- 1Janecka Genomics Laboratory, Duquesne University, Bayer School of Natural and Environmental Sciences, Pittsburgh, PA, United States
- 2Biotechnology Program, Duquesne University, Bayer School of Natural and Environmental Sciences, Pittsburgh, PA, United States
Hippopotamid phylogeny has proven difficult to resolve with proposed relationships between extant and fossil species receiving mixed levels of support. Of particular interest is the divergence between the two extant hippopotamid species, the well-known common hippopotamus (Hippopotamus amphibius) and the enigmatic pygmy hippopotamus (Choeropsis liberiensis). Previous studies have relied on morphological and fossil evidence to identify the ancestral species at the core of this divergence and its timing. In this study, we assembled a molecular matrix of 26 nuclear gene sequences from 11 ungulate species with two primates as an outgroup. We used a Bayesian relaxed molecular clock approach to reconstruct a calibrated time tree for Cetartiodactyla and estimate the divergence date between the common and pygmy hippopotamus. While previous morphological studies have estimated this event to have occurred sometime during the Late Miocene (between 11.6 and 5.3 million years ago), our nuclear gene-based estimates suggest a more recent split of about 4.04 Ma (95% confidence interval: 8.31–1.97 Ma) via RelTime-ML or 2.4 Ma (95% confidence interval: 3.1–1.6 Ma) via MCMCTree. These more recent estimates correspond with the Early Pliocene – Early Pleistocene sub-epochs and align most closely with the results of previous genomic studies. We discuss how our results compare with previous estimates based on both morphological and molecular studies, some of which extend the predicted range of this divergence date even further back in time. Our results suggest a different path of evolution for the understudied pygmy hippopotamus and reveal that morphological evidence alone may not resolve the correct hippopotamid phylogenetic and time trees. We suggest that the common and pygmy hippopotamus may be phylogenetically closer than once believed. Our results also call for further studies to develop a combined approach incorporating both molecular and morphological evidence to reach a consensus on the evolutionary patterns and timing that led to modern hippopotamid evolution.
1. Introduction
The endangered pygmy hippopotamus (Choeropsis liberiensis) is one of only two extant hippopotamid species, the other being the massive common hippopotamus (Hippopotamus amphibius) (Fisher et al., 2007; Boisserie et al., 2011). While H. amphibius can be found near lakes and rivers in many sub-Saharan African nations, the range of the pygmy hippopotamus is now restricted to the West African nation of Liberia and its neighbors (Flacke and Decher, 2019). Several key morphological and ecological differences separate these two species beyond geographical distribution and size (i.e., the pygmy hippopotamus averages 160–270 kg compared with the common hippopotamus averaging 1,400–1,500 kg) (Burnie and Wilson, 2005; Flacke and Decher, 2019). In both species, males are often slightly larger than females (Burnie and Wilson, 2005). C. liberiensis has a more lightweight and slender body for a mostly terrestrial lifestyle in dense rainforests (Boisserie, 2005; Burnie and Wilson, 2005; Flacke and Decher, 2019). The eyes of the pygmy hippopotamus are placed more laterally on their skulls while the eyes of the semiaquatic common hippopotamus are elevated slightly above the roof of their cranium (Boisserie, 2005; Boisserie et al., 2011). Both H. amphibius and C. liberiensis possess four upper incisors, but H. amphibius has four lower incisors (tetraprotodont) while C. liberiensis has only two (diprotodont) (Boisserie, 2005). It is cranial and dental characteristics like these that have guided most previous studies of hippopotamid phylogeny (Boisserie, 2005; Boisserie et al., 2005; Boisserie and Lihoreau, 2006; Orliac et al., 2010).
Molecular studies have determined that Hippopotamidae, the family containing H. amphibius, C. liberiensis, and several extinct hippopotamid genera, are nested deeply within the clade Cetartiodactyla as the sister group of Cetacea (whales, dolphins, and porpoises) (Theodor, 2004; Meredith et al., 2011; Zurano et al., 2019; Springer et al., 2021). Furthermore, molecular evidence suggests that all of the major cetartiodactyl subgroups diverged from one another within a relatively narrow window of time between 66 and 52 million years ago (Ma), between the latest Cretaceous and earliest Eocene (Figure 1) (Meredith et al., 2011; O’Leary et al., 2013; Zurano et al., 2019). While hippopotamids are believed to have diverged from cetaceans between 61.1 and 52.5 Ma, the oldest fossil hippopotamids have been dated to only about 20.6 Ma, during the Early Miocene, leaving an evolutionary gap of at least 30 million years (Orliac et al., 2010). Several groups have been suggested as the potential origin of Hippopotamidae, including Suina (which has been consistently rejected by molecular studies) and Bothriodontinae, a clade within the paraphyletic family Anthracotheriidae (Boisserie et al., 2005; Orliac et al., 2010; Boisserie et al., 2011).
