
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 20 April 2023
Sec. Coevolution
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1131203
This article is part of the Research TopicImpact of Anthropogenic Environmental Changes on Animal MicrobiomesView all 11 articles
The depletion of oxygen as a result of increased stratification and decreased oxygen solubility is one of the most significant chemical changes occurring in aquatic ecosystems as a result of global environmental change. Hence, more aquatic organisms will be exposed to hypoxic conditions over time. Deciphering the effects of hypoxia on strong ecological interactors in this ecosystem’s food web is critical for predicting how aquatic communities can respond to such an environmental disturbance. Here (sub-)lethal effects of hypoxia and whether these are genotype specific in Daphnia, a keystone species of freshwater ecosystems, are studied. This is especially relevant upon studying genetic responses with respect to phenotypic switches upon environmental stress. Further, we investigated the effect of hypoxia on the Daphnia microbial community to test if the microbiome plays a role in the phenotypic switch and tolerance to hypoxia. For this, two Daphnia genotypes were exposed for two weeks to either hypoxia or normoxia and host performance was monitored together with changes in the host associated and free-living microbial community after this period. We detected phenotypic plasticity for some of the tested Daphnia performance traits. The microbial community of the bacterioplankton and Daphnia associated microbial community responded via changes in species richness and community composition and structure. The latter response was different for the two genotypes suggesting that the microbiome plays an important role in phenotypic plasticity with respect to hypoxia tolerance in Daphnia, but further testing (e.g., through microbiome transplants) is needed to confirm this.
The depletion of oxygen is one of the most significant chemical changes currently occurring in freshwater ecosystems as a result of global environmental change. While hypoxia is common as a seasonal disturbance, the duration, spatial scale and frequency have increased in the past few decades and are expected to increase as a result of climate change (Gilbert et al., 2005; Diaz and Rosenberg, 2008; Paerl et al., 2011; Goto et al., 2012). This is concerning as hypoxia has a strong effect on these freshwater ecosystems. The main drivers of deoxygenation are rising water temperature, stratification and anthropogenically induced eutrophication. Warmer surface water holds less soluble oxygen, which in its turn results in increased thermal stratification due to the increasing density difference with deeper colder water depths. This increasing separation reduces circulation between different depths of the water column. Reduced oxygen exchange between the atmosphere and the water causes a further decrease in oxygen amount. Excess nutrient runoff, primarily nitrogen and phosphorus, that enters the water column often leads to plankton blooms, which upon death sink to the bottom and increased oxygen consuming decomposition activity leading to hypoxic conditions in the deeper water depths. This reduction in dissolved oxygen promotes nutrient release from bottom sediments into the surface water (North et al., 2014), enabling the production of phytoplankton blooms. As hypoxic waters and phytoplankton blooms act as reinforcing factors of each other, their co-occurrence is often reported and reinforces that nutrient excess is one of the major causes of increasing oxygen depletion (Zhang et al., 2011). The effect of increasing deoxygenation is 2.75 to 9.3 times larger in freshwater lakes than in oceans, with losses in dissolved oxygen (D.O.) concentration of 5.5% in upper regions and 18.6% in lower regions of 393 studied freshwater bodies between 1980 and 2017 (Jane et al., 2021). Water is considered hypoxic when the dissolved oxygen levels are less than 2 mg/L, since these levels are linked with harmful effects on fish and zooplankton (Vanderploeg et al., 2009).
Deciphering the effects of hypoxia on strong ecological interactors in the food web of freshwater ecosystems, such as the zooplankter Daphnia magna, is thus essential to predict if and how aquatic communities can respond to such a disturbance. When dissolved oxygen becomes depleted in water ecosystems below organismal physiological tolerances, it can (in)directly impact aquatic communities and natural resources. Daphnia is a well-known eco-(toxico)logical model system that has proven to be especially well-suited for studying the interaction between genotype and environment due to their high phenotypic plasticity in response to environmental stressors, short generation time, small size, large number of eggs per clutch, easy manipulation, absence of ethical concerns in experiments and fully sequenced genome (Ebert, 2005; Miner et al., 2012; Ebert, 2022). Mobile organisms, like Daphnia, are less prone to direct lethal effects compared to sessile organism (Díaz and Rosenberg, 1995), but they risk indirect effects of hypoxia when fleeing to more oxygenated regions. Therefore, Daphnia is adapted to hypoxia via a series of physiological (Pirow et al., 2001) and biochemical (Gerke et al., 2011) adaptations, each with their own advantages and costs (Larsson and Lampert, 2011; Galic et al., 2019). Hypoxic conditions induce growth reduction (Kobayashi, 1982; Eby and Crowder, 2002; Seidl et al., 2005) and depending on the Daphnia species, hypoxia can impact reproduction impairments (Seidl et al., 2005; Lyu et al., 2013a). Over a wide range of atmospheric oxygen concentrations, Daphnia can control their oxygen metabolism and metabolic phenotype (Weider and Lampert, 1985; Lee et al., 2022). For example, to avoid fish predation, Daphnia uses vertical migration to seek refuge into hypoxic regions with D.O. concentrations lethal for fish (Hanazato et al., 1985; Decaestecker et al., 2002; Larsson and Lampert, 2011). During hypoxia exposure, Daphnia upregulate hemoglobin synthesis resulting in a higher hypoxia tolerance (Pirow et al., 2001) and a red phenotype (Gorr et al., 2004; Seidl et al., 2005; Zeis et al., 2013), making Daphnia lose their transparent appearance and visual advantage for predation (Larsson and Lampert, 2011). The amount of dissolved oxygen has a significant impact on the population composition as not all genotypes are as effective at surviving in hypoxic conditions, which leads to selection and shifts in the genotype composition of the Daphnia population (Hutchinson, 1957). While respiration rate is genotype independent (Weider and Lampert, 1985), hemoglobin synthesis is genotype dependent (Weider and Lampert, 1985). Genotypes with a lower hypoxia tolerance have an enlarged vulnerability to predation causing changes in community structure, variations in the distribution of species and reduction in biodiversity. During periods of hypoxia, low hypoxia tolerant genotypes may be forced to move to oxygen regions, which are still tolerant for fish (Weider and Lampert, 1985), but the hypoxia-induced red phenotype makes them more visible and prone to predation. Further research is needed into the physiological or ecological mechanisms underlying a Daphnia population tolerance to hypoxia. Such as the mediation of hypoxia tolerance through the microbiome.
The number of studies on the Daphnia-associated microbiota has substantially increased in recent years. The Daphnia microbial composition is dynamic and differs between gut and outer body parts (Qi et al., 2009; Sison-Mangus et al., 2015; Callens et al., 2018). Across studies, bacterial groups such as Comamonadaceae, Flavobacteriacea, Burkholderiaceae, Aeromonas, Limnohabitans, Pedobacter, Ideonella and Pseudomonas have been shown to dominate the gut microbial community of Daphnia (Qi et al., 2009; Macke et al., 2017a; Akbar et al., 2020, 2022; Cooper and Cressler, 2020). Both the environment and host genotype influence the microbial community composition and functionality to achieve a healthy balanced state of the microbiota. Interactions between these factors and/or between these factors and the host-associated microbiota can be temporary and susceptible to selection (Macke et al., 2020; Akbar et al., 2022). It has been shown that depriving Daphnia of its microbiota is detrimental to its fitness and the association between microbial imbalance and disease states is becoming clear, also in Daphnia (Callens et al., 2016; Bulteel et al., 2021; Rajarajan et al., 2022). The Daphnia gut microbial community is known to respond to environmental stressors (Houwenhuyse et al., 2021), such as toxic cyanobacteria (Macke et al., 2017a), antibiotics (Callens et al., 2018; Akbar et al., 2020) and parasites (Bulteel et al., 2021; Rajarajan et al., 2022). Moreover, studies show that the flexible Daphnia microbiome can increase Daphnia tolerance upon environmental stress (Macke et al., 2017b; Akbar et al., 2022).
Hypoxia-induced changes in host-associated microbiota and physiology has been shown in humans and rodents (Han et al., 2021). However, little is known about how hypoxia affects the microbiota of freshwater organisms, like Daphnia. Toxic cyanobacterial blooms (cyanoHABS), which are often co-occurring with depleted oxygen concentrations are associated with reduced fitness of D. magna and a changed gut microbial community (Macke et al., 2017a). We hypothesize that microbial communities change upon hypoxia and may indirectly affect Daphnia metabolism or phenotypic effects upon selective uptake or rejection of microbiota by the host. Microbial data in combination with host performance effects are needed for a comprehensive understanding of hypoxia induced effects on hosts and their microbiomes.
We here investigated whether the Daphnia associated microbial community is affected by hypoxia and if so, whether the effect on the microbiome composition differs between genotypes that differ in their tolerance to hypoxia. Therefore, we investigated the (sub)lethal effects of hypoxia in Daphnia and whether these are genotype specific by exposing two Daphnia genotypes for two weeks to either hypoxia or normoxia and monitored survival, growth and reproduction. To determine the effect of hypoxia on the microbial community of Daphnia, the microbial composition of the gut and body of two genotypes was characterized at the end of the experiment via amplicon sequencing. In addition, bacterioplankton samples of the medium were taken to investigate whether free-living microbial communities released by the Daphnia host also reflected a shift with decreasing oxygen levels and if responses were different than the host associated microbial communities (body and gut). We here thus investigated if the microbiome is relevant in phenotypic responses toward hypoxia. This research provides knowledge needed for further research in microbiome-mediated responses to hypoxia with a particular focus on genotype x microbial community x environmental interactions, as these may mediate microbiome mediated evolutionary responses for D. magna populations during times of hypoxia in the environment.
