- 1Department of Biology, College of Science, United Arab Emirates University, Al-Ain, Abu-Dhabi, United Arab Emirates
- 2Khalifa Center for Genetic Engineering and Biotechnology, United Arab Emirates University, Al Ain, United Arab Emirates
- 3National Water and Energy Center, United Arab Emirates University, Al Ain, United Arab Emirates
Date palms are widely cultivated in arid agroecosystems, where knowledge of irrigation water effect on their soil and root-associated bacterial communities is limited. Using 16S rDNA metabarcoding, we studied soil and root-associated bacterial communities of date palms growing in United Arab Emirates. Overall, 12.18% of bacterial operational taxonomic units (OTUs) were unique to roots, whereas 41.55% were specific to soil. The bacterial diversity was lower in root and community patterns were distinct between compartments, wherein irrigation water pH was a key structuring factor in both compartments, while salinity (electrical conductivity) was important only in the soil. Co-occurrence network analysis revealed a decrease in complexity in the soil–root continuum, and specific taxa/modules also varied with water pH. We observed a higher abundance of endophyte–saprotroph (Bacillus, Streptomyces and Dongia) dual-role OTUs in both compartments, possibly involved in nutrient mobilisation and plant growth. Based on PICRUSt and trait-based analyses, we showed that these OTUs are putatively involved in the nitrogen cycle (nitrification, denitrification, and assimilatory nitrate reduction). Taken together, we suggest that irrigation water pH, not soil pH, transiently affects belowground bacterial communities and selects bacteria in specific pH ranges, which may be important for nutrient cycling (i.e. nitrogen cycle) in arid agroecosystems.
1 Introduction
Phoenix dactylifera L., commonly known as date palm, is a perennial tree species of significant economic and cultural importance. This species is renowned for its exceptional ability to thrive under harsh environmental conditions characterised by high temperatures, low rainfall, and high soil salinity and therefore widely cultivated in arid agroecosystems. Arid agroecosystems are distinct from other agroecosystems in terms of nutrient availability (nitrogen (N), carbon, phosphorus, and other micronutrients) due to prevailing harsh environmental conditions. The nutrients present in soil are lost in the form of gases (carbon dioxide, methane, and dinitrogen) or leach into groundwater through percolation due to agricultural practices (Nielsen and Ball, 2015). A major challenge in arid agroecosystems is the availability and quality of irrigation water because the use of unsuitable groundwater (saline and alkaline) can cause changes in soil pH and salinity, which lead to land degradation (Chen et al., 2019; Batarseh et al., 2021). Soil pH variations can affect soil multifunctionality (potassium, calcium, magnesium, and sodium) (Wei et al., 2022) and crop health. Similarly, salinisation can affect nutrient availability by altering soil properties (flocculation and dispersion), and levels of soil organic matter (SOM) (Zhou et al., 2020), thereby making nutrients unavailable for plants. Irrigation water characteristics are also known to influence edaphic characteristics (soil organic carbon (SOC), electrical conductivity (EC) and pH), which can act as a filter for microbial selection depending on their adaptation capability in belowground compartments.
The microbiota of soil and root compartments perform various roles and work in unison to alleviate stress of their host plant. Alkaline soil pH is one of the important stressors in desert ecosystems (Guo et al., 2021) and soil bacteria from such ecosystems are adapted to alkaline pH through modifications of membrane proteins, accumulation of cationic amino acids (lysine, arginine, and histidine), increased utilisation of H+ for ATP synthase activity, and activation of antiporters (Na+/H+ and Na+(K+)/H+), which are important for maintaining cytosolic pH (Padan et al., 2005). Soil bacteria are also known to produce organic acids which react with alkaline mineral ions and form neutral compounds (Wu et al., 2019), thereby help mitigating negative effects (i.e., nutrient availability and salinity) of soil alkalinity. Similarly, soil bacteria (i.e. Bacillus and Streptomyces) accumulate osmolytes (starch, choline, and glycine betaine-“organic-in”), salts (potassium-“salt-in”), and antioxidant enzymes (catalase and superoxide dismutase) for survival under salinity-stress conditions (Gregory and Boyd, 2021). In addition to these adaptations, bacterial members (i.e. Bacillus) also produce biofilms through the secretion of extracellular polysaccharides under drought conditions, which helps in aggregating soil particles (Tsotetsi et al., 2022), thereby improving nutrient mobilisation (iron and phosphate) and promoting plant growth in arid agroecosystems. A comprehensive understanding of soil and root-associated bacterial functionalities in arid agroecosystems is crucial for enhancing soil health and augmenting crop productivity in these challenging environments.
Bulk soil serves as the starting material for the selection of bacteria in the roots of Arabidopsis thaliana (Schlaeppi et al., 2014). The selection of bacteria in root mainly depends on the adaptation of bacteria to stress or resource availability (root exudates and rhizodeposits), which causes variation in bacterial diversity and communities between endophytic and soil (rhizosphere/bulk) communities. The relative abundance of Actinobacteria is generally high in rhizosphere/bulk soil and is followed by Proteobacteria, Cyanobacteria, and Planctomycetes in arid ecosystems (Vásquez-Dean et al., 2020), whereas the root shows a higher abundance of Acidobacteria, Planctomycetes, Chloroflexi, and Verrucomicrobia (Trivedi et al., 2022). The abundance of bacterial taxa and diversity are known to decrease in a gradient manner in the soil–rhizosphere–endosphere continuum (Zhang et al., 2022). Higher soil pH can alter root-associated bacterial communities directly or spill-over effects through changes in the plant host (stunted growth, altered root morphology, and function (root exudates)) (Guo et al., 2020). Similarly, soil salinity is also known to influence bacterial communities in barley and cotton-cultivated soils (Li et al., 2021). In addition to edaphic characteristics, climate-related factors, such as mean annual temperature and mean annual precipitation, have also been reported as strong predictors of bacterial diversity in both local (Cherif et al., 2015) and global (Vásquez-Dean et al., 2020) levels in arid ecosystems. However, the effect of irrigation water characteristics and associated edaphic changes on diversity and communities in the root–soil continuum is poorly studied.
Bacterial communities in belowground compartments perform various complex functions such as nutrient cycling, plant growth promotion, and biocontrol, which can be inferred comprehensively using co-occurrence network patterns. Co-occurrence indicates the simultaneous presence of two bacterial taxa in a given system but does not necessarily point to actual physical interactions. Studying microbial interactions through co-occurrence networks provides clues related to microbial diversity and community assembly. Network analysis also provides insights into bacterial traits (mutualism (positive) and competition (negative)), which are important for understanding the stability, resistance, and resilience of bacterial communities. In addition, the environmental impact (i.e. drought, salinity etc) on bacterial functions can be inferred from changes in co-occurrence patterns and keystone OTUs (de Vries et al., 2018). Knowledge of co-occurrence patterns and keystone taxa of arid agroecosystems and their changes in response to perturbations in environmental variables is not well documented, and a thorough investigation is required to better understand ecosystem functioning in arid environments.
The overall well-being of agroecosystems depends on the close interaction between the roots and the surrounding soil and their impact on biogeochemical cycles and ecosystem functions. Soil bacteria enhance SOM and N accumulation under alkaline conditions (Zhang et al., 2021), thereby improving the nutrient status of the soils. Saprotrophic soil-dwelling bacteria degrade SOM through hydrolytic enzymes and release sugars, which are fermented to organic acids and used as precursors for methanogenesis (methane and carbon dioxide) in the micro-anoxic niches of the soil matrix (Angel et al., 2011). Similarly, nitrogen fixation is carried out by root-associated and free-living bacteria, while ammonia oxidation and denitrification is generally carried out in the soil (Ling et al., 2022). Changes in edaphic characteristics such as soil salinity and moisture can affect nitrification and denitrification in arid soils (Meng et al., 2020). Similarly, pH also affects nitrogen cycle-related pathways, namely, nitrification (Wang et al., 2019b), denitrification (Čuhel et al., 2010), and nitrate reduction (Rafrafi et al., 2015). Therefore, belowground soil and root compartments can act as a source of climate-active gases such as methane, carbon dioxide, and nitrous oxide (Hall et al., 2012) and lead to critical nutrient loss in nutrient-limited arid soils. The effect of irrigation water characteristics on belowground bacterial communities is a crucial aspect of agroecosystems, as it has the potential to affect biogeochemical cycles and ecosystem functioning (Delgado-Baquerizo et al., 2016; Xing et al., 2022).
In this study, we aimed to elucidate the effect of irrigation water characteristics and concomitant changes in soil chemistry on bacterial diversity and communities of root and bulk soil of date palm (Phoenix dactylifera L.), which are commonly cultivated in arid agroecosystems of United Arab Emirates (UAE). We also investigated the co-occurrence network patterns of bacterial communities and prediction-based functional analyses to understand the implications of irrigation water characteristics on putative microbial interactions and nutrient cycling in soil and root compartments of date palms. Overall, unravelling the influence of irrigation water on bacterial diversity, communities, their interactions, and potential functions will further deepen our understanding of belowground bacterial communities, which has enormous impact improving plant survival, plant health and productivity in challenging arid agroecosystems.
