
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 22 March 2023
Sec. Evolutionary and Population Genetics
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1142810
Understanding phylogeographic patterns of widespread species can provide insights into their speciation processes and guide the conservation and management measures. In the present study, Cyt b sequences were used to investigate the phylogeographic structure of the Opsariichthys acutipinnis-evolans complex in southeastern China. The gene tree revealed six major lineages (lineage A-F) which were distributed allopatrically, with lineage B distributed in the western part (middle Yangtze and Pearl River) and the other lineages in the eastern part (lower Yangtze and coastal waters of southeastern China). Dating of the lineage diversification revealed the early eastward-westward divergence separating lineage A, B, and C during the late Pliocene and early Pleistocene (3.00, 2.61, and 2.12 Ma, respectively), possibly due to the uplift of the Qinghai-Tibetan Plateau and subsequent orogenies in southeastern China. The following northward-southward diversification resulted in the separation of lineage D, E, and F in the early-middle Pleistocene (1.33 and 0.95 Ma), likely associated with the enhanced succession of glacial cycles during the Early-Middle Pleistocene transition. Although the genetic divergence of 0.017–0.070 among lineages indicated possible different species, morphological characters failed to separate them. Therefore, they were treated as a species complex. Given the distinct genetic divergence of the various lineages, they were suggested as different evolutionary significant units.
Understanding the spatial arrangements of genetic variations within populations or among closely related species is a major objective of phylogeography (e.g., Avise et al., 1987; Avise, 2009). Interpretations of phylogeographic patterns and genetic structures provide insights into speciation processes and guidance for conservation and management decisions (Avise, 2000). For example, population genetic and phylogeographic studies revealed that the widespread but endangered Chinese giant salamander (Andrias davidianus) actually harbors seven distinct genetic lineages that might represent at least five different species (Murphy et al., 2000; Yan F. et al., 2018; Liang et al., 2019; Chai et al., 2022), and the lack of awareness of the existence of these cryptic lineages misguided conservation efforts in the past decades. In contrast, genetic assessment of the economically important Lake Tanganyika sprat Stolothrissa tanganicae indicates this species lacks population structure and suggests an integrated stock management strategy (De Keyzer et al., 2019). Understanding the genetic variations and structures of many other species also enhanced our knowledge of the divergence patterns and offered management suggestions (e.g., Van Rossum et al., 2018; Guo et al., 2019; Rincon-Sandoval et al., 2019; Tumendemberel et al., 2019). These findings further emphasize the necessity of genetic evaluations to determine the taxonomic status and identify evolutionary significant units (ESUs) in conservation initiatives, especially for seemingly widespread species (e.g., Taylor et al., 2011; Larson et al., 2014; Yan F. et al., 2018; Li et al., 2020; Li C. et al., 2023).
Opsariichthys acutipinnis and O. evolans are two small riverine fishes widely distributed throughout southeastern China, including the Yangtze River basin, Pearl River basin and independent coastal river basins like the Qiantangjiang River, Minjiang River, and Hanjiang River. These fishes prefer lotic environments with gravel and pebble substrates, and are most common in mountain streams and small tributaries (Chen and Chu, 1998). Opsariichthys acutipinnis was first described by Bleeker in 1871 as Barilius acutipinnis based on a specimen from the Yangtze River (Bleeker, 1871), and then being transferred to the genus Opsariichthys without any explanation (Günther, 1873). Later in 1902, Jordan and Evermann established the genus Zacco to include fishes originally assigned to the genus Opsariichthys, but without conspicuous notched jaws (Jordan and Evermann, 1902). They also described Zacco evolans from the Taiwan Island. Later studies generally treated Opsariichthys acutipinnis and Opsariichthys evolans as synonyms of Opsariichthys platypus (Rendahl, 1928) or Zacco platypus (Oshima, 1919; Nichols, 1943; Yang and Huang, 1964), but did not reach any consensus. In 1982, Chen (1982) revised the taxonomy of opsariichthyine fishes and agreed that the notched jaws could distinguish the genus Opsariichthys and Zacco, thus he considered both O. acutipinnis and O. evolans should be member of the genus Zacco and be synonymized with Zacco platypus. However, some later studies based on osteological or molecular evidence suggest that the notched jaws might not be appropriate to serve as diagnostic feature (Howes, 1980; Ashiwa and Hosoya, 1998; Chen and Chang, 2005; Ma et al., 2006). Chen et al. (2009) suggested that the genus Zacco can be differed from Opsariichthys by vertical bars on flanks fused into patches (versus distinct vertical bars not fused into patches in Opsariichthys) and nuptial tubercles on cheeks in male fused into a plate basally (versus tubercles small and not fused into a plate in Opsariichthys). Thus the species previously identified as Zacco platypus from southern China with such features should be regarded as members of Opsariichthys acutipinnis-O. evolans complex. Later studies generally treated O. acutipinnis and O. evolans as two closely related but distinct species based on deep divergence in mitochondrial gene (s), allopatric distribution, and morphological differences (Huynh and Chen, 2013; Wang et al., 2019). A recent work conducted by Wang et al. (2019) revealed a deeply divergent mitochondrial lineage that related to O. acutipinnis and O. evolans (“O. sp. A”), indicating the existence of previously ignored cryptic diversity. In the past decade our extensive sampling work throughout southeastern China has yielded plenty of morphologically similar specimens that could be identified as either O. acutipinnis and O. evolans. However, we found out that morphological identification based on diagnostic characteristics proposed by Huynh and Chen (2013) and Wang et al. (2019) would be problematic, e.g., specimens with features of O. acutipinnis and O. evolans co-existed in most populations. As a consequence, we tentatively treated these fishes as the Opsariichthys acutipinnis-evolans complex in the current study.
Plenty of studies have suggested that primary freshwater fishes with resident lifestyles always exhibit highly structured genetic patterns between different drainage basins or even different stretches of a river because of their typically low mobility and preference for certain habitats (e.g., Chen et al., 2007; Lin et al., 2008; Huang et al., 2019; Won et al., 2020). For example, populations of the cold-water fish Rhynchocypris oxycephalus in China exhibited deep genetic structure, aside from the geological factors, the low dispersal ability and preference for headwater streams are important contributors to the current divergence patterns (Yu et al., 2014). Prominent genetic differentiation was detected from the stream goby populations within fine-scale regions, and that both paleodrainage alternations and the sedentary lifestyle of this species are major factors in generating the strong genetic structure (Wu et al., 2016). Cases like these reinforce the impression that primary freshwater fishes are ideal candidates to investigate the biological consequences of complex geological changes. The O. acutipinnis-evolans complex is a group of small riverine fishes widely distributed throughout southeastern China, therefore they can be great models to explore how geological and climatic changes affect their genetic makeups and distribution patterns. However, few studies (Perdices et al., 2004; Perdices and Coelho, 2006; Lin, 2008; Yin, 2015) have addressed the genetic differentiation and phylogeographic structure of this species complex due to long term neglect in taxonomy, which hindered us from understanding the diversity and evolutionary history of these small stream fishes, as well as making proper management decisions. In the present study, we used both mitochondrial Cyt b sequences and 25 morphological traits to assess the genetic and phenotypic differentiation among populations of O. acutipinnis-evolans complex in southeastern China. Then we aim to investigate the phylogeographic structure and explore the potential factors that have shaped the patterns of genetic variability. Finally, we wish our phylogeographic evaluation could offer some implications for the conservation and management of this species.
A total of 821 specimens were collected from 36 localities in southeastern China, covering the Yangtze River basin, Pearl River basin, and several small coastal river basins such as the Qiantangjiang River, the Minjiang River and the Hanjiang River (Supplementary Table S1). Fishes were captured using dip net, gill net, and electrofishing between 2009 and 2020. Due to the lack of a generally acceptable classification system, we treated all our Opsariichthys specimens with the following characteristics as Opsariichthys acutipinnis-evolans complex: (1) mouth without conspicuous notch; (2) lateral line scale count lower than 45; (3) tubercles round and small, usually arranged in one row on the lower jaw. Voucher specimens were preserved in 95% ethanol at 4°C. All voucher specimens were deposited at the Institute of Hydrobiology, Chinese Academy of Sciences.
Total genomic DNA was extracted either from a small piece of flesh or pelvic fin clip taken from the right side of each specimen, following the salt-extraction methods (Aljanabi, 1997). The complete mitochondrial cytochrome b (Cyt b) gene was amplified for all the samples with the universal primers L14724 and H15915 (Xiao et al., 2001). Amplification was conducted in a 30-μL reaction volume, containing 11 μl of dd H2O, 15 μl of 2 × MIX, 1 μl of each primer, and 2 μl of template DNA. The thermocycling program for the polymerase chain reaction (PCR) consisted of an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 54°C for 45 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. PCR products were sent to commercial sequencing company (Sangon Biotech, Shanghai) for purifying and sequencing with the same PCR primers.
