
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 30 June 2023
Sec. Ecophysiology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1122304
This article is part of the Research Topic Insect Pollinators in the Anthropocene: How Multiple Environmental Stressors Are Shaping Pollinator Health View all 11 articles
Virus spillover from managed to unmanaged bees and vice versa may be one mechanism driving colony losses of the former and declines of the latter. There is clear evidence that the ubiquitous Deformed wing virus (DWV) is a major driver of honey bee (Apis mellifera) colony mortality. Although DWV has been detected in the solitary bee Osmia bicornis, data on DWV infectivity and virulence from solitary bees are scarce. Here, we used microinjection to investigate whether DWV genotype A (DWV-A) obtained from honey bees can replicate in O. bicornis. DWV-A titers and intermediate strand analyses suggest that DWV-A does not replicate in O. bicornis and thus is probably not infectious for this solitary bee species. Interestingly, the data demonstrate that DWV-A recovered from O. bicornis 16 days post-microinjection remains infectious for A. mellifera. Therefore, despite the lack of apparent virulence of DWV in this solitary bee species, O. bicornis has the potential to act as a virus spillover host and may contribute to increased colony losses of managed honey bees and declines in populations of other managed or unmanaged bee species.
Emerging infectious diseases (EIDs) are of considerable concern to both human and animal health and may pose a risk to ecosystem services such as insect pollination, ultimately leading to potentially detrimental consequences for economic gains and food security (Potts et al., 2010; Vanbergen and Initiative, the I.P, 2013; Potts, et al. 2016a; Potts et al., 2016b). The managed Western honey bee (Apis mellifera), the most economically valuable managed pollinator species, is well described to suffer from a multitude of such emerging diseases, pests, and parasites (Ellis and Munn, 2005; Rosenkranz et al., 2010; Goulson et al., 2015; Neumann et al., 2016). There is clear evidence that these pathogens are key players of unsustainably high losses of managed colonies over the last decades (Neumann and Carreck, 2010). However, other bee populations, managed or unmanaged, are also of dire concern (Potts, et al. 2016a; Tehel et al., 2016). Not only because pathogen spillover from managed honey bees may contribute to reported declines of other populations, but also because these may act as pathogen reservoirs and thus potential hosts for spillback to managed honey bees (Proesmans et al., 2021). This may thereby perpetuate a vicious cycle of colony losses and pollinator population declines (Rhyan and Spraker, 2010; Fürst et al., 2014; Graystock et al., 2016; Potts et al., 2016b).
Pathogen transmission among bee populations can occur through multiple routes, but recently attention has increased regarding spread through the shared use of flowers (Durrer and Schmid-Hempel, 1994; Chen et al., 2006a; Chen et al., 2006b; Burnham et al., 2021; Proesmans et al., 2021). Unsurprisingly, a wide range of pathogens reported from managed honey bees has been detected in other bees’ populations, underlining the commonality of pathogens jumping from one host species to another (host shift) (Fürst et al., 2014; Tehel et al., 2016; McMahon et al., 2018; Alger et al., 2019; Figueroa et al., 2019). This process can be unidirectional or bidirectional: from an initial host spilling over to a novel host, and/or from a novel host spilling back to the initial host, i.e. reverse spillover, or “spill-back” (Woolhouse, 2001). Although the susceptibility of a novel host can be driven by pathogens previously infecting other closely related phylogenetic hosts, another avenue for the development of novel EIDs may be through environmental pressures, such as widespread pesticide use and habitat loss and degradation, that serve as concurrent stressors and contribute to increased susceptibility to pathogens (Longdon et al., 2014; Retschnig et al., 2015; McMahon et al., 2018; Straub et al., 2019; Straub et al., 2022). However, empirical data on the relative importance and interplay of different drivers remain scarce and direct evidence on the type of spillover scenario is lacking (Rigaud et al., 2010; Evison et al., 2012; Straub et al., 2022). In all cases, whether or not a host species resists the infection depends on multiple interacting factors involved in the triad of “host–pathogen–environment” (Rhyan and Spraker, 2010). The emergence of such infectious diseases within bee communities has been suggested as an important driver contributing to wild bee decline (Potts et al., 2010; Manley et al., 2015; Graystock et al., 2016; Potts et al., 2016a; Tehel et al., 2016; Grozinger and Flenniken, 2019).
The worldwide-distributed Deformed wing virus (DWV) is among the most harmful pathogens associated with managed honey bees (Martin et al., 2012; Neumann et al., 2012). DWV is a positive-sense, single-stranded RNA virus (family Iflaviridae, genus Iflavirus) that causes notable clinical symptoms (deformed wings), as well as deleterious effects on foraging and life span in honey bees (de Miranda and Genersch, 2010; Möckel et al., 2011; Dainat et al., 2012; Benaets et al., 2017). It is considered to be among the major drivers of honey bee colony collapses, especially because of its efficient vectoring by the ubiquitous ectoparasitic mite Varroa destructor, which can generate disease epidemics within individual colonies (de Miranda and Genersch, 2010; Neumann et al., 2012; Wilfert et al., 2016; Yañez et al., 2020). This efficient vector activity is based on the injection of DWV particles directly into bee pupae or adult bees by the feeding mite, which is the most efficient method of horizontal transmission known so far (Chen et al., 2006b; Möckel et al., 2011; Yañez et al., 2020). Furthermore, V. destructor has been demonstrated to serve as a biological vector for DWV genotype B (DWV-B) based on viral titers (Di Prisco et al., 2016) as well as DWV intermediate strand analyses, both used as tokens of positive strand RNA virus replication (Ongus et al., 2004; Gisder et al., 2009; Posada-Florez et al., 2019; Posada-Florez et al., 2020). The increased presence of DWV in honey bees due to efficient vectoring thus poses a threat of cross-species virus transmission (Martin and Brettell, 2019).
Deformed wing virus has been detected in a wide range of other species, including several species of bumble bees (Bombus spp.) and solitary mason bees (Osmia cornuta and O. bicornis) (Mazzei et al., 2014; Ravoet, 2014; Martin and Brettell, 2019; Yañez et al., 2020). However, the mere detection of a virus is nonsynonymous with actual replication within its host; it may rather reflect that an individual has ingested or carries viral particles that are not actively replicating (Evison et al., 2012; Tehel et al., 2016). In fact, the production of minus-strand intermediates in positive-strand RNA viruses has been regarded as a prerequisite of pathogenicity for overt DWV infections (Yue and Genersch, 2005; Gisder et al., 2009). Replication of DWV was detected via intermediate strand analyses in several bumble bee species (Bombus spp.), and several studies have demonstrated experimentally that DWV can exploit bumble bees as a host (Singh et al., 2010; Li et al., 2011; Levitt et al., 2013; Fürst et al., 2014; Radzevičiūtė et al., 2017). Thus, DWV is increasingly considered to be a multihost pathogen (Zhang et al., 2012; Fürst et al., 2014). In solitary bees, few studies have similarly demonstrated prevalence of DWV as well as its minus-strand intermediate (Ravoet, 2014; Radzevičiūtė et al., 2017). However, controlled experimental infection scenarios with unequivocal evidence that DWV can exploit solitary bees as a host is largely lacking. Furthermore, the role of a potential reverse shift scenario for viruses from other bee species back to managed honey bees remains largely unexplored. Such research is needed to expand on the potential role of virus spillover for the health of both managed and unmanaged bees.
Here, we investigated through a series of virus infectivity assays whether DWV that has been propagated in honey bee pupae can replicate in the solitary bee Osmia bicornis after abdominal microinjection directly into hemolymph. Presence of replication was assessed by viral titers measured by quantitative polymerase chain reaction (qPCR) and performance of intermediate strand assays. In addition, we tested whether inocula taken from previously microinjected O. bicornis remain infectious to honey bees by reintroduction of inocula into the hemolymph of honey bee pupae. Our results demonstrate that although DWV does not appear to overtly replicate in O. bicornis, inocula harvested 16 days post-microinjection from O. bicornis remained infectious once reintroduced into the hemolymph of honey bees. Our results demonstrate that O. bicornis has the potential to act as a virus spillover or spillback host for DWV despite the lack of obvious replication, and may contribute to increased colony losses of managed honey bees and declines in populations of other wild bee species.
Sealed Western honey bee (Apis mellifera) worker brood combs were sampled from a local colony at the Institute of Bee Health in Bern-Liebefeld, Switzerland. Then, Deformed wing virus (DWV) treatment and control solutions were prepared via standard propagation in pink-eyed honey bee pupae (de Miranda et al., 2013). Five honey bee pupae per treatment were microinjected with 2 μl of a DWV (107 viral copies) or PBS buffer (Phosphate Buffered Saline; pH 7.4), respectively, and incubated at 34.5°C for five days. Afterwards, DWV treatment and control solutions were prepared by homogenization of pupae in 500 µl PBS buffer. 100 µl of chloroform was added and the solution was centrifuged at 13,000 rpm for 10 minutes. Supernatants were collected and stored at −20°C until use. To inactivate possible DWV particles in control PBS solution, it was incubated for 15 min at 65°C (Lelie et al., 1987). DWV titers in the solutions were quantified with standard quantitative polymerase chain reaction (qPCR) as detailed below (de Miranda et al., 2013; Evans et al., 2013). The bees used (N = 5) to create the DWV treatment solution for subsequent infections in Osmia bicornis were tested for variant identity using specific PCR assays (see Section 2.4) for two dominant European variants (DWV genotype A (DWV-A) and genotype B (DWV-B)) (Kevill et al., 2017).
