
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 12 May 2023
Sec. Chemical Ecology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1150532
This article is part of the Research Topic Chemosensory Receptor Systems of Invertebrates, From Expression to Function: Protein Targeting, Interactions with Ligands, Pharmacology, and Pest Management Strategies View all 7 articles
Insects perceive countless odors with the help of odorant receptors (ORs) expressed by olfactory sensory neurons (OSNs). OR complexes are composed of odor-specific proteins OrX and olfactory co-receptor proteins Orco, with both extracellular C-terminus and intracellular N-terminus. Here, we generated a genetically modified Drosophila melanogaster fly line that expresses Orco proteins with an N-terminally fused green fluorescent protein (GFP). We generated four genotypes (homozygous and heterozygous) for which we performed antennal cryosectioning and immunolabeling to determine which genotype would show high GFP labeling intensity in the dendritic areas of OSNs. We found that fluorescent intensity was the highest in homozygous flies. As Orco is known to guide OrX proteins to the OSN dendrites, we checked whether the fusion of GFP to the Orco N-terminus may affect such trafficking and thereby OR function. Using single sensillum recording (SSR), we found no difference in the olfactory capability of homozygous flies compared with wild-type flies. As future application, we want to use this transgenic fly line to resolve the structure of OR complexes using Cryo electron microscopy (cryo-EM).
The fruit fly, Drosophila melanogaster, is one of the most popular model organisms used in scientific and medical research (Stephenson and Metcalfe, 2013). Unique features, such as short life cycle, rapid generation time, cost-effectiveness, and an excellent genetic toolbox, have contributed to this status (Yamaguchi and Yoshida, 2018). These features have also made D. melanogaster a key contributor in studies of molecular mechanisms of olfaction. The Drosophila olfactory organs are the antenna and the maxillary palps (Keil, 1999; Shanbhag et al., 1999). The antenna is divided into three segments: scape, pedicel, and funiculus. The whole surface of the funiculus is packed with chemosensory hairs called sensilla. Depending on the structural features, sensilla are commonly classified into four types: basiconic, trichoid, intermediate, and coeloconic (Shanbhag et al., 1999). Each sensillum contains the dendrites of one-to-four olfactory sensory neurons (OSNs) that detect volatile chemicals from the surrounding environment. The OSNs are thus the first link in the olfactory pathway guiding an insect to forage, mate, escape danger, reproduce, and survive.
The OSNs express various classes of chemosensory receptors: ionotropic receptors (IRs), gustatory receptors (GRs), and odorant receptors (ORs) in their dendrites (Wicher and Miazzi, 2021). Here, we focus on ORs. Insect OR proteins comprise seven transmembrane domains with an intracellular N-terminus and an extracellular C-terminus domain and thus display an inverted topology as compared to vertebrate ORs (Benton et al., 2006; Lundin et al., 2007). Insect ORs are heteromers composed of a highly conserved co-receptor protein (Orco) and a specific tuning receptor protein OrX. ORs form ligand-gated non-selective cation channels that allow the flow of cations, including Ca2+ into the OSN (Larsson et al., 2004; Neuhaus et al., 2005; Sato et al., 2008; Wicher et al., 2008; Vosshall and Hansson, 2011). Orco proteins form homomers in the absence of an OrX subunit and operate as ion channels (Neuhaus et al., 2005; Mukunda et al., 2014). The co-receptor protein also serves as a chaperone to transport OR proteins to the ciliary OSN dendrites (Benton et al., 2006). In two recent studies, the molecular structure of Orco from the wasp Apocrypta bakeri (Butterwick et al., 2018) and OrX constructs from the jumping bristletail M. hrabei (del Mármol et al., 2021) have been elucidated. In both cases, the channels form homo tetramers with a central ion-conducting pore.
With the establishment of cultured cell lines and the Drosophila empty neuron expression system, it has been possible to characterize ORs according to their ligands (Sato et al., 2008; Wicher et al., 2008; Wang et al., 2010; German et al., 2013). Likewise, one of the genetic toolboxes, the GAL4-UAS binary expression system, has been extensively used in studies of Drosophila olfaction to address a wide range of biological questions (Brand and Perrimon, 1993; Busson and Pret, 2007). The GAL4-UAS system allows the expression of genetically labeled fluorescent markers in a specific neuron of interest. Typically, fluorescent markers can be green fluorescent protein (GFP), genetically encoded calcium indicators (GECI), FRET sensors (Förster resonance energy transfer), or several other types (Mank and Griesbeck, 2008; Miazzi et al., 2016). For example, in the open antenna preparation, GECIs, like GCaMP, report changes in the intracellular cellular calcium concentration of OSNs or supporting cells to odor stimulation (Mukunda et al., 2014; Halty-deLeon et al., 2018; Jain et al., 2021; Miazzi et al., 2022; Prelic et al., 2022; Wiesel et al., 2022). The development of these sophisticated systems has made it possible to visualize specific neurons of interest and to perform Ca2+ imaging, patch-clamp recordings (Sato et al., 2008; Wicher et al., 2008), and single sensillum recordings (SSR) (Hallem et al., 2004; Pellegrino et al., 2010) from these.
In the present study, we developed a genetically manipulated D. melanogaster fly line that expresses GFP fused to the Orco N-terminus in Orco null mutant background (orco−/−) using the GAL4-UAS system. The approach was based on our previous study performed with heterologously expressed OR proteins (Wicher et al., 2008). There, we used N-terminal GFP fusion proteins (Orco or Or22a) to demonstrate OR localization in the plasma membrane of HEK293 cells. In the present approach, we produced a D. melanogaster fly that expresses GFP-fused protein at the Orco N-terminus to visualize membrane-tagged OR proteins. We produced four genotypes (see Table 1) and investigated their expression pattern. Finally, we checked whether the olfactory capability of N-GFP-Orco fly was altered due to the GFP insertion.
