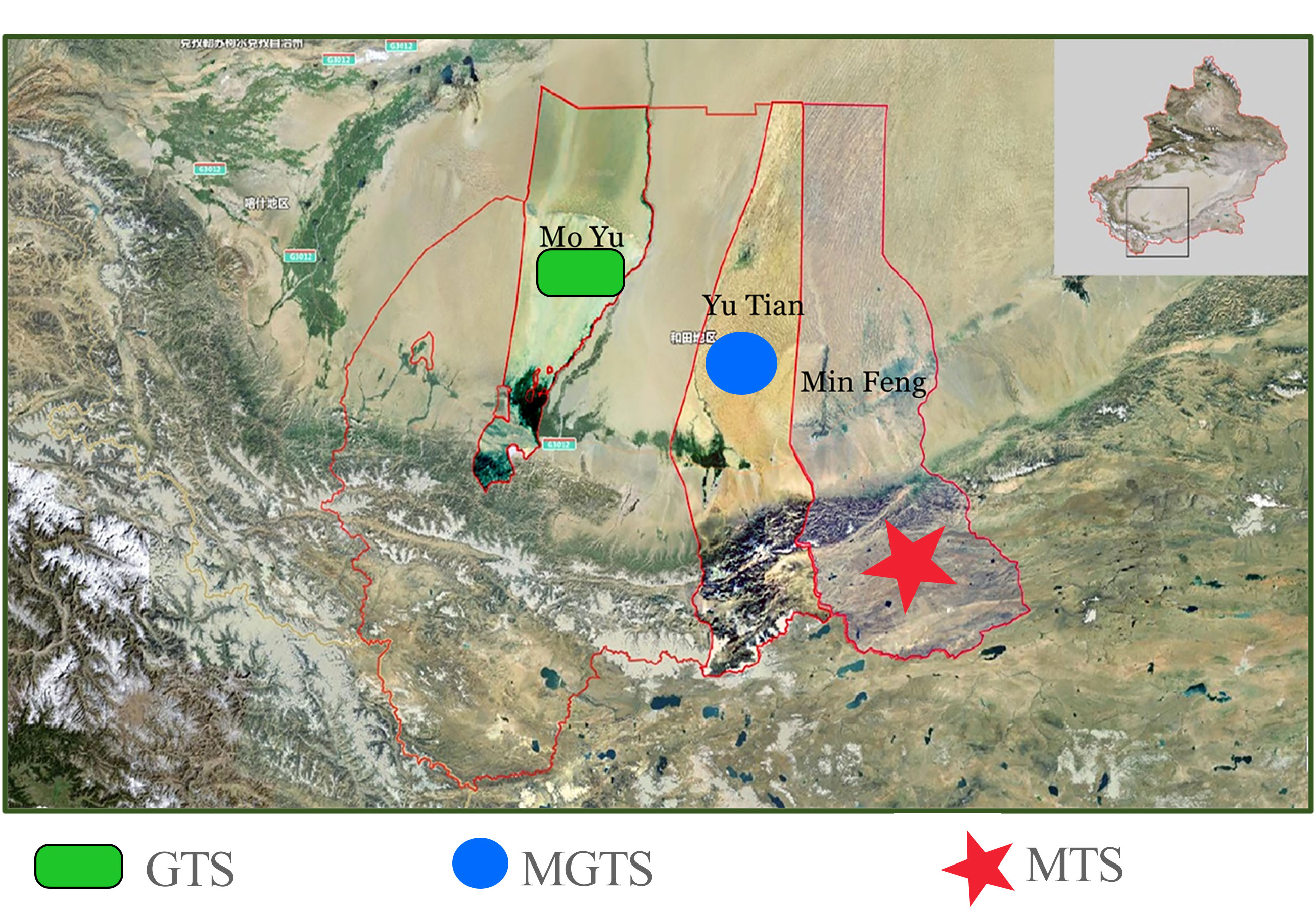
Hetian sheep primarily inhabit the Hetian region in southern Xinjiang, nestled between the Taklimakan Desert to the north and the northern foothills of the Kunlun Mountains to the south. This region experiences minimal rainfall and year-round drought conditions. Hetian sheep are renowned for their resilience in such harsh ecological environments and their prized carpet wool (Doyle et al., 2021). The geographical landscape in the Hetian sheep production area displays distinct topographical differences. Mountains (MTS) are located in the high-altitude mountainous regions in the south, while grasslands (GTS) are found in the low-altitude grasslands in the north, and mountain–grasslands (MGTS) are distributed in areas between high and low altitudes. Additionally, the size and genetic characteristics of Hetian sheep also vary due to their growth environment and feeding methods (Wang et al., 2019). Therefore, gaining a precise understanding of the genetic traits of Hetian sheep is crucial for the preservation of this valuable genetic resource.
Illumina Ovine SNP BeadChip can analyze the variation in single-nucleotide polymorphisms (SNPs) among different individuals. Its emergence provides a powerful tool for population genetics research, allowing us to gain a deeper understanding of the genetic differences and evolutionary history among various subpopulations (Pareek et al., 2011). Adam Abied et al. (2021) used the Ovine Infinium 577K BeadChip to analyze five archetypes of thin-tailed Desert Sheep in Sudan and four breeds of Chinese sheep at the genomic level. They found that thin-tailed Desert Sheep in Sudan exhibited high genetic diversity but low genetic differentiation. Additionally, the study identified potential candidate genes associated with traits such as coat color (PDGFRA), fat deposition (CAMK5), desert environmental adaptability (PSMB1 and SOCS2), and reproductive characteristics (NR5A1 and AP1G1). Zewdu Edea et al. (Edea et al., 2017) used high-density genome-wide SNP to unveil the clustering of five Ethiopian sheep populations based on tail phenotype (short fat-tailed, long fat-tailed, and fat-rumped) and geographic origin (high altitude, mid to high altitude, and low altitude). Tatiana E. Deniskova et al. (Deniskova et al., 2018) utilized Ovine SNP50K BeadChip data to investigate the population structure of 396 Russian sheep from 25 different breeds, concluding that tail type predominantly defined Russian coarse wool cluster. In a comprehensive genome-wide analysis, Jie Jie Cheng et al. (Cheng et al., 2020) investigated the genetic structure of Chaka sheep, uncovering significant genetic divergence between Chaka sheep and Bayinbuluke, Tan, and Oula sheep. This suggests that Chaka sheep have descended from a distinct ancestral lineage. Furthermore, the study identified several genes associated with muscle development (IGF1, GDF3, HDAC9, TGFBR2, CAPN3NF1, and MYOM1). Adam Abied et al. (Abied et al. (2020)), using the Ovine Infinium HD SNP BeadChip, divided five Chinese local sheep breeds (Hetian, Karakul, Yabuyi, Hu, and Wadi sheep) into three genetic clusters based on their tail types and geographical locations. This analysis led to the identification of candidate genes associated with immune response and disease resistance (HERC2, CYFIP1, C5orf30, and ITCH), body weight and digestive metabolism (GJB2, GJA2, PKD2, OMG, and CPAP), and reproductive traits (BMPR1B, TSHR, NF1, and BMP2). In addition to high-density SNP chips, runs of homozygosity (ROH) are also commonly employed in livestock genetics research to evaluate the genetic diversity and kinship among livestock. ROH refers to a continuous DNA sequence in the genome in which an individual inherits the same genotype from both parental copies. ROH length can vary, with longer segments typically indicating more recent common ancestors, while shorter segments suggest more distant common ancestors. M. A. used Illumina Ovine SNP50K BeadChip data to analyze the genetic structure of 5,952 wild Soay sheep under close-kin mating inhibition through ROH analysis. They found that the degree of inbreeding depression decreased with age, suggesting that many alleles with weakly deleterious effects or low frequencies likely contribute to inbreeding depression affecting survival. E. F. Dzomba et al. (2021) utilized Illumina Ovine SNP50K BeadChip data to analyze 14 sheep breeds native to South Africa and 17 sheep breeds of African (6), Asian (2), and European (9) origin to investigate the number and distribution of ROH. The findings revealed that, across all breeds, the majority of detected ROH fell within the smallest 1–6 Mb length category (88.2%). Furthermore, the study identified genes associated with tissue and embryonic growth (SPP1 and LCORL), wool growth regulation (PGFS), adaptive and innate immunity (ADRA4C, GRK3, and SH2BH100), thermogenesis (PLIN199), and selection of horn absence (RXFP2).
Hetian sheep is a renowned breed of wool-producing sheep in China (Doyle et al., 2021). However, our understanding of the genetic evolutionary relationships of Hetian sheep living in different ecological environments remains limited. In this study, based on Illumina Ovine SNP50K BeadChip data and utilizing population genetic diversity and homozygosity analysis methods, we conducted an in-depth investigation into the genetic mechanisms and evolutionary relationships of Hetian sheep inhabiting three distinct ecological environments. This research not only elucidates the genetic differences and evolutionary history among Hetian sheep populations in different ecological environments but also provides fresh insights into the conservation and sustainable utilization of Hetian sheep’s genetic resources.
2 Materials and methods
2.1 Animal ethics
This work was conducted in accordance with the standards set by the Ethics Committee of the College of Animal Science and Technology of Tarim University (SYXK2020-009). Written informed consent was obtained from the owners for the participation of their animals in this study.
2.2 Hetian sheep samples and DNA extraction
For this experiment, we selected 84 adult female Hetian sheep from three distinct ecological regions. These Hetian sheep were of similar age, exhibited good physical health, and had no familial relationships (Figure 1, Table 1, Table S1). Specifically, there were 32 sheep (MTS) from Minfeng County, Xinjiang, 23 sheep (MGTS) from Yutian County, Xinjiang, and 29 sheep (GTS) from Moyu County, Xinjiang. We extracted genomic DNA from ear tissue samples of Hetian sheep using the standard phenol-chloroform method (Green and Sambrook, 2001).