Throughout this study, two D. magna genotypes were used: the KNO 15.04 and the F genotype. Genotype KNO15.04 originates from a small (350 m2), fishless, mesotrophic pond in Knokke, at the Belgian coast (51°20′05.62″ N, 03°20′53.63″ E). The F clone is a standard genotype used in ecotoxicological tests, obtained from the Barrata lab in Barcelona and originally isolated in Scotland (Barata et al., 2017). All D. magna stock genotypes were cultured and maintained for many generations in the Aquatic Biology lab (IRF life sciences lab, KU Leuven department Kortrijk, Belgium). For the experiment, three maternal lines of these stock lines were set up per genotype to exclude maternal effects between individuals of the same genotype. The maternal lines were established by collecting and continuing every second (or third) brood during two or more generations. D. magna used in the experiments were obtained from eggs of the second or third brood, given that these are better quality than first brood offspring. Cultures were kept at a density of one D. magna individual per 50 mL in filtered tap water (Greenline e1902 filter) at a constant room temperature of 19 ± 1°C and under a 16:8 h light–dark cycle. They were fed three times a week with 200.103 cells/mL of the unicellular green algal species Chlorella vulgaris. Chlorella vulgaris cultures were cultured under sterile conditions in 2 L jars with Wright’s Cryptophyte medium (Guillard and Lorenzen, 1972) at a constant room temperature of 20 ± 2°C and under a light–dark cycle of 16:8 h. To prevent bacterial contamination, a 0.22 μm filter was present at the in- and output of the aeration system to supply CO2 and to remove oxygen. Magnetic stirrers were used to ensure a constant mixing in order to avoid precipitation of the algae. Fluorescence-activated cell sorting was used to measure the cell density of the algal cultures (using FACS Verse, Biosciences).
To unravel if different D. magna genotypes show a different sensitivity toward hypoxia exposure, the two D. magna genotypes (KNO 15.04 and F) were exposed to a hypoxic and normoxic exposure. Female D. magna carrying parthenogenetic eggs in their brood pouch, at a stage of ≥48 h after egg laying, were isolated for three maternal lines per genotype. Once the D. magna juveniles were released from the brood pouch, they were individually transferred to a 50 ml Falcon tube containing autoclaved filtered tap water. Per maternal line, ten D. magna juveniles for each of the three maternal lines used per genotype were exposed to either a hypoxic or a normoxic exposure for 14 days. The hypoxic exposure was achieved using Biospherix C-chambers with a ProOx controllers, where a 2% air oxygen level was reached in the chambers using regulated inflow of nitrogen gas under pressure (1.7 mbar). Dissolved oxygen content (mg/L), oxygen saturation (%), temperature (°C) and pressure (hPa) were measured daily to check the stability of the hypoxic exposure using the Hach HQ40d multi-meter and optical dissolved oxygen sensor. The dissolved oxygen in the medium decreased gradually, reaching a stable concentration of 1.83 ± 1.08 mg/L after 2 days. The normoxic conditions had an average dissolved oxygen concentration of 8.7 ± 1.25 mg/L. The experimental exposures were kept constant throughout the experiment with a temperature of 19 ± 1°C and a 16-8 h light–dark cycle. To account for differing light incidence and potential differences between the hypoxia chambers, falcons were randomized daily. After the transfer to the normoxic or hypoxic exposure, survival and fecundity (day of first and second brood, and brood amount) were monitored daily. Body size of 5 D. magna individuals per combination of maternal line and exposure was measured at days 3, 7, 10, and 14 of the exposures using sterile materials, a BMS microscope camera and BMS PIX software. The length of a D. magna was measured from the top of the eye to the base of the apical spine. Starting from day two of the experiment, 200.103 cells/mL of autoclaved Chlorella vulgaris were administered every other day.
Five surviving individuals from each maternal line were collected at the end of the experiment, except for hypoxia exposed KNO 15.04 as only four D. magna survived until the end of the experiment. Individuals were dissected and D. magna guts and bodies were collected separately to determine the microbial community composition of each genotype and body part via amplicon sequencing. In addition, bacterioplankton samples of the medium were taken by filtering 200 ml of the medium over a 0.22 μm filter. Samples were collected on ice (to prevent microbial community shifts and to preserve DNA) in an Eppendorf tube containing 10 μL sterile Milli-Q. For each exposure, individuals were pooled per genotype and maternal line. Amplicon sequencing was performed according to Houwenhuyse et al. (2021). DNA was extracted by the Qiagen PowerSoil DNA isolation kit and dissolved in 20 μL MilliQ water. To determine the total DNA yield, 1 μL of sample was used in an Invitrogen Qubit dsDNA HS assay. Increased specificity and amplicon yield were obtained by using nested PCR. The entire 16S rRNA gene was amplified for 30 cycles (98°C for 10 s; 50°C for 45 s; 72°C for 30 s) with the Life Technologies SuperFi high fidelity polymerase and the EUB8F and 1492R primers on 10 ng of template. The PCR product was purified with the QIAquick PCR purification kit. In a second amplification round, 5 μL (20–50 ng) of the PCR product was amplified for 30 cycles (98°C for 10 s; 50°C for 5 s; 72°C for 30 s) with the 515F and 806R primers to obtain amplicons of the V4-region with a dual index. The latter two primers contained an 8-nucleotide barcode, as well as an Illumina adapter at their 5′-end. PCRs were performed in triplicate and were pooled and gel-purified for each sample with the QIAquick gel extraction kit. To prepare an equimolar library, an Applied Biosystems SequalPrep Normalization Plate was used to normalize amplicon concentrations, after which the library was pooled. Amplicon sequencing was performed using a v2 PE500 kit with custom primers on the Illumina Miseq platform, which resulted in two 250-nucleotide paired-end reads for each of the 36 samples.
All data analysis was performed using R 4.0.4. To select the models with the best combination of variables, the Akaike information criterion (AIC) was used. For survival data, A log-rank test was performed using the ‘survdiff’ function (survival package in R) to determine whether there was a significant difference in survival probability between groups. Survival probability over time for different groups was visualized by plotting Kaplan–Meier curves using the ‘ggsurvplot’ function (survminer package in R). For body size, differences between groups were determined by performing an Analysis of Variance (ANOVA) using the ‘Anova’ function (car package in R) on a generalized linear model (GLM) and contrasts between specific groups was analyzed with a Tukey post-hoc test. When taking maternal lines into account as a random factor, a linear mixed-effects model (lmer function, lme4 package in R) was used on normally distributed data or a generalized linear mixed effects model (glmer function, lme4 package in R) was used when the data was not normally distributed. Body size was compared over time and for each time point separately. The best model was determined by having the lowest AIC. Differences in body size between groups was visualized by making boxplots using the ‘ggplot’ function (ggplot2 package in R). Total fecundity was analyzed with a linear mixed-effects model, controlling for an unbalanced design with a restricted maximum likelihood estimation.
We processed DNA sequences in accordance with Callahan et al. (2016b). Sequences were trimmed on both paired ends (the first 10 nucleotides and starting at position 180) and filtered (maximum of 2 expected errors per read). The high-resolution DADA2 method, which relies on a parameterized model of substitution errors to discriminate sequencing errors from actual biological variation, was used to predict sequence variations (Callahan et al., 2016a). After that, chimeras were removed from the data set. Using the SILVA v138 training set, a naïve Bayesian classifier was used to assign taxonomy. ASVs that were classified as “chloroplast” or “cyanobacteria” or that had no taxonomic assignment at the phylum level were eliminated from the data set. Following filtering, a total of 738,198 reads—an average of 19951.3 reads per sample—were obtained, with the majority of the samples having more than 9,000 reads. ASVs were pooled at the order level, and orders accounting for less than 1% of the readings were disregarded in order to visualize the bacterial orders that varied between the treatments. Measures for alphadiversity of the microbial communities within the different exposures, genotypes and sample types (ASV richness and Shannon Index) were calculated using the vegan package in R (Bellier, 2012). Prior to analyzing alphadiversity, all samples were rarified to a depth of 9,500 reads, based on the number of reads per sample. A generalized linear model (GLM), assuming a Poisson distribution of the data, was used to investigate the effects of sample type (gut, body, or bacterioplankton), oxygen exposure (normoxia or hypoxia), genotype (KNO 15.04 or F), and any possible interactions on ASV richness with maternal line as random factor. The ‘emmeans’ function with a ‘Tukey’ adjustment from the emmeans R package was used to perform pairwise comparisons between significant variables and their interactions. Principal Coordinates Analysis with the phyloseq package in R was used to calculate and plot weighted and unweighted Unifrac distance matrices in order to compare variations in microbial community composition and structure (beta diversity) between variables. Using the Adonis2 function in the vegan package in R, the effect of oxygen exposure, genotype, sample type, and all potential interactions on β-diversity were evaluated through a permutation MANOVA. Obtained p-values were adjusted for multiple comparisons through the control of the false discovery rate (FDR). To identify which bacterial classes significantly differed between the exposures and sample types, ASVs were grouped at the class level, and classes representing <1% of the reads were removed. Differential abundance analyses were then performed with the Bioconducter package DESeq2 (Love et al., 2014).