2 Materials and methods
2.1 Study site and sampling description
We selected seven study sites that are located across the oasis agroecosystem of Al Ain (Abu Dhabi Emirates), UAE. According to the Köppen climate classification, the climate of the study sites was classified as “Bwh”, which is indicative of a tropical and subtropical desert climate (Table S1). These sites were characterized by a higher mean annual temperature (MAT) ranging from 25.3 to 27.8°C and a lower mean annual precipitation (MAP) ranging from 75 to 109 mm, which are typical for arid agroecosystems. At each study site, we have collected triplicate soil and root samples at 20–30 cm depth from date palms separated approximately 50 m away from each other in March 2020. The height and age of the date palm trees were similar across sites and they were irrigated with freshwater at a rate of 50–60 liters per tree once every three days. In total, we collected 42 samples (7 farms × 2 compartments × 3 replicates) from seven different sites. Soil and root samples were transported to the laboratory under cooling conditions and stored at −20°C until DNA isolation. In addition, we have also collected irrigation water samples from each site for chemical analyses (pH and EC). Detailed information regarding the chemical analysis (water and soil) has been provided in previous studies (Loganathachetti et al., 2022; Shamim et al., 2022) and briefly included in Table S1. The irrigation water pH (6.96–7.99) and EC (0.32–3.22 dS.m−1) of the freshwater varied widely between the sampling sites. Among the study sites, the irrigation water pH was the highest in S3 and S4, while irrigation water EC was the highest in S7 (Figure S1). Soil pH (6.74–7.74), EC (0.002–0.503 dS.m−1), and OM (1.7–6.08%) also fluctuated between sites. We found a weak significant correlation (Spearman’s ρ = −0.45; P < 0.05) between water pH and water EC. Soil pH also showed a significant weak correlation (Spearman’s ρ = −0.46; P < 0.05) with soil OM (Figure S1).
2.2 DNA isolation and V3–V4 amplicon sequencing
Soil DNA isolation was performed according to the manufacturer’s instructions using E.Z.N.A soil DNA kit. We added approximately 250 mg of soil sample to a bead-beating tube, along with a lysis buffer (SLSX and DS buffer) and a series of mechanical and chemical treatments to lyse cells and remove contaminants using silica column. Similarly, 1 g of root samples were ground with liquid nitrogen to obtain a ground biomass. This was used for lysis (AP1 buffer) with proteinase K (1.2 mg/mL), followed by binding and purification of DNA as per the DNeasy Plant Mini Kit (Qiagen, USA) protocol. The purity of the DNA was determined using Nanodrop before performing PCR. We used 341F (CCTACGGGNGGCWGCAG) and 805R (GACTACHVGGGTATCTAATCC) (Herlemann et al., 2011) primers for the amplification of V3–V4 16S rRNA genes. PCR conditions and purification of barcoded amplicon libraries were based on a previous study (Loganathachetti et al., 2022). The amplicon libraries were sequenced using Illumina MiSeq at the Integrated Microbiome Resource (IMR) lab, Nova Scotia, Canada (https://www.imr.bio.com) based on standard procedures. The demultiplexed sequence files from this study have been deposited in the Zenodo repository (https://zenodo.org/) (10.5281/zenodo.60782 92 and 10.5281/zenodo.6371857).
2.3 Bioinformatics analyses
We analysed 907,507 raw sequence reads using the Divisive Amplicon Denoising Algorithm 2 v1.12 DADA2 R package (Callahan et al., 2016, accessed on 20 December 2021). We performed primer removal, trimming, error learning, denoising, merging, amplicon sequence variant generation, and removal of chimeric reads using appropriate R packages, as described by Loganathachetti et al. (2022). An additional clustering step was performed at 97% sequence similarity using vsearch v2.15 (Rognes et al., 2016) and a total of 6,279 Operational Taxonomic Units (OTUs) were obtained. We performed a series of steps, namely, clustering, singleton, and chimera removal, using vsearch v2.15, which finally yielded 5,504 OTUs (425,521 reads) after the removal of 1,245 OTUs (12,523 reads). The taxonomic classification of OTUs (5,504 OTUs: 425,521 reads) was carried out by comparing them with the Silva database v138.1 using assignTaxonomy function of the DADA2 R package (Quast et al., 2012). We removed 15 archaeal (45 reads), 11 chloroplast (94,833 reads), 5 mitochondrial (20201 reads), 1 non-bacterial (2 reads), 2 unclassified kingdom level OTUs (36 reads), and 1091 OTUs containing less than five reads (6,852 reads) from the count table. After completing the aforementioned processes, the OTU table consisted of 3487 OTUs (310,404 reads) from 38 samples (read range: 1,147–20,298). We normalised the OTU table based on the sample with the lowest sequence count (1,147), which was used for diversity and community analyses using the rrarefy function of the Vegan R package (Oksanen et al., 2020).
We predicted putative functional pathways of the OTUs using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) for both the soil and root samples. Initially, OTUs were assigned to KEGG orthologues (KO), and then MetaCyc pathway abundance was calculated based on KO. The abundance percentage of putative assimilatory nitrate reduction to ammonium (ANRA) OTUs in the soil and root samples was calculated by comparing the genus information obtained from nirA and narG databases. These databases were prepared after downloading the bacterial taxa information against “nirA” and “narG” search in the Genbank, NCBI (https://www.ncbi.nlm.nih.gov/genbank/, accessed on 29 November 2022) followed by manual curation (Tables S2, S3).
2.4 Statistical analyses
We used R v4.0.3 (R Core Development Team, 2020) for all statistical analyses unless stated otherwise. The OTU abundance table was arcsine-transformed to minimise heterogeneity. The environmental variables (water pH, water EC, soil pH, soil EC, and soil OM) were Z-transformed to Z-scores (0–1) to increase homogeneity. We applied the analyses of variance (ANOVA) test and Tukey’s HSD post-hoc test to statistically analyse the significant differences in bacterial diversity (richness, diversity, and evenness) between soil and root compartments using the Agricolae R package. We performed Non-Metric Multi-Dimensional Scaling (NMDS) analysis to identify the differences in the community structure between soil and root compartments and Permutational Analysis of Variance (PERMANOVA) for identifying environmental variables important for community structuring using Vegan R package. A forward selection approach was applied for the environmental variables, and the model with the lowest Akaike information criterion was considered the final model. Briefly, we initially tested the significance of individual factors, and the final model was prepared by ranking them based on their R2 values. NMDS and PERMANOVA analyses were performed separately for the soil and root compartments, respectively. In functional prediction analyses, the significantly different MetaCyc pathways between the soil and root compartments were identified by t-test using Prism 6 software and visualised using the Pheatmap R package.
Co-occurrence network analysis was performed separately for OTUs detected in soil and root using the iNAP workflow (Feng et al., 2022, accessed on 15 September 2022), which was based on a previous method by Deng et al. (2012). First, we performed majority selection which filtered OTUs based on occurrence in at least 80% of samples. The filtered OTU table was subjected to SparrCC correlation and an adjacency matrix was generated (Friedman and Alm, 2012). The OTUs in the adjacency matrix that passed the correlation threshold (<−0.6 to >0.6) significantly (P < 0.05) were used for calculating network centralities (within module: Zi and between module: Pi) using “Greedy modularity optimization” method (Newman, 2004) using the iNAP workflow. We utilized Gephi v0.9 to calculate topological characteristics, including average path length, average degree, and modularity, and to visualise the co-occurrence network patterns. The average degree is defined as the number of connections in relation to the total number of nodes, and the average path length points are the mean distances of all pairs of nodes. Higher modularity indicates denser interactions of nodes within modules and fewer interactions among the nodes of other modules. The nodes in the co-occurrence networks were categorised as module hubs (Zi > 2.5, Pi ≤ 0.62), network hubs (Zi > 2.5, Pi > 0.62), connector hubs (Zi ≤ 2.5, Pi > 0.62), and peripheral OTUs (Zi ≤ 2.5, Pi ≤ 0.62) (Olesen et al., 2007). Except for peripheral OTUs, all types of nodes (connector-, module-, and network hubs) are called as keystone OTUs (Deng et al., 2012).
