- 1The Swire Institute of Marine Science and Area of Ecology and Biodiversity, School of Biological Sciences, The University of Hong Kong, Pokfulam, Hong Kong SAR, China
- 2The Joint Laboratory for Marine Ecology and Environmental Sciences (JLMEES), The Swire Institute of Marine Science, The University of Hong Kong, Pokfulam, Hong Kong SAR, China
- 3Department of Ecoscience, Aarhus University, Roskilde, Denmark
- 4Simon F.S. Li Marine Science Laboratory, School of Life Sciences, The Chinese University of Hong Kong, Shatin, Hong Kong SAR, China
- 5Department of Geography, University of Victoria, Victoria, BC, Canada
- 6Institute for Climate and Carbon Neutrality, The University of Hong Kong, Pokfulam, Hong Kong SAR, China
- 7LabEx ICONA International CO2 Natural Analogues Network, Shimoda, Japan
Introduction: Reef building oysters provide vast ecological benefits and ecosystem services. A large part of their role in driving ecological processes is mediated by the microbial communities that are associated with the oysters; together forming the oyster holobiont. While changing environmental conditions are known to alter the physiological performance of oysters, it is unclear how multiple stressors may alter the ability of the oyster holobiont to maintain its functional role.
Methods: Here, we exposed oysters to acidification and hypoxia to examine their physiological responses (molecular defense and immune response), changes in community structure of their associated microbial community, and changes in water nutrient concentrations to evaluate how acidification and hypoxia will alter the oyster holobiont’s ecological role.
Results: We found clear physiological stress in oysters exposed to acidification, hypoxia, and their combination but low mortality. However, there were different physiological trade-offs in oysters exposed to acidification or hypoxia, and the combination of stressors incited greater physiological costs (i.e., >600% increase in protein damage and drastic decrease in haemocyte counts). The microbial communities differed depending on the environment, with microbial community structure partly readjusted based on the environmental conditions. Microbes also seemed to have lost some capacity in nutrient cycling under hypoxia and multi-stressor conditions (~50% less nitrification) but not acidification.
Discussion: We show that the microbiota associated to the oyster can be enriched differently under climate change depending on the type of environmental change that the oyster holobiont is exposed to. In addition, it may be the primary impacts to oyster physiology which then drives changes to the associated microbial community. Therefore, we suggest the oyster holobiont may lose some of its nutrient cycling properties under hypoxia and multi-stressor conditions although the oysters can regulate their physiological processes to maintain homeostasis on the short-term.
Introduction
Reef building oysters provide numerous ecological functions in coastal marine systems. Along with building structural complexity on the benthos which provides habitat for many marine species, oysters also drive crucial ecological processes such as biogeochemical cycling and benthic-pelagic coupling (Ray and Fulweiler, 2021). Key to these processes are the microbial communities associated with oysters; together forming the oyster holobiont. The activity of the microbial communities associated with oysters are key for the oysters themselves and for the ecological processes performed by the oyster holobiont as a whole (Green et al., 2012; Dubé et al., 2019; Stevick et al., 2021). For example, the oyster holobiont contributes to reduction of eutrophication through simultaneous bio-deposition from the oyster and denitrification stimulated by microbial conversion of biologically reactive nitrogen to nitrogen gas (Ray and Fulweiler, 2021). However, it remains unclear how changing environments, especially driven by climate change, can impact the ecological performance of the oyster holobiont.
Rapid changes in coastal marine environments, directly or indirectly driven by anthropogenic activity, can cause strong physiological responses in fauna. These responses influence how organisms perform their ecological roles within their ecosystem (Vaquer-Sunyer and Duarte, 2008; Hughes et al., 2020). The decline in the capability of organisms to perform their ecological roles can subsequently manifest into broader consequences, such as the loss of some ecological processes within the ecosystem (Diaz and Rosenberg, 1995; Lemasson et al., 2017; Vizzini et al., 2017; Ullah et al., 2018). In the case of sessile habitat forming species, such as oysters, stressful environmental change like acidification or hypoxia can cause physiological impairment that includes oxidative stress, reduced metabolic activity, or haemocyte apoptosis (Boyd and Burnett, 1999; David et al., 2005; Timmins-Schiffman et al., 2014; Wang et al., 2016). These physiological impairments can lead to broader consequences including changes in energy homeostasis, reduced growth, increased susceptibility to diseases, or mortality (Wang et al., 2016; Lutier et al., 2022), and therefore impact the ecological functions of oysters (Lemasson et al., 2017). On the other hand, the extent of alteration to oyster associated microbes in relation to changing environments remains debatable with some studies indicating limited changes (Zhou et al., 2016; Ge et al., 2021; Garner et al., 2022), while others showing significant changes, especially with longer exposure to environmental change (King et al., 2019; Scanes et al., 2021a,b; Unzueta-Martínez et al., 2021).
Among the multiple environmental stressors that can affect oysters and their associated microbial communities, acidification and hypoxia are likely to have profound impact on the oyster holobiont because the already large pH variability within coastal systems (especially around estuaries) could become more extreme with climate change (Rivest et al., 2017). In addition, hypoxic events have become more frequent because of increased coastal eutrophication (Vaquer-Sunyer and Duarte, 2008). Finally, given the simultaneous increase in eutrophication and warming of coastal systems, the co-occurrence of acidification and hypoxia will become more prevalent as overall increases in system metabolic demands will consume more oxygen and release more carbon dioxide (Cai, 2011; Gobler and Baumann, 2016). Therefore, examining how acidification and hypoxia will impact the oyster holobiont and its function can provide important information for oyster reef restoration or oyster aquaculture activities, both of which attract large efforts and financial investments globally (Bayraktarov et al., 2016).
One of the key ecological roles filled by the oyster holobiont is as an intermediary that links biogeochemical cycles in the sediment and water column through filtration and bio-deposition (Smyth et al., 2013; Ray and Fulweiler, 2021). Organic matter (generally from phytoplankton) from the water column is filtered and digested, and substantial denitrification (conversion of NO3− to N2 gas) can by the microbiome within the oysters’ digestive system (the microbial community that naturally occurs within the oyster) (Ray et al., 2019). Unassimilated matter released as bio-deposits (faeces and pseudo-faeces) that contain a large amount of ammonium (NH4+) is then cycled, through nitrification, to NO2− and NO3− by microbes on the oysters’ shell or in the sediment/water column (Ray et al., 2019). These biologically reactive nitrogen species (NO2− and NO3−) are further denitrified by microbes in the sediment or water column, or utilised by epiphytes (Ray et al., 2019). Importantly, changes in water conditions can alter the nutrient cycling capacity of oysters (Hoellein et al., 2015) by influencing the oysters’ physiology, and hence the amount of nitrogen that is processed (Jeppesen et al., 2018), and by changing the microbial community associated with oysters (King et al., 2019; Unzueta-Martínez et al., 2021; Scanes et al., 2021a,b).
Here, we tested the impact of acidification, hypoxia, and their combination on the oyster holobiont; the physiological traits of the oyster, associated microbial community, and the resulting changes in nutrient cycling. While is it well documented that acidification or hypoxia can drive deteriorative physiological responses in oysters (Timmins-Schiffman et al., 2014), the impact of stressful environmental conditions on the whole oyster holobiont remains unclear. We hypothesised that acidification and hypoxia would drive physiological trade-offs in oysters, which would shift the microbial communities associated with them and thus alter the biogeochemical cycling capacity of the oyster holobiont. To test this hypothesis, we exposed oyster holobionts to hypoxia, acidification, or their combination. We then quantified the level of protein damage, oxidative stress, and immune response of oysters exposed to each condition. Finally, we identified the microbial community associated with the oysters and measured the concentration of different nutrients within experimental treatments to estimate the relationship between oysters, microbes and nutrient cycle. Together, these results provide a broader understanding of how climate driven changes in coastal environmental conditions may impact the oyster holobiont as a whole and affect their nutrient cycling capacity.
Methods
Oyster collection
Oysters were sampled from a natural community at Shui Hau Wan, Hong Kong, during low tide at salinity 33 psu and temperature 23°C. This community is comprised of Magallana angulata and Magallana sikamea. Visual separation of species before the experiment was attempted, however, the exact separation of species remained uncertain because of the morphological similarity of these two species and their co-occurrence at the site (Lau et al., 2020). DNA barcoding using mantle tissues of about 50% of the total oysters used in the experiment showed that the community was composed of 60% M. sikamea and 40% M. angulata (only oysters that were used for tissue sampling as described below were used for DNA barcoding). Oysters were brought to the laboratory and kept in 100 L holding tanks for 10 days for acclimation and fed daily with Shellfish Diet 1800 (Reed Mariculture Inc., United States). During this period, oysters were cleaned to remove all fouling from their shells and half the seawater in the holding tanks was changed daily to maintain water quality. Salinity in the holding tanks was 35 psu and temperature was maintained at 23°C using 300-watt submersible aquarium water heaters.
