
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 30 March 2023
Sec. Evolutionary and Population Genetics
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1146561
Extreme climate change may lead to a decline in biodiversity and species extinction. And it also results in different population structure and genetic diversity of sheep. Therefore, studying the effects of extreme climate change on the distribution pattern of sheep is of great value. We used Illumina Ovine SNP 50K BeadChip to study the sheep around in southern Xinjiang of China, living around the Taklimakan Desert. Among them, 6 sheep breeds were from Xinjiang, China, and 3 sheep breeds were from other countries (Indian, Iran and Bangladeshi). We examined their genetic diversity and population structure and Genes related to year-round estrous in sheep were obtained by selective sweep. The principal component analysis, Admixture and Neighbor-Joining tree showed that a distinct pattern of population structure was observed in 9 sheep breeds. The candidate genes related to year-round estrous were ZO2, IGF1, TSHB and DLG1, and the candidate genes related to sheep reproductive performance were BMP4, LRP8, NF1, and INSL3. This study contributes to a better understanding of the genetic structure and population history of sheep populations in the Taklimakan Desert environment, as well as the regulatory mechanisms of year-round estrous in sheep in the desert environment, which has important implications for the global livestock industry to cope with the challenges of rapidly changing climate and to explore important economic traits.
Sheep have played an important economic and cultural role in the history of human development, not only as a source of human food and ancillary products, but also in the spread of human farming civilization (Deniskova et al., 2018). The geomorphology of southern Xinjiang in China is complex, with the Southern Tianshan Mountains in the north and the Karakoram Mountains in the south, forming an obliterated ring, and the Taklimakan desert is in the east, forming sand touch arid climate. The Taklimakan Desert is a warm temperate arid desert with sufficient light, little precipitation and large temperature difference between day and night. Under the extreme environmental conditions, part of the native sheep in Southern Xinjiang have the characteristics of strong stress resistance, high reproductive capacity and perennial estrous, such as Duolang sheep and Yecheng sheep. The average lambing rate of Duolang sheep is about 130%. Duolang sheep is the main local breed with both meat and fat in Kashgar area of Xinjiang. The average lambing rate of Yecheng sheep is 121.04%, and the central producing area is located in Yecheng County on the western edge of Taklimakan Desert. Both Duolang sheep and Yecheng sheep have excellent genetic characters such as prolific and year-round estrous. Therefore, studying the origin and evolution of sheep, and the advantages of its formation in extreme environments will not only help to understand its breed formation process, but also help to deal with the future challenges of animal husbandry under extreme environmental changes.
Although the native sheep in Southern Xinjiang have many excellent production traits due to its diverse ecosystem (Ma et al., 2019). Many sheep breeds still remain in the state of self-breeding by farmers, which leads to the lag of breeding work and a small number of offspring. Now it is necessary to selectively intervene the breeding work through genomic selection technology, so that the excellent production traits sheep can be better inherited to future generations. Compared with traditional selection methods, genomics can be evaluated at an early stage with higher accuracy (Hayes et al., 2012). Illumina Ovine SNP 50K BeadChip has been widely used in the study of sheep genetic diversity, genetic relatedness and the exploitation of important economic traits (Deniskova et al., 2019; Paim et al., 2021), which allows us to have a deeper understanding of the origin, scale and economic traits of sheep in the process of domestication, as well as understanding the differentiation and intra-breed variation of sheep (Hiendleder et al., 2002). Using Ovine SNP 50K BeadChip, Kijas found that 74 sheep breeds in the world contained high genetic diversity, indicating that domestication occurred on a wide genetic basis; Gene selective sweep revealed 31 regions containing genes for fur pigmentation, growth and reproduction (Kijas et al., 2012). Lv analyzed the Ovine 50K BeadChip data of SNPs from 32 ancient local sheep breeds living in different environments around the globe, and detected 230 SNPs that were selected by environmental selection pressure (Lv et al., 2015). The estrous character of sheep is mainly affected by the factors such as light, temperature and genes. As a result, the majority of sheep in the world are seasonal estrous breeds (Thimonier, 1981; Wayne et al., 1989). Zhang confirmed that FGF3, DNM2 and USP25 were associated with annual estrous in sheep by comparing sheep breeds with different traits (Zhang C. L. et al., 2022). The year-round estrous sheep can break through the restrictions of seasonal breeding, and the production performance is much better than that of seasonal estrous sheep, which can greatly improve the production efficiency. It is of great significance to explore the year-round estrous character of sheep in desert environment by using Ovine SNP 50K BeadChip.
In this study, the Zhang C. L. et al., 2022 Ovine SNP 50K BeadChip was used to analyze the genomes of nine sheep breeds from China, India, Bangladeshi, and Iran. The genetic background, genetic characteristics and genetic mechanism of year-round estrous of native sheep breeds in Taklimakan Desert were studied. Identifying the interspecific genetic structure of Xinjiang native sheep and understanding their genetic differentiation can help prevent the loss of genetic resources of sheep under the changing climate extremes, develop strategies to protect endangered breeds and develop new varieties in a targeted way.
This work was conducted in accordance with the specifications of the Ethics Committee of Tarim University of Science and Technology (SYXK 2020-009).
We used 341 samples of nine sheep breeds from China, India, Bangladeshi and Iran (Table 1). To be specific, we used five sheep breeds from Xinjiang, China, inculding 46 Duolang sheep (DUL), 48 Bayanbroek sheep (BYK), 46 Tashkurgan sheep (TSK), 50 Yecheng sheep (YEC), 45 Kazakh sheep (HSK) and 24 Tibetan sheep (TIB) from Tibet, China. The blood samples were all from ewes. The data of 24 Deccani sheep (IDC) from Indian, 34 Moghani sheep (MOG) from Iran, and 24 Bangladesh Carole (BGA) from Bangladeshi, were included in this study. The data of IDC, MOG, TIB and BGA was from International Sheep Genomics Consortium (ISGC)1.
