- 1Department of Biomedical and Veterinary Sciences, Virginia-Maryland College of Veterinary Medicine, Blacksburg, VA, United States
- 2Department of Fish and Wildlife Conservation, Virginia Tech, Blacksburg, VA, United States
- 3Department of Food Science and Technology, Virginia Tech, Blacksburg, VA, United States
- 4Centre for the Conservation of African Resources, Animals, Communities, and Land Use, Kasane, Botswana
Introduction: Campylobacter spp. infections are responsible for significant diarrheal disease burden across the globe, with prevalence thought to be increasing. Although wild avian species have been studied as reservoirs of Campylobacter spp., our understanding of the role of wild mammalian species in disease transmission and persistence is limited. Host factors influencing infection dynamics in wild mammals have been neglected, particularly life traits, and the role of these factors in zoonotic spillover risk is largely unknown.
Methods: Here, we conducted a systematic literature review, identifying mammalian species that had been tested for Campylobacter spp. infections (molecular and culture based). We used logistic regression to evaluate the relationship between the detection of Campylobacter spp. in feces and host life traits (urban association, trophic level, and sociality).
Results: Our analysis suggest that C. jejuni transmission is associated with urban living and trophic level. The probability of carriage was highest in urban-associated species (p = 0.02793) and the most informative model included trophic level. In contrast, C. coli carriage appears to be strongly influenced by sociality (p = 0.0113) with trophic level still being important. Detection of Campylobacter organisms at the genus level, however, was only associated with trophic level (p = 0.0156), highlighting the importance of this trait in exposure dynamics across host and Campylobacter pathogen systems.
Discussion: While many challenges remain in the detection and characterization of Camploybacter spp., these results suggest that host life traits may have important influence on pathogen exposure and transmission dynamics, providing a useful starting point for more directed surveillance approaches.
Introduction
Campylobacter spp. are a diverse group of gastrointestinal zoonotic pathogens that are responsible for an estimated 96 million cases of foodborne illness in humans annually (Kirk et al., 2015) an important cause of human illness in both economically developed and developing nations (Devleesschauwer et al., 2017). This pathogen group infects a wide range of animal hosts, although domestic species such as cattle and chickens are thought to be the largest known reservoirs of infection (Man, 2011; Wagenaar et al., 2013). Campylobacter jejuni and C. coli are the primary species associated with clinical disease in humans, but a growing number of Campylobacter spp. have been recognized as emerging human pathogens (Man, 2011). Risk factors for Campylobacter infection include consumption of poultry, household water from wells, and exposure to animals (Levesque et al., 2013; MacDonald et al., 2015). This exposure has typically focused on occupational contact with livestock, owning a dog, or allowing animals access to sleeping and food preparation areas (Levesque et al., 2013; Mughini Gras et al., 2013; Kirk et al., 2015; MacDonald et al., 2015; Osbjer et al., 2016). Although consumption of poultry is the most common risk factor in urban environments, increased odds ratios of campylobacteriosis in rural populations are associated with poultry husbandry (Potter et al., 2003), high ruminant density (Arsenault et al., 2012), and contact with wild bird feces (Kapperud et al., 2003). Attribution studies have been performed to examine genetic similarities between clinical cases and animal isolates, with some evidence of contribution from wildlife (French et al., 2009; Strachan et al., 2009). In some studies, wildlife has also been demonstrated to be a competent reservoir of Campylobacter, including important human pathogens such as C. jejuni and C. coli (Weis et al., 2016; Newell et al., 2017). Wildlife may be an important source of Campylobacter spillover to humans, but it is unclear which species present the greatest public health risk. Challenges in both detection and characterization continue to complicate progress in this area of research (Couturier et al., 2013; Frasao et al., 2017).
Life traits and species interactions with conspecifics and socio-ecological systems
Host life traits can have important influence on the way a particular host species interfaces with conspecifics, animal communities, humans, and the environment, creating or eliminating opportunities for pathogen transmission. For example, a study carried out in Northern Botswana found that antibiotic resistance is higher in carnivores and water-associated species, demonstrating that the way an organism interacts with its environment can alter pathogen transmission and carriage (Alexander et al., 2010; Jobbins and Alexander, 2015).