Experimental setup
After acclimation, oysters were transferred into 5 L experimental containers. Five replicate containers per treatment condition (n = 5) were used, each containing 12 individual oysters of mixed sizes. We used 12 oysters per replicate to allow for enough individuals for sampling tissue to measure molecular defence and for sampling haemolymph for quantifying immune response (different individuals were used) in case this sampling created stress or mortality in the oysters. Treatment conditions for the experiments were ambient control (C), hypoxia (H), acidification (A), and hypoxia × acidification (HA) (n = 5 containers per treatment). Treatments were achieved by bubbling pre-mixed gases through the seawater in each experimental container. Hypoxia conditions were created by adding N2 to air to achieve 5% O2 (equivalent to 2 mgO2/L) and acidification conditions were created by mixing CO2 to air to achieve 0.1% CO2 (equivalent to 1,000 ppm). To create HA conditions CO2 was added directly to the hypoxia mixture, while control conditions involved bubbling air into the seawater. All gases were mixed using electronic multi-gas flow controllers (Cole Parmer Masterflex Gas Mass Flowmeter) and manual air flow controllers (Cole Parmer Masterflex Variable-Area Flowmeter). To maintain stable treatment conditions, each gas was constantly bubbled in the appropriate experimental containers and a thin sheet of styrofoam was placed on the surface of the water to limit air/water gas exchange. At the beginning of the experiment, containers were filled with seawater pre-adjusted to the treatment conditions, then placed in a water bath at 23°C, controlled by an external water chiller (Hailea, Model HC.2200BH). Oysters were fed ad libitum with Shellfish Diet 1800 and a two-thirds water change was performed daily with seawater pre-adjusted to the treatment conditions. The experiment was maintained for 16 days, and oyster mortality recorded daily during water change.
Water chemistry
Water quality (pH and DO) was monitored three times each day (in the morning, after the water change, and in the afternoon). Water pH and temperature were monitored with a Mettler Tolledo Seven2Go pH meter. Oxygen concentrations were monitored with a Fibox 4 (PreSens) to which an oxygen dip probe was connected, and salinity wat monitored using a refractometer. To determine the exact pCO2 levels, water samples (70 mL) for total alkalinity were taken once daily. Total alkalinity was measured using a Metrohm Titrator. Finally, pCO2 levels were calculated using CO2Sys_v2.1 (Supplementary Table S1).
Oyster molecular defence
To estimate the effects of hypoxia, acidification, or HA on oyster physiology (i.e., protein damage and oxidative stress), the concentration of Hsp-70, total antioxidant capacity (TAC), and malondialdehyde (MDA) were measured from the gill tissue of oysters. At the end of the experiment, one oyster from each container was dissected to remove the gills, which were immediately frozen in liquid nitrogen and stored at −80°C until further use. Prior to measuring Hsp-70, TAC and MDA, gill tissue samples were thawed and immediately cut and weighed separately for each assay. For all assays, gill tissue samples were homogenised using a TissueLyser 2 at 30 hertz for 1 min. Total protein was measured using a BCA total protein kit (Pierce) following manufacturer’s protocols. Hsp 70 was measured using a Mouse Heat Shock Protein 70 m Hsp-70 Elisa Kit (CUSABIO) following manufacturer’s protocols. TAC and MDA were measured using an OxiSelect Total Antioxidant Capacity (TAC) Assay Kit (Cell Biolabs) and an OxiSelect TBARS (MDA Quantification) Assay Kit (Cell Biolabs), respectively, following manufacturer’s protocols.
Oyster immune response
To quantify the haemocyte populations and measure the level of phagocytosis in oysters from different treatments, haemolymph samples were collected from the pericardial cavity of two new individuals from each experimental container (total of 10 individuals per treatment) at the end of the experiment using a one millilitre syringe and fine needle. Haemolymph samples were immediately placed on ice to prevent coagulation. For cell counts and identification, haemolymph samples were prepared following Wang et al. (2016). In brief, 0.5 mL haemolymph from each individual was fixed with 4% paraformaldehyde, then cells counted and identified using microscopy. Preserved cells were counted and separated as granulocytes and agranulocytes using a haemocytometer mounted on a light microscope. Phagocytic activity was measured immediately after haemolymph sampling using undiluted haemolymph samples following Goedken and De Guise (2004). In brief, 0.5 mL fresh haemolymph was mixed with one micrometre fluorescent latex beads at a ratio of approximately 100 beads/haemocyte. Samples were incubated for 1 hour at 20°C, then analysed on a FACS Aria 2 flow cytometer (Becton Dickinson). 10,000 events were set as a limit and the number of haemocytes that had engulfed zero, one, two or three beads were quantified based on their fluorescence peaks. Filtered seawater, fresh haemolymph samples without added beads, and filtered seawater with beads only were used as controls to clearly separate different populations. The percentage composition of haemocytes that had engulfed zero, one, two, and three beads were calculated as a function of the total haemocyte population.
Microbial community
To identify the effects of treatment conditions on the microbial communities associated with oysters, we sampled the microbes after 12 days of experimental exposure. Microbial samples were taken after day 12 rather than 16 (end of whole experiment) in order to have time to process all samples and ensure no additional sampling was required. We collected 1 litre of water from each oyster container, before water change and feeding. To differentiate between microbes that were already present in the water from microbes that were associated with the oysters, we also collected 1 litre of water from the control containers that did not contain any oysters (n = 3 per treatment). These containers had the same water change regime as the containers with oysters and therefore water in both the controls (no oyster) and treatment containers (with oyster) had been incubated for the same time period between water change and sampling (approximately 20 h). Water samples were filtered on autoclaved 0.2 μm mixed cellulose esters membrane filter papers (Millipore) using a vacuum pump and filtration system (Rocker Scientific). Filter papers were immediately frozen in liquid nitrogen and stored at −80°C until analysis. DNA was extracted from the membrane filter papers using a Qiagen PowerSoil kit following manufacturer’s protocol. DNA concentrations were checked with a nanodrop (Thermo Scientific) and quality checked on an agarose gel. Two extraction blanks were included to quantify the level of contamination that originated from the extraction kit or the extraction process.
Prior to sequencing library preparation, DNA concentrations were measured with a Qubit dsDNA BR Assay Kit on a Qubit 2.0 fluorometer (Thermo Scientific); the concentrations of DNA samples were subsequently normalized to 20 ng/μL by diluting with nuclease free water (Thermo Scientific). Amplicon sequencing libraries targeting the 16S rRNA V4-V5 region were prepared following the TaggiMatrix protocol (Glenn et al., 2019). The targeted locus was amplified with the primer pairs 515F-Y/926R (Parada et al., 2016); all primers included a variable length internal index (5–8 bp). The genes were amplified in 25 μL reactions with NEBNext® Ultra™ II Q5® Master Mix (New England Biolabs), which combined 12.5 μL of the PCR master mix, 1.25 μL of each primer, 5 μL of DNA template, and 5 μL of nuclease free water. The cycling conditions were as follows: 30 s at 98°C; 20 cycles of 10 s at 98°C, 15 s at 62°C, 20 s at 72°C; 2 min at 72°C. PCR products were purified with AMPure XP magnetic beads (Beckman Coulter) in 1:1 ratio and inspected on 1.5% agarose gel. Two PCR blanks were included for each amplicon to monitor contamination originated from the PCR process.
Purified amplicons were pooled together in equal concentrations for a second round of PCR with iTru indexed primers (Glenn et al., 2019). The second PCR was performed in triplicate, in 50 μL reaction volume with NEBNext® Ultra™ II Q5® Master Mix (New England Biolabs) following manufacturer’s instructions. The reaction conditions were as follows: initial denaturation of 30 s at 98°C; 8 cycles of 10 s at 98°C and 75 s at 65°C; and a final extension step at 65°C for 5 min. PCR products were purified with AMPure XP magnetic beads (Beckman Coulter) in 1:1 ratio and inspected on 1.5% agarose gel. The amplicon libraries were then pooled with other amplicon sequencing libraries and sequenced on Illumina MiSeq with MiSeq Reagent Kit version 3 (2 × 300 bp) in the Centre for PanorOmic Sciences, LKS Faculty of Medicine, HKU.
Raw sequencing reads were adapter trimmed and demultiplexed with cutadapt v3.1 (Martin, 2011), and subsequently imported into Qiime2 2020.11 (Bolyen et al., 2019). Read pairs were denoised and merged using DADA2 (Callahan et al., 2016), with the forward and reverse reads truncated to 290 bp and 210 bp, respectively. In order to perform taxonomic classification of the sequencing reads, we used the Silva 138 database as recommended in Qiime2. A feature classifier was trained on the Silva 138 99% OTUs full length sequences dataset; to improve the accuracy of the Naïve Bayes classifier, the dataset was subset so that only the region of the target sequences was used for training the classifier (Bokulich et al., 2018). This yielded a spreadsheet of taxonomic classifications and the number of hits based on our sequences.