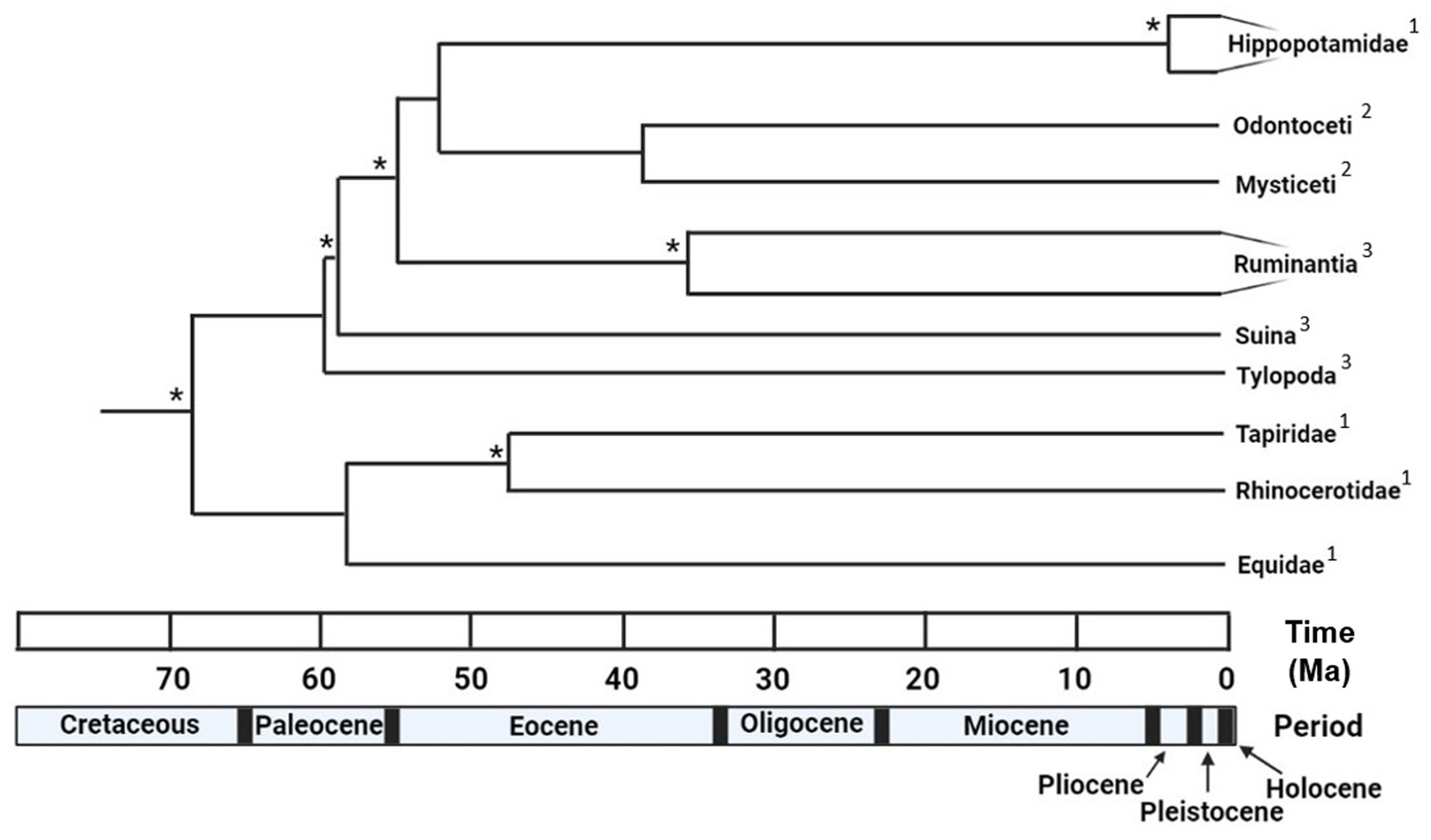
Figure 1. Time tree summarizing the divergence dates of several major ungulate lineages, encompassing both Cetartiodactyla and Perissodactyla. Note that of the ten nodes displayed, five occur within a relatively narrow period of between 60 and 50 million years ago. *Indicates divergence dates estimated from this study, the remaining divergence dates were obtained from Meredith et al. (2011). 1Indicates branches summarized at the taxonomic rank of family. 2Indicates branches summarized at the taxonomic rank of parvorder. 3Indicates branches summarized at the taxonomic rank of suborder. Designed in BioRender.
Taxonomic relationships within Hippopotamidae have proven just as difficult to resolve partly because the majority of species in this family are extinct (Figure 2). With only two extant species, most studies of this nature have relied solely on morphological evidence, specifically cranial and dental characters (Boisserie, 2005; Boisserie and Lihoreau, 2006; Orliac et al., 2010). A morphological analysis by Boisserie (2005) found the pygmy hippopotamus to form a lineage with the primitive genus Saotherium, establishing a sister group to the remaining genera of subfamily Hippopotaminae that diverged during the Late Miocene (Boisserie et al., 2005; Boisserie, 2017). However, Boisserie (2005, 2007) noted that a Saotherium-Choeropsis lineage was only weakly supported morphologically and should be interpreted with caution.
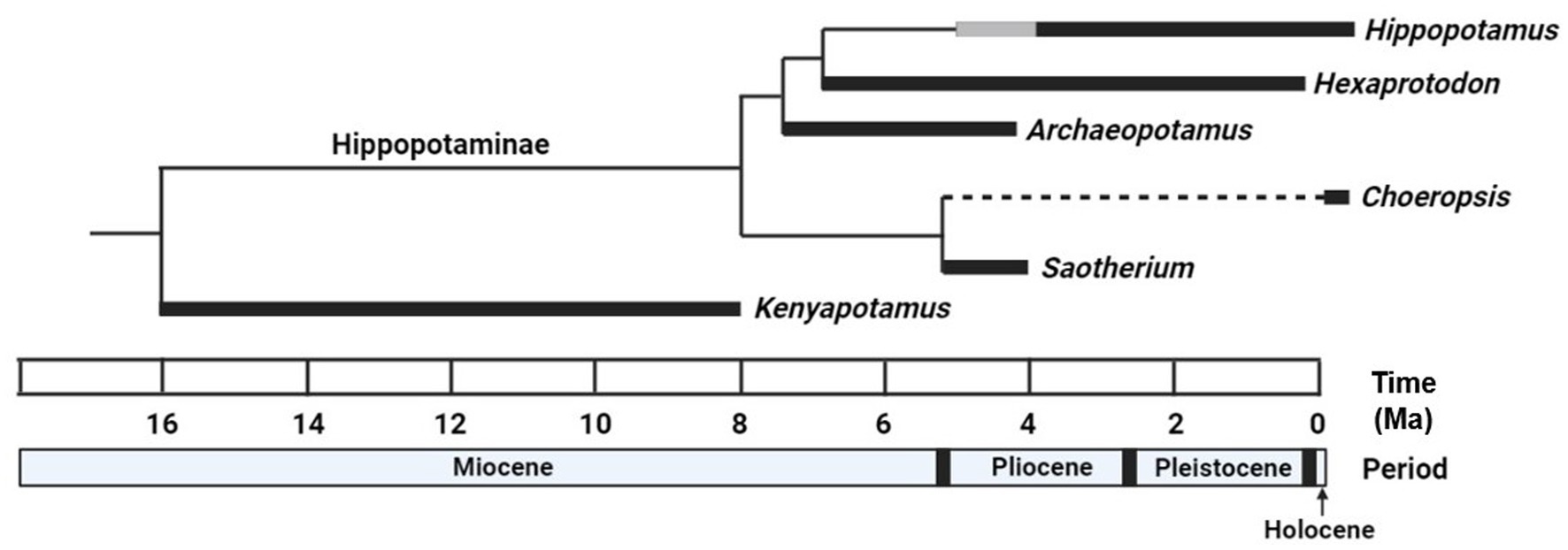
Figure 2. Time tree summarizing the evolutionary history of Hippopotamidae inferred from morphological analyses (i.e., Boisserie, 2005) at the level of genera. The five genera currently placed within subfamily Hippopotaminae are depicted with their more primitive ancestor Kenyapotamus (of Kenyapotaminae) as an outgroup. Thicker, solid black lines indicate the temporal range of each genus from the fossil record. Thicker, solid gray lines indicate potential extended temporal ranges for genera if fossils currently attributed to those genera remain classified as such. Dashed black lines indicate the gap in the fossil record of the pygmy hippopotamus. Temporal ranges for each genus and tree topology were corroborated by Boisserie (2005, 2017), Boisserie et al. (2011), Weston (2000). Designed in BioRender.