3 Results
3.1 Bacterial diversity and community structure of soil and root compartments
Of the total OTUs, 41.55% were found only in the soil, 12.18% were found only in the roots, and 46.27% were recorded in both soil and root samples (Figure S2). Bacterial richness, Shannon diversity index, and evenness were significantly higher in the soil than in the roots (Figures 1A–C). The rarefaction curves appeared to reach a plateau for both the soil and root compartments, indicating a sufficient sequencing depth (Figure S2). We found distinct bacterial community structures in the soil and roots in the NMDS ordination space (Figures 1D–F) (R2 = 0.1635; P = 0.0001). At the individual compartment level, PERMANOVA analyses showed that irrigation water pH was a major factor influencing bacterial community structure in both the soil (R2 = 0.2157; P < 0.0001) and root (R2 = 0.12375; P = 0.0018) (Table 1), whereas soil EC was a significant additional factor in the soil affecting bacterial community structure (R2 = 0.0935; P = 0.0359).
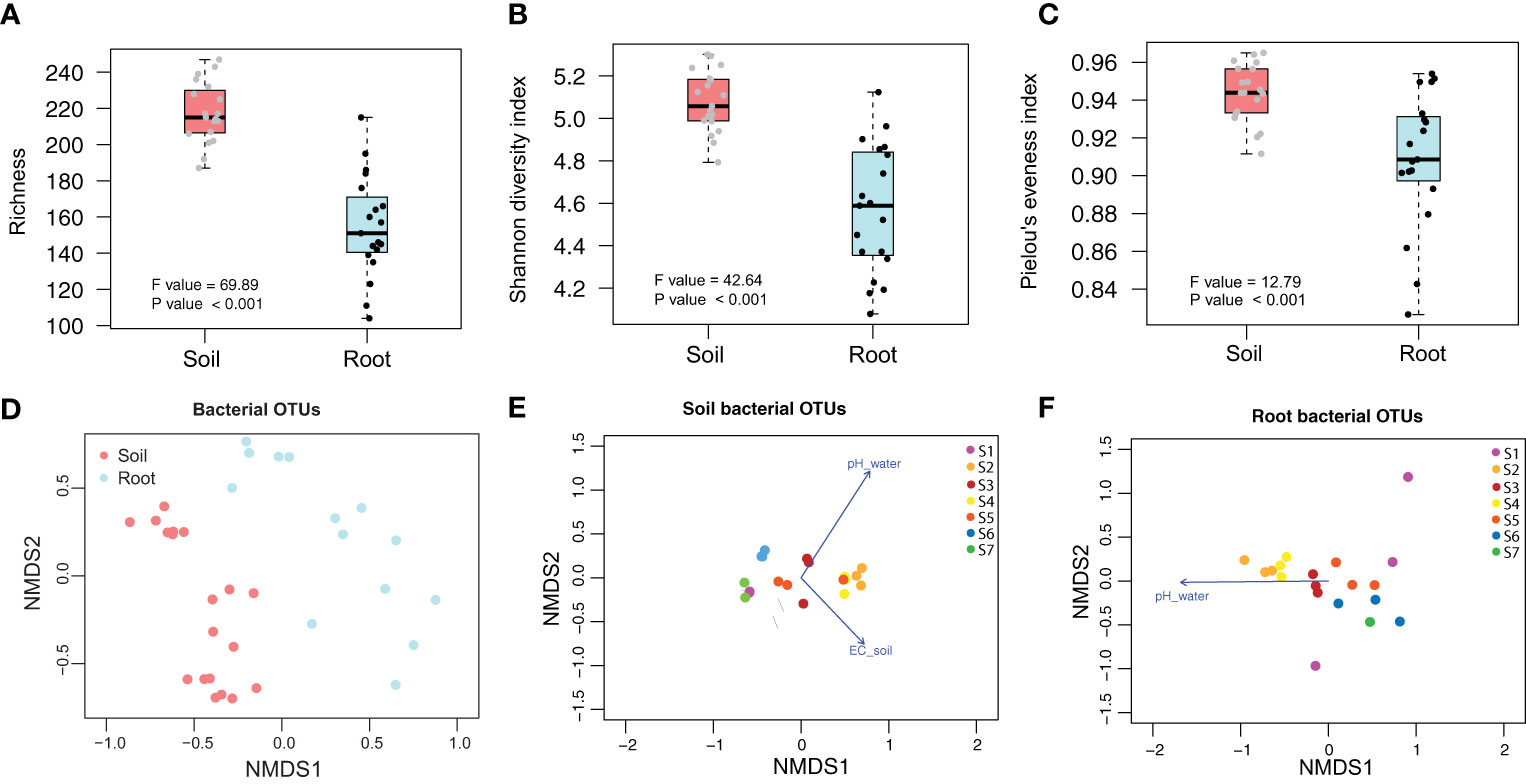
Figure 1 Bacterial diversity and community patterns of soil and root compartments. The boxplots show (A) richness, (B) Shannon’s diversity index, and (C) evenness tested using ANOVA followed by Tukey’s HSD post-hoc test. Statistical inference is highlighted within the plot (P < 0.05). The box spans the interquartile range (IQR; first quartile to the third) with the median indicated by a dark horizontal line, the whiskers show the 1.5xIQR. The bacterial community structure in the NMDS ordination for (D) soil and root compartments, (E) soil compartment, and (F) root compartment. The arrows point in the direction of maximum increase of individual vector variables and had significant effects (P < 0.05) on the NMDS ordination. The S1–S7 points site numbers in (E, F).
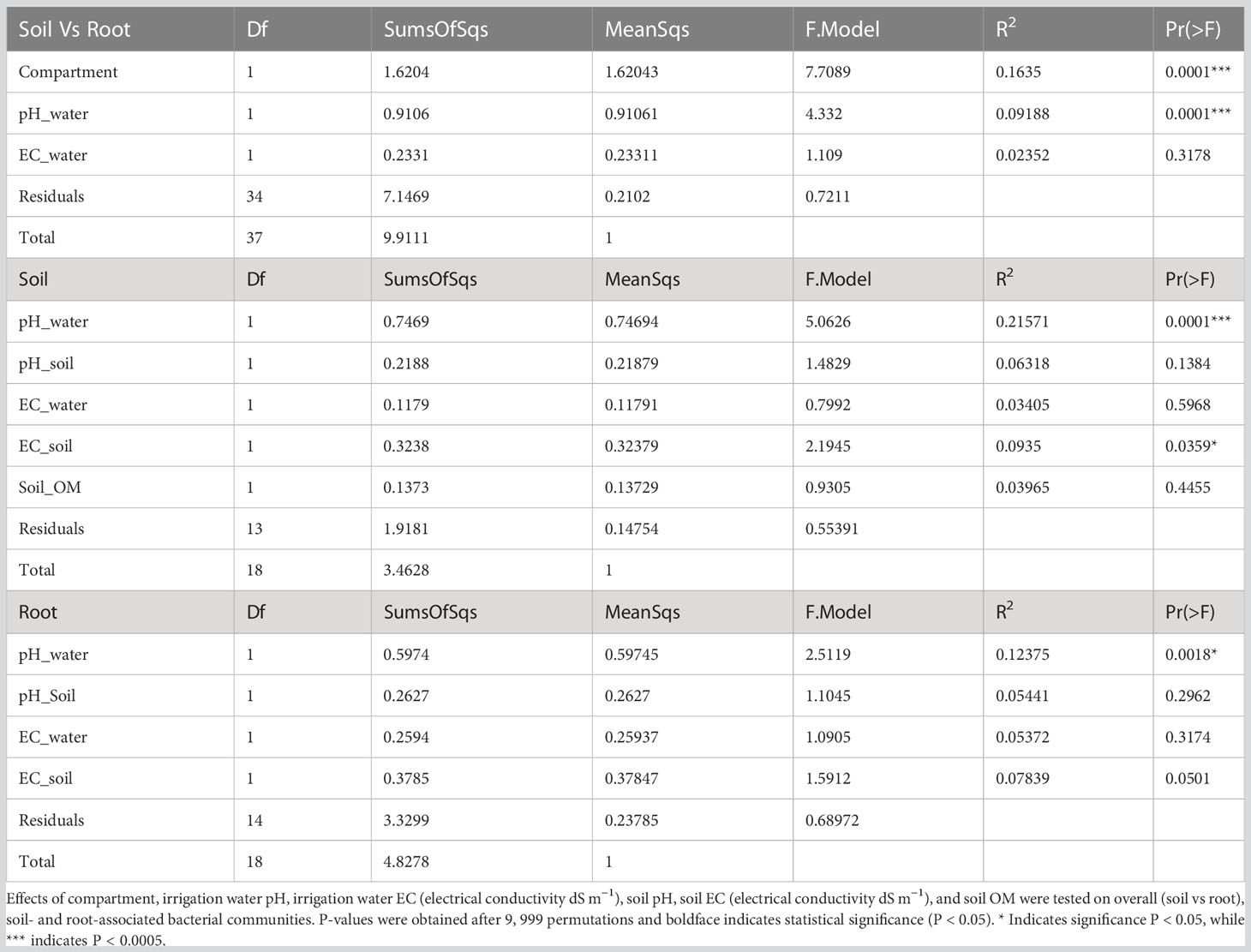
Table 1 PERMANOVA analysis showing effects of environmental variables on date palm bacterial community structure.