Nutrient cycling
After sampling for microbial communities, water samples for measuring nutrient concentrations were taken by filtering 50 mL of water from all experimental treatments (control, hypoxia, acidification, and HA), with and without oysters, into a sterile centrifuge tube using a syringe-mounted 0.22 μm filter (Millipore). Water for nutrient samples were collected immediately after sampling for microbial communities, and from the same experimental container, to capture the microbial community and nutrient concentrations in the water as closely as possible. Filtered water samples were stored at −20°C until analysis. Ammonium concentration was analysed by spectrophotometry on a 96 well-plate following Grasshoff et al. (2009). A gradient of standards was prepared using ammonium sulphate dissolved in MilliQ water (100, 40, 20, 10, 5, 1, and 0.5 μmol/L). 250 μL of standard and sample were then pipetted in a 96 well-plate followed by addition of 10 μL phenol, 5 μL citrate buffer, and 10 μL hypochlorite. The solutions were mixed using the “shake” function on the spectrophotometer and incubated at 37°C for 30 min followed by cool down for 30 min and measurement at 630 nm on the spectrophotometer. Concentrations of nitrate and nitrite were measured following García-Robledo et al. (2014). Standards were prepared using sodium nitrate and sodium nitrite dissolved in MilliQ water (100, 40, 20, 10, 5, 1, and 0.5 μmol/L). 200 μL of standard and sample were then pipetted in a 96 well-plate followed by addition of 20 μL of Greiss reagent. The solutions were mixed using the ‘shake’ function on the spectrophotometer and incubated at 60°C for 20 min followed by cool down for 30 min and measurement at 540 nm on the spectrophotometer. Standards were used to plot a calibration curve based on known nutrient concentrations and spectrophotometer readings, which was then used to determine nutrient concentrations in samples.
Data analyses
Prior to formal analysis, all data were tested for normality and homoscedasticity using a Shapiro–Wilk test and a Bartlett test, respectively. To identify differences in Hsp-70, MDA, TAC, haemocyte community, phagocytic activity, and nutrient concentrations among treatments (control, hypoxia, acidification, and HA), we undertook two-factor permutational-ANOVAs (factors: Hypoxia, Acidification, two levels in each) on Euclidean distance matrices (3 < n < 15 per treatment depending on the variable measured, see Supplementary Tables S2, S3) followed by pairwise tests for significant terms in PERMANOVA. Microbial community data were grouped by phylum and were log (X + 1) transformed prior to analysis because of the large number of zero values within the data set. To identify differences in community structures among treatments, we performed a two-factor permutational-MANOVA (factor: Hypoxia, Acidification) on Bray–Curtis similarity matrix (n = 4). When significant effects were detected, post-hoc pairwise tests were used to determine which treatments differed. We then used an Analysis of Similarity (ANOSIM) to identify the amount of overlap (similarity/dissimilarity) in the community between treatments. Specific microbial phyla that contributed to the differences between treatments were identified using a Similarity Percentage test (SIMPER). The microbial community data were then plotted on a heatmap and a dendrogram was used for hierarchical clustering of microbial phyla. All permutational-MANOVA, ANOSIM, and SIMPER tests were performed on PRIMER+PERMANOVA (version 7). All normality tests, homoscedasticity tests, and hierarchical clustering of microbial phyla for heatmap were performed on R (version 4.12) using the packages “stats” and “gplots.”
Results
Oyster molecular defence
Throughout the experiment, less than 10% of oysters died across all experimental treatments (total 14 individuals died out of 240: 3 in control, 2 in acidification, 4 in hypoxia, and 5 in HA). However, experimental treatments drove differences in the physiological responses of oysters (Figure 1; Supplementary Table S2). Hsp-70 production was independently influenced by hypoxia and acidification (Supplementary Table S2). Oysters exposed to HA produced the highest concentrations of Hsp-70 (Figure 1A); approximately 600% more than oysters in the control (t7 = 3.1738, p = 0.023; Supplementary Table S2), and 500% more than those exposed to acidification (t8 = 3.0245, p = 0.012). Oysters from HA conditions also seemed to produce higher concentrations of Hsp-70 than those from hypoxic conditions, however these were not significant, likely because of large variability (Figure 1A; t7 = 2.0412, p = 0.078). Oysters from hypoxic conditions produced approximately 120% more Hsp-70 than those from control conditions (Figure 1A; t6 = 2.3148, p = 0.029), but were within the same range as those from acidified conditions (t7 = 1.2772, p = 0.247). Finally, oysters from acidification conditions did not produce significantly higher Hsp-70 concentrations than those from control conditions (Figure 1A; t7 = 1.2131, p = 0.317).
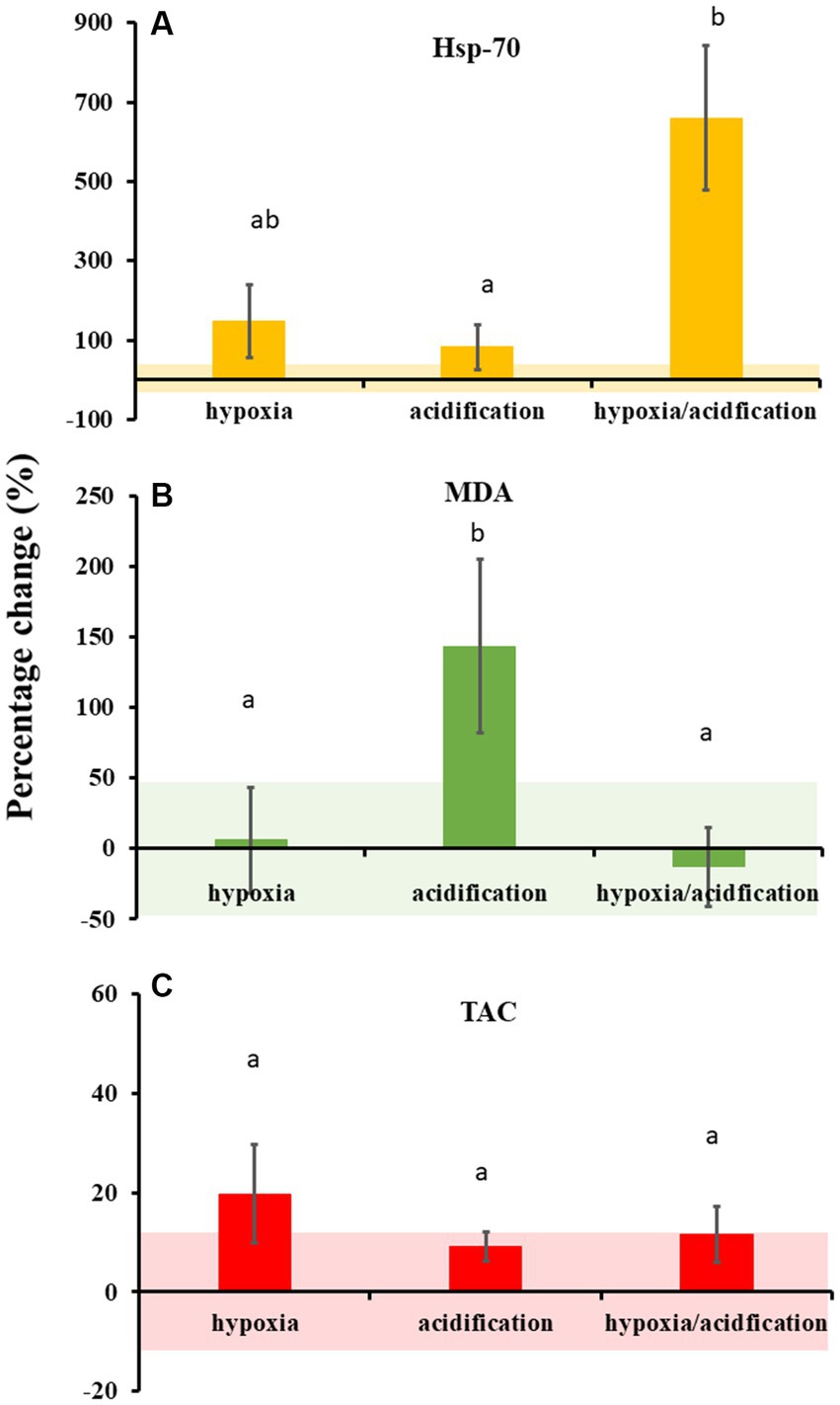
Figure 1. Percentage change relative to control (mean ± SE) in heat shock protein 70 (Hsp-70; (A)), malondialdehyde (MDA; (B)), and total oxidative capacity (TAC; (C)) measured from oysters exposed to hypoxia, acidification, or both. Coloured bands around the x-axis denote the variation (mean ± SE) in oysters exposed to control conditions. Concentrations of Hsp-70, MDA and TAC in control conditions were 0.0014 (±0.0007) ng/mg, 0.071 (±0.03) μM/mg, and 0.0014 (±0.0002) mM/mg, respectively. Note the difference in scale for each graph. Letters denote significant differences.
Overall, oysters from the different conditions produced similar concentrations of MDA (Figure 1B; Supplementary Table S2), except for acidification, meaning that oxidative stress was high in oysters exposed to acidification. Oysters exposed to acidification produced a higher concentration of MDA compared to those exposed to control and HA conditions (Figure 1B; t5 = 3.6523, p = 0.029, t6 = 3.4242, p = 0.043, respectively), but not compared to hypoxia (t5 = 2.5796 p = 0.101: Supplementary Table S1). Finally, no difference in TAC was observed across treatments (Figure 1C; Supplementary Table S2).