We sequenced 622 samples and retrieved 199 sequences from our previous study (Wang et al., 2019). We also included another 193 sequences from GenBank submitted by previous authors (Perdices et al., 2004; Perdices and Coelho, 2006; Lin, 2008; Zheng et al., 2015; Yin et al., 2016; Wang et al., 2019; Supplementary Table S1). Thus a total of 1014 Cyt b sequences from 62 localities were used in the present study, covering most of its distribution in southeastern China, including the Yangtze River basin, Pearl River basin, and several small coastal river basins (Figure 1A, Table 1, Supplementary Table S1). As the information on the taxonomic status of some sequences from previous studies have not been updated, those downloaded from GenBank were verified using sequences from Wang et al. (2019) as reference to confirm their identity prior to the analysis. Opsariichthys macrolepis (OQ428185, OQ428186), O. hainanensis (OQ428183, OQ428184), and O. chengtui (KT725244)were used as outgroup taxa. All the newly determined sequences were deposited in GenBank under the accession numbers OQ214215 to OQ214836.
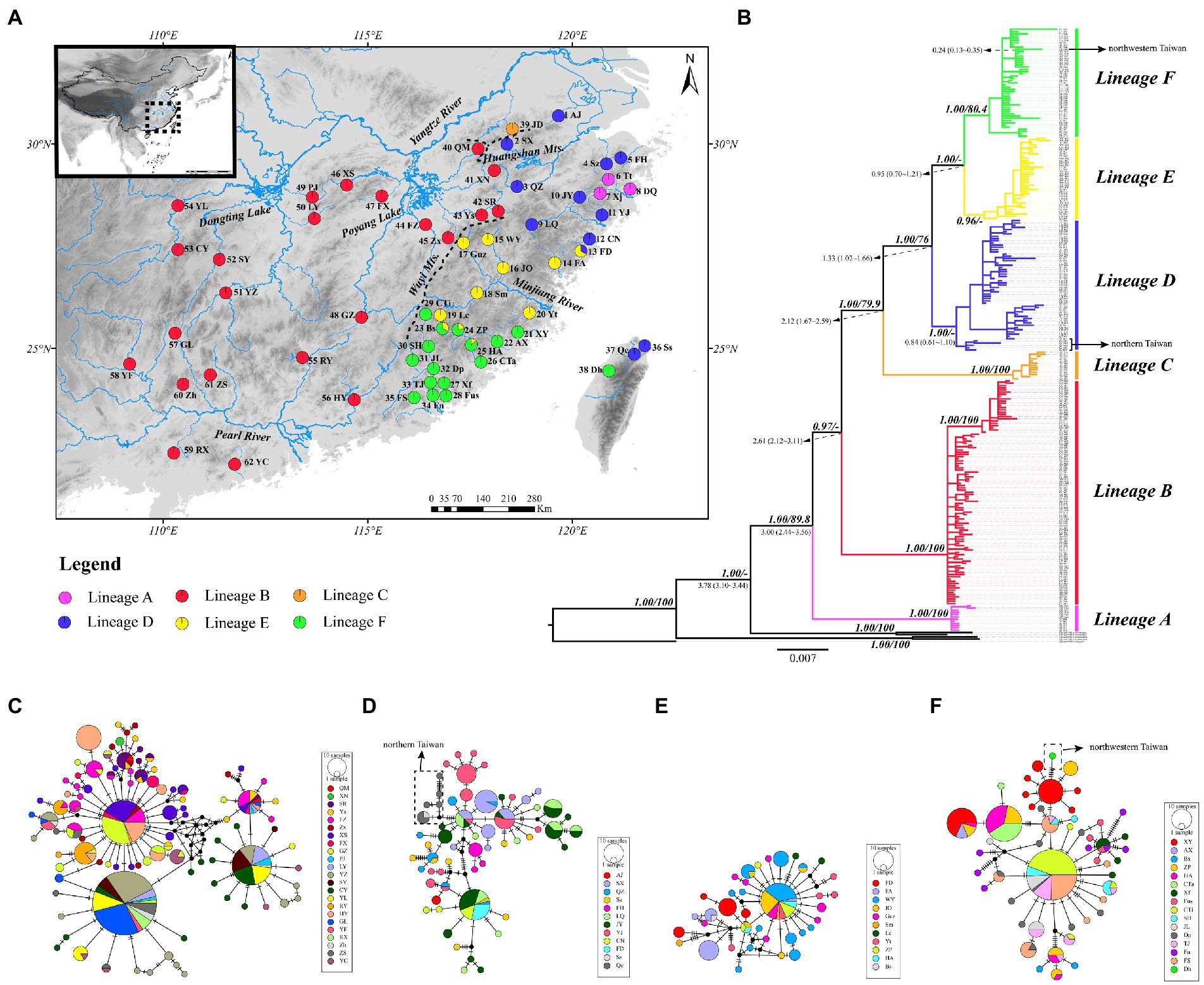
Figure 1. (A) Map of sampling localities of the O. acutipinnis-evolans complex. Codes for sampling localities are given in Table 1 and Supplementary Table S1. Populations are present as pie charts with slice size proportional to the frequency of the six major lineages identified by the Cyt b gene tree as in (B). (B) Cyt b gene tree from Bayesian inference. Numbers in bold and italics at nodes indicate Bayesian posterior probabilities (PP) and bootstrap values (BS) from maximum likelihood analysis, respectively. Only well-supported numbers (PP > 0.95, BS > 70) are shown. Numbers in regular at nodes indicate divergence time estimate of major lineages. Colored branches denote different lineages as in (A). (C–F) Median-joining (MJ) networks of the O. acutipinnis-evolans complex concerning lineage (B, D-F). The mutation steps are shown as hatch marks on the branches. Missing (or unsampled) haplotypes are represented by black dots. The area of each circle is proportional to the number of individuals for each haplotypes. Different colors denote different populations.
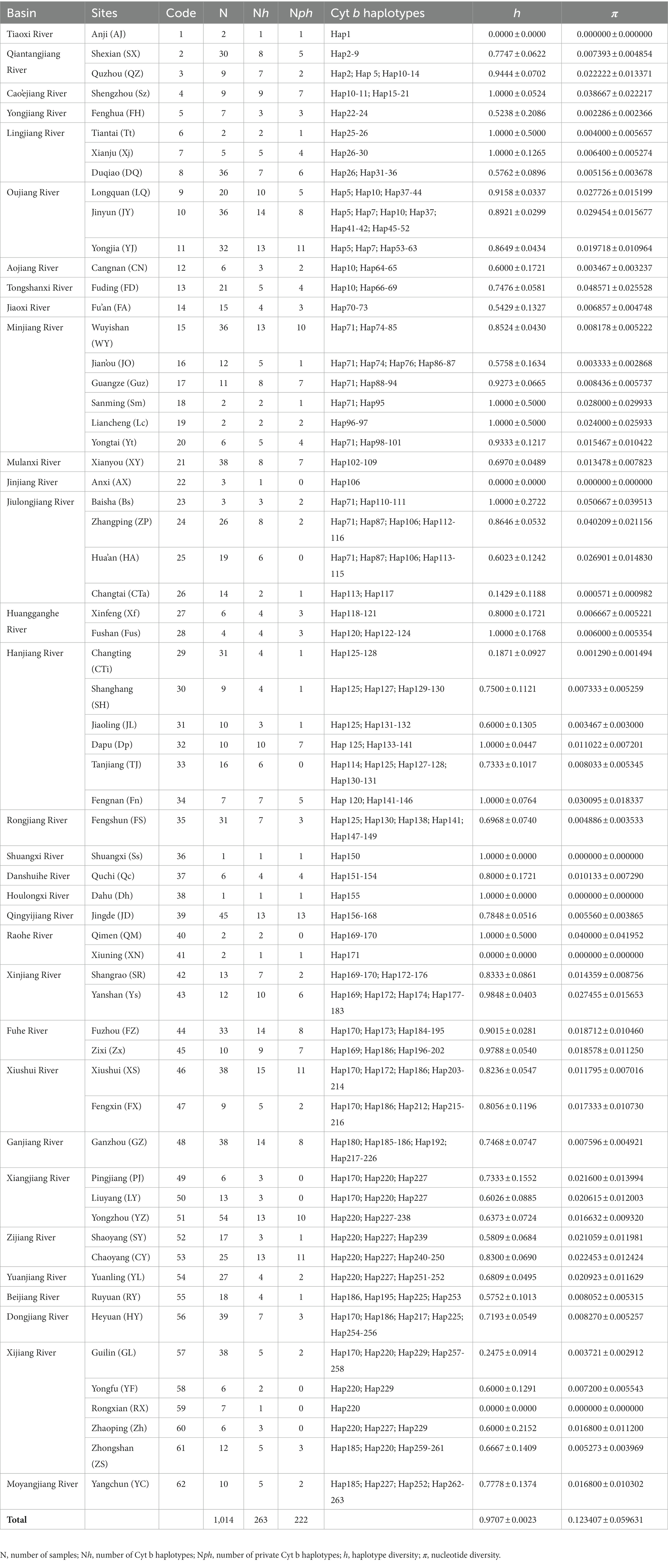
Table 1. Sampling information for the 62 populations of O. acutipinnis-evolans complex used in this study.
Multiple sequence alignment was performed using MAFFT (Katoh and Standley, 2013) implemented in PhyloSuite (Zhang et al., 2020) under default parameters and subsequently checked by eye in SeaView (Gouy et al., 2010). Characteristics and variations of sequences were analyzed in MEGA 7.0 (Kumar et al., 2016). Unique haplotypes were collapsed using ALTER (Glez-Peña et al., 2010). Sequence diversity indices were estimated for both pooled and individual populations using Arlequin 3.5 (Excoffier and Lischer, 2010), including the number of haplotypes (n), haplotype diversity (h) and nucleotide diversity (π).