Multi-host pathogens, such as Campylobacter, can have altered transmission dynamics in urban landscapes increasing pathogen prevalence (Woolhouse et al., 2001; Bradley and Altizer, 2007) in relation to changes in interspecific contact rates, a key determinant of multi-host pathogen transmission (Woolhouse et al., 2001; Renteria-Solis et al., 2014). Indeed, urbanization is increasingly identified as a potential driver of infectious disease emergence as it can change host spatial dynamics and pathogen transmission patterns in ways that are unpredictable (Neiderud, 2015; Hassell et al., 2017). For example, urban landscapes may alter host-pathogen dynamics directly by changing the way pathogens move through a system, or indirectly through changes to host immunocompetence or behavior (Brearley et al., 2013; Brunton et al., 2020). Urbanization has been shown to increase individual dispersal between troops in the banded mongoose (Mungos mungo), which may heighten the transmission risk of pathogens that spread through close contact (Verble et al., 2021). Evidence of increased stress (typically measured by fecal cortisol) and immunosuppression in urban-associated wildlife, however, is mixed. Some species associated with urban landscapes have in fact demonstrated lower stress levels and more robust immune responses, likely associated with consistent resource availability (Audet et al., 2015; Lyons et al., 2017; Iglesias-Carrasco et al., 2020). This was demonstrated in a population of banded mongoose in northern Botswana, where access to anthropogenic food sources was associated with lower levels of circulating glucocorticoids (Laver et al., 2012).
While Campylobacter occurs in many wildlife species, clinical signs of gastrointestinal illness or adverse health effects have rarely been observed. Two noctule bats (Nyctalus noctula; Hazeleger et al., 2018), a dozen rhesus monkeys (Macaca mulatta; Kalashnikova et al., 2002), and several vervet monkeys (Chlorocebus pygerythrus; Ngotho et al., 2006) infected with Campylobacter spp. presented with signs of acute gastrointestinal disease. These infections have the potential to cause mortality, with one wild vervet monkey dying from suspected antibiotic-resistant Campylobacter infection during laboratory quarantine, while several other monkeys in poor condition were euthanized (Ngotho et al., 2006). This outcome suggests that some free-ranging wildlife may be susceptible to acute Campylobacter infections, potentially affecting the quantity of pathogen shed into the environment and subsequent transmission to other animals and humans (Chaban et al., 2010). Even in the absence of acute signs of disease, however, evidence suggests that Campylobacter spp. infection in wildlife can affect body mass and overall health (Taff and Townsend, 2017).
Campylobacter transmission at the human-wildlife interface is also likely influenced by the intrinsic properties of the pathogen itself, including its persistence within infected animals across time (Inglis et al., 2004). In a study of clinical Campylobacter isolates, it was found that there is extensive evolutionary potential due to frequent recombination events, and that genetic exchange occurs regularly between C. coli and C. jejuni, important human pathogens (Wilson et al., 2009). This exchange may be accelerated by human activity and could increase the host range of these organisms (Sheppard et al., 2008; Waldenstrom et al., 2010; Lawton et al., 2018; Wei et al., 2019). For example, C. jejuni sequence type (ST)-137 was found to be widespread across wild bird taxa and has been implicated more recently in sporadic cases of campylobacteriosis (Wei et al., 2019). Hypervariable regions have also been discovered in C. jejuni that likely contribute to rapid evolutionary change, host range expansion, and antibiotic resistance (Parkhill et al., 2000).
A worrying trend is the emergence of multi-drug resistant (MDR) Campylobacter. In a study that took place in the United Kingdom, MDR C. jejuni was found to be widespread in poultry (Lopes et al., 2019). As livestock waste is considered an important reservoir of antibiotic resistance genes (ARGs; He et al., 2020), the risk of ARGs spreading to wildlife through this route is substantial. However, even in the absence of antibiotic use, high levels of resistance have been found in Campylobacter isolated from wild birds (Molina-Lopez et al., 2011; Du et al., 2019). Examples of this phenomenon are also observed in domestic species. For instance, 15% of Campylobacter isolates from a population of Australian chickens were fluoroquinolone-resistant without any history of exposure to this antibiotic class (Abraham et al., 2020). It has been demonstrated that gyrA mutations conferring fluroquinolone resistance are particularly stable and unlikely to incur fitness costs in Campylobacter (Luo et al., 2005; Luangtongkum et al., 2009). There are additional mechanisms that may allow for the persistence of antibiotic resistance genes in the absence of selective pressure, such as fitness-enhancing compensation from other regions of the genome and co-selection with genes that confer an evolutionary advantage (Melnyk et al., 2015). Monitoring antibiotic resistance in Campylobacter will remain key in reducing its spread and preserving the therapeutic agents available to treat disease.