Oyster immune response
The number of granulocytes in oysters were influenced by both acidification and hypoxia (Supplementary Table S3) demonstrating that the immune system was impaired. Oysters exposed to HA and acidification produced fewer granulocytes compared to control or hypoxia oysters (Figure 2A; Supplementary Table S3). The number of agranulocytes in oysters, were also influenced by acidification and hypoxia, and their interaction (Supplementary Table S3). Agranulocyte numbers in oysters from HA conditions were lower than those exposed to hypoxia or acidification (Figure 2A; t33 = 3.6789, p = 0.002, and t32 = 2.3893, p = 0.026, respectively), with counts in the latter two being similar (t31 = 1.0456, p = 0.314), and lower than those in the control (t27 = 5.2238, p = 0.001, and t28 = 5.8149, p = 0.001, respectively).
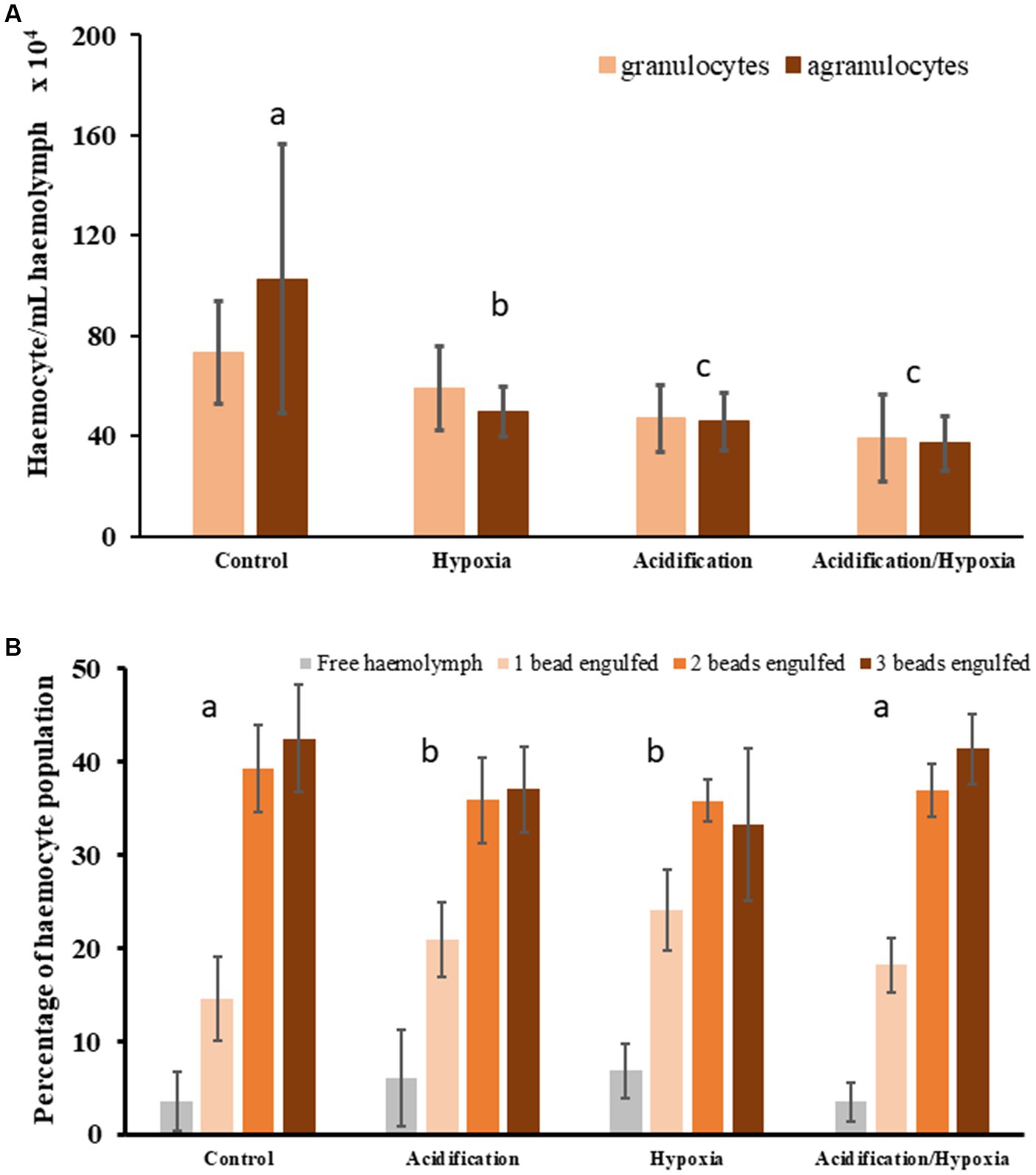
Figure 2. Oyster haemocyte counts (mean ± SE) differentiated as granulocytes and agranulocytes (A) and oyster phagocytic activity measured as the number of fluorescent beads engulfed by haemocytes (mean ± SE) (B). Letters and circles denote significantly different treatment groups.
The overall communities of haemocyte (both granulocytes and agranulocytes) were significantly influenced by hypoxia, acidification, or their combination (HA), with the proportion of granulocyte to agranulocyte also differing (1:1.5 granulocyte to agranulocyte in controls; Figure 2A; Supplementary Table S3). The haemocyte community in oysters exposed to hypoxia were different from those exposed to other experimental treatments in terms of counts and proportion of granulocyte to agranulocyte (more granulocyte than agranulocyte; ratio = 1:0.8; Supplementary Table S3). Oysters exposed to HA and acidification had similar haemocyte community, that is, similar counts and proportion of granulocyte to agranulocyte (1:1) to each other (Figure 2A; t33 = 1.7916 p = 0.059), but also the lowest counts among all treatments (Figure 2A).
The phagocytic activity, that is the percentage of haemocytes that engulfed one, two, three, or no beads, was influenced by hypoxia and acidification which each independently reduced overall phagocytic activity relative to control (fewer haemocytes engulfed 2 or 3 beads: Figure 3; Supplementary Table S3). However, HA produced similar phagocytic activity as control conditions as suggested by the proportion of haemocytes engulfing 2 or 3 beads (Figure 2B; Supplementary Table S3). Although oysters exposed to HA had lower counts of haemocytes, their activity in terms of engulfment of beads, was similar to oysters from control conditions (Figure 2B; t16 = 1.227, p = 0.212). Overall, a larger proportion of haemocytes from both control and HA engulfed two or three beads, with very few engulfing none. The phagocytic activity in oysters exposed to hypoxia and acidification were similar to each other (Figure 2B; t17 = 1.4234, p = 0.148). While the majority of haemocytes from these treatments still engulfed two or three beads, a large proportion also engulfed only one or no beads (Figure 2B).
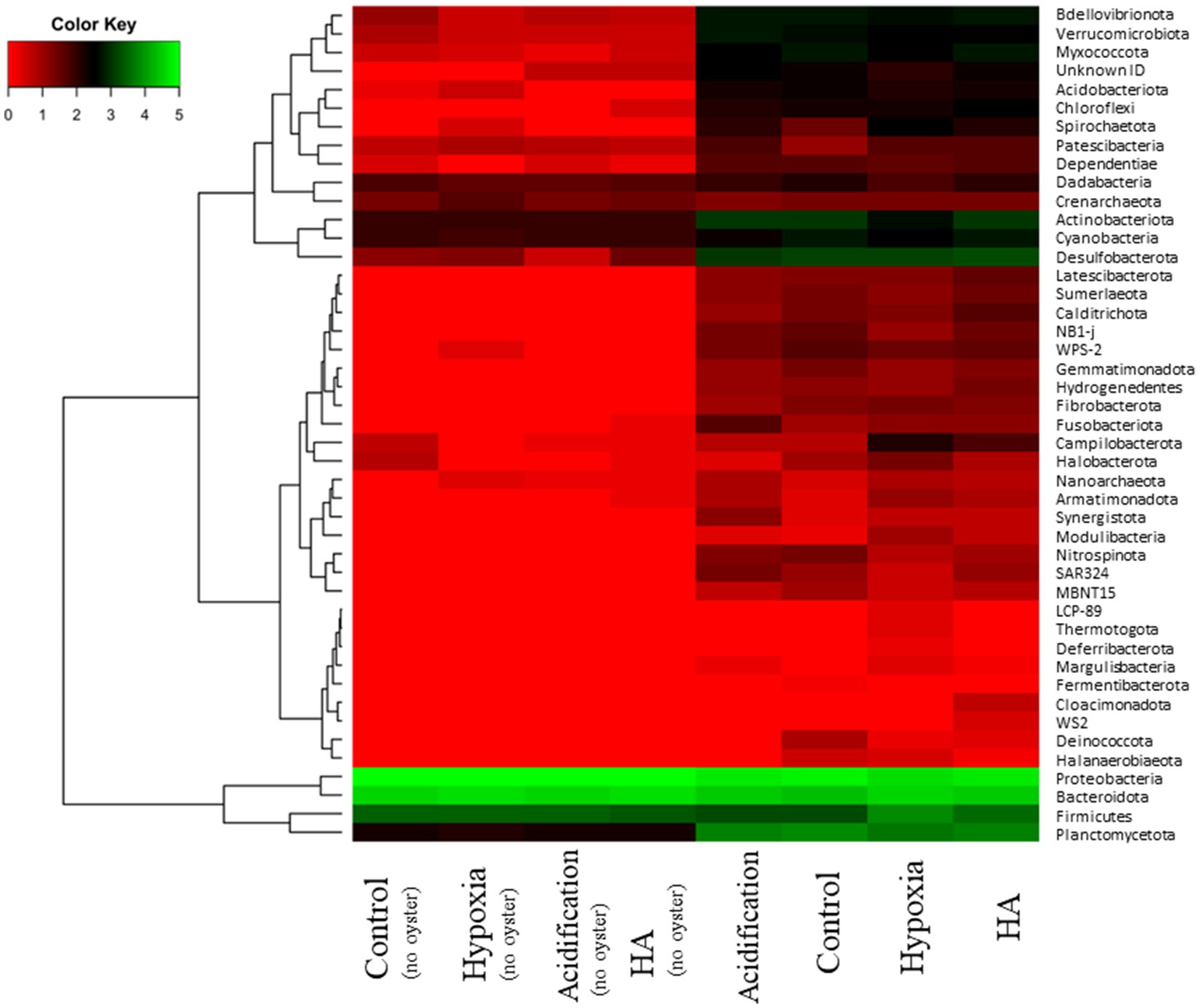
Figure 3. Heatmap comparing abundance of microbial phyla (log x + 1) in the presence or absence of oysters under control, acidification, hypoxia, and hypoxia/acidification (HA) conditions.