O. bicornis cocoons were purchased from WAB-Mauerbienenzucht, Konstanz, Germany, and stored at 4°C until experimental start. Adult bees emerged individually in 1.3 L round plastic cylinders (Ø = 110 mm, height = 160 mm) sealed with multifilament netting (Lanz-Anliker AG, Rohrbach, Switzerland) in a climate-controlled room at 25° C equipped with a sunlight simulation system at the research station of Agroscope, Zürich, Switzerland (Sandrock et al., 2014). Each cage was supplied with sugar water (50% (w/v)) in 0.2 ml Eppendorf® tubes and pollen (Sonnentracht Imkerei GmbH, Bremen, Germany, Petri dish, Ø = 30 mm) ad libitum. Pollen was gamma ray irradiated (Leoni Studer Hard AG, Däniken, Switzerland) prior to use to limit potential pathogen interference via contamination (Sandrock et al., 2014). Two days after emergence, 90 bees (34 males and 56 females) were randomly assigned to each of the two treatments (“O. bicornis DWV”; N = 46; 17 males and 29 females or “O. bicornis PBS Control”; N = 44; 17 males and 27 females). This time span from emergence and treatment enabled the bees to adapt to the cage, recover from eclosion, defecate and feed (Dmochowska-Ślęzak et al., 2015). For each treatment, bees were microinjected between the third and the fourth tergite with 2µl of DWV treatment (107 viral copies) or PBS control solutions that were the product of the procedure mentioned in Section 2.1 (de Miranda et al., 2013) using a Hamilton syringe with 26-gauge needle (0.45 mm) (Human et al., 2013). To facilitate the handling of bees, they were cooled for two hours at 4° C, then ice-chilled for 3 minutes prior to microinjection (Ebadi et al., 1980; Frost et al., 2011; Chen et al., 2014). After microinjection, individuals were returned to their cages in the climate chamber. Pollen and sugar water provisions were refilled every three days. Mortality was assessed daily. The experiment was terminated 16 days post-treatment, and all surviving individuals were freeze-killed and stored at −20° C until molecular analysis was conducted (Chen et al., 2007). Three individuals from each treatment were tested for DWV variant identity using specific PCR assays (see Section 2.4) for two dominant European variants (DWV-A and DWV-B) (Kevill et al., 2017). Twelve O. bicornis bees were frozen immediately after microinjection with DWV treatment solution to confirm the baseline concentration of the virus-inoculated bees (“DWV inoculum”; Supplementary Table 1).
Prior to experimental infection, a total of 19 untreated A. mellifera pupae were selected from two local colonies in Liebefeld, Switzerland and molecularly screened to determine naturally occurring baseline levels of DWV (“A. mellifera No Treatment,” Supplementary Table 2).
To confirm the infectivity of the DWV particles in the DWV treatment solution administered to O. bicornis, pink-eyed worker pupae (A. mellifera), which were not infested by the parasitic mite Varroa destructor, were then collected and randomly assigned to treatment groups (A. mellifera DWV, N = 21); A. mellifera PBS control, N = 20). Pupae were microinjected intraabdominally with the solutions prepared as previously described (de Miranda et al., 2013). Six individuals from both treatment groups were freeze-killed immediately after microinjection at −20° C until molecular processing to assess initial virus levels following microinjection (see Supplementary Table 2).
To test whether DWV particles recovered from O. bicornis remain infectious for A. mellifera, a new treatment solution was prepared by extracting virus particles from previously DWV-treated O. bicornis bees (N = 3) 16 days post-injection as previously described. Inoculation by microinjection in additional pink-eyed worker pupae (“A. mellifera DWV from Osmia”) was performed as detailed before using pupae (N = 21) from the same two colonies as previously mentioned (de Miranda et al., 2013). Six individuals were freeze-killed immediately after microinjection at −20° C until molecular processing to assess initial virus levels (see Supplementary Table 2).
All remaining A. mellifera pupae from the treatment groups “A. mellifera DWV” (N =15) and “A. mellifera DWV from Osmia” (N = 15) were incubated at 34.5°C and ≥ 50% relative humidity and darkness for five days, then stored at −20°C until molecular analysis was conducted. DWV quantification was performed by quantitative PCR as detailed below (de Miranda et al., 2013; Evans et al., 2013).
Individual bees were crushed in 2 ml Eppendorf® tubes with 5 mm metal beads in a TN buffer (100 mM Tris, 100 mM NaCl, pH 7.6) and homogenized for 1 min by 25 1/s frequency using a Retsch® MM 300 mixer mill (Evans et al., 2013). TN buffer volume added depended on the weight of the bee to achieve a concentration of 0.5 mg/μl (Supplementary Tables 1, 2). Fifty μl of homogenate were used for RNA extraction. RNA was extracted with the NucleoSpin® RNA II kit (Macherey−Nagel) following the manufacturer’s recommendations. The extracted RNA was eluted in 60 μl of RNase-free water and stored at −80°C until further processing (Evans et al., 2013). An exogenous internal RNA reference, the Tobacco Mosaic Virus (TMV), was introduced into each sample during the RNA extraction to monitor the efficiency of RNA purification and cDNA synthesis steps (Tentcheva et al., 2006). Reverse transcription was performed by using M-MLV RT enzyme (Promega) with 2.4 μg of RNA template, 1 μg of random hexamer primers, 200 Units of reverse transcriptase, in 25 μl of final reaction volume (de Miranda et al., 2013). The cDNA synthesis was performed in a Thermocycler (Biometra) with a PCR cycling profile of 5 min incubation at 70°C and 60 min at 37°C. The quantitative PCR reactions were prepared with the KAPA SYBR® FAST Universal qPCR kit (KAPA Biosystems) with 3 μl of tenfold-diluted cDNA, 0.24 μl (0.2 µM) of forward and reverse specific primers, 6 μl of 2X KAPA SYBR® green reaction mix, in a total of 12 μl final reaction volume (de Miranda et al., 2013). Primers used for DWV and TMV are detailed in Table 1. Each sample was performed in duplicate. Each plate included no-template negative controls and four positive controls per primer pair obtained from ten-fold serial dilutions of purified PCR products that function as standard curves (Bustin et al., 2009). The reaction was processed in an ECO™ Real-Time PCR machine (Illumina) and the qPCR cycling profile consisted of 3 min incubation at 95°C and 40 cycles of 3 sec at 95°C for denaturation, 30 sec at 57°C for annealing and extension, and data collection. To verify the specificity of the qPCR products, the amplification was followed by a melting curve analysis by reading the fluorescence at 0.5°C increments from 55°C to 95°C.
Viral titers were calculated from qPCR output data and standard curves adjusted by the various experimental dilution factors to arrive at an estimated DWV genome copies per microgram of RNA (Yañez et al., 2012), which were then log-transformed (Supplementary Table 1). All DWV-A titers are reported as log-transformed means ± standard deviations (see Section 3). For DWV variant identity, the relative number of DWV-A or DWV-B genome copies per microgram of RNA are reported as a percentage of the sum of DWV-A and DWV-B genome copies per microgram of RNA (Table 2).
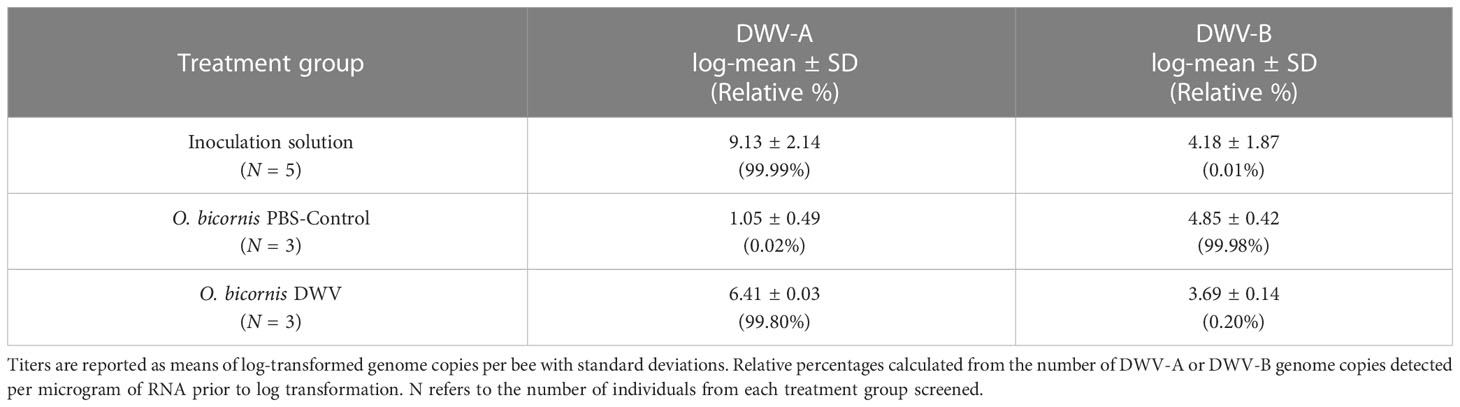
Table 2 Viral variant identification by relative viral titers detected by quantitative PCR for DWV-A and DWV-B in inoculation solution, PBS-Control or DWV-treated O. bicornis.