Blood samples were collected using the Dried Blood Spots (DBS), and NucliSens easyMag instrument was used to extract DNA. The DNA of all samples was quantified by agarose gel electrophoresis and Nanodrop ND-2000 (Thermo Scientific) and the concentration of gDNA was adjusted to 50 ng/μL. Genome-wide amplification was then performed on all samples. The gDNA was fragmented, precipitated, and resuspended in a hybridization buffer after 20–24 h of amplification at 37°C. The resuspended DNA fragments were loaded onto the chip and hybridized for 16–24 h by incubation at 48°C. After hybridization, non-specific binding DNA was removed by washing, and the remaining specific binding sites were extended by a single base. After staining, the genotype files were scanned by Illumina iScan Reader.
Plink V. 1.90 (Purcell et al., 2017) software was used for quality control and unqualified SNP sites were eliminated. The quality control criteria of this study were as follows: (1) individual detection rate > 0.95, (2) SNPs detection rate > 0.95, and (3) Hardy–Weinberg equilibrium (HWE) p value ≥10−6.
Plink V. 1.90 software was used to calculate the observed heterozygosity (HO), expected heterozygosity (He), minor allele frequency (MAF) and inbreeding coefficient.
The genotypic data after quality control was subjected to PCA analysis using GCTA (Yang et al., 2011) software, which identified the main components representing population structure based on genetic correlations among individuals.
The ontogenetic distances and evolutionary relationships of the five sheep breeds were represented by an adjacency evolutionary tree. The VCF2Dis V. 1.092 was used to calculate the P distance matrix, and then the NJ-tree was constructed by ATGC: FastME3 program, and the drawing was done by network tool iTOL (Letunic and Bork, 2021).
Genetic admixture calculations were performed using Admixture (Alexander et al., 2009). In the Admixture analysis of all populations, an Admixture analysis was performed for K values ranging from 2 to 8.
The fixation index (Fst) analysis was performed on the SNP data of YEC and DUL (year-round estrous), and BYK and TSK (seasonal estrous), to analyze the trait of year-round estrous in sheep under an extreme environment. The snp data of DUL and YEC were, respectively, analyzed by Pi to provide information for sheep breeding against future changeable extreme environment. Fst is used to measure the degree of population differentiation and can reflect the level of species population differentiation. This method is suitable for selective signal detection of multiple populations and as follows:
