
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 18 July 2023
Sec. Population, Community, and Ecosystem Dynamics
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1177446
 Erin E. Grabarczyk1,2
Erin E. Grabarczyk1,2 Marina Querejeta3,4
Marina Querejeta3,4 P. Glynn Tillman1
P. Glynn Tillman1 Rebekah D. Wallace5,6
Rebekah D. Wallace5,6 Brittany F. Barnes7
Brittany F. Barnes7 Colton D. Meinecke7
Colton D. Meinecke7 Caterina Villari7
Caterina Villari7 Kamal J. K. Gandhi7
Kamal J. K. Gandhi7 Joseph LaForest5,6
Joseph LaForest5,6 Matt Elliott8
Matt Elliott8 Jason M. Schmidt6*
Jason M. Schmidt6*Introduced species alter established trophic interactions and molecular analysis can resolve changes in community structure and associated foraging links. Joro spiders (Trichonephila clavata) were recently introduced to the United States and their range is rapidly expanding across the east coast. Here, we used DNA metabarcoding of fecal samples, prey remains from webs, and dissected guts to compare diet composition of female Joro spiders in the southeastern United States. We amplified DNA from three material types using arthropod-targeted COI primers and sequenced with IIlumina MiSeq. Prey remains from webs had the highest diversity, richness, as well as the highest proportion of prey reads relative to Joro spider reads. Recovery of prey reads from fecal samples and dissected gut content was low and both were overwhelmed by Joro spider DNA. Although fecal samples and gut content had high proportions of Joro spider reads, fecal samples had higher prey diversity and richness. Moreover, we detected prey DNA from fecal samples several days after capture from the field, which reveals initial gut retention time estimates for fecal samples collected from web-building spiders. Combined, our results offer a first glimpse at the complexity of trophic associations for an introduced web-building spider and identify a viable material, prey remains from webs, as a source of prey DNA for estimates of biodiversity associated with web-building spiders.
The introduction of new species may alter established patterns of trophic interactions, with cascading effects on entire ecosystems (David et al., 2017). DNA collected from foraging animals can serve as biodiversity capsules, leading to a better understanding of trophic interactions as well as serving as a means to understand how community structure has changed as a result of the introduction (Clare, 2014; Boyer et al., 2015). For example, molecular gut content analysis can illustrate dietary niche overlap of native species with that of introduced or invasive species, which may inform conservation management strategies (Brown et al., 2014; Zarzoso-Lacoste et al., 2019). Alternatively, analysis of introduced species diets may reveal consumption of invasive prey, agricultural pests, or disease vectors, suggesting that not all trophic interactions result in negative outcomes for an ecosystem (Martins et al., 2022). As such, molecular diet analysis of introduced species can uncover changes in community structure, either through direct consumption or as competition between native and introduced species.
Despite many advantages, information obtained through molecular diet analysis varies depending on how samples are collected as well as the type of material evaluated. For some arthropod predators, particularly spiders, amplification of focal consumer DNA from gut extractions can overwhelm prey species (Piñol et al., 2014; Cuff et al., 2023). This may potentially lead to under-reporting or missed detections of rare and degraded prey reads. The type of dietary sample also affects study design. Oftentimes in arthropod diet studies, individuals are captured, killed, and destroyed through the DNA extraction process, however this approach does not allow for individuals to be resampled. Destructive sampling is synonymous with scientific harvesting, which is less than ideal when studying endangered or endemic species as well as ecologically important (e.g., keystone) species in vulnerable environments (Lefort et al., 2022).
Alternative materials, such as predator fecal samples, regurgitates, or DNA for foraging sites, may provide DNA evidence to establish tropic links. Collection of fecal material or regurgitate is a minimally disruptive approach, which allows for repeated sampling of arthropod predators in order to understand prey preference, metabolic processing, or seasonal foraging patterns (Clare, 2014; Lefort et al., 2022; Neidel and Traugott, 2023). For web-building spiders, DNA collected from webs, a form of environmental DNA or eDNA, can be used to estimate biodiversity of arthropods as well as other organisms caught in webs (Deiner et al., 2017; Corse et al., 2019; Gregorič et al., 2022). Prey found in webs may serve as an additional source of DNA, which may be used to infer tropic interactions as well as identify web by-catch (Xu et al., 2015; Gregorič et al., 2022). Therefore, diet analysis of arthropod regurgitates, fecal samples, or DNA collected from foraging sites in addition to whole body extractions may be advantageous, particularly if predators are large or if repeated sampling of individuals is of interest.
The Joro spider (Trichonephila clavata) is native to Japan, China, Korea, and Taiwan and was first detected in Georgia, USA in 2014 (Hoebeke et al., 2015; Wallace et al., 2022); however, their range has expanded across parts of the eastern United States (Davis and Frick, 2022). Currently, Joro spiders have been confirmed in Georgia, Alabama, South Carolina, North Carolina, Tennessee, West Virginia, Maryland, and Oklahoma (jorowatch.org; Chuang et al., 2023). Joro spiders spin large webs with golden colored silk to capture prey. Female Joro spiders save remains of their prey, forming a localized collection of arthropod body parts that cluster or clump in webs, thus providing a potential record of prey captured. In Japan, female Joro spiders that consume greater amounts of food are larger and density increases with increased food availability (Miyashita, 1992b). Additionally, high feeding rates of juveniles positively correlates with greater survival and increased dispersal distances (Miyashita, 1992a), suggesting diet plays an important role in the success of Joro spiders in their native range. As a newly introduced species to the United States, we must understand the impact of Joro spider foraging patterns on local ecosystems and community interactions.