The presence of the DWV-A intermediate strand RNA was assessed as a token of viral replication in O. bicornis and A. mellifera by strand-specific RT-PCR (Yue and Genersch, 2005). The analyses were conducted following standard procedures (de Miranda et al., 2013) for all three DWV treatment groups (“A. mellifera DWV”, “O. bicornis DWV”, “A. mellifera DWV from Osmia”) in two separate reactions by first tagging the RNA intermediate strand during the cDNA synthesis using a “Tagged” primer, then by specifically amplifying it using a “Tag” primer (Table 3). Intermediate strand validation controls, labelled as “No Tag,” were run in parallel for the detection of potential unspecific strand amplification (false positives). Those controls do not include “Tag” primers in the PCR reactions and ensure the effective removal of “Tagged” primer during the purification process. RNA was converted to cDNA using a Superscript® III reverse transcriptase (Invitrogen) following the manufacturer’s recommendations with 1 μl of DWV 3F tagged primer (Table 3), 1 μl of 0.01M dNTP mix (Bioline), 4 μl of 5X first strand buffer, 1 μl of 0.1M DTT, 1 μl (200 Units) of reverse transcriptase, in 20 μl of final reaction volume (de Miranda et al., 2013). The reaction was processed in a thermocycler (Biometra) with the following PCR cycling profile: 5 min at 65°C; then 10 min at 25°C and 60 min at 50°C, followed by 15 min at 70°C. The high temperature used for the reverse transcription improves specific strand amplification by reducing secondary structures. The resulting cDNA was purified results using the NucleoSpin® Gel & PCR Clean-up kit (Macherey-Nagel) and eluted in 30 μl of elution buffer. Purified tenfold-diluted cDNA was amplified by using the same conditions as conventional PCR (see above) with MyTaq™ kit (Bioline). A Tag oligonucleotide was used as forward primer and DWV4-R1 was used as reverse primer (Table 3). In addition, other PCR reactions without the Tag primer (“No Tag”) were run as a control for inactivation and efficient removal of the excess of DWV 3F tagged primer via purification after reverse transcription. The thermal cycling profile consisted in 2 min incubation at 95°C and 35 cycles of 20 sec at 95°C for denaturation, 20 sec at 42°C for annealing, and 30 sec at 72°C for extension. The PCR products were purified and analysed by electrophoresis on a 1.2% agarose gel, stained in 30% GelRed® Nucleic Acid Gel Stain bath for 30 min, and visualized under UV light.

Table 3 Primers used for the detection of intermediate strand DWV-A in O. bicornis and A. mellifera.
All statistical analyses and figure preparation were performed using R Statistical Software (v4.2.1, R Core Team, 2022).
Survival analyses of O. bicornis bees were performed using the packages “survival” (Therneau and Grambsch, 2000) and “survminer” (Kosinski et al., 2020) to calculate and create Kaplan-Meier cumulative survival curves for each treatment group. Bees that survived until the experimental endpoint and were freeze-killed were censored from the survival analysis. Significant variation between treatment groups and sex were performed separately using the survdiff() function (rho = 0). Pairwise testing for any significant different variation was done using the pairwise_survdiff() function (rho = 0) with Bonferroni corrected p-values (Bonferroni, 1936; Figure 1).
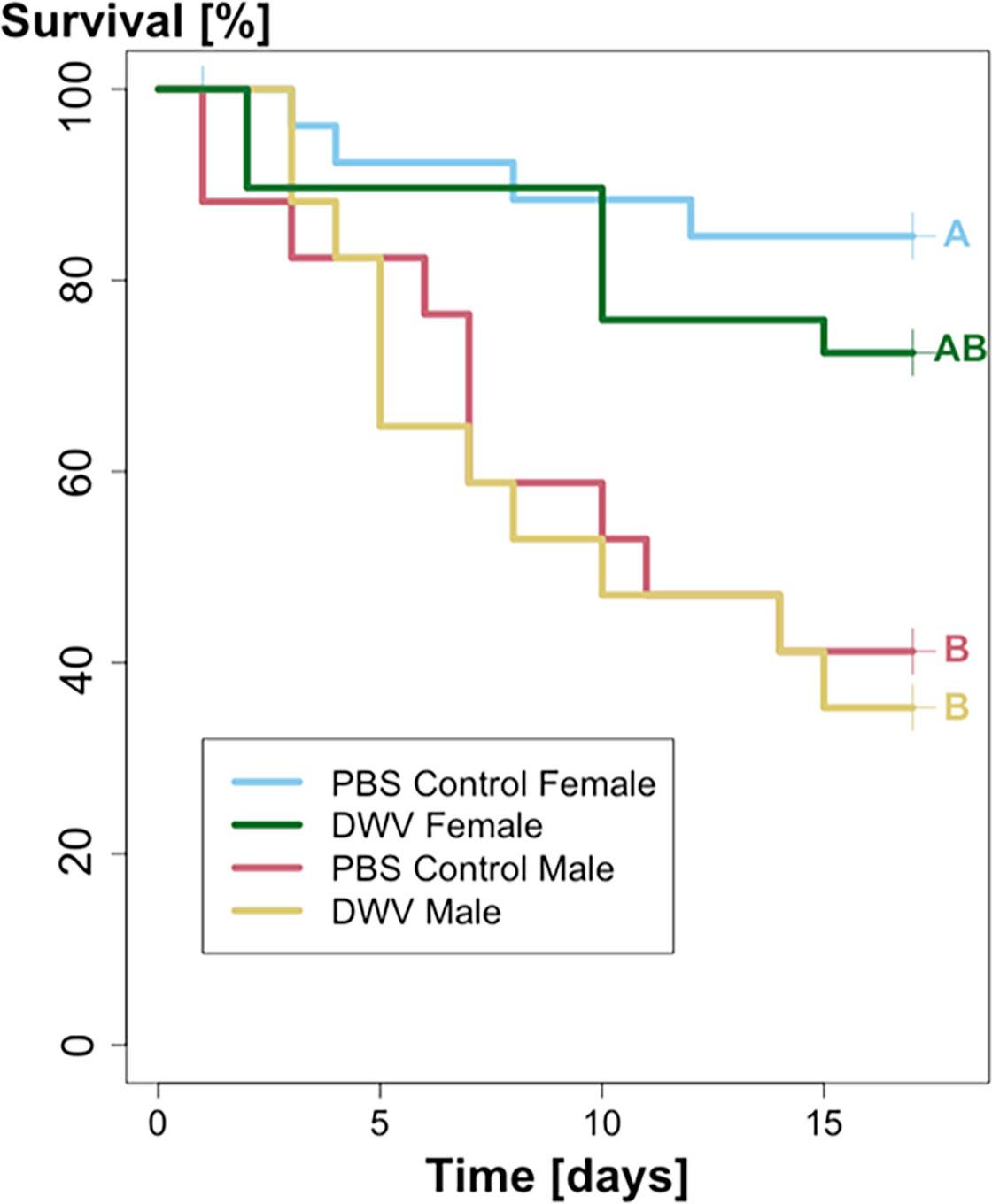
Figure 1 Kaplan-Meier survival analysis of Osmia bicornis. Both male PBS Control (N = 17) and DWV (N = 17) treatment groups showed significantly higher mortality than PBS control females (N = 27), whereas DWV treated females (N = 29) showed no significant differences from other treatment groups. Significant differences are marked by different letters (A, B); Kaplan-Meier log-rank test, p < 0.05).
To test whether viral titers could be explained by sex in O. bicornis, a simple linear regression model (lm) from the “stats” package (R Core Team, 2022) was conducted with log-transformed viral titers as a dependent variable and sex (male/female) as an independent variable for O. bicornis treatment groups (PBS control and DWV). Titer data for O. bicornis was pooled by treatment group regardless of sex. Then, a Shapiro-Wilk’s test was performed on the log-transformed viral titers for all O. bicornis and A. mellifera treatment groups (Figure 2; “DWV inoculum,” “O. bicornis PBS control,” “O. bicornis DWV,” “A. mellifera PBS Control,” “A. mellifera DWV,” and “A. mellifera DWV from Osmia”) using the shapiro.test() function and indicated non-normality (p < 0.05). As such, a Kruskal-Wallis test was done using the kruskal.test() function. A post-hoc pairwise Wilcoxon test for significant differences between all possible pairwise combinations was done with the pairwise.wilcox.test() function, with Bonferroni adjusted p-values (Figure 2).