Where, MSG is the mean square of error within the population, MSP is the mean square of error between the populations, and nc is the average sample size between the populations after correction. By using a sliding window with a window size of 50 Kb and a sliding step size of 25 Kb, the Fst value of each sliding window SNP is calculated. Vcftools was used to calculate the Fst value for each window, and then CMplot was used to plot Manhattan. In both analyses, the top 1% was considered as selection footprints.
The Pi and Fst results were selected for intersection analysis, with annotations referencing the sheep genome Ovis Oar_v4.0. Gene functional annotation was performed referencing the NCBI databases4 and OMIM database.5 The g:Profiler6 was used for autosomal enrichment of candidate genes for GO and Reactome/KEGG pathway analysis.
After genotypic quality control, there was carried out on the SNPs of 341 sheep used in the experiment. After the unqualified SNPs were removed, there were 45,139 informative SNPs in these nine sheep breeds population.
Results of genetic diversity indices for sheep breeds in this study were given in Table 2. The observed and expected heterozygosity ranged from 0.1806 (TSK) to 0.3988 (MOG) and 0.2880 (TIB) to 0.3693 (MOG), respectively. Inbreeding coefficient ranged from −0.08 (HSK) to 0.4557 (TSK), and minor allele frequency ranged from 0.2101 (TIB) to 0.2780 (MOG).

Table 2. The observed heterozygosity (HO), expected heterozygosity (He), minor allele frequency (MAF), and inbreeding coefficient in each population.
PCA results showed that all Xinjiang indigenous sheep breeds could be well distinguished from other sheep breeds (Figure 1). At PC1 and PC2 levels, BGA, MOG and IDC come together and the remaining sheep breeds come together. TIB and TSK extend outwards and were partially mixed, distinguished from other sheep breeds. At all three levels, Xinjiang indigenous sheep breeds are clustered together, with some parts of the YEC extending outward.
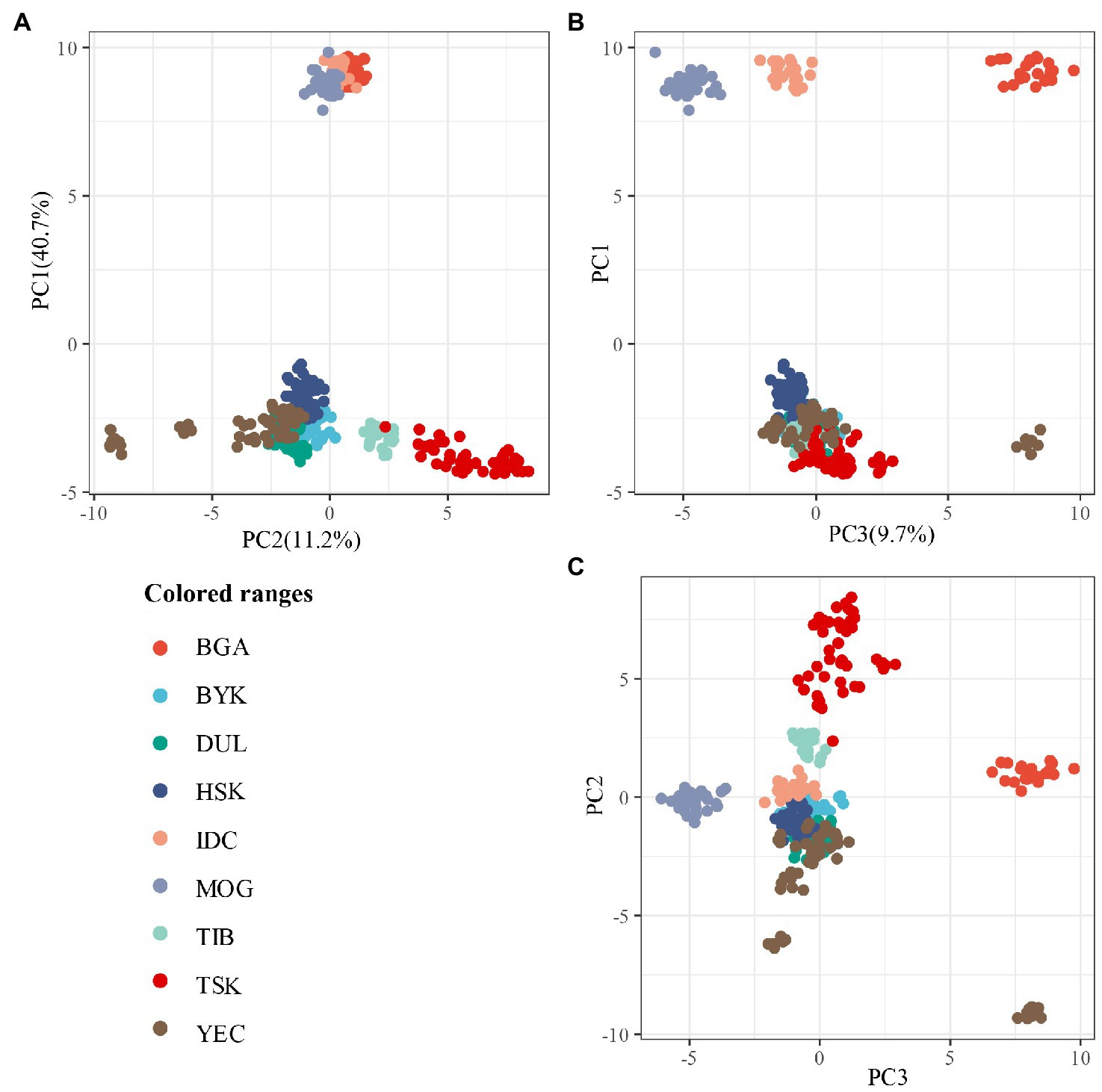
Figure 1. Principal component analysis of nine sheep breeds. The analysis was performed for the first two components (C1 and C2) (A), for the first and third component (C1 and C3) (B) and for the second and third component (C2 and C3) (C).
The Admixture result showed that the sheep in the Tarim Basin had different ancestral components from those in other countries (Figure 2). IDC, MOG and BGA had unique genetic backgrounds different from Chinese indigenous sheep. HSK, BYK, DUL and YEC had similar genetic backgrounds. When K = 2–8, TSK had different genetic backgrounds from other Xinjiang sheep breeds. When K = 3, 4, and 7, TSK and TIB had similar genetic backgrounds.
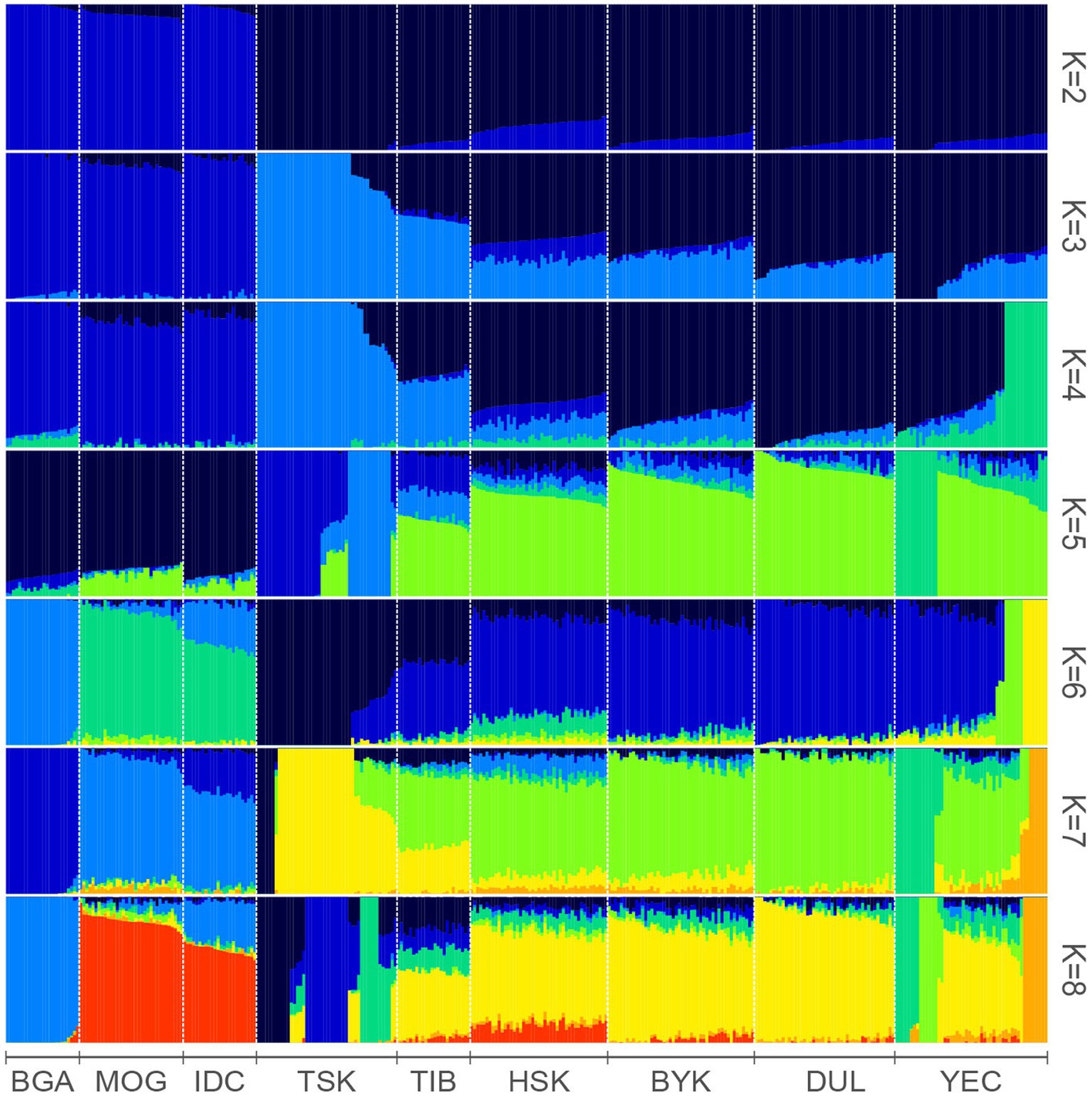
Figure 2. Admixture analysis of nine sheep breeds. Results for inferred numbers of clusters k = 2–8 were shown. Different colors represent different ancestral components.
