
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 16 June 2023
Sec. Population, Community, and Ecosystem Dynamics
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1173750
This article is part of the Research Topic The Effects of Climate Change and Anthropogenic Activities on Patterns, Structures and Functions of Terrestrial Ecosystems View all 18 articles
 Zhongqing Yan1,2,3
Zhongqing Yan1,2,3 Meng Li1,2,3
Meng Li1,2,3 Yanbin Hao4
Yanbin Hao4 Yong Li1,2,3
Yong Li1,2,3 Xiaodong Zhang1,2,3
Xiaodong Zhang1,2,3 Liang Yan1,2,3
Liang Yan1,2,3 Enze Kang1,2,3
Enze Kang1,2,3 Xiaodong Wang1,2,3
Xiaodong Wang1,2,3 Ao Yang1,2,3
Ao Yang1,2,3 Yuechuan Niu4
Yuechuan Niu4 Xiaoshun Yu1,2,3
Xiaoshun Yu1,2,3 Xiaoming Kang1,2,3*
Xiaoming Kang1,2,3* Kerou Zhang1,2,3*
Kerou Zhang1,2,3*Diverse microorganisms drive biogeochemical cycles and consequently influence ecosystem-level processes in alpine peatlands, which are vulnerable to extreme drought induced by climate change. However, there are few reports about the effects of extreme drought on microbial function. Here we identify microbial functional genes associated with carbon and nitrogen metabolisms of extreme drought experiments that occurred at different periods of plant growth, the results show that early extreme drought reduces the abundance of functional genes involved in the decomposition of starch and cellulose; midterm extreme drought increases the abundance of lignin decomposition functional genes; late extreme drought reduces the hemicellulose but increases cellulose decomposition functional genes. In the carbon fixation pathway, extreme drought mainly changes the abundance of functional genes involved in the reductive citrate cycle process, the 3-hydroxy propionate bi-cycle, the dicarboxylate-hydroxybutyrate cycle and the incomplete reductive citrate cycle. Among the nitrogen cycling functional genes, amoA involved in oxidizing ammonia to hydroxylamine significantly increases under early extreme drought; midterm extreme drought reduces nrtC and nifD genes, which participate in nitrate assimilation and nitrogen fixation, respectively; late extreme drought significantly increases hcp genes involved in ammonification. pH and TN had the largest effects on the carbon degradation, fixation and nitrogen cycling functional genes. The composition of microbial community structures involved in carbon fixation differed between treatments in early extreme drought. There is a good linear fit between the diversity of gene abundance and corresponding microbial communities in the reductive citrate cycle, hydroxy propionate-hydroxybutyrate cycle, dicarboxylate-hydroxybutyrate cycle and nitrogen cycling, which suggests that the functional genes and community composition of microorganisms involved in these processes are consistent in response to extreme drought. This study provides new insights into the adaptability and response characteristics of microbial communities and functional genes in plateau peatland ecosystems to extreme drought events.
Peatland is a carbon-rich ecosystem covering 185-423 million hectares of the earth’s surface (Ribeiro et al., 2021). It is an important organic carbon (C) and nitrogen (N) pool and its C and N reserves account for 30% (ca. 644 Gt C) and 10% (8 – 15 Gt N) of the total reserves of the whole terrestrial ecosystem, respectively (Limpens et al., 2006; Wang et al., 2014). A large amount of greenhouse gases is produced and discharged due to C and N migration and transformation in peatlands (Chen et al., 2014; Yao et al., 2022). Global warming is affecting precipitation patterns in a complex way, as heating promotes evaporation, leading to soil surface drying, thus increasing the duration and intensity of drought events (Pokhrel et al., 2021). The global climate model predicts that the frequency and intensity of extreme drought events will increase in the future (Hoover and Rogers, 2016). Extreme drought has seriously affected soil biogeochemical C and N cycling and related greenhouse gas fluxes (Deng et al., 2021), leading to the loss of ecosystem functions (Du et al., 2018; Kang et al., 2018). Therefore, the fate of the large amount of C and N stored in peatland and the response of peatland to extreme drought is very important for the future climate. It is becoming increasingly apparent that in addition to drought intensity, the timing of drought has important effects on the ecosystem C and N cycling (Dietrich and Smith, 2016). Some studies have shown that drought events, occurring at different plant growth stages (i.e., rapid growth, full bloom, and decline stages), have differently influenced ecosystem production and functioning (Knapp et al., 2008; Dietrich and Smith, 2016). However, the ecological implications of greater intra-season variability of rainfall extremes have received minimal notice, especially concerning variable timing of extreme drought at different plant growth stages, which might have the largest ecological consequences.
Extreme drought events mainly lead to water stress in plants, which can alter the distribution pattern of plant biomass (Jentsch et al., 2011), cause a decrease in photosynthetic, transpiration rates (Lefi et al., 2004) and water use efficiency, then decrease in plant suitability (Loik, 2007). Extreme drought events not only directly affect plant growth, but also indirectly affect the supply of soil nutrients through changes in soil moisture content. Studies have shown that extreme drought in summer significantly reduces soil C and N mineralization, leading to a decrease in soil nutrient supply (Borken & Matzner, 2009). Soil microorganisms play an intermediary role in the key steps of all biogeochemical cycles and maintain ecosystem functions (Bell et al., 2005; Zhang et al., 2012; Bastida et al., 2021). Therefore, the effect of extreme drought events on soil water and nutrients regime is highly likely to cause changes in soil microbial activity. As the soil dries, water membranes form on soil particles, concentrating water-containing pore water components (e.g., dissolved nutrients, solutes, toxins), limiting the diffusion of stroma and extracellular enzymes and increasing interactions between microbial populations (Malik and Bouskill, 2022). There is overwhelming evidence that microorganisms will respond to drought in terms of microbial community composition and diversity (Zhou et al., 2012; Guo et al., 2020; Yuan et al., 2021). In addition, changes in microbial communities can in turn regulate soil nutrient regimes through metabolism and decomposition. Under drought conditions, microorganisms can mediate soil C sequestration and alleviate the impact of reduced soil water availability caused by climate change by increasing soil carbon sequestration (Canarini et al., 2016). Drought can reduce the decomposition of soil organic matter by reducing the characteristics of microorganisms (e.g., enzyme activity and functional gene abundance) (Alster et al., 2013; Vogel et al., 2013). Therefore, the understanding of soil microbial ecology is crucial to our ability to evaluate terrestrial C and N cycling, but the complexity of soil microbial communities and the multiple ways that they may be affected by extreme drought hinder our ability to draw clear conclusions on this topic and the metabolic kinetics and phenotypic characteristics of microbial communities are still unclear (Bardgett et al., 2008; de Vries et al., 2018), though microbial communities drive biogeochemical cycles through their specific metabolic activities.
Litter decomposition is a key link between carbon budget and nutrient cycling. In general, drought can cause a decrease in the rate constant of litter decomposition, and the decomposition of C and N in various plant litter leaves has decreased (Sardans and Penuelas, 2010). Sudden drought may have a greater effect on litter decomposition than long-term drought (García-Palacios et al., 2016). In addition, the decomposition rate of different carbon components is also inhibited to varying degrees by drought. For example, the proportion of cellulose in litter under drought treatment in the Mediterranean region increased by 10.9% compared to the ambient conditions, while the lignin content increased by 17.5% (Tu et al, 2017). Some studies suggest that drying conditions have a negative effect on soil microorganisms. Drought can lead to a decrease in soil microbial biomass, diversity and relative abundance, thereby affecting the litter decomposition rates (Zhou et al., 2018). Therefore, understanding the decomposition, transformation, and sequestration of organic matter in soil requires a further understanding of how microbial physiology regulates the processes that control biogeochemical cycling, climate change, and ecosystem sustainability (Bardgett et al., 2008; Roth et al., 2019). Liang et al. (2017) defined two pathways (i.e., ex vivo modification and in vivo turnover) to jointly explain the dynamics of soil C driven by microbial catabolism and/or anabolism. However, it has been difficult to link specific taxa to these ecosystem processes and consequently predict the functional responses of soil microorganisms (Fierer, 2017). Thus, a better understanding of the diversity and abundance of microbial functional genes that predict the functional potential of soil microorganisms can link the knowledge of microbial communities to their key ecosystem functions. Soil microbial genes coding for specific enzymes related to particular ecosystem processes can help us to establish links between genetic diversity, community structure and further to ecosystem functions, such as carbon cycling through studying carbohydrate-active enzymes (CAZy) (Yang et al., 2014; Manoharan et al., 2017).
N cycling is a collection of important biogeochemical pathways mediated by microbial communities. In most northern peatlands, the slow decomposition of dead plant material returns organic N to the soil and causes peat accumulation (Moore, 2002). Microbial conversion of N is usually described as a cycling consisting of denitrification, nitrification, nitrogen fixation and dissimilatory nitrate reduction to ammonium (DNRA) (Glaze et al., 2022). Drought stresses directly or indirectly affect the ecosystem N cycling (Deng et al., 2021). The effect of drought on net N mineralization and flux in soils of different climates, soil, and ecosystem types varies (Hartmann et al., 2013). The increase in duration and intensity of drought is usually related to N mineralization, mobility, and a decrease in the plant uptake rate of inorganic nutrients (Deng et al., 2021). Recent efforts have focused on characterizing functional genes involved in multiple N cycling processes using genetic approaches and relating genetic information to ecosystem functioning. It has been made clear that amoA genes regulate the availability of inorganic N and the production of N2O through nitrification, while nirK and nirS genes are involved in NO3- consumption and N2O production, nosZ genes mediate the conversion of N2O to N2 during denitrification (Dai et al., 2020). Tu et al. (2017) have also correlated soil N cycling with microbial genetic data, which were mainly concentrated in a few gene families by PCR. Petersen et al. (2012) analyzed nitrification and denitrification processes across a vegetation gradient in Alaska by qPCR amplification of gene families including amoA, nirK/S, and nosZ, and suggested that the abundance of these functional genes can be used as good predictors for biogeochemical process rates. Drought was reported to affect the diversity and composition of microbial communities, and therefore the abundance of functional genes related to N cycling in arable land (Banerjee et al., 2016) and pasture soils (Radl et al., 2015). However, comprehensive surveys of genes involved in all N cycling processes in peatland ecosystems under extreme drought events have rarely been carried out.
The Qinghai-Tibet Plateau has large soil C and N stocks that turn over at low rates (Chen et al., 2013; Fu et al., 2021). In alpine peatland soils, microbial metabolism is limited by harsh environments, such as low temperatures or oxygen levels, and microorganisms are less able to decompose and recover soil organic material. The purpose of this study is to conduct a prospective study on whether the functional genes involved in the process of C and N cycling changed significantly after extreme drought events using metagenomics methods and to explore which specific processes and microbial groups are most affected. To address these objectives, an in-situ field extreme drought simulation experiment was conducted for five years in an alpine peatland ecosystem was performed and the functional genes of soil C and N cycling were screened in detail. Finally, we explore the role of drought-affected microbial communities in the C and N cycling at the genetic level.
The location of the peatland investigated in this research is at an altitude of 3430 m above sea level, located in the Zoige plateau (33°47′56.61″ N, 102°57′28.43″ E) on the northeast edge of the Qinghai-Tibet Plateau. The largest alpine peatland in the world is found in this area due to the region’s unique climatic and hydrological regimes and its geomorphologic and soil conditions (Ma et al., 2016). The mean annual temperature and mean annual precipitation are -1.7°C to 3.3°C and 650 mm to 750 mm, respectively and 90% of the annual precipitation occurs from April to September. The soil is highland peat soil. For this study, the vertical depth of peat at the study site was approximately 1.2 m. The SOC content was between 179 g·kg-1 to 276 g·kg-1 and the soil pH was between 6.8 to 7.2. The vegetation was primarily composed of Carex meyeriana, Koeleria tibetica, Carex muliensis, Eriophorum gracile, Blysmus sinocompressus, and Carex secbrirostris.