To generate the UAS –N-GFP -Orco fly, GFP was ligated to the N-terminus of Orco. The genetically encoded green fluorescent protein (GFP) prepared from the plasmid PBac-3XP3-EGFP-hsp-Gal4-sv40-attB was kindly provided by B. Prud’homme (Institut de Biologie du Développment de Marseille, France). A 717-bp GFP fragment without the stop codon was amplified via PCR using the following primer set: GFP-Forward: 5’ATGGTGAGCAAGGGCGAGGAG 3′; GFP-Reverse 5’AGGTTGTCATCTTGTACAGCTCGTCCATGC 3′. Total RNA from D. melanogaster antennae was prepared and cDNA synthesized to PCR-amplify Orco using the gene-specific primers: Orco-Forward: 5’ATGACAACCTCGATGCAGCCGA 3′; Orco-Reverse: 5’ GCTGTACAAGATGACAACCTCGATGCAG 3′. The resulting two constructs of the PCR reactions (GFP 717 bp and Orco 1,461 bp) were sequenced and purified. Subsequently, a third PCR was performed to fuse GFP at the N-terminus of Orco using the primers GFP-forward and Orco-reverse and the two constructs of GFP and Orco as a template. The resultant construct N-GFP-Orco of 2,178 bp was T:A cloned into pCR™8/GW/TOPO (Thermo Fisher Scientific, Schwerte, Germany). Sanger sequencing confirmed all sequences (Eurofins Genomics Germany GmbH, Ebersberg, Germany, and the Department of Insects Symbiosis, Max Planck Institute for Chemical Ecology, Jena, Germany). The transfer of the construct to the destination vector pUASg.attB (kindly provided by J. Bischof, FlyORF Injection, Zürich, Switzerland) was performed as described in Gonzalez et al. (2016). Next, the D. melanogaster germline transformation with the prepared plasmid was carried out by FlyORF Injection (Zürich, Switzerland) using the PhiC31 integration system. The attP40 target landing site was used. FlyORF used the Bloomington fly line number 25709. The vector was inserted into chromosome II to produce the genotype +; UAS-N-GFP-Orco/Cyo; +. Transgenic D. melanogaster flies were bred on conventional cornmeal agar medium under a 12-h light and 12-h dark cycle at 25°C and 70% humidity. To generate the experimental fly line: yw; UAS-N-GFP-Orco/Cyo; Orco-Gal4, orco1/TM2, following parental lines were used: (+; UAS-N-GFP-Orco/Cyo; +), (yw; BL/Cyo; TM2/TM6b), and (yw/w[1118]; UAS-Orco; Orco-Gal4,orco1). The parental lines: (yw; BL/Cyo; TM2/TM6b) was obtained from Bloomington Stock Center (no. BL3704), and (yw/w[1118]; UAS-Orco; Orco-Gal4,orco1) was created in our laboratory using the orco1 (Larsson et al., 2004). The experimental fly line (yw; UAS-N-GFP-Orco/Cyo; Orco-Gal4,orco1/TM2) expresses N-GFP-Orco under the regulation of Orco in an Orco knockout background. All the experimental fly lines used in the experiments had no endogenous Orco.
We used 4- to 6-day-old female flies to perform immunofluorescence experiments from the genotypes given below.
A total of antennae were prepared from fly heads and mounted in OCT (optimal cutting temperature) mounting media (Lot: 03816567; VWR Chemicals, Leuven, Belgium). Microm HM560 was used to slice thin sections of antennae (14 μm) on Fisher SuperFrost Plus slides. In the next step, sections were fixed with 4% paraformaldehyde (PFA) for 10 min (1 mL per slide) in a humidified chamber. Then, the slides were washed with phosphate-buffered saline (PBS) two times 10 min each (1 mL per slide) on a shaker at room temperature. After washing, samples were blocked in the blocking solution (PBS+ 5% NGS (normal goat serum)) for 30 min and covered with the coverslip (#1). Blocking solution was removed after 30 min, and then, 100 μL of freshly prepared primary antibody anti-Orco (kindly provided by Leslie. Vosshall) (1:1000) was added to each slide. Slides were covered with the coverslip (#1) and were incubated overnight at 4°C. The next day, slides were washed with PBS four times 10 min each and blocked for 30 min. Later, 100 μL of secondary fluorescent antibody and goat anti-rabbit Alexa 546 of 1:250 was added to each slide and incubated for 2 h at room temperature in dark conditions. After the incubation time, the samples were washed 4 times 5 min each in PBS in dark on a shaker. Finally, the samples were mounted in Vectashield, covered with coverslips (#0), and stored for further use at 4°C.
Parental control: (+; UAS-N-GFP-Orco; +) was also used to test the presence or the absence of GFP or Orco in our antennal cross-sections (Supplementary Figure S1). We performed double immune labeling for these experiments. Primary antibodies: anti-Orco (kindly provided by Leslie. Vosshall) (1:1000) and Anti-GFP () (1:500) and secondary fluorescent antibody: goat anti-rabbit Alexa 568 of 1:250 and goat anti-mouse Alexa 488 of 1:500 were used.
Zeiss cLSM880 confocal microscope (Carl Zeiss, Oberkochen, Germany) equipped with a 10, 20, or 40×/1.20 water immersion objective (C-Apochromat, Carl Zeiss) was used to image the antennal cross-sections. Samples were excited with 488 nm and 543 nm wavelengths. Laser and detector gain for two channels were optimized to avoid saturation, and all the images were acquired with the same confocal settings for contrast and brightness. Z- Stack images were captured at 1024 × 1024-pixel resolution at 0.12 μm intervals for the antennal cross-sections. All images used for the figure representation were of 8 bits.