The two-way interaction between Daphnia genotype and oxygen exposure (comparison normoxia versus hypoxia) was significant for survival (Supplementary Figure S1; Cox proportional hazard model: interaction Daphnia genotype x oxygen exposure: X2 = 11; df = 3; p = 0.01). When the two genotypes were pooled, the Daphnia individuals which were subjected to normoxia had a higher survival rate compared to the ones in hypoxia (Supplementary Figure S2B; Cox proportional hazard analysis: treatment: X2 = 7.1; df = 1; p = 0.008). Consistent with the significant interaction between genotype and exposure treatment, the tendency was that survival was slightly more reduced in the KNO 15.04 than in the F genotype in hypoxia versus normoxia (Figure 1). A two-way interaction between genotype and oxygen exposure over time could also be seen for body size (p < 0.01). The hypoxia exposure had a significant overall negative effect on Daphnia growth over time (Figure 2, p < 0.001). The general increasing effect of hypoxia on growth over time was mainly attributed to the stronger limiting effect of hypoxia on the growth of the KNO 15.04 genotype (Figure 2A right panel, p < 0.0001) compared to the F genotype Daphnia individuals (Figure 2A left panel, p = 0.5). At day 14, KNO 15.04 Daphnia individuals that were exposed to hypoxia were 15.27% smaller compared to Daphnia of the same genotype who were exposed to normoxia. For the F genotype, there was only a reduction in size of 0.28% between normoxia and hypoxia at day 14 (Figure 2). Notable, there was no differential growth between the two Daphnia genotypes in normoxia (Figure 2B right panel: p = 0.28). The F genotype Daphnia individuals became larger than the KNO 15.04 genotype Daphnia individuals in hypoxia over time (Figure 2B left panel: p < 0.01). Both genotypes had a clear red phenotypic appearance in hypoxia (Supplementary Figure S3) and of the Daphnia surviving until day 14, the proportion of hypoxia exposed reproducing Daphnia (5%) was significantly lower than normoxia exposed Daphnia (40%) (Figure 3A; p < 0.01). Also, the number of eggs in the first clutch was significantly lower in hypoxia compared to normoxia exposed Daphnia (Figure 3B; p < 0.05). Genotype did not influence the number of reproducing Daphnia, nor the amount of produced first clutch eggs. However, the KNO 15.04 genotype had a higher percentage of Daphnia individuals carrying a second clutch compared to the F genotype in normoxia (Figure 3C; p < 0.05). Clutch size did not differ between the genotypes. Not a single Daphnia individual reproduced a second time in hypoxia (Figures 3C,D).
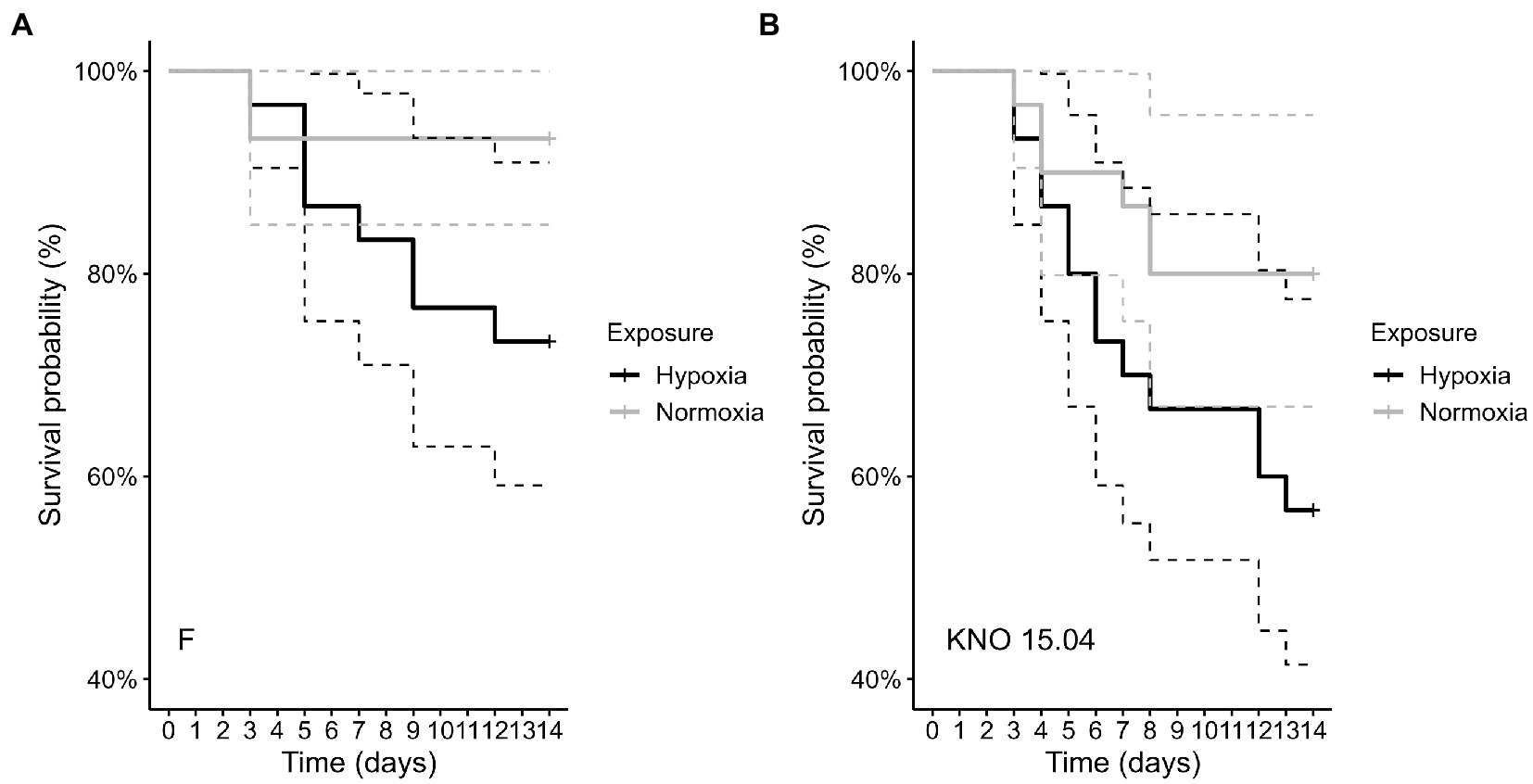
Figure 1. Comparison of survival of the F genotype (A) and KNO 15.04 genotype (B) in the hypoxia (black line) or normoxia (grey line) exposure. Dashed lines represent the 95% confidence intervals. Sample size was n = 30 (10 individuals * 3 independent replicates) for each genotype * exposure combination.
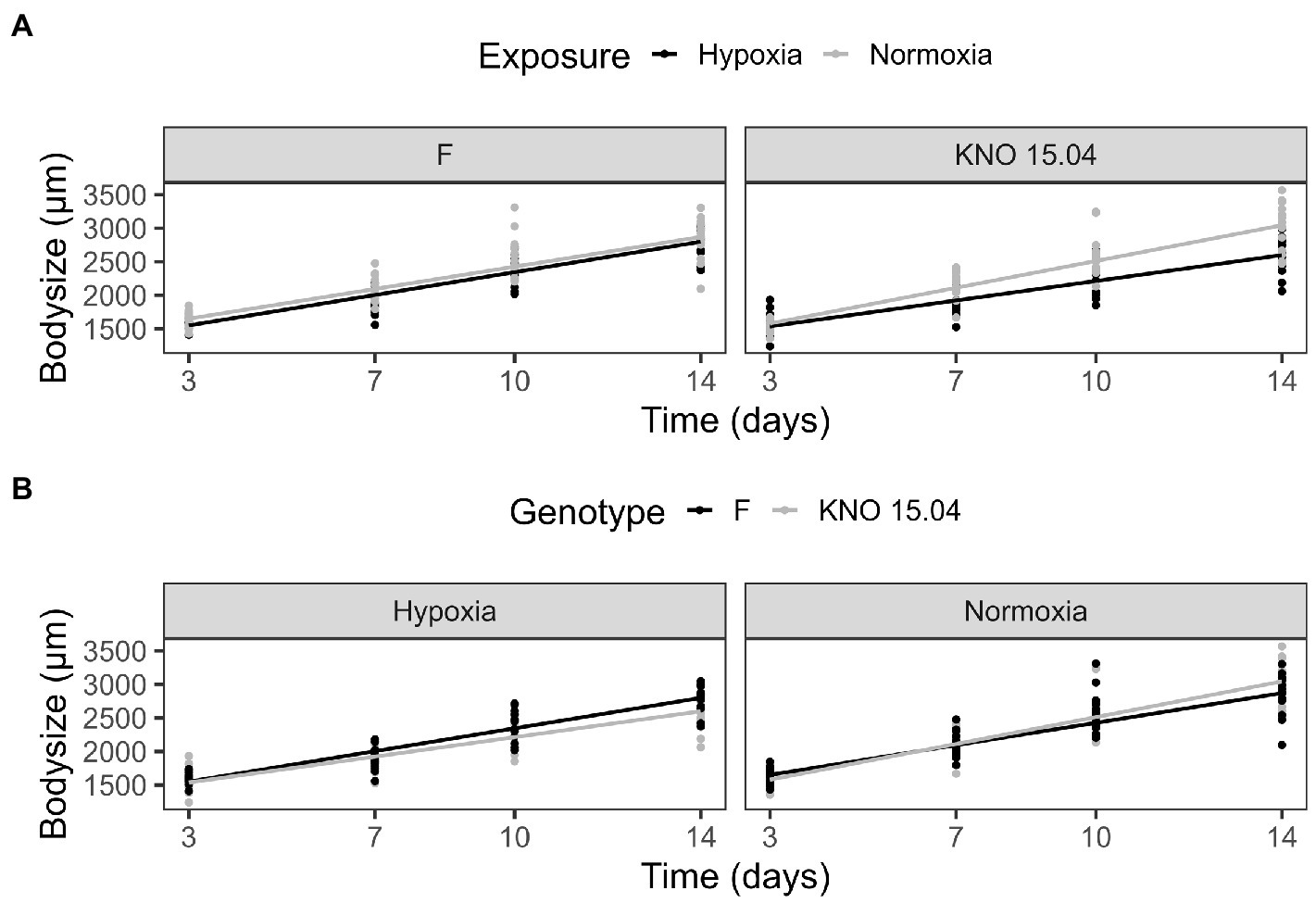
Figure 2. Body size comparison of the F and KNO 15.04 genotype in hypoxia and normoxia. (A) Exposure effect on the body size of the F (left) and KNO 15.04 (right) genotype during hypoxia (black line) or normoxia (grey line) exposure over 14 days. (B) Genotype effect in the hypoxia and normoxia exposure: Black lines correspond to body size of the F genotype and grey lines to body size of the KNO 15.04 genotype. Dots represent individual Daphnia body size data.
