
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 17 February 2023
Sec. Behavioral and Evolutionary Ecology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1085993
This article is part of the Research Topic Adaptations to Subterranean Environments View all 14 articles
 Olga Bondareva1*
Olga Bondareva1* Tatyana Petrova1
Tatyana Petrova1 Semen Bodrov1
Semen Bodrov1 Maria Gavrilo2,3
Maria Gavrilo2,3 Antonina Smorkatcheva4
Antonina Smorkatcheva4 Natalia Abramson1
Natalia Abramson1Life under the earth surface is highly challenging and associated with a number of morphological, physiological and behavioral modifications. Subterranean niche protects animals from predators, fluctuations in environmental parameters, but is characterized by high levels of carbon dioxide and low levels of oxygen and implies high energy requirements associated with burrowing. Moreover, it lacks most of the sensory inputs available above ground. The current study describes results from RNA-seq analysis of four subterranean voles from subfamily Arvicolinae: Prometheomys schaposchnikowi, Ellobius lutescens, Terricola subterraneus, and Lasiopodomys mandarinus. Original RNA-seq data were obtained for eight species, for nine species, SRA data were downloaded from the NCBI SRA database. Additionally assembled transcriptomes of Mynomes ochrogaster and Cricetulus griseus were included in the analysis. We searched for the selection signatures and parallel amino acid substitutions in a total of 19 species. Even within this limited data set, we found significant changes of dN/dS ratio by free-ratio model analysis for subterranean Arvicolinae. Parallel substitutions were detected in genes RAD23B and PYCR2. These genes are associated with DNA repair processes and response to oxidative stress. Similar substitutions were discovered in the RAD23 genes for highly specialized subterranean Heterocephalus glaber and Fukomys damarensis. The most pronounced signatures of adaptive evolution related to subterranean niche within species of Arvicolinae subfamily were detected for Ellobius lutescens. Our results suggest that genomic adaptations can occur very quickly so far as the amount of selection signatures was found to be compliant with the degree of specialization to the subterranean niche and independent from the evolutionary age of the taxon. We found that the number of genomic signatures of selection does not depend on the age of the taxon, but is positively correlated with the degree of specialization to the subterranean niche.
There are approximately 250 species of rodents that spend most of their lives in burrows (Begall et al., 2007). Underground tunnels protect animals from predators, fluctuations in environmental parameters and extreme conditions. Subterranean rodents live in darkness, lack most of the sensory inputs available above ground, face low food supplies and expend a lot of energy digging. Therefore, this lifestyle is highly challenging and associated with a number of morphological, physiological and behavioral modifications (McNab, 1966; Nevo, 1979, 1999; Buffenstein, 2000; Busch et al., 2000; Begall et al., 2007). That’s why they for a long time served as a model for studying adaptive convergence and parallel evolution (Nevo, 1979; Lacey, 2000; Begall et al., 2007).
The molecular studies of adaptation to subterranean lifestyle started with the mitochondrial DNA analyses. First, in analysis of cytochrome b (CYTB) in subterranean rodent families Geomyidae and Bathergidae, Da Silva et al. (2009) found a significantly higher ratio (dN/dS) in each of the subterranean groups with respect to their non-subterranean counterparts. Moreover, the subterranean mole rats and tucu-tucus showed more sites whose amino acid properties may be under positive selection in the CYTB gene than their non-subterranean relatives, and some of the sites identified to be under selection exclusively in subterranean taxa were shared among all subterranean taxa (Da Silva et al., 2009). Later on these results were confirmed in analysis of both CYTB and cytochrome c oxidase subunit 2 (COX2) (Tomasco and Lessa, 2014) and at least three sites under positive selection have been detected in subterranean rodents. Positive selection signals have been detected in various organisms related to shifts in their ecology that imply changes in the metabolic needs (Tomasco and Lessa, 2014). Hence, the variability of mitochondrial genomes may reflect adaptations for demanding lifestyles (Di Rocco et al., 2006; Da Fonseca et al., 2008; Hassanin et al., 2009; Yu et al., 2011; Luo et al., 2013; Wang et al., 2016).
Studies of the mitochondrial genome adaptations were followed by studies of nuclear genes. Results of the Spalax galili Nevo, Ivanitskaya, and Beiles, 2001 genome and transcriptome analyses show high rates of ω values in genes associated with RNA/DNA editing, reduced chromosome rearrangements and over-representation of short interspersed elements (SINEs). These may probably be linked to degeneration of vision and progression of photoperiodic perception, tolerance to hypercapnia and hypoxia and resistance to cancer (Fang et al., 2014a). Another comparative analyses including DNA-seq and RNA-seq data of phylogenetically related subterranean species from Bathergidae family revealed possible molecular adaptations for subterranean conditions and factors of extreme longevity, including a divergent insulin peptide, expression of oxygen-carrying globins in the brain, prevention of high CO2-induced pain perception, and enhanced ammonia detoxification (Fang et al., 2014b). Large-scale analysis of coding DNA sequences of four representatives of independent subterranean mammal lineages from three superorders indicate positive selection and convergent occurrence of substitutions in the same sites for 53 genes (Davies et al., 2018). While African mole-rats and blind mole-rats have been studied extensively in relation to resistance to oxidative stress, toxin-resistance, and circadian biology (Rado et al., 1991; Oosthuizen et al., 2003; Fang et al., 2014a,b), other groups of subterranean rodents have so far been largely overlooked.
The subfamily Arvicolinae (voles, lemmings, and muskrats) is the evolutionary young rodent taxon that emerged the fastest documented adaptive radiation among recent mammals. The number of extant arvicoline rodents is eight times greater than in the sister subfamily Cricetinae, the most recent common ancestors of both are known in the fossil record from the Late Miocene, ca 10 Ma both in Eurasia and North America (Martin, 2003; Fejfar et al., 2011). Nowadays they occupy a variety of landscapes, habitats, and ecological niches in the Northern Hemisphere, from above ground and even arboreal [e.g., tree voles, Arborimus Taylor, 1915 (Corn and Bury, 1988; Swingle and Foreman, 2009) to highly specialized subterranean—Ellobius Fischer, 1814 and Prometheomys Satunin, 1901 (Ognev, 1950; Kryštufek and Shenbrot, 2022)]. Moreover, the transition to the subterranean lifestyle within this subfamily occurred more than once. Our previous studies showed that selection pressure is relaxed at the mitochondrial CYTB gene in particular (Bondareva et al., 2021a), and in mitochondrial genomes for most subterranean Arvicolinae species (Bondareva et al., 2021b). The recent and phylogenetically independent transition of Arvicolinae voles to subterranean lifestyle makes it possible to trace adaptive events at a smaller evolutionary scale, in contrast to the analysis of classical ancient subterranean species such as Heterocephalus Rüppell, 1842, Nannospalax Palmer, 1903, and Fukomys Ogilby, 1838. Thus, this model group may allow insights into the very beginning of the adaptive process at the genome level, comparison with the data obtained in the studies of ancient subterranean taxa may show whether these changes have a convergent character or individual nature in each case.
In the current study, we evaluated the selection signals in nuclear genes of the subterranean Palaearctic voles. Similarly to the previously carried mitochondrial genome analysis, we expect to find the magnitude of selective pressure that occurred in the subterranean Arvicolinae lineages using RNA-seq data. We also aimed to test the hypothesis that convergent molecular signatures of selection may occur in unrelated subterranean mammals.