Fiji ImageJ software (Schindelin et al., 2012) was used to create some slice projections of antenna cross-section z-stacks. After generating projections, region of interests (somata and sensilla) were marked from each cross-section of N-GFP-Orco flies, and then, the fluorescence intensity was measured. The mean fluorescence intensity of soma or sensilla was corrected for the background noise and given as F in arbitrary units (a.u.). Statistical analysis was performed using the Prism 9.4.0 software (GraphPad Software Inc., La Jolla, CA, United States), and Kruskal–Wallis test was performed. Data are presented as mean ± SEM (standard error of the mean). 26 ≤ n ≤ 31 soma and 29 ≤ n ≤ 40 sensilla were selected from 6 ≤ n ≤ 11 antennal cross-sections from all the genotypes.
For N-GFP Orco expression analysis, the plot profile method was used from the Fiji ImageJ software (Schindelin et al., 2012). Three sensilla that were adjacent to each other were marked for both anti-Orco and native GFP signals from each N-GFP-Orco homozygous cross-section and also from the parental line (Supplementary Figure S2). Once the fluorescent intensity values were obtained, they were normalized and plotted on Prism 9.4.0 software. Parametric, unpaired two-tailed t-test was performed. n = 15 sensilla for five cross-sections.
Single sensillum recording (SSR) was performed on 4- to 8-day-old adult female Canton-S and homozygous N-GFP-Orco flies as described in Pellegrino et al. (2010). A single female fly was inserted into the narrow end of 200-μL pipette tip with the help of a fly aspirator and was then immediately placed onto the glass slide with the help of dental wax. The antenna was raised upward and positioned on a coverslip. The tip of a glass capillary was used to stabilize the position of the antenna in the dorsomedial direction (Clyne et al., 1997; Lin and Potter, 2015) This orientation allows one to observe large basiconic sensilla (ab3 sensilla type) present on the third antennal segment. Ab3 sensilla house the ab3A neuron that expresses the Or22a receptor and the ab3B neuron expressing Or85b. Ab3 sensilla were visualized using a compound microscope (Olympus BX51W1) with 1,000x magnification. Meanwhile, the recording and ground electrode, made of tungsten wire, were electrochemically sharpened using 3 M KOH. Neurons action potential was recorded by inserting the ground electrode into the eye and recording electrode into the base of ab3 sensilla using a micromanipulator system (Luigs and Neumann SM-10). AC signals were amplified (Syntech Universal AC/DC Probe), sampled (96,000samples/s) and filtered (3 kHz high-300 Hz low, 50/60 Hz suppression), and transferred to computer using a USB-IDAC system. Neuronal activity was recorded using AutoSpike software (v3.7) for 10s, starting 1 s before stimulation, and action potentials were counted 1 s period prior to the stimulation and for 1 s after the onset of the stimulation.
Odor was freshly prepared by pipetting 10 μL of ethyl hexanoate and 2-heptanone, dissolved in hexane (solvent), and then added onto the filter paper (diameter 10 mm). Ethyl hexanoate (99% purity) was obtained from Sigma-Aldrich (Steinheim, Germany). 2-Heptanone (≥98%) was obtained from Sigma-Aldrich (Steinheim, Germany). N-hexane (≥95%) was obtained from Carl Roth GmbH (Karlsruhe, Germany). The filter paper was then inserted into a glass Pasteur pipette and kept for 30 min for equilibrating the pipette headspace. The glass Pasteur pipette was attached to a stimulus controller (Stimulus Controller CS-55, Syntech) to puff odor for 500 ms to a prehumidified air, which was continuously delivered on the fly at a rate of 0.6 LPM using a flowmeter (Cole-Parmer, www.coleparmer.com). Only ab3 sensilla were recorded for each replicate from the Canton-S and N-GFP-Orco homozygous flies. Ab3A neurons show spikes of higher and ab3B with lower amplitude.
SSR traces were analyzed using AutoSpike32 software 3.7 version (Syntech NL 1998). The differences in spike count (action potential) were calculated by subtracting the number of spikes (spontaneous activity) one second prior to the stimulus from those after the onset of stimulus. Changes in spikes patterns were then compared using a two-tailed unpaired, non-parametric t-test with Mann–Whitney comparison, n = 5 flies. The entire statistical test was performed in GraphPad Prism 9.4.0.
We generated a transgenic D. melanogaster fly line that expresses a modified Orco protein in which a GFP is coupled to its N-terminus region. The main reason to establish this fly line was to provide a permanent marker for OR-expressing OSNs directly linked to an OR protein. Using gene transformation and the GAL4-UAS system, we produced four genotypes upon crossing male and female flies (Homo, Hetero1, Hetero2, and Hetero3) and checked which genotype would show a high level of GFP expression in the sensillum type housing the dendrite. We performed cryo sectioning and immunohistochemistry for all genotypes to check the coincidence of GFP- and anti-Orco fluorescence as well as the differences in GFP expression. Representative results for antennal images are given in Figures 1A–L; they show that GFP fluorescence is co-localized with anti-Orco staining. An analysis of GFP fluorescence intensity is presented for somata in Figure 1M and sensilla in Figure 1N. The GFP signal in homozygous flies is stronger in both soma (Figure 1M) and sensilla (Figure 1N) compared with the heterozygous genotypes (except for similar expression in somata of Hetero3 flies) (Figure 1M). We also observed differences in the N-GFP-Orco expression (Figure 2) pattern in the somata among heterozygous phenotypes (Hetero1, Hetero2, and Hetero3) (Figure 1M), which could be due to the number of copies of GAL4 and UAS expressed in these phenotypes. Given the high GFP expression in the sensilla of homozygous flies (Figure 1N), we concluded that this genotype was the optimal candidate for further experiments.