Spiders present both opportunities and challenges for metabarcoding analysis. For large web building spiders, a variety of material types, including fecal samples (Sint et al., 2015), DNA from webs (Xu et al., 2015; Gregorič et al., 2022), and whole-body extractions can be used to evaluate trophic interactions. However, predator DNA oftentimes amplifies extremely well in analysis of spider diets, and therefore may reduce the quality or read depth of prey DNA (Piñol et al., 2014; Krehenwinkel et al., 2019; Cuff et al., 2023). Therefore, as a model to advance knowledge of web-building spider trophic interactions, our objective was to compare prey composition, diversity, richness, and the proportion of predator reads relative to prey reads from three types of materials collected from female Joro spiders. The three material types analyzed included dissected guts, fecal samples, and DNA of prey remains from webs. We used DNA metabarcoding to characterize Joro spider diets and calculated diversity and richness of prey consumed.
A total of 213 females were collected from their webs for gut content analysis at 38 sites in Georgia and South Carolina, USA from October 2, 2021 to October 24, 2021 (Table 1; Supplemental Materials Figure 1). Sites were selected from public data sources tracking Joro spider presence (iNaturalist.org) and included 35 public access parks and three private residences. At each of the 38 sites, we collected samples of prey remains (i.e. clusters of arthropod body parts found in webs) from a web along with the resident Joro spider, for a total of 93 prey remains collected from webs. Captured females were placed in 20 mL sterile vials and covered with 100% chilled ethanol and placed immediately on ice. Prey remains were placed in sterile vials and put on ice. Upon return to the lab, spiders and prey remains were stored at −20°C. The intestinal track of Joro spiders was dissected and kept at −20°C in 100% ethanol until DNA extractions. During dissections, individuals were positioned on a sterilized petri dish and dissected over ice.
Two adult female Joro spiders were collected from 16 sites around northeast Georgia USA, for a total of 32 females held in captivity. Sites included 14 public access parks and two private residences. Females were removed from webs between October 8, 2021 and October 24, 2021, placed in a sterile plastic container (washed with 96% ethanol and 10% bleach), and provided water. Following capture from the field, we collected a fecal sample from each Joro spider at 24–48 hours post-capture and again at 72–96 hours post-capture. Spider fecal samples were small, around 3–5 mm in diameter, and sterile forceps were used to scoop a single specimen from the bottom of containers. All samples were stored in 1.5 mL microcentrifuge tubes that contained RNALater (ThermoFisher) and kept at −20°C. Following each fecal sample collection, females were transferred to a new, sterilized plastic container.
We extracted DNA from spider fecal samples and prey remains with the DNeasy PowerSoil Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Fecal samples were transferred directly into extraction tubes, whereas prey remains were dried and sterilized scissors used to break apart larger portions of the web remains, until all remains could fit into the extraction tube (<25 mg of dry tissue). A blank, negative control extraction that did not contain any materials was included during PCR plate preparation. We extracted dissected spider guts with the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
We followed a two-step, nested DNA metabarcoding approach to detect arthropod DNA in Joro spider dietary samples (Kitson et al., 2019). In the first round of PCR, DNA was amplified with the primer pair ZBJ-ArtF1c/ZBJ-ArtR2c (Zeale et al., 2011), which targets a 157 base pair fragment of mitochondrial Cytochrome Oxidase subunit I (COI) gene. Each sample was dual-tagged with a unique combination of eight forward and eight reverse base pair tags (Kitson et al., 2019). Following this round, PCR products were cleaned with magnetic beads using the MagBio HighPrep™ PCR System following the manufacturer’s instructions based on sample volume (MagBio Genomics Inc. MD, USA). During the second round of PCR, Illumina adapters and molecular tags were added to the amplified COI region. Each plate of 96 samples received forward and reverse tags that consisted of a unique eight nucleotide sequence, which enabled demultiplexing of plates and individual identification of samples post-sequencing. PCR products were again cleaned with MagBio HighPrep™ PCR System. We estimated the concentrations of amplicons (ng/uL) on a Qiaxcel Advanced System (Qiagen, Hilden, Germany) and pooled PCR products based on their concentrations, which were standardized across samples and plates. Samples were processed by the Georgia Genomics and Bioinformatics Core lab (GCBU-UGA) for sequencing on an Illumina MiSeq (Illumina, San Diego, CA, USA) with V3 chemistry and 300 cycles.
Multiplexed library quality was checked using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) on forward and reverse reads separately. Libraries were demultiplexed and primers trimmed using cutadapt (Martin, 2011). Trimmed reads were assembled using PEAR (Zhang et al., 2014). Subsequent analyses were performed using VSEARCH v2.8.2 (Rognes et al., 2016). We filtered sequences by quality (fastq_maxee = 1) and dereplicated. After, singletons, insertions and deletions (indels), and chimeras were removed. At this point, we obtained a set of sequences that correspond to Amplicon Single Variants (ASVs), which were clustered into Operational Taxonomic Units (OTUs) applying the greedy algorithm for clustering with a cut-off threshold of 97%. Next, we generated a FASTA file with all the OTUs and a by-sample OTU table. Taxonomic classification of the OTUs was performed against the NCBI Genbank nr/nt using the BLAST algorithm (Johnson et al., 2008) and R package taxonomizr (Sherrill-Mix, 2023) in order to infer species level classifications when available. A complementary taxonomic classification was obtained against the Barcode of Life Data System (BOLD) database using the python package bold_identification (https://github.com/linzhi2013/bold_identification). Based on OTU tables for taxa reads recovered from each sample material, we further filtered our data to include all identified reads at the Genus level with >95% identity and >120bp length overlap for the primer pair ZBJ-ArtF1c/ZBJ-ArtR2c. We removed rare reads by setting a threshold for detection of taxa to >10 reads. Any taxa with less than ten reads was adjusted to zero and considered absent (e.g., Deagle et al., 2019). All reads were combined by Family for an overall summary (Supplemental Table 1) and combined by Order for visual displays.