3.2 Co-occurrence network patterns and topological characteristics of soil and root compartments
The co-occurrence networks were generated from 436 soil and 129 root associated bacterial OTUs. These networks were used for the calculation of topological characteristics (average degree, network diameter, average path length, modularity, modules, nodes, co-occurrence, and co-exclusion) and identification of keystone OTUs (connector, module and network hubs), which are shown in Figure 2 and Table S4. The complexity of the co-occurrence patterns (higher average degree, higher nodes, lower modularity, and lower average path length) was relatively higher in the soil than in the roots. The soil samples consisted of four modules (SM1–4) while the root contained four modules (RM1–3). Soil modules (SM3 and SM4) were positively correlated to irrigation water pH (P < 0.05). Similarly, specific root modules also showed positive correlation to irrigation water pH (RM2 and RM3) and soil pH (RM2) respectively (P < 0.05) (Figure 2), while RM1 module was negatively correlated to soil OM (P < 0.05). The correlation and the significance of the module–trait (water pH, water EC, soil pH, soil EC, and soil OM) relationships of both soil and root are shown in Figure 2. List of keystone OTUs detected in both compartments are presented in Table S4.
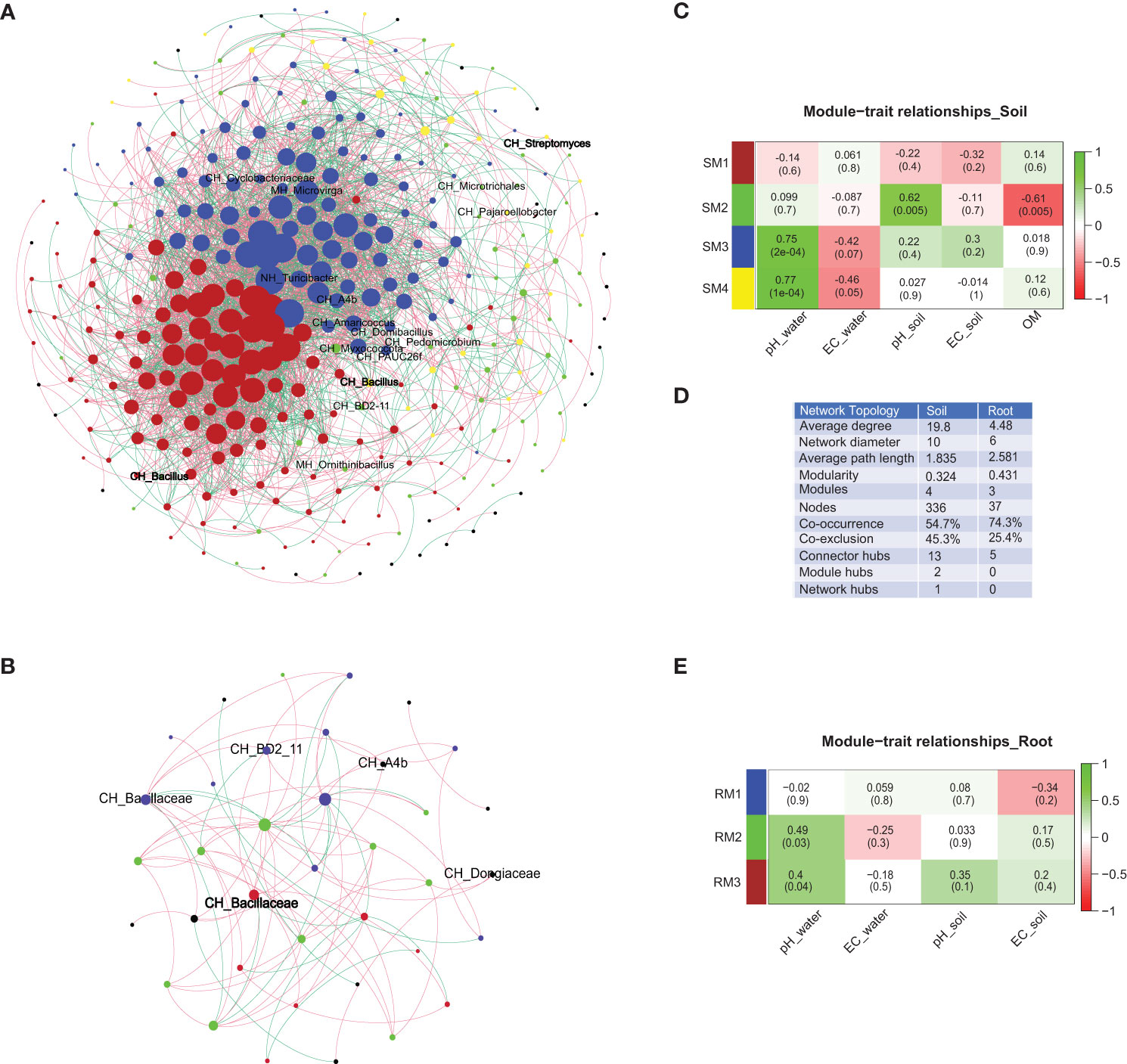
Figure 2 Bacterial co-occurrence and co-exclusion network patterns and module-trait relationships of soil and root compartments. Co-occurrence and co-exclusion network patterns of (A) soil, (B) root, (C) module–trait analysis of root, (D) topological characteristics, and (E) module–trait analysis of soil. The edges indicating co-occurrence are coloured in green, while co-exclusion is coded in red. The colour of the nodes in the networks and the first column of the module–trait relationship indicate respective modules. The direction of the correlation (scale 1 to −1) and statistical significance (P-value) of module–trait analysis of soil and root modules is given in the tiles. Connector hubs (CH), module hubs (MH) and network hubs (NH) are labelled within the networks.
3.3 Bacterial composition and putative pathways of soil and root compartments
Soil samples contained the following dominant phyla: Actinobacteria (23.9%), Firmicutes (22.3%), Proteobacteria (22%), Chloroflexi (10.4%), Acidobacteria (8%), and Myxococcota (2.3%), whereas root samples predominantly contained Proteobacteria (36.9%), Actinobacteria (25%), Firmicutes (12.3%), Bacteroidetes (5.6%), Chloroflexi (5.4%), Acidobacteria (4.75%), and Myxococcota (3.3%) (Table S5; Figure 3A). Proteobacteria were represented by Rhizobiales (10.8% and 18.8%), Burkholderiales (1.5% and 2.7%), and Gammaproteobacterales (0.04% and 2.2%) in the root and soil samples, respectively. In contrast, Actinobacteria consisted of Streptomycetales (0.5%, 5.7%), Actinomarinales (7%, 3.6%), Micromonosporales (1.2%, 3.5%), and Gaiellales (2.4%,0.8%), with increased abundance in soil compared to roots (Figure 3B). Acidobacteria order Vicinamibacterales (4.2%, 2.1%) was higher in bulk soil than in root samples. The Bacteroidota orders Bacillales (17.2%, 9.4%) and Paenibacillales (2.4%, 0.3%) were also higher in the bulk soil samples.