To further investigate the genetic structure of nine sheep breeds, we constructed Neighbor-Joining tree for all individuals. The results showed that Neighbor-Joining tree could separate sheep breeds from China and other countries according to geographical distance. It can be clearly observed that BYK can be divided into two populations, and the genetic distance from TIB and HSK was relatively close. YEC and DUL have the closest genetic distance, which was consistent with the results of PCA and Admixture (Figure 3).
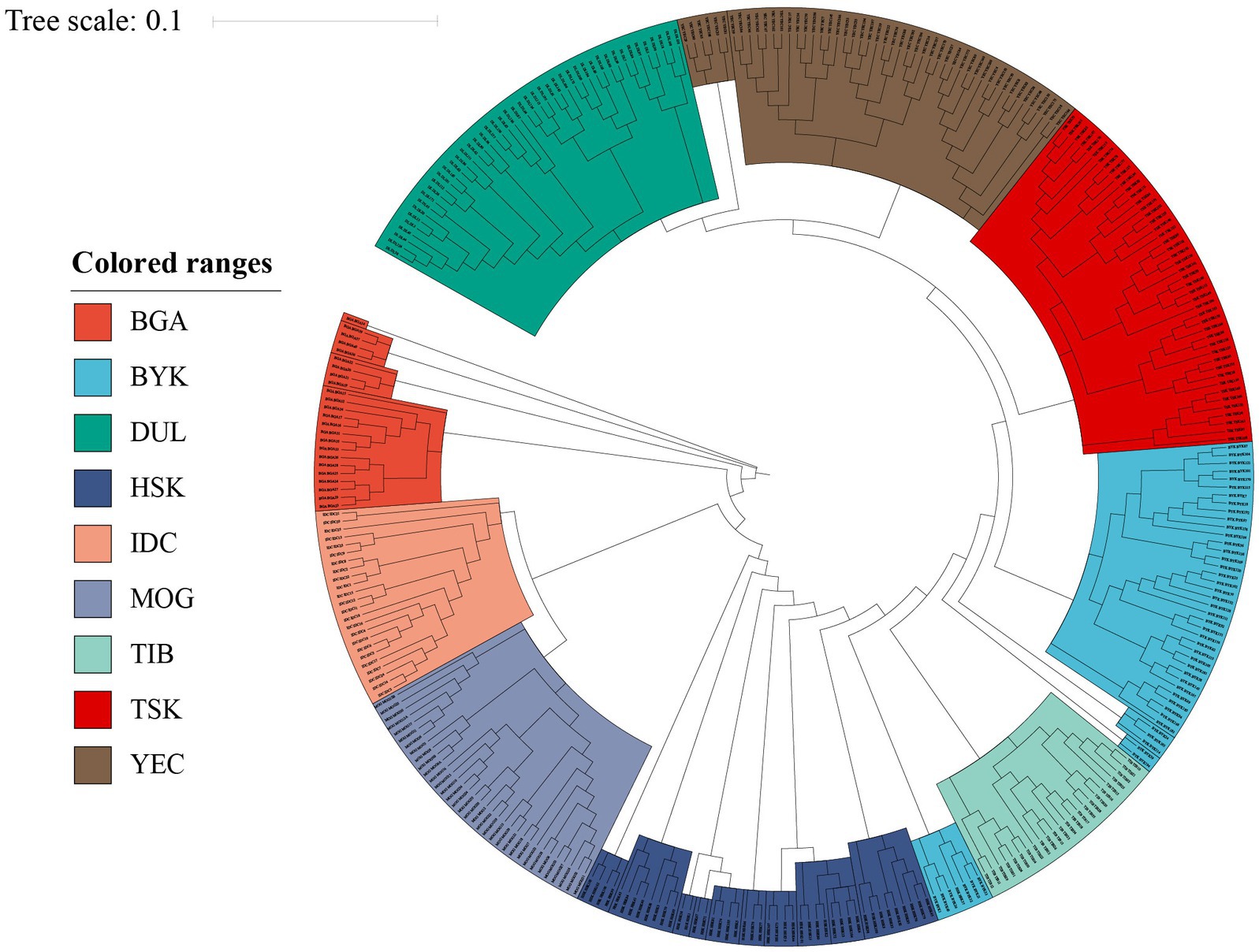
Figure 3. The result of Neighbor-Joining tree of nine sheep breeds. The indigenous sheep breeds in Xinjiang can be distinguished from those in other countries, and the relationships of YEC and DUL are closest.
The Fst values were calculated, and the Fst values were arranged in descending order (Figure 4A). The first 5% was regarded as the significant window, and a total of 953 genes were obtained. The enrichment results of the KEGG pathway of the candidate genes and their relationship were shown in Figure 4B. We identified some genes associated with perennial estrous and reproduction in sheep by GO analysis and pathway enrichment combined with references. For example, TSHB and ZO2 regulate circadian rhythms, IGF1 regulates GnRH expression, and all play important roles in the estrous cycle. DLG1, BMP4 and NF1 are important candidate genes for increasing litter size in sheep. NYAP1 and INSL3 play a role in follicular development, ovulation and luteal formation. The Pi results of YEC and DUL showed that 526 and 523 genes were obtained, respectively, under the last 5% threshold, and 60 and 66 genes were obtained by intersection with Fst results (Figure 5). GO analysis was performed on the candidate genes obtained by the intersection (Figure 6).