We fit generalized linear mixed effects models (GLMM) and linear mixed effects models (LMM) in R program software verision 4.3.0 (R CoreTeam 2023) to compare prey read recovery, diversity, and richess of the three material types. To analyze the overall proportion of prey reads by material type and to compare the proportion of prey reads between time points for fecal samples, we used the package lme4 (function: glmer, family: binomial; Bates et al., 2015) specifying site or date collected (for time post feeding) as a random effect. For analysis of prey diversity and richness, we employed an iterative process and fit rarefaction curves to maximize the number of samples included in our comparisons with the package Rarefy (function: GUniFrac; Chen et al., 2018), while standardizing read depth across samples (see Deagle et al., 2019; Grabarczyk et al., 2022). Due to variable read recovery, we evaluated prey read depth for each material type seperately. For example, a rarefaction threshold of 1,000 reads allowed 88% of all prey remains samples to be included in diversity and richness estimates; whereas for fecal samples, a rarefaction threshold of 100 reads retained 100% of samples (Supplemental Materials Figure 2). However, for gut content samples, Joro spider reads overwhelmed non-Joro reads, and despite a rarefaction threshold set to 100 reads, we only retained 13 out of 213 samples for our analyses (Supplemental Materials Figure 2). For each material type, we estimated two dietary metrics at the sample level based on the rarefied relative read data matrix (read depth set at 100 for all materials): Shannon diversity index (H) and species richness (S) with the vegan package (function: diversity; Oksanen et al., 2022). Next, we fit linear mixed effects models (function: lmer; Bates et al., 2015) with site as a random effect to assess differences in diversity (H) and richness (S) by material type. Likewise, we fit linear mixed effects models to test whether (H) and (S) in fecal samples differed between time points with date collected specificed as a random effect. To account for an unbalanced number of samples by material type, we evaluated signficance with a type “III” sums of squares and report the Wald F-tests or χ2. For all analyses, model adequacy was assessed with residual plots. We evaluated pair-wise contrasts for significant main effects with emmeans (function: pairs; Lenth, 2023). Finally, to gauge potential change in prey taxa composition between material types or fecal sample collection time, we fit PERMANOVA on the rarefied read matrix (Permutaional Multivariate Analysis of Variance using distance matrices) with the vegan package (function: adonis2; Oksanen et al., 2022). All analyses were conducted at the level of Family (pooled reads for Families by samples). For ease of creating interpretable graphical displays, we pooled rarefied reads by Order.
A total of 340 samples across three materials were sequenced and provided an average sequencing depth of 22,227 (± 1 SD 9,923) reads recovered per sample. Of the 32 female Joro spiders held in captivity, we collected a fecal sample from all females 24–48 hours post-capture and collected fecal samples from 26 females at 72–96 hours post-capture. Of the 58 fecal samples, 54 amplified both prey and Joro spider DNA. Reads were recovered from all 93 collected prey remains and 213 gut contents. However, despite dissecting the digestive tract of females to limit Joro spider DNA, most reads recovered from gut samples were of Joro spiders.
Among material types, prey remains collected from Joro spider webs contained the highest proportion of non-Joro reads (χ2 = 4,661, df = 2, P < 0.0001; Figure 1A). At the sample level, we recovered on average 17,159 (±1SE 1,038) of non-Joro reads from prey remains, 3,468 (±1SE 1,016) from fecal samples, and 88 (±1SE 37) from Joro gut content samples. The total number of reads after quality filtration was 17,656,770.