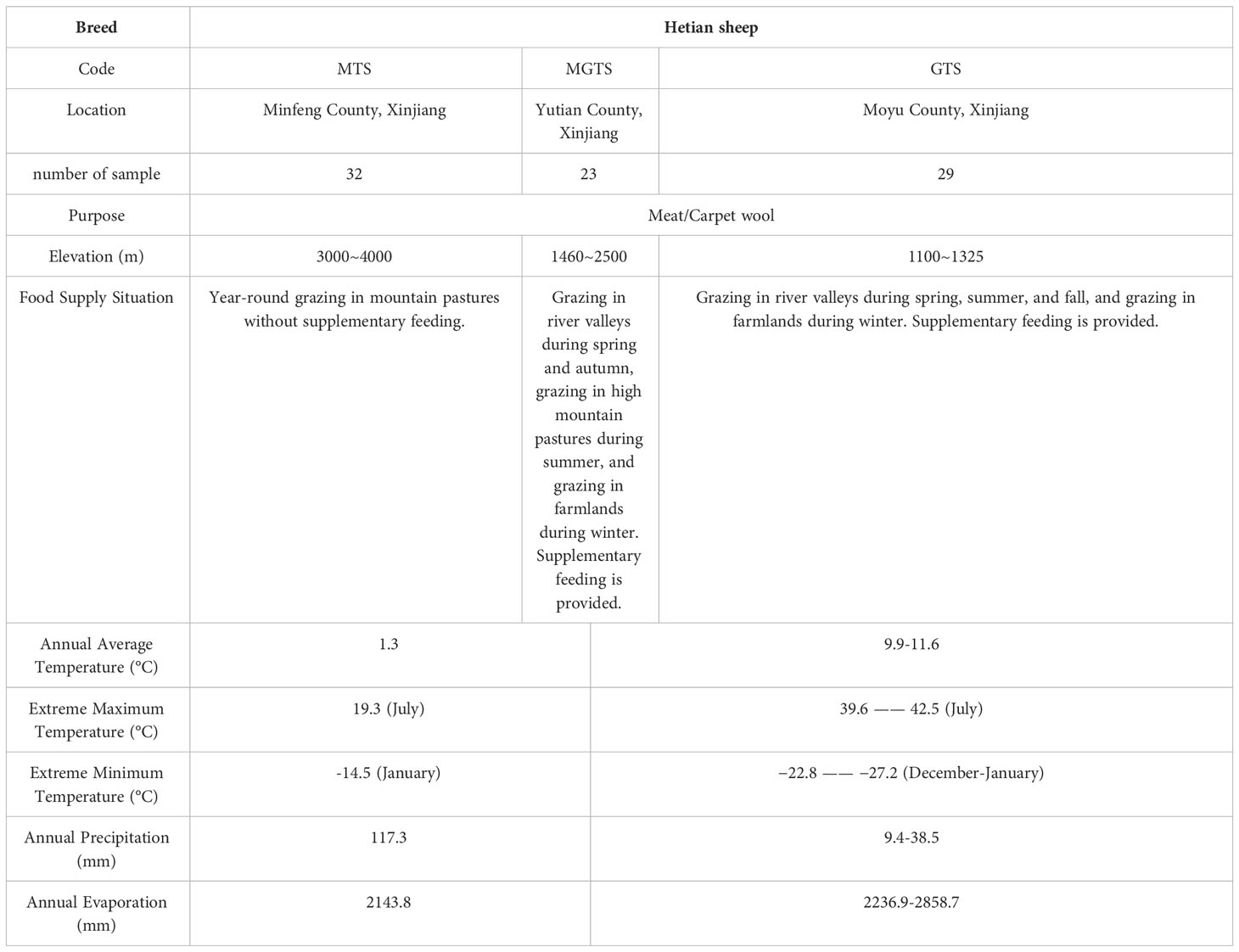
Table 1 Information on the location of the experimental animals and the environmental conditions in which they are found.
2.3 Genotyping and quality control
We genotyped DNA samples using the Illumina Ovine SNP50 BeadChip (Compass Biotechnology Co., Ltd., Beijing, China). The quality of the genotyped SNP data was evaluated using Plink V.1.90 software (Purcell et al., 2007), and any SNPs that did not meet the quality criteria were excluded. The quality control criteria employed in this study were as follows: (1) SNPs with a call rate > 95%; (2) animals with over 5% missing genotypes were excluded; and (3) p-value for Hardy–Weinberg equilibrium ≤0.00001.
2.4 Principal component analysis
Conduct PCA analysis on quality-controlled genotype data using the “-pca” command in PLINK software v.1.905 (Patterson et al., 2006; Gauch et al., 2019), and then create a PCA plot using R software.
2.5 Phylogenetic analysis
We constructed an Identity By State (IBS) matrix using Plink V.1.90 software to quantify genetic similarity between individuals and measure pairwise genetic distance between varieties (Rhodes, 2020). Subsequently, we established the neighbor-joining (NJ) phylogenetic tree using the NJ method in Mega 6.0 software.
2.6 Admixture analysis
For admixture analysis, we assumed Hardy–Weinberg equilibrium, complete linkage equilibrium, and no prior information about individual population origin. We analyzed K values (representing the number of ancestral populations) from 2 to 4 using ADMIXTURE software to determine ancestral origins, infer species origins, and quantify population admixture values (Alexander et al., 2009). We also calculated respective cross-validation (CV) errors.
2.7 LD analysis
In linkage disequilibrium (LD) analysis, we used r2 as a measure of LD, which represents the degree of statistical and genetic correlation between two loci (0 < r2 < 0.6) and remains stable and insensitive to changes in gene frequency (Hill, 1974; Ardlie et al., 2002; Zhao et al., 2007). We employed PopLDdecay software to detect LD levels and attenuation of SNP loci within a 300-kb range in three populations. Subsequently, we generated LD decay plots using the “perlPlot_OnePop.pl” command based on the statistical results.
2.8 ROH analysis
We conducted ROH analysis on the three populations using the homozyg command in PLINK software version 1.905. The analysis parameters were as follows: (1) One missing SNP and a maximum of one potential heterozygous genotype were allowed in ROH. (2) The minimum ROH length was set to 1,000 kb to exclude short ROH segments caused by LD. (3) A minimum SNP density of 100 was required for each ROH segment. (4) The maximum gap between two consecutive SNPs in ROH was set to 250 kb. (5) To reduce false positives, the minimum number of SNPs constituting an ROH was determined using the method proposed by Lentz et al (Lencz et al., 2007).
Among them, IROH represents the minimum number of SNPs constituting an ROH, α is the percentage of false positive ROH (set to 0.05 in this study), nS is the number of SNPs for each individual, ni is the number of individuals, and is the average heterozygosity of all SNPs.
To compare the distribution of ROH in each population, we classified the length of ROH fragments into three groups: 1–5 Mb, 5–10 Mb, and ≥10 Mb. We recorded the number of ROH for each length category and separately calculated the coverage of ROH on each chromosome (ROH%, which is the number of ROH on each chromosome divided by the total number of ROH), as well as the coverage of long ROH on each chromosome (MROH%, which is the number of ROHs ≥10 Mb on each chromosome divided by the total number of ROHs).
2.9 Genome region selection and candidate gene identification
We analyzed the distribution of ROH within the chromosomes of Hetian sheep across three different regions. Specifically, we considered only overlapping ROH segments between MTS and MGTS, as well as between GTS and MGTS, to identify genomic segments potentially inherited by MGTS (Stoffel et al., 2021). These segments were then annotated in the sheep genome Oar_v4.0 based on their physical locations. For gene cluster analysis, we utilized the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/). Additionally, we used Quick Gene Ontology (GO, https://www.ebi.ac.uk/QuickGO/) and the Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/kegg2.html) to retrieve relevant information and functions of annotated genes (Ashburner et al., 2000). Finally, we conducted a literature search on the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/pmc/) to identify candidate genes associated with environmental adaptation.
3 Results
3.1 SNP quality control
Genotypic quality control was performed for 46,327 SNPs from 84 sheep in the experiment. After removing the sex chromosomes and SNPs that did not meet quality control criteria, 44,274 SNPs were left for subsequent analysis.
3.2 Population genetic structure and linkage disequilibrium
The PCA plot results indicate that Hetian sheep from three different environmental regions are clearly distinguished at PC1 and PC2 levels (Figure 2A). At PC1 and PC3 levels, MGTS and GTS cluster together (Figure 2B). At PC2 and PC3 levels, Hetian sheep from three different environmental regions intermingle (Figure 2C).
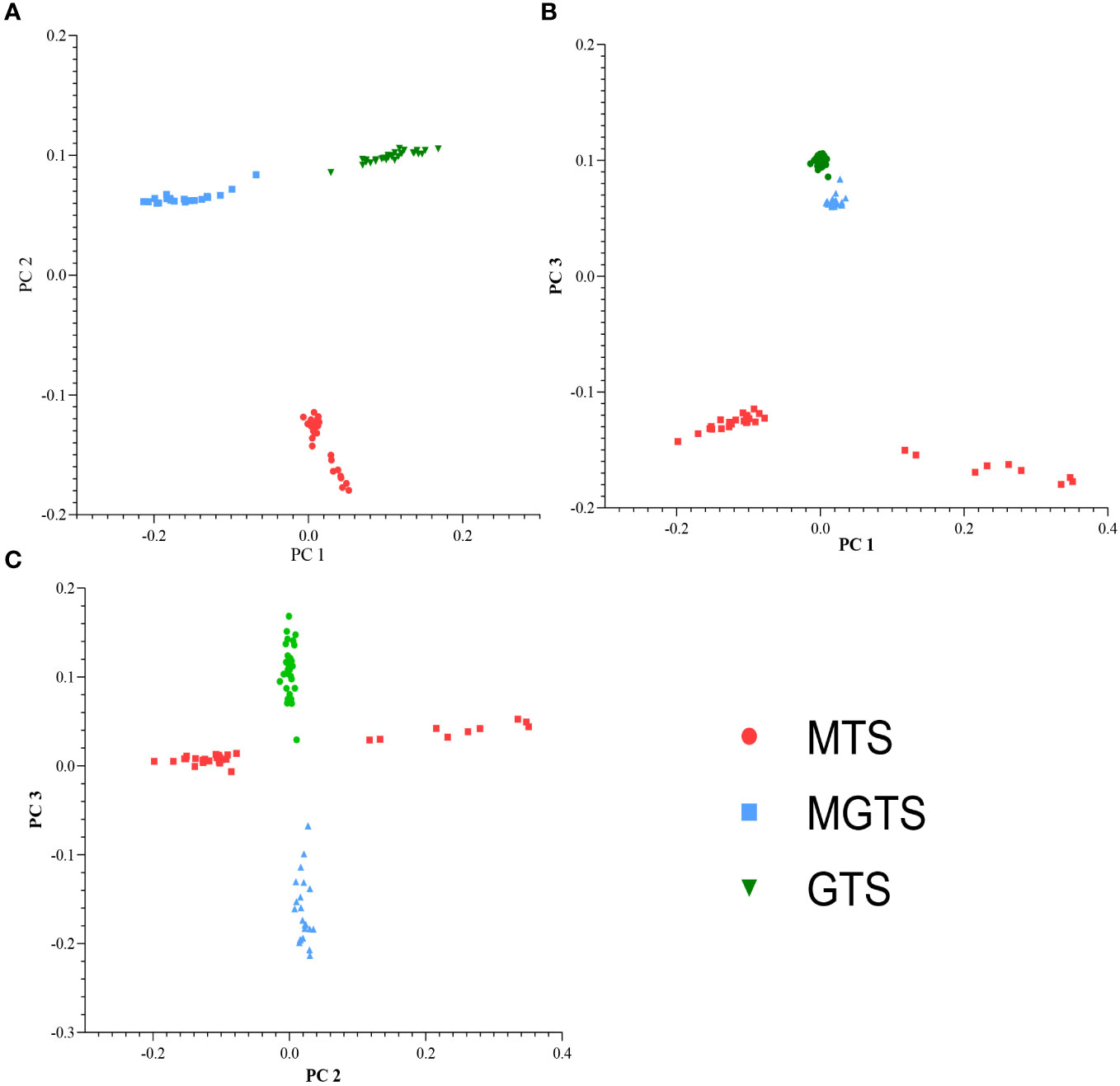
Figure 2 Principal component analysis of Hetian sheep. (A) The analysis was performed for the first two components (PC1 vs. PC2), (B) the first and third components (PC1 vs. PC3), and (C) the second and third components (PC2 vs. PC3).