Morphological and molecular evidence often, but certainly not always, predict similar phylogenies (Naylor and Adams, 2001; Springer et al., 2017). For instance, morphological and molecular data were at odds regarding the phylogenetic relationship between cetaceans and artiodactyls prior to the 2000s (O’Leary, 1999; Boisserie, 2007). In this case, molecular data provided the first evidence that hippopotamids and cetaceans are closer phylogenetically than was once believed, a hypothesis that has since gained the support of complementary morphological evidence (Geisler et al., 2007). Similarly, the snow leopard (Panthera uncia) was once thought to be the most basal member of Panthera based on morphological characters until molecular studies found this species to be a sister species of the tiger (Panthera tigris) (Davis et al., 2010). Insectivora represents one more example, a now-defunct clade that once grouped members of Afrotheria and Laurasiatheria together (Burnie and Wilson, 2005; Symonds, 2005).
Phylogenetic analyses should incorporate both molecular and morphological evidence whenever possible, but this may be difficult in the case of hippopotamids with only two surviving species to provide molecular data. Phylogenetic studies of Hippopotamidae are further complicated by a nonexistent fossil record for the pygmy hippopotamus, making the task of identifying its direct ancestor especially difficult (Boisserie et al., 2005; Boisserie, 2007). Previous studies of hippopotamid evolution have almost exclusively used morphological data, leaving the potential contribution of molecular evidence underrepresented.
Molecular studies that did include C. liberiensis were primarily focused on other species (McGowen et al., 2020; Figuet et al., 2021). With so few molecular studies of the pygmy hippopotamus, we sought to generate a phylogenetic time tree of Cetartiodactyla from nuclear gene sequences to estimate the divergence date between H. amphibius and C. liberiensis. We then consider the implications of our divergence date on current hypotheses of hippopotamid evolution.
2. Materials and methods
2.1. Gene sequences and alignments
Sequences of 26 nuclear gene segments were downloaded from NCBI for eight cetartiodactyls, three perissodactyls, and two primates for an outgroup (Supplementary Table S1). Among cetartiodactyls, suborder Tylopoda was represented by Lama glama, suborder Suina by Sus scrofa, suborder Ruminantia by Bos taurus and Tragulus napu, family Hippopotamidae by H. amphibius and C. liberiensis, and infraorder Cetacea by Tursiops truncatus and Caperea marginata. Sequences for C. liberiensis were obtained from whole genome sequencing data (Accession: SRX9926581), downloaded from NCBI SRA and analyzed in CLC Genomics Workbench v.10. Sequence reads were mapped to reference files of H. amphibius gene sequences, the closest living relative of C. liberiensis (Springer et al., 2021), at a similarity fraction of 0.9. Consensus sequences were extracted with a low coverage threshold of four reads per position with Ns inserted in positions of low coverage. The noise threshold and minimum nucleotide count values were kept at their default settings of 0.1 and 1, respectively. Once all sequences were obtained, multiple sequence alignments were performed in MEGA-X. Each gene was aligned as DNA using the MUSCLE algorithm and regions that did not have sequences for a majority of species (if present) were trimmed by eye.
2.2. Bayesian analysis
Aligned files for each gene segment were used to assemble a molecular matrix as a Nexus file [available in DRYAD (doi:10.5061/dryad.cjsxksnbg)]. A Maximum Clade Credibility (MCC) tree was generated in MrBayes 3.2.7a using a General Time Reversible (GTR) evolutionary model with gamma-distributed rate variation and a proportion of invariable sites. The analysis was run for 1 × 106 generations and sampled every 1 × 103 generations. In total, 31,890 nucleotides were analyzed for each species.
2.3. Divergence date estimations
The consensus tree from the Bayesian analysis was used to estimate divergence dates with six fossil calibrations in MEGA-X using the RelTime-ML algorithm. Fossil calibrations were entered with normal distributions and 95% confidence intervals set to the upper and lower bounds of each calibration (Supplementary Table S2). Calibrated nodes (and corresponding branches) included Ungulata (Cetartiodactyla + Perissodactyla), Cetartiodactyla (Artiofabula + Tylopoda), Cetruminantia (Ruminantia + Cetancodonta (Whippomorpha)), Cetancodonta (Hippopotamidae + Cetacea), Cetacea (Mysticeti + Odontoceti), and Perissodactyla (Equidae + Tapiridae with Rhinocerotidae). Divergence dates were also estimated with an MCMC tree generated in PAML using the Hasegawa, Kishino and Yano 1985 Model (HKY85). Parameters included 26 gene partitions, gamma value of 0.5, rate of 4, five rate categories, burn-in of 2,000 chains, sampling frequency of 10, and 20,000 samples with a prior on the root less than 100 Ma. 95% confidence intervals for each divergence date were generated in PAML as well.
3. Results
3.1. Phylogenetic analysis
The MCC tree generated by the Bayesian analysis received clade credibility values of 100 for all nodes (Figure 3). Nodes receiving strong levels of support worthy of note include Cetartiodactyla (Node 4), Artiofabula (Node 5), Cetruminantia (Node 6), Cetancodonta (Node 7), and Ruminantia (Node 10). The topology of Cetartiodactyla generated in this study is consistent with the most widely accepted phylogenetic studies of this clade based on the species included (Price et al., 2005; Meredith et al., 2011; Springer et al., 2019; Figuet et al., 2021).
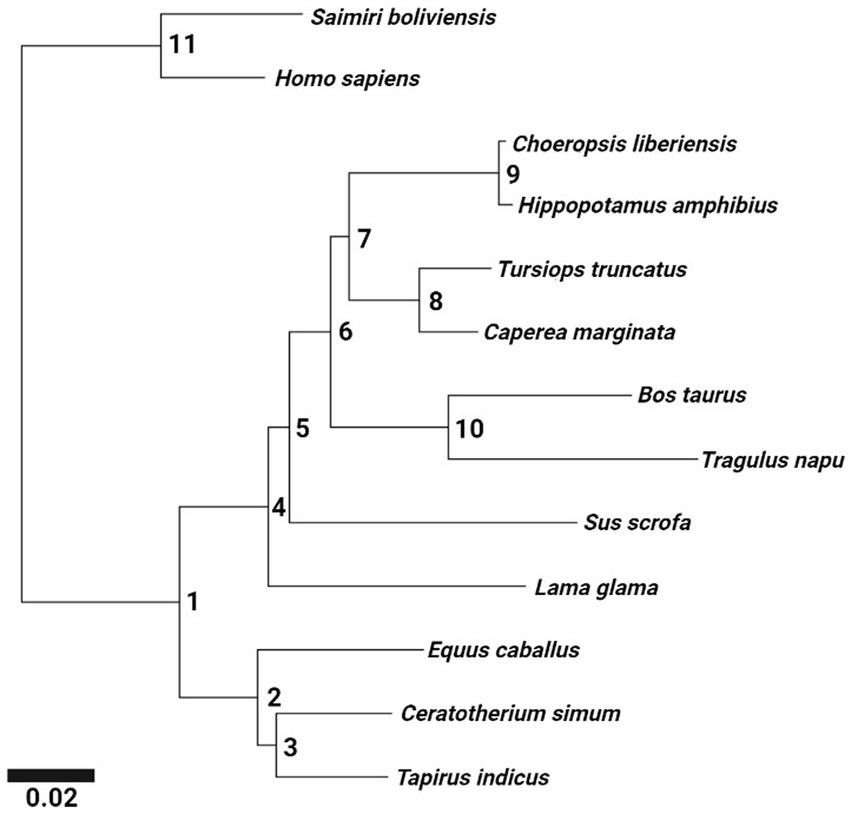
Figure 3. Ungulate (Cetartiodactyla + Perissodactyla) phylogeny generated from the Bayesian analysis (MrBayes 3.2.7a) displaying relative branch lengths. Two primates (Saimiri boliviensis and Homo sapiens) were included as an outgroup. All nodes received clade credibility values of 100. Nodes are numbered to facilitate referencing in the text. Exported from MEGA-X and edited in BioRender.