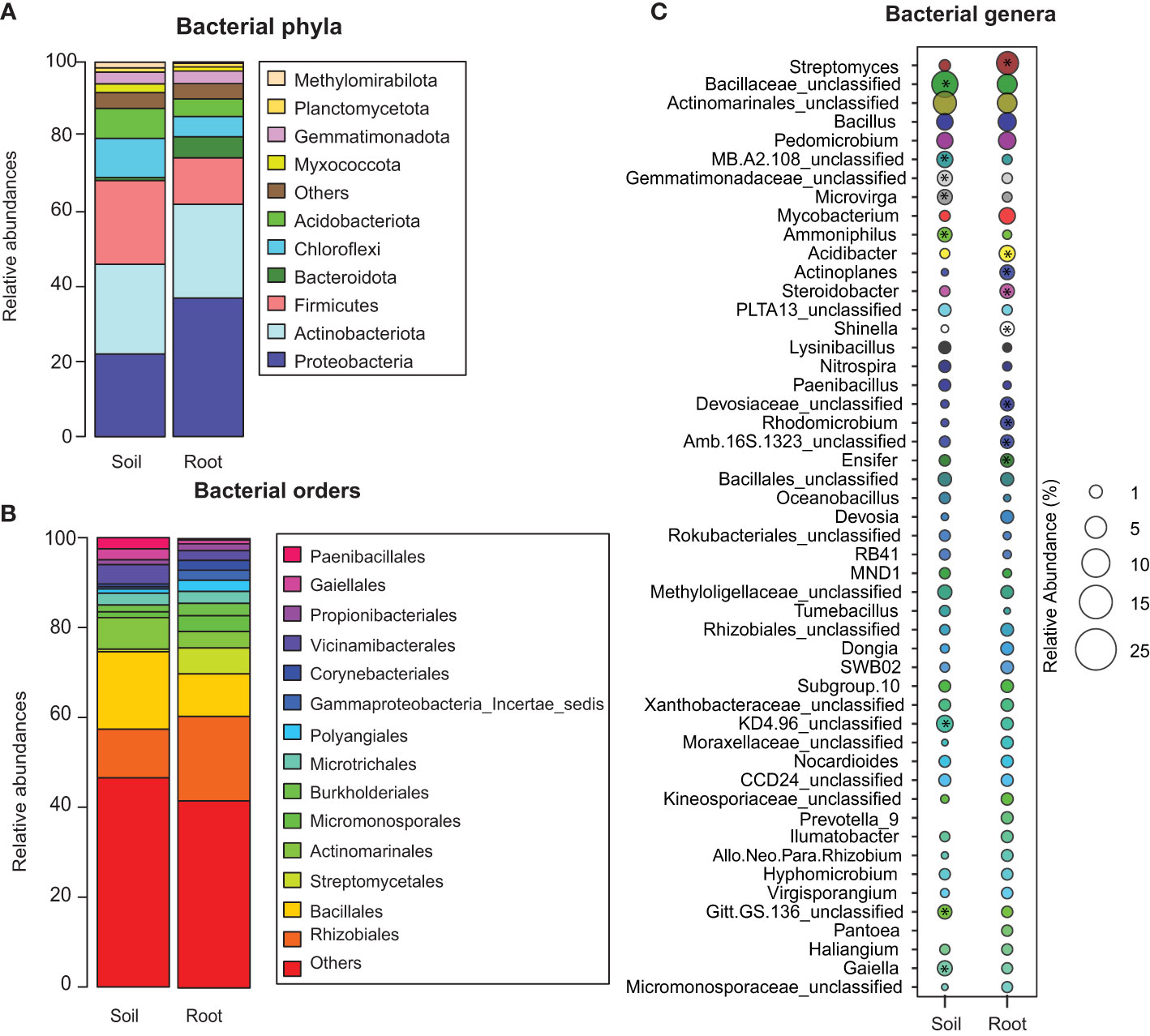
Figure 3 The relative abundance of bacterial taxa in the soil and root compartments of date palm. Relative abundance percentage of bacteria at the (A) phylum, (B) order, and (C) genus levels. * Indicates significantly higher operational taxonomic units (OTUs; P < 0.05) in the marked compartment (root/soil).
Soil compartment was enriched with Microvirga, Ammoniphilus, and Gaiella, whereas root showed enhancement of Streptomyces, Mycobacterium, Devosia, Allo_Neo_Para_Rhizobium, and Steroidobacter, while Bacillus and Pedomicrobium were highly abundant in both compartments (Table 2; Figure 3C). We performed heatmap clustering to identify taxa that differed significantly between soil and root compartments. Bulk soil samples showed a higher abundance of Bacillaceae_unclassified, KD4_96_unclassifed, Gitt_GS-136_unclassifed, Gaiella, Dongia, Gemmatimonadaceae_unclassifed, MB-A2-108_unclassified, Microvirga, and Ammoniphilus. Similarly, root samples showed an increased abundance of Streptomyces, Acidibacter, Actinoplanes, Steroidobacter, Shinella, Rhodomicrobium, Devosiacease unclassified, Amb-16S-1323_unclassified, and Ensifer (Figure 3C). Nitrification- and denitrification-related OTUs were generally dominant in the soil (Nitrospira and Gaiella), whereas roots consisted of genera involved in nitrate reduction (Nitrosomonadaceae) (data not shown).
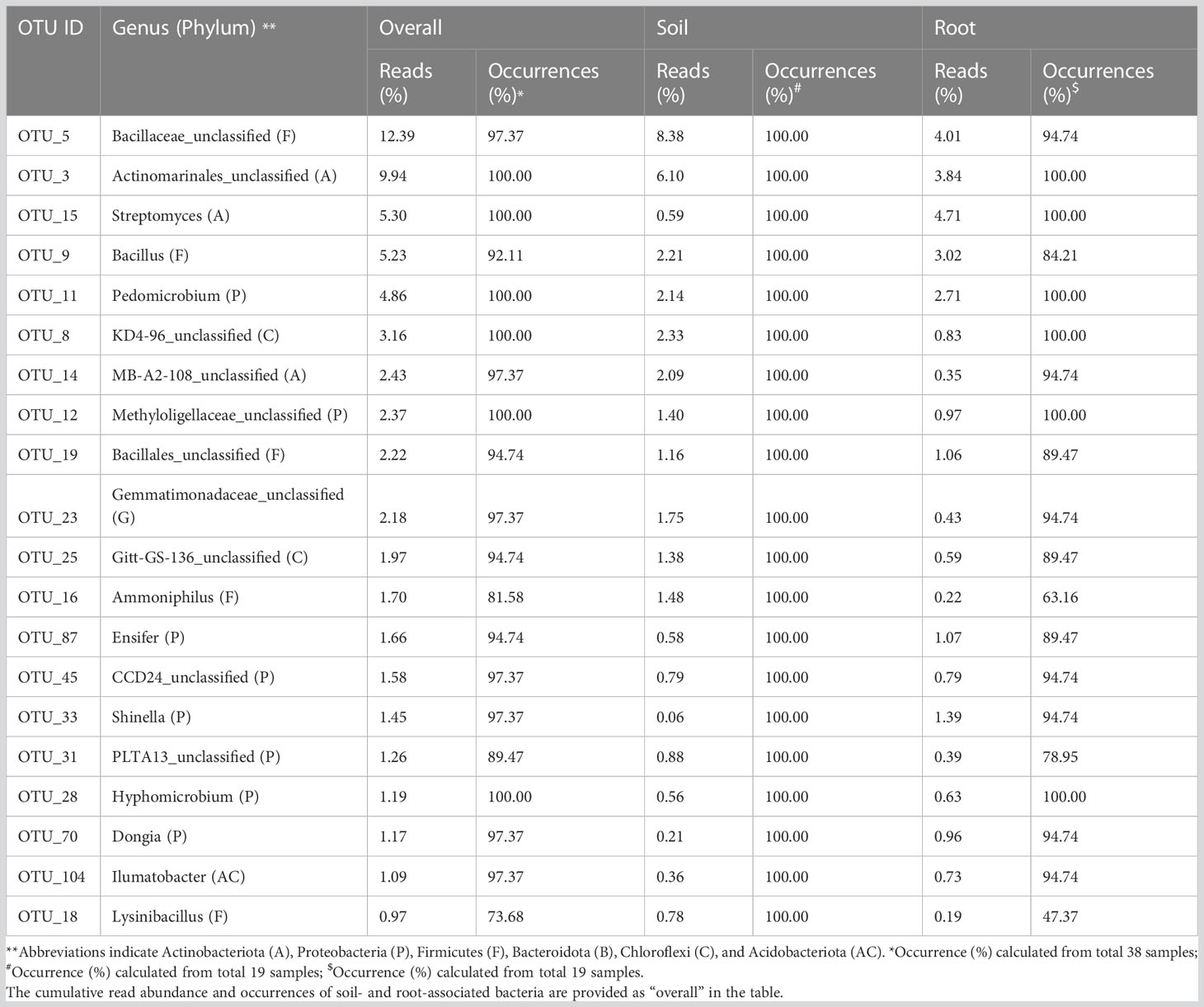
Table 2 Taxonomic identity, sequence read abundance, and occurrences of the top 20 operational taxonomic units (OTUs) detected from soil and root compartments of date palm.
Of the 426 MetaCyc pathways, 84 were found to be significantly (P < 0.05) different between the root and soil. Biogeochemical pathways related to anaerobic degradation, fermentation, methanogenesis, and nitrifier denitrification-related pathways were enriched in the soil (Figure 4). Similarly, amino acid synthesis- and nitrate reduction-related pathways were enriched in the roots. The percentage of taxa capable of assimilatory nitrate reduction was higher in the root than in the soil (Figure S3).