Phylogenetic relationships among mitochondrial Cyt b haplotypes were reconstructed using Bayesian inference (BI) and maximum likelihood (ML) approaches. TN + F + G4 was selected as the best-fit nucleotide substitution model based on the corrected Akaike information criterion (AICc) in Modelfinder (Kalyaanamoorthy et al., 2017).
BI was conducted in MrBayes 3.2.7 (Ronquist et al., 2012) with two independent runs of Metropolis-coupled Markov Chain Monte Carlo analyses, each comprising one cold chain and three heated chains at a default temperature of 0.1. The chains were run for 10 million generations and sampled every 1,000 generations. The average standard deviation of split frequencies were required to drop below 0.01, and the convergence diagnostic for branch length posterior probabilities (potential scale reduction factor) to approach 1. The first 25% of the trees were discarded as burn-in and the remaining trees were used to generate a consensus tree. ML analysis was performed in PhyML 3.0 (Guindon et al., 2010). The robustness of each branch was estimated with 1000 non-parametric bootstrap replicates. To visualize the relationships among haplotypes in each lineages, the median-joining (MJ) networks were generated using PopArt (Leigh and Bryant, 2015) on lineages that contained multiple populations.
A dated gene tree using the Cyt b haplotypes was reconstructed using BEAST 2.6.0 (Bouckaert et al., 2019) to estimate divergence time among major lineages. The hypothesis of a strict molecular clock was first tested using the sequence-based likelihood ratio test in DAMBE 7(Xia, 2018). The hypothesis of a strict molecular clock cannot be rejected at the significance level of 0.05 (p = 0.8798). We calibrated the Cyt b gene tree with 1.05% per site per million year under the strict clock model to obtain the absolute divergence time estimates. This substitution rate was previously inferred for several opsariichthyine fishes from Lake Biwa based on geological event calibration (Tabata et al., 2016). Analyses were performed for 10 million generations while sampling every 1000 generations under the Yule speciation tree prior. The BI tree was used as the starting tree. Three independent runs were carried out. Tracer 1.7 (Rambaut et al., 2018) was used to check for adequate effective sample sizes (ESS > 200) of the posterior distribution of each parameter and to access convergence among independent runs. The runs were then combined using LogCombiner (Bouckaert et al., 2019) after removing the first 25% samples as burn-in. A maximum clade credibility tree (MCC tree) was generated using TreeAnnotator in the BEAST package.
A total of 273 specimens representing each lineages were randomly selected for morphological examination. Twenty morphometric measurements and three meristic characters were used in this study (Supplementary Table S2). Besides, the position of the posterior end of pectoral fin and maxilla were check visually. Measurements and counts were made on the left side of each individual following Armbruster (2012). Measurements were taken point to point with a digital caliper (precision of 0.01mm). The pectoral fin was removed from the selected specimen, cleared and stained following Taylor and van Dyke (1985), and then inspected under a stereoscope.
Morphometric measurements were subject to principal component analysis (PCA) to check body shape differentiations among major lineages. Prior to the analysis, the method of Reist (1985) was used to normalize all the measurements except for standard length to eliminate the influence of allometry of body parts. Normalization was conducted in R and PCA was run with SPSS 21 (SPSS, Chicago, IL, USA). The PCA loadings were presented in Supplementary Table S3.
The final alignment of Cyt b consisted of 912 base pairs (bp), of which 174 bp (19.1%) were parsimony informative. The base composition was unequal (A = 24.29%, T = 30.75%, G = 16.44%, C = 28.52%), with strong compositional biases against G existed at the second and especially at the third codon position (just an average of 10.5%). We translated all sequences into amino acids, and no internal stop codon was detected. A total of 263 haplotypes were identified among the 1014 Cyt b sequences (Table 1), of which 84.4% (222) were unique to certain populations. The Cyt b haplotype diversity (h) and nucleotide diversity (π) varied among populations (Table 1). The overall h in all samples was 0.9655 ± 0.0028, and the overall π was 0.157542 ± 0.076141.
The ML and BI trees based on Cyt b haplotypes yielded similar gene tree topologies and congruently revealed six evolutionary lineages clearly correlating with their geographical distribution (Figure 1B). Lineage A located at the basal part of the tree, comprised all samples from population 6–8 (Tt, Xj and DQ) belonging to the Lingjiang River basin on China’s southeastern coast. Lineage B, sister to the remaining four lineages, consisted of samples from population 40–62 belonging to the middle Yangtze River basin and the Pearl River basin. Lineage C consisted of samples from population 39 (JD) of the Qingyijiang River belonging to the lower Yangtze. Lineage D contained samples from northern Taiwan island (population 36–37) and northern populations in China’s southeastern coast (population 2–5, 9–12) except for one population (population 1, AJ), which originated from Tiaoxi River of the lower Yangtze River. Lineage E and F contained samples from middle and southern populations (population 13–35) in the southeastern coastal region, respectively. Population 38 (Dh) from western Taiwan island clustered within lineage F. The monophyly and relationships of lineages was well supported (PP>0.95, BS>75) unless mentioned otherwise. The relationships among haplotypes in lineage B, D, E, and F were clearly shown by the MJ networks (Figures 1C-F). It was clearly depicted that the northern Taiwan populations (population 36, Ss, and population 37, Qc) were derived from Qiantangjiang/Oujiang River poopulations, and the northwestern Taiwan population (population 38, Dh) originated from Mulanxi River population. The inter-lineage genetic distances ranged from 0.0177 (between lineage E and F) to 0.0700 (between lineage A and C) (Table 2).
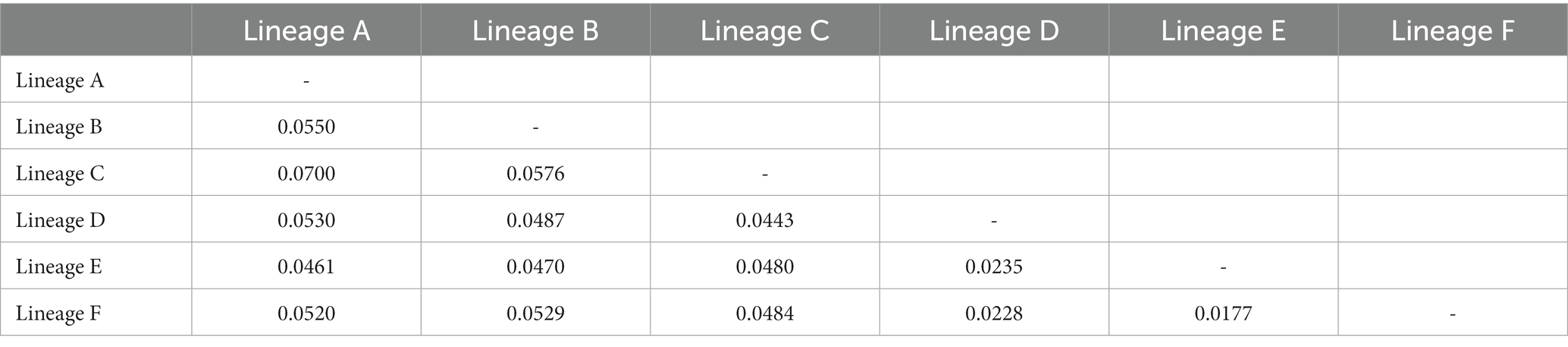
Table 2. Genetic distance between major lineages of O. acutipinnis-evolans complex based on Cyt b sequences.
The divergence time estimates of major lineages are provided in Figure 1B. The dating analysis suggested that the most recent common ancestor (TMRCA) of O. acutipinnis-evolans complex was dated back to 3.78 Ma (95% HPD 3.10 ~ 4.44 Ma). TMRCA of lineage A occurred at 3.00 Ma (95% HPD 2.44 ~ 3.56 Ma). TMRCA of lineage B was dated to 2.61 Ma (95% HPD 2.12 ~ 3.11 Ma). TMRCA of lineage C diverged at 2.12 Ma (95% HPD 1.67 ~ 2.59 Ma), and TMRCA of lineage D diverged at 1.33 Ma (95% HPD 1.02–1.66 Ma). The split between the continental and northern Taiwan island populations occurred at 0.84 Ma (95% HPD 0.61 ~ 1.10 Ma). Time since the split of lineage E and lineage F was estimated to be 0.95 Ma (95% HPD 0.70 ~ 1.21 Ma). The northwestern Taiwan population was isolated from the continental populations at around 0.24 Ma (95% HPD 0.13 ~ 0.35 Ma).
The scatter plot of the first two principal components indicated no clear separation between the six evolutionary lineages (Figure 2A). The number of scales and branched pectoral fin rays counts showed subtle differences and significant overlap among different lineages (Figures 2B–D; Supplementary Table S2). The most frequently observed predorsal scales ranged from 15 to 16, and circumpeduncular scales ranged from 16 to 18. The clear and stain procedure revealed that the branched pectoral fin rays ranged from 13 to 16, with 14 and 15 being the most frequent counts. Besides, the pectoral fin length varied among individuals, sex and life stages. Longer pectoral fins reaching or extending beyond the origin of the pelvic fin were only observed in sexually active males. The posterior end of pectoral fin never extends to pelvic fin origin in females and juveniles (Figures 2E,F; Supplementary Table S2). The position of the posterior end of maxilla also varied among individuals and did not exhibit specific patterns between lineages (Figure 2G; Supplementary Table S2).