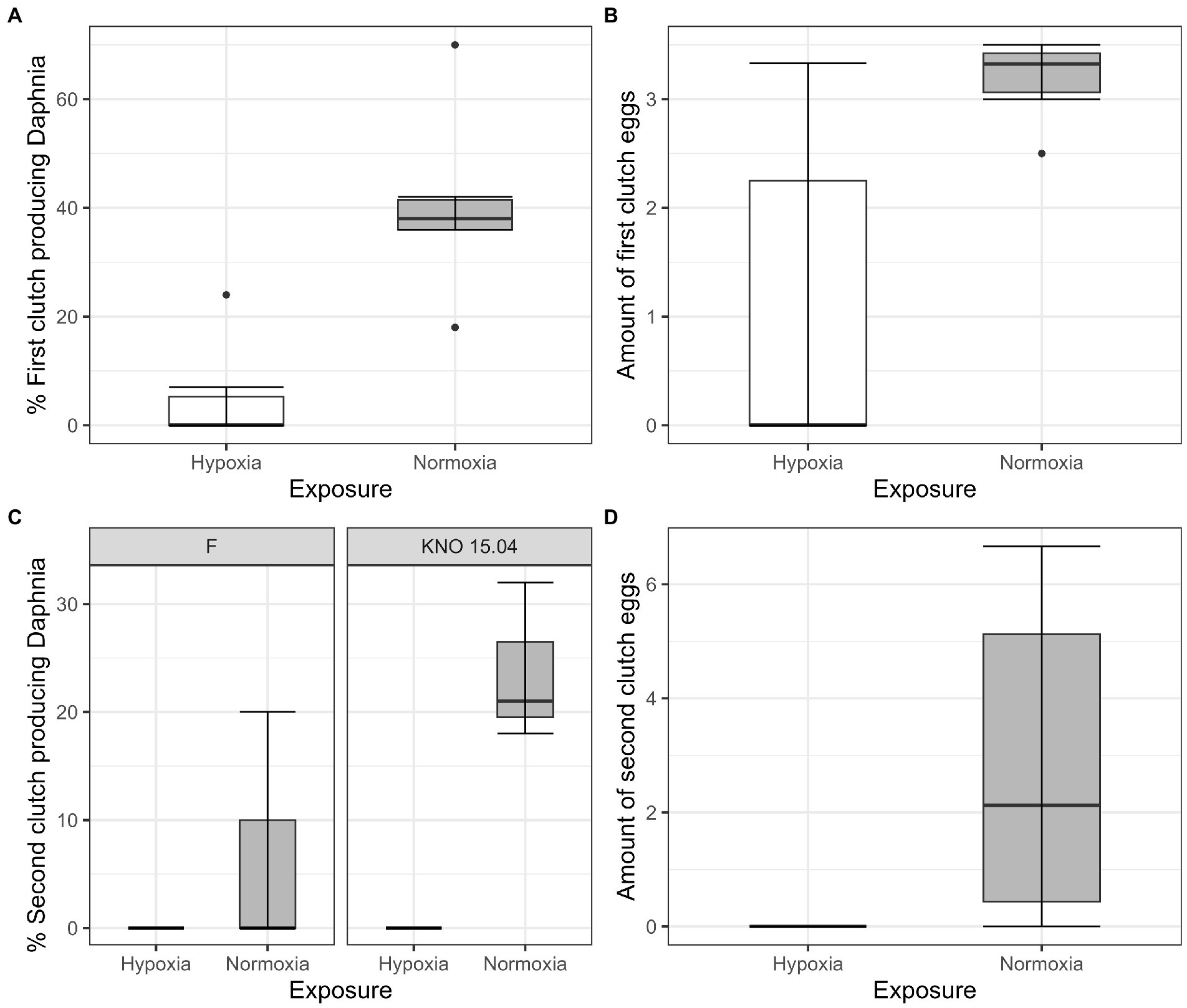
Figure 3. Fecundity of Daphnia genotypes F and KNO 15.04 under hypoxia (white boxplots) and normoxia (grey box plot). (A) Percentage of first clutch producing individuals of the Daphnia surviving up to 14 days. (B) Size of first clutch in the two exposures. (C) Percentage of second clutch producing individuals of the Daphnia surviving up to 14 days of the F and KNO 15.04 genotype, left and right panel, respectively. (D) Size of second clutch in the two exposures. The box plot includes 50% of the data from the first to the third quartile, the whiskers extend to the minimum and maximum data within the 1.5 interquartile range and the dots represent single outlier data points outside that range.
The species richness (SR) of the complete dataset showed a two-way interaction between exposure and sample type (p < 0.05), with a higher SR in bacterioplankton compared to body and gut samples (bacterioplankton-body samples: p < 0.0001, bacterioplankton-gut samples: p < 0.001, ANOVA) when pooling the two genotypes in hypoxia. This was reflected in a higher amount of Actinobacteria (Wald test: BPK-body: padj<0.0001; BPK-gut: padj<0.05) and a lesser amount of Gammaproteobacteria (Wald test: BPK-body: padj<0.0001; BPK-gut: padj<0.0001) in the bacterioplankton samples compared to the Daphnia body and gut in hypoxia (Figure 4). The difference in SR between bacterioplankton and the other sample types was not present in normoxia (p = 0.09). However, also in normoxia Gammaproteobacteria were less present in the bacterioplankton compared to the gut samples (Wald test: padj<0.05). In addition, the bacterioplankton contained more Bacteroidiota than the Daphnia gut samples (Figure 4, Wald test: padj<0.05) and more Verrucomicrobiae than the Daphnia body (Wald test: padj<0.01) and gut samples (Wald test: padj<0.05) in normoxia (Figure 4). When taken relative abundances of species into account, the two-way interaction between sample type and exposure disappeared, as the Shannon Index (SI) of the bacterioplankton samples differed significantly not only from gut (p < 0.001) and body (p < 0.001) samples in hypoxia, but also from gut samples in normoxia (p < 0.05).
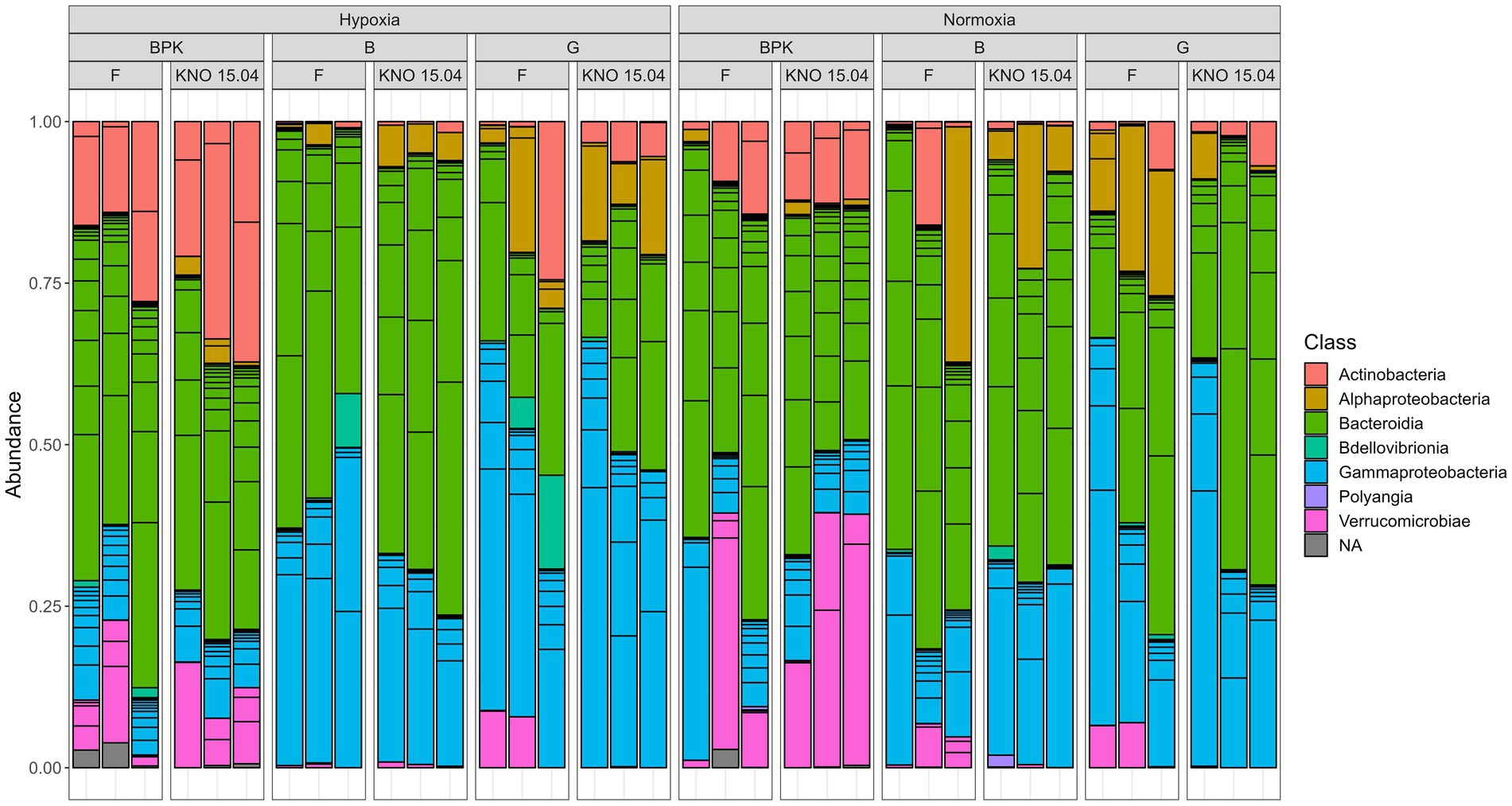
Figure 4. Overview of the relative abundance of different classes of bacteria present in three sample types (Bacterioplankton – BPK, Body – B, Gut – G of the two D. magna genotypes) (F versus KNO 15.04).