The extreme drought events were designed by statistical extremity concerning a historical reference period (extreme value theory) independent of biological effects. We focused on one extreme drought event, not a long-term drought, so local rainfall statistics at the study site have been collected over the past 50 years (China Meteorological Data Network). Daily rainfall of ≤ 3 mm is defined as non-effective rainfall and the minimum duration of non-effective rainfall during the study period was set to 32 days (where the days without effective rainfall are the duration of the drought) (Jentsch et al., 2011), which has the same design idea with the previous research(Zhou et al., 2019). Different periods of extreme drought events were applied during the growing season. The experiment consisted of a control (CK) with ambient precipitation and then different periods of extreme drought. The names and dates of three drought periods were: early drought (ED) from June 18 to July 20, 2019; midterm drought (MD) from July 20 to August 23, 2019; and late drought (LD) from August 23 to September 25, 2019, following 5 years of continuous extreme drought events from 2014 to 2018.
Precipitation was excluded from the study site using a transparent awning (length × width × height: 2.5 m × 2.5 m × 1.8 m). The light transmittance of the shelter material was more than 90%. Each drought treatment consisted of three replicate plots of 2 m × 2 m each, and these were selected randomly in the study field. The iron plates around each experimental community are smashed into the ground for 1m to prevent the lateral flow of water. During rain, roof water was collected with gutters along the shelter awning and transported 50 m downhill of the experimental site, and after each extreme drought event, we removed the shelters and then allow the extreme drought plots to receive natural precipitation. Three soil cores of 0–20 cm in depth were collected and mixed from each plot at the end of each simulated extreme drought period (i.e., ED, MD and LD). One part of the soil samples was transported to the laboratory in coolers with dry ice and then were stored at −80°C for later DNA extraction, the other part was passed through a 2–mm sieve to remove the plant roots and determine soil biochemical properties.
The soil pH was determined in a 1:2.5 soil/water solution using a pH meter. soil water content (SWC) was determined using the oven-drying method. Soil ammonium (NH4+) and nitrate (NO3−) concentrations were determined by the colorimetric method. Soil organic carbon (SOC) was determined by the rapid dichromate oxidation-titration method. Dissolved organic carbon (DOC) was measured using a continuous-flow analyzer. Soil total nitrogen (TN) was determined by the Kjeldahl procedure. These have already been demonstrated in our previous research (Kang et al., 2022; Yan et al., 2022).
Total genomic DNA was extracted from the soil samples using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to the manufacturer’s instructions. The concentration and purity of extracted DNA were determined using TBS-380 and NanoDrop2000, respectively. DNA extract quality was checked using 1% agarose gel.
DNA extract was fragmented to an average size of about 400 bp using Covaris M220 (Gene Company Limited, China) for paired-end library construction. A paired-end library was constructed using NEXTFLEX Rapid DNA-Seq (Bioo Scientific, Austin, TX, USA). Adapters containing the full complement of sequencing primer hybridization sites were ligated to the blunt-end of fragments. Paired-end sequencing was performed on an Illumina NovaSeq (Illumina Inc., San Diego, CA, USA) at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) using NovaSeq Reagent Kits according to the manufacturer’s instructions (www.illumina.com). Sequence data associated with this project have been deposited in the NCBI Short Read Archive database (Accession Number: SRP368675).
The data were analyzed on the free online platform, Majorbio Cloud Platform (www.majorbio.com). The paired-end Illumina reads were trimmed of adaptors, and low-quality reads (length<50 bp or with a quality value <20 or having N bases) were removed by fastp (Chen et al., 2018) (https://github.com/OpenGene/fastp, version 0.20.0). Metagenomics data were assembled using MEGAHIT (Li et al., 2015) (https://github.com/voutcn/megahit, version 1.1.2), which makes use of succinct de Bruijn graphs. Contigs with a length being or over 300 bp were selected as the final assembling result, and then the contigs were used for further gene prediction and annotation.
Open reading frames (ORFs) from each assembled contig were predicted using MetaGene (Noguchi et al., 2006) (http://metagene.cb.k.u-tokyo.ac.jp/). The predicted ORFs with length being or over 100 bp were retrieved and translated into amino acid sequences using the NCBI translation table (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=tgencodes#SG1. A non-redundant gene catalog was constructed using CD-HIT (Fu et al., 2012) (http://www.bioinformatics.org/cd-hit/,version4.6.1) with 90% sequence identity and 90% coverage. Reads after quality control were mapped to the non-redundant gene catalog with 95% identity using SOAPaligner (Li et al., 2008) (http://soap.genomics.org.cn/, version 2.21), and gene abundance in each sample was evaluated.
Representative sequences of non-redundant gene catalog were aligned to the NCBI and NR databases with an e-value cutoff of 1e-5 using Diamond (Buchfink et al., 2015) (http://www.diamondsearch.org/index.php, version 0.8.35) for taxonomic annotations. The KEGG annotation was conducted using Diamond (Buchfink et al., 2015) (http://www.diamondsearch.org/index.php, version 0.8.35) against the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/keeg/) with an e-value cutoff of 1e-5.
Independent sample t-tests were used to analyze the functional genes related to C and N affected by extreme drought in different periods. Linear discriminant analysis (LDA) effect size analysis (LEfSe) was conducted using the Galaxy server (http://huttenhower.sph.harvard.edu/galaxy) to analyze the microbial communities at genus level affected by extreme drought in different periods, and the linear fitting variance was used to test the consistency of microbial community and functional gene diversity. Constrained ordination (redundancy analysis, RDA) was used to investigate the relationships between soil factors and the functional genes involved in C degradation, C fixation and N cycling. The differences of microbial community structure between the control and extreme drought treatments were determined using non-metric multidimensional scaling analysis (NMDS) in the “vegan” package of R software. Analysis of variance was performed by SPSS 26.0, and the figures were drawn with OriginPro.
In this study, a total of 65 functional genes (Table S1) responsible for the process of soil C decomposition were selected and 116 functional genes involved in the soil C sequestration pathway were detected (Table S2).
Extreme drought significantly affected many important microbial functional genes involved in C decomposition (Figure 1). Specifically, the functional genes involved in Chitin, Pectin and Aromatics decomposition were not significantly affected by ED. nplT significantly decreased (P < 0.05) by 23.73% in the ED, and the abundance of the PGM3 gene changed from 0.38% to 0, which participated in the decomposition of starch, and the abundance of bglB gene involved in cellulose decomposition was significantly reduced (P < 0.05) by 6.16%; In the MD, pmm-pgm and xynB were significantly reduced (P < 0.05) by 5.08% and 7.52%, while treS and gmuG were significantly increased (P < 0.05) by 4.58% and 42.28%, they participated in the decomposition of starch and hemicellulose, and the glx gene involved in the decomposition of Lignin was changed from 0 to 4.55%. The abf1 gene significantly decreased (P < 0.05) by 56.89%, but the glucan 1, 3-beta-glucosidase (EC: 32.1.58) significantly increased (P < 0.05) by 84.94%. These are involved in the decomposition of hemicellulose and cellulose, respectively. In general, the ED reduced the abundance of functional genes involved in the decomposition of starch and cellulose, and the MD increased the abundance of genes involved in the decomposition of Lignin, but had different effects on the functional genes involved in the different processes of the decomposition of starch and cellulose. The LD reduced the functional genes involved in the decomposition of hemicellulose, but increased the functional genes involved in the decomposition of cellulose.
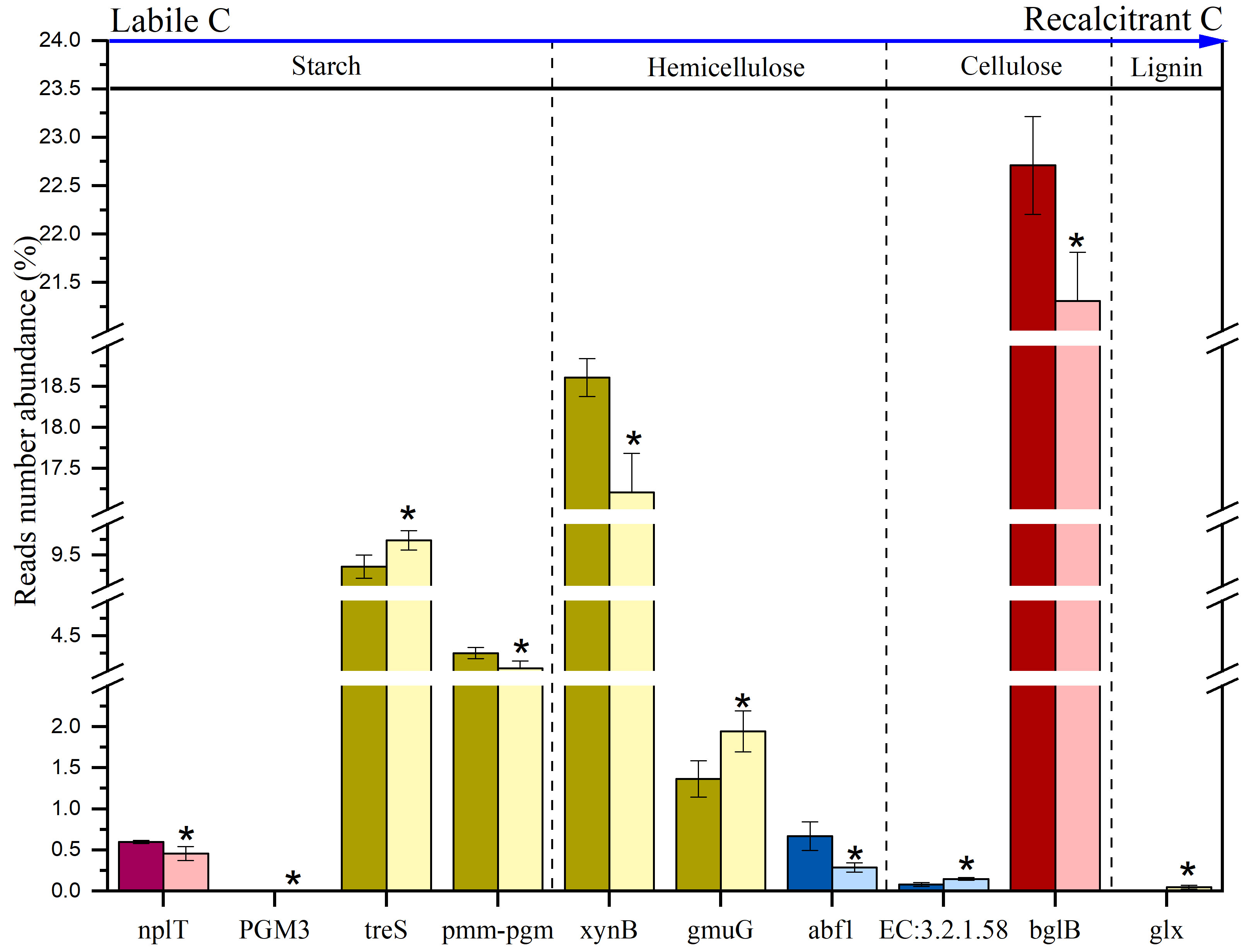
Figure 1 Significantly changed C degradation functional genes are compared between extreme drought and control sites. CK: light color; Extreme drought: dark color; Red: ED; Yellow: MD; Blue: LD. Error bars represent standard error (n = 3). The differences between extreme drought and control sites were analyzed by two-tailed paired t-tests. *P < 0.05.