In the current study we obtained new material for eight Arvicolinae species (Alticola lemminus Miller, 1898, Chionomys nivalis Martins, 1842, Terricola subterraneus de Selys-Longshamps, 1836, Dicrostonyx torquatus Pallas, 1778, Lemmus sibiricus Kerr, 1792, Myopus schisticolor Lilljeborg, 1844, Ondatra zibethicus Linnaeus, 1766, and Prometheomys schaposchnikowi Satunin, 1901), see details for sampling localities in Biosamples description. Tissue mix (muscles, liver, heart, lungs, and testes for males) was fixed in an intactRNA buffer (Evrogen, Moscow, Russia) to avoid RNA degradation directly at the moment of catching the animal in the field. The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Zoological Institute Russian Academy of Sciences (permission #2-17/14-07-2022).
For nine species, SRA data were downloaded from the NCBI SRA database. Additionally, two already assembled transcriptomes of Mynomes ochrogaster Wagner, 1842 and Cricetulus griseus Milne-Edwards, 1867 were included in the analysis. Thus, in total, we analyzed transcriptomes of 19 Cricetidae species (see Supplementary Table 1 for details).
We categorized species as either subterranean or above ground based on their behavior and morphological traits by classical description of subterranean rodents (Lacey, 2000; Lessa et al., 2008) as previously (Bondareva et al., 2021b). A species was considered subterranean if it is known to spend the most time underground, perform regular digging activities, and come to the surface only incidentally and within the vicinity of burrow openings (Lessa et al., 2008; Smorkatcheva and Lukhtanov, 2014). Two species fit these definitions in our dataset: Ellobius lutescens Thomas, 1897 and Prometheomys schaposchnikowi. Following the data available in literature, Terrícola subterraneus and Lasiopodomys mandarinus Milne-Edwards, 1871 also could be assigned to subterranean or semi-subterranean species (Ognev, 1950; Kryštufek and Shenbrot, 2022). Taking this into account we categorized all four species as subterranean. They are marked by color on Figure 1. We were limited only by these species being unable to consider other subterranean Arvicolinae forms (ex gr. Microtus pinetorum Le Conte, 1830) in the analyses due to the lack of data in the SRA database at the period of analysis and the poorly described ecology habits for several potential subterranean species.
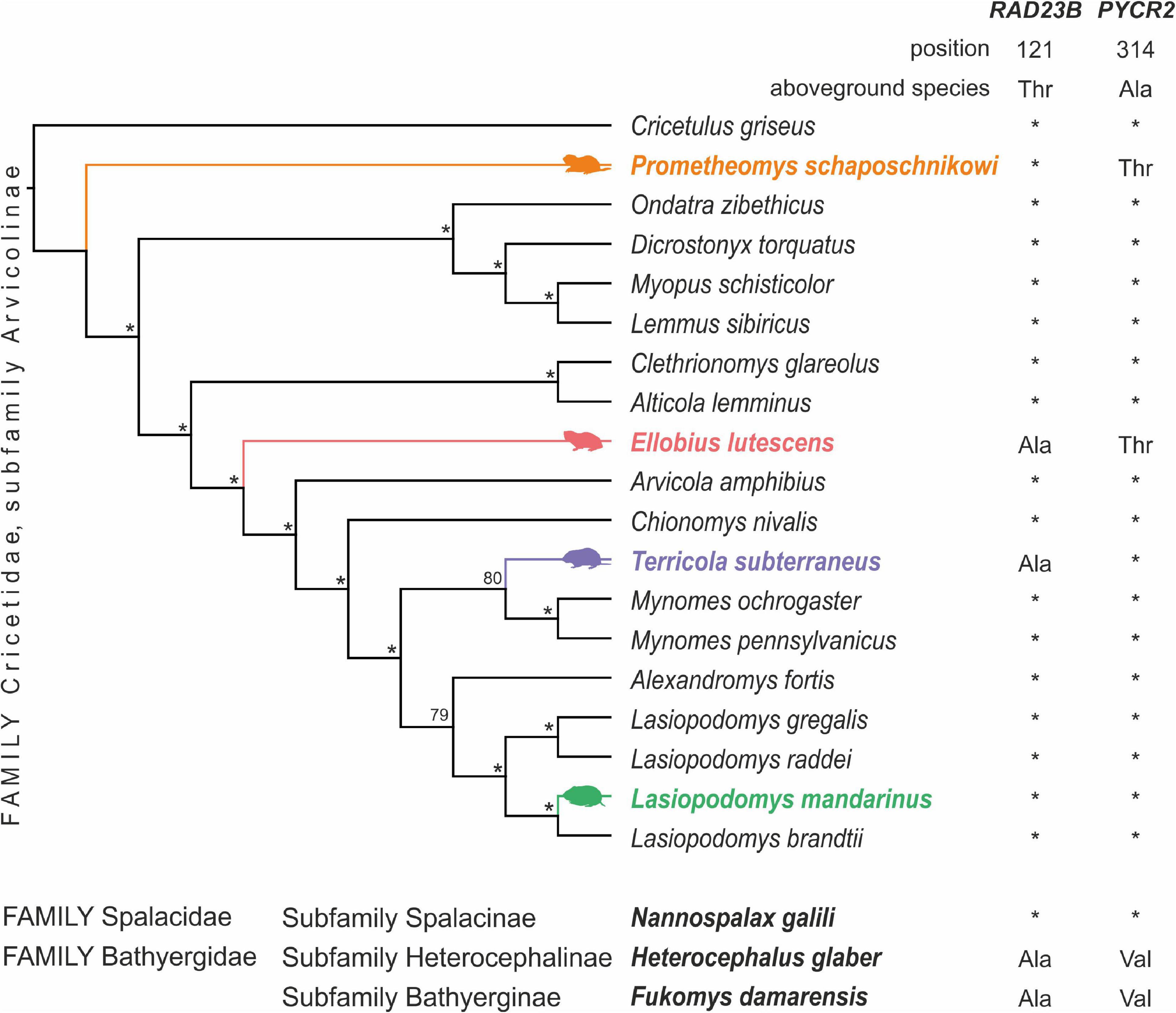
Figure 1. Phylogenetic tree of Arvicolinae subfamily based on transcriptome data with observed substitutions unique for subterranean species. Subterranean Arvicolinae species are shown in color. Nodes with 98–100 ML support are marked with an asterisk. On the right, the amino acid substitutions in certain positions are shown. Below the tree, the amino acid substitutions in the same sites in specialized subterranean rodents from other families are shown.
Total RNA was isolated with an RNeasy mini kit (Qiagen) under animal cells/spin protocol. RNA quality was quantified using a Bioanalyzer 2100 (Agilent Genomics) with a minimum of 7.0 for the RNA Integrity Number score. The combined protocol of the NEBNext Poly(A) mRNA Magnetic Isolation Module and the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina1 was used to isolate polyA-RNA from the total RNA fraction and prepare DNA libraries. Preparation was carried out according to the protocol with the following modifications: (1) sample fragmentation was conducted at 94°C for 10 min; (2) the synthesis of the first chain was hold using the program with the following parameters: 25°C - 10 min, 42°C - 30 min, 70°C - 15 min; (3) DNA purification and concentration was carried out according to the protocol using Ampure XP magnetic particles. Elution was transferred with bidistilled water; (4) the number of cycles in PCR consisted of 10 or 15, depending on the initial RNA concentration.