When looking at the exposure effect on the separate sample types, hypoxia only affected the bacterioplankton community significantly, where there was an enrichment in Actinobacteria (Wald test: padj<0.05) and a reduction in Gammaproteobacteria (Wald test: padj<0.05) and Verrucomicrobiae (Wald test: padj<0.05) (Supplementary Figure S4). No significant exposure effect could be found within the body and gut samples community using the Deseq2 analysis, but when looking at the rare classes (representing less than 1% of the data), it can be seen that the class Polyangia was no longer represented in hypoxia in both the gut and body samples compared to normoxia (Supplementary Table S1). For the gut samples, the classes Acidimicrobiia, Armatimonadia and Kapabacteria were no longer present in hypoxia (Supplementary Table S1). While there were no differences in species richness (p = 0.98) nor Shannon Index (p = 0.99) between gut and body samples, it should be noted that when investigating rare bacterial classes, the classes Bacilli, Desulfitobacteriia, Acidimicrobiia, Armatimonadia, and Kapabacteria were not present in the Daphnia body samples, while they were present in the Daphnia gut samples (Supplementary Table S1).
The three-way interaction between exposure, sample type and genotype in SI (p < 0.05) can be explained by the fact that the two-way interaction between exposure and sample type differed for the different genotypes. This three-way interaction was only borderline significant for SR (p = 0.08). In the F genotype there was a two-way interaction between exposure and sample type: sample types did not differ from each other in normoxia, but they did in hypoxia: bacterioplankton samples differed from both body (p < 0.05 and p < 0.01, for SR and SI, respectively) and gut (p < 0.05 and p < 0.05, for SR and SI, respectively) samples (Figure 5). In the KNO 15.04 genotype, there was no two-way interaction: the SR of bacterioplankton samples differed from both body and gut samples in normoxia as well as in hypoxia (Normoxia: bacterioplankton-body samples: p < 0.0001, bacterioplankton-gut samples: p = 0.0001; Hypoxia: bacterioplankton-body samples: p < 0.0001, bacterioplankton-gut samples: p < 0.001, ANOVA). For SI, the difference between bacterioplankton samples and both body and gut samples in normoxia became smaller in hypoxia to a point were none of the sample types differed significantly from each other when the relative abundances of the species were taken into account (Figure 5: SI: Normoxia: bacterioplankton-body samples: p < 0.01, bacterioplankton-gut samples: p < 0.05; Hypoxia: bacterioplankton-body samples: p = 0.06, bacterioplankton-gut samples: p = 0.12, ANOVA). Within the gut samples, there was a significant two-way interaction between exposure and genotype in SR. Within the F genotype, gut samples in hypoxia had a significant lower species richness compared to normoxia (p < 0.05). This trend toward a lower species richness and Shannon index upon hypoxia, although not significant, could also be seen in the body samples of the two genotypes, but not in the gut samples of KNO 15.04. An opposite trend could be seen in the bacterioplankton samples, where hypoxia exposure resulted in a higher SR and SI than in normoxia in the F genotype and a higher SR in the KNO 15.04 genotype (Figure 5). In normoxia, the gut of the KNO 15.04 genotype had a significantly lower species richness compared to the F genotype (p < 0.05).
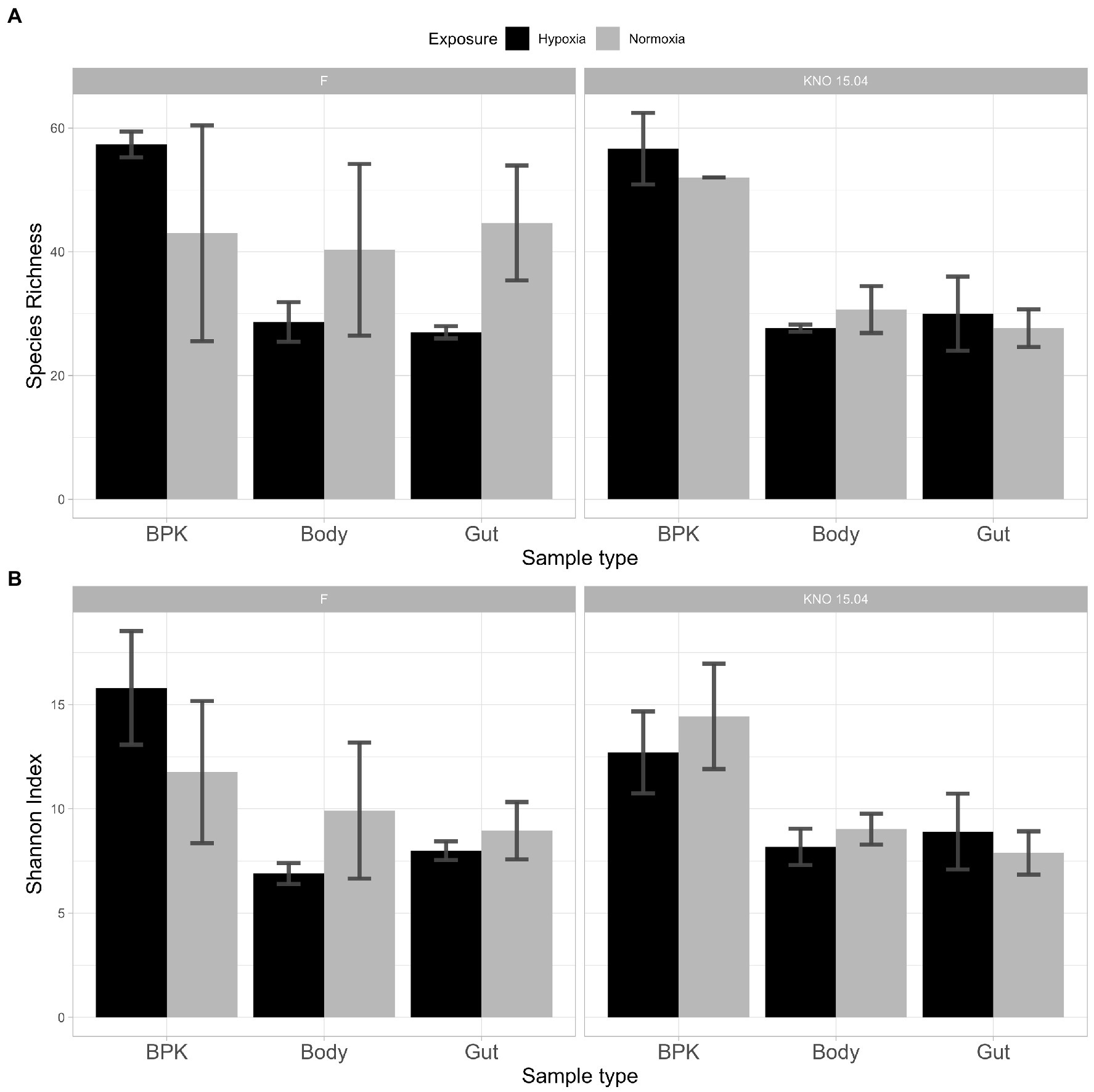
Figure 5. Alpha diversity: (A) Species Richness and (B) Shannon Index of bacterial communities between genotypes and sample types in hypoxia (black) and normoxia (grey). Left panels: F-clone; right panels: KNO 15.04.
For beta-diversity, the composition of the sample types did not differ from one another in normoxia (p = 0.19), but hypoxia exposure made the bacterial communities of the different sample types more distinct from each other causing bacterioplankton, body and gut samples to form three distinct clusters, when pooling genotypes (p = 0.0001). This distinction between sample types was present in the structure of the bacterial microbiota community in normoxia (p < 0.01), but was more pronounced in hypoxia (p = 0.0001). The strongest effect could be seen in bacterioplankton samples in which both structure (p < 0.05; Supplementary Figure S5A left panel) and composition (p < 0.05. Supplementary Figure S5B left panel) differed between normoxia and hypoxia. In the Daphnia body samples, the composition of the bacterial community differed between normoxia and hypoxia (p < 0.05; Supplementary Figure S5B middle panel), but the overall structure was the same (p = 0.35; Supplementary Figure S5A middle panel). There was no difference in composition (p = 0.18; Supplementary Figure S5B right panel) or structure (p = 0.25; Supplementary Figure S5A right panel) of the gut bacterial communities between hypoxia and normoxia.
When looking at the genotypes separately, the bacterial microbiota of the different sample types did not differ in their overall structure (p = 0.29; Figure 6A upper right panel) and composition (p = 0.89; Figure 6B upper right panel) in normoxia, but hypoxia significantly impacted both the overall structure (p < 0.01; Figure 6A upper left panel) and composition (p < 0.01; Figure 6B upper left panel) of the bacterial microbiota causing a more distinct bacterial community per sample type in the F genotype. This change of going from a shared (more common) structure and composition between the sample types in normoxia toward a distinct structure and composition per sample type in hypoxia is less pronounced in the KNO 15.04 genotype in which this is only true for composition (p = 0.09 in normoxia and p < 0.05 in hypoxia; Figure 6B lower right and left panel, respectively). The overall structure of the sample types of the KNO 15.04 genotype was not significantly impacted by hypoxia as the sample types already significantly differed from each other in normoxia (p < 0.01; Figure 6A lower right panel) and remained to differ in hypoxia (p < 0.01; Figure 6A lower left panel).