Extreme drought also significantly affected many important functional genes involved in different C fixation pathways (Figure 2), including the reductive citrate cycle, the 3-hydroxypropionate bi-cycle, the dicarboxylate-hydroxybutyrate cycle and the incomplete reductive citrate cycle, while the hydroxy propionate-hydroxybutyrate cycle, the reductive acetyl-CoA pathway, and the phosphate acetyltransferase-acetate kinase pathway were almost unchanged. In the ED, korC decreased by 30.78% and 28.39% (P < 0.05) in the Reductive citrate cycle and in the Incomplete reductive citrate cycle, respectively. The abundance of E4.2.1.2B, participating in the 3-Hydroxypropionate bi-cycle process, increased significantly by 3.18%. In the MD, sdhC in the reductive citrate cycle, the 3-hydroxy propionate bi-cycle and the dicarboxylate-hydroxybutyrate cycle significantly increased (P < 0.05) by 7.63%, 9.42%, and 6.57%, respectively. mch and smtA1 decreased significantly (P < 0.05) by 18.58% and 19.78%, respectively, while K15052 increased significantly (P < 0.05) by 38.46% in the 3-Hydroxypropionate bi-cycle. In the LD, sucC significantly decreased (P < 0.05) by 4.61% in the Dicarboxylate-hydroxybutyrate cycle.
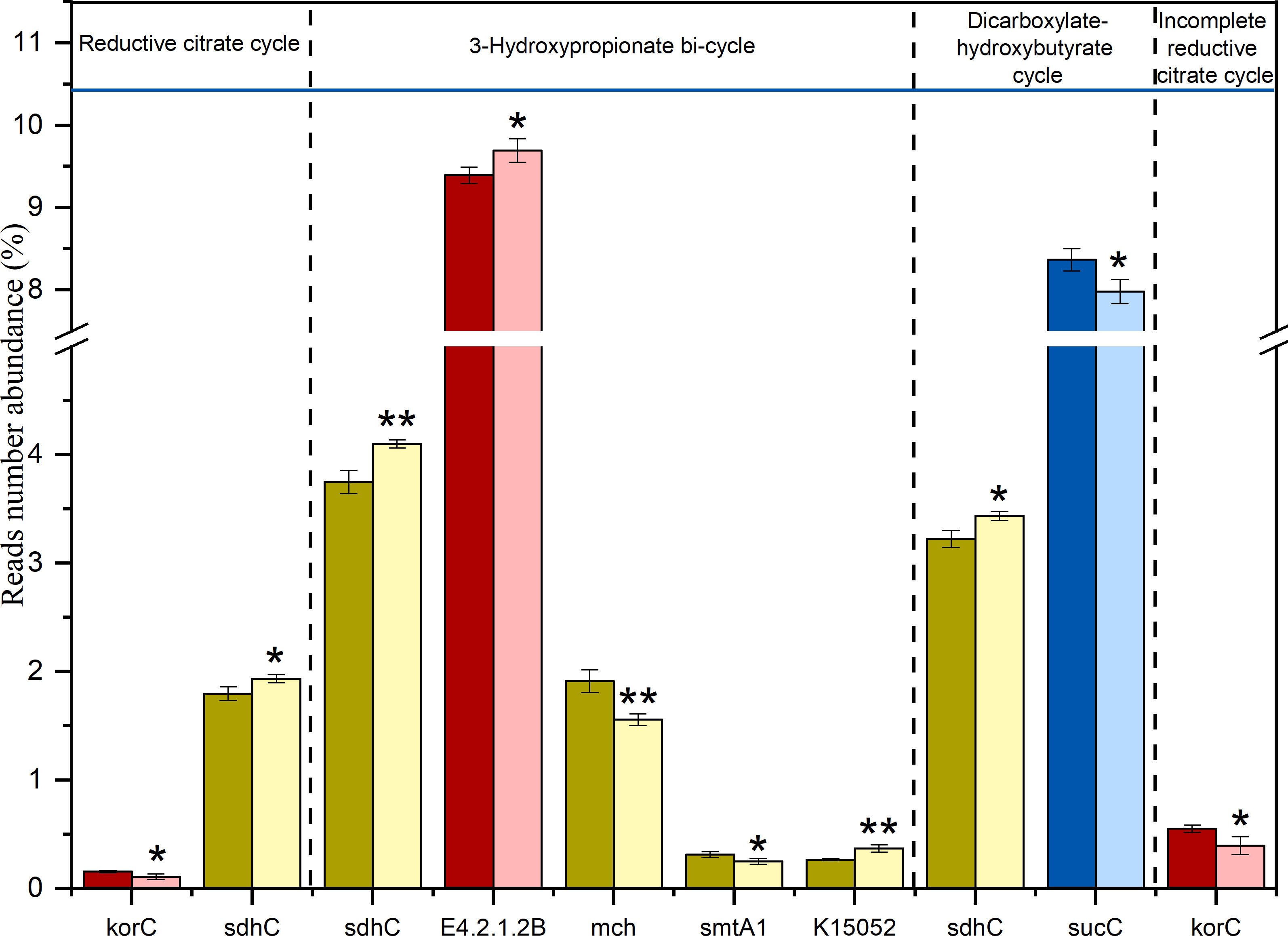
Figure 2 Significantly changed C degradation functional genes compared between extreme drought test plots and control test plots. CK: light color; Extreme drought: dark color Red: ED; Yellow: MD; Blue: LD. Error bars represent standard error (n = 3). The differences between extreme drought and control sites were analyzed using two-tailed paired t-tests. **P < 0.01, *P < 0.05.
Results of the RDA indicated that microbial functional genes involved in C degradation and fixation across the different treatments could be distinguished by soil factors (Figure 3). pH and TN had the largest effects on the C degradation and fixation functional genes. The first axis described 52.38% of the variation in microbial C degradation functional genes, and the second axis explained 4.26% of the variation. The first axis described 37.21% of the variation in microbial C fixation functional genes, and the second axis explained 4.58% of the variation.
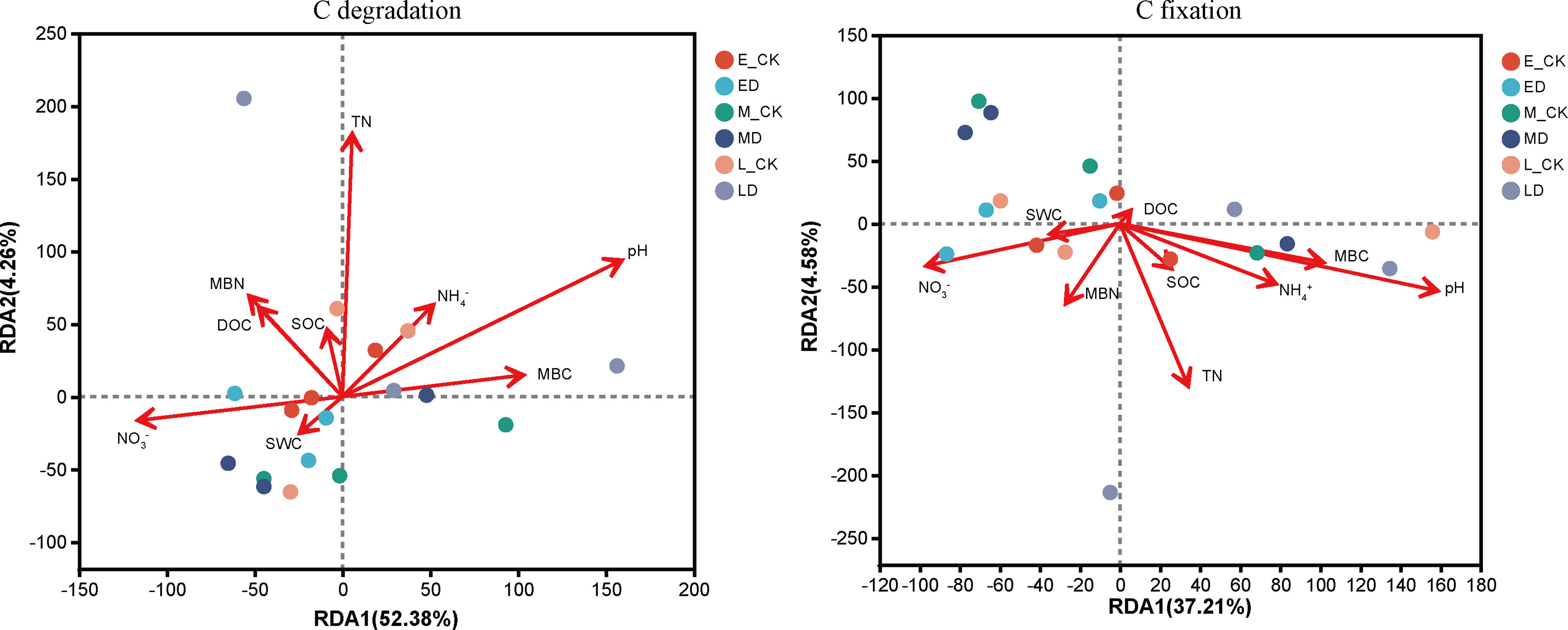
Figure 3 Ordination biplots of redundancy analysis (RDA) showing the relationship of C degradation and fixation functional genes to soil characteristics at 0–20 cm soil layers.
In this study, a total of 50 genes (Table S3) related to the N cycling were detected, including those associated with ammonification, nitrate assimilation, assimilatory N reduction, dissimilatory N reduction, nitrification, denitrification and N fixation. In different periods of extreme drought, the affected functional genes and their response processes were different (Figure 4). When the ED event occurred, the abundance of the ammonia oxidation key gene of nitrification (amoA) in the extreme drought soil increased significantly by 55.32% compared with the control (P < 0.05). This gene is responsible for oxidizing ammonia to hydroxylamine. The abundance of nrtC and nifD genes of soil microorganisms significantly decreased by 13.51% and 25.21%, respectively, compared with the control under the MD (P < 0.05), they participate in the process of nitrate assimilation and N fixation respectively. In the LD, the abundance of hcp genes involved in the soil amelioration process was significantly reduced by 57.78% compared with the control (P < 0.05). Results of the RDA indicated that pH, TN and NO3- had the largest effects on the N cycling functional genes. The first axis described 20.62% of the variation in microbial C degradation functional genes, and the second axis explained 12.59% of the variation (Figure 5).
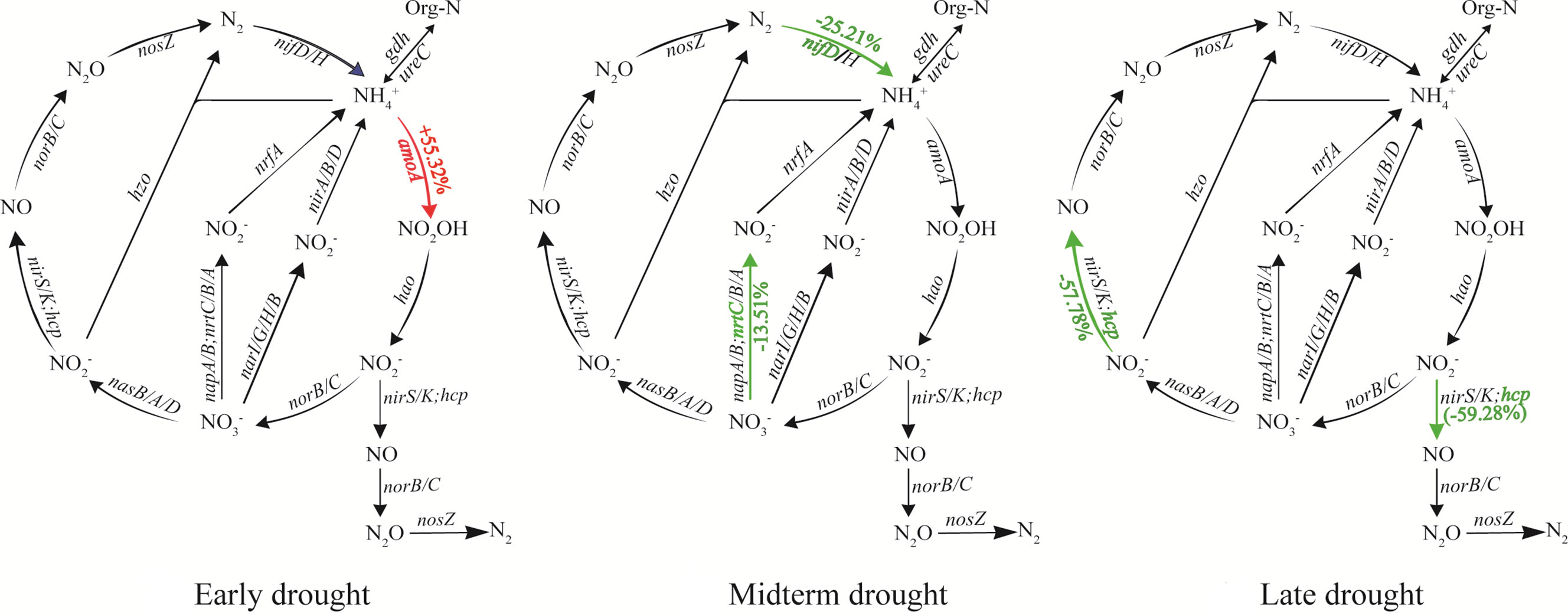
Figure 4 The extreme drought effect on N cycling genes. The percentages in brackets indicate changes in average abundances of functional genes between extreme drought and control sites. Red and green represent the increase and decrease in the average abundance by extreme drought, respectively.