The results of the N-J tree analysis indicate that Hetian sheep from three different environmental regions can be clearly distinguished based on geographical distance. It is clear that the 84 Hetian sheep can be divided into three groups, with MGTS and GTS showing relatively close genetic distances (Figure 3).
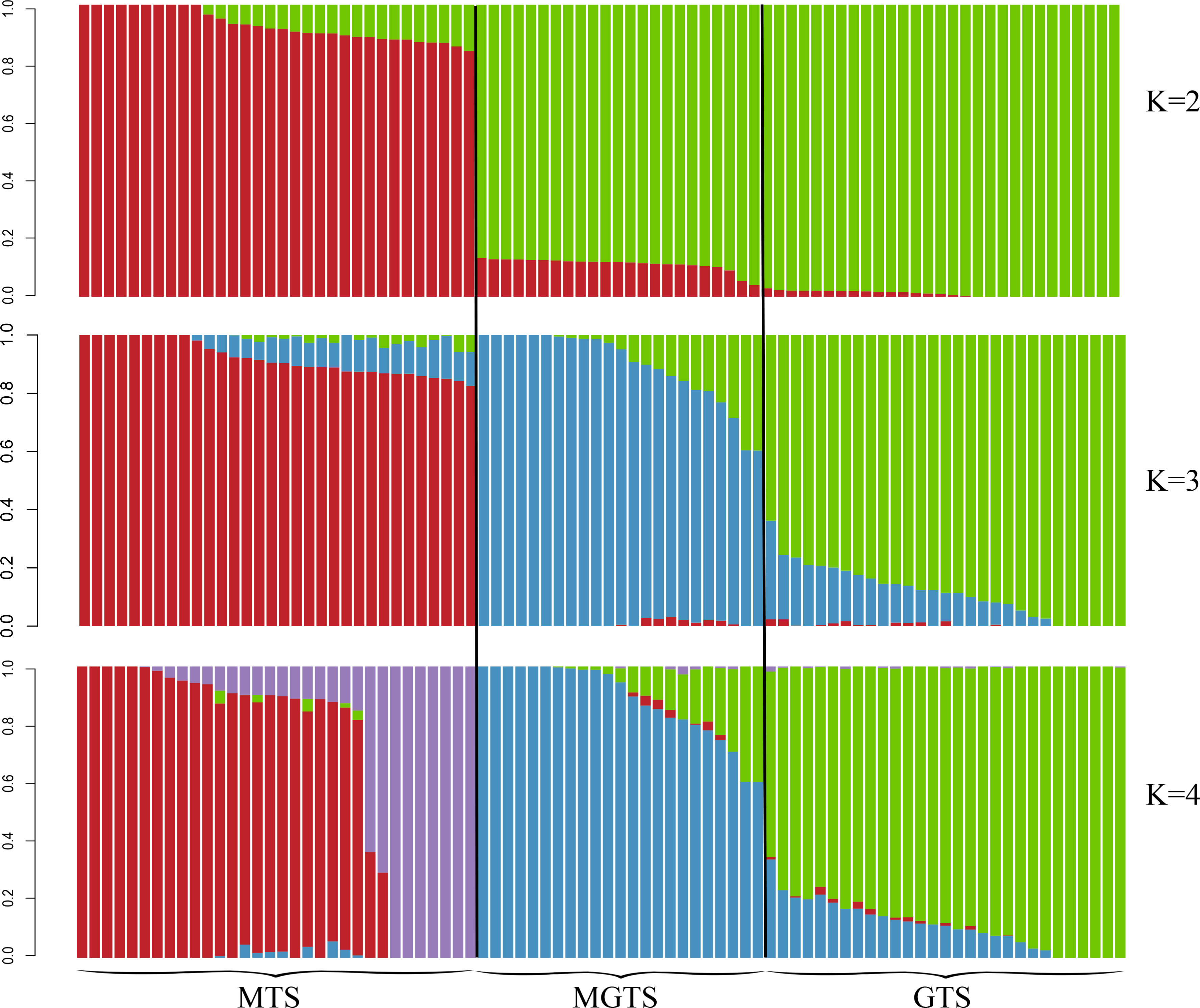
To further investigate the genetic structure of Hetian sheep from different environmental regions, we conducted an Admixture analysis for all individuals. Each column represents a sample. The values on the left, ranging from 0.0 to 1.0, represent the proportion of specific genes, and the values on the right, represented by K, represent the assumed number of ancestors. The results indicate that at K = 2, MGTS and GTS show similar genetic backgrounds. At K = 3, Hetian sheep from the three different environmental regions each have their unique genetic backgrounds (Figure 4). This is consistent with the results from PCA and the N-J tree analysis.
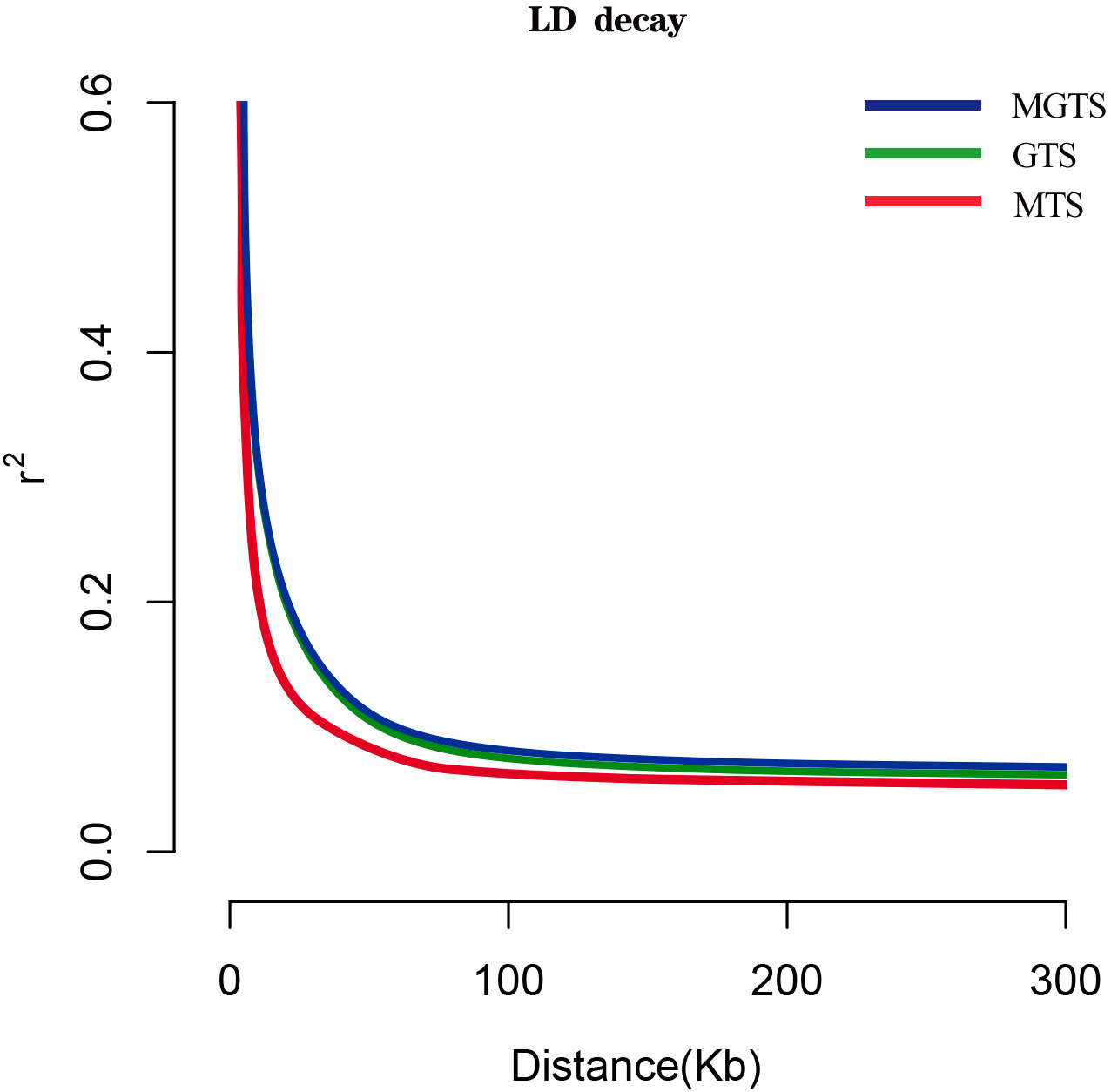
Our observations revealed that the average LD coefficient r2 among Hetian sheep from three different survival environments rapidly decreases within 30 kb, with MTS showing the most rapid decay. Eventually, the LD coefficient r2 levels of GTS and MGTS are higher than those of MTS (Figure 5).
3.3 Runs of homozygosity
A total of 739 ROHs were identified in the genomes of 84 Hetian sheep. Among them, MTS detected 342 ROHs, with 204 in the range of 1–5 Mb, 40 in the range of 5–10 Mb, and 58 exceeding 10 Mb. In GTS, 313 ROHs were observed, with 227 in the 1–5 Mb range, 63 in the 5–10 Mb range, and 23 exceeding 10 Mb. In MGTS, 84 ROHs were detected, with 68 in the 1–5 Mb range, 13 in the 5–10 Mb range, and 3 exceeding 10 Mb. It is worth noting that the majority of ROHs are concentrated within the 10 Mb range (Figure 6A, Tables S2–S4).