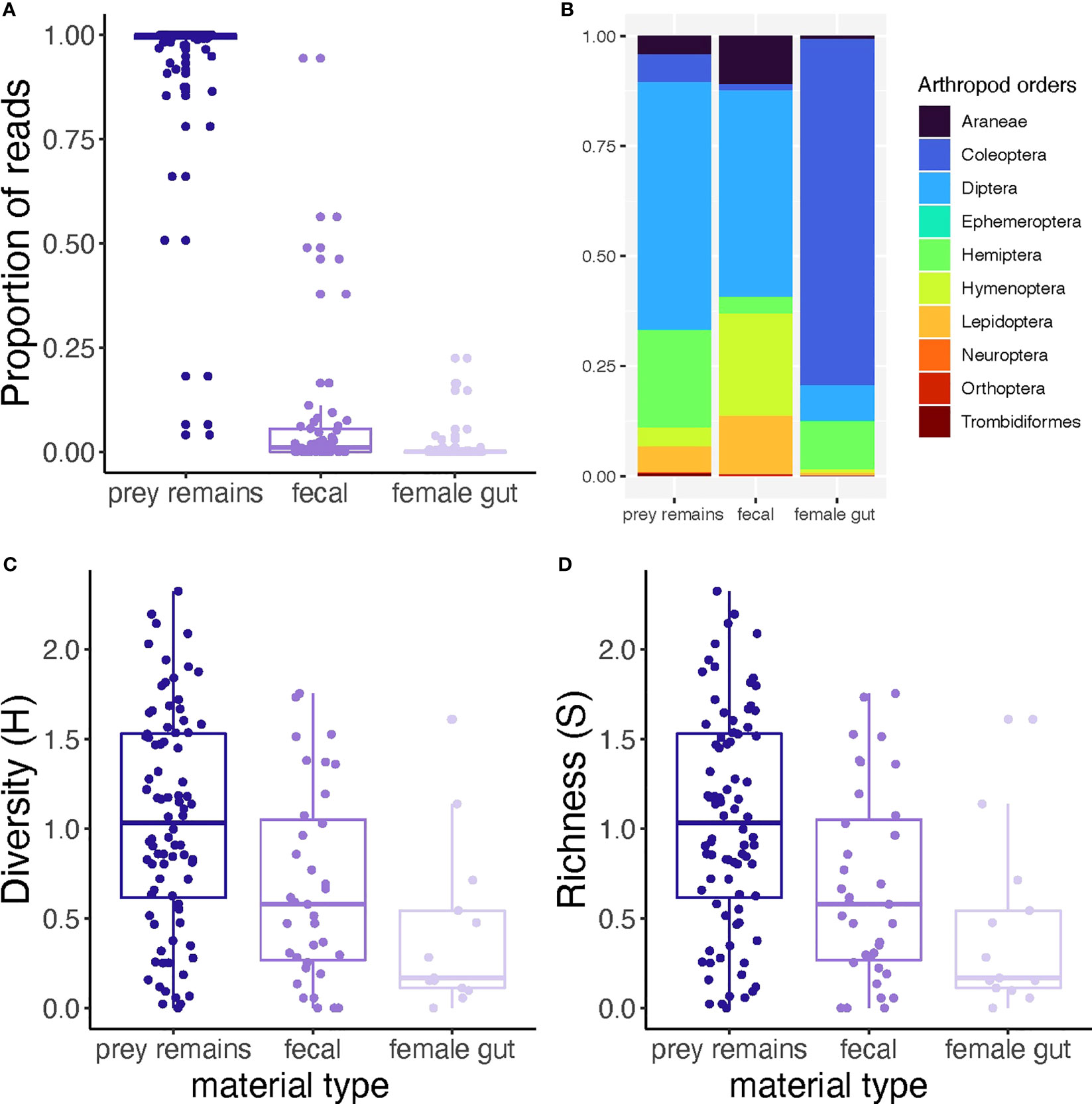
Figure 1 Summary of recovered read distribution relative to Joro spider material type. (A) All non-Joro prey reads were retained in relation to total reads recovered for each sample, and compared between materials. (B) Rarified prey reads were used to standardize read depth across samples, then the distribution of prey orders recovered was presented according to material type. (C, D) Rarefied prey reads per sample were used to estimate (C) Shannon diversity index (H) and (D) species richness (S) of prey found in three types of material collected from female Joro spiders or their webs. Lower-case letters indicate the results of linear contrasts between sample materials, where different letters represent significant differences (α = 0.05).
Data sets were individually rarefied to standardize read depth prior to visualizing or calculating diversity metrics of non-Joro reads recovered from each material type. To simplify display complexity, reads were binned by arthropod Orders (Figure 1B). Overall, we observed10 arthropod Orders across material types and recovered a total of 138 taxa (Supplemental Table 1). For prey remains and fecal samples, Diptera (flies) amplified in high proportions. For gut contents, Coleoptera (beetles) was the most prevalent order (Figure 1B). Diversity and richness of arthropod taxa detected among materials differed significantly (F2, 135 = 5.245, P < 0.0001, F2, 135 = 6.219, P = 0.003, Figures 1C, D, respectively); prey remains had the highest diversity and species richness (Figures 1C, D). Differences in diversity and richness between materials were also related to composition dissimilary in prey taxa observed (PerMANOVA, F2,135 = 2.765, P < 0.0001, Figure 1B; Supplemental Materials Table 1).
We assessed whether DNA from fecal samples collected at two time intervals produced identifiable prey reads from 32 female Joro spiders held in captivity. We recovered a significantly higher proportion of prey reads from the 48-hour than the 96-hour time point (χ2 = 4.6, df = 1, P = 0.03, Figure 2). Despite a greater number of reads from the 48-hour collection, we found no difference in prey diversity (F1, 33 = 2.6, P = 0.1) or richness (F1, 33 = 0.9, P = 0.3) for fecal sample by time point. In addition, we found no significant difference in compositional changes between time points (PerMANOVA, F1,34 = 2.1, P = 0.098). However, at the 96-hour point, two common prey orders were not detected, which included Coleoptera (beetles) and other Araneae (spiders).