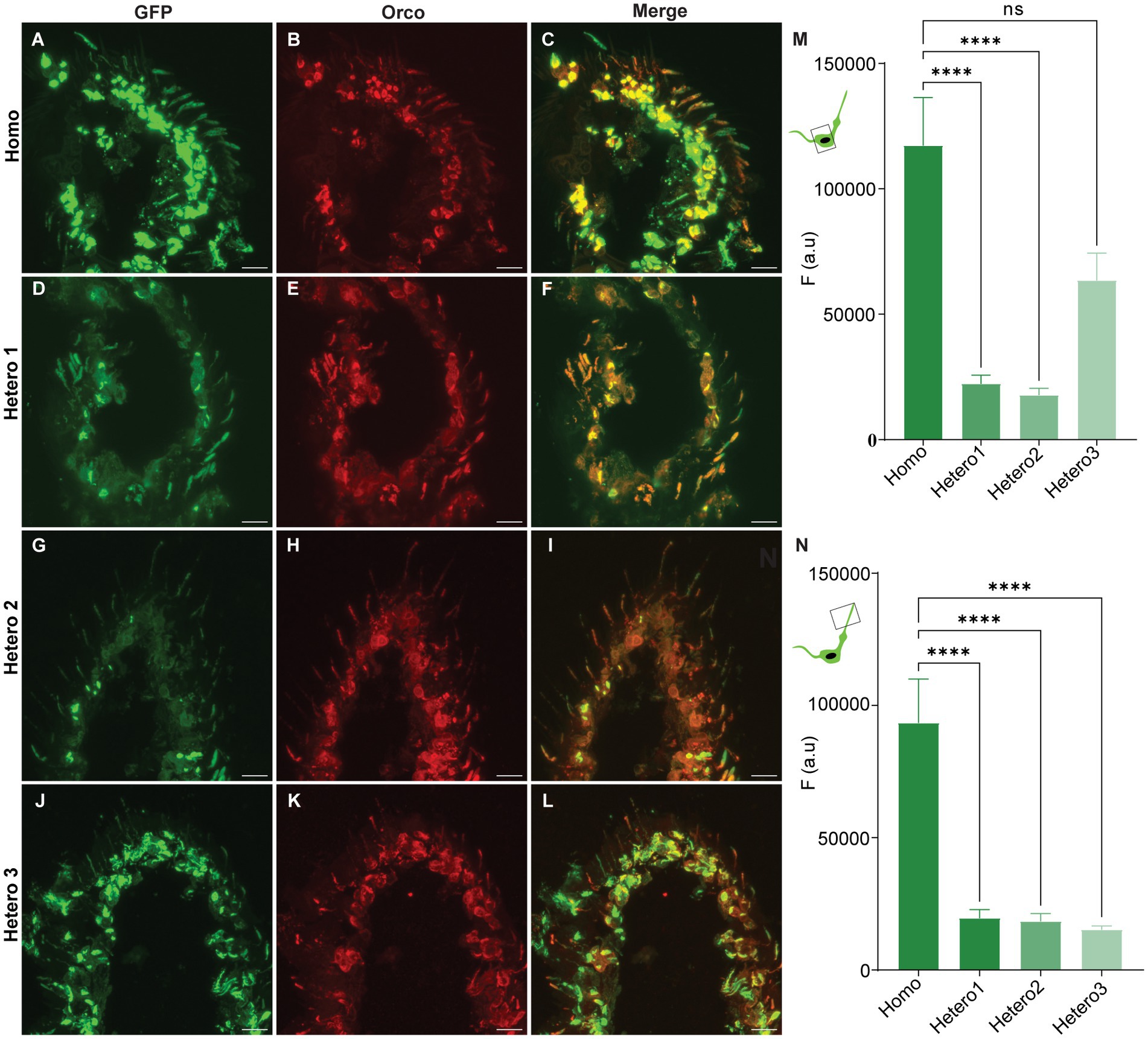
Figure 1. Confocal images of N-GFP-Orco expression pattern in antennal sections from the genotypes (Homo, Hetero1, Hetero2, and Hetero3) and their mean fluorescent intensity values. (A,D,G,J) GFP fluorescence, (B,E,H,K) anti-Orco fluorescence, (C,F,I,L) merged GFP and anti-Orco signal in individual genotypes. Mean fluorescence intensity responses (F (a.u.)) from the N-GFP-Orco signal in soma (M) and sensilla (N) of Homo, Hetero1, Hetero2, and Hetero3 flies. Kruskal–Wallis test (***p < 0.0001, ns = not significant) 6 ≤ n ≤ 14. Scale bar: 11 μm.
We investigated the co-localization of GFP and anti-Orco signals in detail for sensilla of homozygous flies (Figure 2). Representative images of three sensilla are shown for GFP fluorescence in Figure 2A, anti-Orco fluorescence in Figure 2B, and merged signals in Figure 2C. For quantitative comparison, we normalized GFP and anti-Orco fluorescence intensities as described in Materials and Methods section. Plots for the example shown in Figures 2A–C are given in Figures 2D–F. The maxima of normalized GFP and anti-Orco fluorescence intensities were not significantly different (Figure 2G), which confirmed the co-localization of GFP and Orco protein in the OSN dendrites.