While wildlife is considered an important reservoir of Campylobacter spp., our understanding of infection and transmission dynamics is mostly limited to avian species. The current body of evidence of Campylobacter among mammalian wildlife hosts is inadequate to fully utilize a One Health approach to disease control strategies. Here, we review and summarize the literature concerning Campylobacter infection in wildlife hosts, identifying and expanding our current understanding of Campylobacter in mammalian wildlife species. Using infection detection data, we assess the potential influence of host life history traits such as trophic level, urban association, and sociality on the probability of Campylobacter carriage in wildlife species. We discuss these results and study limitations as well as implications to Campylobacter transmission dynamics and surveillance and management approaches.
Methods
Literature review
This study followed guidelines provided under the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA), a framework for the standardization of reviews and meta-analyses (Page et al., 2021). Briefly, published studies of Campylobacter in wildlife were retrieved from online scientific search engines (Web of Science, PubMed, Google Scholar) using the key words: ‘wildlife + Campylobacter’ and ‘wild mammals + Campylobacter.’ Only the first 300 records from each query in Google Scholar were screened, as this resource was considered a secondary database to capture articles not located on Web of Science or PubMed (Haddaway et al., 2015; Gusenbauer and Haddaway, 2020). Duplicate records were removed automatically using EndNote 20, a reference manager (The EndNote Team, 2013). Reference lists of pertinent publications were searched for additional related articles. Information was retrieved on the location of sample collection, order, genus, and species studied, animal status (captive, wild feces or wild carcass), detection of Campylobacter spp. and particular Campylobacter species identified, and method of testing used (culture and PCR or other molecular methods).
Classification of life traits
Mammals that had Campylobacter detection data were categorized by trophic level (herbivore, omnivore, and carnivore) and sociality using the PanTHERIA database (Jones et al., 2009) and the Animal Diversity Web (ADW; Myers et al., 2019). In cases where species are occasionally omnivores but are primarily herbivores or carnivores, the predominant trophic level was chosen for analysis. Sociality of the species was defined as either social or solitary, depending on whether a species lived alone or within a social group (Myers et al., 2019). Mammals were also classified based on their association with urban areas. The classification system from Santini et al. (Santini et al., 2019) was used to classify species into urban ‘dwellers’ and urban ‘visitors’, with species meeting more than one criterion labeled as both (i.e., ‘dwellers\visitors’). Per this classification system, species that are classified as urban ‘dwellers’ have been demonstrated to utilize urban areas for all resources, while ‘visitors’ may exist in urban areas for some parts of their life cycle, but still obtain most resources from natural environments. Urban association for species that were not included in the Santini et al. (Santini et al., 2019) review were searched for using ‘taxon name (genus and species) + urban, urbanization or humans’ (Supplementary Table S1). An additional category was added for purposes of this study to classify species that had no urban interaction, which were labeled as urban ‘avoiders’ based on an absence of urban interaction in the literature. Several marine and alpine species were assumed to be avoiders due to habitat needs that did not overlap with urban areas. Information on consumption of species by humans was collected from the IUCN Red List (IUCN, 2019) and Mildenstein et al. (2016) for some bat species.