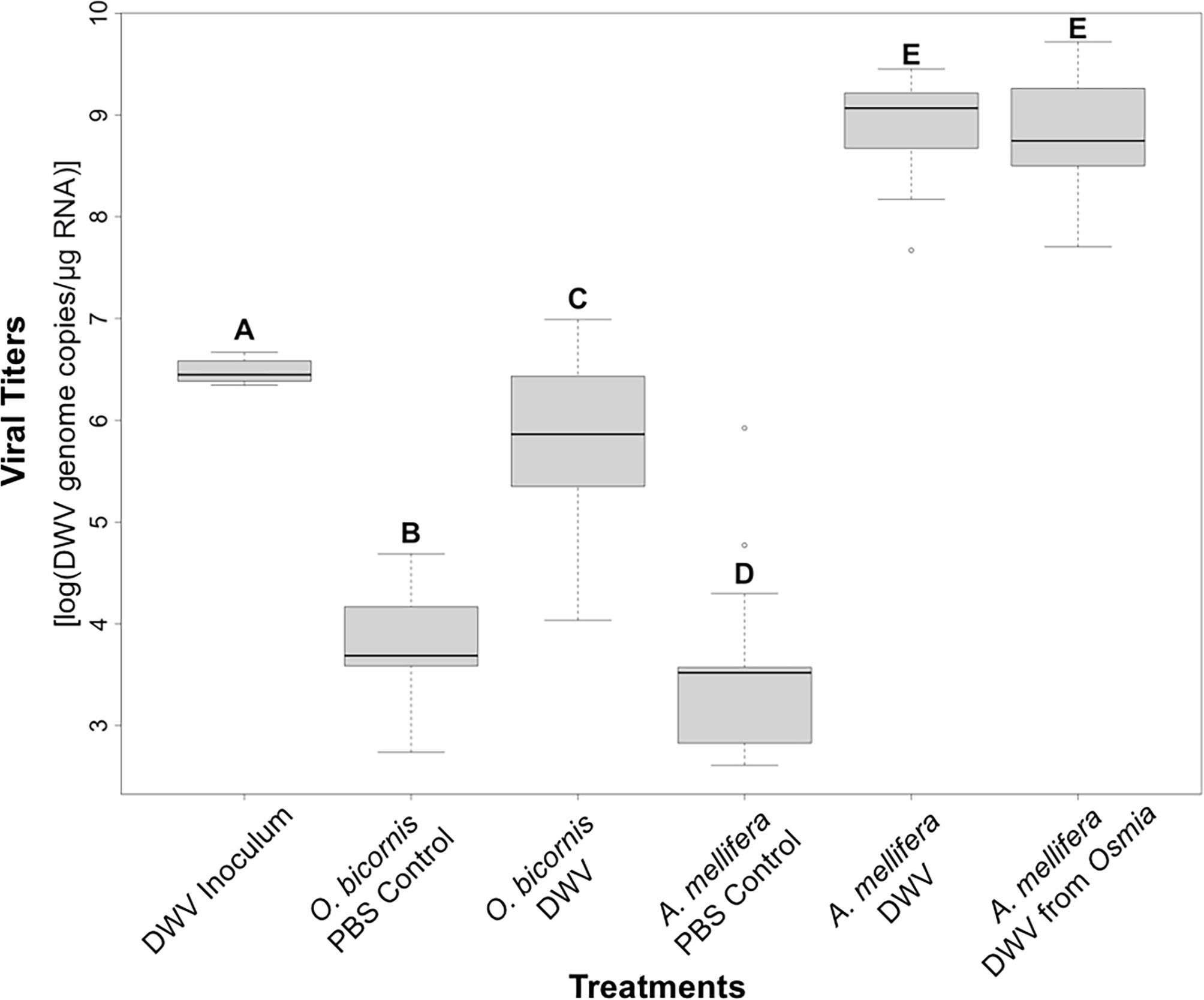
Figure 2 DWV-A titers expressed as log transformed DWV-A genome copies per microgram of RNA across the six treatment groups. Different letters (A–E) indicate significant differences between treatment levels (Post-hoc pairwise Wilcoxon test, p < 0.05; “DWV inoculum”: the initial microinjected DWV-A titer in O. bicornis, N = 12; “O. bicornis PBS control”: O. bicornis microinjected with DWV-free PBS solution, N = 44; “O. bicornis DWV”: Osmia bicornis microinjected with DWV, N = 46; “A. mellifera PBS Control”: A. mellifera microinjected with DWV-free PBS solution N = 14; “DWV A. mellifera”: Apis mellifera microinjected with DWV, N = 15; “A. mellifera DWV Osmia”: Apis mellifera microinjected with DWV originating from previously infected O. bicornis, N = 15). A five number summary is visually displayed in each box whisker plot: 1) minimum value, 2) first quartile, 3) median, 4) third quartile, and 5) maximum value.
The virus-strain specific PCR for two predominant European variants (DWV genotypes A (DWV-A) and B (DWV-B)) showed that DWV inoculation solution propagated in Apis mellifera and further used for infection assays in Osmia bicornis consisted integrally of DWV-A (99.99%; Table 2).
In O. bicornis PBS-injected controls, very low titers of DWV-A were detected (log mean 1.05 ± 0.49), whereas DWV-B comprised 99.98% of the variant composition (log mean 4.85 ± 0.42). Oppositely, DWV-treated O. bicornis had relatively high titers of DWV-A (log mean 6.41 ± 0.3, N = 3) compared to DWV-B (log mean 3.69 ± 0.14), thus representing 99.80% of the variant composition as DWV-A (Table 2), similar to the inoculation solution.
A total of 56 O. bicornis survived until experimental end, with 29 PBS-Control individuals (7 males, 22 females) and 27 DWV-treated individuals (6 male, 21 female). O. bicornis males treated with DWV showed significantly higher mortality compared to O. bicornis PBS Control treated females (Figures 1A, B; Kaplan-Meier log-rank test, p = 0.0046). However, PBS Control females and PBS Control males demonstrated significantly different lifespan outcomes (Kaplan-Meier log-rank test, p = 0.0023), suggesting that sex (male vs. female), rather than treatment (PBS vs. DWV), contributed to differences in mortality.
The screening for background levels of DWV-A in the two local colonies selected to serve as the source of A. mellifera individuals for the present study demonstrated low titers in screened, untreated individuals expressed at the logarithmic scale of genome copies per µg of RNA with log means of 4.05 ± 0.36 (N = 19) (Supplementary Table 2; “A. mellifera No Treatment”). Individuals from both colonies were then used for subsequent creation of inoculation solution and for further experimental virus transmission assays.
For O. bicornis, viral titers between males and females were not significantly different (lm, t-value = −1.076, p = 0.287), indicating that sex did not influence viral titer outcomes. As such, males and female O. bicornis were pooled based on treatment group for further analyses. DWV-A was detected in DWV-treated O. bicornis bees (“DWV inoculum”) that were frozen immediately after treatment at a log-mean of 6.48 ± 0.11 (N=12), demonstrating successful inoculation with DWV by the procedure of microinjection (Figure 2). Furthermore, microinjection with PBS control solution in O. bicornis (“O. bicornis PBS Control”) and A. mellifera (“A. mellifera PBS Control”) resulted in low levels of DWV-A with titer log means of 3.68 ± 0.42 (N = 44) and 3.55 ± 0.92 (N = 14), respectively, suggesting that the stress of microinjection did not contribute to the development of infection.
O. bicornis treated with DWV (“O. bicornis DWV”) showed a significantly higher titer with a log mean of 5.55 ± 0.70 (N = 46) (Figures 2B, C) of DWV-A than those treated with PBS control solution (“O. bicornis PBS Control”). However, this value was significantly lower than the initial inoculum level (Figures 2A–C), suggesting a lack of overt infection with DWV-A. In comparison, A. mellifera treated with DWV showed significantly higher levels of DWV-A than both the initial inoculum (“DWV inoculum”) and A. mellifera treated with PBS control solution (“A. mellifera PBS Control”), with a log mean of 8.89 ± 0.52 (N = 15), consistent with establishment of infection (Figures 2A–E). Furthermore, A. mellifera treated with DWV that had been recovered from O. bicornis (“A. mellifera DWV from Osmia”) showed similar levels of DWV-A suggestive of infection with a log mean of 8.80 ± 0.54 (N = 15).
Strong, clear bands representing DWV-A were visualized using gel electrophoresis following the intermediate strand-specific RT-PCR assay in A. mellifera pupae five days after microinjection with DWV treatment solution, indicating that the microinjected virus particles were infective (Figure 3). In contrast, O. bicornis bees 16 days after microinjection with DWV treatment solution showed only very faint bands. For those A. mellifera pupae microinjected with DWV particles recovered from previously DWV-microinjected O. bicornis, the intermediate strand DWV-A RNA was clearly detectable, indicating active replication. Intermediate strand validation controls (“No Tag,” Figure 3), displayed no visible bands, indicating an efficient removal of the DWV 3F tagged primer, ruling out the possibility false-positive results.