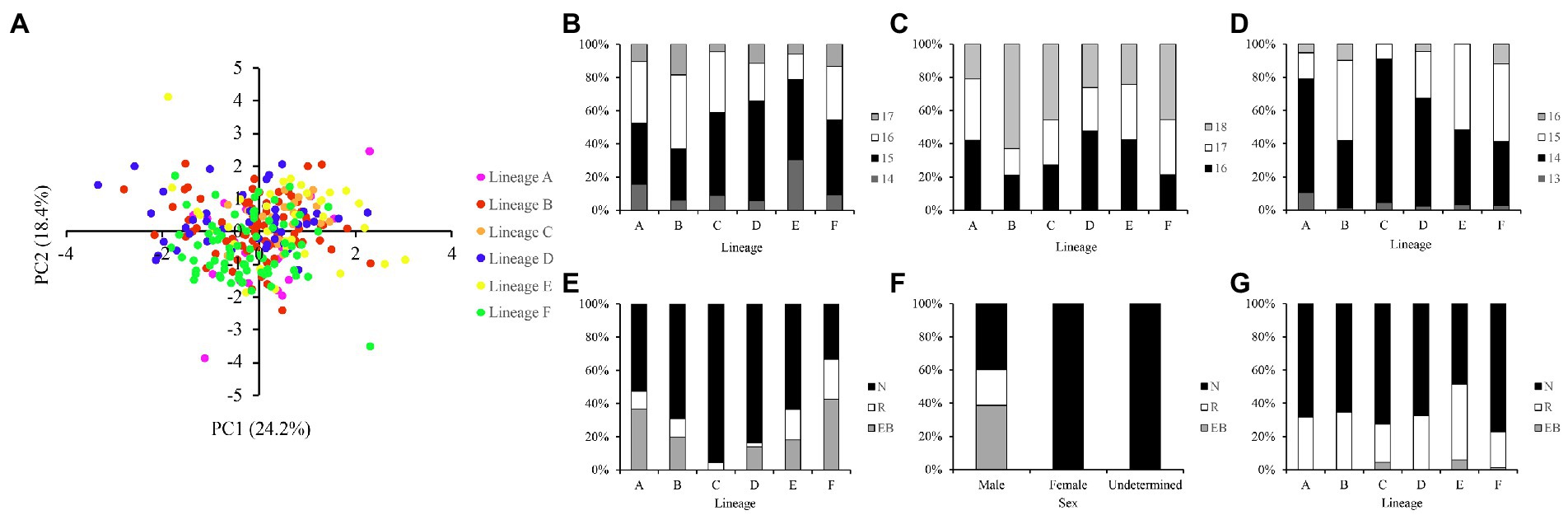
Figure 2. (A) Scatter plot of the first two principal components based on 20 measurements. The meristic counts differences between different lineages in (B) predorsal scales, (C) circumpeduncular scales, and (D) branched pectoral fin rays. The differences in the position of posterior end of pectoral fin against ventral fin origin between different (E) lineages and (F) sexes. (G) The differences in the position of the posterior end of maxilla against vertical of anterior margin of orbit. N, not reaching; R, reaching; EB, extend beyond.
One of the major goals of phylogeography is to decipher the spatial arrangements of genetic variations within populations or among closely related species (Avise, 2000). Our results clearly revealed that phylogeographic pattern of the O. acutipinnis-evolans complex in southeastern China corresponded to category I defined by Avise (2000), which was allopatric lineages with deep differentiation. This pattern could be explained by the existence of long-term biogeographic barriers. While lineage A, C, D, E, and F originated from the relatively eastern part of their range, including the coastal area of southeastern China, lineage B occupied the relatively western part, covering the middle Yangtze River basin and the Pearl River basin. The most conspicuous geographic break between the eastern and western lineages is the Huangshan Mountains and the Wuyi mountains. These mountains serve as the natural watershed between the Yangtze River basin and the coastal river basins, and are usually perceived as an important zoogeographic boundary (Zhang and Chen, 1997; Tang W. Q. et al., 2022) and barriers to dispersal for many fish species (e.g., Cao et al., 2013; Chiang et al., 2013; Zheng et al., 2015). Geological evidence have indicated that the large-scale uplifts of the Huangshan Mountains and the Wuyi Mountains took place during the Quaternary (Chao, 1981; Zhang et al., 1990; Wang and Zhu, 1997; Huang et al., 1998). This period correlated with the rapid uplifting stages of the Qinghai-Tibetan Plateau (QTP) since the late Pliocene at around 3.6 Ma (Li and Fang, 1999). The intensive uplifting of the QTP has been assumed to act as a dominant driving force of tectonic movements in eastern China (Xu and Zhou, 2012), resulting in a terrain gradually sloping from west to east and new landscape features (Shi et al., 1998; Zhang et al., 2000; Yan Y. et al., 2018). The early diversification of lineage A, B, and C could be considered as eastward-westward divergence of the ancestral population. The temporal diversification was dated back to the late Pliocene and early Pleistocene (3.00, 2.61, and 2.12 Ma), largely coincide with the orogeny of mountains in southeastern China (like Wuyi Mountains and Huangshan Mountains) and the eastern margin of the QTP during this period. Despite that lineage A is locally endemic to a small river basin in the present, it could be once widespread but only eventually survived in this small region after its isolation. The uplift of mountain ranges likely acted as a natural divide between formerly connected rivers, impeding dispersal and gene flow and resulting in the isolation of populations on both slopes. As a small resident stream fish, the pale chub might be prone to the alternation in landscapes and water systems, resulting in the formation of current phylogeographical structures. Recent phylogeographic studies on two gudgeon species have congruently revealed similar scenarios, in which the significant genetic differences between the coastal river populations and Yangtze River populations were attributed to the long-term isolation between these river drainages as a result of the orogenic events in southeastern China (Yang et al., 2022; Li M. et al., 2023). These findings, along with ours, emphasized the important role of the low mountains and hills in structuring the phylogeographic patterns of small freshwater fishes in southeastern China.
The modern lineage D, E and F occupy the southeastern coastal area of China, and a southward diversification pattern along the coast could be clearly observed. The divergence time between lineage D and lineage E was estimated to be 1.33 Ma and the split between lineage E and F were estimated to be 0.95 Ma, both correspond to the timing of the Early-Middle Pleistocene transition (EMPT, approximately 0.8 ~ 1.2 Ma). In this period an intensification in amplitude and duration of climatic cycles marked the onset of a pronounced succession of glacial and interglacial which characterized the late Pleistocene (e.g., Clark et al., 2006; Ehlers et al., 2018). Geological evidence has suggested that during the EMPT, 25-50m and 20-30m changes in the sea-level took place between 1.2 ~ 2.8 Ma and 0.9 ~ 1.0 Ma, respectively, (Kitamura and Kawagoe, 2006; Berends et al., 2021). The amplitude in eustatic sea level changes of the EMPT were incomparable to those of the LGM, which were 120–130 m lower than the present (Lambeck and Chappell, 2001). However, given that the water depth of inner continental shelf in the East China Sea was usually lower than 60m (Yi et al., 2014), one could still suspect that the alternations in water course due to sea level changes could exert a great impact on the evolution of freshwater fish. The coastal area was impacted by lower sea levels during glacial periods, exposing large areas of continental shelf and paleo-rivers. These paleo-rivers likely increased population connectivity through river anastomoses, which would promote dispersal and gene flow and result in the colonization into new habitats. During the subsequent interglacial periods, sea-level highstands might induce a fragmentation of the formerly connected river systems and result in the isolation of populations. The cyclic changes in water courses could have enabled the southward colonization of the ancestral population and independent evolution after isolations. Drainage alternations associated with sea-level fluctuations during the Quaternary may be of great importance to the range dynamics of freshwater species in coastal China, as many previous phylogeographic studies have suggested (Yang and He, 2008; Yang, 2012; Yu et al., 2014; Yang et al., 2016).
Our findings also shed lights on the origin of primary freshwater fish fauna of the Taiwan Island. Previous studies had widely assumed that rivers in Zhejiang and Fujian region (e.g., Qiantangjiang River and Minjiang River) possibly act as major dispersal routes from continental East Asia to the Taiwan Island, especially for freshwater fishes that restricted to the northern or northwestern part of the island (e.g., Wang et al., 2004; Xu et al., 2005; Lin et al., 2010; Yang, 2012; Ju et al., 2018). The natural distribution of O. acutipinnis-evolans complex in Taiwan was confined to the northern part of the island (Lin, 2008). Our results agree fairly well with this hypothesis. The phylogeographic analysis indicated that populations in Taiwan derived from the Zhejiang-Fujian region via two independent colonization events in different period. Samples from the northern part of the island, including the Danshuihe River and Shuangxi River, showed a close relationship with those sampled from the Qiangtangjiang/ Oujiang River (Figure 1D), while samples from northwestern part of the island derived from those sampled from Mulanxi River (Figure 1F). Geological and phylogeographical evidence have shown that a land bridge recurrently connected the Taiwan Island with the continental East Asia during the Quaternary glacial cycles, facilitating biotic interchanges between both sides (e.g., Wang et al., 2007; Chiang et al., 2010; Lin et al., 2022). The periodic emergence of the land bridge could be a plausible explanation for the multiple colonization routes at different time stages as some previous studies suggested (Yu, 1995; Wang et al., 2004; Chang et al., 2016).