Microbial community
There was a clear difference in the microbial community structure when oysters were present oysters, irrespective of the treatment, compared when oysters were absent (Figure 3; Pseudo-F3,30 = 101.01, p = 0.001). The microbial community associated with oysters was characterised by microbes that are commonly observed in the environment (benthos, water column) or as part of the microbiome of organisms (e.g., the phyla Latescibacterota and Acidobacteriota), but also multiple other phyla associated with organic matter degradation (e.g., Hydrogenedentes, Sumerlaeota) or inorganic nutrient cycling (e.g., Desulfobacterota, Nitrospinota, ‘uncultured’ NB1-j, Planctomycetota). The community structure of microbes associated with oysters also differed among the four treatments (Supplementary Table S4), albeit with some overlap in the number of phyla (ANOSIM: 0.2 < R statistic <0.5). Therefore, differences among the communities were driven by turnover in a few taxa rather than wholesale community change. The community in acidified conditions was distinguished by the presence of bacteria resistant to extreme environments (e.g., phylum Deinococcota), organic carbon fermenting bacteria (e.g., phylum Fermentibacterota), and nitrite-oxidizing bacteria (e.g., phylum Nitrospinota) (Figure 3). On the other hand, the community in hypoxic conditions was primarily distinguished by the presence of bacteria from multiple phyla that thrive in anaerobic conditions (e,g., Deferribacterota, Halanaerobiaeota, Modulibacteria, and Thermotogota). In addition, pathogenic bacteria, such as from the phyla Firmicutes and Spirochaetoses, contributed more to the hypoxic community compared to all other treatments. Finally, in the HA treatment, the microbial community was characterised by anaerobic bacteria and organic carbon degraders (e.g., phyla Calditrichota, Cloacimonadota, and Hydrogenedentes).
Nutrient cycling
The concentration of ammonium and nitrite were very low (close to zero) in the absence of oysters under all treatments (Figure 4). In contrast, the concentrations of both were high (> 15 μmol/L for ammonium and > 35 μmol/L for nitrite) in all experimental conditions when oysters were present. The concentrations of ammonium was higher in control and acidification treatments when oysters were present compared to when they were absent (up to 25 μmol/L g; Figure 4; t6 = 5.0176, p = 0.016 and t6 = 3.4189, p = 0.047, respectively), but these concentrations were low compared to the concentration of nitrite in both control and acidification conditions in the presence of oysters (both over 40 μmol/L; t6 = 7.8128, p = 0.019 and t6 = 13.198, p = 0.018, respectively). Finally, nitrate concentrations remained unchanged in the presence or absence of oysters or between different treatments (Figure 4). The concentrations of ammonium were two times higher under hypoxia and HA conditions compared to acidification (Figure 4; t7 = 3.3436, p = 0.009 and t8 = 2.5076, p = 0.036). The concentrations of ammonium were higher under hypoxia compared to control conditions (Figure 4; t7 = 2.9393, p = 0.024), and while the concentrations under HA seemed higher, they were not statistically significant (t8 = 2.1081, p = 0.057). No differences were found in nitrite concentrations between treatments, except for acidification where nitrite was lower compared to control conditions (Figure 4; t8 = 2.2273, p = 0.027). Finally, the concentration of nitrate under hypoxia and HA were both less than that in acidification (Figure 4; t7 = 3.2422, p = 0.014 and t8 = 32.5526, p = 0.007, respectively).
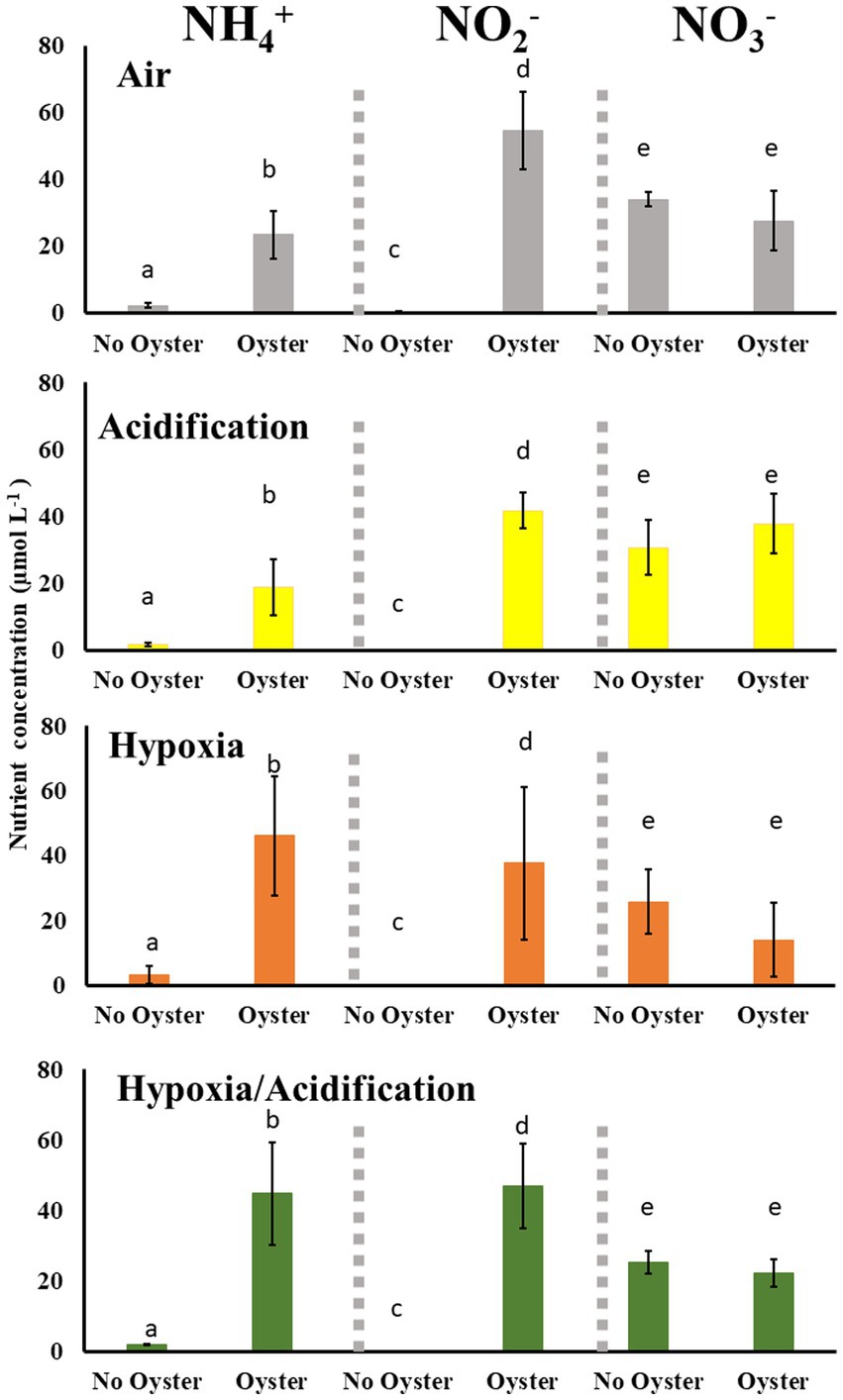
Figure 4. Ammonium (NH4+), Nitrite (NO2−) and Nitrate (NO3−) concentrations (mean ± SE) measured from seawater in the presence and absence of oysters under control, acidification, hypoxia, and Hypoxia/acidification conditions. Letters denote significantly different concentrations for each nutrient in the presence and absence of oysters within each treatment.
Discussion
Climate change is projected to drive more extreme fluctuations in environmental conditions, potentially exposing organisms to periods of physiologically stressful conditions, especially with compound changes in two or more environmental conditions. For habitat forming organisms (e.g., oysters, mussels, seagrasses, or corals), physiological stress can quickly cascade into ecosystem-wide consequences (Wernberg et al., 2013) through changes in performance of the organism that result in changes in their function (e.g., loss of habitat forming capacity or changes in ecological processes in which they are involved). Here, we show that acidification, and its combination with hypoxia (HA), were particularly stressful to oysters and caused a large molecular response. Simultaneously, acidification, hypoxia, and HA impaired the immune response of oysters. More importantly, however, changes to the oyster-associated microbial community under altered conditions drove a loss in efficiency of nutrient cycling in hypoxic and HA conditions.