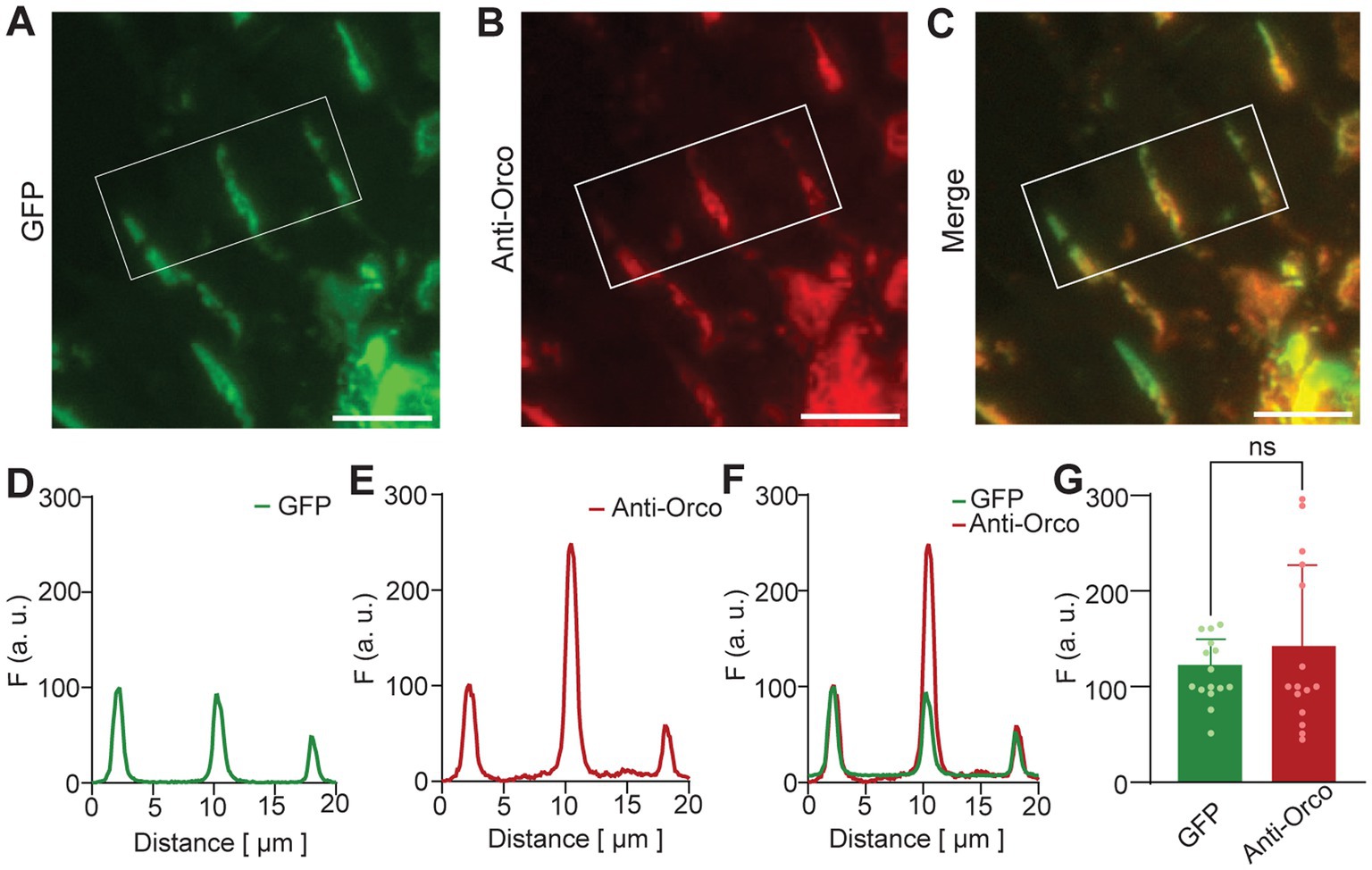
Figure 2. N-GFP-Orco expression in OSN sensilla. Confocal images of antennal sections from Homo genotype. (A) GFP fluorescence, (B) anti-Orco fluorescence, (C) merged GFP and anti-Orco signal. Sensilla in the white box were taken for analysis as shown in D-F. (D–F) Sensilla fluorescence intensity distribution, (D) GFP fluorescence, (E) anti-Orco fluorescence, (F) merged GFP and anti-Orco signals, (G) normalized maxima of GFP and anti-Orco fluorescence from 15 sensilla. Unpaired two-tailed t-test, ns: not significant, scale bar: 9 μm.
Finally, we asked whether the function of tagged OSNs in the N-GFP-Orco homozygous fly might be affected due to the presence of GFP at the Orco N-terminus. To check this, we performed single sensillum recordings (SSRs) (Figure 3A), which allow extracellular monitoring of OSN action potentials (Clyne et al., 1997; Hallem et al., 2004; Hallem and Carlson, 2006; Pellegrino et al., 2010). We targeted ab3 sensilla as they are easily accessible and house two well-characterized OSNs, namely ab3A and ab3B (Dobritsa et al., 2003). The ab3A OSN expresses the broadly tuned receptor protein Or22a and is activated by fruit odors such as ethyl hexanoate and ethyl butyrate (Dobritsa et al., 2003). The ab3B OSN expresses Or85b and is activated by 2-heptanone (Dobritsa et al., 2003). We performed our experiments by inserting a recording electrode into the base of ab3 sensilla and stimulated with ethyl hexanoate and 2-heptanone at 10−4 dilution for 500 ms. We found no difference in the number of action potentials (spikes) elicited between control Canton-S flies (Figure 3B) and N-GFP-Orco homozygous flies (Figure 3C). This was supported by analyzing the changes in action potential spike count after odor stimulation in both neurons, ab3A (Or22a) and ab3B (Or85b) in Canton-S and N-GFP-Orco flies (Figures 3D–E). These results showed that OSN function remained unaffected in the genetically modified fly.