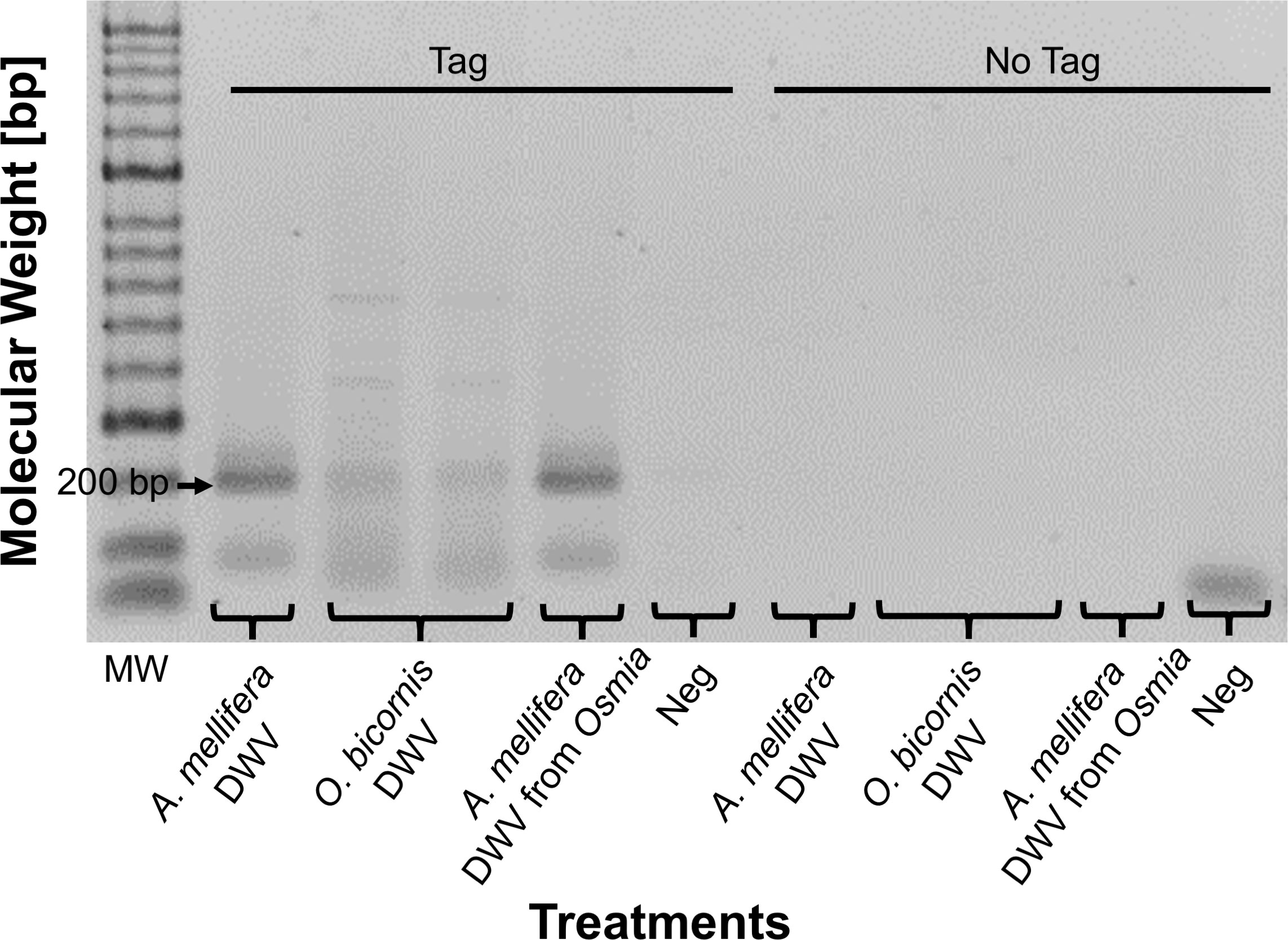
Figure 3 Intermediate strand assay for replication of DWV-A in three treatment groups “A. mellifera DWV,” “O bicornis DWV” and “A. mellifera DWV from Osmia,” as well as a no-template negative control “Neg.” Positive results are shown with the presence of a 200 bp sized band representing DWV-A. Validation controls (“No Tag”) for the detection of potential unspecific strand amplification (false positives) are displayed. (MW = molecular weight size marker; bp = base pairs).
The viral infectivity assay employed in our study offers the stimulation of an extreme infection scenario for the studied solitary bee species, Osmia bicornis, through an artificial microinjection with a high number of Deformed wing virus genotype A (DWV-A) copies of genome (106 genome copies per microgram of RNA). This viral infectivity assay offers two important advantages to test for infectivity of a virus within a new host by (1) overcoming of the natural physical barriers and physiological antiviral defenses to viral infections, thereby enabling the virus to rapidly spread into the host’s body and (2) negatively affecting the expression of immune response genes (Yang and Cox-Foster, 2005; Möckel et al., 2011; Yañez et al., 2012; Ryabov et al., 2014).
In support of apparent infection, as was demonstrated by DWV microinjected A. mellifera in our study (“A. mellifera DWV”), an increase in viral titers for DWV-A (Figure 2) and the presence of strong, clear bands by intermediate strand analysis representing replication of DWV-A (Figure 3) are expected. In contrast, DWV treated O. bicornis (“O. bicornis DWV”) showed slightly yet significantly lower viral titers compared to starting levels (“DWV Inoculum”), and only faint bands were present by intermediate strand analysis. In the case of active viral replication, we would expect viral titers to increase after inoculation, which was not observed. Instead, the viral titers in microinjected O. bicornis were detected at a slightly yet significantly lower level to the original virus inoculum, suggesting that DWV-A particles visualized by the intermediate strand assay may be residual post-microinjection. However, a very low rate of DWV-A replication in O. bicornis cannot be excluded.
Although pupae of A. mellifera were used as positive controls to confirm the infectivity of the experimental DWV particles according to the standard method of virus propagation, newly emerged adults were used for experimental transmission of DWV to O. bicornis (de Miranda et al., 2013). Data from the literature in adult A. mellifera suggest an inoculation range of 104 to 108 DWV particles for covert, low-level infections, and 1010 to 1011 DWV particles for overt, high-level infections (Highfield et al., 2009; Zioni et al., 2011; de Miranda et al., 2013; McMahon et al., 2018). DWV-treated A. mellifera in the present study demonstrated titers within this range, which is consistent with previous findings (Ryabov et al., 2014). Interestingly, our results for O. bicornis demonstrate a DWV particle range that matches the range described for an asymptomatic honey bee with a covert infection (104 to 108 DWV copies of genome), which is likely due to experimental DWV injection. These findings are similarly consistent with reported DWV low-level infections in Bombus spp., with an estimated range between 104 to 106 DWV particles (McMahon et al., 2018).
Interestingly, A. mellifera treated with DWV sourced from previously microinjected O. bicornis (“A. mellifera DWV from Osmia”) showed similarly high titers of DWV-A as A. mellifera injected with DWV that had been propagated in honey bee pupae (“A. mellifera DWV”). This suggests that although DWV did not cause an overt infection in O. bicornis, the virus remains viable and infectious to A. mellifera, even sixteen days post-microinjection. Although the virus particles do not appear to actively replicate in O. bicornis, residual particles from microinjection may remain in a latent state and become virulent upon microinjection into an optimized host, i.e., A. mellifera. Furthermore, the presence of only faint bands representing DWV-A for DWV-treated O. bicornis (“O. bicornis DWV”) on the intermediate strand assay could be potentially explained by the presence of inoculum remnants, whose source was DWV-A propagated in honey bee pupae. The mechanism of this ability of DWV-A to not cause overt infection in O. bicornis, yet become infective upon inoculation into its optimized host, A. mellifera, up to sixteen days post-inoculation, remains to be understood. Should a natural scenario occur in which O. bicornis may serve as a source of latent DWV-A particles that have the potential to become infectious to an optimized host such as A. mellifera, there may be implications for repercussions on other managed or unmanaged bee communities in terms of pathogen transmission.
How can a solitary bee species such as O. bicornis become infected with DWV in nature? Although this remains unclear, it has been suggested that virus uptake occurs per os via a food-borne transmission, likely via shared flowers (Chen et al., 2006a; Chen et al., 2006b; Singh et al., 2010; Ravoet, 2014; Radzevičiūtė et al., 2017; Burnham et al., 2021; Keller et al., 2021). Though DWV is a key pathogen in managed honey bees and spillover to other species, e.g. bumble bees, has been reported repeatedly, more data are required before deriving general conclusions on the role of spillover of viruses contributing to solitary bee decline (Fürst et al., 2014; Alger et al., 2019; Gusachenko et al., 2019; Tehel et al., 2020; Burnham et al., 2021; Cilia et al., 2021). Our results are in line with data of field survey study, in which intermediate strand RNA of DWV was detected in only one solitary bee species (Andrena haemorrhoa) (Fabricius, 1778), but not in a range of other analyzed solitary bee species, including O. bicornis (Radzevičiūtė et al., 2017). In contrast, several Bombus spp. have displayed intermediate strand RNA of DWV (Radzevičiūtė et al., 2017; Alger et al., 2019). A lower infectivity and virulence of DWV in solitary bees compared to social bees could be explained by several factors.