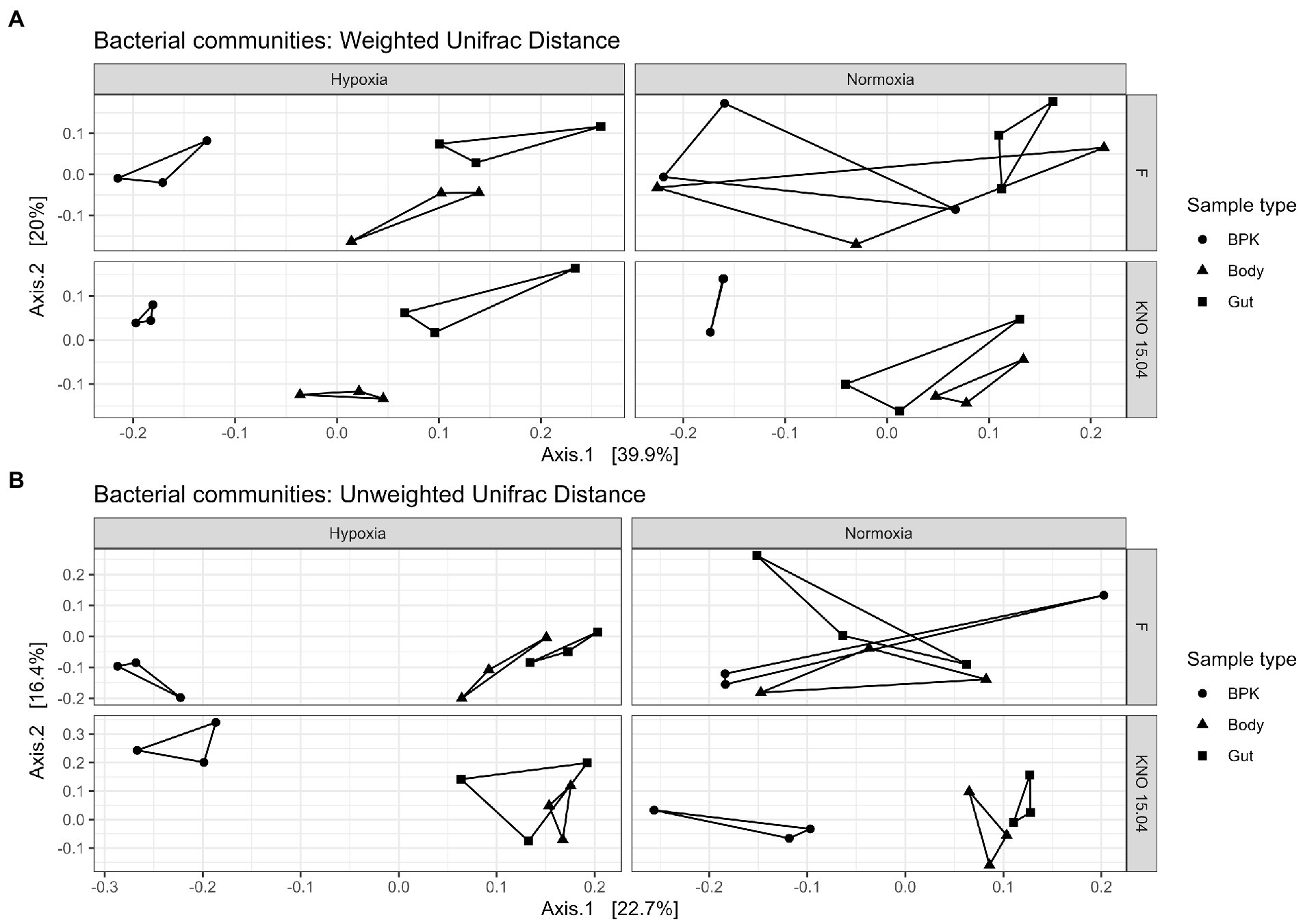
Figure 6. Beta diversity: (A) Weighted and (B) Unweighted Unifrac distance of the bacterial communities for the different genotypes in hypoxia and normoxia. Circles correspond to bacterioplankton (BPK) samples, triangles to body samples and squares to gut samples.
In this study we investigated the effect of a long-term hypoxia exposure on D. magna performance and its associated microbial community. The microbial communities studied were these from Daphnia body and gut and the free-living bacterioplankton released by Daphnia. As hypothesized, hypoxia at an environmentally relevant concentration reduced survival, fecundity and growth of the D. magna individuals tested. These results are in line with previous research in D. magna (Homer and Waller, 1983) and another Daphnia species, D. similis, that has intensively been investigated under hypoxia (Lyu et al., 2013a). In comparison with D. similis, the D. magna genotypes here tested had high survival percentages, which are in line with high survival percentages of D. magna in earlier studies (Homer and Waller, 1983; Lyu et al., 2013a). This suggests that Daphnia tolerance to hypoxia is interspecific even for Daphnia species with a similar body size. Hypoxia here affected mainly reproduction via the amount of first and second clutch producing D. magna individuals and the amount of the first clutch eggs produced, which is in line with earlier effect in D. similis (Lyu et al., 2013a) and reproduction impairments in D. magna exposed to D.O. levels below 2.7 mg/L (Homer and Waller, 1983). But our results contradict (Seidl et al., 2005), where clutch size was unchanged in the first five clutches in D. magna acclimated to hypoxia. Hypoxia induced reproduction impairments can affect future generations and the population structure in the aquatic environment. In this study, hypoxia had the largest effect on body size, where a genotype x exposure two-way interaction showed a genotype dependent effect of hypoxia on body size with the KNO 15.04 genotype growing slower compared to the F genotype in hypoxia. Reduction in body size as a result of hypoxia exposure is consistent with other studies in Daphnia spp. (Homer and Waller, 1983; Hanazato, 1996; Seidl et al., 2005; Lyu et al., 2013a). On the one hand, hypoxia-induced growth reduction (Kobayashi, 1982; Eby and Crowder, 2002; Seidl et al., 2005) is beneficial for diffusive oxygen transport pathways (Pirow et al., 2004; Pirow and Buchen, 2004; Seidl et al., 2005), but on the other hand, it can be detrimental since the smaller body size implies a diffusive bypass in the haemolymph circulation (Pirow et al., 2004; Pirow and Buchen, 2004) which reduces the effectiveness of the circulatory system’s ability to resist oxygen overload in body tissues in regions with high D.O. concentrations (Seidl et al., 2005). Daphnia individuals divert energy from their development, growth and reproduction toward producing hemoglobin to facilitate oxygen uptake under hypoxic stress (Hanazato and Dodson, 1995). The fact that we did not find strong genotype x environment interactions in survival and reproduction may be because we tested two genotypes that showed a relative high hypoxia tolerance. Body size was more affected than survival and fecundity, suggesting that different D. magna performance traits are differentially responsive to hypoxia. A similar phenomenon was found for nitrite presence, another environmental stressor linked with water pollution, where reproduction was more affected than survival and molting (Lyu et al., 2013b).
Diet and antibiotics are known traditional environmental factors that shape the gut microbial composition in a way that can be associated with changes in performance traits, also in D. magna (Akbar et al., 2020). We here showed that also hypoxia is an environmental factor that affects Daphnia associated microbial communities and especially the bacterioplankton that surrounded the Daphnia individuals. To determine the effect of hypoxia on the microbial community of D. magna, the microbial composition of the gut and body of the two genotypes was investigated. In addition, bacterioplankton samples of the medium were taken to investigate whether microbial communities in the medium experienced a shift with low oxygen levels. Two patterns with respect to selective uptake of microbial strains to obtain tolerance toward a toxic cyanobacterial diet upon the exposure of Daphnia to microbial inocula have been proposed by Houwenhuyse et al. (2021): (1) selection of specific beneficial and/or adapted strains, and/or (2) selection for a high strain diversity with complementary gene functions. While support for both these patterns was found for the tolerance of D. magna to the toxic cyanobacteria (Houwenhuyse et al., 2021), our results only showed evidence of the first pattern. Important to note is that in our study no extra inocula were added, so the microbial community in the bacterioplankton are the bacterial strains that were shedded from the Daphnia and were growing in the experimental Daphnia medium. In our results, there was a trend towards a reduced alpha diversity in terms of species richness and Shannon index in D. magna body tissue and gut upon hypoxia, which can be seen especially in the gut samples of the F genotype where species richness was significantly lower in hypoxia compared to normoxia. This trend was associated with a trend toward an increased alpha diversity in the bacterioplankton samples, causing the bacterioplankton community to differ strongly from gut and body samples in hypoxia while the bacterioplankton community was similar to gut and body bacterial communities in normoxia. Our results imply that hypoxia is structuring the bacterioplankton community. However, we do not see that effect independently of the Daphnia host. Hypoxia is selecting for microbial strains well adapted to these hypoxic conditions and Daphnia is strengthening this effect by up concentrating and shedding these strains. In the longer term and upon exposing Daphnia population experiments under hypoxic conditions, we expect that the Daphnia and bacterioplankton community’s merge. Hypoxia significantly reduced the alpha diversity of the gut microbiota of the F-genotype, but in general, hypoxia had only a minor effect on the gut or body of Daphnia. Nevertheless, hypoxia significantly altered the bacterioplankton community and significantly affected Daphnia life history. Microbial priority effects may have masked effects, given that we did not use germ-free individuals here. It is also possible that microbiome influencing factors mediate host fitness and the host associated microbiota in a hypoxic environment in a time dependent way. The chronic hypoxia exposure here may be the result of host mediated stabilizing effects which masks shorter term responses. Although the F genotype was clearly more responsive to hypoxia than KNO 15.04 with respect to responses in the microbial community, there was no significant host genotype specific response on microbial alpha diversity. Studies where Daphnia gut and bacterioplankton differed in their microbial community demonstrate the impact of the environmental conditions of Daphnia on their interaction with microbial symbionts (Freese and Schink, 2011; Callens et al., 2016). On the one hand, Actinobacteria were found to be more abundant in the bacterioplankton compared to body and gut in hypoxia which reflects a potential expelling effect under hypoxia by D. magna. Bacteria of the class Verrucomicrobiae, on the other hand, seem to thrive well under hypoxia as they are more abundant in bacterioplankton in normoxia compared to body and gut samples, while in hypoxia the amount does not significantly differ between sample types. This theory that Verrucomicrobiae are only partially secreted in the medium in hypoxia is supported by the fact that they are less than half as abundant in the bacterioplankton in hypoxia compared to normoxia. The sample type differences in the alpha diversity were translated in the beta diversity. When looking at the difference between communities, hypoxia caused the composition and structure of the bacterial communities from the bacterioplankton, gut and body to change from more similar communities to three distinct community clusters in the F genotype. This could only be seen in the composition of the sample types in the KNO 15.04 genotype, while the differences between the microbial structure of the sample types became larger in hypoxia. The F genotype showed thus a stronger microbial response to hypoxia for both beta- and alpha-diversity. Nevertheless, it should be noted that the observed microbial effects can also be a direct effect of hypoxia on the bacteria rather than a genotype dependent effect. Future research should build further on alpha- and beta diversity analyses and focus on low-level taxonomic ranks to describe abundance or reduction of aerobic to anaerobic bacterial proportions in hypoxic aquatic conditions.