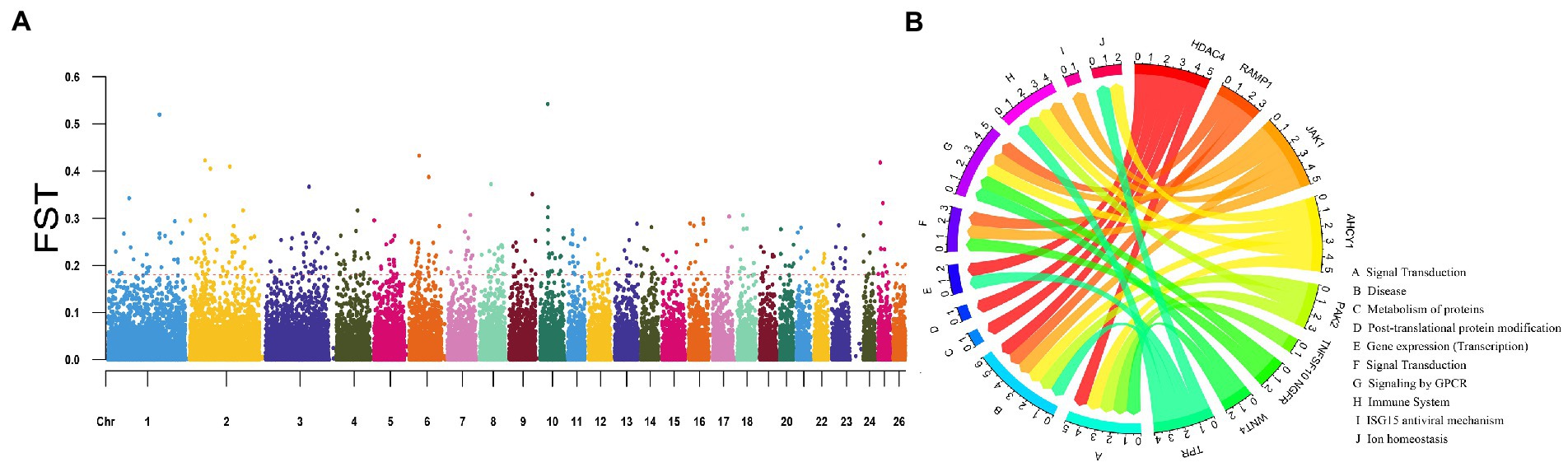
Figure 4. Fst detection results of four sheep breeds. (A) The red line represents the 5% threshold line. (B) Enrichment results of candidate gene pathways.

Figure 5. Pi analysis results of YEC and DUL. The intersection with Fst results obtained 66 (A) and 60 (B) genes, respectively.
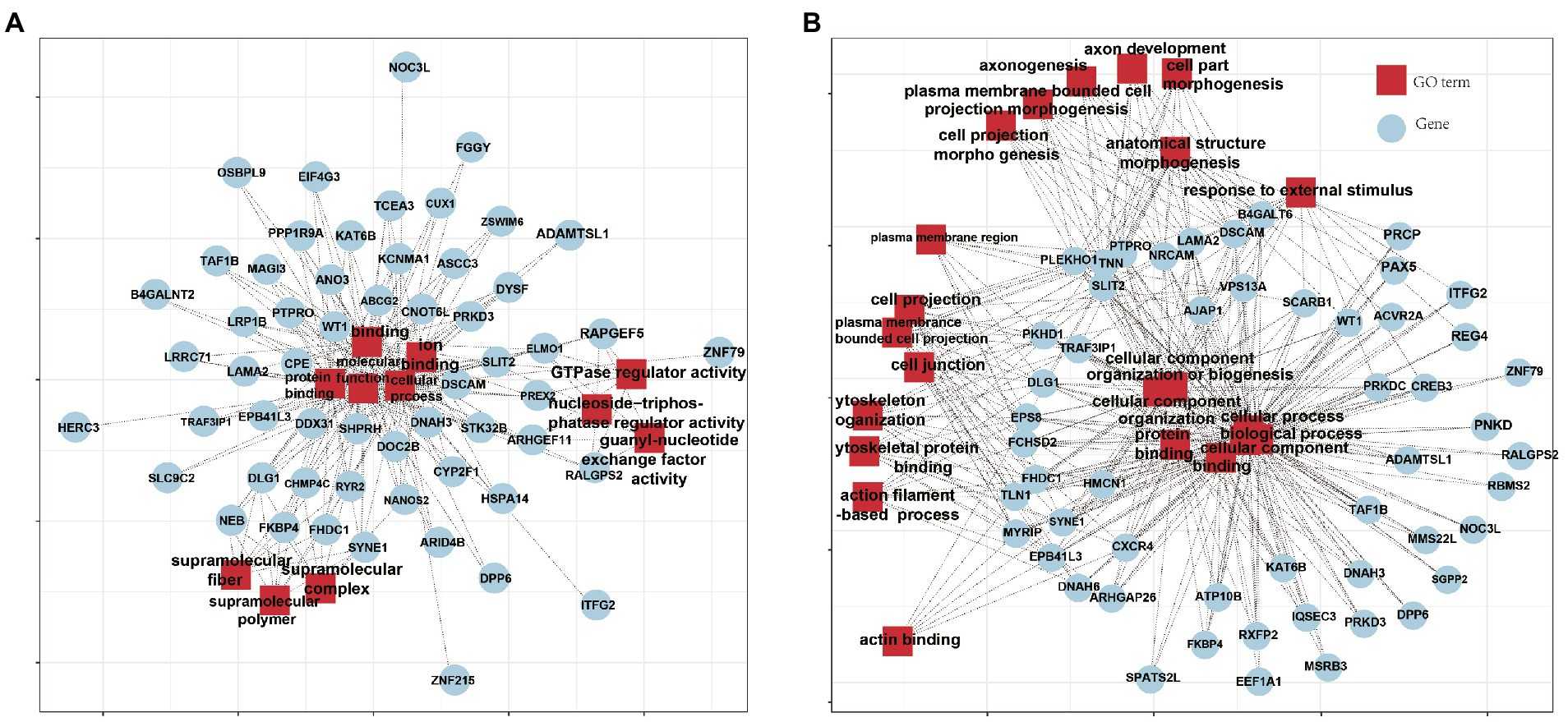
Figure 6. Selective sweep gene GO analysis at the last 5% threshold of YEC (66) and DUL (60). (A) GO analysis was performed on 66 candidate genes obtained by selective sweep of YEC, and they mainly focused on molecular function, protein binding and ion binding. (B) GO analysis was performed on 60 candidate genes obtained by selective sweep of DUL, and they mainly focused on anatomical structure morphogenesis, plasma membrane bounded cell projection and cell projection.