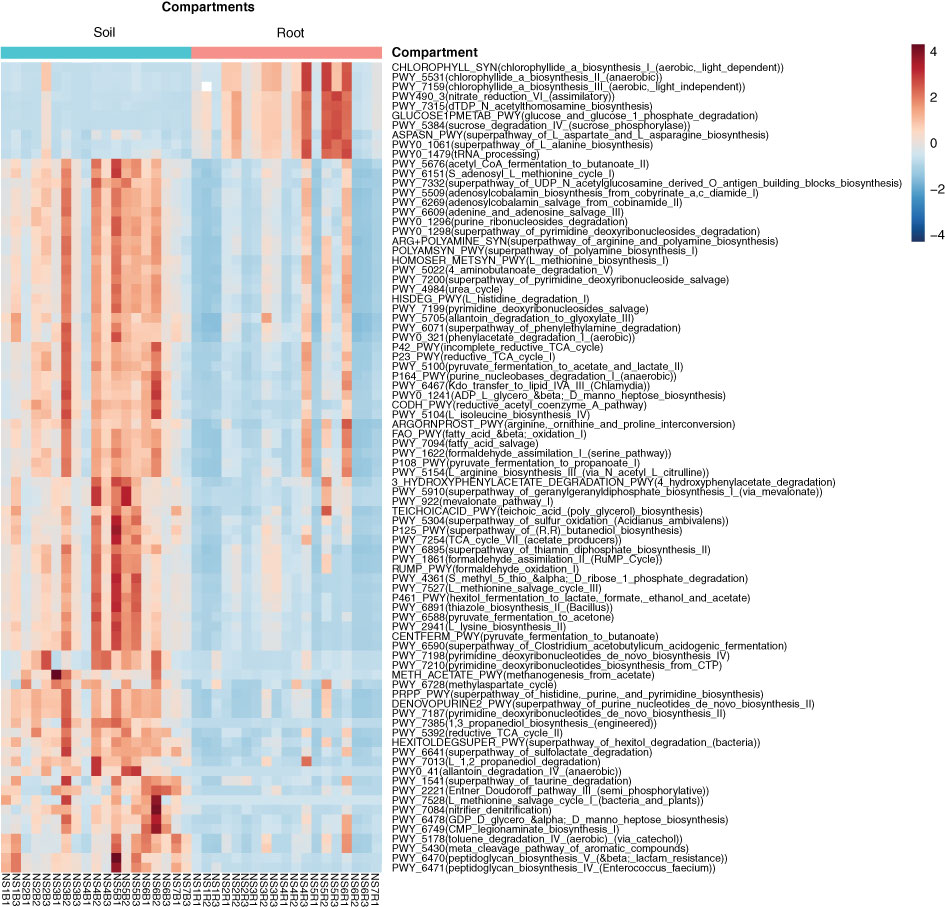
Figure 4 Heatmap showing putative MetaCyc pathway abundance of bacterial OTUs. The intensity of the colours in the heatmap indicates the relatively higher/lower abundance (scale of −4 to 4) of the MetaCyc pathways (P < 0.05) in the soil and root compartments calculated using PICRUSt2.
4 Discussion
We find a lower bacterial diversity in root and distinct bacterial communities between compartments (soil and root) of date palms across sampling sites, in which irrigation water pH was an important structure factor for both compartments, while soil EC is an additional factor for soil bacterial communities. The complexity and stability of the network was higher in soil compared to root, and the abundance of OTUs in specific network modules which are known to participate in nitrogen cycle (nitrification, denitrification, and assimilatory nitrate reduction) increases according to water pH in both compartments. We demonstrate that water pH, and not soil pH, plays a major role in altering bacterial communities and modulating specific modules/taxa involved in the nitrogen cycling in arid agroecosystem.
4.1 Soil and root-associated bacterial community structure based on irrigation water pH
We attribute lower bacterial diversity (richness, Shannon diversity index and evenness) and selection of only 12.8% of total OTUs in the root to the “root effect”, since date palm roots act as “gatekeepers” by selecting specific bacteria from the surrounding soil (Mosqueira et al., 2019) through possible modulations in root exudates and rhizodeposits (Bulgarelli et al., 2012). In this study, we found distinct bacterial communities between soil and root compartments, which were shaped mainly by irrigation water pH. Soil pH is known to structure bacterial communities directly by selecting specific bacteria depending on pH preference or indirectly through changes in nutrient availability (Lammel et al., 2018), especially SOC by altering its dissolution (Anderson et al., 2018). Soil pH has also been reported to structure bacterial communities distinctly according to gradients of agroecosystems (Rousk et al., 2010) and arid (Zeng et al., 2019) ecosystems. It is also known as a global predictor for bacterial diversity (Bahram et al., 2018) and communities. Therefore, we expect an irrigation water pH-dependent filtering effect in roots (Lopes et al., 2021; Xiong et al., 2021), which possibly amplifies host selection pressure and transiently select specific OTUs (i.e. Gaiella–soil and Streptomyces–root), thereby distinctly structure the bacterial communities between compartments. In addition, the higher percolation rate of water through soil (aridsol) in the study regions (Abdelfattah and Shahid, 2007) may also play an important role in transient filtering in arid agroecosystems, while the water pH variations may be attributed to the possible increase in carbonate and bicarbonate (Batarseh et al., 2021) levels. In this study, apart from water pH, soil bacterial communities were also structured based on soil EC, wherein salinity negatively affects bacterial growth through plasmolysis and selectively allows colonisation of salinity-tolerant OTUs (i.e. Bacillus), thereby shaping bacterial communities (O’Brien et al., 2019; Zhang et al., 2019).
4.2 Soil bacterial co-occurrence patterns are more complex and affected by water chemistry and soil OM
Higher resource availability (i.e. root exudates, rhizodeposits, and organic acids) in roots possibly alter the bacterial co-occurrence pattern from a highly complex (higher average degree, higher nodes, lower modularity, and lower average path length) to a less complex pattern in the soil–root continuum. This, along with higher bacterial diversity in soil, indicates higher network stability compared to roots in this study since complex networks were reported to be more resistant to changes in environmental conditions compared to less-complex simple networks (Santolini and Barabási, 2018; Wang et al., 2019a). We observed relatively dense connections within modules than between modules (modularity) in the root than in the soil, which indicates niche partitioning of root-associated bacterial communities of date palm due to selection pressure exerted by the plant host (Qian et al., 2019). In addition, we found a higher percentage of co-occurrence interactions among OTUs in root, pointing a possible enhancement of mutualistic interactions useful for survival and growth under arid conditions (Mosqueira et al., 2019). We detected a higher abundance of putative dual-role root-associated bacteria (Hardoim et al., 2008), Bacillus (biofilm) (Khan et al., 2021), Streptomyces (biocontrol) (Saeed et al., 2017) and Dongia (heat stress), whose modules (RM2 and RM3) correlated positively to irrigation water pH, indicating their versatile role in plant homeostasis and growth promotion. Similarly, we found enrichment of specific OTUs, Hyphomicrobium (denitrification) (Martineau et al., 2015), Romboutsia (acetogenesis), Novibacillus (biocontrol), Gracilibacillus (biocontrol), Pedomicrobium (iron cycle), and Gemmatimonadaceae_unclassified (nitrous oxide reduction) (Oshiki et al., 2022) and their respective modules (SM3 and SM4) to water pH in soil, which perhaps help in regulating soil health and nutrient cycling in arid soil. We also found a module (SM2), which correlates negatively with soil OM concentrations and positively with soil pH, which indicates the preference of bacteria (i.e. Microvirga) for oligotrophic conditions (Zi et al., 2022) and neutral–alkaline pH (Li et al., 2020). Detailed prediction-based functional analysis of OTUs is useful for understanding the irrigation water characteristics associated changes in nutrient cycling of agroecosystems.
4.3 Modulation of nitrogen cycle related OTUs and putative pathways in soil and root compartments
The enrichment of aerobic denitrification (nitrifier denitrification)-related MetaCyc pathways along with a higher abundance of Nitrosomonadaceae OTUs indicate that aerobic denitrification is predominantly carried out in the soil. In this study, the enrichment of specific OTUs (i.e. Nitrosomonadaceae) in relation to water pH increase indicate that water pH is an important factor for denitrification in soil instead of soil pH (Čuhel et al., 2010). We also observed a higher percentage of root-associated bacterial OTUs (Bacillus, Streptomyces, Mycobacterium, Actinoplanes and Hyphomicrobium), which are putatively involved in assimilatory nitrate reduction to ammonium (ANRA) (Fahrbach et al., 2008; Xu et al., 2017; Zhang et al., 2021). In addition, the enhancement of ANRA MetaCyc pathways suggest the importance of root-associated bacteria in preventing nitrogen loss, in which nitrate is converted to ammonium and incorporated into biomass through glutamine biosynthesis (Bernard and Habash, 2009). This is further supported by the presence of OTUs in roots putatively containing nitrite reductase (nirA) and nitrate reductase (narG) at higher abundance.