Sample concentrations were measured on a Qubit fluorometer. The quality of the obtained libraries was checked using the Bioanalyzer 2100 Agilent using the DNA High Sensitivity kit. RNA extraction and library preparation were performed using resources of the Skoltech Genomics Core Facility2.
Sequencing was performed on the Illumina HiSeq4000 with read length 75 bp and paired-end tracks. Demultiplexing and conversion of data to the fastq format was performed using the bcl2fastq2 Illumina software. Sequencing was performed using resources of the Skoltech Genomics Core Facility (see text footnote 2).
The quality of the obtained paired readings was assessed by the FastQC (Andrews, 2010). Trimmomatic 0.39 (Bolger et al., 2014) was used to remove low quality reads and Illumina adapters. We took only reads with a quality higher than 28–29.
Transcriptomes were assembled with a standard Trinity package (Haas et al., 2013). Base transcriptome statistics were also estimated with Trinity Stats. The ORFs in assembled transcriptomes were predicted by Transdecoder3. The resulting sequences were purified from chimeric ones using the DIAMOND program (Buchfink et al., 2015) with the reference from the NCBI protein base (nr). Transcriptome completeness was evaluated by BUSCO (Manni et al., 2021). Orthologous genes were determined using Proteinortho (Lechner et al., 2011). We searched for single-copy orthologs that were present in all analyzed transcriptomes for future analyzes.
Separate orthological genes were aligned using prank (Löytynoja, 2014) with the correction for the protein-coding genes tripletity. Aligned orthologs were concatenated into one meta-alignment with Python-based script.
Phylogenetic reconstruction for 19 species was based on found orthological genes and carried out in the IQ-tree4. Conserved blocks from multiple alignments were previously removed by Gblocks (Castresana, 2000).
Phylogenetic trees used for tracing signatures of the selection pressure on each subterranean species separately (Figure 2) were obtained with MrBayes 3.2.2 (Ronquist et al., 2012) using seven nuclear genes (6,421 bp), see methods in Bondareva et al. (2021a). The following analysis parameters were: nst = mixed and the distribution of the substitution rates between sites, the dataset was divided into partitions by genes. Each analysis was started with a random tree and had two replicates with four Markov chains (MCMC) and 1 million generations, with the results recorded every hundredth generation. Stationarity and convergence of separate runs were assessed using ESS statistics in Tracer v1.6 (Rambaut et al., 2014). The trees were visualized using the FigTree v1.6 program (accessed on 26 November 2021)5.
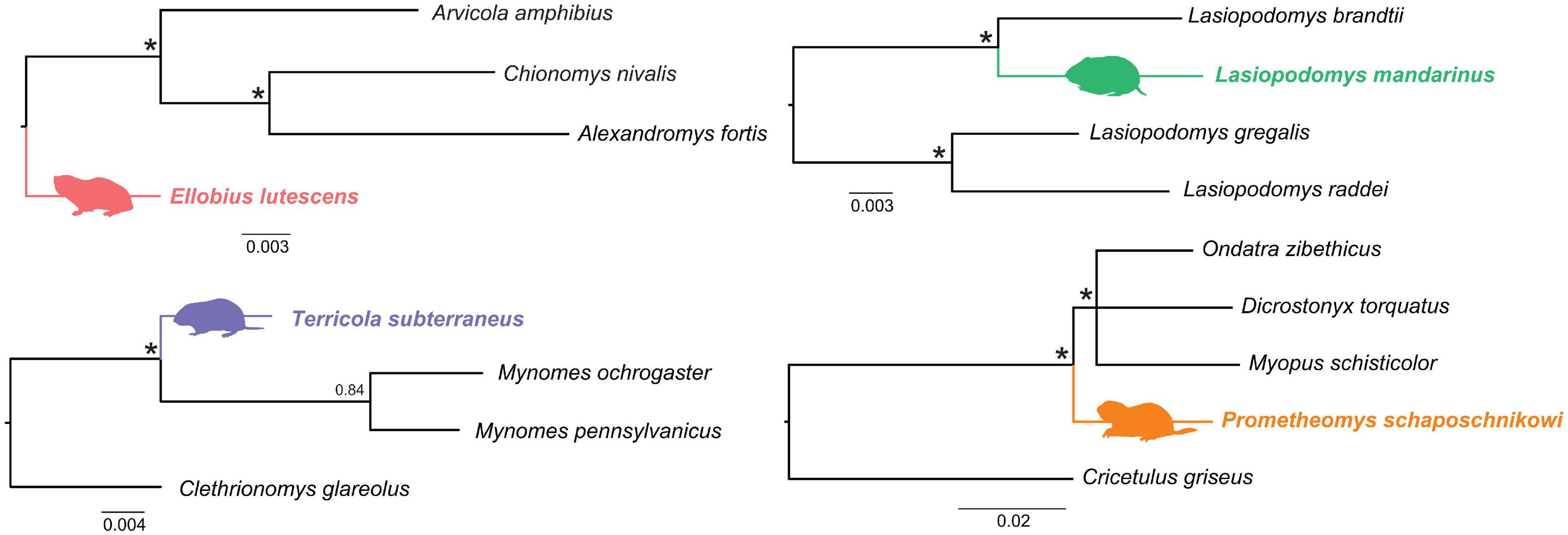
Figure 2. Trees used for tracing signatures of the selection pressures on the subterranean vole species. Fifty-percent majority rule consensus trees from Bayesian inference analysis are given. Subterranean species are shown in color. Nodes with 0.99–1.0 BI support are indicated with asterisks.
The evaluation of both convergent and parallel amino acid substitutions was carried out by the ProtParCon program (Yuan et al., 2019) with a further search for amino acids characteristic only for subterranean rodents: we looked for replacements occurring at least in two subterranean taxa using a script on Python 3. The significance of the substitution frequency was estimated with the Evolver program included in the PAML4 (Yang, 2007) using Monte Carlo simulation algorithm to obtain event probability of the substitution in each position. Simulated sequences evolving according to the following parameters: an evolutionary model, the branching pattern and branch lengths of the tree, amino acid frequencies and sequence length. Additionally Holm multiple adjustment used to obtain p-value in R software v.3.4.4 (R Core Team, 2020). Detected significant amino acid substitutions were subsequently checked for influencing protein structure by the Sift program6 and modeled in SWISS-MODEL (Waterhouse et al., 2018). Visualization and comparison of modeled structures was carried out in PyMol 2.2.0 (Schrödinger and DeLano, 2020).
To test for possible convergent occurrence of the detected substitutions in other subterranean rodents, we used protein sequences of homologous genes from NCBI Protein database (Heterocephalus glaber—XP_004848222.1, XP_021097910.1; Nannospalax galili—XP_008820604.1, XP_008852875.1; Fukomys damarensis—XP_010611512.1, XP_010637758.1).
Selective pressure was estimated as variation in the levels of non-synonymous (dN) and synonymous (dS) substitutions, as well as ω (dN/dS ratio) using several approaches. We applied the classic codeml program (Yang et al., 2000; Yang, 2007) with the comparisons between free-branch and M0, and free-branch and neutral-branch models with the ete-toolkit interface (Huerta-Cepas et al., 2016).