3.2. Divergence date estimations
As mentioned previously, most major cetartiodactyl lineages are believed to have diverged within a relatively short window of about 14 million years (Figure 4) (Meredith et al., 2011; O’Leary et al., 2013; Zurano et al., 2019). For example, fossil calibrations used in this study for Cetartiodactyla (Node 4 in Figure 3) and Cetruminantia (Node 6 in Figure 3) were nearly identical at 65.8–52.5 and 66.0–52.4 million years ago, respectively. The divergence date for Artiofabula (Node 5 in Figure 3) would therefore also be expected to fall within this same range as Suina diverged from Cetruminantia after Tylopoda (Price et al., 2005; Meredith et al., 2011). Based on the molecular matrix and fossil calibrations used in this study, a divergence date of 4.04 Ma (95% confidence interval: 8.31–1.97 Ma) was estimated using RelTime-ML. The estimated divergence date from the MCMC tree was younger still at roughly 2.4 Ma (Supplementary Figure S1). The 95% confidence interval for this estimate ranged from 3.1–1.6 Ma (Supplementary Figure S2).
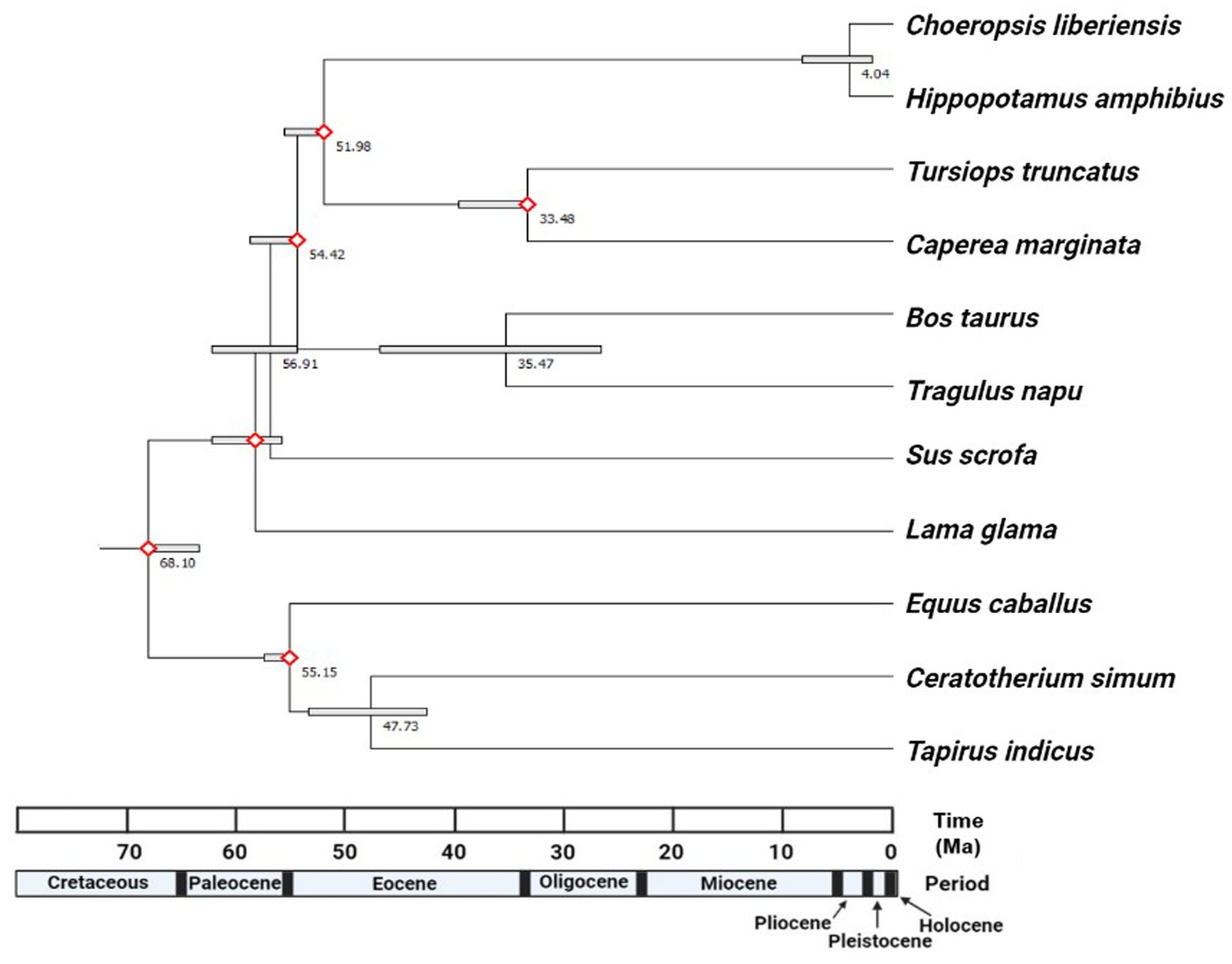
Figure 4. Time tree generated via RelTime-ML (in MEGA-X) from the results of our phylogenetic analysis with six fossil calibrations. Divergence dates for each ungulate node are listed in millions of years ago (Ma). Nodes calibrated with fossils are indicated by hollow red diamonds. Bars around each node represent 95% confidence intervals for divergence date estimates. Exported from MEGA-X and edited in BioRender.