5 Conclusion
We show a distinct variation in bacterial diversity and community structure of belowground compartments (root and soil) and changes in water and soil chemistry were the important factors affecting the community. Soil bacterial communities were complex, connected and stable, while root-associated bacterial communities were relatively less complex and stable. Root-associated bacterial communities show a higher amount of mutualistic interactions compared to soil indicating their critical role in plant homeostasis. The perturbations in bacterial communities and modules to irrigation water pH provide insights into the possible adaptation mechanisms (endophytic–saprotrophic dual-role) of bacterial OTUs (Bacillus, Streptomyces, Mycobacterium, Dongia and Gaiella) in the belowground compartments of arid agroecosystems. Irrigation water pH may modulate specific modules/OTUs (Bacillus, Streptomyces, Mycobacterium, Actinoplanes and Hyphomicrobium) and possibly increase the abundance of aerobic denitrification (soil) and ANRA pathways (root). Taken together, our findings suggest that irrigation water pH, rather than soil pH, alters bacterial diversity and communities in both soil and root compartments by allowing the growth of selected bacteria in specific pH ranges, which may be important for nutrient cycling (i.e. the nitrogen cycle) in arid agroecosystems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://zenodo.org/record/6078292#.Y73QrezP2HE, 10.5281/zenodo.6078292; https://zenodo.org/record/6371857#.Y73QwezP2HE, 10.5281/zenodo.6371857.
Author contributions
DSL: Performed bioinformatic, statistical analyses and drafted the article; SM: Conceived the project idea, performed fieldwork and funding acquisition. Both authors contributed to the article and approved the submitted version.
Funding
This research work is supported by Asian University Alliance (AUA) grant, G00003654 and #12R133.
Acknowledgments
We thank Subha Chandran for helping in DNA extraction and also thank the Asian University Alliance for providing funding.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1142073/full#supplementary-material
References
Abdelfattah M. A., Shahid S. A. (2007). A comparative characterization and classification of soils in Abu Dhabi coastal area in relation to arid and semi-arid conditions using USDA and FAO soil classification systems. Arid L. Res. Manage. 21, 245–271. doi: 10.1080/15324980701426314
Anderson C. R., Peterson M. E., Frampton R. A., Bulman S. R., Keenan S., Curtin D. (2018). Rapid increases in soil pH solubilise organic matter, dramatically increase denitrification potential and strongly stimulate microorganisms from the firmicutes phylum. PeerJ 6, e6090. doi: 10.7717/peerj.6090
Angel R., Matthies D., Conrad R. (2011). Activation of methanogenesis in arid biological soil crusts despite the presence of oxygen. PLoS One 6, e20453. doi: 10.1371/journal.pone.0020453
Bahram M., Hildebrand F., Forslund S. K., Anderson J. L., Soudzilovskaia N. A., Bodegom P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560, 233–237. doi: 10.1038/s41586-018-0386-6
Batarseh M., Imreizeeq E., Tilev S., Al Alaween M., Suleiman W., Al Remeithi A. M., et al. (2021). Assessment of groundwater quality for irrigation in the arid regions using irrigation water quality index (IWQI) and GIS-zoning maps: case study from Abu Dhabi emirate, UAE. Groundw. Sustain. Dev. 14, 100611. doi: 10.1016/j.gsd.2021.100611
Bernard S. M., Habash D. Z. (2009). The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 182, 608–620. doi: 10.1111/j.1469-8137.2009.02823.x
Bulgarelli D., Rott M., Schlaeppi K., Ver Loren van Themaat E., Ahmadinejad N., Assenza F., et al. (2012). Revealing structure and assembly cues for arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Čuhel J., Šimek M., Laughlin R. J., Bru D., Chèneby D., Watson C. J., et al. (2010). Insights into the effect of soil pH on N2O and N2 emissions and denitrifier community size and activity. Appl. Environ. Microbiol. 76, 1870–1878. doi: 10.1128/AEM.02484-09
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen L. J., Li C. S., Feng Q., Wei Y. P., Zhao Y., Zhu M., et al. (2019). An integrative influence of saline water irrigation and fertilization on the structure of soil bacterial communities. J. Agric. Sci. 157, 693–700. doi: 10.1017/S002185962000012X
Cherif H., Marasco R., Rolli E., Ferjani R., Fusi M., Soussi A., et al. (2015). Oasis desert farming selects environment-specific date palm root endophytic communities and cultivable bacteria that promote resistance to drought. Environ. Microbiol. Rep. 7, 668–678. doi: 10.1111/1758-2229.12304
Delgado-Baquerizo M., Grinyer J., Reich P. B., Singh B. K. (2016). Relative importance of soil properties and microbial community for soil functionality: insights from a microbial swap experiment. Funct. Ecol. 30, 1862–1873. doi: 10.1111/1365-2435.12674
Deng Y., Jiang Y.-H., Yang Y., He Z., Luo F., Zhou J. (2012). Molecular ecological network analyses. BMC Bioinf. 13, 113. doi: 10.1186/1471-2105-13-113
de Vries F. T., Griffiths R. I., Bailey M., Craig H., Girlanda M., Gweon H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9, 3033. doi: 10.1038/s41467-018-05516-7
Fahrbach M., Kuever J., Remesch M., Huber B. E., Kampfer P., Dott W., et al. (2008). Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 58, 2215–2223. doi: 10.1099/ijs.0.65342-0
Feng K., Peng X., Zhang Z., Gu S., He Q., Shen W., et al. (2022). iNAP: an integrated network analysis pipeline for microbiome studies. iMeta 1-11. doi: 10.1002/imt2.13
Friedman J., Alm E. J. (2012). Inferring correlation networks from genomic survey data. PloS Comput. Biol. 8, e1002687. doi: 10.1371/journal.pcbi.1002687
Gregory G. J., Boyd E. F. (2021). Stressed out: bacterial response to high salinity using compatible solute biosynthesis and uptake systems, lessons from vibrionaceae. Comput. Struct. Biotechnol. J. 19, 1014–1027. doi: 10.1016/j.csbj.2021.01.030
Guo H., Shi X., Ma L., Yang T., Min W. (2020). Long-term irrigation with saline water decreases soil nutrients, diversity of bacterial communities, and cotton yields in a gray desert soil in China. Pol. J. Environ. Stud. 29, 4077–4088. doi: 10.15244/pjoes/120158
Guo J., Zhou Y., Guo H., Min W. (2021). Saline and alkaline stresses alter soil properties and composition and structure of gene-based nitrifier and denitrifier communities in a calcareous desert soil. BMC Microbiol. 21, 246. doi: 10.1186/s12866-021-02313-z
Hall S. J., Silver W. L., Amundson R. (2012). Greenhouse gas fluxes from atacama desert soils: a test of biogeochemical potential at the earth’s arid extreme. Biogeochemistry 111, 303–315. doi: 10.1007/s10533-011-9650-7
Hardoim P. R., van Overbeek L. S., van Elsas J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. doi: 10.1016/j.tim.2008.07.008
Herlemann D. P., Labrenz M., Jürgens K., Bertilsson S., Waniek J. J., Andersson A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Khan M. A., Hamayun M., Asaf S., Khan M., Yun B.-W., Kang S.-M., et al. (2021). Rhizospheric bacillus spp. rescues plant growth under salinity stress via regulating gene expression, endogenous hormones, and antioxidant system of oryza sativa l. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.665590
Lammel D. R., Barth G., Ovaskainen O., Cruz L. M., Zanatta J. A., Ryo M., et al. (2018). Direct and indirect effects of a pH gradient bring insights into the mechanisms driving prokaryotic community structures. Microbiome 6, 106. doi: 10.1186/s40168-018-0482-8
Li Y. Q., Chai Y. H., Wang X. S., Huang L. Y., Luo X. M., Qiu C., et al. (2021). Bacterial community in saline farmland soil on the Tibetan plateau: responding to salinization while resisting extreme environments. BMC Microbiol. 21, 119. doi: 10.1186/s12866-021-02190-6
Li J., Gao R., Chen Y., Xue D., Han J., Wang J., et al. (2020). Isolation and identification of microvirga thermotolerans hr1, a novel thermo-tolerant bacterium, and comparative genomics among microvirga species. Microorganisms 8, 101. doi: 10.3390/microorganisms8010101
Ling N., Wang T., Kuzyakov Y. (2022). Rhizosphere bacteriome structure and functions. Nat. Commun. 13, 836. doi: 10.1038/s41467-022-28448-9
Loganathachetti D. S., Alhashmi F., Chandran S., Mundra S. (2022). Irrigation water salinity structures the bacterial communities of date palm (Phoenix dactylifera)-associated bulk soil. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.944637
Lopes L. D., Hao J., Schachtman D. P. (2021). Alkaline soil pH affects bulk soil, rhizosphere and root endosphere microbiomes of plants growing in a sandhills ecosystem. FEMS Microbiol. Ecol. 97, fiab028. doi: 10.1093/femsec/fiab028
Martineau C., Mauffrey F., Villemur R. (2015). Comparative analysis of denitrifying activities of hyphomicrobium nitrativorans, hyphomicrobium denitrificans, and hyphomicrobium zavarzinii. Appl. Environ. Microbiol. 81, 5003–5014. doi: 10.1128/AEM.00848-15
Meng Y., He Z., Liu B., Chen L., Lin P., Luo W. (2020). Soil salinity and moisture control the processes of soil nitrification and denitrification in a riparian wetlands in an extremely arid regions in northwestern China. Water 12, 2815. doi: 10.3390/w12102815
Mosqueira M. J., Marasco R., Fusi M., Michoud G., Merlino G., Cherif A., et al. (2019). Consistent bacterial selection by date palm root system across heterogeneous desert oasis agroecosystems. Sci. Rep. 9, 4033. doi: 10.1038/s41598-019-40551-4
Newman M. E. J. (2004). Fast algorithm for detecting community structure in networks. Phys. Rev. E 69, 66133. doi: 10.1103/PhysRevE.69.066133
Nielsen U. N., Ball B. A. (2015). Impacts of altered precipitation regimes on soil communities and biogeochemistry in arid and semi-arid ecosystems. Glob. Change Biol. 21, 1407–1421. doi: 10.1111/gcb.12789
O’Brien F. J. M., Almaraz M., Foster M. A., Hill A. F., Huber D. P., King E. K., et al. (2019). Soil salinity and pH drive soil bacterial community composition and diversity along a lateritic slope in the Avon river critical zone observatory, Western Australia. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01486
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2020) Vegan: community ecology package. Available at: https://cran.r-project.org/package=vegan.