A branch models assumes significant differences between ω values for marked branches (foreground or ωfrg) and the rest branches of the tree (background branches or ωbkg). For branch analysis the total set of species was subdivided into several subsets so that each subset contained a subterranean species and its phylogenetically closest or sister aboveground species (Figure 2). For each subterranean species analysis was implemented with free-branch model (b_free, where ωfrg, and ωbkg are free), neutral-branch model (b_neut, where ωfrg is fixed to one) and M0 model, where all branches evolve at the same rate. The comparison between free-branch and M0 shows whether foreground branches have ω significantly different from the rest of the tree. The comparison between free-branch and neutral-branch models detects if the ωfrg value is significantly higher than 1. The values 999 and 0.001 were regarded as errors. The calculated p-values of all likelihood-ratio tests (LRTs) were corrected using Holm multiple adjustment in R software v.3.4.4.
In addition, we calculate a selection level for each species by free-ratio method. Free-ratio method does not imply the selection of branches and calculates across the whole three giving a unique value for each species. The Wilcoxon test was used to compare the differences between the mean values of dN/dS ratio for subterranean and terrestrial species groups. Test was conducted using R v.3.4.4.
Newly obtained raw sequence RNA-data for nine Arvicolinae species listed above (see “section 2 Materials and Methods,” Supplementary Table 1) are available at the NCBI Sequence Read Archive: SRR21996854—SRR21996864, SRR21996867—SRR21996869 (BioProject No. PRJNA590630).
After the de novo assembly, from 34,750 to 371,688 “Trinity genes” per species were generated (Supplementary Table 2). The N50 length was 1,644–3,571 bp, whereas the mean contig length was about 525 bp. Complete BUSCO values (C in Supplementary Table 2) varied from 48 to 80.1% except already assembled Mynomes ochrogaster and Cricetulus griseus with mean values 98 and 93%, respectively. Since we did not make a functional comparison of transcriptomes, even such a low percentage (48%) of complete BUSCO for several species was sufficient for further analysis.
The search for orthologs by Proteinortho resulted in more than 1,000 orthologous genes for the studied species. But only 112 single-copy orthologs were present in all analyzed transcriptomes. They were used in further analyzes. Such a small number of intersections is explained by our assembly’s incompleteness and limited amount of initial raw data. Since the already assembled and published transcriptomes of Mynomes and Cricetulus were also included in the analysis, we were confident that detected universal single-copy genes were not chimeric or erroneously generated by Trinity annotation.
The total length of the concatenated single-orthologs alignment was 214,696 nucleotides. The 98,595 nucleotides remained after cleaning with the Gblocks (45% of the original alignment). The topology of trees for nuclear (Figure 1) and previously published mitochondrial gene trees (Abramson et al., 2021) practically does not contradict each other. The only difference concerns the position of the taxa in the first basal radiation. According to the data on nuclear genes the earliest split is represented by the Prometheomys schaposchnikowi, whereas the tribe Lemmini is the earliest derivative according to mitochondrial genes (Abramson et al., 2021).
The calculation of free dN/dS ratio model values across the whole tree for each taxa was carried out for each of 112 single-copy orthologs independently. Free dN/dS ratio model values across all genes showed that median dN/dS meanings turned out to be significantly (p-value = 0.002) higher for subterranean rodents as compared to above ground forms: 0.078 and 0.037, respectively.
To test for branch-specific positive selection in each of the four subterranean rodents, we ran a branch model for each of these species independently in comparison only with phylogenetically close terrestrial taxa (Figure 2). For each of 112 single-copy orthologs we failed to find any genes with significant changes in selection level. Despite the observed trend of higher dN/dS values for subterranean species and possible selection relaxation for these genes (Supplementary Table 3), all LRT p-values were insignificant after Holm correction. This tendency is especially pronounced for Ellobius lutescens, where for several genes p-values were significant before Holm correction, in contrast to other subterranean Arvicolinae analysis.
We compared respective sets of substitutions generated by ProtParCon on 112 single-copy orthologous protein sequences in search of parallel molecular selective signatures among the four subterranean species. Parallel amino acid substitutions shared in at least two subterranean species were found in several genes: ERP29, RAD23B, HIKESHI, ZADH2, MRPS14, PYCR2, CCDC86, GTPBP2, SNAPC2, and TTLL12 (Table 1). We additionally checked manually that there was no variability in these positions for above ground rodents sequences. Subsequent verification by the Evolver simulation process showed the significance of non-random occurrence of substitutions in the RAD23 and PYCR2 genes. These genes are associated with DNA repair processes (Pohjoismäki et al., 2012) and response to oxidative stress (Kuo et al., 2016), respectively.
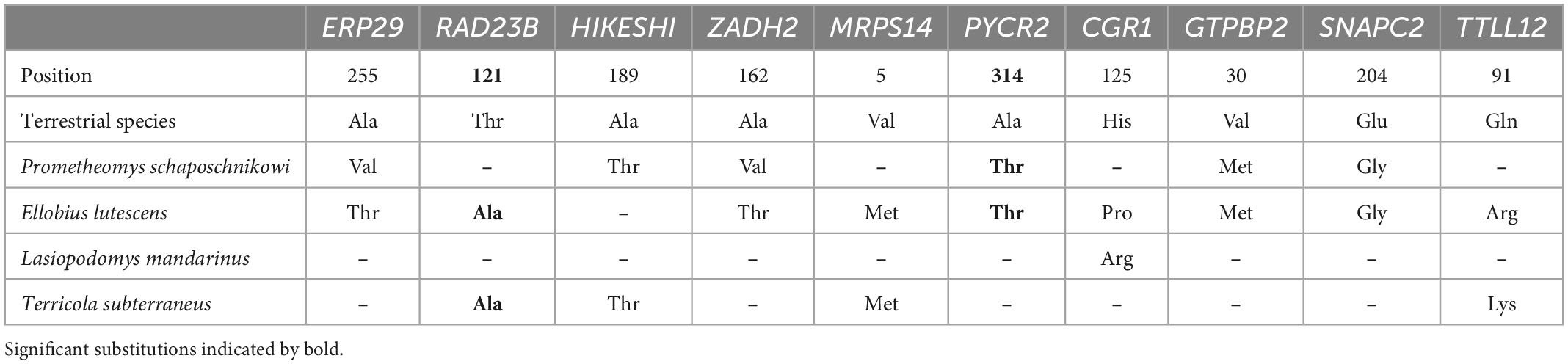
Table 1. Obtained genes with parallel amino acid substitutions shared in at least two subterranean species.
Substitution Ala314Thr in PYCR2 gene was predicted as affecting the protein function with a significant score of 0.00 by Sift program, while Thr143Ala substitution in RAD23 gene was shown as tolerated with a score of 0.81. The RAD23 tertiary structure modeling showed that the replacement site in the protein is located at a non-structural fragment and therefore does not have visible effect on the protein (Figure 3). It was not possible performing such modeling for the PYCR2 gene, because the length of the template turned out to be less than the length of our original sequence. By coincidence, the site of interest was located in the lacking part of the template. Other templates offered by the SWISS-MODEL program were much worse and could not be used.