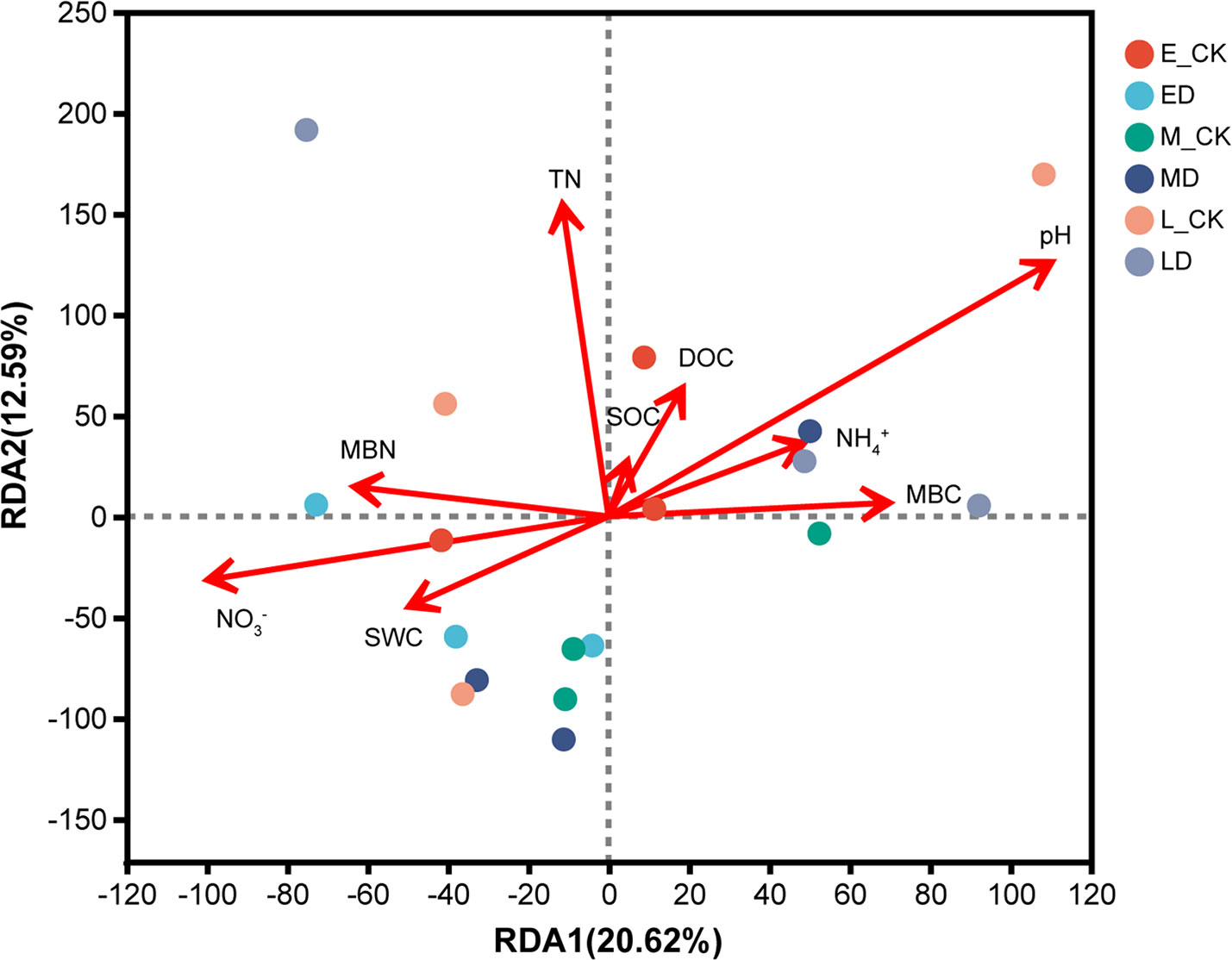
Figure 5 Ordination biplots of redundancy analysis (RDA) showing the relationship of N cycling functional genes to soil characteristics at 0–20 cm soil layers.
In all the samples in this study, the detection numbers at different community classification levels were that Domain: 4; Kingdom: 7; Phylum: 87; Class: 161; Order: 293; Family: 501; Genus: 1631; Species: 6269. The co-occurrence relationship between the sample and the microbial community (Phylum and Genus level) is shown in Figure 6. At the Phylum level, the dominant communities were mainly Proteobacteria, Actinobacteria, Acidobacteria and Chroroflexi. They accounted for about 40%, 20%, 9% and 3% of all communities in the different drought treatments.
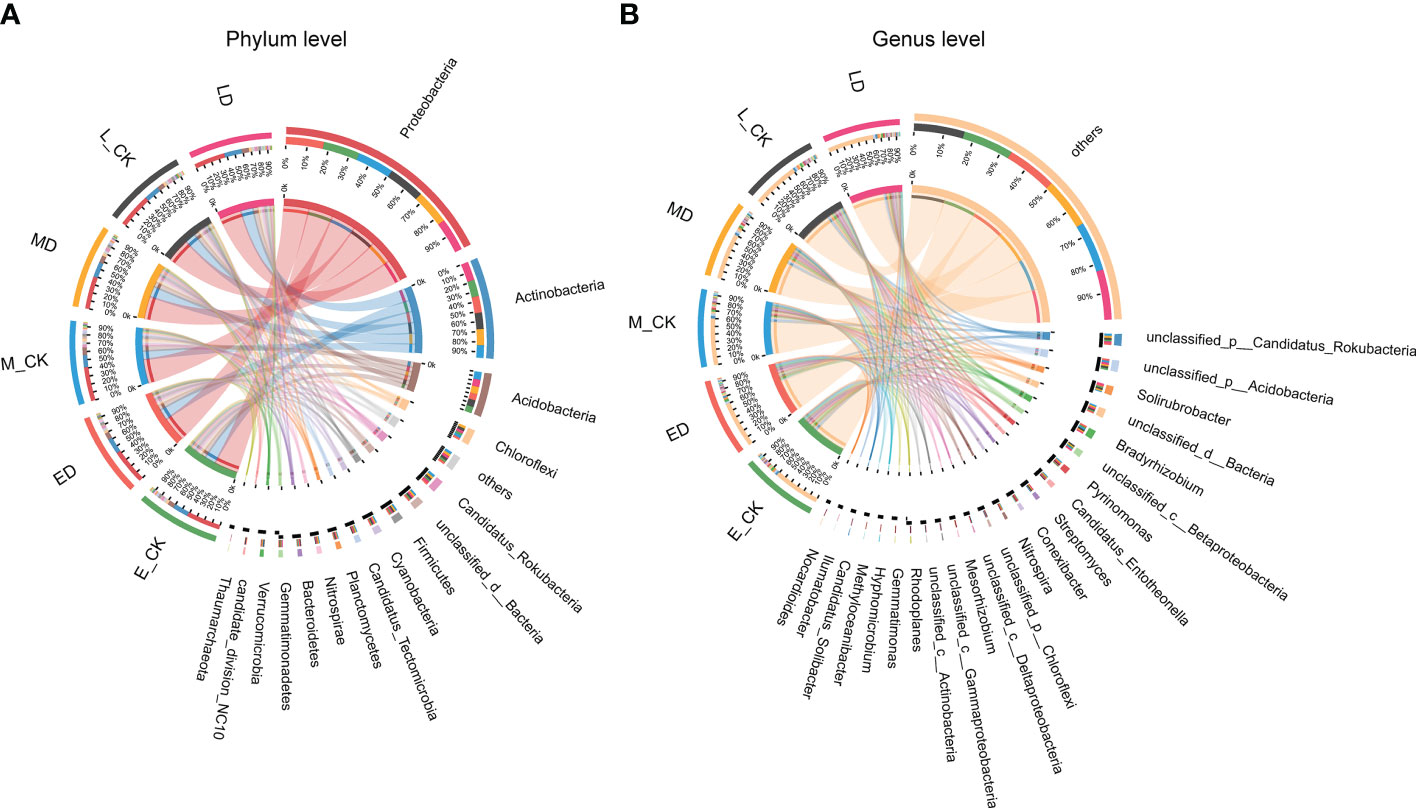
Figure 6 Co-occurrence relationship between samples and species. (A) Microbial community at phylum level; (B) Microbial community at genus level.
Extreme drought also significantly affected many microbial groups at the genus level (Figure 7). In the ED, MD and LD, there were 32, 41 and 2 microbial communities at the genus level that changed significantly, indicating that the impact of ED and MD on microbial community structure was stronger than that of LD. Among microbial communities that have undergone significant changes, extreme drought during different periods leads to different directions of change in microbial communities. ED reduced the abundance of 90.63% of microbial communities, while MD increased the abundance of 95.12% of microbial communities. LD significantly reduced the community abundance of Thermobacterium and Gallionella. In addition, only ED significantly reduced the composition of the Mycobacteriaceae, which belongs to Archaea. In other treatments, there were no significant changes in the Archaea or Fungal categories, indicating that extreme drought mainly changed the composition of bacterial groups.