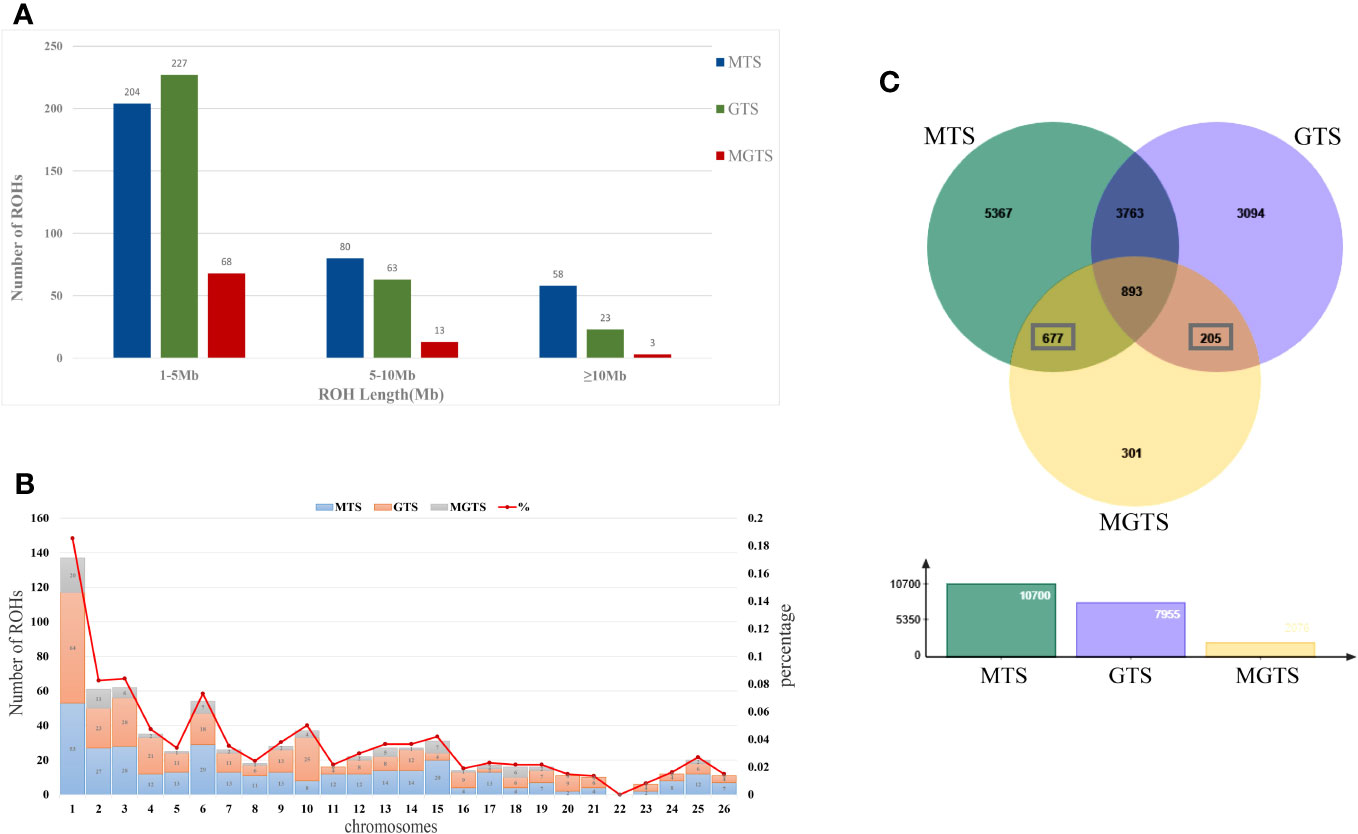
Figure 6 Distribution of runs of homozygosity. (A) Total number of ROHs in each length category. (B) Number of ROHs per autosome and ROH coverage per autosome (ROH%). (C) Three population candidate gene Wayne diagrams.
Among the three groups of Hetian sheep, significant variations were observed in the number of ROHs. MGTS exhibited a notably lower count of ROHs when compared to MTS and GTS. Additionally, within MTS, certain individuals displayed unusually lengthy ROH segments. We assessed the ROH coverage for each autosomal chromosome and observed a non-uniform distribution with a concentration on chromosomes 1, 2, 3, 4, 6, 10, and 15. Notably, chromosome 1 exhibited the highest number of ROHs in all three Hetian sheep groups, while chromosome 22 had the least ROH coverage. Intriguingly, MGTS exhibited a complete absence of ROH segments on chromosomes 11, 20, 21, 22, 23, 24, and 26 (Figure 6B, Tables S2–S4).
3.4 Identification of ROH islands and gene annotation
To elucidate the distinct genetic characteristics of Hetian sheep across three diverse environments, we conducted an annotation of the identified ROH fragments using the Oar_v4.0 reference genome. Specifically, 10,700 specific genes were identified in MTS, 2,076 specific genes in MGTS, and 7,955 specific genes in GTS. To identify shared genetic elements between MTS and MGTS, we conducted an intersection analysis, removing the specific genes also found in GTS. This process left us with 677 candidate genes unique to MTS and MGTS. Similarly, when we performed an intersection analysis between GTS and MGTS, eliminating genes shared with MTS, 205 candidate genes specific to GTS and MGTS were identified (Figure 6C, Table S5).
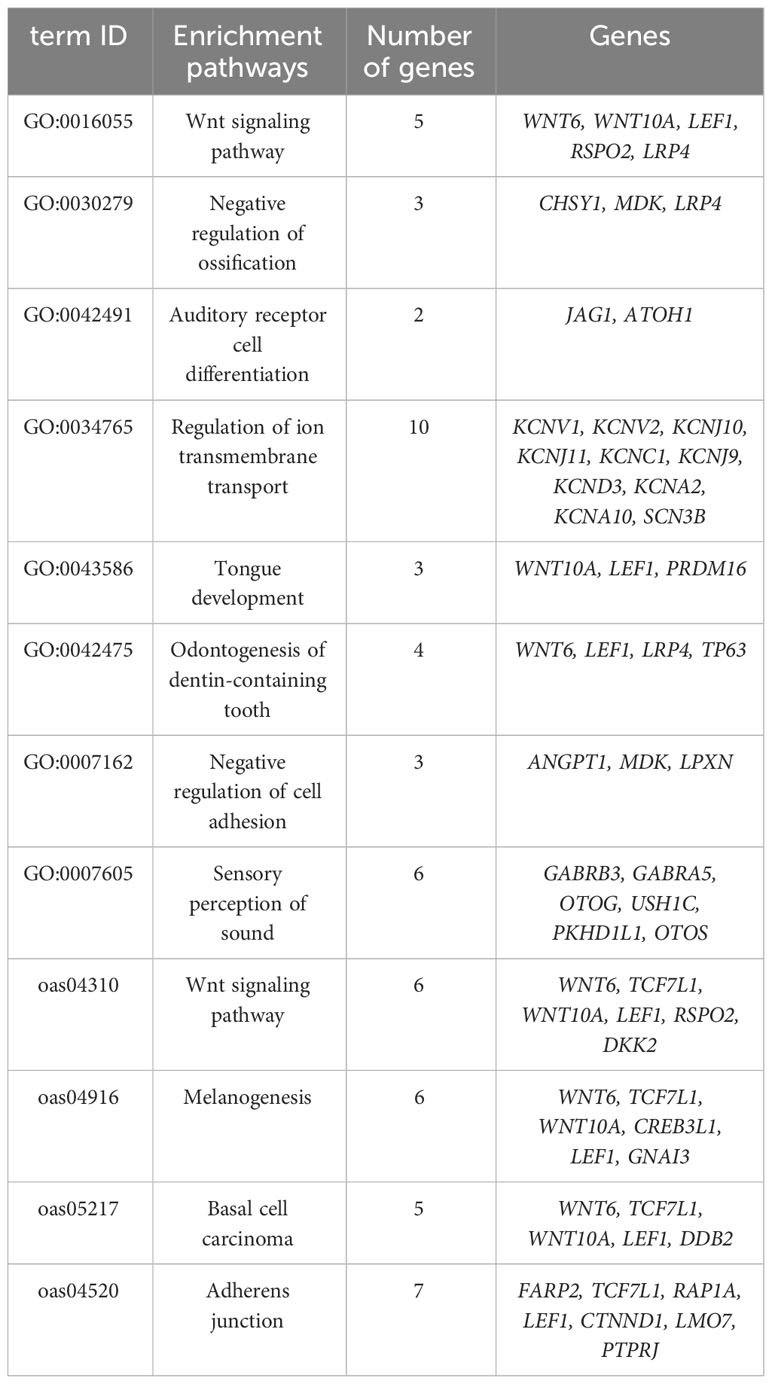
To gain insights into the pathways linked to these genes, we conducted GO and KEGG functional enrichment analyses on MTS- and MGTS-specific genes, resulting in 677 candidate genes enriched across 72 pathways (Table S6). Among these pathways, the top three in significance included GO:0016021 (integral component of membrane), oas04740 (olfactory conductance), and GO:0005886 (plasma membrane). Furthermore, we identified 12 crucial pathways tailored for the high-altitude environment, encompassing pivotal genes associated with neural development, skeletal morphogenesis, tooth production, tongue development, and phenotypes related to vision and hearing (Table 2). Uncovering the genetic changes underpinning the expression of these traits will contribute to our understanding of the genetic mechanisms governing the adaptation of Hetian sheep to the high-altitude environment in natural conditions.
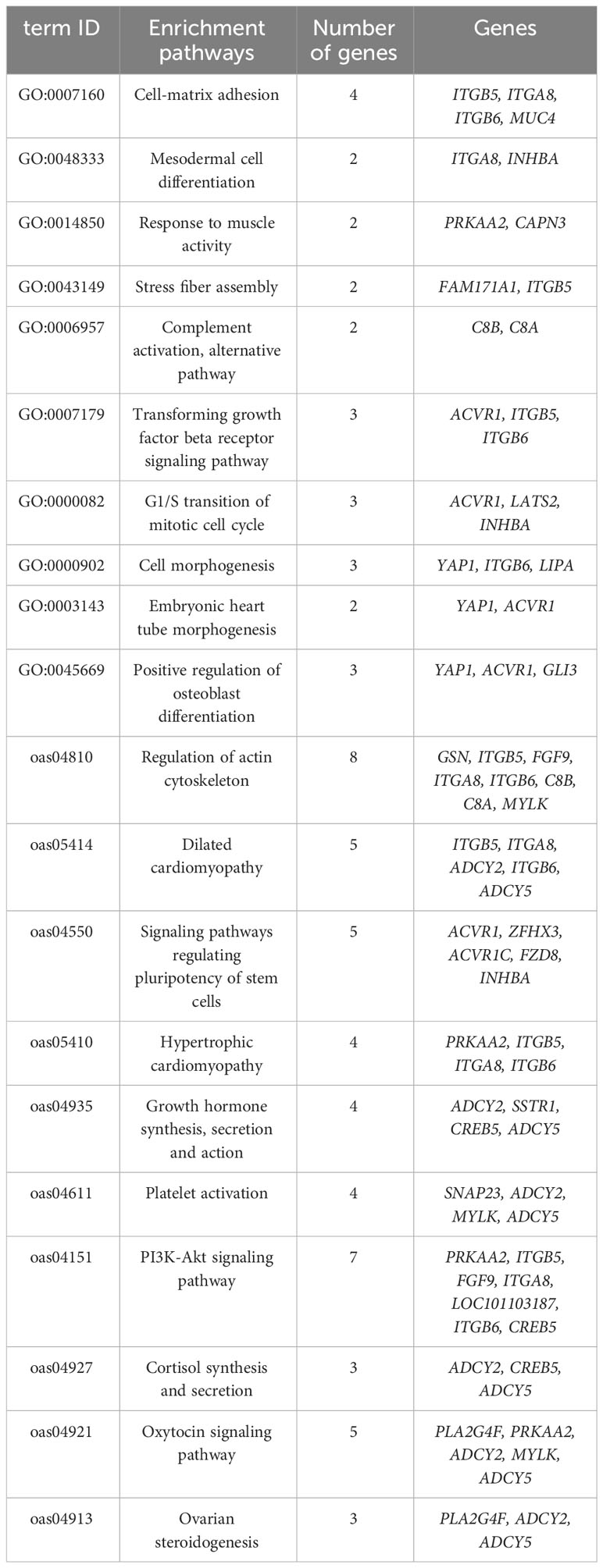
Similarly, we conducted GO and KEGG functional enrichment analyses on GTS- and MGTS-specific genes, yielding 205 candidate genes enriched across 47 pathways (Table S7). Among these, the top three significant pathways included GO:0007160 (cell–matrix adhesion), GO:0048333 (mesodermal cell differentiation) and GO:0014850 (response to muscle activity). Additionally, we identified 20 crucial pathways associated with economic traits, encompassing important genes enriched in reproductive, growth, and immune-related pathways (Table 3). Investigating the genetic changes responsible for these phenotypic alterations will not only enhance our understanding of the genetic mechanisms associated with multiparous traits but also provide valuable breeding strategies to enhance traits productivity.