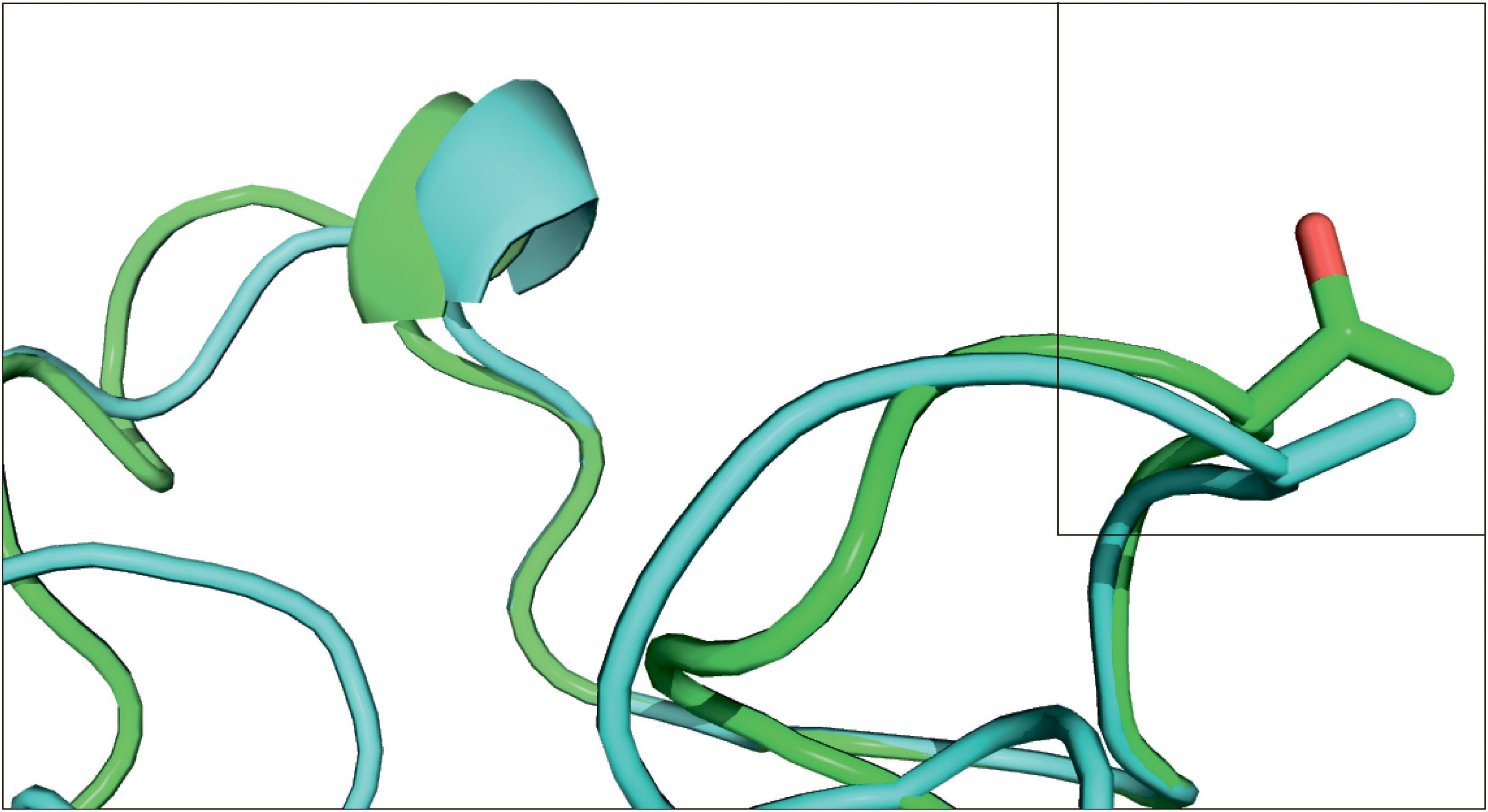
Figure 3. Structural model of substitutions in the RAD23 gene. E. lutescens (subterranean) model is green, L. gregalis Pallas, 1779 (terrestrial) is cyan. Position 143 is framed.
Search of convergent protein substitution across ancient and highly specialized subterranean species revealed substitution in RAD23 gene of Heterocephalus glaber and Fukomys damarensis. For the same species, we found a substitution in the position 314 of the PYCR2 gene, but with an amino acid different from those found in subterranean Arvicolinae: Val instead of Thr, respectively. We were unable to find similar substitutions in another subterranean rodent subfamily Spalacidae—Nannospalax galii. Amino acids in the studied positions coincided for this species with those in terrestrial Arvicolinae rodents.
In the current study, we test for molecular adaptations signs in protein-coding nuclear genes across the subterranean lineages of vole species. Specifically we aimed to check whether the tendencies revealed in studies on mitochondrial genes (Bondareva et al., 2021b) may also be observed across transcriptome data.
Within our transcriptome-based data set we uncover several genes with parallel amino acid substitutions. Verification showed the reliable substitutions in two genes: the RAD23B (Nucleotide Excision Repair Protein) and PYCR2 (pyrroline-5-carboxylate reductase 2). These substitutions were found in both genes in the genome of Ellobius lutescens, in PYCR2 in Prometheomys schaposchnikowi and in the RAD23B in Terricola subterraneus respectively (Table 1).
The found genes were not previously reported in studies of molecular adaptations of subterranean rodents. At the present stage it is impossible directly to link the functional impact of these substitutions to adaptive processes. However, the biochemical pathways and processes in which these genes are involved may play a certain role in the adaptation to subterranean lifestyle. The active work of the mitochondria potentially causes the formation of reactive oxygen forms that represent a damaging factor for the cell (Turrens, 2003). The RAD23B and PYCR2 genes are associated with DNA repair processes (Pohjoismäki et al., 2012) and response to oxidative stress (Kuo et al., 2016), respectively. The RAD23 homolog gene was found in the study of drought adaptation in plants, since clear signs of positive selection were observed (Zhang et al., 2013). The change in expression level of this gene was also revealed in the analysis of cold tolerance of Thinopyrum intermedium (Jaikumar et al., 2020). Moreover, the presence of the same amino acid substitutions in highly specialized subterranean rodent species from the family Bathyergidae may also indicate the importance of these substitutions in the adaptive processes.
The small number of genes with parallel substitutions typical for subterranean forms is not surprising. Even the wide scale study of Kalina Davies revealed (Davies et al., 2018) only 35 genes with parallel substitutions in all analyzed species which was less than 1% of the total number of analyzed genes. Another wide-scale study performed on subterranean rodents (Fang, 2015) discovered parallel substitutions for Heterocephalus glaber and Fukomys damarensis only in two genes: ARG1 and Na(V)1.7 across the whole-genome comparison. However, interesting to note that in the analysis of CYTB gene sequences across more than 60 Arvicolinae species we identify only three parallel substitutions occurring in subterranean forms (Bondareva et al., 2021a).
The previously observed tendency of the selection relaxation in mitochondrial genomes was not fully confirmed for nuclear data. The median dN/dS value is significantly higher across all analyzed genes for subterranean voles compared with above ground using free ratio model. However, branch-model analysis failed to detect genes with significant changes in selection level for individual subterranean lineage compared to its sister one at the tree. Opposite to this in the analogous branch-model analysis of mitochondrial genomes we observed dramatic differences in a number of genes with clear selection signatures in subterranean lineages compared to sister above ground ones (Bondareva et al., 2021b).
This difference between mitochondrial and nuclear genes may be explained both by the slower rate of nuclear genes evolution (Lin and Danforth, 2004) compared to mitochondrial ones and the insufficient number of analyzed genes. It is worth mentioning that Arvicolinae is the youngest group among order Rodentia and even its oldest subterranean lineage, Prometheomys schaposchnikowi, is not older than 7 Ma (Abramson et al., 2021). However even in the very old and highly specialized subterranean groups from different mammal superorders signs of positive selection were revealed only in 10% out of all protein-coding nuclear genes studied (over 8,000) (Davies et al., 2018).