The physiological response of oysters differed based on the type of environmental stressor they were exposed to, but, overall, both molecular processes and immune response were impacted. Oysters exposed to acidification produced a higher concentration of MDA, suggesting an increased lipid peroxidation and oxidative stress (Tsikas, 2017), a response commonly observed in oysters exposed to acidification (Tomanek et al., 2011; Mackenzie et al., 2014; Wang et al., 2016, 2020). Simultaneously, oysters exposed to acidification produced fewer haemocytes and the phagocytic capacity of these haemocytes was also reduced. The reduction of haemocytes and phagocytosis is generally representative of stressful conditions (Mackenzie et al., 2014) and here it could be indicative of two responses. First, energy may have been redistributed towards production of MDA for reduction of oxidative stress within the organism, at the expense of the immune system (trade-off mechanism; Rauw, 2012). Second, oxidative stress within the haemolymph may have reduced phagocytic capacity of haemocytes by impacting cell membrane strength (e.g., Xie et al., 2017; Cao et al., 2018). The low haemocyte production and phagocytic activity means that extended exposure to acidification can severely impact oyster immune capacity, both through impaired molecular response and immune response, and, therefore, increase their susceptibility to pathogens.
Oysters exposed to hypoxia did not respond through changes to molecular defence, but their immune capacity was significantly lowered. Specifically, we found that both the number of granulocytes and agranulocytes in oysters were reduced indicating a reduction in innate immune capacity (Jiang et al., 2016). Depressed immune capacity (haemocyte counts and phagocytic activity) under hypoxic conditions has been observed in a range of organisms (Wang et al., 2012; Chen et al., 2017; Wang et al., 2017; Shen et al., 2019; Zhan et al., 2022), and is often considered a major contributor to mortality because of the associated susceptibility to pathogenic load (Ellis et al., 2011). In our experiment, exposure to hypoxic conditions for 16 days did not have major lethal consequences likely because of the controlled conditions that organisms were reared in (that is natural pathogens were not as prevalent as they could be in the natural environment). However, considering that hypoxic conditions on the benthos can last for days up to months (Levin et al., 2009; Qian et al., 2018), and oysters inhabit dynamic coastal environments where pathogens are more prevalent (either occurring in the natural environment or resulting from anthropogenic discharge), it is likely that prolonged hypoxia will drive higher mortality rates of oysters in natural ecosystems.
The coupled impact of acidification and hypoxia tends to be synergistically negative on organismal physiology (Gobler et al., 2014; Gobler and Baumann, 2016) and can cause rapid mortality (Chan et al., 2019). In our experiment, oysters produced substantially more Hsp-70 under combined hypoxic and acidified conditions and their haemocyte counts decreased considerably. Yet, the phagocytic activity (proportion of cells that engulf beads) remained similar to that of oysters from the control conditions (i.e., most cells engulfed three and few engulfed less than two beads) and, most importantly, mortality remained low. This combination of responses suggests that oysters under HA conditions likely experienced acute physiological stress. As the production and chaperoning action of HSPs are ATP-dependent processes that require high levels of aerobic metabolism and come at a high energetic cost (Feder and Hofmann, 1999; Sokolova et al., 2012), our results suggest that a large amount of energy may have been directed towards the production of Hsp-70 chaperones that serve to maintain protein integrity (Mayer and Bukau, 2005). The increased Hsp-70 production suggests that the organisms experienced protein damage, a condition commonly driven by excessive oxidative stress that rapidly affects the structure and activity of proteins (Reichmann et al., 2018). Therefore, rather than allocating energy towards reduction of oxidative stress (e.g., MDA), oysters may have allocated energy towards maintaining the structure of the rapidly deteriorating proteins (by producing Hsp-70 chaperones). Finally, the low counts of haemocytes but strong phagocytic capacity indicates that energy may have also been invested towards efficient immune response, such as through repair of haemocyte cell membrane (Xie et al., 2017), rather than production of more haemocytes. The investment in repairing haemocyte cell membranes could be driven by an increasing pathogenic load or deteriorating tissue that required haemocytes to respond (Bachère et al., 2015), but the large amount of energy already allocated to Hsp-70 production meant that production of new haemocyte may be too costly, and maintenance of already existing haemocytes provided a better trade-off. Overall, these results suggest that the combined effects of hypoxia and acidification are physiologically costly to oysters and can rapidly increase their susceptibility to pathogens.
The microbial community associated with the oysters was affected by exposure to different environmental stressors. Considering that the microbial community structure was modified (enriched differently) in different environmental conditions, it is conceivable that short-term rapid environmental change will drive some extent of oyster holobiont restructuring in natural systems (e.g., some microbes that were more suited to exploit or survive in the acidification, hypoxia, or both conditions, and, therefore, became more prevalent in these conditions). While oyster holobiont research remains limited, similar changes in some microbial taxonomic groups within the holobiont have been observed when exposed to similar environmental stressors for the Sydney rock oysters (Saccostrea glomerata; Scanes et al., 2021a,b), some sponges (Ribes et al., 2016; Posadas et al., 2022) and some corals (Bourne et al., 2016; Meunier et al., 2021). However, complete/broader turnover of associated microbial communities is not often observed (Meron et al., 2012; Bourne et al., 2016; Zhou et al., 2016; Ge et al., 2021). Broader host associated microbial community turnover is more likely to occur after long-term exposure to changed environments, such as in oysters exposed to 2,800 ppm pCO2 for 80 days (Unzueta-Martínez et al., 2021) or clams exposed to several years of increased temperature (Neu et al., 2021). Our results, along with findings from other studies, therefore, suggest that host associated microbial communities can be subject to important shifts in enrichment that cause compositional changes driven by alterations to their environment.
The two times lower concentration of ammonium than nitrite (ratio of approximately 1:2) suggest perhaps low ammonium release through bio-deposits by oysters but, importantly, efficient nitrification rates by microbes in both control and acidification. In contrast, the doubled ammonium concentration in hypoxic and HA conditions compared acidification, and to some extent, control conditions suggest more ammonium release by oysters in hypoxic and HA conditions. A probable explanation of for the increased ammonium concentrations is that increased stress induced higher filtration rates (energy acquisition), which lead to more ammonium excretion. While we did not directly measure filtration rates, the linkage between physiological stress, filtration rates and ammonium releases has previously been established (e.g., Guzmán-Agüero et al., 2013). The 1:1 ratio of ammonium to nitrite in hypoxic and HA conditions also suggest approximately 50% less efficient nitrification compared to control or acidification conditions. Therefore, the changes in ammonium and nitrite concentrations measured in our study suggest high nitrification capacity by microbes in the acidification treatment, that is a possible increase in nitrification by microbes in acidified conditions compared to control or other treatments. Under hypoxia and the combination of stressors, however, the high concentration of both ammonium and nitrite measured in the experiment suggest ammonium release by oysters (e.g., through excretion) may be higher and that although nitrification was maintained, there was loss of nitrification efficiency/capacity under hypoxia and HA conditions.
Overall, our study shows that the oyster holobiont can maintain some reduced nutrient cycling capacity under acidification or hypoxic conditions, or their combination, as long as the host itself can regulate its physiological processes. The enrichment of microbial communities seemed to change mostly in an exploitive manner based on the environmental conditions. However, it is also clear that processes such as nitrification become less efficient in hypoxic and combined hypoxic and acidified conditions. On the other hand, oyster molecular defence and immune capacity was impacted under both acidification and hypoxia, and even more so under the combined stressors. While mortality was low under all altered conditions, the decreased immune capacity coupled with other physiological impairments would likely render oysters vulnerable to pathogens and other future environmental stressors. If there was a resultant increase in mortality and decline in oyster populations this would drive a drastic loss of the functions associated with the oyster holobiont. The negative impact of climate change on oyster physiology and holobiont functions, therefore, has implications for the future of oyster reefs whereby the increase hypoxic and acidified conditions in coastal environments could render conservation and restoration efforts less able to achieve their full potential.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: University of Hong Kong (HKU) Data Repository, accession 10.25442/hku.22717084.v1; NCBI BioProject, accession PRJNA903533.