Our results support the theory that environmental stress can alter host-microbial community relationships and the pattern seems to be host genotype dependent (Akbar et al., 2020). We assume that this effect affects Daphnia performance but further testing, e.g., through microbiome transplant experiments, for this is needed. No general hypoxia-associated microbiome was found and although the F genotype showed a stronger response to hypoxia in terms of microbial responses and survival, it was the KNO 15.04 that showed the strongest effects on body size and kept its microbial community equal between the different sample types tested. Also, non-adaptive symbiont loss due to hypoxic stress, similar to the process of symbiont loss in coral bleaching (Johnson et al., 2021), is a hypothesis that we cannot exclude here. Another possibility is that the detected changes in host performance and shifts in microbial communities are due to a change in Daphnia metabolism and filtration(feeding) rates (Hanazato and Dodson, 1995; Lee et al., 2022). The metabolic phenotype of Daphnia in response to hypoxia was found earlier to be influenced by both micro-evolutionary differences and spatial and temporal environmental heterogeneity of the aquatic environment (Weider and Lampert, 1985; Lee et al., 2022). Daphnia subjected to hypoxia as a result of thermal stratification are also exposed to variations in pH, temperature, salinity, conductivity, nutrients and food levels. Studies that consider the interacting impacts of these factors that may also affect metabolism and life history traits are required in order to effectively estimate the effects of lower DO in natural aquatic environments. It has already been shown that further reductions in life history traits arose in hypoxia, if it is accompanied by food shortage as food shortage and oxygen deficiency cause synergistic effects on the life history of D. magna (Hanazato, 1996).
In conclusion, hypoxia reduced survival, body size and reproduction in D. magna differentially with the strongest genotype specific effect reflected in Daphnia body size. Alongside impairements in D. magna performance traits, hypoxia induced the composition and structure of the bacterioplankton, and the Daphnia associated microbial communities (gut and body) to shift into distinct communities. Within the gut and body samples a trend toward a lower alpha diversity in hypoxia was found, which was associated with a higher alpha diversity in the surrounding bacterioplankton and this effect was different for the two genotypes. This finding is relevant in the context of host acclimatization and evolutionary potential upon climate change, which is the primary cause of hypoxia and is predicted to worsen over the next decades, inducing effects in the zooplankton and its associated microbial community and in turn affecting the biodiversity of the natural freshwater systems with effects for the quality of drinking water.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA922487.
MC and ED designed the experiment. MC performed the experiment and IV performed extractions and sample processing for sequencing. MC analyzed the data, with help from SH and ED. MC drafted the manuscript and managed revisions with input from ED and CV. All authors contributed to the article and approved the submitted version.
Funding was provided by the KU Leuven research project C16/17/002.
We are grateful for the assistance of Dzhamilyat Kurbanova and Alec De Buyck during the experimental work. We thank Amruta Rajarajan, Luc De Meester, Robby Stoks en Koenraad Muylaert for the stimulating discussions and Eline Beert and Jonas Blockx for the technical help with the hypoxia chambers.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1131203/full#supplementary-material
Akbar, S., Gu, L., Sun, Y., Zhang, L., Lyu, K., Huang, Y., et al. (2022). Understanding host-microbiome-environment interactions: insights from daphnia as a model organism. Sci. Total Environ. 808:152093. doi: 10.1016/j.scitotenv.2021.152093
Akbar, S., Gu, L., Sun, Y., Zhou, Q., Zhang, L., Lyu, K., et al. (2020). Changes in the life history traits of Daphnia magna are associated with the gut microbiota composition shaped by diet and antibiotics. Sci. Total Environ. 705:135827. doi: 10.1016/j.scitotenv.2019.135827
Barata, C., Campos, B., Rivetti, C., Leblanc, G. A., Eytcheson, S., Mcknight, S., et al. (2017). Validation of a two-generational reproduction test in Daphnia magna: an Interlaboratory exercise. Sci. Total Environ. 579, 1073–1083. doi: 10.1016/j.scitotenv.2016.11.066
Bellier, E. (2012). D. Borcard, F. Gillet, P. Legendre: numerical ecology with R. J. Agric. Biol. Environ. Stat. 17, 308–309. doi: 10.1007/s13253-012-0094-x
Bulteel, L., Houwenhuyse, S., Declerck, S. A. J., and Decaestecker, E. (2021). The role of microbiome and genotype in Daphnia magna upon parasite re-exposure. Genes (Basel), 12 12:10070. doi: 10.3390/genes12010070
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016a). Dada2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Callahan, B. J., Sankaran, K., Fukuyama, J. A., Mcmurdie, P. J., and Holmes, S. P. (2016b). Bioconductor workflow for microbiome data analysis: from raw reads to community analyses [version 1; peer review: 3 approved]. F1000 Res. 5:1492. doi: 10.12688/f1000research.8986.2
Callens, M., Macke, E., Muylaert, K., Bossier, P., Lievens, B., Waud, M., et al. (2016). Food availability affects the strength of mutualistic host-microbiota interactions in Daphnia magna. ISME J. 10, 911–920. doi: 10.1038/ismej.2015.166
Callens, M., Watanabe, H., Kato, Y., Miura, J., and Decaestecker, E. (2018). Microbiota inoculum composition affects Holobiont assembly and host growth in daphnia. Microbiome 6:56. doi: 10.1186/s40168-018-0444-1
Cooper, R. O., and Cressler, C. E. (2020). Characterization of key bacterial species in the Daphnia magna microbiota using shotgun Metagenomics. Sci. Rep. 10:652. doi: 10.1038/s41598-019-57367-x
Decaestecker, E., De Meester, L., and Ebert, D. (2002). In deep trouble: habitat selection constrained by multiple enemies in zooplankton. Proc. Natl. Acad. Sci. 99, 5481–5485. doi: 10.1073/pnas.082543099
Díaz, R. J., and Rosenberg, R. (1995). Marine benthic hypoxia: a review of its ecological effects and the behavioural responses of benthic macrofauna. Oceanogr. Mar. Biol. 33, 245–303.
Diaz, R. J., and Rosenberg, R. (2008). Spreading dead zones and consequences for marine ecosystems. Science 321, 926–929. doi: 10.1126/science.1156401
Ebert, D. (2005). Ecology, Epidemiology, and Evolution of Parasitism in Daphnia., Bethesda, MD: National Library of Medicine (Us), National Center for Biotechnology Information.
Ebert, D. (2022). Daphnia as a versatile model system in ecology and evolution. EvoDevo 13, 1–16. doi: 10.1186/s13227-022-00199-0
Eby, L. A., and Crowder, L. B. (2002). Hypoxia-based habitat compression in the Neuse River estuary: context-dependent shifts in behavioral avoidance thresholds. Can. J. Fish. Aquat. Sci. 59, 952–965. doi: 10.1139/f02-067
Freese, H. M., and Schink, B. (2011). Composition and stability of the microbial community inside the digestive tract of the aquatic crustacean Daphnia magna. Microb. Ecol. 62, 882–894. doi: 10.1007/s00248-011-9886-8
Galic, N., Hawkins, T., and Forbes, V. E. (2019). Adverse impacts of hypoxia on aquatic invertebrates: a meta-analysis. Sci. Total Environ. 652, 736–743. doi: 10.1016/j.scitotenv.2018.10.225
Gerke, P., Börding, C., Zeis, B., and Paul, R. J. (2011). Adaptive Haemoglobin gene control in daphnia Pulex at different oxygen and temperature conditions. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 159, 56–65. doi: 10.1016/j.cbpa.2011.01.017
Gilbert, D., Sundby, B., Gobeil, C., Mucci, A., and Tremblay, G. H. (2005). A seventy-two-year record of diminishing deep-water oxygen in the St. Lawrence estuary: the Northwest Atlantic connection. Limnol. Oceanogr. 50, 1654–1666. doi: 10.4319/lo.2005.50.5.1654
Gorr, T. A., Cahn, J. D., Yamagata, H., and Bunn, H. F. (2004). Hypoxia-induced synthesis of hemoglobin in the crustacean Daphnia magna is hypoxia-inducible factor-dependent. J. Biol. Chem. 279, 36038–36047. doi: 10.1074/jbc.M403981200
Goto, D., Lindelof, K., Fanslow, D., Ludsin, S., Pothoven, S., Roberts, J. J., et al. (2012). Indirect consequences of Hypolimnetic hypoxia on zooplankton growth in a large eutrophic Lake. Aquat. Biol. 16, 217–227. doi: 10.3354/ab00442
Guillard, R. R. L., and Lorenzen, C. J. (1972). Yellow-green algae with Chlorophyllide C1,2. J. Phycol. 8, 10–14.