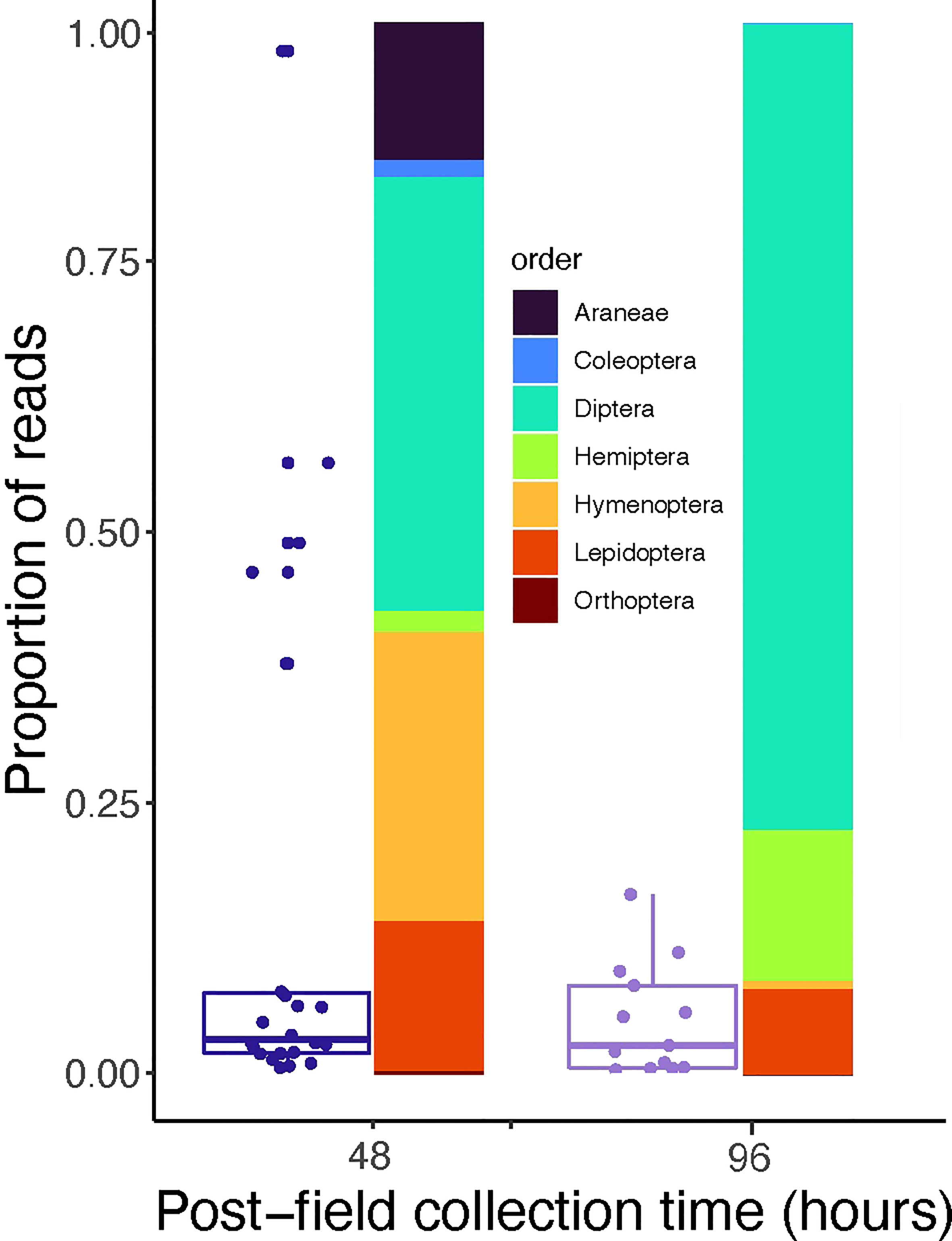
Figure 2 Summary of fecal sample read recovery of female Joro spiders held in captivity post-field collection. Box and whisker plots show the proportion of non-Joro spider reads recovered in relation to time, and the stacked bars represent the relative read recovery of detected arthropod Orders. Araneae reads exclude the Joro spider.
Analysis of female Joro spider dietary samples provided a diverse snapshot of prey DNA linked to three material types, and for fecal samples, an initial view of prey retention in metabarcoding studies. For web-building spiders, prey remains are a potential source of DNA that allows for repeated sampling of individuals (Xu et al., 2015; Gregorič et al., 2022). Reads from Joro spider prey remains had high diversity, richness, and minimal contamination from predator reads. In contrast, fecal samples and dissected guts had high levels of predator reads, which likely reduced detection of prey DNA. Due to poor read recovery from gut samples, we are unable to answer the question: how closely do the diets of females match prey remains in webs? Moreover, what is the function of prey remains for the Joro spider – food cache or garbage dump? Prior studies indicate that web prey remains or mainlining caches of arthropod materials can supplement web-building spider diets (Champion de Crespigny et al., 2001; Griffiths et al., 2003). However, other predators, such as insectivorous birds, may be attracted to and feed from webs with prey remains or caches (Schronce and Davis, 2022). Therefore, the value of prey remains as an alternative food source and whether prey remains accurately represent resident female foraging patterns is unknown. Regardless, the diversity of species recovered from this material suggests Joro spider webs trap an array of arthropods.
We confirm all three material types collected from female Joro spiders yield amplifiable arthropod sequences via DNA metabarcoding, which can be used to characterize their diet. The most common arthropod Orders among samples included Coleoptera, Diptera, Hemiptera, and Hymenoptera, however, Araneae, Ephemeroptera, Lepidoptera, Neuroptera, Orthoptera, and Trombidiformes also amplified (Supplemental Materials Table 1). Combined reads from all materials resulted in 138 taxa at Family level. Prey remains captured the highest taxonomic diversity among materials tested (Figure 1B, Supplemental Materials Table 1). However, prey remains may also include web by-catch, and therefore does not necessarily imply that all taxa found were consumed by resident spiders. Nevertheless, many taxa were found in all three material types, such as Carabidae (ground beetles), Chironomidae (non-biting midges), a diversity of Diptera, Aphididae (aphids), Aphrophoridae (spittlebugs), and Cicadellidae (leafhoppers), which provides stronger evidence of consumption. Moreover, nuisance species, including Culicidae (mosquitos) and Pentatomidae (stink bugs) were found in both fecal samples and prey remains, which suggests Joro spiders catch and eat both species. In terms of overall proportion of reads among material types, Diptera was the most prevalent Order for both prey remains and fecal samples (Supplemental Materials Table 1). For gut samples, Coleoptera accounted for the highest proportion of prey reads, however, based on total number, Coleoptera reads were much higher in prey remains than gut samples (Supplemental Materials Table 1).