4. Discussion
4.1. Current understanding of hippopotamid phylogeny
Before discussing the results and implications of our study, we first describe our current understanding of the phylogeny of Hippopotamidae in greater detail. This family is commonly divided into two subfamilies, the more ancient and primitive Kenyapotaminae and the more recent and derived Hippopotaminae (Pickford, 1983; Harris et al., 2008; Orliac et al., 2010; Boisserie et al., 2011). By the end of the 20th century, only two of the five genera currently placed within Hippopotaminae were recognized: Hippopotamus (H.) and Hexaprotodon (Hex.) (Boisserie, 2005). While the pygmy hippopotamus had been discovered and named Choeropsis liberiensis more than a century earlier, it had been reclassified as a species of Hexaprotodon (Hex. liberiensis) in 1977 based on similarities in skull anatomy between the two genera (Coryndon, 1977). While later examinations found enough differences to revalidate Choeropsis as a distinct genus, some authors still refer to the pygmy hippopotamus as Hex. liberiensis (Boisserie, 2005). By 2005, at least one dozen extinct species of Hexaprotodon had been described, and the genus was believed to be paraphyletic, with Hippopotamus nested within Hexaprotodon (Weston, 2000; Boisserie and White, 2004; Boisserie, 2005). However, reevaluation of some of these species by Boisserie (2005) led to the reclassification of three Hexaprotodon species into two newly established genera: Archaeopotamus and Saotherium. Proposed relationships and temporal distributions between these five genera along with Kenyapotamus of Kenyapotaminae were consolidated from the sources referenced in this paragraph (Figure 2).
Today, the African H. amphibius is the sole surviving Hippopotamus species, but extinct members of this genus had spread into Eurasia as well (Petronio, 1995; Martino et al., 2021). Hippopotamus likely first appeared around 4 Ma during the Early Pliocene of east Africa, though the exact date is difficult to determine based on fragmentary remains from the earliest Pliocene (Boisserie, 2005). Hexaprotodon now refers mainly to extinct species of Asian hippopotamids, some of which could have survived into the earliest Holocene roughly 10,000 years ago (Jukar et al., 2019). This genus likely originated at least 7 Ma with the discovery of Hex. garyam in Central Africa and had reached southern Europe by about 6 Ma with the discovery of material currently described as a species of Hexaprotodon (Martino et al., 2021). Most of the remaining African species of Hexaprotodon were reclassified as species of Hippopotamus by Boisserie (2005), but at least one species (Hex. bruneti) returned to Africa from Asia around 2.5 Ma (Boisserie and White, 2004). Unlike the common hippopotamus, fossils of which date back to roughly 2 Ma, the pygmy hippopotamus is completely absent from the fossil record (Boisserie, 2005). Fossils of Saotherium mingoz (the only species of this genus) ranging from about 5.3–4 million years old have been discovered in the central African nation of Chad (Boisserie, 2005). Archaeopotamus first appeared in Kenya at least 7.5 Ma and survived into the Early Pliocene, spreading as far away as Abu Dhabi in the Middle East (Boisserie et al., 2005; Boisserie, 2007).
Boisserie (2005) was essentially the only study to suggest a Saotherium-Choeropsis lineage, a cautious interpretation that appears in many subsequent analyses but is rarely discussed. The tentative grouping of these species as a sister group to the remaining members of Hippopotaminae is based on similarities in cranial and dental anatomy, including primitive traits such as large orbits placed laterally on their skulls and a lack of strong canine processes in both Saotherium and the pygmy hippopotamus (Boisserie, 2005). Beyond these, however, Boisserie (2005) noted that there is little other evidence that specifically supports a Saotherium-Choeropsis lineage. Ultimately, intrafamilial relationships within Hippopotamidae are vastly understudied and the phylogeny of this family lacks a clear consensus. The phylogenetic placement of C. liberiensis has not been a major focus of morphological studies since Boisserie (2005) and remains difficult to assess with confidence. Hippopotamidae has never been the focus of a rigorous phylogenetic analysis combining both morphological and molecular evidence. A study of this nature could, for the first time, integrate molecular data with morphological evidence in a study of hippopotamid phylogeny while incorporating morphological discoveries since 2005.
The taxonomic position of C. liberiensis is complicated by the mixture of primitive and derived features of this animal (Boisserie, 2005; Flacke and Decher, 2019). The pygmy hippopotamus has a diprotodont lower jaw and a short mandibular symphysis, both of which are considered derived characteristics (Boisserie, 2005). Due to the lack of pygmy hippopotamus fossils, it remains unclear as to whether the primitive characteristics of this species are relics from the earliest hippopotamids or have evolved convergently to suit the secluded lifestyle that might have been shared by C. liberiensis and the earliest hippopotamids. Boisserie discussed these two potential evolutionary histories, suggesting that the former is more generally accepted but the latter is possible and carries major implications on hippopotamid phylogeny if evidence is found in support of it (Boisserie, 2007; Boisserie et al., 2011). The more recent divergence date estimated in our study gives reason to further investigate Boisserie’s second hypothesis, that C. liberiensis adapted secondarily to life in tropical rainforests and is a younger specialized hippopotamid as opposed to an ancient genus (Boisserie et al., 2011). While the pygmy hippopotamus is terrestrial, the sensitive skin and watertight nose and ears of this species represent physiological traits shared with the semiaquatic common hippopotamus (Boisserie, 2007). These traits suggest a semiaquatic common ancestor to Hippopotamidae and provide more evidence that the terrestrial nature of C. liberiensis is a derived rather than ancestral trait (Boisserie et al., 2011).
4.2. Comparison of estimated divergence dates with previous studies
The results of our study estimated the divergence date between H. amphibius and C. liberiensis to about 4.04 Ma (95% confidence interval: 8.31–1.97 Ma) via RelTime-ML or 2.4 Ma (95% confidence interval: 3.1–1.6 Ma) via MCMCTree. Tree topology was identical in every analysis performed and congruent with our current understanding of ungulate phylogeny (Meredith et al., 2011). The more ancient divergence date (and broader confidence interval) estimated in RelTime-ML is likely due to the fossil calibrations used to generate the RelTime-ML estimate. Of six calibrations, five range from 60–56 Ma (confidence intervals extend this range to 66–50 Ma). These dates are substantially older than the expected divergence date range for the common and pygmy hippopotamus (i.e., no older than 10 Ma) and likely pulled the estimate for this divergence date back in time slightly, making it appear more ancient than it actually is. The influence of using different fossil calibrations will be another major component of future studies incorporating both extinct and extant taxa. With only two extant hippopotamids, the most recent fossil calibration for this lineage used in this study was for the divergence between Hippopotamidae and Cetacea c. 57 Ma. The incorporation of extinct taxa in a combined analysis would allow the hippopotamid lineage to be better calibrated than it could be using only extant taxa. However, both the RelTime-ML and MCMCTree estimates from this study are distinctly younger than most previous estimates and support a an Early Pliocene – Early Pleistocene divergence date for H. amphibius and C. liberiensis, giving reason for future studies to evaluate Boisserie’s (2007) hypothesis that the pygmy hippopotamus may have secondarily evolved for a terrestrial lifestyle more recently than expected.
Boisserie (2005) (and all subsequent morphological studies referencing the Saotherium-Choeropsis lineage tentatively proposed therein) predicted a divergence date for the two extant hippopotamids of roughly 8–5.3 Ma during the Late Miocene. This general range spans the time between the early radiation of Hippopotaminae (roughly 8–7.5 Ma) and the first appearance datum (FAD) of Saotherium of about 5.3 Ma (Boisserie, 2005; Boisserie et al., 2011). This range reflects the hypothesis that the primitive traits of C. liberiensis are due to an ancient divergence from the rest of Hippopotaminae rather than a more recent secondary adaptation to terrestrial life as predicted by Boisserie’s (2007) alternative hypothesis and the results of this study.