4 Discussion
4.1 Correlation of population genetic structure
In this study, we used the SNP50K BeadChip to analyze SNP loci in the Hetian sheep genome. PC2 and PC3 displayed a clustering pattern, with all three populations at the same level (Figure 2C), implying a shared pedigree and ancestry among the populations. The phylogenetic tree shows a common “root node”, consistent with the PCA results (Figure 3). These findings indicate minimal genetic distance and strong genetic similarity among the three populations. Importantly, these results align with the known genetic background of Hetian sheep, providing strong evidence for the validity and feasibility of our experimental approach.
4.2 Diversity of population genetic structure
According to PCA, the cluster maps for PC1 and PC3 show GTS and MGTS forming a cohesive cluster, indicating a close genetic relationship (Figure 2B). Typically, this clustering pattern reflects the geographical distribution of the varieties, aligning with the analysis of the shared “root node” between MGTS and GTS in the phylogenetic tree, indicating a common ancestral lineage (Figure 3). Admixture analysis at K = 2–3 confirms MGTS’s origin from GTS (Figure 4). This leads to the hypothesis that MGTS evolved from GTS under selection pressure. The PC1 and PC2 clustering plot clearly demonstrates distinct separation among the three sheep strains (Figure 2A). This separation is also evident in the phylogenetic tree, with each sheep strain forming a unique branch. Notably, Admixture analysis at K = 4 indicates a lack of gene flow between MGTS and GTS. In conclusion (Frichot et al., 2014), at the genomic level, Hetian sheep can be classified into three distinct types: MTS, GTS, and MGTS, which can be clearly distinguished based on geographical location.
4.3 Population linkage genetic degree
In the context of LD, the decay rate is at its lowest when the genetic diversity within the population decreases, resulting in more tightly linked loci (McRae et al., 2002).
Regarding LD attenuation, the minimum LD attenuation distance in MTS corresponds to the maximum attenuation rate, possibly due to its high genetic diversity, which reflects less domestication selection (Figure 5). In contrast, MGTS shows a more advanced stage of evolution, with the maximum LD attenuation distance attributed to significant artificial domestication pressures. The study suggests that MTS is genetically closer to Hetian sheep ancestors, while MGTS has the greatest genetic distance from them. GTS falls between MTS and MGTS, implying that it likely evolved from MTS. Additionally, as agriculture developed in the Grass District, increased fertilizer usage and a staged downhill sheep-shifting method for manure production laid the foundation for MTS evolving into GTS.
4.4 Gene region selection situation
ROHs are genomic regions with complete homozygosity across genetic loci, inherited from parents to offspring. The length and frequency of ROH segments offer valuable insights into the genetic history of a population (Purfield et al., 2012; Joaquim et al., 2019). Longer ROH segments signal high inbreeding, suggesting a closer link to ancestral genetics due to limited chromosome recombination over generations. Conversely, in populations with more distant genetic relationships, lengthy ROH segments are rare (Sams and Boyko, 2019).
In this study, we observed distinct patterns of ROH fragments among the MTS, GTS, and MGTS populations. MTS had fewer ROH fragments in the 0–5 Mb range than GTS but more exceeding 5 Mb, indicating recent intragroup inbreeding and a closer genetic relationship with Hetian sheep ancestors. In contrast, MGTS had significantly fewer and shorter ROH fragments than both MTS and GTS, suggesting prolonged intragroup inbreeding and a more distant genetic connection to Hetian sheep ancestors, which is consistent with LD results. We propose that MTS evolved from mountainous to grassland habitats, giving rise to GTS, while MGTS may represent a distinct lineage resulting from artificial breeding or natural selection within the GTS population.
4.5 Exploring the regulatory mechanisms of high-altitude adaptive pathways
For further investigation into the adaptation mechanism of MGTS and MTS performance traits under high-altitude conditions, we identified 677 genes specific to MGTS and MTS using GO and KEGG enrichment, associated with 72 pathways. Notably, these genes were not expressed in GTS. Bioaccumulation results established a causal link between changes in gene expression and alterations in physiological traits, forming the foundation for adaptive physiological changes at high elevations (Table 2, Table S6).
GO:0034765, related to ion transmembrane transport regulation, includes 10 key genes. Among these, inwardly rectifying potassium channel-related genes are KCNJ9, KCNJ10, and KCNJ11, while voltage-gated potassium channel-related genes consist of KCNA2, KCNA10, KCNC1, KCNC2, KCNC4, KCNV1, and KCNV2. GO:0034765 is fundamental in neural electrophysiology. Notably, potassium channels are central to neuronal excitability and the establishment of a negative resting membrane potential (Qu et al., 2017). Furthermore, they are vital for cellular expression within the central nervous system, spinal cord, kidney, heart, skeletal muscle, and more (Wulff et al., 2009). These genes provide the physiological underpinning that enables Hetian sheep to sense and adapt to various internal and external environmental cues at high altitudes. Subsequently, they control and regulate the activities of diverse bodily systems and organs, coordinate movements, and transmit information via peripheral nerves to maintain a relative balance between the body and its external surroundings.
The genes WNT6, WNT10A, LEF1, RSPO2, and LRP4 are prominently enriched in the GO: 0016055 pathway (Wnt signaling pathway). These genes are closely associated with cell proliferation and differentiation, playing pivotal roles in various biological processes such as bone cell generation, embryonic development, organ formation, hair and tooth development, and regeneration. Additionally, they play integral roles in wound healing and cardiovascular system functioning. WNT6 plays a critical role in multiple aspects of normal development. It contributes not only to embryonic morphogenesis and decidualization but also to tooth formation and the maintenance of mesenchymal stem cell function. Furthermore, WNT6 plays an essential role in post-natal homeostasis (Wei et al., 2020). WNT10A, aside from its signaling function, plays a pivotal role in wound healing by regulating collagen expression and synthesis (Wang et al., 2018). Moreover, it is vital for the development and regulation of ectodermal derivatives, which, in turn, impact the normal formation and regeneration of hair, teeth, skin, and nails (Adaimy et al., 2007). The synergistic effect of WNT6 and WNT10A also promotes osteoblast formation and mutual support (Cawthorn et al., 2012). LEF1, as a key member of the LEF effector protein family, exerts crucial regulatory control over skeletal development. The LEF1 transcription factor is instrumental in both normal osteogenesis and the prevention of bone loss (Luo et al., 2019). RSPO2 not only regulates osteoblast activity and facial morphogenesis but also plays a pivotal role in controlling limb development (Yue et al., 2022). LRP4, on the other hand, influences not only the number, morphology, and growth of teeth but also plays a vital role in maintaining bone homeostasis (Ahn et al., 2017; Martínez-Gil et al., 2022).The Gene Ontology term GO:0030279 (negative regulation of ossification) predominantly includes CHSY1, MDK, and LRP4. CHSY1 is indispensable for skeletal development and the promotion of bone length (Wilson et al., 2012). MDK and LEF1 assume vital roles in cartilage development, skeletal growth, and osteoblast differentiation and regeneration (Haffner-Luntzer et al., 2016; Yan et al., 2022). In the context of GO:0042475 (odontogenesis of dentin-containing tooth), the key genes WNT6, LEF1, LRP4, and TP63 play pivotal roles in dental cell differentiation, skeletal development, and osteoblast differentiation (Ahn et al., 2017; Luo et al., 2019; Wei et al., 2020; Gong et al., 2021). GO:0043586 (tongue development) was primarily enriched with WNT10A, LEF1, and PRDM16. PRDM16 is essential for normal palatogenesis and plays critical and multifaceted roles during embryonic development (Bjork et al., 2010).
The expression patterns of these genes can be elucidated by the challenging conditions of high-altitude environments, which are characterized by sparse vegetation, food scarcity, and limited water sources. In contrast to sheep raised on plains, Hetian sheep in high-altitude regions must actively seek food and water to meet their physiological requirements for survival, thus requiring excellent physical fitness. The expression of genes such as WNT6 and WNT10A serves as the foundation for the formation, proliferation, and differentiation of osteocytes, as well as hair development. Genes like TCFL1, along with others, contribute to the versatile regulation of the cardiovascular system, playing a pivotal role in long-distance travel and adaptation to hypoxia in high-altitude environments. Simultaneously, genes like LEF1 and PRDM16 facilitate the development of teeth and the tongue, providing the necessary conditions for chewing on vegetation and accessing water sources. These characteristics are primarily linked to their ability to thrive in high-elevation environments, including energy expenditure, thermoregulation, and foraging strategies, among other factors.
GO:0007162, associated with the negative regulation of cell adhesion, exhibits significant enrichment, primarily linked to MDK. MDK plays a pivotal role in influencing the proliferation of glial cells and the formation of Muller glial-derived progenitor cells. Furthermore, MDK is notably expressed in the embryonic retina, where it plays a critical role in promoting the survival of retinal neurons (Campbell et al., 2021). On the other hand, GO:0007605, focused on sound perception, is predominantly enriched in OTOG and OTOS. Deficiency in OTOG and OTOS has been associated with hearing loss, particularly when the auditory organ structures remain intact (Delprat et al., 2005; Ganaha et al., 2019). While USH1C is expressed in both the retina and the ear, it is essential to note that its expression level in the retina is comparatively lower than in the ear (Tian et al., 2010). Additionally, PKHD1L1 exhibits specific expression in the apical region of stereocilia, particularly within the high-frequency cochlear region, and is indispensable for maintaining normal hearing function (Wu et al., 2019). GO:0042491, which is associated with auditory receptor cell differentiation, demonstrated significant enrichment primarily related to JAG1 and ATOH1. JAG1 exhibits expression in the neurosensory domain and plays a pivotal role in determining the sensory and neural components of the ear (Pan et al., 2010). Additionally, the reactivation of ATOH1 expression has a direct impact on the number of sensory hair cells within the inner ear, enhancing the ability to detect and balance sounds (Gómez-Dorado et al., 2021). The expression of these genes suggests that Hetian sheep inhabiting high-altitude environments possess superior vision and heightened hearing compared to sheep raised in lowland areas. Consequently, this enhances their alertness, attributes crucial for foraging and evading natural predators within the high-altitude habitat.