Han, N., Pan, Z., Liu, G., Yang, R., and Yujing, B. (2021). Hypoxia: the “invisible pusher” of gut microbiota. Front. Microbiol. 12:690600. doi: 10.3389/fmicb.2021.690600
Hanazato, T. (1996). Combined effects of food shortage and oxygen deficiency on life history characteristics and filter screens of daphnia. J. Plankton Res. 18, 757–765. doi: 10.1093/plankt/18.5.757
Hanazato, T., and Dodson, S. I. (1995). Synergistic effects of low oxygen concentration, predator Kairomone, and a pesticide on the Cladoceran daphnia Pulex. Limnol. Oceanogr. 40, 700–709. doi: 10.4319/lo.1995.40.4.0700
Hanazato, T., Yasuno, M., and Hosomi, M. (1985). Significance of a low oxygen layer for a daphnia population in Lake Yunoko, Japan. Hydrobiologia 185, 19–27.
Homer, D. H., and Waller, W. T. (1983). Chronic effects of reduced dissolved oxygen on Daphnia magna. Water Air Soil Pollut. 20, 23–28. doi: 10.1007/BF00279493
Houwenhuyse, S., Stoks, R., Mukherjee, S., and Decaestecker, E. (2021). Locally adapted gut microbiomes mediate host stress tolerance. ISME J. 15, 2401–2414. doi: 10.1038/s41396-021-00940-y
Jane, S. F., Hansen, G. J. A., Kraemer, B. M., Leavitt, P. R., Mincer, J. L., North, R. L., et al. (2021). Widespread Deoxygenation of Temperate Lakes. Nature (London) 594, 66–70. doi: 10.1038/s41586-021-03550-y
Johnson, M. D., Scott, J. J., Leray, M., Lucey, N., Bravo, L. M. R., Wied, W. L., et al. (2021). Rapid ecosystem-scale consequences of acute Deoxygenation on a Caribbean coral reef. Nat. Commun. 12:4522. doi: 10.1038/s41467-021-24777-3
Kobayashi, M. (1982). Influence of body size on Haemoglobin concentration and resistance to oxygen deficiency in Daphnia magna. Comp. Biochem. Physiol. A Comp. Physiol. 72, 599–602.
Larsson, P., and Lampert, W. (2011). Experimental evidence of a low-oxygen refuge for large zooplankton. Limnol. Oceanogr. 56, 1682–1688. doi: 10.4319/lo.2011.56.5.1682
Lee, T. M., Westbury, K. M., Martyniuk, C. J., Nelson, W. A., and Moyes, C. D. (2022). Metabolic phenotype of daphnia under hypoxia: macroevolution, microevolution, and phenotypic plasticity. Front. Ecol. Evol. 10:2935. doi: 10.3389/fevo.2022.822935
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for Rna-Seq data with Deseq2. Genomebiology.Com 15:550. doi: 10.1186/s13059-014-0550-8
Lyu, K., Cao, H., Chen, R., Wang, Q., and Yang, Z. (2013a). Combined effects of hypoxia and ammonia to daphnia Similis estimated with life-history traits. Environ. Sci. Pollut. Res. Int. 20, 5379–5387. doi: 10.1007/s11356-013-1555-7
Lyu, K., Wang, Q., Chen, R., Lu, Q., and Yang, Z. (2013b). Inter-specific differences in survival and reproduction of Cladocerans to nitrite gradient and the ecological implications. Biochem. Syst. Ecol. 48, 151–156. doi: 10.1016/j.bse.2012.12.002
Macke, E., Callens, M., De Meester, L., and Decaestecker, E. (2017a). Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat. Commun. 8:1608. doi: 10.1038/s41467-017-01714-x
Macke, E., Callens, M., Massol, F., Vanoverberghe, I., De Meester, L., and Decaestecker, E. (2020). Diet and genotype of an aquatic invertebrate affect the composition of free-living microbial communities. Front. Microbiol. 11:380. doi: 10.3389/fmicb.2020.00380
Macke, E., Tasiemski, A., Massol, F., Callens, M., and Decaestecker, E. (2017b). Life history and eco-evolutionary dynamics in light of the gut microbiota. Oikos 126, 508–531. doi: 10.1111/oik.03900
Miner, B. E., De Meester, L., Pfrender, M. E., Lampert, W., and Hairston, N. G. Jr. (2012). Linking genes to communities and ecosystems: daphnia as an Ecogenomic model. Proc. R. Soc. B Biol. Sci. 279, 1873–1882. doi: 10.1098/rspb.2011.2404
North, R. P., North, R. L., Livingstone, D. M., Köster, O., and Kipfer, R. (2014). Long-term changes in hypoxia and soluble reactive phosphorus in the Hypolimnion of a large temperate Lake: consequences of a climate regime shift. Glob. Chang. Biol. 20, 811–823. doi: 10.1111/gcb.12371
Paerl, H. W., Hall, N. S., and Calandrino, E. S. (2011). Controlling harmful Cyanobacterial blooms in a world experiencing anthropogenic and climatic-induced change. Sci. Total Environ. 409, 1739–1745. doi: 10.1016/j.scitotenv.2011.02.001
Pirow, R., Bäumer, C., and Paul, R. J. (2001). Benefits of Haemoglobin in the Cladoceran crustacean Daphnia magna. J. Exp. Biol. 204, 3425–3441. doi: 10.1242/jeb.204.20.3425
Pirow, R., Bäumer, C., and Paul, R. J. (2004). Crater landscape: two-dimensional oxygen gradients in the circulatory system of the microcrustacean Daphnia magna. J. Exp. Biol. 207, 4393–4405. doi: 10.1242/jeb.01318
Pirow, R., and Buchen, I. (2004). The dichotomous Oxyregulatory behaviour of the planktonic crustacean Daphnia magna. J. Exp. Biol. 207, 683–696. doi: 10.1242/jeb.00812
Qi, W., Nong, G., Preston, J. F., Ben-Ami, F., and Ebert, D. (2009). Comparative Metagenomics of daphnia Symbionts. BMC Genomics 10:172. doi: 10.1186/1471-2164-10-172
Rajarajan, A., Wolinska, J., Walser, J.-C., Mader, M., and Spaak, P. (2022). Infection by a eukaryotic gut parasite in wild daphnia Sp. associates with a distinct bacterial community. FEMS Microbiol. Ecol. 98:97. doi: 10.1093/femsec/fiac097
Seidl, M. D., Paul, R. J., and Pirow, R. (2005). Effects of hypoxia acclimation on Morpho-physiological traits over three generations of Daphnia magna. J. Exp. Biol. 208, 2165–2175. doi: 10.1242/jeb.01614
Sison-Mangus, M. P., Mushegian, A. A., and Ebert, D. (2015). Water fleas require microbiota for survival, growth and reproduction. ISME J. 9, 59–67. doi: 10.1038/ismej.2014.116
Vanderploeg, H. A., Ludsin, S. A., Ruberg, S. A., Höök, T. O., Pothoven, S. A., Brandt, S. B., et al. (2009). Hypoxia affects spatial distributions and overlap of pelagic fish, zooplankton, and phytoplankton in Lake Erie. J. Exp. Mar. Biol. Ecol. 381, S92–S107. doi: 10.1016/j.jembe.2009.07.027
Weider, L. J., and Lampert, W. (1985). Differential response of daphnia genotypes to oxygen stress: respiration rates, hemoglobin content and low-oxygen tolerance. Oecologia 65, 487–491. doi: 10.1007/BF00379661
Zeis, B., Becker, D., Gerke, P., Koch, M., and Paul, R. J. (2013). Hypoxia-inducible Haemoglobins of daphnia Pulex and their role in the response to acute and chronic temperature increase. Biochimica et Biophysica Acta. Proteins Proteomics 1834, 1704–1710. doi: 10.1016/j.bbapap.2013.01.036
Zhang, M., Wang, Z., Xu, J., Liu, Y., Ni, L., Cao, T., et al. (2011). Ammonium, microcystins, and hypoxia of blooms in eutrophic water cause oxidative stress and C–N imbalance in submersed and floating-leaved aquatic plants in Lake Taihu, China. Chemosphere 82, 329–339. doi: 10.1016/j.chemosphere.2010.10.038
Keywords: Daphnia magna , hypoxia, genotype x environment interaction, host-associated microbiome, global change
Citation: Coone M, Vanoverberghe I, Houwenhuyse S, Verslype C and Decaestecker E (2023) The effect of hypoxia on Daphnia magna performance and its associated microbial and bacterioplankton community: A scope for phenotypic plasticity and microbiome community interactions upon environmental stress? Front. Ecol. Evol. 11:1131203. doi: 10.3389/fevo.2023.1131203
Received: 24 December 2022; Accepted: 27 March 2023;
Published: 20 April 2023.
Edited by:
Kai Lyu, Nanjing Normal University, ChinaReviewed by:
Clay Cressler, University of Nebraska-Lincoln, United StatesCopyright © 2023 Coone, Vanoverberghe, Houwenhuyse, Verslype and Decaestecker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manon Coone, bWFub24uY29vbmVAa3VsZXV2ZW4uYmU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.