Molecular studies of the pygmy hippopotamus have been few in number up to this point. We are aware of three studies that incorporated mitogenomic data (Montgelard et al., 1997; Hassanin et al., 2012; Zurano et al., 2019), one that utilized transcriptomic data (Figuet et al., 2021), one that focused on ten nuclear genes of interest (Springer et al., 2021), and one that utilized whole genomic data (McGowen et al., 2020). Among these, only Springer et al. (2021) placed special emphasis on the pygmy hippopotamus.
Montgelard et al. (1997) used cytochrome b and 12S rRNA mitochondrial gene sequences to predict cetartiodactyl divergence dates using a local molecular clock as opposed to the relaxed molecular clock approach used in this study. Their analysis predicted a divergence date of roughly 5.7 Ma for the common and pygmy hippopotamus (Montgelard et al., 1997). However, 12S rRNA gene sequences failed to support a monophyletic Cetancodonta (Montgelard et al., 1997). Hassanin et al. (2012) and Zurano et al. (2019) used complete cetartiodactyl mitogenomes to calculate divergence dates. Hassanin et al. (2012) estimated the divergence date for H. amphibius and C. liberiensis to range from roughly 9–7 Ma, while Zurano et al. (2019) estimated a date of 8.7–7.6 Ma. Both analyses recovered support for Cetruminantia and Cetancodonta, but not Artiofabula, finding Suina to diverge before Tylopoda (Hassanin et al., 2012; Zurano et al., 2019). Artiofabula monophyly seems to receive support from analyses using nuclear DNA, but is rejected by analyses using mitochondrial DNA (Zurano et al., 2019).
McGowen et al. (2020) estimated a divergence date of roughly 7–4 Ma for H. amphibius and C. liberiensis based on how the data set was partitioned and what evolutionary models were applied (i.e., autocorrelated or independent rate models), with most estimates falling near the earlier end of this range. The overwhelming majority of taxa surveyed in this study (i.e., at least 70 of roughly 80 species) are cetaceans. Given the slower evolutionary rate of Cetacea compared to Hippopotamidae, the inclusion of so much cetacean data and fossil calibrations could predict a more ancient divergence date for the common and pygmy hippopotamus (as with the RelTime-ML estimate in this study) (McGowen et al., 2020). Springer et al. (2021) referenced the time tree published in McGowen et al. (2020) and did not perform any new divergence date analyses in their study. McGowen et al. (2020) used roughly 3,200 protein-coding genes while we used only 26. While a much smaller number, our set of 26 gene segments was specifically selected based on Meredith et al. (2011) for their stable rates of evolution across all mammalian orders, producing a strong phylogenetic signal capable of predicting divergence dates with a high degree of accuracy. However, substantial overlap exists between the range of McGowen et al.’s estimates and the confidence intervals of the RelTime-ML analysis of this study for the common and pygmy hippopotamus divergence, with most divergence dates estimated by McGowen et al. (2020) falling within the Late Miocene – Early Pliocene while the estimates of this study fall within the Early Pliocene – Early Pleistocene sub-epochs.
Divergence date estimates for H. amphibius and C. liberiensis based on a transcriptomic analysis by Figuet et al. (2021) are also more ancient, ranging from 8.5–7.1 Ma (coevol analysis) to as old as 9.6 Ma (TimeTree). The span of mitogenome/transcriptome-based estimates extend the range of the H. amphibius – C. liberiensis divergence date even further back in time to almost 10 Ma, older than the radiation of Hippopotaminae estimated to begin around 8 Ma based on morphological evidence and the fossil record (Boisserie et al., 2011). The range of nuclear gene-based estimates incorporates dates more recent than the FAD of Saotherium, spanning from roughly 4 Ma (McGowen et al., 2020) to younger still with our results. Our estimates align most closely with those of McGowen et al. (2020), the only other study we are aware of that used genomic data to estimate this divergence date. The estimate of 5.7 Ma by Montgelard et al. (1997) is the next closet molecular estimate to our results, despite the inability of the 12S rRNA gene to recover a monophyletic Cetancodonta without incorporation of the cytochrome b gene. The variation in estimates and tree topology across these studies based on the type of sequence data used (i.e., mitochondrial genes, whole mitogenomes, large genomic, small genomic, or transcriptomic) underscores the need for future studies to evaluate the effects of using different types of data and methods of data partitioning on estimating the H. amphibius – C. liberiensis divergence date.
4.3. Influence of types of data used in phylogenetic analyses
Some studies have noted incongruences in generating phylogenies or estimating divergence dates when using nuclear versus mitogenomic sequences (DeBry and Seshadri, 2001; Springer et al., 2001; Kjer and Honeycutt, 2007; Zhou et al., 2011). In particular, Artiofabula monophyly is often supported by analyses of nuclear gene sequences (e.g., Meredith et al., 2011) but rejected by analyses of mitochondrial gene sequences (e.g., Hassanin et al., 2012). In most cases, nuclear data seems to better encapsulate the most widely accepted topology for Cetartiodactyla (Hassanin and Douzery, 2003; Willows-Munro et al., 2005; Velazco et al., 2022). However, this provides another example of the influence of the type of data used to generate phylogenies on resulting tree topologies. A critical future direction will be to further characterize this variation when using different types of data to predict cetartiodactyl relationships and divergence dates.
We argue that the most comprehensive approach to estimate the divergence date for the common and pygmy hippopotamus would incorporate molecular evidence for extant species as well as morphological evidence for both extant and extinct species. A matrix with multiple sources of data for as many extant cetartiodactyls and their extinct relatives as possible would fill gaps in our understanding of cetartiodactyl evolution. Our study may therefore be limited by the inclusion of so few cetartiodactyl species (only eight in total) with a dataset lacking morphological data and extinct cetartiodactyls. However, the inclusion of 26 nuclear gene segments totaling 31,890 nucleotides for each species should represent a robust dataset that establishes the significance of our study.
Our results call for future studies to give hippopotamid evolution the same treatment as cetacean phylogenetic relationships, i.e., the production of dedicated data matrices comprising both molecular and morphological data to evaluate the relative contributions and methods of partitioning for both (O’Leary, 1999). If morphological traits are under strong selection in the pygmy hippopotamus, this study could be the first evidence of accelerated morphological evolution in C. liberiensis and a closer relationship with H. amphibius than previously predicted. This would support the hypothesis of Boisserie (2007) and Boisserie et al. (2011) that C. liberiensis is a specialized hippopotamid that has secondarily adapted to a terrestrial lifestyle, though this could not be confirmed without morphological evidence in the form of pygmy hippopotamus fossils.