The populations of O. acutipinnis-evolans complex in southeastern China exhibit a high level of genetic diversity. Over 80% of the haplotypes were unique to specific populations, which suggests the possibility of population isolation. This phenomenon may be attributed not only to geographic factors, but also to intrinsic properties of this group of fishes. The O. acutipinnis-evolans complex is a group of small resident fish that prefers lotic habitats, typically found in small montane streams and tributaries. The low dispersal potentials and ecological preference of these fishes could render them vulnerable to the alternation in landscape and hydrological patterns, leading to population isolation or restricted gene flow among populations. Similar patterns have also been observed in other small freshwater fishes in southeastern China (e.g., Yu et al., 2014; Yang et al., 2022).
Grant and Bowen (1998) have proposed four categories of population histories based on haplotype and nucleotide diversity. The first category includes populations with both low haplotype and nucleotide diversity (h < 0.5, π < 0.5%), which may have underwent recent population bottleneck or founder event. The second category comprises population with high haplotype diversity (h > 0.5) but low nucleotide diversity (π < 0.5%), which could be attributed to rapid population growth after population bottleneck. The third category represents populations that have low haplotype diversity (h < 0.5) but high nucleotide diversity (π > 0.5%), which may have experienced bottleneck in a previously large and stable population, or secondary contact. The forth category includes populations that have both large haplotype and nucleotide diversity (h > 0.5, π > 0.5%), which is likely associated with large, stable populations or secondary contact of previously isolated populations. The nucleotide and haplotype diversity were both high (h = 0.9655 ± 0.0028, π = 0.157542 ± 0.076141) in pooled and most individual populations of the O. acutipinnis-evolans complex, confirming the classification in category four. These fishes are typically found in large, stable populations. In general, effective population size and molecular diversity are positively correlated, as observed in many freshwater fishes (e.g., Chiang et al., 2013; Wang et al., 2022).
According to Grant and Bowen’s study (1998), geographic co-occurrence between previously allopatric lineages could lead to large nucleotide diversity. Our study found that several coastal river populations (population 13, FD; population 23, Bs; population 24, ZP; population 25, HA) exhibited higher nucleotide diversity than other populations due to the admixture of two major lineages (Figure 1A), while the remaining populations had a single lineage. Some previous studies focusing on both animals (e.g., Zhao et al., 2008; Li M. et al., 2023) and plants (e.g., Nettel et al., 2008; Havrdová et al., 2015) have attributed the elevated nucleotide diversity to a result of secondary contact. In our case, population 13 (FD) was originated from a small coastal stream (Tongshanxi River). It is plausible to attribute this phenomenon to secondary contact that arise as the consequences of the drainage alternation due to sea level changes or river capture events. Though the relative role of them requires further investigation. Population 23 (BS), population 24 (ZP) and population 25 (HA) all contained haplotypes from both lineage E and F. These populations were all derived from upper or middle reaches of the Jiulongjiang River. Since the lower reach population 26 (CTi) did not exhibit admixture, secondary contact via river capture could possibly contribute to this scenario.
Phylogeographic analysis can provide insights into the speciation processes as it elucidates the presence of incipient subpopulations on the way to becoming new species or that may already represent cryptic species (e.g., Marshall et al., 2011). Our Cyt b gene tree revealed six distinct, almost allopatric evolutionary lineages. Large genetic distances can be observed between some lineages, which could be considered as subspecies or even species level divergence as some previous studies have suggested (e.g., Yu et al., 2014; Chen et al., 2017; Li et al., 2020). However, our morphological analysis concerning both morphometrics and meristic counts failed to distinguish all the six major mitochondrial lineages recovered by the Cyt b gene tree, suggesting that O. acutipinnis-evolans complex could be morphologically conserved. The presence of cryptic species has been suggested to explain the high intraspecific divergence in the opsariichthyine fishes (Wang et al., 2019). Morphologically cryptic lineages could have different adaptations in other aspects, such as behaviors, ecology and physiology (e.g., Braune et al., 2008; Feckler et al., 2014; Taylor and Friesen, 2017). For example, the allochrony in migration and breeding has been assumed to act as an prezygotic isolation mechanism in maintaining the cryptic diversity of the morphologically conserved sand martin subspecies (Tang Q. et al., 2022). Therefore, we still cannot resist that cryptic species might occur. Given the genetic distinctness and the morphological conservatism among lineages, it is still more appropriate to consider this group of fishes as a species complex.
According to Huynh and Chen (2013) and Wang et al. (2019), the two currently described species, O. acutipinnis and O. evolans, can be distinguished by: (1) two distinct mitochondrial lineages with genetic divergence of 2.5%–2.7% in D-loop sequences or 5.2% in Cyt b sequences; (2) allopatric distribution, with O. acutipinnis occupies the Yangtze River basin and Pearl River basin and O. evolans inhabits coastal drainages in southeastern China; and (3) variation of a single branched pectoral fin ray and a single scale above the lateral line, differences in predorsal scales (18–20 in O. acutipinnis vs. 16–17 in O. evolans), and the relative position of the posterior end of maxilla and pectoral fin. Compared to the previous works, our Cyt b gene tree did recovered two monophyletic groups that correspond to these two previously defined species: lineage D + E + F correspond to O. evolans and lineage B corresponds to O. acutipinnis. However, the results of morphological examination disagree with the previous studies that these lineages could be separated by the aforementioned morphological features. The differences of merely a single branched pectoral fin rays, a single scale above the lateral line can be attributed to intra-population variations. They might not be appropriate to act as diagnostic features for distinct species. The length of pectoral fin varied between individuals, age, and sex, with longer pectoral fin that extending way beyond ventral fin origin observed mostly in sexually active males. Therefore, distinguishing O. acutipinnis and O. evolans with the previously proposed morphological diagnostic features would be problematic. Besides, the results of our phylogeographic analysis detected lineage A from the coastal populations and lineage C out of the Yangtze populations. Arbitrarily assigning the coastal lineages as O. evolans and Yangtze-Pearl lineages as O. acutipinnis would also be a hasty decision. Thus distribution as an identification key for both species should also be used with care. Since morphological characteristics and distribution cannot effectively distinguish these two species, DNA barcoding becomes a safer way in their identification. Still, the taxonomy of the O. acutipinnis-evolans complex should be revised carefully, addressing not only morphological differences but also other aspects like ecology and behavior. In addition, the present work, as well as the former ones, only used mitochondrial markers to infer the evolutionary history of the O. acutipinnis-evolans complex. The mitochondrial DNA can only represent a tiny fraction of the whole genome and its limitations have long been addressed (e.g., Galtier et al., 2009). In future studies, it is recommended that additional data, especially from nuclear genome, be incorporated to produce more robust results.
Evolutionary significant units (ESUs) and management units (MUs) are important concepts in conservation genetics (e.g., Moritz, 1994; Fraser and Bernatchez, 2001). According to Avise (2005), an evolutionary significant unit (ESU) is one or a set of conspecific populations with relatively long term evolutionary history mostly separate from other such units. Our phylogeographic analysis demonstrated six allopatrically distributed lineages that underwent long geographical isolation. Given this phylogeographic distinctiveness and evolutionary independence, each major lineage should be regarded as a different evolutionary significant unit (ESU). Neglecting the uniqueness of different lineages will certainly increase the chance of genetic homogenization and contamination of the gene pool, which may increase the risk of extinction of local populations. The aforementioned Chinese giant salamander serves as a tragic example (Yan F. et al., 2018). Previous research on the Japanese pale chub Zacco platypus revealed that a distinct lineage native to the Lake Biwa was introduced to other locations in Japan, and resulted in the successful establishment of this lineage in the new environments and widely hybridization with the endemic populations throughout most of the areas in the Japanese islands (Takamura and Nakahara, 2015). Similar situations of other fishes also call for attention to the phylogeographic structures and genetic uniqueness of populations (e.g., Jang-Liaw et al., 2019; Watanabe et al., 2020). Thus, genetic and phylogeographic assessment are crucial for the seemingly widespread but morphologically conserved species, especially when it comes to implementing conservation actions such as translocation or reintroduction.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was reviewed and approved by Animal Care and Use Committee of Institute of Hydrobiology, Chinese Academy of Sciences.
HL conceived the research and provided financial support. JG collected samples, conducted laboratory work, and performed data analysis. DY contributed to the sampling and data analysis. JG wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by the National Key R & D Program of China (2018YFD0900806), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31000000), and National Natural Science Foundation of China (31872234).