Sheep belong to Artiodactyla, Bovidae, Capovinae and Ovis in zoological taxonomy, and the origin of sheep has been controversial. Ovis gmelinii, Ovis ammon, and Ovis vignei, which are generally believed to be distributed in Eurasia, are most closely related to domestic sheep (Hiendleder et al., 2002), among which Ovis gmelinii is considered to be the most likely ancestor of domestic sheep. Therefore, in order to reveal the origin of sheep breeds around Tarim Basin, we selected sheep breeds from the Asian mainland for analysis. Our results showed that the level of genetic variability around Tarim Basin was comparable to that demonstrated in other worldwide populations (Dossybayev et al., 2019; Adeniyi et al., 2022). However, they had high levels of inbreedingcoefficient, indicating an urgent need to establish a conservation program to reduce inbreeding and increase genetic diversity. Through the population structure, we have observed that the genetic distance between Xinjiang native sheep and other sheep was far away. Chinese indigenous sheep were classified geographically and morphologically into three groups: Mongolian, Kazakh, and Tibetan. The population structure showed that the germplasm resources of all sheep breeds in southern Xinjiang were close to each other, which was obviously different from other sheep breeds of other countries. PCA and N-J tree can distinguish sheep breeds in different regions according to geographical distance. The Admixture results showed that TIB and TSK may be the ancestral groups of sheep in Xinjiang. These results suggested that Xinjiang in China may also have been the first place to domesticate sheep (Wei et al., 2015). The population structure showed that DUL and YEC were closely related and had similar ancestral components. The results of N-J tree revealed that DUL and HSK were separated, which was consistent with the results of Wei’s study on 10 native sheep breeds in China (Wei et al., 2015). Studies have also shown that the genetic background of domestic sheep, especially Western sheep, is similar to that of Ovis ammon polii living in Xinjiang, suggesting that domestic sheep may have originated from Ovis ammon living in the Pamir Plateau, rather than Ovis gmelinii.
DUL and YEC are geographically close to each other in the desert environment, and have good traits such as strong stress resistance, multiple parities and year-round estrous. Given the increasing threat of climate changes, the effective conservation of local genetic resources for livestock has become urgent. It is of great significance to understand the mechanism of year-round estrous, as an important and unique trait to DUL and YEC, on the genomic level.
Breeding activities are carried out under short day conditions, which is one of the important factors limiting the production efficiency of sheep (Ebling, 2010). Most sheep in the world are in seasonal estrous. However, DUL and YEC living in the desert environment have the trait of year-round estrous, which may be caused by the longer light time in the desert environment. DUL and YEC live in the area with the longest sunshine duration in China, which can reach 2,550 to 3,300 h a year. Sheep estrous is a very complicated physiological process (Habeeb and Anne Kutzler, 2021). After receiving light stimulation, the retina of sheep converts it into nerve impulse to promote the pulsing release of gonadotropin-releasing hormone (GnRH) in the hypothalamus, thus promoting the secretion of follicle-stimulating hormone (FSH) and luteinizing hormone (LH) in the pituitary gland and acting on the gonadal axis (Chen et al., 2017; Voliotis et al., 2019; Zhang C. L. et al., 2022). The modality of hypothalamic–pituitary-gonadal (HPG) axis affects reproductive function in animals, especially the regulation of the estrous cycle (Schaub et al., 2008). Zonula occludens protein 2 (ZO2) is an important protein that makes up the blood-cerebrospinal fluid barrier and regulates its permeability. The photo period regulates the expression of ZO2 and thus the permeability of the barrier, thereby inducing the reproductive activity of sheep (Lagaraine et al., 2011). For example, short periods of light can cause a surge in ZO2, but longer periods of light can decrease the ZO2 protein and increase the permeability of the blood-cerebrospinal fluid barrier, allowing high concentrations of proteins (progesterone, estradiol, and leptin) to enter (Lagaraine et al., 2011). Thyroid stimulating hormone (TSH) plays a role in controlling thyroid structure and metabolism, and is required for endogenous shifts in the seasonal reproductive state of animals 21 (Pierce and Parsons, 1981). The expression of thyroid Stimulating Hormone Subunit Beta (TSHB) in the pituitary can induce the expression of deiodinase 2 (DIO2), which ultimately stimulates the release of GnRH and triggers long-term photoinduced seasonal reproduction, which may be associated with year-round estrous (Huang et al., 2013). Insulin-like growth factor 1 (IGF1) also plays an important role in regulating many hormones that are critical to the reproductive system (Miller and Gore, 2001). As a regulator of reproductive neuroendocrine, IGF1 is believed to play a role in regulating GnRH release, including follicular growth and ovulation, and is an important candidate gene for influencing sheep fertility (He et al., 2012).