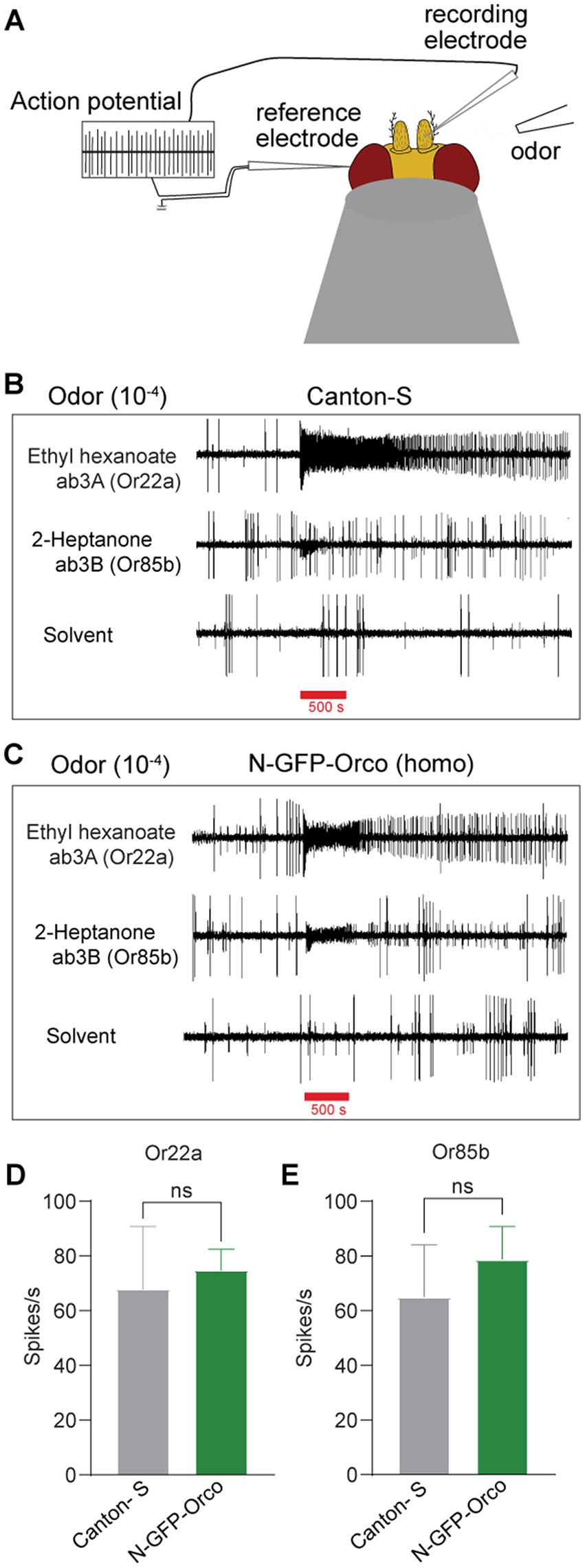
Figure 3. Olfactory performance of the N-GFP-Orco Homo flies. (A) Schematic diagram of SSR principle. The recording electrode is inserted into the base of sensillum, the reference electrode is placed into the eye, and the odor is presented via a tube. Ab3 sensilla have two neurons, the ab3A neuron which expresses the Or22a receptor that is activated by ethyl hexanoate and the ab3B neuron expressing Or85b that is activated by 2-heptanone. (B,C) SSR traces from ab3A (Or22a) and ab3B (Or85b) with its diagnostic odorants and hexane as solvent control in both Canton-S (B) and N-GFP-Orco Homo flies (C) at 10−4 odor dilution with an odor delivery of 500 ms. (D,E) Spike count in ab3A (D) and ab3B sensilla (E). Spike count was calculated by subtracting the number of spikes 1 s before the odor presentation and then after 1 s after the onset of the stimulus in both Canton-S and N-GFP-Orco Homo flies. Two-tailed unpaired, non-parametric t-tests with Mann–Whitney comparison test, ns = not significant, n = 5 antenna.
Labeling olfactory receptors may help in investigating the details of the sensory machinery such as the distribution of specific receptors in OSN dendrites. To label ORs in general, the ubiquitary co-receptor Orco (for recent reviews, (see Stengl and Funk, 2013; Fleischer et al., 2018; Wicher and Miazzi, 2021)) is a good candidate to be tagged. For D. melanogaster Orco, an N-terminal GFP-coupled modification was seen to be incorporated in the plasma membrane in heterologous cells (Wicher et al., 2008). We thus generated a transgenic D. melanogaster fly line that expressed GFP at the N-terminal region of Orco as a new method to genetically label OR-expressing OSNs.
We produced four genotypes of N-GFP-Orco fly lines (Homo, Hetero1, Hetero2, and Hetero3) and investigated the expression pattern, thereby observing pronounced differences among these genotypes. For example, in Hetero1 (Figures 1D–F,M) and Hetero2 (Figures 1G–I,M) flies, we observed only weak GFP signals in the somata. This might be due to the presence of only one copy of the N-GFP-Orco gene in the second chromosome and one copy of the Orco-Gal4, orco1 gene in the third chromosome. By contrast, there was no significant difference in fluorescence intensity between Homo and Hetero3 fly somata (Figures 1J–L,M), which might be due to the presence of two copies of the N-GFP-Orco genes in the second chromosome and only one copy of the Orco-Gal4, orco1 gene in the third chromosome. Most importantly, we observed a much weaker fluorescence intensity in the sensilla of all Hetero genotypes than in the Homo genotype (Figure 1N). The differences in GFP fluorescence in both somata and dendrites could be due to the presence of balancer chromosomes. These are used to maintain deleterious mutations and prevent crossovers in the flies (Miller et al., 2019), but we do not know whether the balancer chromosome and N-GFP-Orco are interacting in a different way and may affect the gene expression of N-GFP-Orco in heterozygous flies. With the given good GFP expression in the sensilla of Homo flies, we therefore used only homozygous flies in our experiments.
A prerequisite for labeling ORs is a functional membrane insertion of Orco and OrX proteins in the dendritic membrane. In the soma membrane, there is a high probability to find homomeric Orco complexes (Benton et al., 2006). Furthermore and most pronounced in Figure 1A, a major portion of N-GFP-Orco may get stuck in the cytoplasm. Inspection of GFP- and anti-Orco fluorescence signals in the sensilla (Figure 2) confirmed the co-localization of GFP and Orco proteins. Orco plays an important role in guiding OrX proteins to the dendritic membrane (Larsson et al., 2004). Flies lacking Orco or bearing mutations in Orco show impaired OrX transport, which leads to behavioral deficits (Larsson et al., 2004; Benton et al., 2006; Getahun et al., 2016; Jain et al., 2021). We thus tested whether the Orco modification might have affected OSN olfactory functionality in N-GFP-Orco Homo flies. Recording the neuronal activity with SSR measurements did not reveal any significant differences in spike counts between N-GFP-Orco Homo and Canton-S OSNs (Figures 3D,E). The tested neurons are thus functional and respond to odors, unlike in the case of Orco mutant flies, where the OSN activity is completely abolished (Larsson et al., 2004). Our results add to a previous study demonstrating that membrane-targeted GFP did not affect OR function (Lin and Potter, 2015).