For example, solitary bees cannot rely on social immunity and must therefore entirely rely on individual immune responses (Wilson-Rich et al., 2009; Meunier, 2015). These individual immune responses may be better developed compared to workers in social insects, which can be considered analogous to somatic cells. Therefore, losses of individual workers can be compensated for as long as the germ line remains intact (Evans, et al., 2006; Straub et al., 2015). Furthermore, differential gut microbiota enabling the host to fight against pathogens may also play a role (Engel et al., 2016; Keller et al., 2021). In any case, there appear to be significant differences between host species and their susceptibility to DWV infections (McMahon et al., 2018). Furthermore, variations arise both between and within RNA viruses due to the high mutation rates and ample opportunity for local strains to adapt to novel hosts (Daszak et al., 2000; Parrish et al., 2008; Gisder et al., 2018; Paxton et al., 2022).
For example, intermediate strand RNA of Black queen cell virus (BQCV), another RNA virus, has been detected in Anthophora plumipes, several Bombus spp., Xylocopa spp., Vespa velutina, the stingless bee Melipona colimana, and in O. bicornis (Radzevičiūtė et al., 2017; Mazzei et al., 2019; Morfin et al., 2021). Since BQCV transmission is not attributed to an efficient biological vector, as is largely the case with DWV in honey bees and the ubiquitous ectoparasitic mite Varroa destructor, exploring transmission dynamics in BQCV and other RNA viruses that are not associated with an efficient vector may be a better predictor for their possible role in novel hosts (Neumann et al., 2012).
Nonetheless, these results indicate that DWV has the potential to cause overt infection in A. mellifera when sourced from O. bicornis, potentially posing an additional threat to other managed and unmanaged bees in terms of DWV transmission. This could cause a spillback scenario for DWV and possibly for the other honey bee viruses detected in O. bicornis (Radzevičiūtė et al., 2017). Interestingly, the variant identity of DWV that predominated in the present study was DWV genotype A (DWV-A). A recent study by Paxton et al. (2022) has highlighted the worldwide replacement of DWV-A by DWV genotype B (DWV-B). Thus, further research is needed to investigate to which extent results regarding infectivity of O. bicornis and its potential role with regard to spillover obtained in here for DWV-A may differ for the recently increasingly DWV-B. Given the abundance of RNA viruses identified in populations of both managed and unmanaged bees and the subsequent potential of virus spillbacks, it is prudent to take such a scenario also into account for managed honey bee health.
Because the mere detection of DWV in managed or unmanaged bee species is not a reliable sign of a host shift, survey data should ideally be accompanied by controlled infection scenarios. In our study, we demonstrate for the first time through experimental transmission that Deformed wing virus genotype A (DWV-A) does not obviously replicate in Osmia bicornis as a novel host. Nevertheless, DWV-A particles-maintained infectivity for A. mellifera within O. bicornis up to sixteen days post microinjection. Therefore, this solitary bee species has the potential to serve as a transient spillover host, which may ultimately contribute to colony losses and diminishing populations of wild bee species more detrimentally affected by DWV infections, as is observed for several bumblebee species. More survey and controlled infection data are required from a range of species and viruses to draw general conclusions on the role of virus spillover and spillback for the health of both managed and unmanaged bees.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethical approval was not required for the study on animals in accordance with the local legislation and institutional requirements.
Conceptualization, PN and OY. Data curation, AS, NB and OY. Formal analysis, AS, NB, AB and OY. Funding acquisition, PN. Investigation, AS and NB. Methodology, NB and OY. Project administration, OY, MA and PN. Supervision, OY, MA and PN. Validation, AS and OY. Visualization, AS and AB. Writing – original draft, AS, NB, OY and PN. Writing – review and editing, AS, NB, OY, AB, MA and PN. All authors contributed to the article and approved the submitted version.
Financial support was granted to PN via the Vinetum Foundation. Open access funding by University of Bern.
Appreciation is expressed to Monika Haueter, Kaspar Roth, Stephan Bosshart and Mario Wald-burger for technical assistance and for introduction into laboratory methods. A special thank you is paid to Robert Paxton for providing advice for the contents of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1122304/full#supplementary-material
Alger S. A., Burnham P. A., Boncristiani H. F., Brody A. K. (2019). RNA virus spillover from managed honeybees (Apis mellifera) to wild bumblebees (Bombus spp.). PloS One 14 (6), e0217822. doi: 10.1371/journal.pone.0217822
Benaets K., Van Geystelen A., Cardoen D., De Smet L., de Graaf D. C., Schoofs L., et al. (2017). Covert deformed wing virus infections have long-term deleterious effects on honeybee foraging and survival. Proc. R. Soc. B: Biol. Sci. 284 (1848), 20162149. doi: 10.1098/rspb.2016.2149
Bonferroni C. E. (1936). Teoria statistica delle classi e calcolo delle probabilità (8: Pubblicazioni Del R Isti tuto Superiore Di Scienze Economiche e Commer ciali Di Firenze).
Burnham P. A., Alger S. A., Case B., Boncristiani H., Hébert‐Dufresne L., Brody A. K. (2021). Flowers as dirty doorknobs: deformed wing virus transmitted between Apis mellifera and Bombus impatiens through shared flowers. J. Appl. Ecol. 58 (10), 2065–2074. doi: 10.1111/1365-2664.13962
Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi: 10.1373/clinchem.2008.112797
Chen Y. P., Pettis J. S., Collins A., Feldlaufer M. F. (2006b). Prevalence and transmission of honeybee viruses. Appl. Environ. Microbiol. 72 (1), 606–611. doi: 10.1128/AEM.72.1.606-611.2006
Chen Y., vans J., Hamilton M., Feldlaufer M. (2007). The influence of RNA integrity on the detection of honey bee viruses: molecular assessment of different sample storage methods. J. Apicultural Res. 46 (2), 81–87. doi: 10.1080/00218839.2007.11101372
Chen Y.-M., Fu Y., He J., Wang J.-H. (2014). Effects of cold narcosis on memory acquisition, consolidation and retrieval in honeybees (Apis mellifera). Zoological Res. 35 (2), 118–123. doi: 10.11813/j.issn.0254-5853.2014.2.118
Chen Y., Evans J., Feldlaufer M. (2006a). Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebrate Pathol. 92 (3), 152–159. doi: 10.1016/j.jip.2006.03.010
Cilia G., Zavatta L., Ranalli R., Nanetti A., Bortolotti L. (2021). Replicative deformed wing virus found in the head of adults from symptomatic commercial bumblebee (Bombus terrestris) colonies. Veterinary Sci. 8 (7), 117. doi: 10.3390/vetsci8070117
Dainat B., Evans J. D., Chen Y. P., Gauthier L., Neumann P. (2012). Dead or alive: deformed wing virus and Varroa destructor reduce the life span of winter honeybees. Appl. Environ. Microbiol. 78 (4), 981–987. doi: 10.1128/AEM.06537-11
Daszak P., Cunningham A. A., Hyatt A. D. (2000). Emerging infectious diseases of wildlife– threats to biodiversity and human health. Science 287, 8. doi: 10.1126/science.287.5452.443
de Miranda J. R., Bailey L., Ball B. V., Blanchard P., Budge G. E., Chejanovsky N., et al. (2013). Standard methods for virus research in Apis mellifera. J. Apicultural Res. 52 (4), 1–56. doi: 10.3896/IBRA.1.52.4.22
Di Prisco G., Annoscia D., Margiotta M., Ferrara R., Varricchio P., Zanni V., et al. (2016). A mutualistic symbiosis between a parasitic mite and a pathogenic virus undermines honey bee immunity and health. Proc. Natl. Acad. Sci. 113 (12), 3203–3208. doi: 10.1073/pnas.1523515113
Dmochowska-Ślęzak K., Giejdasz K., Fliszkiewicz M., Żóltowska K. (2015). Variations in antioxidant defense during the development of the solitary bee Osmia bicornis. Apidologie 46 (4), 432–444. doi: 10.1007/s13592-014-0333-y
Durrer S., Schmid-Hempel P. (1994). Shared use of flowers leads to horizontal pathogen transmission. Proc. R. Soc. London. Ser. B: Biol. Sci. 258 (1353), 299–302. doi: 10.1098/rspb.1994.0176
Ebadi R., Gary N. E., Lorenzen K. (1980). Effects of carbon dioxide and low temperature narcosis on honey bees, Apis mellifera. Environ. Entomology 9 (1), 144–150. doi: 10.1093/ee/9.1.144
Ellis J. D., Munn P. A. (2005). The worldwide health status of honey bees. Bee World 86 (4), 88–101. doi: 10.1080/0005772X.2005.11417323
Engel P., Kwong W. K., McFrederick Q., Anderson K. E., Barribeau S. M., Chandler J. A., et al. (2016). The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. MBio 7 (2), e02164–15.
Evans J. D., Schwarz R. S., Chen Y. P., Budge G., Cornman R. S., De la Rua P., et al. (2013). Standard methods for molecular research in Apis mellifera. J. Apicultural Res. 52 (4), 1–54. doi: 10.3896/IBRA.1.52.4.11
Evans J. D., Aronstein K., Chen Y. P., Hetru C., Imler J. L., Jiang H., et al. (2006). Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol Biol. 15 (5), 645–56.