4.4. Closing remarks on hippopotamid phylogeny
Our results contradict the idea of a Saotherium-Choeropsis lineage (based on the first appearance datum of Saotherium around 5.3 Ma) and reveal that H. amphibius and C. liberiensis are closer phylogenetically than previously hypothesized. These findings echo one of Boisserie’s hypotheses that the pygmy hippopotamus has secondarily evolved a terrestrial lifestyle after diverging from a semiaquatic ancestor, and warrant further investigation of this potential evolutionary path (Boisserie et al., 2011). However, the H. amphibius – C. liberiensis divergence date and the phylogeny of Hippopotamidae at large remain difficult to resolve. The Early Pliocene – Early Pleistocene divergence date estimates for the two extant hippopotamid species based on molecular data in this study are more recent than most previous estimates. We propose that the two extant hippopotamids are closer phylogenetically than once believed based on greater than expected sequence similarity. However, without fossil evidence of a direct pygmy hippopotamus ancestor or a consensus of molecular and morphological studies, these questions remain unanswered. Limited morphological evidence due to the gap in the fossil record for C. liberiensis means that the last common ancestor of the two extant hippopotamid species still cannot be identified with confidence. It is also possible that a repeat of this study incorporating more cetartiodactyl species and morphological data could generate a different divergence date and support or conflict with the conclusions of this or other studies. More phylogenetic studies including molecular data for the pygmy hippopotamus will be of the utmost importance in resolving the taxonomic relationships and placement of this species. It is our hope that this study inspires a new era of hippopotamid research characterized by combined matrices of morphological and molecular data with both extinct and extant taxa to evaluate the evolutionary history of this family.
Data availability statement
The original contributions presented in the study are included in the Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MK extracted consensus sequences from the C. liberiensis whole genome sequence, performed the phylogenetic analyses and wrote the first and revised drafts of the manuscript. SV collected and organized genomic data from NCBI. JJ conceived the project, contributed to compiling sequences, phylogenetic analyses, and writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by NSF DEB Award #1654964 to JJ, John Wible, Thomas Williamson, Steve Brusatte, and Michelle Spaulding.
Acknowledgments
We would like to thank John Wible (Carnegie Museum of Natural History), SV, Manuel Frontera, David Negron, Maya Joiner, and Melanie Quain for assistance with this project. We would also like to thank the reviewers of this manuscript, whose thoughtful and thorough feedback greatly improved the quality of early drafts.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1144711/full#supplementary-material
References
Boisserie, J.-R. (2005). The phylogeny and taxonomy of hippopotamidae (mammalia: artiodactyla): a review based on morphology and cladistic analysis. Zool. J. Linnean Soc. 143, 1–26. doi: 10.1111/j.1096-3642.2004.00138.x
Boisserie, J. R. (2007). “Family Hippopotamidae” in The evolution of artiodactyls. eds. D. R. Prothero and S. E. Foss (Baltimore: The Johns Hopkins University Press).
Boisserie, J.-R. (2017). Hippopotamidae (Cetartiodactyla, Hippopotamoidea) from Kanapoi, Kenya, and the taxonomic status of the early Pliocene hippopotamids from the Turkana Basin. J. Hum. Evol. 140:102377. doi: 10.1016/j.hevol.2017.07.017
Boisserie, J.-R., Fisher, R. E., Lihoreau, F., and Weston, E. M. (2011). Evolving between land and water: key questions on the emergence and history of the Hippopotamidae (Hippopotamoidea, Cetancodonta, and Cetartiodactyla). Biol. Rev. 86, 601–625. doi: 10.1111/j.1469-185X.2010.00162.x
Boisserie, J.-R., and Lihoreau, F. (2006). Emergence of hippopotamidae: new scenarios. (2005). C. R. Palevol. 5, 749–756. doi: 10.1016/j.crpv.2005.11.004
Boisserie, J.-R., Lihoreau, F., and Brunet, M. (2005). The position of hippopotamidae within cetartiodactyla. PNAS 102, 1537–1541. doi: 10.1073/pnas.0409518102
Boisserie, J.-R., and White, T. D. (2004). A new species of Pliocene hippopotamidae from the middle awash. Ethiopia. J. Vertebr. Paleontol. 24, 464–473. doi: 10.1671/2475
Davis, B. W., Li, G., and Murphy, W. J. (2010). Supermatrix and species tree methods resolve phylogenetic relationships within the big cats, Panthera (Carnivora: Felidae). Mol. Phylogenet. Evol. 56, 64–76. doi: 10.1016/j.ympev.2010.01.036
DeBry, R. W., and Seshadri, S. (2001). Nuclear intron sequences for phylogenetics of closely related mammals: an example using the phylogeny of Mus. J. Mammal. 82, 280–288. doi: 10.1644/1545-1542(2001)082<0280:NISFPO>2.0.CO;2
Figuet, E., Ballenghien, M., Lartillot, N., and Galtier, N. (2021). Reconstruction of body mass evolution in the Cetartiodactyla data. PCI. Evol. Biol. 1:e42. doi: 10.24072/pcjournal.55
Fisher, R. E., Scott, K. M., and Naples, V. L. (2007). Forelimb myology of the pygmy hippopotamus (Choeropsis liberiensis). Anat. Rec. 290, 673–693. doi: 10.1002/ar.20531
Flacke, G. L., and Decher, J. (2019). Choeropsis liberiensis (Artiodactyla: Hippopotamidae). Mamm. Species 51, 100–118. doi: 10.1093/mspecies/sez017
Geisler, J. H., Theodor, J. M., Uhen, M. D., and Foss, S. E. (2007). “Phylogenetic relationships of cetaceans to terrestrial artiodactyls” in The evolution of artiodactyls. eds. D. R. Prothero and S. E. Foss (Baltimore: The Johns Hopkins University Press).