Olesen J. M., Bascompte J., Dupont Y. L., Jordano P. (2007). The modularity of pollination networks. Proc. Natl. Acad. Sci. 104, 19891–19896. doi: 10.1073/pnas.0706375104
Oshiki M., Toyama Y., Suenaga T., Terada A., Kasahara Y., Yamaguchi T., et al. (2022). N2O reduction by gemmatimonas aurantiaca and potential involvement of gemmatimonadetes bacteria in N2O reduction in agricultural soils. Microbes Environ. 37, ME21090. doi: 10.1264/jsme2.ME21090
Padan E., Bibi E., Ito M., Krulwich T. A. (2005). Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta - Biomembr. 1717, 67–88. doi: 10.1016/j.bbamem.2005.09.010
Qian X., Li H., Wang Y., Wu B., Wu M., Chen L., et al. (2019). Leaf and root endospheres harbor lower fungal diversity and less complex fungal co-occurrence patterns than rhizosphere. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01015
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team. (2020). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
Rafrafi Y., Ranaivomanana H., Bertron A., Albrecht A., Erable B. (2015). Surface and bacterial reduction of nitrate at alkaline pH: conditions comparable to a nuclear waste repository. Int. Biodeterior. Biodegradation 101, 12–22. doi: 10.1016/j.ibiod.2015.03.013
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Rousk J., Bååth E., Brookes P. C., Lauber C. L., Lozupone C., Caporaso J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Saeed E. E., Sham A., Salmin Z., Abdelmowla Y., Iratni R., El-Tarabily K., et al. (2017). Streptomyces globosus UAE1, a potential effective biocontrol agent for black scorch disease in date palm plantations. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01455
Santolini M., Barabási A.-L. (2018). Predicting perturbation patterns from the topology of biological networks. Proc. Natl. Acad. Sci. 115, E6375–E6383. doi: 10.1073/pnas.1720589115
Schlaeppi K., Dombrowski N., Oter R. G., Ver Loren van Themaat E., Schulze-Lefert P. (2014). Quantitative divergence of the bacterial root microbiota in arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. 111, 585–592. doi: 10.1073/pnas.1321597111
Shamim A., Sanka Loganathachetti D., Chandran S., Masmoudi K., Mundra S. (2022). Salinity of irrigation water selects distinct bacterial communities associated with date palm (Phoenix dactylifera l.) root. Sci. Rep. 12, 12733. doi: 10.1038/s41598-022-16869-x
Trivedi P., Batista B. D., Bazany K. E., Singh B. K. (2022). Plant–microbiome interactions under a changing world: responses, consequences and perspectives. New Phytol. 234, 1951–1959. doi: 10.1111/nph.18016
Tsotetsi T., Nephali L., Malebe M., Tugizimana F. (2022). Bacillus for plant growth promotion and stress resilience: what have we learned? Plants 11, 2482. doi: 10.3390/plants11192482
Vásquez-Dean J., Maza F., Morel I., Pulgar R., González M. (2020). Microbial communities from arid environments on a global scale. a systematic review. Biol. Res. 53, 29. doi: 10.1186/s40659-020-00296-1
Wang Z., Meng Y., Zhu-Barker X., He X., Horwath W. R., Luo H., et al. (2019b). Responses of nitrification and ammonia oxidizers to a range of background and adjusted pH in purple soils. Geoderma 334, 9–14. doi: 10.1016/j.geoderma.2018.07.038
Wang W., Wang J., Ye Z., Zhang T., Qu L., Li J. (2019a). Soil property and plant diversity determine bacterial turnover and network interactions in a typical arid inland river basin, Northwest China. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02655
Wei Y., Jing X., Su F., Li Z., Wang F., Guo H. (2022). Does pH matter for ecosystem multifunctionality? an empirical test in a semi-arid grassland on the loess plateau. Funct. Ecol. 36, 1739–1753. doi: 10.1111/1365-2435.14057
Wu J., Liu W., Zhang W., Shao Y., Duan H., Chen B., et al. (2019). Long-term nitrogen addition changes soil microbial community and litter decomposition rate in a subtropical forest. Appl. Soil Ecol. 142, 43–51. doi: 10.1016/j.apsoil.2019.05.014
Xing W., Lu X., Ying J., Lan Z., Chen D., Bai Y. (2022). Disentangling the effects of nitrogen availability and soil acidification on microbial taxa and soil carbon dynamics in natural grasslands. Soil Biol. Biochem. 164, 108495. doi: 10.1016/j.soilbio.2021.108495
Xiong C., He J. Z., Singh B. K., Zhu Y. G., Wang J. T., Li P. P., et al. (2021). Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environ. Microbiol. 23, 1907–1924. doi: 10.1111/1462-2920.15262
Xu L., Zhang Y., Read N., Liu S., Friman V.-P. (2017). Devosia nitraria sp. nov., a novel species isolated from the roots of nitraria sibirica in China. Antonie Van Leeuwenhoek 110, 1475–1483. doi: 10.1007/s10482-017-0901-z
Zeng Q., An S., Liu Y., Wang H., Wang Y. (2019). Biogeography and the driving factors affecting forest soil bacteria in an arid area. Sci. Total Environ. 680, 124–131. doi: 10.1016/j.scitotenv.2019.04.184
Zhang Z., Chai X., Gao Y., Zhang B., Lu Y., Huang C., et al. (2022). Dynamics in diversity, co-occurrence pattern, and community assembly of a perennial desert plant root-associated bacteria. Rhizosphere 22, 100526. doi: 10.1016/j.rhisph.2022.100526
Zhang H., Ma B., Huang T., Shi Y. (2021). Nitrate reduction by the aerobic denitrifying actinomycete streptomyces sp. XD-11-6-2: performance, metabolic activity, and micro-polluted water treatment. Bioresour. Technol. 326, 124779. doi: 10.1016/j.biortech.2021.124779
Zhang K., Shi Y., Cui X., Yue P., Li K., Liu X., et al. (2019). Salinity is a key determinant for soil microbial communities in a desert ecosystem. mSystems 4, e00225–e00218. doi: 10.1128/mSystems.00225-18
Zhou W., Han G., Liu M., Zeng J., Liang B., Liu J., et al. (2020). Determining the distribution and interaction of soil organic carbon, nitrogen, ph and texture in soil profiles: a case study in the lancangjiang river basin, southwest China. Forests 11, 532. doi: 10.3390/f11050532
Keywords: arid agroecosystems, date palms (Phoenix dactylifera L.), irrigation water, nitrogen cycle, root and soil bacterial community, co-occurrence network
Citation: Sanka Loganathachetti D and Mundra S (2023) Water pH, not soil pH, alters bacterial community structural pattern and nitrogen cycling pathways in date palm (Phoenix dactylifera L.) roots and bulk soil under freshwater irrigation regime. Front. Ecol. Evol. 11:1142073. doi: 10.3389/fevo.2023.1142073
Received: 12 January 2023; Accepted: 01 June 2023;
Published: 15 June 2023.
Edited by:
Arnaldo Marín, University of Murcia, SpainReviewed by:
David Correa Galeote, National Autonomous University of Mexico, MexicoMohamed Anli, Centre Universitaire De Patsy, Comoros
Copyright © 2023 Sanka Loganathachetti and Mundra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunil Mundra, c3VuaWxtdW5kcmFAdWFldS5hYy5hZQ==
 Dinesh Sanka Loganathachetti
Dinesh Sanka Loganathachetti Sunil Mundra
Sunil Mundra