The periodic increase in the level of related hormones in the body is a crucial part of year-round estrous. The FSH and LH secreted by the pituitary reach the ovary along with the blood circulation to stimulate follicle development and maturation, and further produce E2, and eventually lead to ovulation (Zhang J. et al., 2022). Subsequently, under the action of LH, ovarian granulosa cells become luteinized and secrete progesterone, and the hypothalamus pituitary gland is negatively feedback, and the body enters a hypoestrous period. Discs Large MAGUK Scaffold Protein 1 (DLG1) is expressed at a high level in oocytes and granulosa cells, which can regulate the activation of primordial follicles in the Hippo signaling pathway and increase the birth rate, and is an important candidate gene for increasing the litter size of sheep (Hernández-Montiel et al., 2020). LDL receptor related protein 8 (LRP8) is widely present in mammalian ovarian granulosa cells and luteal cells, and can control follicular growth and development and ovulation (Fayad et al., 2007) The expression of follicular bone Morphogenetic Protein 4 (BMP4) was significantly higher in FecB carrying sheep, indicating their importance in governing the fecundity in FecB carrier (Goyal et al., 2017). As an important candidate gene for increasing the number of lambs in Texel sheep, neurofibromin 1 (NF1) plays an important role in follicular genesis and the development of LH signaling (Xu et al., 2018). Different expression levels of neuronal tyrosine phosphorylated phosphoinositide-3-Kinase adaptor 1 (NYAP1) in sheep may contribute to increased reproductive rate by regulating target genes related to the oxytocin signaling pathway (Miao et al., 2016). Insulin Like 3 (INSL3) and its receptor relaxin Family Peptide Receptor 2 (RXFP2) were expressed in ovarian luteal cells, and INSL3 showed cyclic changes in the estrous cycle, and the change pattern was consistent with that of estrogen, both increased after luteal lysis, and then sharply decreased after LH surge and may play an important role in sheep year-round estrous (Satchell et al., 2013).
The year-round estrous is the result of a complex, multi-system, multi-gene collaboration. It is regulated by the HPG axis and is closely related to hormone levels in the body. The results of Fst showed that many genes related to perennial estrous can affect the regulation of HPG axis and hormone levels in animals, so as to show year-round estrous. These genes may also be important candidate genes for influencing litter size in sheep.
The datasets presented in this study can be found in online repositories. The name of the repository and a link to the data can be found below: Figshare; http://doi.org/10.6084/m9.figshare.21900612.
This work was conducted in accordance with the specifications of the Ethics Committee of Tarim University of Science and Technology (SYXK 2020-009).
JZ and SL contributed to the study for the experimental design, collection, laboratory analysis and statistical analysis of the data, and writing of the manuscript. MT and CZ contributed to the experimental design and revising of the manuscript. All authors contributed to the article and approved the submitted version.
This study was financially supported through grants from the National Natural Science Fund Project in China (32060743) and Bintuan Science and Technology Program (2022CB001-09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1146561/full#supplementary-material
1. ^ https://www.sheephapmap.org
2. ^ https://github.com/BGI-shenzhen/VCF2Dis
3. ^ http://www.atgc-montpellier.fr/fastme
4. ^ http://www.ncbi.nlm.nih.gov/gene
Adeniyi, O. O., Simon, R., Bytyqi, H., Kugler, W., Mehmeti, H., Berisha, K., et al. (2022). Capturing genetic diversity and selection signatures of the endangered Kosovar Balusha sheep breed. Genes 13:866. doi: 10.3390/genes13050866
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Chen, W. Y., Du, Y. Q., Guan, X., Zhang, H. Y., and Liu, T. (2017). Effect of GnRHR polymorphisms on in vitro fertilization and embryo transfer in patients with polycystic ovary syndrome. J. Hum. Genet. 62, 1065–1071. doi: 10.1038/jhg.2017.85
Deniskova, T., Dotsev, A., Lushihina, E., Shakhin, A., Kunz, E., Medugorac, I., et al. (2019). Population structure and genetic diversity of sheep breeds in the Kyrgyzstan. Front. Genet. 10:1311. doi: 10.3389/fgene.2019.01311
Deniskova, T. E., Dotsev, A. V., Selionova, M. I., Kunz, E., Medugorac, I., Reyer, H., et al. (2018). Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. 50:29. doi: 10.1186/s12711-018-0399-5
Dossybayev, K., Orazymbetova, Z., Mussayeva, A., Saitou, N., Zhapbasov, R., Makhatov, B., et al. (2019). Genetic diversity of different breeds of Kazakh sheep using microsatellite analysis. Arch. Anim. Breed. 62, 305–312. doi: 10.5194/aab-62-305-2019
Ebling, F. J. (2010). Photoperiodic regulation of puberty in seasonal species. Mol. Cell. Endocrinol. 324, 95–101. doi: 10.1016/j.mce.2010.03.018
Fayad, T., Lefebvre, R., Nimpf, J., Silversides, D. W., and Lussier, J. G. (2007). Low-density lipoprotein receptor-related protein 8 (LRP8) is upregulated in granulosa cells of bovine dominant follicle: Molecular characterization and spatio-temporal expression studies. Biol. Reprod. 76, 466–475. doi: 10.1095/biolreprod.106.057216
Goyal, S., Aggarwal, J., Dubey, P. K., Mishra, B. P., Ghalsasi, P., Nimbkar, C., et al. (2017). Expression analysis of genes associated with prolificacy in FecB carrier and noncarrier Indian sheep. Anim. Biotechnol. 28, 220–227. doi: 10.1080/10495398.2016.1262869
Habeeb, H. M. H., and Anne Kutzler, M. (2021). Estrus synchronization in the sheep and goat. Vet. Clin. North Am. Food Anim. Pract. 37, 125–137. doi: 10.1016/j.cvfa.2020.10.007