Harris, J. M., Cerling, T. E., Leakey, M. G., and Passey, B. H. (2008). Stable isotope ecology of fossil hippopotamids from the Lake Turkana Basin of East Africa. J. Zool. 275, 323–331. ISSN: 0952-8369. doi: 10.1111/j.1469-7998.2008.00444.x
Hassanin, A., and Douzery, E. J. P. (2003). Molecular and morphological phylogenies of ruminantia and the alternative position of the moschidae. Syst. Biol. 52, 206–228. doi: 10.1080/10635150390192726
Hassanin, A., Delsuc, F., Ropiquet, A., Hammer, C., Jansen van Vuuren, B., Matthee, C., et al. (2012). Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C. R. Biol. 335, 32–50. doi: 10.1016/j.crvi.2011.11.002
Jukar, A. M., Patnaik, R., Chauhan, P. R., Li, H. C., and Lin, J. P. (2019). The youngest occurrence of Hexaprotodon falconer and Cautley, 1836 (Hippopotamidae, Mammalia) from South Asia with a discussion on its extinction. (2019). Quat. Int. 528, 130–137. doi: 10.1016/j.quaint.2019.01.005
Kjer, K. M., and Honeycutt, R. L. (2007). Site specific rates of mitochondrial genomes and the phylogeny of eutheria. BMC Evol. Biol. 7:8. doi: 10.1186/1471-2148-7-8
Martino, R., Pignatti, J., Rook, L., and Pandolfi, L. (2021). Hippopotamid dispersal across the Mediterranean in the latest Miocene: a re-evaluation of the Gravitelli record from Sicily. Italy. Acta Palaeontol. Pol. 66, S67–S78. doi: 10.4202/app.00838.2020
McGowen, M. R., Tsagkogeorga, G., Álvarez-Carretero, S., Dos Reis, M., Struebig, M., Deaville, R., et al. (2020). Phylogenomic resolution of the cetacean tree of life using target sequence capture. Syst. Biol. 69, 479–501. doi: 10.1093/sysbio/syz068
Meredith, R. W., Janečka, J. E., Gatesy, J., Ryder, O. A., Fisher, C. A., Teeling, E. C., et al. (2011). Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524. doi: 10.1126/science.1211028
Montgelard, C., Catzeflis, F. M., and Douzery, E. (1997). Phylogenetic relationships of artiodactyls and cetaceans as deduced from the comparison of cytochrome b and 12S rRNA mitochondrial sequences. Mol. Biol. Evol. 14, 550–559. doi: 10.1093/oxfordjournals.molbev.a025792
Naylor, G. J., and Adams, D. C. (2001). Are the fossil data really at odds with the molecular data? Morphological evidence for cetartiodactyla phylogeny reexamined. Syst. Biol. 50, 444–453. doi: 10.1080/10635150118184
O’Leary, M. A. (1999). Parsimony analysis of total evidence from extinct and extant taxa and the cetacean – artiodactyl question (Mammalia, Ungulata). Cladistics 15, 315–330. doi: 10.1111/j.1096-0031.1999.tb00269.x
O’Leary, M. A., Bloch, J. I., Flynn, J. J., Gaudin, T. J., Giallombardo, A., Giannini, N. P., et al. (2013). The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339, 662–667. doi: 10.1126/science.1229237
Orliac, M., Boisserie, J.-R., MacLatchy, L., and Lihoreau, F. (2010). Early miocene hippopotamids (cetartiodactyla) constrain the phylogenetic and spatiotemporal settings of hippopotamid origin. PNAS 107, 11871–11876. doi: 10.1073/pnas.1001373107
Pickford, M. (1983). On the origins of hippopotamidae together with descriptions of two new species, a new genus and a new subfamily from the Miocene of Kenya. Geobios 16, 193–217. doi: 10.1016/S0016-6995(83)80019-9
Price, S. A., Bininda-Emonds, O. R. P., and Gittleman, J. L. (2005). A complete phylogeny of the whales, dolphins, and even-toed hoofed mammals (cetartiodactyla). Biol. Rev. 80, 445–473. doi: 10.1017/S1464793105006743
Springer, M. S., DeBry, R. W., Douady, C., Amrine, H. M., Madsen, O., de Jong, W. W., et al. (2001). Mitochondrial versus nuclear gene sequences in deep-level mammalian phylogeny reconstruction. Mol. Biol. Evol. 18, 132–143. doi: 10.1093/oxfordjournals.molbev.a003787
Springer, M. S., Emerling, C. A., Meredith, R. W., Janečka, J. E., Eizirik, E., and Murphy, W. J. (2017). Waking the undead: implications of a soft explosive model for the timing of placental mammal diversification. Mol. Phylogenet. Evol. 106, 86–102. doi: 10.1016/j.ympev.2016.09.017
Springer, M. S., Foley, N. M., Brady, P. L., Gatesy, J., and Murphy, W. J. (2019). Evolutionary models for the diversification of placental mammals across the KPg boundary. Front. Genet. 10:1241. doi: 10.3389/fgene.2019.01241
Springer, M. S., Guerrero-Juarez, C. F., Huelsmann, M., Collin, M. A., Danil, K., McGowen, M. R., et al. (2021). Genomic and anatomical comparisons of skin support independent adaptation to life in water by cetaceans and hippos. Curr. Biol. 31, 2124–2139.e3. doi: 10.1016/j.cub.2021.02.057
Symonds, M. (2005). Phylogeny and life histories of the ‘Insectivora’: controversies and consequences. Biol. Rev. 80, 93–128. doi: 10.1017/S1464793104006566
Theodor, J. M. (2004). Molecular clock divergence estimates and the fossil record of Cetartiodactyla. J. Paleontol. 78, 39–44. doi: 10.1666/0022-3360(2004)078<0039:MCDEAT>2.0.CO;2
Velazco, P. M., Buczek, A. J., Hoffman, E., Hoffman, D. K., O’Leary, M. A., and Novacek, M. J. (2022). Combined data analysis of fossil and living mammals: a Paleogene sister taxon of Placentalia and the antiquity of Marsupialia. Cladistics 38, 359–373. doi: 10.1111/cla.12499
Weston, E. M. (2000). A new species of hippopotamus hexaprotodon lothagamensis (mammalia: hippopotamidae) from the late Miocene of Kenya. J. Vertebr. Paleontol. 20, 177–185. doi: 10.1671/0272-4634(2000)020[0177:ANSOHH]2.0.CO;2
Willows-Munro, S., Robinson, T. J., and Matthee, C. A. (2005). Utility of nuclear DNA intron markers at lower taxonomic levels: phylogenetic resolution among nine Tragelaphus spp. Mol. Phylogenet. Evol. 35, 624–636. doi: 10.1016/j.ympev.2005.01.018
Zhou, X., Xu, S., Yang, Y., Zhou, K., and Yang, G. (2011). Phylogenomic analyses and improved resolution of Cetartiodactyla. Mol. Phylogenet. Evol. 61, 255–264. doi: 10.1016/j.ympev.2011.02.009
Keywords: Cetartiodactyla, divergence date estimation, Hippopotamidae, Hexaprotodon, Choeropsis liberiensis, Hippopotamus, Early Pliocene – Early Pleistocene
Citation: Kardos MC, Velmurugan S and Janecka JE (2023) Molecular data suggests a Pliocene – Early Pleistocene divergence date for the common and pygmy hippopotamus. Front. Ecol. Evol. 11:1144711. doi: 10.3389/fevo.2023.1144711
Edited by:
Daniele Salvi, University of L’Aquila, ItalyReviewed by:
Robert William Meredith, Montclair State University, United StatesJean-Renaud Boisserie, CNRS, France
Copyright © 2023 Kardos, Velmurugan and Janecka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan E. Janecka, amFuZWNrYWpAZHVxLmVkdQ==
 Michael C. Kardos
Michael C. Kardos Sindhu Velmurugan
Sindhu Velmurugan Jan E. Janecka
Jan E. Janecka