For many arthropod metabarcoding studies, focal predator DNA constitutes a significant portion of total reads for a given sample, which results in fewer prey reads (Cuff et al., 2023). Here, we used a set of commonly used arthropod-targeted primers, which in previous studies showed lower amplification of some spider taxa (Krehenwinkel et al., 2018). Unfortunately, we found that Joro spider DNA amplified exceedingly well with ZBJ-ArtF1c/ZBJ-ArtR2c primers. Recent studies suggest some spider species amplify much better than others, when predator DNA over amplifies, the read depth for prey species in gut content samples may be significantly reduced. For example, Krehenwinkel et al. (2019) found that the abundance of spider predator DNA reads relative to prey reads was species-specific; some spider species over-amplified whereas others did not. Although more complicated and costly, the use of multiple primers may help balance biases among taxa detected in dietary samples (Corse et al., 2019; Krehenwinkel et al., 2019; Forsman et al., 2022; Cuff et al., 2023). In addition, limiting the amount of predator tissue included in an extraction may cut down on predator reads (Krehenwinkel et al., 2017). Here, we dissected females to isolate the digestive tract, yet still amplified an overwhelming amount of Joro spider DNA with the Zeale COI primer. Thus, dissection alone was insufficient to adequately enrich prey reads for downstream analysis of gut contents. For Joro spiders, future studies should use multiple primers to achieve balanced amplification of taxa in samples and to better approximate prey DNA in spider guts (Elbrecht et al., 2019).
Analysis of fecal samples or regurgitates is common in vertebrate diet research, however fewer studies have explored arthropod fecal samples or regurgitates to understand trophic interactions (but see Waldner and Traugott, 2012; Scriven et al., 2013; Sint et al., 2015; Kamenova et al., 2018; Neidel and Traugott, 2023). For some spiders, digestion is slow, and prey may take as long as 140 hours to completely breakdown (Greenstone et al., 2014). A timed post-feeding test with regurgitates of beetles shows that prey are detected over long periods of time, but individual metabolic processing times vary significantly (Neidel and Traugott, 2023). In female Joro spiders, we detected prey DNA from fecal samples at both 48- and 96-hour collection intervals. However, for this type of material, smaller, degraded DNA fragments isolated from consumer feces may limit the scope of prey detection, simply due to digestion time in the gut (Kamenova et al., 2018). Despite many constraints, we recovered a diverse range of prey DNA in Joro spider fecal samples. In Lycosid spiders, analysis of fecal samples amplified similar types of prey as whole-body spider extractions (Sint et al., 2015). Thus, if regurgitates or fecal samples provide similar results, this may be an advantageous approach, especially if working with a species of conservation concern (Scriven et al., 2013). For questions that seek to understand how individual foraging patterns vary over time, regurgitates or fecal samples could be used to understand how long a prey is detected in diets or potentially link diet analysis at multiple time points to individual fitness (Grabarczyk et al., 2022).
Invasive species are a leading cause of biodiversity loss (Gurevitch and Padilla, 2004; Mollot et al., 2017; Crystal-Ornelas and Lockwood, 2020). Understanding whether introduced species show the potential to negatively affect community structure or ecosystem function is essential for mitigating adverse effects. Although Joro spiders may be considered a nuisance by homeowners, we found that resident spiders prey on and capture a diversity of insects, including pests such as mosquitos and stink bugs. By considering three types of materials collected from female Joro spiders, we show that dissected guts, fecal samples, and prey remains all amplify prey DNA. The overwhelming amount of predator DNA amplified from dissected guts suggests that a deeper understanding of the biological significance of prey remains collected from webs is warranted for Joro spiders, as this material type may be an alternative to gut content analysis. Furthermore, this approach could be readily applied to other web-building species that collect prey in webs. Whether prey remains are an accurate representation of Joro spider diets or whether only certain prey types (e.g., hard exoskeletons versus soft-bodied prey) vary in persistence in webs requires further attention. In addition, fecal samples collected from captive females show that prey DNA can be detected in these samples upwards of 96 hours post capture, which in combination with prey remains, offers clues regarding Joro spider trophic links and may be key to uncovering their impact on ecosystems in the southeastern United States.
The original contributions presented in the study are publicly available. This data can be found here: NCBI BioProject PRJNA938034.
Methodology, analysis, and writing – original draft preparation: EG, MQ, and JS. Conceptualization and writing – review and editing: EG, MQ, PT, RW, BB, CM, CV, KG, JL, ME, JS. All authors have approved the submitted version of this manuscript.
Funding for MQ was provided by FICYT and “Plan de Ciencia, Tecnologia e Innovacion 2018–2022” from the Government of Asturias (Grant AYUD/2021/58607).
We thank Magdy Alabady and Georgia Genomics and Bioinformatics Core lab at the University of Georgia (GGBC-UGA) for sequencing services.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the University of Georgia, Valdosta State University, or the United States Department of Agriculture, Agricultural Research Service.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1177446/full#supplementary-material
Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Software 67. doi: 10.18637/jss.v067.i01
Boyer S., Cruickshank R. H., Wratten S. D. (2015). Faeces of generalist predators as “biodiversity capsules”: a new tool for biodiversity assessment in remote and inaccessible habitats. Food Webs 3, 1–6.