We thank Siqing Liu, Huaming Hu, Mingzheng Li, and other colleagues for their assistance with specimen collection. We also thank Ping Yang, Lan Zhu, Xianglong Jiang, Qiaoling Deng, Tingyue Yang, Xiaojuan Zhao, and Yahan Wu for their help with laboratory work and data analysis. Thanks also to Tereza Mchura Magati for improving the languages.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1142810/full#supplementary-material
Aljanabi, S. (1997). Universal and rapid salt-extraction of high quality genomic DNA for PCR- based techniques. Nucleic Acids Res. 25, 4692–4693. doi: 10.1093/nar/25.22.4692
Armbruster, J. W. (2012). Standardized measurements, landmarks, and meristic counts for cypriniform fishes. Zootaxa 3586:8. doi: 10.11646/zootaxa.3586.1.3
Ashiwa, H., and Hosoya, K. (1998). Osteology of Zacco pachycephalus, sensu Jordan & Evermann (1903), with special reference to its systematic position. Environ. Biol. Fish 18, 163–171. doi: 10.1007/978-94-015-9016-7_10
Avise, J. C. (2000). Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press.
Avise, J. C. (2005). “Phylogenetic units and currencies above and below the species level” in Phylogeny and conservation. eds. A. Purvis, J. L. Gittleman, and T. Brooks. 1st ed (Cambridge, United Kingdom: Cambridge University Press), 76–100.
Avise, J. C. (2009). Phylogeography: retrospect and prospect. J. Biogeogr. 36, 3–15. doi: 10.1111/j.1365-2699.2008.02032.x
Avise, J. C., Arnold, J., Ball, R. M., Bermingham, E., Lamb, T., Neigel, J. E., et al. (1987). Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 18, 489–522. doi: 10.1146/ANNUREV.ES.18.110187.002421
Berends, C. J., de Boer, B., and van de Wal, R. S. W. (2021). Reconstructing the evolution of ice sheets, sea level, and atmospheric CO2 during the past 3.6 million years. Clim. Past 17, 361–377. doi: 10.5194/cp-17-361-2021
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., et al. (2019). BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15:e1006650. doi: 10.1371/journal.pcbi.1006650
Braune, P., Schmidt, S., and Zimmermann, E. (2008). Acoustic divergence in the communication of cryptic species of nocturnal primates (Microcebus ssp.). BMC Biol. 6:19. doi: 10.1186/1741-7007-6-19
Cao, L., Liang, X. F., Tang, W., and Zhao, J. (2013). Phylogeography of Coreoperca whiteheadi (Perciformes: Coreoperca) in China based on mitochondrial and nuclear gene sequences. Biochem. Syst. Ecol. 50, 223–231. Available at:. doi: 10.1016/j.bse.2013.04.007
Chai, J., Lu, C. Q., Yi, M. R., Dai, N. H., Weng, X. D., Di, M. X., et al. (2022). Discovery of a wild, genetically pure Chinese giant salamander creates new conservation opportunities. Zool. Res. 43, 469–480. Available at:. doi: 10.24272/j.issn.2095-8137.2022.101
Chang, H. Y., Wang, W. K., Chen, K. N., Su, J. K., Hsin, C. Y., Li, J., et al. (2016). Phylogeography and genetic structure of the endemic cyprinid fish Microphysogobio brevirostris in northern Taiwan. Biochem. Syst. Ecol. 65, 176–184. doi: 10.1016/j.bse.2016.02.020
Chao, C. P. (1981). An investigation of the causes of the richness of the biological resources in the Wuyishan natural reserve. Wuyi Sci. J. 1, 223–227.
Chen, Y. Y. (1982). A revision of opsariichthine cyprinid fishes. Oceanol. Limnol. Sin. 13, 293–299.
Chen, I. S., and Chang, Y. C. (2005). A photographic guide to the inland-water fishes of Taiwan. Keelung: The Sueichan Press.
Chen, X. L., Chiang, T. Y., Lin, H. D., Zheng, H. S., Shao, K. T., Zhang, Q., et al. (2007). Mitochondrial DNA phylogeography of Glyptothorax fokiensis and Glyptothorax hainanensis in Asia. J. Fish Biol. 70, 75–93. Available at:. doi: 10.1111/j.1095-8649.2007.01370.x
Chen, Y. Y., and Chu, X. L. (1998) Fauna Sinica, Osteichthyes, Cypriniformes II: Danioninae. Beijing: Science Press.
Chen, I. S., Wu, J.-H., and Huang, S.-P. (2009). The taxonomy and phylogeny of the cyprinid genus Opsariichthys Bleeker (Teleostei: Cyprinidae) from Taiwan, with description of a new species. Environ. Biol. Fish 86, 165–183. doi: 10.1007/s10641-009-9499-y
Chen, W., Zhong, Z., Dai, W., Fan, Q., and He, S. (2017). Phylogeographic structure, cryptic speciation and demographic history of the sharpbelly (Hemiculter leucisculus), a freshwater habitat generalist from southern China. BMC Evol.Biol. 17:216. Available at:. doi: 10.1186/s12862-017-1058-0
Chiang, T. Y., Lin, H. D., Shao, K. T., and Hsu, K. C. (2010). Multiple factors have shaped the phylogeography of Chinese spiny loach Cobitis sinensis in Taiwan as inferred from mitochondrial DNA variation. J. Fish Biol. 76, 1173–1189. Available at:. doi: 10.1111/j.1095-8649.2010.02589.x
Chiang, T. Y., Lin, H. D., Zhao, J., Kuo, P. H., Lee, T. W., and Hsu, K. C. (2013). Diverse processes shape deep phylogeographical divergence inCobitis sinensis(Teleostei: Cobitidae) in East Asia. J. Zool. Syst. Evol. Res. 51, 316–326. doi: 10.1111/jzs.12030
Clark, P. U., Archer, D., Pollard, D., Blum, J. D., Rial, J. A., Brovkin, V., et al. (2006). The middle Pleistocene transition: characteristics, mechanisms, and implications for long-term changes in atmospheric pCO2. Quat. Sci. Rev. 25, 3150–3184. Available at:. doi: 10.1016/j.quascirev.2006.07.008
de Keyzer, E. L. R., de Corte, Z., van Steenberge, M., Raeymaekers, J. A. M., Calboli, F. C. F., Kmentová, N., et al. (2019). First genomic study on Lake Tanganyika sprat Stolothrissa tanganicae: a lack of population structure calls for integrated management of this important fisheries target species. BMC Evol. Biol. 19:6. Available at:. doi: 10.1186/s12862-018-1325-8
Ehlers, J., Gibbard, P. L., and Hughes, P. D. (2018). ‘Quaternary glaciations and chronology’, in past glacial environments. Netherland: Elsevier, 77–101.
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Feckler, A., Zubrod, J. P., Thielsch, A., Schwenk, K., Schulz, R., and Bundschuh, M. (2014). Cryptic species diversity: an overlooked factor in environmental management? J. Appl. Ecol. 51, 958–967. Available at:. doi: 10.1111/1365-2664.12246
Fraser, D. J., and Bernatchez, L. (2001). Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol. Ecol. 10, 2741–2752. doi: 10.1046/j.1365-294X.2001.t01-1-01411.x
Galtier, N., Nabholz, B., Glemin, S., and Hurst, G. D. D. (2009). Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18, 4541–4550. doi: 10.1111/j.1365-294X.2009.04380.x
Glez-Peña, D., Gómez-Blanco, D., Reboiro-Jato, M., Fdez-Riverola, F., and Posada, D. (2010). ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 38, W14–W18. doi: 10.1093/nar/gkq321
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user Interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Grant, W., and Bowen, B. W. (1998). Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J. Hered. 89, 415–426. doi: 10.1093/jhered/89.5.415
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Günther, A. (1873). Report on a collection of fishes from China. J. Natl. Hist. 12, 239–250. doi: 10.1080/00222937308680749
Guo, X. Z., Zhang, G. R., Wei, K. J., Ji, W., Yan, R. J., Wei, Q. W., et al. (2019). Phylogeography of the threatened tetraploid fish, Schizothorax waltoni, in the Yarlung Tsangpo River on the southern Qinghai-Tibet plateau: implications for conservation. Sci. Rep. 9:2704. doi: 10.1038/s41598-019-39128-y
Havrdová, A., Douda, J., Krak, K., Vít, P., Hadincová, V., Zákravský, P., et al. (2015). Higher genetic diversity in recolonized areas than in refugia of Alnus glutinosa triggered by continent-wide lineage admixture. Mol. Ecol. 24, 4759–4777. doi: 10.1111/mec.13348
Howes, G. J. (1980). The anatomy, phylogeny and classification of bariliine cyprinid fishes. Bull. Brit. Mus. (Natural History) Zool. 37, 129–198.
Huang, P., Diffendal, R. F., Yang, M., and Helland, P. E. (1998). Mountain evolution and environmental changes of Huangshan (Yellow Mountain), China. J. Geogr. Sci. 9, 25–34.