Our screening of nuclear genes showed despite the small number of them in the analysis we nevertheless may see not strong but clear adaptive signs. These signs can be seen in the increase of dN/dS ratio in the free ratio analysis and in the presence of parallel substitutions in subterranean forms. These may from the first look indicate adaptive signals to subterranean life, but we cannot exclude any other explanation for these findings. Analogous results detected in other subterranean rodents were linked to a variation in metabolic rate, body mass, population size, and generation time among lineages (Martin and Palumbi, 1993; Martin, 1995; Bromham et al., 1996). However, we assume that in case of decrease of effective population size, that is characteristic for most subterranean forms, we would observe, in the first turn, the decrease of overall genetic diversity but not the increase of non-synonymous substitutions as we see in our case. Similarly, the other factors listed above may explain the difference in overall genetic diversity between subterranean forms and above ground ones, but could not be responsible for the significant increase in non-synonymous substitutions.
We aimed first to test the hypothesis on convergent molecular signatures of selection in several phylogenetically distant lineages of subterranean Arvicolinae and second, compare if found these signs with published data on other subterranean mammals. According to published data, we can identify two main trends: selection relaxation and occurrence of parallel amino acid substitutions. Combined results of analysis of mitochondrial genomes and transcriptomes showed both these tendencies for subterranean Arvicolinae (Figure 4).

Figure 4. Graphic representation of possible molecular adaptations in response to subterranean life. Genes are colored as follows: Red—under positive selection in at least one subterranean lineage; yellow—with parallel substitution in at least one pair of subterranean taxa; green—both.
Despite the limited number of nuclear genes in the analysis we have found parallel substitutions for subterranean forms in RAD23B and PYCR2 genes in the current work. Studies performed on representatives of the families Bathyergidae and Spalacidae revealed parallel substitutions in completely different genes, e.g., Na(V)1.7 and ARG1 (Fang et al., 2014b), EPAS1 and AJUBA (Shao et al., 2015). Thirty-five loci with parallel substitutions were common across subterranean pairwise comparisons (Golden moles, African mole-rats, blind mole rats, Star-nosed mole) by Davies et al. (2018). They include FREM1, LAMA3, MFI2, and PIEZO2 (Safran et al., 2010). Additionally, at least three of the loci (SLC26A8, TDRD6, and TEX15) have roles in sperm development, and at least eight (e.g., C3, CLCA1, and TLR4) are involved with an immune response (Safran et al., 2010). As mentioned above, RAD23B and PYCR2 genes were not listed in previous studies, but their function could be associated with adaptive processes. Our own analysis of the protein sequences of these genes in highly specialized Heterocephalus glaber and Fukomys damarensis from Bathyergidae family shows convergent substitution in the same position with Arvicolinae subterranean rodents.
Several previous studies have investigated convergent molecular evolution based on dN/dS ratio estimation in subterranean mammals. Da Silva et al. (2009) found a significant difference for its values in CYTB sequences of subterranean rodents from families Ctenomyidae, Geomyidae and Bathyergidae compared to their aboveground closely related species. Later, signatures of selection were found in several mitochondrial genes of subterranean rodents (Tomasco and Lessa, 2011, 2014; Tavares and Seuánez, 2018). These have revealed convergent evolution in genes relating to hypoxia between plateau zokors (Spalacidae) and naked mole-rats (Bathyergidae), and comparisons of subterranean and closely related terrestrial rodents found significant differences in substitution rates of protein-coding genes between the two groups (Shao et al., 2015). Additionally, two genome-wide screens for changes in evolutionary rates identified enrichment in vision- and skin-related loci (Prudent et al., 2016; Partha et al., 2017). Our previous studies on the Arvicolinae subfamily also showed the relaxation of selection strength in CYTB and mitochondrial protein-coding genes reflecting an increased ω ratio in subterranean lineages compared to aboveground ones (Bondareva et al., 2021a,b). In this work we observed the trend to selection relaxation by free-ratio analysis, but didn’t find individual genes with significant changes in selection level. We can associate it with the fact that Arvicolinae is the youngest group among order Rodentia and even its oldest subterranean lineage, Prometheomys schaposchnikowi, is not older than 7 Ma (Abramson et al., 2021). It is quite likely that this time span is insufficient for the pronounced selection change in slowly evolving nuclear genes. In addition, our sample dataset was limited, which could also affect the result of sensitive branch-based analysis.
The greatest number of selection signs within Arvicolinae subterranean species was found in the genus Ellobius both in mitochondrial genes (Bondareva et al., 2021a,b) and in the nuclear protein-coding genes. This genus is younger than the oldest representative of the subfamily, subterranean P. schaposchnikowi, by at least ca. 2 Ma. The oldest fossil remains of the true Ellobius are known from the Late Pliocene (Lytchev and Savinov, 1974; Zazhigin, 1988; Tesakov, 2004) but the morphological features in all species of the genus are very similar with those observed in the “model” subterranean rodents of the families Bathyergidae and Spalacidae: protruding incisors, very small eyes, and isolation of the mouth region by lips (Gromov and Polyakov, 1977).
So, on RNA-seq data we additionally confirmed our previous conclusion that the number of selection signatures is independent of the evolutionary age of the lineage but fits the degree of specialization to the subterranean niche and adaptive mechanisms to the subterranean lifestyle are convergent subterranean rodents from different families.
We indicate the signs of positive selection in nuclear genes for voles during colonization of the subterranean environment. Also we found two genes with parallel substitutions: RAD23B and PYCR2. Similar substitutions were found in representatives of the highly specialized family Bathyergidae. Our combined results of mitochondrial and nuclear genes analyses show that the number of adaptive traits does not depend on the evolutionary age of the lineage, but coincides well with the degree of specialization to the subterranean niche. The obtained data suggests that these signatures are convergent across distant subterranean rodents and mammals.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, SRR21996854—SRR21996864 and SRR21996867—SRR21996869.
This animal study was reviewed and approved by Ethics Committee of Zoological Institute Russian Academy of Sciences (permission #1-17, April 07, 2022).
OB: methodology, software, investigation, formal analysis, data curation, writing—original draft, and writing—review and editing. TP: data curation, formal analysis, and visualization. SB: data curation and writing—review and editing. MG: data curation. AS: data curation and writing—original draft. NA: conceptualization, data curation, methodology, resources, writing—review and editing, project administration, and funding acquisition. All authors read and approved the final manuscript.
This work was supported by the Russian Science Foundation (RSF) N 19-74-20110 (for OB, SB, TP, and NA) and Russian Foundation for Basic Research (RFBR) N 19-04-00538a (for AS).
We are grateful to colleagues Malikov V. G. (ZIN RAS, Saint Petersburg), Okhlopkov I. M. and Mamayev N. V. (Institute for Biological Problems of Cryolithozone, Yakutsk), Genelt-Yanovskiy E. A. (ZIN RAS, Saint Petersburg) who shared the material for the study, and Rasskazov D. A. (The Kurchatov Genome Center of the Institute of Cytology and Genetics SB RAS, Novosibirsk) for help with server management. The material for D. torquatus was obtained during the expedition “Open Ocean: Arctic Archipelagos–2019, Severnaya Zemlya”.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1085993/full#supplementary-material
Abramson, N. I., Bodrov, S. Y., Bondareva, O. V., Genelt-Yanovskiy, E. A., and Petrova, T. V. (2021). A mitochondrial genome phylogeny of voles and lemmings (Rodentia: Arvicolinae): Evolutionary and taxonomic implications. PLoS One 16:e0248198. doi: 10.1371/journal.pone.0248198
Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. 2010. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [accessed April 10, 2020].