Brown D. S., Burger R., Cole N., Vencatasamy D., Clare E. L., Montazam A., et al. (2014). Dietary competition between the alien Asian musk shrew (Suncus murinus) and a re-introduced population of Telfair's skink (Leiolopisma telfairii). Mol. Ecol. 23, 3695–3705.
Champion de Crespigny F. E., Herberstein M. E., Elgar M. A. (2001). Food caching in orb-web spiders (Araneae: Araneoidea). Naturwissenschaften 88, 42–45.
Chen J., Zhang X., Zhou H. (2018). GUniFrac: generalized UniFrac distances, distance-based multivariate methods and feature-based univariate methods for microbiome data analysis (R package version, 1).
Chuang A., Deitsch J. F., Nelsen D. R., Sitvarin M. I., Coyle D. R. (2023). The Jorō spider (Trichonephila clavata) in the southeastern US: an opportunity for research and a call for reasonable journalism. Biol. Invasions 25, 17–26.
Clare E. L. (2014). Molecular detection of trophic interactions: emerging trends, distinct advantages, significant considerations and conservation applications. Evol. Appl. 7 (9), 1144–1157.
Corse E., Tougard C., Archambaud-Suard G., Agnèse J. F., Messu Mandeng F. D., Bilong Bilong C. F., et al. (2019). One-locus-several-primers: a strategy to improve the taxonomic and haplotypic coverage in diet metabarcoding studies. Ecol. Evo. 9, 4603–4620.
Crystal-Ornelas R., Lockwood J. L. (2020). Cumulative meta-analysis identifies declining but negative impacts of invasive species on richness after 20 yr. Ecology 101, e03082.
Cuff J. P., Kitson J. J., Hemprich-Bennett D., Tercel M. P., Browett S. S., Evans D. M. (2023). The predator problem and PCR primers in molecular dietary analysis: swamped or silenced; depth or breadth? Mol. Ecol. Resour. 23, 41–51.
David P., Thébault E., Anneville O., Duyck P.-F., Chapuis E., Loeuille N. (2017). “Impacts of invasive species on food webs,” in Advances in ecological research (Elsevier), 1–60. doi: 10.1016/bs.aecr.2016.10.001
Davis A. K., Frick B. L. (2022). Physiological evaluation of newly invasive Jorō spiders (Trichonephila clavata) in the southeastern USA compared to their naturalized cousin, Trichonephila clavipes. Physiol. Entomol. 47, 170–175.
Deagle B. E., Thomas A. C., McInnes J. C., Clarke L. J., Vesterinen E. J., Clare E. L., et al. (2019). Counting with DNA in metabarcoding studies: how should we convert sequence reads to dietary data? Mol. Ecol. 28, 391–406.
Deiner K., Bik H. M., Mächler E., Seymour M., Lacoursière-Roussel A., Altermatt F., et al. (2017). Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895.
Elbrecht V., Braukmann T. W. A., Ivanova N. V., Prosser S. W. J., Hajibabaei M., Wright M., et al. (2019). Validation of COI metabarcoding primers for terrestrial arthropods. PeerJ 7, e7745. doi: 10.7717/peerj.7745
Forsman A. M., Hoenig B. D., Gaspar S. A., Fischer J. D., Siegrist J., Fraser K. (2022). Evaluating the impacts of metabarcoding primer selection on DNA characterization of diet in an aerial insectivore, the purple martin. Auk 139, ukab075.
Grabarczyk E. E., Gill S. A., Vonhof M. J., Alabady M. S., Wang Z., Schmidt J. M. (2022). Diet composition and diversity does not explain fewer, smaller urban nestlings. PloS One 17, e0264381.
Greenstone M. H., Payton M. E., Weber D. C., Simmons A. M. (2014). The detectability half-life in arthropod predator-prey research: what it is, why we need it, how to measure it, and how to use it. Mol. Ecol. 23, 3799–3813. doi: 10.1111/mec.12552
Gregorič M., Kutnjak D., Bačnik K., Gostinčar C., Pecman A., Ravnikar M., et al. (2022). Spider webs as eDNA samplers: biodiversity assessment across the tree of life. Mol. Ecol. Resour. 22, 2534–2545.
Griffiths B. V., Holwell G. I., Herberstein M. E., Elgar M. A. (2003). Frequency, composition and variation in external food stores constructed by orb-web spiders: nephila edulis and Nephila plumipes (Araneae: Araneoidea). Aust. J. Zool. 51, 119–128.
Gurevitch J., Padilla D. K. (2004). Are invasive species a major cause of extinctions? Trends Ecol. Evol. 19, 470–474.
Hoebeke E. R., Huffmaster W., Freeman B. J. (2015). Nephila clavata l koch, the Joro spider of East Asia, newly recorded from north America (Araneae: Nephilidae). PeerJ 3, e763.
Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9.
Kamenova S., Mayer R., Rubbmark O. R., Coissac E., Plantegenest M., Traugott M. (2018). Comparing three types of dietary samples for prey DNA decay in an insect generalist predator. Mol. Ecol. Resour. 18, 966–973.
Kitson J. J., Hahn C., Sands R. J., Straw N. A., Evans D. M., Lunt D. H. (2019). Detecting host–parasitoid interactions in an invasive lepidopteran using nested tagging DNA metabarcoding. Mol. Ecol. 28, 471–483.