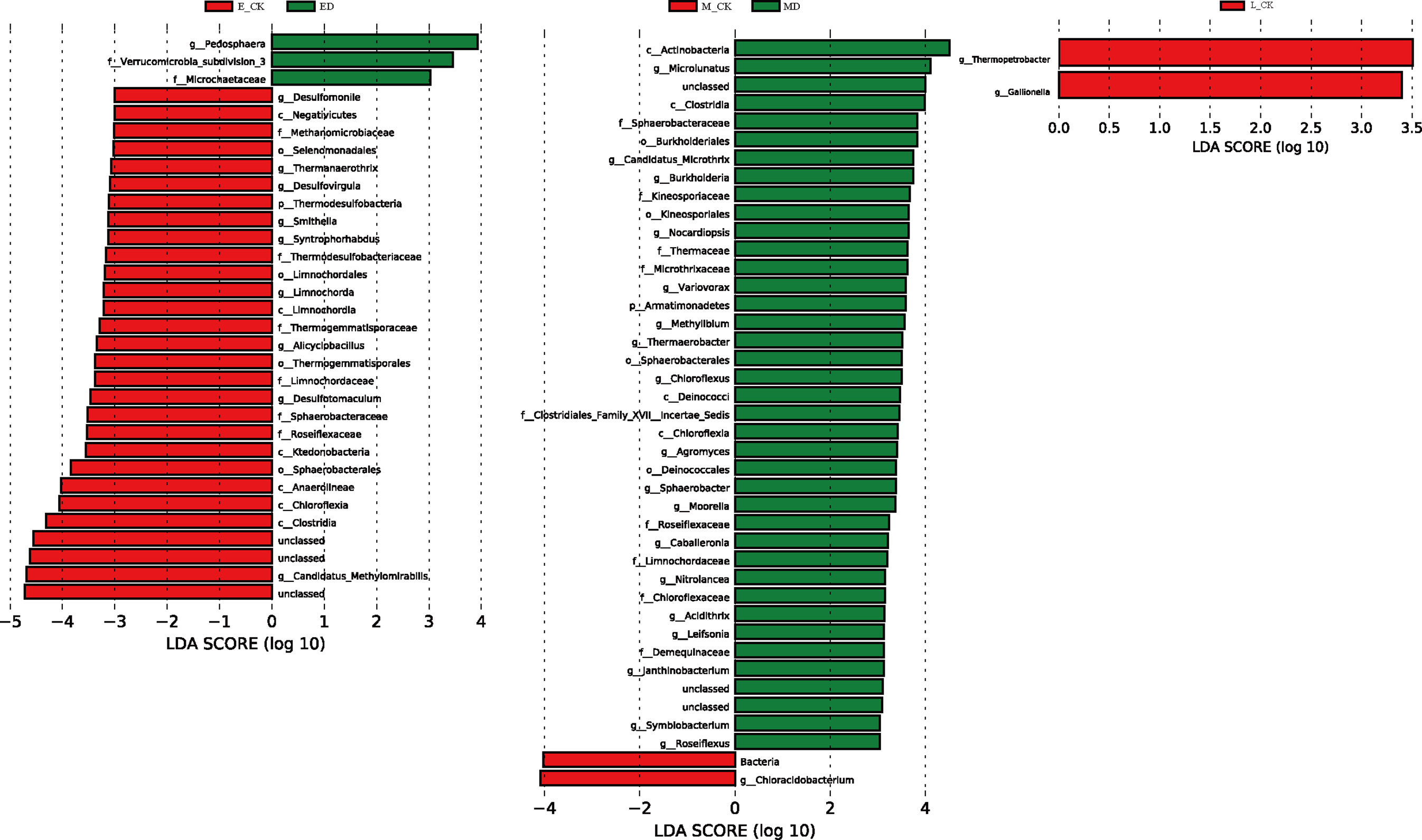
Figure 7 A linear discriminant analysis effect size (LEfSe) method identifies the responses of soil microbial abundant taxa under extreme drought treatment. The threshold on the logarithmic LDA score was 3.0. Red indicates a significant increase in abundance in CK plots, and green indicates a significant increase in abundance in extreme drought plots. The replicate (n = 3).
Based on the abundance information of species and function, α diversity of species and function were calculated, respectively, and linear regression analysis was performed to evaluate the consistency of species and function. In this study, the microbial communities of some C sequestration processes exhibited good consistency with the corresponding functional genes, including the reductive citrate cycle, the hydroxypropionate-hydroxybutylate cycle and the dicarboxylate-hydroxybutyrate cycle (Table 1), The associated linear fits were y=1.643X+0.8031 (R2 = 0.37, P < 0.01), y=0.7463X+4.9030 (R2 = 0.28, P < 0.05) and y=0.6539X+4.2184 (R2 = 0.24, P < 0.05), respectively. The species and functional gene α diversity of the N cycling also showed good consistency, and the linear fit was y=0.3084X+4.8109 (R2 = 0.28, P < 0.05). NMDS analyses showed that the composition of microbial community structures involved in C fixation differed between treatments in early extreme drought (Figure 8), while the microbial community structure involved in C degradation and N cycling has little change under extreme drought treatment in different periods. Furtherly, the relative contributions of the top ten functional genes to microbial communities involved in C degradation, C fixation and N cycling processes (Figure 9) showed that as the abundance of microbial communities decreased, although the composition of related functional genes changed, the functional categories did not show significant changes, indicating a complex relationship between microbial community composition and functional gene abundance. The changes in population abundance were not consistent with the changes in function.
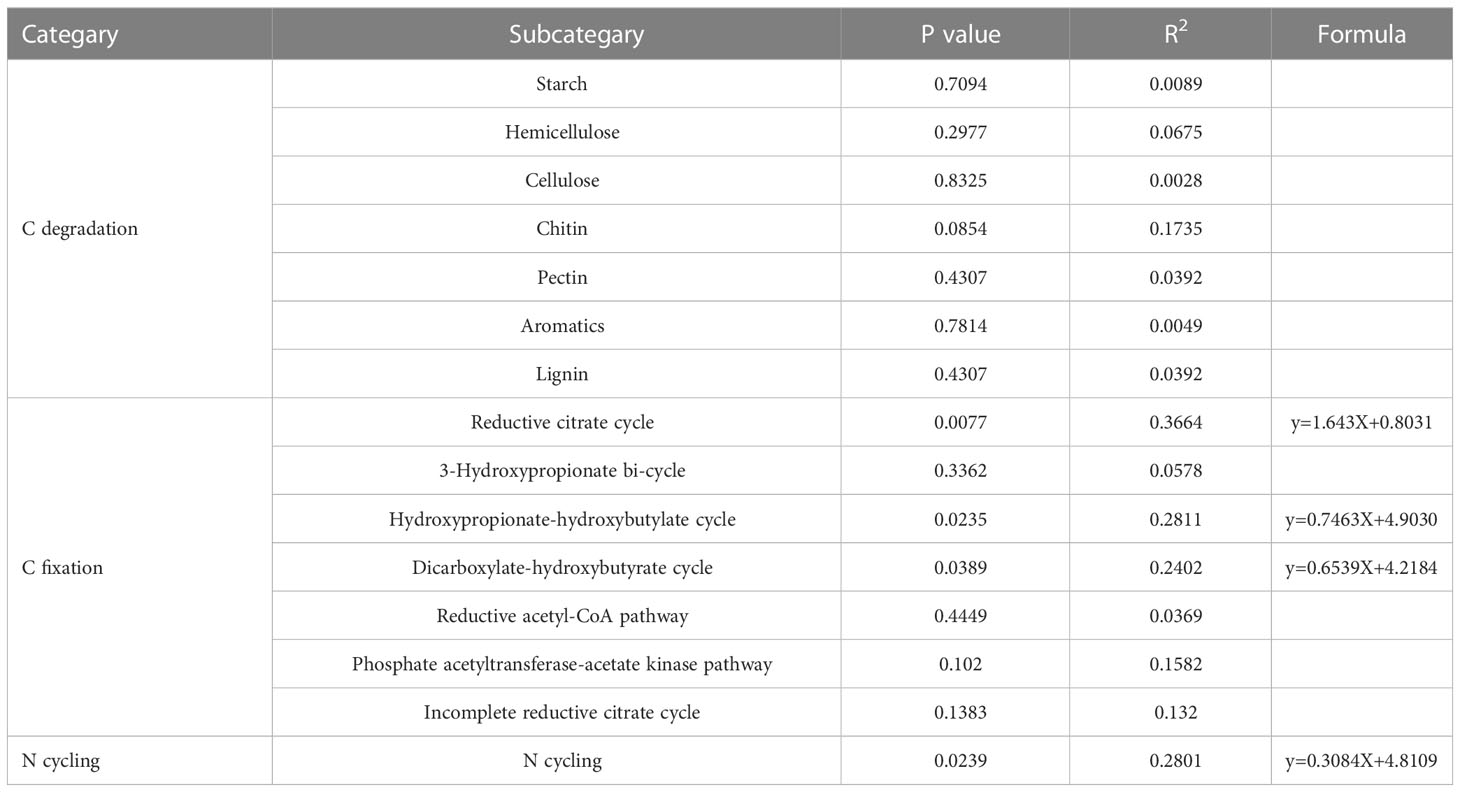
Table 1 Regression relationship of α diversity (Shannon index) for species at the genus level and their corresponding functions in different processes.
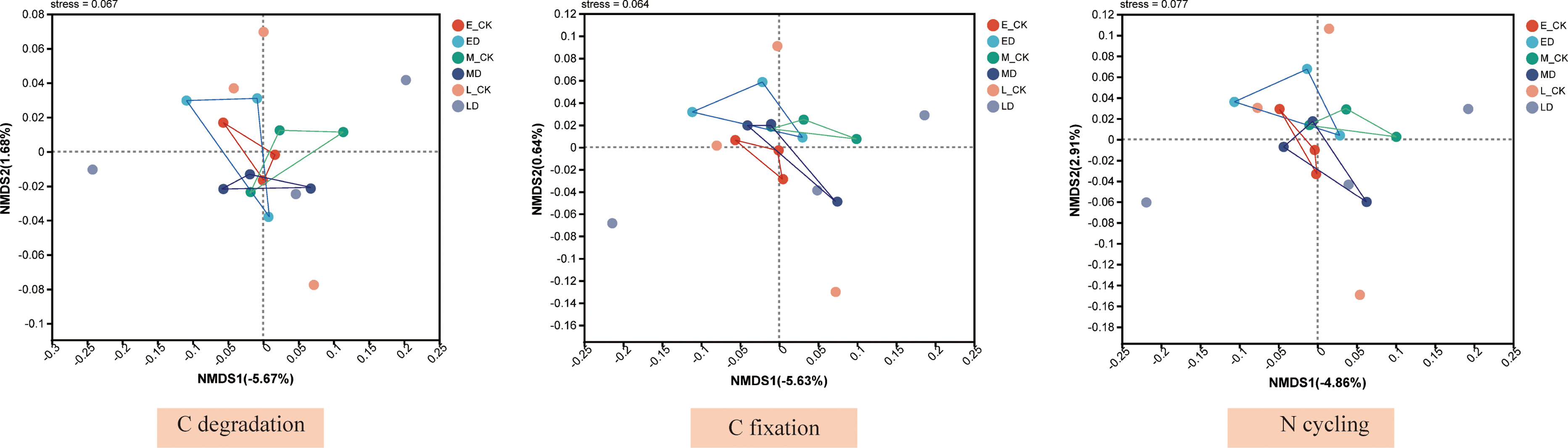
Figure 8 Non–metric multidimensional scaling (NMDS) ordination of the microbial community involved in C degradation, C fixation and N cycling at different treatments. The Bray–Curtis distance matrix based on the abundance of microbial community at genus level was used to determine the compositional variation.