Begall, S., Burda, H., and Schleich, C. E. (2007). Subterranean rodents: News from underground. Berlin: Springer.
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bondareva, O. V., Potapova, N. A., Konovalov, K. A., Petrova, T. V., and Abramson, N. I. (2021a). Searching for signatures of positive selection in cytochrome b gene associated with subterranean lifestyle in fast-evolving arvicolines (Arvicolinae, Cricetidae, Rodentia). BMC Ecol. Evol. 21:92. doi: 10.1186/s12862-021-01819-4
Bondareva, O., Genelt-Yanovskiy, E., Petrova, T., Bodrov, S., Smorkatcheva, A., and Abramson, N. (2021b). Signatures of adaptation in mitochondrial genomes of Palearctic subterranean voles (Arvicolinae, Rodentia). Genes 12:1945. doi: 10.3390/genes12121945
Bromham, L., Rambaut, A., and Harvey, P. H. (1996). Determinants of rate variation in mammalian DNA sequence evolution. J. Mol. Evol. 43, 610–621. doi: 10.1007/BF02202109
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Buffenstein, R. (2000). “Ecophysiological responses of subterranean rodents to underground habitats,” in Life underground: The biology of subterranean rodents, eds E. A. Lacey, J. L. Patton, and G. N. Cameron (Chicago, IL: University of Chicago Press), 63–110.
Busch, C., Antinuchi, C. A., Del Valle, J. C., Kittlein, M. J., Malizia, A. I., Vassallo, A. I., et al. (2000). “Population ecology of subterranean rodents,” in Life underground: The biology of subterranean rodents, eds E. A. Lacey, J. L. Patton, and G. N. Cameron (Chicago, IL: University of Chicago Press), 183–226.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Corn, P. S., and Bury, R. B. (1988). Distribution of the voles arborimus longicaudus and phenacomys intermedius in the central oregon cascades. J. Mammal. 69, 427–429.
Da Fonseca, R. R., Johnson, W. E., O’Brien, S. J., Ramos, M. J., and Antunes, A. (2008). The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics 9:119. doi: 10.1186/1471-2164-9-119
Da Silva, C. C., Tomasco, I. H., Hoffmann, F. G., and Lessa, E. P. (2009). Genes and ecology: Accelerated rates of replacement substitutions in the cytochrome b gene of subterranean rodents. Open Evol. J. 3, 17–30. doi: 10.2174/1874404400903010017
Davies, K. T. J., Bennett, N. C., Faulkes, C. G., and Rossiter, S. J. (2018). Limited evidence for parallel molecular adaptations associated with the subterranean niche in mammals: A comparative study of three superorders. Mol. Biol. Evol. 35, 2544–2559. doi: 10.1093/molbev/msy161
Di Rocco, F., Parisi, G., Zambelli, A., and Vida-Rioja, L. (2006). Rapid evolution of cytochrome c oxidase subunit II in camelids (Tylopoda, Camelidae). J. Bioenerg. Biomembr. 38, 293–297. doi: 10.1007/s10863-006-9048-8
Fang, X. (2015). Convergent and divergent adaptations of subterranean rodents genome wide studies on Heterocephalus, Fukomys and Spalax. Ph.D. thesis. Copenhagen: University of Copenhagen.
Fang, X., Nevo, E., Han, L., Levanon, E. Y., Zhao, J., Avivi, A., et al. (2014a). Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat. Commun. 5:3966. doi: 10.1038/ncomms4966
Fang, X., Seim, I., Huang, Z., Gerashchenko, M. V., Xiong, Z., Turanov, A. A., et al. (2014b). Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes. Cell Rep. 8, 1354–1364. doi: 10.1016/j.celrep.2014.07.030
Fejfar, O., Heinrich, W.-D., Kordos, L., and Maul, L. C. (2011). Microtoid cricetids and the early history of arvicolids (Mammalia, Rodentia). Palaeontol. Electron. 14, 1–38.
Gromov, I. M., and Polyakov, I. Y. (1977). Fauna of the USSR. Mammals. Volume 3. Vyp. 8. Voles (Microtinae). Leningrad: Nauka.
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-Seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hassanin, A., Ropiquet, A., Couloux, A., and Cruaud, C. (2009). Evolution of the mitochondrial genome in mammals living at high altitude: New insights from a study of the tribe Caprini (Bovidae, Antilopinae). J. Mol. Evol. 68, 293–310. doi: 10.1007/s00239-009-9208-7
Huerta-Cepas, J., Serra, F., and Bork, P. (2016). ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638. doi: 10.1093/molbev/msw046
Jaikumar, N. S., Dorn, K. M., Baas, D., Wilke, B., Kapp, C., and Snapp, S. S. (2020). Nucleic acid damage and DNA repair are affected by freezing stress in annual wheat (Triticum aestivum) and by plant age and freezing in its perennial relative (Thinopyrum intermedium). Am. J. Bot. 107, 1693–1709. doi: 10.1002/ajb2.1584
Kryštufek, B., and Shenbrot, G. I. (2022). Voles and lemmings (Arvicolinae) of the palaearctic region. Maribor: University of Maribor Press.
Kuo, M. L., Lee, M. E., Tang, M., Besten, W., Hu, S., Sweredoski, M. J., et al. (2016). PYCR1 and PYCR2 interact and collaborate with RRM2B to protect cells from overt oxidative stress. Sci. Rep. 6:18846. doi: 10.1038/srep18846
Lacey, E. A. (2000). Life underground: The biology of subterranean rodents. Chicago, IL: University of Chicago Press.
Lechner, M., Findeiß, S., Steiner, L., Marz, M., Stadler, P. F., and Prohaska, S. J. (2011). Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics 12:124. doi: 10.1186/1471-2105-12-124
Lessa, E. P., Vassallo, A. I., Verzi, D. H., and Mora, M. S. (2008). Evolution of morphological adaptations for digging in living and extinct ctenomyid and octodontid rodents. Biol. J. Linn. Soc. 95, 267–283. doi: 10.1111/j.1095-8312.2008.01057.x
Lin, C., and Danforth, B. (2004). How do insect nuclear and mitochondrial gene substitution pattern differ? Insight from Bayesian analyses of combined datasets. 2004. Mol. Phylogenet. Evol. 30, 686–702. doi: 10.1016/S1055-7903(03)00241-0
Löytynoja, A. (2014). Phylogeny-aware alignment with PRANK. Methods Mol. Biol. 1079, 155–170. doi: 10.1007/978-1-62703-646-7_10
Luo, Y., Yang, X., and Gao, Y. (2013). Mitochondrial DNA response to high altitude: A new perspective on high-altitude adaptation. Mitochondrial DNA 24, 313–319. doi: 10.3109/19401736.2012.760558
Lytchev, G. F., and Savinov, P. F. (1974). Late Pliocene lagomorphs and rodents from Kiikbai. Mater. Istor. Faune Kazakhstana 6, 39–57.