Furthermore, the KEGG_PATHWAY analysis revealed the enrichment of four additional intriguing genes: WNT6, WNT10A, LEF1, and TCF7L1, which also exhibited increased significance in the GO analysis. The gene expressions of WNT6 and WNT10A play a crucial role in stimulating osteoblast formation and differentiation, essential for maintaining bone homeostasis (Cawthorn et al., 2012). Notably, bone differentiation has a cascading effect, promoting osteocalcin production and regulating tooth formation (Zhang R. et al., 2021). On a different note, WNT signaling pathways are known to boost the proliferation of melanin stem cells and induce their differentiation for pigment production (Infarinato et al., 2020). LEF1 secretes a variety of growth factors (Cho et al., 2020), providing a degree of protection for the heart and concurrently stimulating osteoblast differentiation while promoting tooth development (Kratochwil et al., 2002; Luo et al., 2019). It is also significantly expressed in melanoma and melanin (Infarinato et al., 2020; Kim et al., 2020). TCF7L1, beyond its role in directly regulating cardiomyocyte differentiation and promoting hair follicle development, has an impact on eye development (Ku et al., 2017; Liang and Liu, 2018; Young et al., 2019). The influence of WNT6, WNT10A, LEF1, and TCF7L1 predominantly encompasses somatic growth, heart function, teeth development, and wool production. These attributes form a foundation for Hetian sheep to adapt to the challenges of migration, hypoxia, foraging, and cold in high-altitude environments. Additionally, LEF1’s role in promoting melanin expression provides protection for the skin and eyes against ultraviolet radiation in the high-altitude environment, benefiting Hetian sheep. However, it is important to note that excessive growth of melanocytes, if unchecked, can lead to melanoma, a cancerous condition.
4.6 Exploring the regulatory mechanisms of low-altitude economic pathways
To gain a deeper understanding of the genetic traits and physiological mechanisms of MGTS and GTS in grassy environments, we focused on identifying unique genes within these groups. Through GO and KEGG enrichment, we identified 205 genes specific to MGTS and GTS, associated with 47 pathways, which were not expressed in MTS. The outcomes of our biological enrichment analysis provide valuable insights into identifying genes linked to crucial economic traits. Furthermore, they enhance our comprehension of the biological processes and mechanisms involved in natural or artificial selection within the Hetian sheep population (Table 2, Table S7).
Pathways such as GO:0045669 (positive regulation of osteoblast differentiation), GO:0000082 (mitotic cell cycle G1/S transition), GO:0003143 (embryonic heart tube morphogenesis), GO:0000902 (cell morphogenesis), GO:0007160 (cell–matrix adhesion), GO:0007179 (transforming growth factor beta receptor signaling pathway), GO:0048333 (mesangial cell differentiation), GO:0014850 (muscle activity response), and GO:0043149 (stress fiber assembly) are enriched with genes primarily responsible for regulating osteoblast proliferation and differentiation, cell morphogenesis, matrix adhesion, stimulation of muscle activity, and various states or activities of organismal muscle cells.
YAP1 plays a crucial role in maintaining normal cerebellar morphology and coordinated movement (Rojek et al., 2019). ITGA8 is responsible for inducing the expression of actin filaments (Marek et al., 2020). FGF9 is involved in regulating myogenesis through its signal transduction mechanism (Huang et al., 2019). ITGB6 is expressed in skeletal muscle tissues (Ducceschi et al., 2014). ACVR1 plays a critical role in bone formation and development (Allen et al., 2020). CAPN3 has significant functions in promoting calcium release from skeletal muscle fibers, facilitating sarcoplasmic reticulum calcium uptake, contributing to muscle formation, and participating in muscle remodeling (Chen et al., 2021). GLI3 regulates the activation of muscle stem cells (Brun et al., 2022). Additionally, INHBA and EIF2S2 are involved in crucial biological processes related to male gonad development and reproduction. INHBA, a mesenchyme-specific gene, collaborates with testosterone to facilitate epididymal coiling by promoting epithelial proliferation (Tomaszewski et al., 2007). Beyond this, INHBA also exerts its influence on sperm production and the maintenance of normal testicular morphogenesis, highlighting its multifunctional significance in male reproductive biology (Archambeault et al., 2011). These characteristics are influenced by the constraints imposed by agricultural production methods. When selecting agricultural production and breeding strategies, there is often a greater focus on the growth, development, and reproductive traits of Hetian sheep, sometimes at the expense of genetic traits. Consequently, this emphasis on specific traits may result in a reduction of the sheep’s genetic potential and lead to a wider genetic divergence from its ancestors.
C8A and C8B are found to be enriched in GO:0006957, which corresponds to complement activation via the alternative pathway. C8A and C8B are two subunits that encode the C8 protein, a key component of the complement system. The complement system plays a crucial role in the immune system, aiding the body in combatting various pathogens such as bacteria and viruses (Mannes et al., 2023; Shah et al., 2023). These findings provide fresh insights into the genomic alterations brought about by artificial selection and their implications for aspects such as fertility, growth, development, and immune-related genes.
Furthermore, in the KEGG_PATHWAY analysis, the genes PLA2G4F, ACVR1, PRKAA2, ADCY2, ADCY5, ZFHX3, and INHBA were enriched in the following pathways: oas04921 (Oxytocin signaling pathway), oas04913 (Ovarian steroidogenesis) and oas04550 (Signaling pathways regulating pluripotency of stem cells). These genes are primarily associated with reproduction-related processes, functioning within pathways involving the fallopian tubes, fetal weight regulation, uterine artery diameter, and various other aspects of the reproductive system. PLA2G4F releases arachidonic acid, a precursor of prostaglandin synthesis, which plays a pivotal role in processes such as ovulation, fertilization, implantation, and parturition. Prostaglandins exert dual effects by either contracting or relaxing the muscle layers of the oviduct, facilitating the transportation of gametes and zygotes (Gauvreau et al., 2010; Cerny et al., 2015). ACVR1 is notably present in granulosa cells during mid-estrus and in pre-ovulation follicles, exhibiting significant elevation during testicular proliferation (da Silveira et al., 2014; Alvarez-Rodriguez et al., 2020). PRKAA2 influences uterine artery diameter, fetal growth, birth weight, and metabolic homeostasis (Griffiths et al., 2020). Additionally, estradiol has been shown to significantly enhance the expression of ADCY2 (Zhao et al., 2020). ZFHX3 assumes a critical role in the proper development of lactation in the mammary gland (Zhao et al., 2016). Furthermore, ADCY5 demonstrates robust associations not only with fetal birth weight but also with the regulation of placental glucose transporter expression, vitamin B2 uptake in the placenta, and the architecture and permeability of the materno-fetal placental barrier (Freathy et al., 2010).
ITGB5, ITGB6, ITGA8, ADCY2, ADCY5, PRKAA2, MYLK, PLA2G4F, and SNAP23 are enriched in pathways associated with inflammatory responses, such as oas05414 (dilated cardiomyopathy), oas05410 (hypertrophic cardiomyopathy), and oas04611 (platelet-activating pathway). For instance, SNAP23 plays a pivotal role in the development of platelets and mast cells, particularly in connective tissue mast cells especially during allergic responses (Cardenas et al., 2021). PLA2G4F has been identified as a potential candidate gene associated with inflammatory bowel disease (Esworthy et al., 2012), and MYLK has been found to mediate regulatory pathways in neutrophils during the innate immune response (Xu et al., 2008).
Furthermore, genes such as ITGB5, ITGB6, ITGA8, ADCY2, ADCY5, PRKAA2, MYLK, SSTR1, CREB5, FGF9, and GSN have been implicated in the regulation of the actin cytoskeleton within pathways like oas04810 (regulation of the actin cytoskeleton), oas04151 (the PI3K-Akt signaling pathway), oas04935 (synthesis, secretion, and actions of growth hormone), and oas04927 (cortisol synthesis and secretion). These genes play pivotal roles in governing cellular growth, development, and metabolism, and modulating actin cytoskeleton dynamics, cellular transcription, translation, proliferation, overall growth, and survival in Hetian sheep. Additionally, they are involved in specific regulatory mechanisms that impact overall body growth and development of muscle cells. These pathways indicate the important economic traits of Hetian sheep in reproduction, growth, development and immunity after artificial selection and breeding. The peptide hormone SSTR1 is considered a promising candidate gene associated with livestock growth traits, overseeing various processes such as cell proliferation and growth hormone release (Li et al., 2021). CREB5 contributes to the maintenance of articular chondrocyte expression (Zhang CH. et al., 2021), while FGF9 plays a crucial role in skeletal development and exhibits high expression levels in mature osteoblasts (Wang et al., 2017). Furthermore, GSN is involved in regulating actin cytoskeleton remodeling and more (Wang et al., 2022). Collectively, these pathways underscore the significant economic traits of Hetian sheep, encompassing reproduction, growth, development, and immunity, all cultivated through artificial selection and breeding efforts.
5 Conclusions
Using the data from Illumina Ovine SNP50K BeadChip, we investigated the genetic structure and ROH in the Hetian sheep population. This novel study is the first to classify Hetian sheep into three distinct subtypes at the genomic level: MTS, MGTS, and GTS. Notably, GTS and MGTS are more closely genetically related to each other. In addition, our ROH analysis revealed the presence of genes associated with multiparous traits in GTS and MGTS, which provides valuable insights for the potential development of Hetian sheep lines with enhanced multiparousal traits.
Data availability statement
The datasets presented in this study can be found in online repositories. A link to the data can be found below: https://doi.org/10.6084/m9.figshare.23504796.v1 The following resources were used in preparing this article:PLINK (RRID:SCR_001757)MEGA Software (RRID:SCR_000667)R-pbutils (RRID:SCR_002995)DAVID (RRID:SCR_001881)QuickGO (RRID:SCR_004608)KEGG (RRID:SCR_012773).