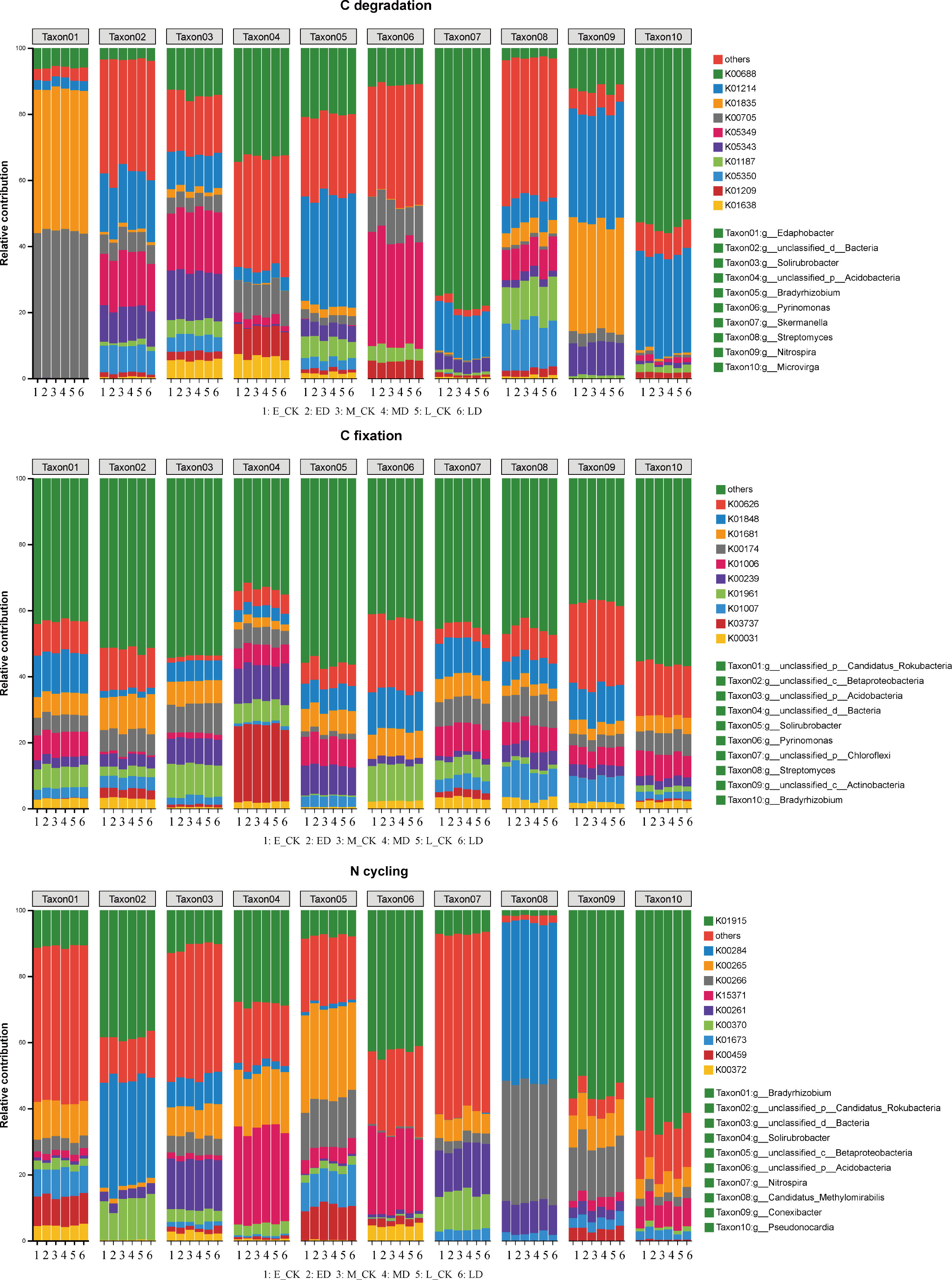
Figure 9 The relative contributions of the top ten functional genes to microbial communities involved in C degradation, C fixation and N cycling processes. The taxonomy of the bacterial communities is shown at the genus level, and the relative contributions are represented by different colors.
Microorganisms have two distinct and critical roles in controlling terrestrial C fluxes i.e., facilitating the release of C into the atmosphere through their catabolic activities, but also blocking it by stabilizing it into a form that is not easily decomposed (Liang and Balser, 2011; Liang et al., 2017). Microbial necromass rather than standing biomass may be a better indicator of microbial contributions to soil C pools, and research increasingly shows a substantial microbial role in the sequestration of C into stable soil C pools (Ludwig et al., 2015).
Our research showed that different periods of extreme drought events have different effects on the functional genes involved in organic C decomposition. After five years of extreme drought, only 15.38% of the selected key functional genes show significant changes. The impact of extreme drought on the different organic matter has certain complexities, and the time of occurrence of extreme drought events within the seasonal cycle plays an important role. In addition, although the changing trend of functional genes is also not consistent, our research showed that the change of functional genes for the decomposition of labile C is relatively complex, and the functional genes for the decomposition of lignin are significantly increased. The production of extracellular degrading enzymes may be reduced under drought conditions because of the lower rate of return of decomposition products due to diffusion limitations (Sardans and Penuelas, 2010). The microbial community is very sensitive to changes in precipitation conditions and even a slight precipitation event can cause rapid microbial response (Nielsen and Ball, 2015). Yue et al. (Yue et al., 2015) used gene chips to study soil microorganisms in alpine meadows and noted that the relative abundance of catabolic genes related to C and N cycling decreased with the increase of temperature, with the most obvious change being in genes related to stable C. Our study also suggests that drought significantly increases the catabolic genes related to lignin and has complex effects on labile C such as cellulose and hemicellulose. This is consistent with research that has indicated that extreme drought promotes more gene expression in microorganisms degrading stable C, and reduces the long-term stability of soil C. Due to the huge diversity of microorganisms and the unclear ecological functions of most species, detection methods for specific functional microbial groups need to be greatly improved (Rinke et al., 2013).
Similar to the decomposition process of soil organic matter, the genes involved in the C fixation process exhibited different changes for different extreme drought periods, although the response trend of related processes was not consistent, indicating that the response of microorganisms to extreme drought is very complex. However, it is difficult to deduce the change direction of the final C fixation capacity from the change of a certain function. Assimilation of CO2 by soil autotrophic microorganisms is an important process of the soil C cycling, particularly in vegetation-constrained ecosystems with environmental stresses such as on the Tibetan Plateau which is characterized by low temperature and high UV radiation levels. Lynn et al. (2017) showed that the C sequestration rate of autotrophic microorganisms in wetland soil was 85.1 mg C m-2 d-1, and that of autotrophic microorganisms in grassland and woodland soil was 21.9 mg C m-2 d-1 and 32.9 mg C m-2 d-1, respectively. Precipitation and latitude together accounted for 76.00% difference in soil C sequestration microbial metabolic pathways in differently vegetated areas. In addition, oxygen sensitivity can also one of the reasons for differences in CO2 sequestration pathways (Liu et al., 2016).
In our study, nirK and nirS genes in denitrifying microbial communities for reducing nitrite (NO2−) to nitric oxide (NO), and nosZ genes in denitrifying bacterial communities for reducing N2O to N2 (Jetten, 2008; Levy-Booth et al., 2014) were not affected by extreme drought. This may greatly influence the nitrification and denitrification rates and associated N2O emissions. Keil et al. (Keil et al., 2015) used qPCR to study microbial communities. Their results showed that the abundance of nirK, nirS and nosZ all changed, while the abundance of the nosZ denitrifying gene decreased by 3.1%, which was greatly affected by drought. Hartmann et al. (Hartmann et al., 2013) studied the response of soil N cycling microorganisms to simulated drought through qPCR. Drought increased the N mineralization rate by 14%, and reduced the relative abundance of denitrification gene nirS by 7%. In an incubation experiment, it was found that under both flooded and non-flooded conditions, warming increased the N mineralization rate, which was reflected in the increase of denitrification rate and the abundance of copper reductase (nirK) (Gao et al., 2009; Fang et al., 2021).
Snider et al. (2015) documented significant changes in denitrification gene abundance (nirS, nosZ), which increased after heavy rainfall events increased WFPS from < 40% to > 70%, consistent with large fluxes of N2O. A meta-analysis found that the effect sizes of nirK and nosZ showed a U-shaped relationship with the effect sizes of soil moisture (Li et al., 2020), contrary to the belief that lower soil moisture and correspondingly higher soil oxygen concentration may inhibit nirK and nosZ genes under drought conditions (Delgado-Baquerizo et al., 2014; Homyak et al., 2017). Although our experiment was carried out continuously for 5 years, rewetting during the period of non-shielding may play a great role in the recovery of microorganisms. Some studies have pointed out that the significant resistance and/or resilience of the functional microbial community participating in the N cycling to extreme weather events may indicate that the microorganisms in silt are better adapted to the expected pressure conditions (Hammerl et al., 2019).
Ammonia oxidation is the first step of nitrification, which is the key process in the global N cycling and forms nitrate through microbial activities (Leininger et al., 2006). We observed an increase in abundance of amoA genes from the ED plots. This suggests that the ammonia oxidation process increases after extreme drought events. Che et al. (2017) used N cycling gene abundance (amoA) to find that a major shift in the N cycling led to a five-fold increase in nitrobacteria during grassland degradation in Tibet. Nitrification produces easily leached nitrates, which are the limiting substrate for denitrification in alpine meadows (Xie et al., 2014). Therefore, nitrates increase during periods of high soil moisture or in moist soil microhabitats. However, it has also been reported that both ammoxidation archaea (AOA) abundance and community composition are unaffected by drought, supporting earlier observations of AOA resistance to drought stress (Gleeson et al., 2010) and AOA’s good adaptation to a wide range of growing conditions and substrate concentrations (Schleper, 2010). Our study did not distinguish between AOA and AOB, but the increase of overall gene abundance indicates that extreme drought events in peatland may enhance the ammonia oxidation process. Previous studies have reported that although total N concentration sharply decreases under drought, the availability of soluble organic N and mineral N significantly increases, thereby promoting plant N absorption in the short term (Rennenberg et al., 2009). The opposite research results suggest that drought does not affect the inorganic N content in the soil, even under long-term drought treatment. Part of the reason for these two results is the difference in drought intensity and timing (Hartmann et al., 2013). We demonstrated that the soil pH, TN and NO3- had the largest effects on the N cycling functional genes (Figure 5).
Most biogeochemical transformations appear to be mediated by a limited set of metabolic pathways in multiple taxa (Louca et al., 2018). We still do not know the appropriate solutions to establish links between microbial diversity and ecosystem processes in order to be able to capture the taxonomic distribution, metabolic capacity and response to environmental perturbations of microorganisms, although the diversity of microbes is incredible (Gans et al., 2005). The relationship between microbial biodiversity and ecosystem function is mainly inferred by measuring the change of species diversity and community composition derived using the marker gene method, but this inference does not explain the causal relationship between microbial diversity and ecosystem function (Krause et al., 2014). Research on the relationship between microbial biodiversity and ecosystem function needs to further characterize the functional performance and role of microorganisms in the ecosystem (Wang et al., 2019).
Our results showed that the composition of microbial community structures involved in C fixation function differed between treatments in early extreme drought (Figure 8). In addition to the derailed ecosystem function and microbial community function mentioned above, microbial taxonomic composition and functional potential may not always be linearly correlated. In our study, microbial community diversity involved in the N cycling process exhibited a good linear relationship with functional gene diversity, and only 3 out of 7 microbial C fixation processes had a good linear relationship. However, the diversity of genes involved in soil C degradation screened by us showed no linear relationship with the corresponding microbial communities, indicating that the decomposition of organic matter was involved in the decoupling between microbial groups and genes. This may be caused by different microbial groups having the same functional genes. This separation of microbial functional and taxonomic composition raises key questions about the relative spatial and temporal value of information on microbial phylogenetic diversity versus metabolic diversity of functional microorganisms for understanding biogeochemical reactions (Wertz et al., 2007).
For microorganisms performing relatively “extensive” functions, such as the decomposition and turnover of organic matter, the recombination of functional gene diversity (i.e. the functional potential of the community) does not necessarily lead to changes in ecosystem functions, such as decomposition rate (Allison and Martiny, 2008; Miki et al., 2014). A recent meta-analysis based on a metagenomic dataset of 365 samples worldwide revealed a consistent pattern in the relative frequencies of eight metabolic pathways associated with N transformation, hinting at the ability of metagenomic approaches to make potential inferences about the role of microorganisms in mediating biogeochemical cycles (Nelson et al., 2016). Studies have shown strong evidence to support the hypothesis of functional redundancy in soil microorganisms, as the degree of variation in microbial taxonomic composition is greater than the functional potential based on metagenomic gene abundance in global terrestrial ecosystems (Chen et al., 2022). In the future, we need to further integrate macro genomics, experimental culture and field measurement technology to link the information on microbial classification, phylogeny and functional genes with the measured phenotypic traits and environmental preferences (Barberan et al., 2017).