Hayes, B. J., Lewin, H. A., and Goddard, M. E. (2012). The future of livestock breeding: Genomic selection for efficiency, reduced emissions intensity, and adaptation. Trends Genet. 29, 206–214. doi: 10.1016/j.tig.2012.11.009
He, J. N., Zhang, B. Y., Chu, M. X., Wang, P. Q., Feng, T., Cao, G. L., et al. (2012). Polymorphism of insulin-like growth factor 1 gene and its association with litter size in small tail Han sheep. Mol. Biol. Rep. 39, 9801–9807. doi: 10.1007/s11033-012-1846-y
Hernández-Montiel, W., Martínez-Núñez, M. A., Ramón-Ugalde, J. P., Román-Ponce, S. I., Calderón-Chagoya, R., and Zamora-Bustillos, R. (2020). Genome-wide association study reveals candidate genes for litter size traits in Pelibuey sheep. Animals 10:434. doi: 10.3390/ani10030434
Hiendleder, S., Kaupe, B., Wassmuth, R., and Janke, A. (2002). Molecular analysis of wild and domestic sheep questions current nomenclature and provides evidence for domestication from two different subspecies. Proc. Biol. Sci. 269, 893–904. doi: 10.1098/rspb.2002.1975
Huang, D. W., Wang, J. X., Liu, Q. Y., Chu, M. X., Di, R., He, J. N., et al. (2013). Analysis on DNA sequence of TSHB gene and its association with reproductive seasonality in goats. Mol. Biol. Rep. 40, 1893–1904. doi: 10.1007/s11033-012-2245-0
Kijas, J. W., Lenstra, J. A., Hayes, B., Boitard, S., Porto Neto, L. R., San Cristobal, M., et al. (2012). Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 10:e1001258. doi: 10.1371/journal.pbio.1001258
Lagaraine, C., Skipor, J., Szczepkowska, A., Dufourny, L., and Thiery, J. C. (2011). Tight junction proteins vary in the choroid plexus of ewes according to photoperiod. Brain Res. 1393, 44–51. doi: 10.1016/j.brainres.2011.04.009
Letunic, I., and Bork, P. (2021). Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lv, F. H., Peng, W. F., Yang, J., Zhao, Y. X., Li, W. R., Liu, M. J., et al. (2015). Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol. Biol. Evol. 32, 2515–2533. doi: 10.1093/molbev/msv139
Miao, X., Luo, Q., Zhao, H., and Qin, X. (2016). Co-expression analysis and identification of fecundity-related long non-coding RNAs in sheep ovaries. Sci. Rep. 6:39398. doi: 10.1038/srep39398
Ma, H., Fang, C., Liu, L., Wang, A., Aniwashi, J., Sulaiman, Y., et al. (2019). Identification of novel genes associated with litter size of indigenous sheep population in Xinjiang, China using specific-locus amplified fragment sequencing technology. PeerJ. 7: e8079. doi: 10.7717/peerj.8079
Miller, B. H., and Gore, A. C. (2001). Alterations in hypothalamic insulin-like growth factor-I and its associations with gonadotropin releasing hormone neurones during reproductive development and ageing. J. Neuroendocrinol. 13, 728–736. doi: 10.1046/j.1365-2826.2001.00686.x
Paim, T. P., Paiva, S. R., de Toledo, N. M., Yamaghishi, M. B., Carneiro, P. L. S., Facó, O., et al. (2021). Origin and population structure of Brazilian hair sheep breeds. Anim. Genet. 52, 492–504. doi: 10.1111/age.13093
Pierce, J. G., and Parsons, T. F. (1981). Glycoprotein hormones: Structure and function. Annu. Rev. Biochem. 50, 465–495. doi: 10.1146/annurev.bi.50.070181.002341
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2017). PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Satchell, L., Glister, C., Bleach, E. C., Glencross, R. G., Bicknell, A. B., Dai, Y., et al. (2013). Ovarian expression of insulin-like peptide 3 (INSL3) and its receptor (RXFP2) during development of bovine antral follicles and corpora lutea and measurement of circulating INSL3 levels during synchronized estrous cycles. Endocrinology 154, 1897–1906. doi: 10.1210/en.2012-2232
Schaub, C. E., Gersting, J. A., Keller-Wood, M., and Wood, C. E. (2008). Development of ER-alpha and ER-beta expression in the developing ovine brain and pituitary. Gene Exp. Patter. 8, 457–463. doi: 10.1016/j.gep.2008.03.001
Thimonier, J. (1981). Control of seasonal reproduction in sheep and goats by light and hormones. J. Reprod. Fertil. Suppl. 30, 33–45.
Voliotis, M., Li, X. F., De Burgh, R., Lass, G., Lightman, S. L., Obyrne, K. T., et al. (2019). The origin of GnRH pulse generation: An integrative mathematical-experimental approach. J. Neurosci. 39, 9738–9747. doi: 10.1523/JNEUROSCI.0828-19.2019
Wayne, N. L., Malpaux, B., and Karsch, F. J. (1989). Social cues can play a role in timing onset of the breeding season of the ewe. J. Reprod. Fertil. 87, 707–713. doi: 10.1530/jrf.0.0870707
Wei, C., Wang, H., Liu, G., Wu, M., Cao, J., Liu, Z., et al. (2015). Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genomics 16:194. doi: 10.1186/s12864-015-1384-9
Xu, S. S., Gao, L., Xie, X. L., Ren, Y. L., Shen, Z. Q., Wang, F., et al. (2018). Genome-wide association analyses highlight the potential for different genetic mechanisms for litter size among sheep breeds. Front. Genet. 9:118. doi: 10.3389/fgene.2018.00118
Yang, J., Lee, S. H., Goddard, M. E., and Visscher, P. M. (2011). GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. doi: 10.1016/j.ajhg.2010.11.011
Zhang, C. L., Liu, C., Zhang, J., Zheng, L., Chang, Q., Cui, Z., et al. (2022). Analysis on the desert adaptability of indigenous sheep in the southern edge of Taklimakan Desert. Sci. Rep. 12:12264. doi: 10.1038/s41598-022-15986-x
Keywords: sheep, genetic diversity, population structure, year-round estrous, desert environment
Citation: Zhang J, Zhang C-l, Tuersuntuohe M and Liu S (2023) Population structure and selective signature of sheep around Tarim Basin. Front. Ecol. Evol. 11:1146561. doi: 10.3389/fevo.2023.1146561
Received: 17 January 2023; Accepted: 13 March 2023;
Published: 30 March 2023.
Edited by:
Yukio Nagano, Saga University, JapanReviewed by:
Moe Lwin, Livestock Breeding and Veterinary Department, MyanmarCopyright © 2023 Zhang, Zhang, Tuersuntuohe and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shudong Liu, bGl1c2h1ZG9uZzYzQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.