Taken together, our study shows that GFP insertion at the N-terminal region of Orco does not affect the localization of OR proteins in the dendritic OSN membrane and that physiological olfactory functions are maintained after this manipulation. A possible application of tagging OR proteins to the OSN membrane is the identification of ORs with cryo-CLEM (correlative light electron microscopy) methods as a preparation to resolve the structure of OR complexes using cryo-EM (electron microscopy). This technique can also be applied to express N-GFP-Orco constructs in a specific population of neurons to determine the structural arrangements of OrX/Orco complexes in dendritic membranes using high-resolution fluorescence microscopy, such as dSTORM (direct stochastic optical reconstruction microscopy) (Heilemann et al., 2008). We believe that the method presented here can unravel many unanswered questions in the field of insect olfaction by increasing our understanding regarding how receptors are composed and arranged.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
This study was performed on the fruit fly, Drosophila melanogaster, in Germany, where research on invertebrates does not require a permit from a committee that approves animal research.
DW formulated the ideas and edited the manuscript for this study. KJ, RS, DW, and BH designed the study. RS designed and generated N-GFP-Orco D. melanogaster flies for the studies. SK helped with immunohistochemistry experiments. KJ performed immunohistochemistry and SSR experiments, analyzed the data and wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
The study was funded by the Max Planck Society (KJ, RS, SK, BH, DW).
The authors thank Ibrahim Alali, Silke Trautheim, Megha Treesa Tom, and Veit Grabe for their technical support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1150532/full#supplementary-material
Benton, R., Sachse, S., Michnick, S. W., and Vosshall, L. B. (2006). Atypical membrane topology and heteromeric function of drosophila odorant receptors in vivo. PLoS Biol. 4:e20. doi: 10.1371/journal.pbio.0040020
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. doi: 10.1242/dev.118.2.401
Busson, D., and Pret, A.-M. (2007). “GAL4/UAS targeted gene expression for studying drosophila hedgehog signaling” in Hedgehog Signaling Protocols. ed. J. I. Horabin (Totowa, NJ: Humana Press), 161–201.
Butterwick, J. A., del Mármol, J., Kim, K. H., Kahlson, M. A., Rogow, J. A., Walz, T., et al. (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452. doi: 10.1038/s41586-018-0420-8
Clyne, P., Grant, A., O'Connell, R., and Carlson, J. R. (1997). Odorant response of individual sensilla on the drosophila antenna. Invertebr. Neurosci. 3, 127–135. doi: 10.1007/BF02480367
del Mármol, J., Yedlin, M. A., and Ruta, V. (2021). The structural basis of odorant recognition in insect olfactory receptors. Nature 597, 126–131. doi: 10.1038/s41586-021-03794-8
Dobritsa, A. A., Van Der Goes van Naters, W., Warr, C. G., Steinbrecht, R. A., and Carlson, J. R. (2003). Integrating the molecular and cellular basis of odor coding in the drosophila antenna. Neuron 37, 827–841. doi: 10.1016/S0896-6273(03)00094-1
Fleischer, J., Pregitzer, P., Breer, H., and Krieger, J. (2018). Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell. Mol. Life Sci. 75, 485–508. doi: 10.1007/s00018-017-2627-5
German, P. F., van der Poel, S., Carraher, C., Kralicek, A. V., and Newcomb, R. D. (2013). Insights into subunit interactions within the insect olfactory receptor complex using FRET. Insect Biochem. Mol. Biol. 43, 138–145. doi: 10.1016/j.ibmb.2012.11.002
Getahun, M. N., Thoma, M., Lavista-Llanos, S., Keesey, I., Fandino, R. A., Knaden, M., et al. (2016). Intracellular regulation of the insect chemoreceptor complex impacts odour localization in flying insects. J. Exp. Biol. 219, 3428–3438. doi: 10.1242/jeb.143396
Gonzalez, F., Witzgall, P., and Walker, W. B. (2016). Protocol for heterologous expression of insect odourant receptors in drosophila. Front. Ecol. Evol. 4:24. doi: 10.3389/fevo.2016.00024
Hallem, E., Hallem EA, C. J., and Carlson, J. R. (2006). Coding of odors by a receptor repertoire. Cell 125, 143–160. doi: 10.1016/j.cell.2006.01.050
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the drosophila antenna. Cells 117, 965–979. doi: 10.1016/j.cell.2004.05.012
Hallem, E., Ho, M., Hallem EA, C. J., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the drosophila antenna. Cell 117, 965–979. doi: 10.1016/j.cell.2004.05.012
Halty-deLeon, L., Hansson, B. S., and Wicher, D. (2018). The Drosophila melanogaster Na+/Ca2+ exchanger CALX controls the Ca2+ level in olfactory sensory meurons at rest and after odorant receptor activation. Front. Cell. Neurosci. 12:186. doi: 10.3389/fncel.2018.00186
Heilemann, M., Van De Linde, S., Schüttpelz, M., Kasper, R., Seefeldt, B., Mukherjee, A., et al. (2008). Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. 47, 6172–6176. doi: 10.1002/anie.200802376
Jain, K., Lavista-Llanos, S., Grabe, V., Hansson, B. S., and Wicher, D. (2021). Calmodulin regulates the olfactory performance in Drosophila melanogaster. Sci. Rep. 11:3747. doi: 10.1038/s41598-021-83296-9
Keil, T. A. (1999). “Morphology and development of the peripheral olfactory organs” in Insect Olfaction. ed. B. S. Hansson (Berlin, Heidelberg: Springer Berlin Heidelberg), 5–47.