Huang, X. X., Hsu, K. C., Kang, B., Kuo, P. H., Tsai, W. H., Liang, C. M., et al. (2019). Population structure of Aphyocypris normalis: phylogeography and systematics. ZooKeys 872, 77–90. doi: 10.3897/zookeys.872.33105
Huynh, T. Q., and Chen, I.-S. (2013). A new species of cyprinid fish of genus Opsariichthys from Ky Cung-bang Giang River basin, northern Vietnam with notes on the taxonomic status of the genus from northern Vietnam and southern China. J. Mar. Sci. and Technol. 21:Article 18. doi: 10.6119/JMST-013-1219-12
Jang-Liaw, N. H., Tominaga, K., Zhang, C., Zhao, Y., Nakajima, J., Onikura, N., et al. (2019). Phylogeography of the Chinese false gudgeon, Abbottina rivularis, in East Asia, with special reference to the origin and artificial disturbance of Japanese populations. Ichthyol. Res. 66, 460–478. doi: 10.1007/s10228-019-00686-w
Jordan, D. S., and Evermann, B. W. (1902). Notes on a collection of fishes from the island of Formosa. Proc. U. S. Natl. Mus. 25, 315–368. doi: 10.5479/si.00963801.25-1289.315
Ju, Y. M., Hsu, K. C., Yang, J. Q., Wu, J. H., Li, S., Wang, W. K., et al. (2018). Mitochondrial diversity and phylogeography of Acrossocheilus paradoxus (Teleostei: Cyprinidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 29, 1194–1202. doi: 10.1080/24701394.2018.1431227
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kitamura, A., and Kawagoe, T. (2006). Eustatic Sea-level change at the mid-Pleistocene climate transition: new evidence from the shallow-marine sediment record of Japan. Quat. Sci. Rev. 25, 323–335. doi: 10.1016/j.quascirev.2005.02.009
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lambeck, K., and Chappell, J. (2001). Sea level change through the last glacial cycle. Science 292, 679–686. doi: 10.1126/science.1059549
Larson, W. A., Seeb, J. E., Pascal, C. E., Tremplin, W. D., and Seeb, L. W. (2014). Single-nucleotide polymorphisms (SNPs) identified through genotyping-by-sequencing improve genetic stock identification of Chinook salmon (Oncorhynchus tshawytscha) from western Alaska. Can. J. Fish. Aquat. Sci. 71, 698–708. doi: 10.1139/cjfas-2013-0502
Leigh, J. W., and Bryant, D. (2015). POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Li, J., and Fang, X. (1999). Uplift of the Tibetan plateau and environmental changes. Chin. Sci. Bull. 44, 2117–2124. doi: 10.1007/BF03182692
Li, C., Jiang, S., Schneider, K., Jin, J., Lin, H., Wang, J., et al. (2020). Cryptic species in white Cloud Mountain minnow, Tanichthys albonubes: taxonomic and conservation implications. Mol. Phylogenet. Evol. 153:106950. doi: 10.1016/j.ympev.2020.106950
Li, C., Xiao, H., Zhang, X., Lin, H., Elmer, K. R., and Zhao, J. (2023). Deep genome-wide divergences among species in white Cloud Mountain minnow Tanichthys albonubes (Cypriniformes: Tanichthyidae) complex: conservation and species management implications. Mol. Phylogenet. Evol. 182:107734. doi: 10.1016/j.ympev.2023.107734
Li, M., Yang, X., Ni, X., and Fu, C. (2023). The role of landscape evolution in the genetic diversification of a stream fish Sarcocheilichthys parvus from southern China. Front. Genet. 13:1075617. doi: 10.3389/fgene.2022.1075617
Liang, Z., Chen, W. T., Wang, D. Q., Zhang, S. H., Wang, C. R., He, S. P., et al. (2019). Phylogeographic patterns and conservation implications of the endangered Chinese giant salamander. Ecol. Evol. 9, 3879–3890. doi: 10.1002/ece3.5014
Lin, H. D. (2008). Phylogeography of Cyprinidae fishes in Taiwan and mainland China. Doctoral thesis. Tainan: National Cheng Kung University.
Lin, H., Dai, C., Yu, H., Tu, J., Yu, J., He, J., et al. (2022). Historical connectivity and environmental filtering jointly determine the freshwater fish assemblages on Taiwan and Hainan Islands of China. Curr. Zool. 1–9. doi: 10.1093/cz/zoac015
Lin, H. D., Hsu, K. C., Shao, K. T., Chang, Y. C., Wang, J. P., Lin, C. J., et al. (2008). Population structure and phylogeography of Aphyocypris kikuchii (Oshima) based on mitochondrial DNA variation. J. Fish Biol. 72, 2011–2025. Available at:. doi: 10.1111/j.1095-8649.2008.01836.x
Lin, C. J., Lin, H. D., Wang, J. P., Chao, S. C., and Chiang, T. Y. (2010). Phylogeography ofHemibarbus labeo(Cyprinidae): secondary contact of ancient lineages of mtDNA. Zool. Scr. 39, 23–35. doi: 10.1111/j.1463-6409.2009.00407.x
Ma, G. C., Tsao, H. S., Lu, H. P., and Yu, H. T. (2006). AFLPs congruent with morphological differentiation of Asian common minnow Zacco (Pisces: Cyprinidae) in Taiwan. Zool. Scr. 35, 341–351. Available at:. doi: 10.1111/j.1463-6409.2006.00232.x
Marshall, D. C., Hill, K. B. R., Cooley, J. R., and Simon, C. (2011). Hybridization, mitochondrial DNA Phylogeography, and prediction of the early stages of reproductive isolation: lessons from New Zealand cicadas (genus Kikihia). Syst. Biol. 60, 482–502. doi: 10.1093/sysbio/syr017
Moritz, C. (1994). Defining “evolutionarily significant units” for conservation. Trends Ecol. Evol. 9, 373–375. doi: 10.1016/0169-5347(94)90057-4
Murphy, R. W., Fu, J., Upton, D. E., de Lema, T., and Zhao, E. M. (2000). Genetic variability among endangered Chinese giant salamanders, Andrias davidianus. Mol. Ecol. 9, 1539–1547. doi: 10.1046/j.1365-294x.2000.01036.x
Nettel, A., Dodd, R. S., Afzal-Rafii, Z., and Tovilla-Hernández, C. (2008). Genetic diversity enhanced by ancient introgression and secondary contact in East Pacific black mangroves: introgression in east pacific black mangroves. Mol. Ecol. 17, 2680–2690. doi: 10.1111/j.1365-294X.2008.03766.x
Nichols, J. T. (1943). The fresh-water fishes of China. New York: American Museum of Natural History.
Oshima, M. (1919). Contributions to the study of the fresh water fishes of the island of Formosa. Ann. Carnegie Mus. 12, 169–328. doi: 10.5962/p.34608
Perdices, A., and Coelho, M. M. (2006). Comparative phylogeography of Zacco platypus and Opsariichthys bidens (Teleostei, Cyprinidae) in China based on cytochrome b sequences. J. Zool. Syst. Evol. Res. 44, 330–338. doi: 10.1111/j.1439-0469.2006.00368.x
Perdices, A., Cunha, C., and Coelho, M. M. (2004). Phylogenetic structure of Zacco platypus (Teleostei, Cyprinidae) populations on the upper and middle Chang Jiang (=Yangtze) drainage inferred from cytochrome b sequences. Mol. Phylogenet. Evol. 31, 192–203. doi: 10.1016/j.ympev.2003.07.001
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in Bayesian Phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Reist, J. D. (1985). An empirical evaluation of several univariate methods that adjust for size variation in morphometric data. Can. J. Zool. 63, 1429–1439. doi: 10.1139/z85-213
Rendahl, H. (1928). Beiträge zur Kenntnis der chinesischen Süßwasserfische. I. Systematischer Teil. Ark. Zool. 20, 1–194.
Rincon-Sandoval, M., Betancur-R, R., and Maldonado-Ocampo, J. A. (2019). Comparative phylogeography of trans-Andean freshwater fishes based on genome-wide nuclear and mitochondrial markers. Mol. Ecol. 28, 1096–1115. doi: 10.1111/mec.15036
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shi, Y., Li, J., and Li, B. (1998). Uplift and environmental changes of Qinghai-Tibetan plateau in the late Cenozoic. Guangzhou, China: Guangdong Science and Technology Press.
Tabata, R., Kakioka, R., Tominaga, K., Komiya, T., and Watanabe, K. (2016). Phylogeny and historical demography of endemic fishes in Lake Biwa: the ancient lake as a promoter of evolution and diversification of freshwater fishes in western Japan. Ecol. Evol. 6, 2601–2623. doi: 10.1002/ece3.2070
Takamura, K., and Nakahara, M. (2015). Intraspecific invasion occurring in geographically isolated populations of the Japanese cyprinid fish Zacco platypus. Limnology 16, 161–170. doi: 10.1007/s10201-015-0450-y
Tang, Q., Burri, R., Liu, Y., Suh, A., Sundev, G., Heckel, G., et al. (2022). Seasonal migration patterns and the maintenance of evolutionary diversity in a cryptic bird radiation. Mol. Ecol. 31, 632–645. doi: 10.1111/mec.16241
Tang, W. Q., Song, X. L., and Zhang, Y. (2022). Species diversity and zoogeographical analysis of freshwater fishes in central and southern mountainous area of East China. Acta Hydrobiol. Sin. 45, 621–629. doi: 10.7541/2022.2021.017
Taylor, R. S., and Friesen, V. L. (2017). The role of allochrony in speciation. Mol. Ecol. 26, 3330–3342. doi: 10.1111/mec.14126
Taylor, E. B., Tamkee, P., Keeley, E. R., and Parkinson, E. A. (2011). Conservation prioritization in widespread species: the use of genetic and morphological data to assess population distinctiveness in rainbow trout (Oncorhynchus mykiss) from British Columbia, Canada. Evol. Appl. 4, 100–115. doi: 10.1111/j.1752-4571.2010.00136.x
Taylor, W. R., and Van Dyke, G. C. (1985). Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium 9, 107–119.