Statistical analyses
Once data had been compiled, duplicate records were removed from the original database through an automated process using EndNote 20 (The EndNote Team, 2013). If the same species was studied in two different regions of the world, and no subspecies are apparent, only one entry for this species was kept in our database. Due to the significant differences in sample sizes from different manuscripts (sample sizes varied from one to 1,168; Sus scrofa), we used a dummy categorical variable for Campylobacter presence in wildlife species, with 1 indicating that Campylobacter spp. had been detected (culture and/or polymerase chain reaction (PCR) based methods) or 0 indicating no Campylobacter spp. detection. This dummy variable was used to create graphs and carry out statistical analyses of the data. If at least one study of a species reported the detection of Campylobacter spp., it was recorded as present for the purposes of analysis. For studies that only identified Campylobacter to the genus level or where the authors did not attempt to detect C. jejuni or C. coli, no data was recorded at the species level and this field was left blank. All statistical analyses were conducted in R 4.1.2, an open source integrated programming environment (R Core Development Team, 2022). To investigate the effect of the categorical input variables (trophic level, urban association, and sociality) on the binary presence/absence of Campylobacter, C. jejuni, and C. coli, a series of logistic regression (LR) models were created in order to determine the ability of a model to predict carriage of either Campylobacter genus or the two species of Campylobacter analyzed in this study (C. jejuni and C. coli). An important assumption of LR models is that multicollinearity does not exist among the predictors. The generalized variance inflation factor (GVIF) was used to assess for multicollinearity in the predictor variables of all models used for analysis. GVIF is generally corrected by ½ degrees of freedom (df) when applied to categorical variables, and a cutoff of 1.73 following (Usio, 2020) was used, with independent variables below this threshold having little-to-no correlation. To obtain the GVIF1/2df value, we used the vif function of the car package (Fox et al., 2012). No independent variables in the models had GVIF1/2df values significantly above 1, indicating that multicollinearity was not present in this dataset.
We use the Akaike information criterion (AIC) to score and select the model with the best fit for the data balanced against the number of model parameters included (Aho et al., 2014). For this analysis we use the AICc, which is a sample size adjusted formula to calculate AIC (Bedrick and Tsai, 1994), due to the relatively low sample size (n = 128). AICc values were generated using AICcmodavg (Mazerolle and Mazerolle, 2017) and the model with the lowest AICc value was chosen as the best fit and used in additional analyses. The predict function in R 4.1.2 was used to estimate the probability of pathogen carriage for each level of the factors included in the in our top model. Data were visualized and graphed using ggplot2 (Wickham et al., 2016). Summary statistics were generated using the janitor package (Firke, 2021).
Results
We identified 65 published papers (1985–2021) and included these in our analysis representing 128 different free-ranging mammal species across 38 families and 9 orders. The plurality of studies was conducted on European wildlife (n = 30), but many world regions, including North America (n = 11), South America (n = 3), the Caribbean (n = 4), the Middle East (n = 2), Africa (n = 6), Asia (n = 6), Oceania (n = 1) and Antarctica (n = 2) were represented (See Supplementary materials S2), having at least one published study on Campylobacter spp. in mammalian wildlife. Campylobacter spp., C. jejuni, and C. coli carriage was detected by culture (n = 56), PCR (or other molecular methods; n = 6), or both (n = 2). One study used microarray for detection of Campylobacter (Jaing et al., 2015). Additionally, two of the studies that utilized culture techniques leveraged matrix-assisted laser desorption/ionization (MALDI-TOF) mass spectrometry for species identification. Four papers were included that investigated Campylobacter spp. in wildlife carcasses destined for human consumption. A full list of species, references, and associated data included in our statistical analyses are listed in Supplementary Table S2. As a species was often covered by multiple studies, there were many cases of redundancy in data that were removed for the analysis. An additional 11 studies analyzed Campylobacter spp. presence in captive wild mammals. These studies were excluded from statistical analysis but included below due to the added value they provide in understanding Campylobacter spp. carriage in wildlife species.