Author contributions
DH: conceptualization, methodology, formal analysis, investigation, and writing – original draft. LF: conceptualization, methodology, writing – review & editing, and funding acquisition. KC: conceptualization, methodology, formal analysis, investigation, and writing – review & editing. LM: investigation and writing – review & editing. AC: conceptualization, methodology, investigation, and writing – review & editing. BR: conceptualization, methodology, writing – original draft, review & editing, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by a Faculty of Science (HKU) Rising Star Fund grant to BR, a University of Hong Kong Post-Doctoral Fellowship to DH, and CUHK funding to LF. The computations were performed using research computing facilities offered by Information Technology Services, the University of Hong Kong.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1083315/full#supplementary-material
References
Bachère, E., Rosa, R. D., Schmitt, P., Poirier, A. C., Merou, N., Charrière, G. M., et al. (2015). The new insights into the oyster antimicrobial defense: cellular, molecular and genetic view. Fish Shellfish Immunol. 46, 50–64. doi: 10.1016/j.fsi.2015.02.040
Bayraktarov, E., Saunders, M. I., Abdullah, S., Mills, M., Beher, J., Possingham, H. P., et al. (2016). The cost and feasibility of marine coastal restoration. Ecol. Appl. 26, 1055–1074. doi: 10.1890/15-1077
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 1–17. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Boyd, J. N., and Burnett, L. E. (1999). Reactive oxygen intermediate production by oyster hemocytes exposed to hypoxia. J. Exp. Biol. 202, 3135–3143. doi: 10.1242/jeb.202.22.3135
Cai, W. J. (2011). Estuarine and coastal ocean carbon paradox: CO2 sinks or sites of terrestrial carbon incineration? Annu. Rev. Mar. Sci. 3, 123–145. doi: 10.1146/annurev-marine-120709-142723
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cao, R., Wang, Q., Yang, D., Liu, Y., Ran, W., Qu, Y., et al. (2018). CO2-induced ocean acidification impairs the immune function of the Pacific oyster against Vibrio splendidus challenge: an integrated study from a cellular and proteomic perspective. Sci. Total Environ. 625, 1574–1583. doi: 10.1016/j.scitotenv.2018.01.056
Chan, F., Barth, J. A., Kroeker, K. J., Lubchenco, J., and Menge, B. A. (2019). The dynamics and impact of ocean acidification and hypoxia. Oceanography 32, 62–71. doi: 10.5670/oceanog.2019.312
Chen, N., Wu, M., Tang, G. P., Wang, H. J., Huang, C. X., Wu, X. J., et al. (2017). Effects of acute hypoxia and reoxygenation on physiological and immune responses and redox balance of Wuchang bream (Megalobrama amblycephala Yih, 1955). Front. Physiol. 8:375. doi: 10.3389/fphys.2017.00375
David, E., Tanguy, A., Pichavant, K., and Moraga, D. (2005). Response of the Pacific oyster Crassostrea gigas to hypoxia exposure under experimental conditions. FEBS J. 272, 5635–5652. doi: 10.1111/j.1742-4658.2005.04960.x
Diaz, R. J., and Rosenberg, R. (1995). Marine benthic hypoxia: a review of its ecological effects and the behavioural responses of benthic macrofauna. Oceanogr. Mar. Biol. 33, 245–203.
Dubé, C. E., Ky, C. L., and Planes, S. (2019). Microbiome of the black-lipped pearl oyster Pinctada margaritifera, a multi-tissue description with functional profiling. Front. Microbiol. 10:1548. doi: 10.3389/fmicb.2019.01548
Ellis, R. P., Parry, H., Spicer, J. I., Hutchinson, T. H., Pipe, R. K., and Widdicombe, S. (2011). Immunological function in marine invertebrates: responses to environmental perturbation. Fish Shellfish Immunol. 30, 1209–1222. doi: 10.1016/j.fsi.2011.03.017
Feder, M. E., and Hofmann, G. E. (1999). Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282. doi: 10.1146/annurev.physiol.61.1.243
García-Robledo, E., Corzo, A., and Papaspyrou, S. (2014). A fast and direct spectrophotometric method for the sequential determination of nitrate and nitrite at low concentrations in small volumes. Mar. Chem. 162, 30–36. doi: 10.1016/j.marchem.2014.03.002
Garner, N., Ross, P. M., Falkenberg, L. J., Seymour, J. R., Siboni, N., and Scanes, E. (2022). Can seagrass modify the effects of ocean acidification on oysters? Mar. Pollut. Bull. 177:113438. doi: 10.1016/j.marpolbul.2022.113438
Ge, R., Liang, J., Yu, K., Chen, B., Yu, X., Deng, C., et al. (2021). Regulation of the coral-associated Bacteria and Symbiodiniaceae in Acropora valida under ocean acidification. Front. Microbiol. 12:767174. doi: 10.3389/fmicb.2021.767174
Glenn, T. C., Nilsen, R. A., Kieran, T. J., Sanders, J. G., Bayona-Vásquez, N. J., Finger, J. W., et al. (2019). Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext). Peer J 7:e7755. doi: 10.7717/peerj.7755
Gobler, C. J., and Baumann, H. (2016). Hypoxia and acidification in ocean ecosystems: coupled dynamics and effects on marine life. Biol. Lett. 12:20150976. doi: 10.1098/rsbl.2015.0976
Gobler, C. J., DePasquale, E. L., Griffith, A. W., and Baumann, H. (2014). Hypoxia and acidification have additive and synergistic negative effects on the growth, survival, and metamorphosis of early life stage bivalves. Plo S one 9:e83648. doi: 10.1371/journal.pone.0083648
Goedken, M., and De Guise, S. (2004). Flow cytometry as a tool to quantify oyster defence mechanisms. Fish Shellfish Immunol. 16, 539–552. doi: 10.1016/j.fsi.2003.09.009
Grasshoff, K., Kremling, K., and Ehrhardt, M. eds., (2009). Methods of Seawater Analysis. John Wiley & Sons, Hoboken, NJ.
Green, D. S., Boots, B., and Crowe, T. P. (2012). Effects of non-indigenous oysters on microbial diversity and ecosystem functioning. PLoS One 7:e48410. doi: 10.1371/journal.pone.0048410
Guzmán-Agüero, J. E., Nieves-Soto, M., Hurtado, M. Á., Pina-Valdez, P., and Garza-Aguirre, M. D. C. (2013). Feeding physiology and scope for growth of the oyster Crassostrea corteziensis (Hertlein, 1951) acclimated to different conditions of temperature and salinity. Aquac. Int. 21, 283–297. doi: 10.1007/s10499-012-9550-4
Hoellein, T. J., Zarnoch, C. B., and Grizzle, R. E. (2015). Eastern oyster (Crassostrea virginica) filtration, biodeposition, and sediment nitrogen cycling at two oyster reefs with contrasting water quality in Great Bay estuary (New Hampshire, USA). Biogeochemistry 122, 113–129. doi: 10.1007/s10533-014-0034-7
Hughes, D. J., Alderdice, R., Cooney, C., Kühl, M., Pernice, M., Voolstra, C. R., et al. (2020). Coral reef survival under accelerating ocean deoxygenation. Nat. Clim. Chang. 10, 296–307. doi: 10.1038/s41558-020-0737-9
Jeppesen, R., Rodriguez, M., Rinde, J., Haskins, J., Hughes, B., Mehner, L., et al. (2018). Effects of hypoxia on fish survival and oyster growth in a highly eutrophic estuary. Estuar. Coasts 41, 89–98. doi: 10.1007/s12237-016-0169-y
Jiang, S., Jia, Z., Zhang, T., Wang, L., Qiu, L., Sun, J., et al. (2016). Functional characterisation of phagocytes in the Pacific oyster Crassostrea gigas. Peer J 4:e2590. doi: 10.7717/peerj.2590
King, W. L., Jenkins, C., Seymour, J. R., and Labbate, M. (2019). Oyster disease in a changing environment: decrypting the link between pathogen, microbiome and environment. Mar. Environ. Res. 143, 124–140. doi: 10.1016/j.marenvres.2018.11.007
Lau, S. C. Y., Thomas, M., Hancock, B., and Russell, B. D. (2020). Restoration potential of Asian oysters on heavily developed coastlines. Restor. Ecol. 28, 1643–1653. doi: 10.1111/rec.13267
Lemasson, A. J., Fletcher, S., Hall-Spencer, J. M., and Knights, A. M. (2017). Linking the biological impacts of ocean acidification on oysters to changes in ecosystem services: a review. J. Exp. Mar. Biol. Ecol. 492, 49–62. doi: 10.1016/j.jembe.2017.01.019
Levin, L. A., Ekau, W., Gooday, A. J., Jorissen, F., Middelburg, J. J., Naqvi, S. W. A., et al. (2009). Effects of natural and human-induced hypoxia on coastal benthos. Biogeosciences 6, 2063–2098. doi: 10.5194/bg-6-2063-2009
Lutier, M., Di Poi, C., Gazeau, F., Appolis, A., Le Luyer, J., and Pernet, F. (2022). Revisiting tolerance to ocean acidification: insights from a new framework combining physiological and molecular tipping points of Pacific oyster. Glob. Chang. Biol. 28, 3333–3348. doi: 10.1111/gcb.16101
Mackenzie, C. L., Lynch, S. A., Culloty, S. C., and Malham, S. K. (2014). Future oceanic warming and acidification alter immune response and disease status in a commercial shellfish species, Mytilus edulis L. PLoS One 9:e99712. doi: 10.1371/journal.pone.0099712