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., and Zdobnov, E. M. (2021). BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654. doi: 10.1093/molbev/msab199
Martin, A. P. (1995). Metabolic rate and directional nucleotide substitution in animal mitochondrial DNA. Mol. Biol. Evol. 12, 1124–1131. doi: 10.1093/oxfordjournals.molbev.a040286
Martin, A. P., and Palumbi, S. R. (1993). Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl. Acad. Sci. U.S.A. 90, 4087–4091. doi: 10.1073/pnas.90.9.4087
Martin, R. A. (2003). Biochronology of latest miocene through pleistocene arvicolid rodents from the central great plains of North America. Coloq. Paleontol. 1, 373–383.
McNab, B. K. (1966). The metabolism of fossorial rodents: A study of convergence. Ecology 47, 712–733.
Nevo, E. (1979). Adaptive convergence and divergence of subterranean mammals. Ann. Rev. Ecol. Syst. 10, 269–308.
Nevo, E. (1999). Mosaic evolution of subterranean mammals: Regression, progression and global convergence. Oxford: Oxford University Press.
Ognev, S. I. (1950). Zveri SSSR I prilezhashhih stran (The mammals of the USSR and adjacent countries). Vol VII. Gryzuny (Rodentia). Cambridge: Academic Publishing.
Oosthuizen, M. K., Cooper, H. M., and Bennett, N. C. (2003). Circadian rhythms of locomotor activity in solitary and social species of African mole-rats (family: Bathyergidae). J. Biol. Rhythms 18, 481–490. doi: 10.1177/0748730403259109
Partha, R., Chauhan, B. K., Ferreira, Z., Robinson, J. D., Lathrop, K., Nischal, K. K., et al. (2017). Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. eLife 6:e25884. doi: 10.7554/eLife.25884
Pohjoismäki, J. L., Boettger, T., Liu, Z., Goffart, S., Szibor, M., and Braun, T. (2012). Oxidative stress during mitochondrial biogenesis compromises mtDNA integrity in growing hearts and induces a global DNA repair response. Nucleic Acids Res. 40, 6595–6607. doi: 10.1093/nar/gks301
Prudent, X., Parra, G., Schwede, P., Roscito, J. G., and Hiller, M. (2016). Controlling for phylogenetic relatedness and evolutionary rates improves the discovery of associations between species’ phenotypic and genomic differences. Mol. Biol. Evol. 33, 2135–2150. doi: 10.1093/molbev/msw098
R Core Team (2020). European environment agency. Available online: https://www.eea.europa.eu/data-and-maps/indicators/oxygen-consuming-substances-in-rivers/r-development-core-team-2006 [accessed February 11, 2021].
Rado, R., Gev, H., Goldman, B. D., and Terkel, J. (1991). “Light and circadian activity in the blind mole rat,” in Photobiology, ed. E. Riklis (Boston, MA: Springer). doi: 10.1007/978-1-4615-3732-8_61
Rambaut, A., Drummond, A. J., and Suchard, M. (2014). Tracer v1. 6. Available online at: http://beast.bio.ed.ac.uk/Tracer (accessed November 26, 2021).
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Safran, M., Dalah, I., Alexander, J., Rosen, N., Iny Stein, T., Shmoish, M., et al. (2010). GeneCards version 3: The human gene integrator. Database 2010:baq020. doi: 10.1093/database/baq020
Schrödinger, L., and DeLano, W. (2020). PyMOL. Available online at: http://www.pymol.org/pymol (accessed November 26, 2021).
Shao, Y., Li, J.-X., Ge, R.-L., Zhong, L., Irwin, D. M., Murphy, R. W., et al. (2015). Genetic adaptations of the plateau Zokor in high-elevation burrows. Sci. Rep. 5:17262. doi: 10.1038/srep17262
Smorkatcheva, A. V., and Lukhtanov, V. A. (2014). Evolutionary association between subterranean lifestyle and female sociality in rodents. Mamm. Biol. 79, 101–109. doi: 10.1016/j.mambio.2013.08.011
Swingle, J. K., and Foreman, E. D. (2009). Home range areas and activity patterns of red tree voles (Arborimus longicaudus) in Western Oregon. Northwest Sci. 83, 273–286. doi: 10.3955/046.083.0310
Tavares, W. C., and Seuánez, H. N. (2018). Changes in selection intensity on the mitogenome of subterranean and fossorial rodents respective to aboveground species. Mamm. Genome 29, 353–363. doi: 10.1007/s00335-018-9748-5
Tesakov, A. S. (2004). Biostratigraphy of middle Pliocene – Eopleistocene of Eastern Europe (based on small mammals). Moscow: Nauka.
Tomasco, I. H., and Lessa, E. P. (2011). The evolution of mitochondrial genomes in subterranean caviomorph rodents: Adaptation against a background of purifying selection. Mol. Phylogenet. Evol. 61, 64–70. doi: 10.1016/j.ympev.2011.06.014
Tomasco, I. H., and Lessa, E. P. (2014). Two mitochondrial genes under episodic positive selection in subterranean octodontoid rodents. Gene 534, 371–378. doi: 10.1016/j.gene.2013.09.097
Turrens, J. F. (2003). Mitochondrial formation of reactive oxygen species. J. Physiol. 552, 335–344. doi: 10.1113/jphysiol.2003.049478
Wang, Y., Shen, Y., Feng, C., Zhao, K., Song, Z., Zhang, Y., et al. (2016). Mitogenomic perspectives on the origin of tibetan loaches and their adaptation to high altitude. Sci. Rep. 6:29690. doi: 10.1038/srep29690
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z., Nielsen, R., Goldman, N., and Pedersen, A.-M. K. (2000). Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449. doi: 10.1093/genetics/155.1.431
Yu, L., Wang, X., Ting, N., and Zhang, Y. (2011). Mitogenomic analysis of Chinese snub-nosed monkeys: Evidence of positive selection in NADH dehydrogenase genes in high-altitude adaptation. Mitochondrion 11, 497–503. doi: 10.1016/j.mito.2011.01.004
Yuan, F., Nguyen, H., and Graur, D. (2019). ProtParCon: A framework for processing molecular data and identifying parallel and convergent amino acid replacements. Genes 10:181. doi: 10.3390/genes10030181
Zazhigin, V. S. (1988). “Paleontological descriptions: Insectivora, Lagomorpha, Rodentia,” in Biostratigraphy of late Pliocene - early pleistocene of Tadzhikistan (Mammalian Fauna), eds E. A. Vangengeim and K. V. Nikiforova (Moscow: Nauka), 18–24.
Keywords: selective pressure, protein-coding genes, subterranean voles, subterranean lifestyle, Arvicolinae, RNA-seq
Citation: Bondareva O, Petrova T, Bodrov S, Gavrilo M, Smorkatcheva A and Abramson N (2023) How voles adapt to subterranean lifestyle: Insights from RNA-seq. Front. Ecol. Evol. 11:1085993. doi: 10.3389/fevo.2023.1085993
Received: 31 October 2022; Accepted: 30 January 2023;
Published: 17 February 2023.
Edited by:
Enrico Lunghi, University of L’Aquila, ItalyCopyright © 2023 Bondareva, Petrova, Bodrov, Gavrilo, Smorkatcheva and Abramson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olga Bondareva,  b2xnYS52LmJvbmRhcmV2YUBnbWFpbC5jb20=
b2xnYS52LmJvbmRhcmV2YUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.