Evison S. E. F., Roberts K. E., Laurenson L., Pietravalle S., Hui J., Biesmeijer J. C., et al. (2012). Pervasiveness of parasites in pollinators. PloS One 7 (1), e30641. doi: 10.1371/journal.pone.0030641
Figueroa L. L., Blinder M., Grincavitch C., Jelinek A., Mann E. K., Merva L. A., et al. (2019). Bee pathogen transmission dynamics: deposition, persistence and acquisition on flowers. Proceedings of the Royal Society B 286(1903), 20190603. doi: 10.1098/rspb.2019.0603
Frost E. H., Shutler D., Hillier N. K. (2011). Effects of cold immobilization and recovery period on honeybee learning, memory, and responsiveness to sucrose. J. Insect Physiol. 57 (10), 1385–1390. doi: 10.1016/j.jinsphys.2011.07.001
Fürst M. A., McMahon D. P., Osborne J. L., Paxton R. J., Brown M. J. F. (2014). Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 506 (7488), 364–366. doi: 10.1038/nature12977
Gauthier L., Tentcheva D., Tournaire M., Dainat B., Cousserans F., Colin M. E., et al. (2007). Viral load estimation in asymptomatic honey bee colonies using the quantitative RT-PCR technique. Apidologie 38 (5), 426–435. doi: 10.1051/apido:2007026
Gisder S., Aumeier P., Genersch E. (2009). Deformed wing virus: replication and viral load in mites (Varroa destructor). J. Gen. Virol. 90 (Pt 2), 463–467. doi: 10.1099/vir.0.005579-0
Gisder S., Möckel N., Eisenhardt D., Genersch E. (2018). In vivo evolution of viral virulence: switching of deformed wing virus between hosts results in virulence changes and sequence shifts. Environ. Microbiol. 20 (12), 4612–4628. doi: 10.1111/1462-2920.14481
Goulson D., Nicholls E., Botías C., Rotheray E. L. (2015). Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347 (6229), 1255957. doi: 10.1126/science.1255957
Graystock P., Blane E. J., McFrederick Q. S., Goulson D., Hughes W. O. H. (2016). Do managed bees drive parasite spread and emergence in wild bees? Int. J. Parasitology: Parasites Wildlife 5 (1), 64–75. doi: 10.1016/j.ijppaw.2015.10.001
Grozinger C. M., Flenniken M. L. (2019). Bee viruses: ecology, pathogenicity, and impacts. Annu. Rev. Entomology 64 (1), 205–226. doi: 10.1146/annurev-ento-011118-111942
Gusachenko O. N., Woodford L., Balbirnie-Cumming K., Ryabov R. E. V., Evans D. J. (2020). Evidence for and against deformed wing virus spillover from honey bees to bumble bees: A reverse genetic analysis. Sci Rep 10 (1), 16847.
Highfield A. C., El Nagar A., Mackinder L. C. M., Noël L. M.-L. J., Hall M. J., Martin S. J., et al. (2009). Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 75 (22), 7212–7220. doi: 10.1128/AEM.02227-09
Human H., Brodschneider R., Dietemann V., Dively G., Ellis J. D., Forsgren E., et al. (2013). Miscellaneous standard methods for Apis mellifera research. J. Apicultural Res. 52 (4), 1–53. doi: 10.3896/IBRA.1.52.4.10
Keller A., McFrederick Q. S., Dharampal P., Steffan S., Danforth B. N., Leonhardt S. D. (2021). (More than) hitchhikers through the network: the shared microbiome of bees and flowers. Curr. Opin. Insect Sci. 44, 8–15. doi: 10.1016/j.cois.2020.09.007
Kevill J. L., Highfield A., Mordecai G. J., Martin S. J., Schroeder D. C. (2017). ABC assay: method development and application to quantify the role of three DWV master variants in overwinter colony losses of European honey bees. Viruses 9 (11), E314. doi: 10.3390/v9110314
Kosinski M., Biecek P, Fabian S. (2020). Survminer: drawing survival curves using “ggplot2”. R package version 0.4, 8, p.556. Available at: https://CRAN.R-project.org/package=survminer.
Lelie P. N., Reesink H. W., Lucas C. J. (1987). Inactivation of 12 viruses by heating steps applied during manufacture of a hepatitis b vaccine. J. Med. Virol. 23 (3), 297–301. doi: 10.1002/jmv.1890230313
Levitt A. L., Singh R., Cox-Foster D. L., Rajotte E., Hoover K., Ostiguy N., et al. (2013). Cross-species transmission of honey bee viruses in associated arthropods. Virus Res. 176 (1–2), 232–240. doi: 10.1016/j.virusres.2013.06.013
Li J., Peng W., Wu J., Strange J. P., Boncristiani H., Chen Y. (2011). Cross-species infection of deformed wing virus poses a new threat to pollinator conservation. J. Economic Entomology 104 (3), 732–739. doi: 10.1603/ec10355
Longdon B., Brockhurst M. A., Russell C. A., Welch J. J., Jiggins F. M. (2014). The evolution and genetics of virus host shifts. PloS Pathog. 10 (11), e1004395. doi: 10.1371/journal.ppat.1004395
Manley R., Boots M., Wilfert L. (2015). Emerging viral disease risk to pollinating insects: ecological, evolutionary and anthropogenic factors. J. Appl. Ecol. 52 (2), 331–340. doi: 10.1111/1365-2664.12385
Martin S. J., Brettell L. E. (2019). Deformed wing virus in honeybees and other insects. Annu. Rev. Virol. 6 (1), 49–69. doi: 10.1146/annurev-virology-092818-015700
Martin S. J., Highfield A. C., Brettell L., Villalobos E. M., Budge G. E., Powell M., et al. (2012). Global honey bee viral landscape altered by a parasitic mite. Sci. (New York N.Y.) 336 (6086), 1304–1306. doi: 10.1126/science.1220941
Mazzei M., Carrozza M. L., Luisi E., Forzan M., Giusti M., Sagona S., et al. (2014). Infectivity of DWV associated to flower pollen: experimental evidence of a horizontal transmission route. PloS One 9 (11), e113448. doi: 10.1371/journal.pone.0113448
Mazzei M., Cilia G., Forzan M., Lavazza A., Mutinelli F., Felicioli A. (2019). Detection of replicative Kashmir bee virus and black queen cell virus in Asian hornet Vespa velutina (Lepelieter 1836) in Italy. Sci. Rep. 9 (1), 10091. doi: 10.1038/s41598-019-46565-2
McMahon D. P., Wilfert L., Paxton R. J., Brown M. J. F. (2018). Emerging viruses in bees: From molecules to ecology. Adv Virus Res 101, 251–291. doi: 10.1016/bs.aivir.2018.02.008
Meunier J. (2015). Social immunity and the evolution of group living in insects. Philos. Trans. R. Soc. B: Biol. Sci. 370 (1669), 20140102. doi: 10.1098/rstb.2014.0102
Möckel N., Gisder S., Genersch E. (2011). Horizontal transmission of deformed wing virus: pathological consequences in adult bees (Apis mellifera) depend on the transmission route. J. Gen. Virol. 92 (Pt 2), 370–377. doi: 10.1099/vir.0.025940-0
Morfin N., Gashout H. A., Macías-Macías J. O., De la Mora A., Tapia-Rivera J. C., Tapia-González J. M., et al. (2021). Detection, replication and quantification of deformed wing virus-a, deformed wing virus-b, and black queen cell virus in the endemic stingless bee, Mlipona colimana, from Jalisco, Mexico. Int. J. Trop. Insect Sci. 41 (2), 1285–1292. doi: 10.1007/s42690-020-00320-7
Neumann P., Carreck N. L. (2010). Honey bee colony losses. J. Apicultural Res. 49 (1), 1–6. doi: 10.3896/IBRA.1.49.1.01
Neumann P., Yañez O., Fries I., de Miranda J. R. (2012). Varroa invasion and virus adaptation. Trends Parasitol. 28 (9), 353–354. doi: 10.1016/j.pt.2012.06.004
Neumann P., Pettis J. S., Schäfer M. O. (2016). Quo vadis aethina tumida? biology and control of small hive beetles. Apidologie 47 (3), 427–466. doi: 10.1007/s13592-016-0426-x
Ongus J. R., Peters D., Bonmatin J.-M., Bengsch E., Vlak J. M., van Oers M. M. (2004). Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 85 (Pt 12), 3747–3755. doi: 10.1099/vir.0.80470-0
Parrish C. R., Holmes E. C., Morens D. M., Park E.-C., Burke D. S., Calisher C. H., et al. (2008). Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72 (3), 457–470. doi: 10.1128/MMBR.00004-08
Paxton R. J., Schäfer M. O., Nazzi F., Zanni V., Annoscia D., Marroni F., et al. (2022). Epidemiology of a major honey bee pathogen, deformed wing virus: potential worldwide replacement of genotype a by genotype b. Int. J. Parasitology: Parasites Wildlife 18, 157–171. doi: 10.1016/j.ijppaw.2022.04.013
Posada-Florez F., Childers A. K., Heerman M. C., Egekwu N. I., Cook S. C., Chen Y., et al. (2019). Deformed wing virus type a, a major honey bee pathogen, is vectored by the mite Varroa destructor in a non-propagative manner. Sci. Rep. 9 (1), 12445. doi: 10.1038/s41598-019-47447-3
Posada-Florez F., Ryabov E. V., Heerman M. C., Chen Y., Evans J. D., Sonenshine D. E., et al. (2020). Varroa destructor mites vector and transmit pathogenic honey bee viruses acquired from an artificial diet. PloS One 15 (11), e0242688. doi: 10.1371/journal.pone.0242688
Potts S. G., Biesmeijer J. C., Kremen C., Neumann P., Schweiger O., Kunin W. E. (2010). Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25 (6), 345–353. doi: 10.1016/j.tree.2010.01.007
Potts S. G., Ngo H. T., Biesmeijer J. C., Breeze T. D., Dicks L.V., Garibaldi L.A., et al. (2016a). The assessment report of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services on pollinators, pollination and food production. Bonn, Germany: Secretariat of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services, 556pp.