Larsson, M. C., Domingos, A. I., Jones, W. D., Chiappe, M. E., Amrein, H., and Vosshall, L. B. (2004). Or83b encodes a broadly expressedodorant receptor essential for drosophila Oolfaction. Neuron 43, 703–714. doi: 10.1016/j.neuron.2004.08.019
Lin, C. C., and Potter, C. J. (2015). Re-classification of Drosophila melanogaster trichoid and intermediate sensilla using fluorescence-guided single sensillum recording. PLoS One 10:e0139675. doi: 10.1371/journal.pone.0139675
Lundin, C., Käll, L., Kreher, S. A., Kapp, K., Sonnhammer, E. L., Carlson, J. R., et al. (2007). Membrane topology of the drosophila OR83b odorant receptor. FEBS Lett. 581, 5601–5604. doi: 10.1016/j.febslet.2007.11.007
Mank, M., and Griesbeck, O. (2008). Genetically encoded calcium indicators. Chem. Rev 108, 1550–1564. doi: 10.1002/chin.200833268
Miazzi, F., Hansson, B. S., and Wicher, D. (2016). Odor-induced cAMP production in Drosophila melanogaster olfactory sensory neurons. J. Exp. Biol. 219, 1798–1803. doi: 10.1242/jeb.137901
Miazzi, F., Jain, K., Kaltofen, S., Bello, J. E., Hansson, B. S., and Wicher, D. (2022). Targeting insect olfaction in vivo and in vitro using functional imaging. Front. Cell. Neurosci. 16:839811. doi: 10.3389/fncel.2022.839811
Miller, D. E., Cook, K. R., and Hawley, R. S. (2019). The joy of balancers. PLOS. Genetics 15:e1008421. doi: 10.1371/journal.pgen.1008421
Mukunda, L., Lavista-Llanos, S., Hansson, B. S., and Wicher, D. (2014). Dimerisation of the drosophila odorant coreceptor Orco. Front. Cell. Neurosci. 8:261. doi: 10.3389/fncel.2014.00261
Mukunda, L., Miazzi, F., Kaltofen, S., Hansson, B. S., and Wicher, D. (2014). Calmodulin modulates insect odorant receptor function. Cell Calcium 55, 191–199. doi: 10.1016/j.ceca.2014.02.013
Neuhaus, E. M., Gisselmann, G., Zhang, W., Dooley, R., Störtkuhl, K., and Hatt, H. (2005). Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat. Neurosci. 8, 15–17. doi: 10.1038/nn1371
Pellegrino, M., Nakagawa, T., and Vosshall, L. B. (2010). Single sensillum recordings in the insects Drosophila melanogaster and Anopheles gambiae. J. Vis. Exp. 36, 1–5. doi: 10.3791/1725-v
Prelic, S., Pal Mahadevan, V., Venkateswaran, V., Lavista-Llanos, S., Hansson, B. S., and Wicher, D. (2022). Functional interaction between drosophila olfactory sensory neurons and their support cells. Front. Cell. Neurosci. 15:789086. doi: 10.3389/fncel.2021.789086
Sato, K., Pellegrino, M., Nakagawa, T., Nakagawa, T., Vosshall, L. B., and Touhara, K. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi: 10.1038/nature06850
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Shanbhag, S., Müller, B., and Steinbrecht, R. A. (1999). Atlas of olfactory organs of Drosophila melanogaster 1. Types, external organization, innervation and distribution of olfactory sensilla. Int. J. Insect Morphol. Embryol. 28, 377–397. doi: 10.1016/S0020-7322(99)00039-2
Stengl, M., and Funk, N. W. (2013). The role of the coreceptor Orco in insect olfactory transduction. J. Comp. Physiol. A 199, 897–909. doi: 10.1007/s00359-013-0837-3
Stephenson, R., and Metcalfe, N. H. (2013). Drosophila melanogaster: a fly through its history and current use. J. R. Coll. Physicians Edinb. 43, 70–75. doi: 10.4997/JRCPE.2013.116
Vosshall, L. B., and Hansson, B. S. (2011). A unified nomenclature system for the insect olfactory coreceptor. Chem. Senses 36, 497–498. doi: 10.1093/chemse/bjr022
Wang, G., Carey, A. F., Carlson, J. R., and Zwiebel, L. J. (2010). Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. 107, 4418–4423. doi: 10.1073/pnas.0913392107
Wicher, D., and Miazzi, F. (2021). Functional properties of insect olfactory receptors: ionotropic receptors and odorant receptors. Cell Tissue Res. 383, 7–19. doi: 10.1007/s00441-020-03363-x
Wicher, D., Schäfer, R., Bauernfeind, R., Stensmyr, M. C., Heller, R., Heinemann, S. H., et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011. doi: 10.1038/nature06861
Wiesel, E., Kaltofen, S., Hansson, B. S., and Wicher, D. (2022). Homeostasis of mitochondrial Ca2+ stores is critical for signal amplification in Drosophila melanogaster olfactory sensory neurons. Insects 13:270. doi: 10.3390/insects13030270
Keywords: insect olfaction, olfactory sensory neuron, antenna, immunohistochemistry, single sensillum recording, Drosophila melanogaster
Citation: Jain K, Stieber R, Kaltofen S, Hansson BS and Wicher D (2023) A new Drosophila melanogaster fly that expresses GFP-tagged Orco. Front. Ecol. Evol. 11:1150532. doi: 10.3389/fevo.2023.1150532
Received: 24 January 2023; Accepted: 31 March 2023;
Published: 12 May 2023.
Edited by:
William Benjamin Walker III, Temperate Tree Fruit and Vegetable Research Unit, Agricultural Research Service (USDA), United StatesReviewed by:
Christopher John Potter, Johns Hopkins University, United States Greg Pask, Middlebury College, United StatesCopyright © 2023 Jain, Stieber, Kaltofen, Hansson and Wicher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dieter Wicher, ZHdpY2hlckBpY2UubXBnLmRl
†These authors shared supervision
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.