In this study, it was shown that under extreme drought conditions, microbial communities with low abundance underwent more significant changes than those with high abundance, and the abundance of functional genes involved in related processes showed a similar pattern, pH and TN had the largest effects on microbial functional genes. Changes in α diversity of microbial communities and functional genes were consistent in some C fixation processes and N cycling. In summary, progress has been made in understanding the potential negative impacts and positive contributions of soil microorganisms to extreme drought by considering the direct and indirect impacts of climate change on microorganisms and the ability of such impacts to amplify or inhibit key processes in the C and N cycling, despite uncertainties in predicting ecosystem function. We believe that in the future it will be necessary to link microbial ecology with whole-ecosystem scale flux measurements and C cycling feedback models. It is possible for ecosystem models to make better predictions of the ecosystem effects of future extreme drought by better evaluating microorganisms’ functional capacity and response.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
ZY, XK, YL, KZ and YH contributed to the conception and design of the study. EK, XZ and XY organized the database. ZY, ML and YL performed the statistical analysis. ZY and LY wrote the first draft of the manuscript. AY, XW, and YN wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 32201410, 32171597, 42041005), the Fundamental Research Funds of the Chinese Academy of Forestry (No. CAFYBB2022SY041) and the Department of Science and Technology of Sichuan Province (2020YFH0131).
We would also like to acknowledge Dr. Willaim R.T. (University of North Wales) for providing language editing in the preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1173750/full#supplementary-material
Allison S. D., Martiny J. B. H. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 105, 11512–11519. doi: 10.1073/pnas.0801925105
Alster C. J., German D. P., Lu Y., Allison S. D. (2013). Microbial enzymatic responses to drought and to nitrogen addition in a southern California grassland. Soil Biol. Biochem. 64, 68–79. doi: 10.1016/j.soilbio.2013.03.034
Banerjee S., Helgason B., Wang L., Winsley T., Ferrari B. C., Siciliano S. D. (2016). Legacy effects of soil moisture on microbial community structure and N2O emissions. Soil Biol. Biochem. 95, 40–50. doi: 10.1016/j.soilbio.2015.12.004
Barberan A., Caceres Velazquez H., Jones S., Fierer N. (2017). Hiding in plain sight: mining bacterial species records for phenotypic trait information. mSphere 2 (4), e00237–17. doi: 10.1128/mSphere.00237-17
Bardgett R. D., Freeman C., Ostle N. J. (2008). Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2 (8), 805–814. doi: 10.1038/ismej.2008.58
Bastida F., Eldridge D. J., García C., Kenny Png G., Bardgett R. D., Delgado-Baquerizo M. (2021). Soil microbial diversity–biomass relationships are driven by soil carbon content across global biomes. ISME J. 15 (7), 2081–2091. doi: 10.1038/s41396-021-00906-0
Bell T., Newman J. A., Silverman B. W., Turner S. L., Lilley A. K. (2005). The contribution of species richness and composition to bacterial services. Nature 436 (7054), 1157–1160. doi: 10.1038/nature03891
Borken W., Matzner E. (2009). Reappraisal of drying and wetting effects on c and n mineralization and fluxes in soils. Glob. Change Biol. 15, 808–824. doi: 10.1111/j.1365-2486.2008.01681.x
Buchfink B., Xie C., Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12 (1), 59–60. doi: 10.1038/nmeth.3176
Canarini A., Carrillo Y., Mariotte P., Ingram L., Dijkstra F. A. (2016). Soil microbial community resistance to drought and links to c stabilization in an Australian grassland. Soil Biol. Biochem. 103, 171–180. doi: 10.1016/j.soilbio.2016.08.024
Che R., Wang F., Wang W., Zhang J., Zhao X., Rui Y., et al. (2017). Increase in ammonia-oxidizing microbe abundance during degradation of alpine meadows may lead to greater soil nitrogen loss. Biogeochemistry 136 (3), 341–352. doi: 10.1007/s10533-017-0399-5
Chen H. H., Ma K. Y., Lu C. Y., Fu Q., Qiu Y. B., Zhao J. Y., et al. (2022). Functional redundancy in soil microbial community based on metagenomics across the globe. Front. Microbiol. 13, 878978. doi: 10.3389/fmicb.2022.878978
Chen H., Yang G., Peng C., Zhang Y., Zhu D., Zhu Q., et al. (2014). The carbon stock of alpine peatlands on the qinghai-Tibetan plateau during the Holocene and their future fate. Quaternary Sci. Rev. 95, 151–158. doi: 10.1016/j.quascirev.2014.05.003
Chen S. F., Zhou Y. Q., Chen Y. R., Gu J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), 884–890. doi: 10.1093/bioinformatics/bty560
Chen H., Zhu Q., Peng C., Wu N., Wang Y., Fang X., et al. (2013). The impacts of climate change and human activities on biogeochemical cycles on the qinghai-Tibetan plateau. Glob. Change Biol. 19 (10), 2940–2955. doi: 10.1111/gcb.12277
Dai Z., Yu M., Chen H., Zhao H., Huang Y., Su W., et al. (2020). Elevated temperature shifts soil n cycling from microbial immobilization to enhanced mineralization, nitrification and denitrification across global terrestrial ecosystems. Glob. Change Biol. 26 (9), 5267–5276. doi: 10.1111/gcb.15211
Delgado-Baquerizo M., Maestre F. T., Escolar C., Gallardo A., Ochoa V., Gozalo B., et al. (2014). Direct and indirect impacts of climate change on microbial and biocrust communities alter the resistance of the n cycle in a semiarid grassland. J. Ecol. 102 (6), 1592–1605. doi: 10.1111/1365-2745.12303
Deng L., Peng C., Kim D.-G., Li J., Liu Y., Hai X., et al. (2021). Drought effects on soil carbon and nitrogen dynamics in global natural ecosystems. Earth Sci. Rev. 214, 103501. doi: 10.1016/j.earscirev.2020.103501
de Vries F. T., Griffiths R. I., Bailey M., Craig H., Girlanda M., Gweon H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9 (1), 3033. doi: 10.1038/s41467-018-05516-7
Dietrich J. D., Smith M. D. (2016). The effect of timing of growing season drought on flowering of a dominant C4 grass. Oecologia 181, 391–399. doi: 10.1007/s00442-016-3579-4
Du L., Mikle N., Zou Z., Huang Y., Shi Z., Jiang L., et al. (2018). Global patterns of extreme drought-induced loss in land primary production: identifying ecological extremes from rain-use efficiency. Sci. Total Environ. 628, 611–620. doi: 10.1016/j.scitotenv.2018.02.114
Fang X., Zheng R., Guo X., Fu Q., Fan F., Liu S. (2021). Yak excreta-induced changes in soil microbial communities increased the denitrification rate of marsh soil under warming conditions. Appl. Soil Ecol. 165, 103935. doi: 10.1016/j.apsoil.2021.103935
Fierer N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fu L. M., Niu B. F., Zhu Z. W., Wu S. T., Li W. Z. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28 (23), 3150–3152. doi: 10.1093/bioinformatics/bts565
Fu B., Ouyang Z., Shi P., Fan J., Wang X., Zheng H., et al. (2021). Current condition and protection strategies of qinghai-Tibet plateau ecological security barrier. Bull. Chin. Acad. Sci. 36 (11), 1298–1306.
Gans J., Wolinsky M., Dunbar J. (2005). Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309 (5739), 1387–1390. doi: 10.1126/science.1112665
Gao J. Q., Ouyang H., Xu X. L., Zhou C. P., Zhang F. (2009). Effects of temperature and water saturation on CO2 production and nitrogen mineralization in alpine wetland soils. Pedosphere 19 (1), 71–77. doi: 10.1016/S1002-0160(08)60085-5
García-Palacios P., Prieto I., Ourcival J. M., Hattenschwiler S. (2016). Disentangling the litter quality and soil microbial contribution to leaf and fine root litter decomposition responses to reduced rainfall. Ecosystems 19 (3), 490–503. doi: 10.1007/s10021-015-9946-x
Glaze T. D., Erler D. V., Siljanen H. M. P. (2022). Microbially facilitated nitrogen cycling in tropical corals. ISME J. 16 (1), 68–77. doi: 10.1038/s41396-021-01038-1
Gleeson D. B., Mueller C., Banerjee S., Ma W., Siciliano S. D., Murphy D. V. (2010). Response of ammonia oxidizing archaea and bacteria to changing water filled pore space. Soil Biol. Biochem. 42 (10), 1888–1891. doi: 10.1016/j.soilbio.2010.06.020
Guo X., Gao Q., Yuan M., Wang G., Zhou X., Feng J., et al. (2020). Gene-informed decomposition model predicts lower soil carbon loss due to persistent microbial adaptation to warming. Nat. Commun. 11 (1), 4897. doi: 10.1038/s41467-020-18706-z
Hammerl V., Kastl E. M., Schloter M., Kublik S., Schmidt H., Welzl G., et al. (2019). Influence of rewetting on microbial communities involved in nitrification and denitrification in a grassland soil after a prolonged drought period. Sci. Rep. 9, 2280. doi: 10.1038/s41598-018-38147-5
Hartmann A. A., Barnard R. L., Marhan S., Niklaus P. A. (2013). Effects of drought and n-fertilization on n cycling in two grassland soils. Oecologia 171 (3), 705–717. doi: 10.1007/s00442-012-2578-3
Homyak P. M., Allison S. D., Huxman T. E., Goulden M. L., Treseder K. K. (2017). Effects of drought manipulation on soil nitrogen cycling: a meta-analysis. J. Geophys. Res. Biogeosci. 122 (12), 3260–3272. doi: 10.1002/2017JG004146
Hoover D. L., Rogers B. M. (2016). Not all droughts are created equal: the impacts of interannual drought pattern and magnitude on grassland carbon cycling. Glob. Change Biol. 22 (5), 1809–1820. doi: 10.1111/gcb.13161
Jentsch A., Kreyling J., Elmer M., Gellesch E., Glaser B., Grant K., et al. (2011). Climate extremes initiate ecosystem-regulating functions while maintaining productivity. J. Ecol. 99 (3), 689–702. doi: 10.1111/j.1365-2745.2011.01817.x
Jetten M. S. M. (2008). The microbial nitrogen cycle. Environ. Microbiol. 10 (11), 2903–2909. doi: 10.1111/j.1462-2920.2008.01786.x
Kang E., Li Y., Zhang X., Yan Z., Zhang W., Zhang K., et al. (2022). Extreme drought decreases soil heterotrophic respiration but not methane flux by modifying the abundance of soil microbial functional groups in alpine peatland. Catena. 212, 106043. doi: 10.1016/j.catena.2022.106043
Kang X., Yan L., Cui L., Zhang X., Hao Y., Wu H., et al. (2018). Reduced carbon dioxide sink and methane source under extreme drought condition in an alpine peatland. Sustainability 10), 4285. doi: 10.3390/su10114285
Keil D., Niklaus P. A., von Riedmatten L. R., Boeddinghaus R. S., Dormann C. F., Scherer-Lorenzen M., et al. (2015). Effects of warming and drought on potential N2O emissions and denitrifying bacteria abundance in grasslands with different land-use. FEMS Microbiol. Ecol. 91 (7), fiv066. doi: 10.1093/femsec/fiv066
Knapp A. K., Beier C., Briske D. D., Classen A. T., Luo Y., Reichstein M. (2008). Consequences of more extreme precipitation regimes for terrestrial ecosystems. BioScience 58, 811–821. doi: 10.1641/B580908
Krause S., Le Roux X., Niklaus P. A., Van Bodegom P. M., Lennon J. T., Bertilsson S., et al. (2014). Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 5, 251. doi: 10.3389/fmicb.2014.00251
Lefi E., Medrano H., Cifre J. (2004). Water uptake dynamics, photosynthesis and water use efficiency in field-grown medicago arborea and medicago citrina under prolonged Mediterranean drought conditions. Ann. Appl. Biol. 144, 299–307. doi: 10.1111/j.1744-7348.2004.tb00345.x
Leininger S., Urich T., Schloter M., Schwark L., Qi J., Nicol G. W., et al. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442 (7104), 806–809. doi: 10.1038/nature04983
Levy-Booth D. J., Prescott C. E., Grayston S. J. (2014). Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 75, 11–25. doi: 10.1016/j.soilbio.2014.03.021
Li R. Q., Li Y. R., Kristiansen K., Wang J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24 (5), 713–714. doi: 10.1093/bioinformatics/btn025
Li D. H., Liu C. M., Luo R. B., Sadakane K., Lam T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 31 (10), 1674–1676. doi: 10.1093/bioinformatics/btv033
Li L. F., Zheng Z. Z., Wang W. J., Biederman J. A., Xu X. L., Ran Q. W., et al. (2020). Terrestrial N2O emissions and related functional genes under climate change: a global meta-analysis. Glob. Change Biol. 26 (2), 931–943. doi: 10.1111/gcb.14847
Liang C., Balser T. C. (2011). Microbial production of recalcitrant organic matter in global soils: implications for productivity and climate policy. Nat. Rev. Microbiol. 9 (1), 75. doi: 10.1038/nrmicro2386-c1
Liang C., Schimel J. P., Jastrow J. D. (2017). The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2 (8), 17105. doi: 10.1038/nmicrobiol.2017.105
Limpens J., Heijmans M. M. P. D., Berendse F. (2006). Boreal Peatland ecosystems. Eds. Wieder R. K., Vitt D. H.. (Berlin, Heidelberg: Springer Berlin Heidelberg), 195–230.