Potts S. G., Imperatriz-Fonseca V., Ngo H. T., Aizen M. A., Biesmeijer J. C., Breeze T. D., et al. (2016b). Safeguarding pollinators and their values to human well-being. Nature 540, 220–229. doi: 10.1038/nature20588
Proesmans W., Albrecht M., Gajda A., Neumann P., Paxton R. J., Pioz M., et al. (2021). Pathways for novel epidemiology: plant–Pollinator–Pathogen networks and global change. Trends Ecol. Evol. 36 (7), 623–636. doi: 10.1016/j.tree.2021.03.006
Radzevičiūtė R., Theodorou P., Husemann M., Japoshvili G., Kirkitadze G., Zhusupbaeva A., et al. (2017). Replication of honey bee-associated RNA viruses across multiple bee species in apple orchards of Georgia, Germany and Kyrgyzstan. J. Invertebrate Pathol. 146, 14–23. doi: 10.1016/j.jip.2017.04.002
Ravoet J., De Smet L., Meeus I., Smagghe G., Wenseleers T., de Graaf D. C. (2014). Widespread occurrence of honey bee pathogens in solitary bees | elsevier enhanced reader. J Invertebr Pathol 122, 55–58. doi: 10.1016/j.jip.2014.08.007
R Core Team (2022) R: a language and environment for statistical computing. Available at: http://www.R-project.org/.
Retschnig G., Williams G. R., Odemer R., Boltin J., Di Poto C., Mehmann M. M., et al. (2015). Effects, but no interactions, of ubiquitous pesticide and parasite stressors on honey bee (Apis mellifera) lifespan and behaviour in a colony environment: stressor effects in honey bee colonies. Environ. Microbiol. 17 (11), 4322–4331. doi: 10.1111/1462-2920.12825
Rhyan J. C., Spraker T. R. (2010). Emergence of diseases from wildlife reservoirs. Veterinary Pathol. 47 (1), 34–39. doi: 10.1177/0300985809354466
Rigaud T., Perrot-Minnot M.-J., Brown M. J. F. (2010). Parasite and host assemblages: embracing the reality will improve our knowledge of parasite transmission and virulence. Proc. R. Soc. B: Biol. Sci. 277 (1701), 3693–3702. doi: 10.1098/rspb.2010.1163
Rosenkranz P., Aumeier P., Ziegelmann B. (2010). Biology and control of Varroa destructor. J. Invertebrate Pathol. 103, S96–S119. doi: 10.1016/j.jip.2009.07.016
Ryabov E. V., Wood G. R., Fannon J. M., Moore J. D., Bull J. C., Chandler D., et al. (2014). A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PloS Pathog. 10 (6), e1004230. doi: 10.1371/journal.ppat.1004230
Sandrock C., Tanadini L. G., Pettis J. S., Biesmeijer J. C., Potts S. G., Neumann P. (2014). Sublethal neonicotinoid insecticide exposure reduces solitary bee reproductive success. Agric. For. Entomology 16 (2), 119–128. doi: 10.1111/afe.12041
Singh R., Levitt A. L., Rajotte E. G., Holmes E. C., Ostiguy N., van Engelsdorp D., et al. (2010). RNA Viruses in hymenopteran pollinators: evidence of inter-taxa virus transmission via pollen and potential impact on non-apis hymenopteran species. PloS One 5 (12), e14357. doi: 10.1371/journal.pone.0014357
Straub L., Williams G. R., Pettis J., Fries I., Neumann P. (2015). Superorganism resilience: eusociality and susceptibility of ecosystem service providing insects to stressors. Curr. Opin. Insect Sci. 12, 109–112. doi: 10.1016/j.cois.2015.10.010
Straub L., Williams G. R., Vidondo B., Khongphinitbunjong K., Retschnig G., Schneeberger A., et al. (2019). Neonicotinoids and ectoparasitic mites synergistically impact honeybees. Sci. Rep. 9 (1), 8159. doi: 10.1038/s41598-019-44207-1
Straub L., Strobl V., Yañez O., Albrecht M., Brown M. J. F., Neumann P. (2022). Do pesticide and pathogen interactions drive wild bee declines? Int. J. Parasitology: Parasites Wildlife 18, 232–243. doi: 10.1016/j.ijppaw.2022.06.001
Tehel A., Brown M. J., Paxton R. J. (2016). Impact of managed honey bee viruses on wild bees. Curr. Opin. Virol. 19, 16–22. doi: 10.1016/j.coviro.2016.06.006
Tehel A., Streicher T., Tragust S., Paxton R. J. (2020). Experimental infection of bumblebees with honeybee-associated viruses: no direct fitness costs but potential future threats to novel wild bee hosts. R. Soc. Open Sci. 7 (7), 200480. doi: 10.1098/rsos.200480
Tentcheva D., Gauthier L., Bagny L., Fievet J., Dainat B., Cousserans F., et al. (2006). Comparative analysis of deformed wing virus (DWV) RNA in Apis mellifera and Varroa destructor. Apidologie 37 (1), 41–50. doi: 10.1051/apido:2005057
Therneau T. M., Grambsch P. M. (2000). “The cox model,” in Modeling survival data: extending the cox model (New York, NY: Springer New York), 39–77. doi: 10.1007/978-1-4757-3294-8_3
Vanbergen A. J., Initiative, the I.P (2013). Threats to an ecosystem service: pressures on pollinators. Front. Ecol. Environ. 11 (5), 251–259. doi: 10.1890/120126
Wilfert L., Long G., Leggett H. C., Schmid-Hempel P., Butlin R., Martin S. J. M., et al. (2016). Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 351 (6273), 594–597. doi: 10.1126/science.aac9976
Wilson-Rich N., Spivak M., Fefferman N. H., Starks P. T. (2009). Genetic, individual, and group facilitation of disease resistance in insect societies. Annu. Rev. Entomology 54, 405–423. doi: 10.1146/annurev.ento.53.103106.093301
Woolhouse M., Taylor L. H., Haydon D. T. (2001). Population biology of multihost pathogens. Science 292 (5519), 1109–1112. doi: 10.1126/science.1059026
Yañez O., Jaffé R., Jarosch A., Fries I., Moritz R. F. A., Paxton R. J., et al. (2012). Deformed wing virus and drone mating flights in the honey bee (Apis mellifera): implications for sexual transmission of a major honey bee virus. Apidologie 43 (1), 17–30. doi: 10.1007/s13592-011-0088-7
Yañez O., Piot N., Dalmon A., de Miranda J. R., Chantawannakul P., Panziera D., et al. (2020). Bee viruses: routes of infection in hymenoptera. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00943
Yang X., Cox-Foster D. L. (2005). Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. United States America 102 (21), 7470–7475. doi: 10.1073/pnas.0501860102
Yue C., Genersch E. (2005). RT-PCR analysis of deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 86 (Pt 12), 3419–3424. doi: 10.1099/vir.0.81401-0
Zhang X., He S. Y., Evans J. D., Pettis J. S., Yin G. F., Chen Y. P. (2012). New evidence that deformed wing virus and black queen cell virus are multi-host pathogens. J. Invertebrate Pathol. 109 (1), 156–159. doi: 10.1016/j.jip.2011.09.010
Keywords: Apis mellifera, host shift, Osmia bicornis, pathogen spillover, pollinators, wild bee
Citation: Schauer A, Bianco N, Yañez O, Brown A, Albrecht M and Neumann P (2023) Deformed wing virus prevalence in solitary bees put to the test: an experimental transmission study. Front. Ecol. Evol. 11:1122304. doi: 10.3389/fevo.2023.1122304
Received: 12 December 2022; Accepted: 05 June 2023;
Published: 30 June 2023.
Edited by:
Fabio Sgolastra, University of Bologna, ItalyReviewed by:
Giovanni Cilia, Council for Agricultural and Economics Research (CREA), ItalyCopyright © 2023 Schauer, Bianco, Yañez, Brown, Albrecht and Neumann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandria Schauer, YWxleGFuZHJpYS5zY2hhdWVyQHVuaWJlLmNo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.