Krehenwinkel H., Kennedy S. R., Adams S. A., Stephenson G. T., Roy K., Gillespie R. G. (2019). Multiplex PCR targeting lineage-specific SNP s: a highly efficient and simple approach to block out predator sequences in molecular gut content analysis. Methods Ecol. Evol. 10, 982–993. doi: 10.1111/2041-210X.13183
Krehenwinkel H., Kennedy S., Pekár S., Gillespie R. G. (2017). A cost-efficient and simple protocol to enrich prey DNA from extractions of predatory arthropods for large-scale gut content analysis by illumina sequencing. Methods Ecol. Evol. 8, 126–134. doi: 10.1111/2041-210X.12647
Krehenwinkel H., Kennedy S. R., Rueda A., Lam A., Gillespie R. G. (2018). Scaling up DNA barcoding – primer sets for simple and cost efficient arthropod systematics by multiplex PCR and illumina amplicon sequencing. Methods Ecol. Evol. 9, 2181–2193. doi: 10.1111/2041-210X.13064
Lefort M.-C., Cruickshank R. H., Descovich K., Adams N. J., Barun A., Emami-Khoyi A., et al. (2022). Blood, sweat and tears: a review of non-invasive DNA sampling. Peer Community J. 2, e16. doi: 10.24072/pcjournal.98
Lenth R. (2023). Emmeans: estimated marginal means, aka least-squares means (R package version 1.8.6). Available at: https://CRAN.R-project.org/package=emmeans.
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12.
Martins B., Silva-Rocha I., Mata V. A., Gonçalves Y., Rocha R., Rato C. (2022). Trophic interactions of an invasive gecko in an endemic-rich oceanic island: insights using DNA metabarcoding. Front. Ecol. Evol. 10, 1197.
Miyashita T. (1992a). Feeding rate may affect dispersal in the orb-web spider Nephila clavata. Oecologia 92, 339–342.
Miyashita T. (1992b). Food limitation of population density in the orb-wed spider, Nephila clavata. Popul. Ecol. 34, 143–153.
Mollot G., Pantel J. H., Romanuk T. N. (2017). “The effects of invasive species on the decline in species richness: a global meta-analysis,” in Advances in ecological research (Academic Press), 61–83.
Neidel V., Traugott M. (2023). Laboratory protocol is important to improve the correlation between target copies and metabarcoding read numbers of seed DNA in ground beetle regurgitates. Sci. Rep. 13, 1995.
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2022 2020). Vegan: community ecology package (R package version 2), 5–7.
Piñol J., San Andrés V., Clare E. L., Mir G., Symondson W. O. C. (2014). A pragmatic approach to the analysis of diets of generalist predators: the use of next-generation sequencing with no blocking probes. Mol. Ecol. Resour. 14, 18–26.
R Core Team (2023). R: a language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584.
Schronce A., Davis A. K. (2022). Novel observation: northern cardinal (Cardinalis cardinalis) perches on an invasive Jorō spider (Trichonephila clavata) web and steals food. Insects 13, 1049.
Scriven J. J., Woodall L. C., Goulson D. (2013). Nondestructive DNA sampling from bumblebee faeces. Mol. Ecol. Resour. 13, 225–229.
Sherrill-Mix S. (2023). _taxonomizr: functions to work with NCBI accessions and taxonomy_ (R package version 0.10.2). Available at: https://CRAN.R-project.org/package=taxonomizr.
Sint D., Thurner I., Kaufmann R., Traugott M. (2015). Sparing spiders: faeces as a non-invasive source of DNA. Front. Zool. 12, 1–5.
Waldner T., Traugott M. (2012). DNA-Based analysis of regurgitates: a noninvasive approach to examine the diet of invertebrate consumers. Mol. Ecol. Resour. 12, 669–675.
Wallace R. D., Hoebeke E. R., Hudson W., Schmidt J. M., Barnes B., Gandhi K., et al. (2022). Joro spider (Trichonephila clavata). Univ. Ga. Press 1273.
Xu C. C. Y., Yen I. J., Bowman D., Turner C. R. (2015). Spider web DNA: a new spin on noninvasive genetics of predator and prey. PloS One 10, e0142503.
Zarzoso-Lacoste D., Bonnaud E., Corse E., Dubut V., Lorvelec O., De Meringo H., et al. (2019). Stuck amongst introduced species: trophic ecology reveals complex relationships between the critically endangered niau kingfisher and introduced predators, competitors and prey. NeoBiota 53, 61–82.
Zeale M. R., Butlin R. K., Barker G. L., Lees D. C., Jones G. (2011). Taxon-specific PCR for DNA barcoding arthropod prey in bat faeces. Mol. Ecol. Resour. 11, 236–244.
Keywords: golden-orb weaver, molecular gut content analysis, introduced species, spider web, trophic interactions
Citation: Grabarczyk EE, Querejeta M, Tillman PG, Wallace RD, Barnes BF, Meinecke CD, Villari C, Gandhi KJK, LaForest J, Elliott M and Schmidt JM (2023) DNA metabarcoding analysis of three material types to reveal Joro spider (Trichonephila clavata) trophic interactions and web capture. Front. Ecol. Evol. 11:1177446. doi: 10.3389/fevo.2023.1177446
Received: 01 March 2023; Accepted: 29 June 2023;
Published: 18 July 2023.
Edited by:
Andrea Sciarretta, University of Molise, ItalyCopyright © 2023 Grabarczyk, Querejeta, Tillman, Wallace, Barnes, Meinecke, Villari, Gandhi, LaForest, Elliott and Schmidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jason M. Schmidt, anNjaG1pZDJAdWdhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.