Ethics statement
The studies involving animals were reviewed and approved by the Ethics Committee of Tarim University of Science and Technology (SYXK 2020-009). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
ZH and SL conceived and supervised the study. ZH analyzed the data with contribution from WZ. LZ, RW, CL, and XB contributed samples or provided help during the sample collection. ZH and SL wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
The study was funded by grants from the National Natural Science Foundation of China (No. 32060743) and the Bingtuan Science and Technology Program (2022CB001-09).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1182966/full#supplementary-material
References
Abied A., Ahbara A. M., Berihulay H., Xu L., Islam R., El-Hag F. M., et al. (2021). Genome Divergence and Dynamics in the Thin-Tailed Desert Sheep From Sudan. Front. Genet. 12, 659507. doi: 10.3389/fgene.2021.659507
Abied A., Bagadi A., Bordbar F., Pu Y., Augustino S. M. A., Xue X., et al. (2020). Genomic Diversity, Population Structure, and Signature of Selection in Five Chinese Native Sheep Breeds Adapted to Extreme Environments. Genes 11 (5), 494. doi: 10.3390/genes11050494
Adaimy L., Chouery E., Megarbane H., Mroueh S., Delague V., Nicolas E., et al. (2007). Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia. Am. J. Hum. Genet. 81 (4), 821–828. doi: 10.1086/520064
Ahn Y., Sims C., Murray M. J., Kuhlmann P. K., Fuentes-Antrás J., Weatherbee S. D., et al. (2017). Multiple modes of Lrp4 function in modulation of Wnt/β-catenin signaling during tooth development. Dev. (Cambridge England) 144 (15), 2824–2836. doi: 10.1242/dev.150680
Alexander D. H., Novembre J., Lange K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19 (9), 1655–1664. doi: 10.1101/gr.094052.109
Allen R. S., Tajer B., Shore E. M., Mullins M. C. (2020). Fibrodysplasia ossificans progressiva mutant ACVR1 signals by multiple modalities in the developing zebrafish. eLife 9, e53761. doi: 10.7554/eLife.53761
Alvarez-Rodriguez M., Martinez C., Wright D., Barranco I., Roca J., Rodriguez-Martinez H. (2020). The Transcriptome of Pig Spermatozoa, and Its Role in Fertility. Int. J. Mol. Sci. 21 (5), 1572. doi: 10.3390/ijms21051572
Archambeault D. R., Tomaszewski J., Childs A. J., Anderson R. A., Yao H. H. (2011). Testicular somatic cells, not gonocytes, are the major source of functional activin A during testis morphogenesis. Endocrinology 152 (11), 4358–4367. doi: 10.1210/en.2011-1288
Ardlie K. G., Kruglyak L., Seielstad M. (2002). Patterns of linkage disequilibrium in the human genome. Nature reviews. Genetics 3 (4), 299–309. doi: 10.1038/nrg777
Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25 (1), 25–29. doi: 10.1038/75556
Bjork B. C., Turbe-Doan A., Prysak M., Herron B. J., Beier D. R. (2010). Prdm16 is required for normal palatogenesis in mice. Hum. Mol. Genet. 19 (5), 774–789. doi: 10.1093/hmg/ddp543
Brun C. E., Sincennes M. C., Lin A. Y. T., Hall D., Jarassier W., Feige P., et al. (2022). GLI3 regulates muscle stem cell entry into GAlert and self-renewal. Nat. Commun. 13 (1), 3961. doi: 10.1038/s41467-022-31695-5
Campbell W. A., Fritsch-Kelleher A., Palazzo I., Hoang T., Blackshaw S., Fischer A. J. (2021). Midkine is neuroprotective and influences glial reactivity and the formation of Müller glia-derived progenitor cells in chick and mouse retinas. Glia 69 (6), 1515–1539. doi: 10.1002/glia.23976
Cardenas R. A., Gonzalez R., Sanchez E., Ramos M. A., Cardenas E. I., Rodarte A. I., et al. (2021). SNAP23 is essential for platelet and mast cell development and required in connective tissue mast cells for anaphylaxis. J. Biol. Chem. 296, 100268. doi: 10.1016/j.jbc.2021.100268
Cawthorn W. P., Bree A. J., Yao Y., Du B., Hemati N., Martinez-Santibañez G., et al. (2012). Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone 50 (2), 477–489. doi: 10.1016/j.bone.2011.08.010
Cerny K. L., Garrett E., Walton A. J., Anderson L. H., Bridges P. J. (2015). A transcriptomal analysis of bovine oviductal epithelial cells collected during the follicular phase versus the luteal phase of the estrous cycle. Reprod. Biol. Endocrinol.: RB&E 13, 84. doi: 10.1186/s12958-015-0077-1
Chen L., Tang F., Gao H., Zhang X., Li X., Xiao D. (2021). CAPN3: A muscle−specific calpain with an important role in the pathogenesis of diseases (Review). Int. J. Mol. Med. 48 (5), 203. doi: 10.3892/ijmm.2021.5036
Cheng J., Zhao H., Chen N., Cao X., Hanif Q., Pi L., et al. (2020). Population structure, genetic diversity, and selective signature of Chaka sheep revealed by whole genome sequencing. BMC Genomics 21 (1), 520. doi: 10.1186/s12864-020-06925-z
Cho H. M., Lee K. H., Shen Y. M., Shin T. J., Ryu P. D., Choi M. C., et al. (2020). Transplantation of hMSCs Genome Edited with LEF1 Improves Cardio-Protective Effects in Myocardial Infarction. Mol. Ther. Nucleic Acids 19, 1186–1197. doi: 10.1016/j.omtn.2020.01.007
da Silveira J. C., Carnevale E. M., Winger Q. A., Bouma G. J. (2014). Regulation of ACVR1 and ID2 by cell-secreted exosomes during follicle maturation in the mare. Reprod. Biol. Endocrinol.: RB&E 12, 44. doi: 10.1186/1477-7827-12-44
Delprat B., Ruel J., Guitton M. J., Hamard G., Lenoir M., Pujol R., et al. (2005). Deafness and cochlear fibrocyte alterations in mice deficient for the inner ear protein otospiralin. Mol. Cell. Biol. 25 (2), 847–853. doi: 10.1128/MCB.25.2.847-853.2005
Deniskova T. E., Dotsev A. V., Selionova M. I., Kunz E., Medugorac I., Reyer H., et al. (2018). Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genetics Selection Evolution: GSE 50 (1), 29. doi: 10.1186/s12711-018-0399-5
Doyle E. K., Preston J. W. V., McGregor B. A., Hynd P. I. (2021). The science behind the wool industry. The importance and value of wool production from sheep. Anim. Frontiers: Rev. Magazine Anim. Agric. 11 (2), 15–23. doi: 10.1093/af/vfab005
Ducceschi M., Clifton L. G., Stimpson S. A., Billin A. N. (2014). Post-transcriptional regulation of ITGB6 protein levels in damaged skeletal muscle. J. Mol. Histol. 45 (3), 329–336. doi: 10.1007/s10735-014-9567-2
Dzomba E. F., Chimonyo M., Pierneef R., Muchadeyi F. C. (2021). Runs of homozygosity analysis of South African sheep breeds from various production systems investigated using OvineSNP50k data. BMC Genomics 22 (1), 7. doi: 10.1186/s12864-020-07314-2
Edea Z., Dessie T., Dadi H., Do K. T., Kim K. S. (2017). Genetic Diversity and Population Structure of Ethiopian Sheep Populations Revealed by High-Density SNP Markers. Front. Genet. 8, 218. doi: 10.3389/fgene.2017.00218
Esworthy R. S., Kim B. W., Rivas G. E., Leto T. L., Doroshow J. H., Chu F. F. (2012). Analysis of candidate colitis genes in the Gdac1 locus of mice deficient in glutathione peroxidase-1 and -2. PloS One 7 (9), e44262. doi: 10.1371/journal.pone.0044262
Freathy R. M., Mook-Kanamori D. O., Sovio U., Prokopenko I., Timpson N. J., Berry D. J., et al. (2010). Variants in ADCY5 and near CCNL1 are associated with fetal growth and birth weight. Nat. Genet. 42 (5), 430–435. doi: 10.1038/ng.567
Frichot E., Mathieu F., Trouillon T., Bouchard G., François O. (2014). Fast and efficient estimation of individual ancestry coefficients. Genetics 196 (4), 973–983. doi: 10.1534/genetics.113.160572
Ganaha A., Kaname T., Yanagi K., Tono T., Higa T., Suzuki M. (2019). Clinical characteristics with long-term follow-up of four Okinawan families with moderate hearing loss caused by an OTOG variant. Hum. Genome Variation 6, 37. doi: 10.1038/s41439-019-0068-4
Gauch H. G., Qian S., Piepho H. P., Zhou L., Chen R. (2019). Consequences of PCA graphs, SNP codings, and PCA variants for elucidating population structure. PloS One 14 (6), e0218306. doi: 10.1371/journal.pone.0218306
Gauvreau D., Moisan V., Roy M., Fortier M. A., Bilodeau J. F. (2010). Expression of prostaglandin E synthases in the bovine oviduct. Theriogenology 73 (1), 103–111. doi: 10.1016/j.theriogenology.2009.08.006
Gómez-Dorado M., Daudet N., Gale J. E., Dawson S. J. (2021). Differential regulation of mammalian and avian ATOH1 by E2F1 and its implication for hair cell regeneration in the inner ear. Sci. Rep. 11 (1), 19368. doi: 10.1038/s41598-021-98816-w
Gong F., Gao L., Ma L., Li G., Yang J. (2021). Uncarboxylated osteocalcin alleviates the inhibitory effect of high glucose on osteogenic differentiation of mouse bone marrow-derived mesenchymal stem cells by regulating TP63. BMC Mol. Cell Biol. 22 (1), 24. doi: 10.1186/s12860-021-00365-7
Green M. R., Sambrook J. F. (2001). Molecular cloning : a laboratory manual. Anal Biochem. 186(1), 182–183. doi: 10.1016/0003-2697(90)90595-Z
Griffiths R. M., Pru C. A., Behura S. K., Cronrath A. R., McCallum M. L., Kelp N. C., et al. (2020). AMPK is required for uterine receptivity and normal responses to steroid hormones. Reprod. (Cambridge England) 159 (6), 707–717. doi: 10.1530/REP-19-0402
Haffner-Luntzer M., Heilmann A., Rapp A. E., Roessler R., Schinke T., Amling M., et al. (2016). Antagonizing midkine accelerates fracture healing in mice by enhanced bone formation in the fracture callus. Br. J. Pharmacol. 173 (14), 2237–2249. doi: 10.1111/bph.13503
Hill W. G. (1974). Estimation of linkage disequilibrium in randomly mating populations. Heredity 33 (2), 229–239. doi: 10.1038/hdy.1974.89
Huang J., Wang K., Shiflett L. A., Brotto L., Bonewald L. F., Wacker M. J., et al. (2019). Fibroblast growth factor 9 (FGF9) inhibits myogenic differentiation of C2C12 and human muscle cells. Cell Cycle (Georgetown Tex) 18 (24), 3562–3580. doi: 10.1080/15384101.2019.1691796
Infarinato N. R., Stewart K. S., Yang Y., Gomez N. C., Pasolli H. A., Hidalgo L., et al. (2020). BMP signaling: at the gate between activated melanocyte stem cells and differentiation. Genes Dev. 34 (23-24), 1713–1734. doi: 10.1101/gad.340281.120