Liu J. F., Mbadinga S. M., Sun X. B., Yang G. C., Yang S. Z., Gu J. D., et al. (2016). Microbial communities responsible for fixation of CO2 revealed by using mcrA, cbbM, cbbL, fthfs, fefe-hydrogenase genes as molecular biomarkers in petroleum reservoirs of different temperatures. Int. Biodeter. Biodegr. 114, 164–175. doi: 10.1016/j.ibiod.2016.06.019
Loik M. E. (2007). Sensitivity of water relations and photosynthesis to summer precipitation pulses for artemisia tridentata and purshia tridentata. Plant Ecol. 191, 95–108. doi: 10.1007/s11258-006-9217-1
Louca S., Polz M. F., Mazel F., Albright M. B. N., Huber J. A., O'Connor M. I., et al. (2018). Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2 (6), 936–943. doi: 10.1038/s41559-018-0519-1
Ludwig M., Achtenhagen J., Miltner A., Eckhardt K. U., Leinweber P., Emmerling C., et al. (2015). Microbial contribution to SOM quantity and quality in density fractions of temperate arable soils. Soil Biol. Biochem. 81, 311–322. doi: 10.1016/j.soilbio.2014.12.002
Lynn T. M., Ge T., Yuan H., Wei X., Wu X., Xiao K., et al. (2017). Soil carbon-fixation rates and associated bacterial diversity and abundance in three natural ecosystems. Microb. Ecol. 73 (3), 645–657. doi: 10.1007/s00248-016-0890-x
Ma K., Liu J. G., Balkovic J., Skalsky R., Azevedo L. B., Kraxner F. (2016). Changes in soil organic carbon stocks of wetlands on china's zoige plateau from 1980 to 2010. Ecol. Model. 327, 18–28. doi: 10.1016/j.ecolmodel.2016.01.009
Malik A. A., Bouskill N. J. (2022). Drought impacts on microbial trait distribution and feedback to soil carbon cycling. Funct. Ecol. 36 (6), 1442–1456. doi: 10.1111/1365-2435.14010
Manoharan L., Kushwaha S. K., Ahrén D., Hedlund K. (2017). Agricultural land use determines functional genetic diversity of soil microbial communities. Soil Biol. Biochem. 115, 423–432. doi: 10.1016/j.soilbio.2017.09.011
Miki T., Yokokawa T., Matsui K. (2014). Biodiversity and multifunctionality in a microbial community: a novel theoretical approach to quantify functional redundancy. Proc. R Soc. Lond B Biol. Sci. 281 (1776), 20132498. doi: 10.1098/rspb.2013.2498
Moore P. D. (2002). The future of cool temperate bogs. Environ. Conserv. 29 (1), 3–20. doi: 10.1017/S0376892902000024
Nelson M. B., Martiny A. C., Martiny J. B. H. (2016). Global biogeography of microbial nitrogen-cycling traits in soil. Proc. Natl. Acad. Sci. U.S.A. 113 (29), 8033–8040. doi: 10.1073/pnas.1601070113
Nielsen U. N., Ball B. A. (2015). Impacts of altered precipitation regimes on soil communities and biogeochemistry in arid and semi-arid ecosystems. Glob. Change Biol. 21 (4), 1407–1421. doi: 10.1111/gcb.12789
Noguchi H., Park J., Takagi T. (2006). MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 34 (19), 5623–5630. doi: 10.1093/nar/gkl723
Pokhrel Y., Felfelani F., Satoh Y., Boulange J., Burek P., Gädeke A., et al. (2021). Global terrestrial water storage and drought severity under climate change. Nat. Clim. Change 11 (3), 226–233. doi: 10.1038/s41558-020-00972-w
Radl V., Kindler R., Welzl G., Albert A., Wilke B.-M., Amelung W., et al. (2015). Drying and rewetting events change the response pattern of nitrifiers but not of denitrifiers to the application of manure containing antibiotic in soil. Appl. Soil Ecol. 95, 99–106. doi: 10.1016/j.apsoil.2015.06.016
Ribeiro K., Pacheco F. S., Ferreira J. W., de Sousa-Neto E. R., Hastie A., Krieger Filho G. C., et al. (2021). Tropical peatlands and their contribution to the global carbon cycle and climate change. Glob. Change Biol. 27 (3), 489–505. doi: 10.1111/gcb.15408
Rinke C., Schwientek P., Sczyrba A., Ivanova N. N., Anderson I. J., Cheng J. F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499 (7459), 431–437. doi: 10.1038/nature12352
Roth V. N., Lange M., Simon C., Hertkorn N., Bucher S., Goodall T., et al. (2019). Persistence of dissolved organic matter explained by molecular changes during its passage through soil. Nat. Geosci. 12 (9), 755. doi: 10.1038/s41561-019-0417-4
Sardans J., Penuelas J. (2010). Soil enzyme activity in a Mediterranean forest after six years of drought. Soil Sci. Soc Am. J. 74 (3), 838–851. doi: 10.2136/sssaj2009.0225
Schleper C. (2010). Ammonia oxidation: different niches for bacteria and archaea? ISME J. 4 (9), 1092–1094. doi: 10.1038/ismej.2010.111
Snider D., Thompson K., Wagner-Riddle C., Spoelstra J., Dunfield K. (2015). Molecular techniques and stable isotope ratios at natural abundance give complementary inferences about N2O production pathways in an agricultural soil following a rainfall event. Soil Biol. Biochem. 88, 197–213. doi: 10.1016/j.soilbio.2015.05.021
Tu Q. C., He Z. L., Wu L. Y., Xue K., Xie G., Chain P., et al. (2017). Metagenomic reconstruction of nitrogen cycling pathways in a CO2- enriched grassland ecosystem. Soil Biol. Biochem. 106, 99–108. doi: 10.1016/j.soilbio.2016.12.017
Vogel A., Eisenhauer N., Weigelt A., Scherer-Lorenzen M.. (2013). Plant diversity does not buffer drought effects on early-stage litter mass loss rates and microbial properties. Glob. Change Biol. 19 (9), 2795–2803. doi: 10.1111/gcb.12225
Wang M., Chen H., Wu N., Peng C., Zhu Q., Zhu D., et al. (2014). Carbon dynamics of peatlands in China during the Holocene. Quaternary Sci. Rev. 99, 34–41. doi: 10.1016/j.quascirev.2014.06.004
Wang J.-T., Egidi E., Li J., Singh B. K. (2019). Linking microbial diversity with ecosystem functioning through a trait framework. J. Biosci. 44 (5), 109. doi: 10.1007/s12038-019-9928-9
Wertz S., Degrange V., Prosser J. I., Poly F., Commeaux C., Guillaumaud N., et al. (2007). Decline of soil microbial diversity does not influence the resistance and resilience of key soil microbial functional groups following a model disturbance. Environ. Microbiol. 9 (9), 2211–2219. doi: 10.1111/j.1462-2920.2007.01335.x
Xie Z., Le Roux X., Wang C., Gu Z., An M., Nan H., et al. (2014). Identifying response groups of soil nitrifiers and denitrifiers to grazing and associated soil environmental drivers in Tibetan alpine meadows. Soil Biol. Biochem. 77, 89–99. doi: 10.1016/j.soilbio.2014.06.024
Yan Z., Kang E., Zhang K., Hao Y., Wang X., Li Y., et al. (2022). Asynchronous responses of microbial CAZymes genes and the net CO2 exchange in alpine peatland following 5 years of continuous extreme drought events. ISME Commun. 2, 115. doi: 10.1038/s43705-022-00200-w
Yang Y., Gao Y., Wang S., Xu D., Yu H., Wu L., et al. (2014). The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J. 8, 430–440. doi: 10.1038/ismej.2013.146
Yao Z., Yan G., Ma L., Wang Y., Zhang H., Zheng X., et al. (2022). Soil C/N ratio is the dominant control of annual N2O fluxes from organic soils of natural and semi-natural ecosystems. Agric. For. Meteorol. 327, 109198. doi: 10.1016/j.agrformet.2022.109198
Yuan M. M., Guo X., Wu L., Zhang Y., Xiao N., Ning D., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11 (4), 343–U100. doi: 10.1038/s41558-021-00989-9
Yue H., Wang M., Wang S., Gilbert J. A., Sun X., Wu L., et al. (2015). The microbe-mediated mechanisms affecting topsoil carbon stock in Tibetan grasslands. ISME J. 9 (9), 2012–2020. doi: 10.1038/ismej.2015.19
Zhang X., Zhao X., Zhang M. (2012). Functional diversity changes of microbial communities along a soil aquifer for reclaimed water recharge. FEMS Microbiol. Ecol. 80 (1), 9–18. doi: 10.1111/j.1574-6941.2011.01263.x
Zhou C., Biederman J. A., Zhang H., Li L., Cui X., Kuzyakov Y., et al. (2019). Extreme-duration drought impacts on soil CO2 efflux are regulated by plant species composition. Plant Soil 439 (1-2), 357–372. doi: 10.1007/s11104-019-04025-w
Zhou S., Huang C., Xiang Y., Tie L., Han B., Scheu S. (2018). Effects of reduced precipitation on litter decomposition in an evergreen broad-leaved forest in Western China. For. Ecol. Manage. 430, 219–227. doi: 10.1016/j.foreco.2018.08.022
Keywords: alpine peatland, extreme drought, carbon cycling, nitrogen cycling, functional genes
Citation: Yan Z, Li M, Hao Y, Li Y, Zhang X, Yan L, Kang E, Wang X, Yang A, Niu Y, Yu X, Kang X and Zhang K (2023) Effects of extreme drought on soil microbial functional genes involved in carbon and nitrogen cycling in alpine peatland. Front. Ecol. Evol. 11:1173750. doi: 10.3389/fevo.2023.1173750
Received: 25 February 2023; Accepted: 01 June 2023;
Published: 16 June 2023.
Edited by:
Cuicui Jiao, Sichuan University of Science and Engineering, ChinaReviewed by:
Hang Wang, Southwest Forestry University, ChinaCopyright © 2023 Yan, Li, Hao, Li, Zhang, Yan, Kang, Wang, Yang, Niu, Yu, Kang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoming Kang, eG1rYW5nQHVjYXMuYWMuY24=; Kerou Zhang, emhhbmdrZXJvdTE5OTFAbndhZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.