Tumendemberel, O., Zedrosser, A., Proctor, M. F., Reynolds, H. V., Adams, J. R., Sullivan, J. M., et al. (2019). Phylogeography, genetic diversity, and connectivity of brown bear populations in Central Asia. PLoS One 14:e0220746. doi: 10.1371/journal.pone.0220746
van Rossum, F., Martin, H., le Cadre, S., Brachi, B., Christenhusz, M. J. M., and Touzet, P. (2018). Phylogeography of a widely distributed species reveals a cryptic assemblage of distinct genetic lineages needing separate conservation strategies. Perspect. Plant Ecol. Evol. Syst. 35, 44–51. doi: 10.1016/j.ppees.2018.10.003
Wang, J. P., Lin, H. D., Huang, S., Pan, C. H., Chen, X. L., and Chiang, T. Y. (2004). Phylogeography of Varicorhinus barbatulus (Cyprinidae) in Taiwan based on nucleotide variation of mtDNA and allozymes. Mol. Phylogenet. Evol. 31, 1143–1156. doi: 10.1016/j.ympev.2003.10.001
Wang, X., Liu, F., Yu, D., and Liu, H. (2019). Mitochondrial divergence suggests unexpected high species diversity in the opsariichthine fishes (Teleostei: Cyprinidae) and the revalidation of Opsariichthys macrolepis. Ecol. Evol. 9, 2664–2677. doi: 10.1002/ece3.4933
Wang, T. Y., Tzang, C., Teng, H. Y., and Chang, T. C. (2007). Phylogeography and identification of a 187-bp-Long duplication within the mitochondrial control region of Formosania lacustre (Teleostei: Balitoridae). Zool. Stud. 46, 569–582.
Wang, J., Zhang, W., Wu, J., Li, C., Ju, Y. M., Lin, H. D., et al. (2022). Multilocus Phylogeography and population genetic analyses of Opsariichthys hainanensis reveal Pleistocene isolation followed by high gene flow around the Gulf of Tonkin. Genes 13:1908. doi: 10.3390/genes13101908
Wang, X., and Zhu, C. (1997). A Model of the Uplift in Huangshan Area Since the Late Cretaceous. Geogr. Res. 16, 75–81.
Watanabe, K., Tabata, R., Nakajima, J., Kobayakawa, M., Matsuda, M., Takaku, K., et al. (2020). Large-scale hybridization of Japanese populations of Hinamoroko, Aphyocypris chinensis, with a. kikuchii introduced from Taiwan. Ichthyol. Res. 67, 361–374. doi: 10.1007/s10228-019-00730-9
Won, H., Jeon, H. B., and Suk, H. Y. (2020). Evidence of an ancient connectivity and biogeodispersal of a bitterling species, Rhodeus notatus, across the Korean peninsula. Sci. Rep. 10:1011. doi: 10.1038/s41598-020-57625-3
Wu, T. H., Tsang, L. M., Chen, I. S., and Chu, K. H. (2016). Multilocus approach reveals cryptic lineages in the goby Rhinogobius duospilus in Hong Kong streams: role of paleodrainage systems in shaping marked population differentiation in a city. Mol. Phylogenet. Evol. 104, 112–122. doi: 10.1016/j.ympev.2016.07.014
Xia, X. (2018). DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 35, 1550–1552. doi: 10.1093/molbev/msy073
Xiao, W., Zhang, Y., and Liu, H. (2001). Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mo. Phylogenet. Evol. 18, 163–173. doi: 10.1006/mpev.2000.0879
Xu, G. J., Cai, K. Z., Shao, G. Z., Wang, J. P., and Lin, H. D. (2005). Phylogeography and population genetic structure of Sinibrama macrops based on mtDNA. BioFormosa 40, 58–67.
Xu, J., and Zhou, B. (2012). The recent tectonic stress field of offshore of China mainland and adjacent areas. Earth Sci. Front. 19, 284–292.
Yan, F., Lü, J., Zhang, B., Yuan, Z., Zhao, H., Huang, S., et al. (2018). The Chinese giant salamander exemplifies the hidden extinction of cryptic species. Curr. Biol. 28, R590–R592. Available at:. doi: 10.1016/j.cub.2018.04.004
Yan, Y., Yao, D., Tian, Z. X., Huang, C. Y., Dilek, Y., Clift, P. D., et al. (2018). Tectonic topography changes in Cenozoic East Asia: a landscape erosion-sediment archive in the South China Sea. Geochem. Geophys. Geosystems. 19, 1731–1750. doi: 10.1029/2017GC007356
Yang, J. Q. (2012). Phylogeographical analysis on Squalidus argentatus recapitulates historical landscapes and drainage evolution on the island of Taiwan and mainland China. Int. J. Mol. Sci. 13, 1405–1425. doi: 10.3390/ijms13021405
Yang, L., and He, S. (2008). Phylogeography of the freshwater catfish Hemibagrus guttatus (Siluriformes, Bagridae): implications for South China biogeography and influence of sea-level changes. Mol. Phylogenet. Evol. 49, 393–398. doi: 10.1016/j.ympev.2008.05.032
Yang, J. Q., Hsu, K. C., Liu, Z. Z., Su, L. W., Kuo, P. H., Tang, W. Q., et al. (2016). The population history of Garra orientalis (Teleostei: Cyprinidae) using mitochondrial DNA and microsatellite data with approximate Bayesian computation. BMC Evol. Biol. 16:73. doi: 10.1186/s12862-016-0645-9
Yang, G. R., and Huang, H. J. (1964). The cyprinids fishes of China (I): Leuciscinae. Shanghai: Shanghai People’s Press.
Yang, X., Ni, X., and Fu, C. (2022). Phylogeographical analysis of the freshwater gudgeon Huigobio chenhsienensis (Cypriniformes: Gobionidae) in southern China. Life 12:1024. doi: 10.3390/life12071024
Yi, L., Ye, X., Chen, J., Li, Y., Long, H., Wang, X., et al. (2014). Magnetostratigraphy and luminescence dating on a sedimentary sequence from northern East China Sea: constraints on evolutionary history of eastern marginal seas of China since the early Pleistocene. Quat. Int. 349, 316–326. doi: 10.1016/j.quaint.2014.07.038
Yin, W. (2015). Studies on phylogeny and biogeography of the opsariichthine fishes. Doctoral thesis. Shanghai: Fudan University.
Yin, W., Cao, K., He, H., and Fu, C. (2016). Four complete mitochondrial genomes of the genera Candidia, Opsariichthys, and Zacco (Cypriniformes: Cyprinidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 4613–4614. doi: 10.3109/19401736.2015.1101582
Yu, H. T. (1995). Patterns of diversification and genetic population structure of small mammals in Taiwan. Biol. J.Linn. Soc. 55, 69–89. doi: 10.1016/0024-4066(95)90029-2
Yu, D., Chen, M., Tang, Q., Li, X., and Liu, H. (2014). Geological events and Pliocene climate fluctuations explain the phylogeographical pattern of the cold water fish Rhynchocypris oxycephalus (Cypriniformes: Cyprinidae) in China. BMC Evol. Biol. 14:225. doi: 10.1186/s12862-014-0225-9
Zhang, E., and Chen, Y. (1997). Fish fauna in northeastern Jiangxi Province with a discussion on the zoogeographical division of East China. Acta Hydrobiol. Sin. 21, 254–261.
Zhang, D., Fengquan, L., and Jianmin, B. (2000). Eco-environmental effects of the Qinghai-Tibet plateau uplift during the quaternary in China. Environ. Geol. 39, 1352–1358. Available at:. doi: 10.1007/s002540000174
Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., et al. (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, H. N., Chen, W. G., Huang, K. R., Li, Z. Q., Zhang, F. L., and Chen, G. Z. (1990). The new geological structures, tectonic movements and geological environment in coastal line of South China. Beijing: Earthquake Press.
Zhao, L., Zhang, J., Liu, Z., Funk, S. M., Wei, F., Xu, M., et al. (2008). Complex population genetic and demographic history of the Salangid, Neosalanx taihuensis, based on cytochrome b sequences. BMC Evol. Biol. 8:201. doi: 10.1186/1471-2148-8-201
Keywords: Opsariichthys acutipinnis-evolans complex, phylogeography, cytochrome b, allopatry, quaternary glaciation, evolutionary significant unit
Citation: Gao J, Yu D and Liu H (2023) Phylogeographic analysis revealed allopatric distribution pattern and biogeographic processes of the widespread pale chub Opsariichthys acutipinnis-evolans complex (Teleostei: Cyprinidae) in southeastern China. Front. Ecol. Evol. 11:1142810. doi: 10.3389/fevo.2023.1142810
Received: 12 January 2023; Accepted: 01 March 2023;
Published: 22 March 2023.
Edited by:
Hung-Du Lin, National Tainan First Senior High School, TaiwanReviewed by:
Chenhong Li, Shanghai Ocean University, ChinaCopyright © 2023 Gao, Yu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanzhang Liu, aHpsaXVAaWhiLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.