Joaquim L. B., Chud T. C. S., Marchesi J. A. P., Savegnago R. P., Buzanskas M. E., Zanella R., et al. (2019). Genomic structure of a crossbred Landrace pig population. PloS One 14 (2), e0212266. doi: 10.1371/journal.pone.0212266
Kim G. H., Fang X. Q., Lim W. J., Park J., Kang T. B., Kim J. H., et al. (2020). Cinobufagin Suppresses Melanoma Cell Growth by Inhibiting LEF1. Int. J. Mol. Sci. 21 (18), 6706. doi: 10.3390/ijms21186706
Kratochwil K., Galceran J., Tontsch S., Roth W., Grosschedl R. (2002). FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1(-/-) mice. Genes Dev. 16 (24), 3173–3185. doi: 10.1101/gad.1035602
Ku A. T., Shaver T. M., Rao A. S., Howard J. M., Rodriguez C. N., Miao Q., et al. (2017). TCF7L1 promotes skin tumorigenesis independently of β-catenin through induction of LCN2. eLife 6, e23242. doi: 10.7554/eLife.23242
Lencz T., Lambert C., DeRosse P., Burdick K. E., Morgan T. V., Kane J. M., et al. (2007). Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc. Natl. Acad. Sci. United States America 104 (50), 19942–19947. doi: 10.1073/pnas.0710021104
Li X., Ding N., Zhang Z., Tian D., Han B., Liu S., et al. (2021). Identification of Somatostatin Receptor Subtype 1 (SSTR1) Gene Polymorphism and Their Association with Growth Traits in Hulun Buir Sheep. Genes 13 (1), 77. doi: 10.3390/genes13010077
Liang R., Liu Y. (2018). Tcf7l1 directly regulates cardiomyocyte differentiation in embryonic stem cells. Stem Cell Res. Ther. 9 (1), 267. doi: 10.1186/s13287-018-1015-x
Luo Y., Ge R., Wu H., Ding X., Song H., Ji H., et al. (2019). The osteogenic differentiation of human adipose-derived stem cells is regulated through the let-7i-3p/LEF1/β-catenin axis under cyclic strain. Stem Cell Res. Ther. 10 (1), 339. doi: 10.1186/s13287-019-1470-z
Mannes M., Halbgebauer R., Wohlgemuth L., Messerer D. A. C., Savukoski S., Schultze A., et al. (2023). Combined Heterozygous Genetic Variations in Complement C2 and C8B: An Explanation for Multidimensional Immune Imbalance? J. Innate Immun. 15 (1), 412–427. doi: 10.1159/000528607
Marek I., Hilgers K. F., Rascher W., Woelfle J., Hartner A. (2020). A role for the alpha-8 integrin chain (itga8) in glomerular homeostasis of the kidney. Mol. Cell. Pediatr. 7 (1), 13. doi: 10.1186/s40348-020-00105-5
Martínez-Gil N., Ugartondo N., Grinberg D., Balcells S. (2022). Wnt Pathway Extracellular Components and Their Essential Roles in Bone Homeostasis. Genes 13 (1), 138. doi: 10.3390/genes13010138
McRae A. F., McEwan J. C., Dodds K. G., Wilson T., Crawford A. M., Slate J. (2002). Linkage disequilibrium in domestic sheep. Genetics 160 (3), 1113–1122. doi: 10.1093/genetics/160.3.1113
Pan W., Jin Y., Stanger B., Kiernan A. E. (2010). Notch signaling is required for the generation of hair cells and supporting cells in the mammalian inner ear. Proc. Natl. Acad. Sci. United States America 107 (36), 15798–15803. doi: 10.1073/pnas.1003089107
Pareek C. S., Smoczynski R., Tretyn A. (2011). Sequencing technologies and genome sequencing. J. Appl. Genet. 52 (4), 413–435. doi: 10.1007/s13353-011-0057-x
Patterson N., Price A. L., Reich D. (2006). Population structure and eigenanalysis. PloS Genet. 2 (12), e190. doi: 10.1371/journal.pgen.0020190
Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81 (3), 559–575. doi: 10.1086/519795
Purfield D. C., Berry D. P., McParland S., Bradley D. G. (2012). Runs of homozygosity and population history in cattle. BMC Genet. 13, 70. doi: 10.1186/1471-2156-13-70
Qu J., Lu S. H., Lu Z. L., Xu P., Xiang D. X., Qu Q. (2017). Pharmacogenetic and case-control study on potassium channel related gene variants and genetic generalized epilepsy. Medicine 96 (26), e7321. doi: 10.1097/MD.0000000000007321
Rhodes J. A. (2020). Topological Metrizations of Trees, and New Quartet Methods of Tree Inference. IEEE/ACM Trans. Comput. Biol. Bioinf. 17 (6), 2107–2118. doi: 10.1109/TCBB.2019.2917204
Rojek K. O., Krzemień J., Doleżyczek H., Boguszewski P. M., Kaczmarek L., Konopka W., et al. (2019). Amot and Yap1 regulate neuronal dendritic tree complexity and locomotor coordination in mice. PloS Biol. 17 (5), e3000253. doi: 10.1371/journal.pbio.3000253
Sams A. J., Boyko A. R. (2019). Fine-Scale Resolution of Runs of Homozygosity Reveal Patterns of Inbreeding and Substantial Overlap with Recessive Disease Genotypes in Domestic Dogs. G3 (Bethesda Md.) 9 (1), 117–123. doi: 10.1534/g3.118.200836
Shah S. M., Joshi D., Chbib C., Roni M. A., Uddin M. N. (2023). The Autoinducer N-Octanoyl-L-Homoserine Lactone (C8-HSL) as a Potential Adjuvant in Vaccine Formulations. Pharm. (Basel Switzerland) 16 (5), 713. doi: 10.3390/ph16050713
Stoffel M. A., Johnston S. E., Pilkington J. G., Pemberton J. M. (2021). Genetic architecture and lifetime dynamics of inbreeding depression in a wild mammal. Nat. Commun. 12 (1), 2972. doi: 10.1038/s41467-021-23222-9
Tian C., Liu X. Z., Han F., Yu H., Longo-Guess C., Yang B., et al. (2010). Ush1c gene expression levels in the ear and eye suggest different roles for Ush1c in neurosensory organs in a new Ush1c knockout mouse. Brain Res. 1328, 57–70. doi: 10.1016/j.brainres.2010.02.079
Tomaszewski J., Joseph A., Archambeault D., Yao H. H. (2007). Essential roles of inhibin beta A in mouse epididymal coiling. Proc. Natl. Acad. Sci. United States America 104 (27), 11322–11327. doi: 10.1073/pnas.0703445104
Wang L., Roth T., Abbott M., Ho L., Wattanachanya L., Nissenson R. A. (2017). Osteoblast-derived FGF9 regulates skeletal homeostasis. Bone 98, 18–25. doi: 10.1016/j.bone.2016.12.005
Wang K. Y., Yamada S., Izumi H., Tsukamoto M., Nakashima T., Tasaki T., et al. (2018). Critical in vivo roles of WNT10A in wound healing by regulating collagen expression/synthesis in WNT10A-deficient mice. PloS One 13 (3), e0195156. doi: 10.1371/journal.pone.0195156
Wang J., Yang R., Cheng Y., Zhou Y., Zhang T., Wang S., et al. (2022). Methylation of HBP1 by PRMT1 promotes tumor progression by regulating actin cytoskeleton remodeling. Oncogenesis 11 (1), 45. doi: 10.1038/s41389-022-00421-7
Wang W., Zhang X., Zhou X., Zhang Y., La Y., Zhang Y., et al. (2019). Deep Genome Resequencing Reveals Artificial and Natural Selection for Visual Deterioration, Plateau Adaptability and High Prolificacy in Chinese Domestic Sheep. Front. Genet. 10. doi: 10.3389/fgene.2019.00300
Wei M., Zhang C., Tian Y., Du X., Wang Q., Zhao H. (2020). Expression and Function of WNT6: From Development to Disease. Front. Cell Dev. Biol. 8. doi: 10.3389/fcell.2020.558155
Wilson D. G., Phamluong K., Lin W. Y., Barck K., Carano R. A., Diehl L., et al. (2012). Chondroitin sulfate synthase 1 (Chsy1) is required for bone development and digit patterning. Dev. Biol. 363 (2), 413–425. doi: 10.1016/j.ydbio.2012.01.005
Wu X., Ivanchenko M. V., Al Jandal H., Cicconet M., Indzhykulian A. A., Corey D. P. (2019). PKHD1L1 is a coat protein of hair-cell stereocilia and is required for normal hearing. Nat. Commun. 10 (1), 3801. doi: 10.1038/s41467-019-11712-w
Wulff H., Castle N. A., Pardo L. A. (2009). Voltage-gated potassium channels as therapeutic targets. Nature reviews. Drug Discovery 8 (12), 982–1001. doi: 10.1038/nrd2983
Xu J., Gao X. P., Ramchandran R., Zhao Y. Y., Vogel S. M., Malik A. B. (2008). Nonmuscle myosin light-chain kinase mediates neutrophil transmigration in sepsis-induced lung inflammation by activating beta2 integrins. Nat. Immunol. 9 (8), 880–886. doi: 10.1038/ni.1628
Yan M., Xiong M., Wu Y., Lin D., Chen P., Chen J., et al. (2022). LRP4 is required for the olfactory association task in the piriform cortex. Cell Biosci. 12 (1), 54. doi: 10.1186/s13578-022-00792-9
Young R. M., Ewan K. B., Ferrer V. P., Allende M. L., Godovac-Zimmermann J., Dale T. C., et al. (2019). Developmentally regulated Tcf7l2 splice variants mediate transcriptional repressor functions during eye formation. eLife 8, e51447. doi: 10.7554/eLife.51447
Yue Z., Niu X., Yuan Z., Qin Q., Jiang W., He L., et al. (2022). RSPO2 and RANKL signal through LGR4 to regulate osteoclastic premetastatic niche formation and bone metastasis. J. Clin. Invest. 132 (2), e144579. doi: 10.1172/JCI144579
Zhang C. H., Gao Y., Jadhav U., Hung H. H., Holton K. M., Grodzinsky A. J., et al. (2021). Creb5 establishes the competence for Prg4 expression in articular cartilage. Commun. Biol. 4 (1), 332. doi: 10.1038/s42003-021-01857-0
Zhang R., Lin J., Liu Y., Yang S., He Q., Zhu L., et al. (2021). Transforming Growth Factor-β Signaling Regulates Tooth Root Dentinogenesis by Cooperation With Wnt Signaling. Front. Cell Dev. Biol. 9, 687099. doi: 10.3389/fcell.2021.687099
Zhao G., Li X., Miao H., Chen S., Hou Y. (2020). Estrogen Promotes cAMP Production in Mesenchymal Stem Cells by Regulating ADCY2. Int. J. Stem Cells 13 (1), 55–64. doi: 10.15283/ijsc19139
Zhao D., Ma G., Zhang X., He Y., Li M., Han X., et al. (2016). Zinc Finger Homeodomain Factor Zfhx3 Is Essential for Mammary Lactogenic Differentiation by Maintaining Prolactin Signaling Activity. J. Biol. Chem. 291 (24), 12809–12820. doi: 10.1074/jbc.M116.719377
Keywords: Illumina OvineSNP50 BeadChip, population genetic structure, evolutionary relationship, Hetian sheep, ROH
Citation: Han Z, Zhou W, Zhang L, Wang R, Liu C, Bai X and Liu S (2023) Genetic diversity and runs of homozygosity analysis of Hetian sheep populations revealed by Illumina Ovine SNP50K BeadChip. Front. Ecol. Evol. 11:1182966. doi: 10.3389/fevo.2023.1182966
Received: 09 March 2023; Accepted: 16 October 2023;
Published: 31 October 2023.
Edited by:
Haipeng Li, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Ottmar Distl, University of Veterinary Medicine Hannover, GermanyRan Li, Northwest A&F University, China
Mei Liu, Hunan Agricultural University, China
Copyright © 2023 Han, Zhou, Zhang, Wang, Liu, Bai and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shudong Liu, bGl1c2h1ZG9uZzYzQDEyNi5jb20=
 Zhipeng Han
Zhipeng Han Wen Zhou1,2
Wen Zhou1,2 Shudong Liu
Shudong Liu