Mayer, M. P., and Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684. doi: 10.1007/s00018-004-4464-6
Meron, D., Rodolfo-Metalpa, R., Cunning, R., Baker, A. C., Fine, M., and Banin, E. (2012). Changes in coral microbial communities in response to a natural pH gradient. ISME J. 6, 1775–1785. doi: 10.1038/ismej.2012.19
Meunier, V., Geissler, L., Bonnet, S., Rädecker, N., Perna, G., Grosso, O., et al. (2021). Microbes support enhanced nitrogen requirements of coral holobionts in a high CO2 environment. Mol. Ecol. 30, 5888–5899. doi: 10.1111/mec.16163
Neu, A. T., Hughes, I. V., Allen, E. E., and Roy, K. (2021). Decade-scale stability and change in a marine bivalve microbiome. Mol. Ecol. 30, 1237–1250. doi: 10.1111/mec.15796
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Posadas, N., Baquiran, J. I. P., Nada, M. A. L., Kelly, M., and Conaco, C. (2022). Microbiome diversity and host immune functions influence survivorship of sponge holobionts under future ocean conditions. ISME J. 16, 58–67. doi: 10.1038/s41396-021-01050-5
Qian, W., Gan, J., Liu, J., He, B., Lu, Z., Guo, X., et al. (2018). Current status of emerging hypoxia in a eutrophic estuary: the lower reach of the Pearl River estuary, China. Estuar. Coast. Shelf Sci. 205, 58–67. doi: 10.1016/j.ecss.2018.03.004
Rauw, W. M. (2012). Immune response from a resource allocation perspective. Front. Genet. 3:267. doi: 10.3389/fgene.2012.00267
Ray, N. E., and Fulweiler, R. W. (2021). Meta-analysis of oyster impacts on coastal biogeochemistry. Nat. Sustain. 4, 261–269. doi: 10.1038/s41893-020-00644-9
Ray, N. E., Henning, M. C., and Fulweiler, R. W. (2019). Nitrogen and phosphorus cycling in the digestive system and shell biofilm of the eastern oyster Crassostrea virginica. Mar. Ecol. Prog. Ser. 621, 95–105. doi: 10.3354/meps13007
Reichmann, D., Voth, W., and Jakob, U. (2018). Maintaining a healthy proteome during oxidative stress. Mol. Cell 69, 203–213. doi: 10.1016/j.molcel.2017.12.021
Ribes, M., Calvo, E., Movilla, J., Logares, R., Coma, R., and Pelejero, C. (2016). Restructuring of the sponge microbiome favors tolerance to ocean acidification. Environ. Microbiol. Rep. 8, 536–544. doi: 10.1111/1758-2229.12430
Rivest, E. B., Comeau, S., and Cornwall, C. E. (2017). The role of natural variability in shaping the response of coral reef organisms to climate change. Curr. Clim. Change Rep. 3, 271–281. doi: 10.1007/s40641-017-0082-x
Scanes, E., Parker, L. M., Seymour, J. R., Siboni, N., King, W. L., Danckert, N. P., et al. (2021a). Climate change alters the haemolymph microbiome of oysters. Mar. Pollut. Bull. 164:111991. doi: 10.1016/j.marpolbul.2021.111991
Scanes, E., Parker, L. M., Seymour, J. R., Siboni, N., King, W. L., Wegner, K. M., et al. (2021b). Microbiome response differs among selected lines of Sydney rock oysters to ocean warming and acidification. FEMS Microbiol. Ecol. 97:fiab 09. doi: 10.1093/femsec/fiab099
Shen, Y., Huang, Z., Liu, G., Ke, C., and You, W. (2019). Hemolymph and transcriptome analysis to understand innate immune responses to hypoxia in Pacific abalone. Comp. Biochem. Physiol. Part D Genomics Proteomics 30, 102–112. doi: 10.1016/j.cbd.2019.02.001
Smyth, A. R., Geraldi, N. R., and Piehler, M. F. (2013). Oyster-mediated benthic-pelagic coupling modifies nitrogen pools and processes. Mar. Ecol. Prog. Ser. 493, 23–30. doi: 10.3354/meps10516
Sokolova, I. M., Frederich, M., Bagwe, R., Lannig, G., and Sukhotin, A. A. (2012). Energy homeostasis as an integrative tool for assessing limits of environmental stress tolerance in aquatic invertebrates. Mar. Environ. Res. 15, 1–18. doi: 10.1016/j.marenvres.2012.04.003
Stevick, R. J., Post, A. F., and Gómez-Chiarri, M. (2021). Functional plasticity in oyster gut microbiomes along a eutrophication gradient in an urbanized estuary. Anim. Microbiome 3, 1–17. doi: 10.1186/s42523-020-00066-0
Timmins-Schiffman, E., Coffey, W. D., Hua, W., Nunn, B. L., Dickinson, G. H., and Roberts, S. B. (2014). Shotgun proteomics reveals physiological response to ocean acidification in Crassostrea gigas. BMC Genomics 15, 1–18. doi: 10.1186/1471-2164-15-951
Tomanek, L., Zuzow, M. J., Ivanina, A. V., Beniash, E., and Sokolova, I. M. (2011). Proteomic response to elevated P CO2 level in eastern oysters, Crassostrea virginica: evidence for oxidative stress. J. Exp. Biol. 214, 1836–1844. doi: 10.1242/jeb.055475
Tsikas, D. (2017). Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: analytical and biological challenges. Anal. Biochem. 524, 13–30. doi: 10.1016/j.ab.2016.10.021
Ullah, H., Nagelkerken, I., Goldenberg, S. U., and Fordham, D. A. (2018). Climate change could drive marine food web collapse through altered trophic flows and cyanobacterial proliferation. PLoS Biol. 16:e2003446. doi: 10.1371/journal.pbio.2003446
Unzueta-Martínez, A., Downey-Wall, A. M., Cameron, L. P., Ries, J. B., Lotterhos, K. E., and Bowen, J. L. (2021). Ocean acidification alters the diversity and structure of oyster associated microbial communities. Limnol. Oceanogr. 6, 348–359. doi: 10.1002/lol2.10214
Vaquer-Sunyer, R., and Duarte, C. M. (2008). Thresholds of hypoxia for marine biodiversity. Proc. Natl. Acad. Sci. 105, 15452–15457. doi: 10.1073/pnas.0803833105
Vizzini, S., Martínez-Crego, B., Andolina, C., Massa-Gallucci, A., Connell, S. D., and Gambi, M. C. (2017). Ocean acidification as a driver of community simplification via the collapse of higher-order and rise of lower-order consumers. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-03802-w
Wang, Q., Cao, R., Ning, X., You, L., Mu, C., Wang, C., et al. (2016). Effects of ocean acidification on immune responses of the Pacific oyster Crassostrea gigas. Fish Shellfish Immunol. 49, 24–33. doi: 10.1016/j.fsi.2015.12.025
Wang, Y., Hu, M., Cheung, S. G., Shin, P. K. S., Lu, W., and Li, J. (2012). Immune parameter changes of hemocytes in green-lipped mussel Perna viridis exposure to hypoxia and hyposalinity. Aquaculture 356-357, 22–29. doi: 10.1016/j.aquaculture.2012.06.001
Wang, X., Huang, W., Wei, S., Shang, Y., Gu, H., Wu, F., et al. (2020). Microplastics impair digestive performance but show little effects on antioxidant activity in mussels under low pH conditions. Environ. Pollut. 258:113691. doi: 10.1016/j.envpol.2019.113691
Wang, Q. F., Shen, W. L., Hou, C. C., Liu, C., Wu, X. F., and Zhu, J. Q. (2017). Physiological responses and changes in gene expression in the large yellow croaker Larimichthys crocea following exposure to hypoxia. Chemosphere 169, 418–427. doi: 10.1016/j.chemosphere.2016.11.099
Wernberg, T., Smale, D. A., Tuya, F., Thomsen, M. S., Langlois, T. J., De Bettignies, T., et al. (2013). An extreme climatic event alters marine ecosystem structure in a global biodiversity hotspot. Nat. Clim. Chang. 3, 78–82. doi: 10.1038/nclimate1627
Xie, J., Zhao, C., Han, Q., Zhou, H., Li, Q., and Diao, X. (2017). Effects of pyrene exposure on immune response and oxidative stress in the pearl oyster, Pinctada martensii. Fish Shellfish Immunol. 63, 237–244. doi: 10.1016/j.fsi.2017.02.032
Zhan, Y., Zha, S., Peng, Z., Lin, Z., and Bao, Y. (2022). Hypoxia-mediated immunotoxicity in the blood clam Tegillarca granosa. Mar. Environ. Res. 177:105632. doi: 10.1016/j.marenvres.2022.105632
Keywords: oyster holobiont, oyster physiology, acidification, hypoxia, oyster reef
Citation: Hemraj DA, Falkenberg LJ, Cheung K, Man L, Carini A and Russell BD (2023) Acidification and hypoxia drive physiological trade-offs in oysters and partial loss of nutrient cycling capacity in oyster holobiont. Front. Ecol. Evol. 11:1083315. doi: 10.3389/fevo.2023.1083315
Edited by:
Jennifer L. Matthews, University of Technology Sydney, AustraliaReviewed by:
Mathias Wegner, Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research (AWI), GermanyLuke P. Miller, San Diego State University, United States
Copyright © 2023 Hemraj, Falkenberg, Cheung, Man, Carini and Russell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bayden D. Russell, YnJ1c3NlbGxAaGt1Lmhr
 Deevesh Ashley Hemraj
Deevesh Ashley Hemraj Laura J. Falkenberg
Laura J. Falkenberg Khan Cheung
Khan Cheung Lauren Man
Lauren Man Alessia Carini
Alessia Carini Bayden D. Russell
Bayden D. Russell