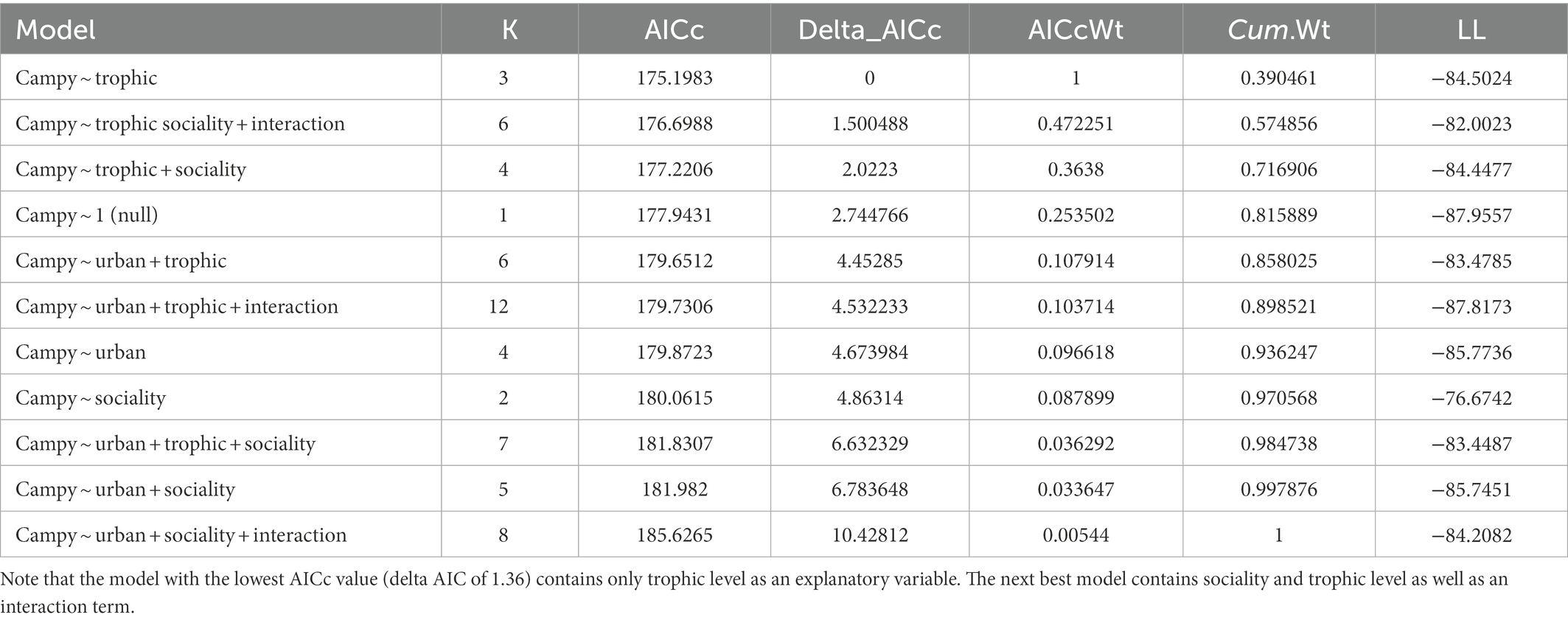
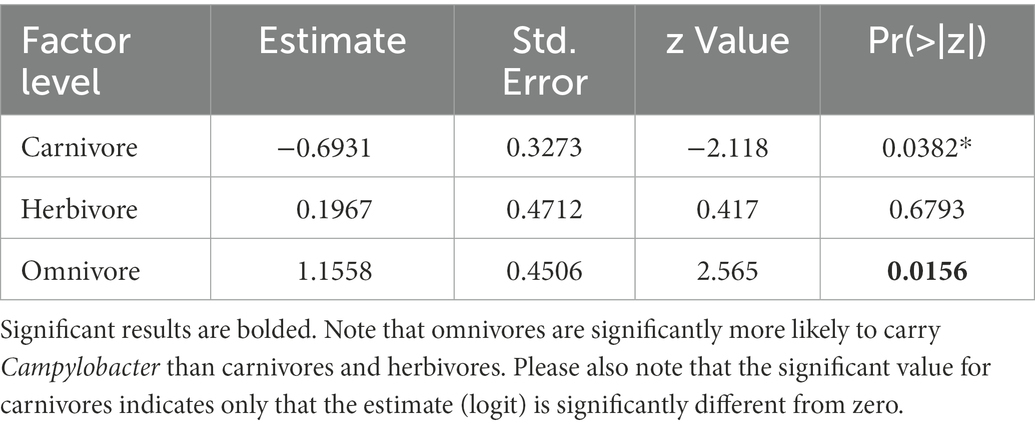
Campylobacter spp. presence was only correlated with trophic level in the model containing the highest explanatory power (lowest AICc, ΔAICc value of 1.5; Table 1). Based on this model, omnivores are significantly more likely to carry the Campylobacter genus than either herbivores or carnivores (p value = 0.0156, Table 2). Given the breath of Campylobacter and host species included in the genus level analysis, this finding suggests that trophic position has an important cross-species influence on exposure dynamics for this pathogen group. Urban association and sociality life traits would appear to have a more heterogenous influence across the various Campylobacter pathogen-wildlife host systems.
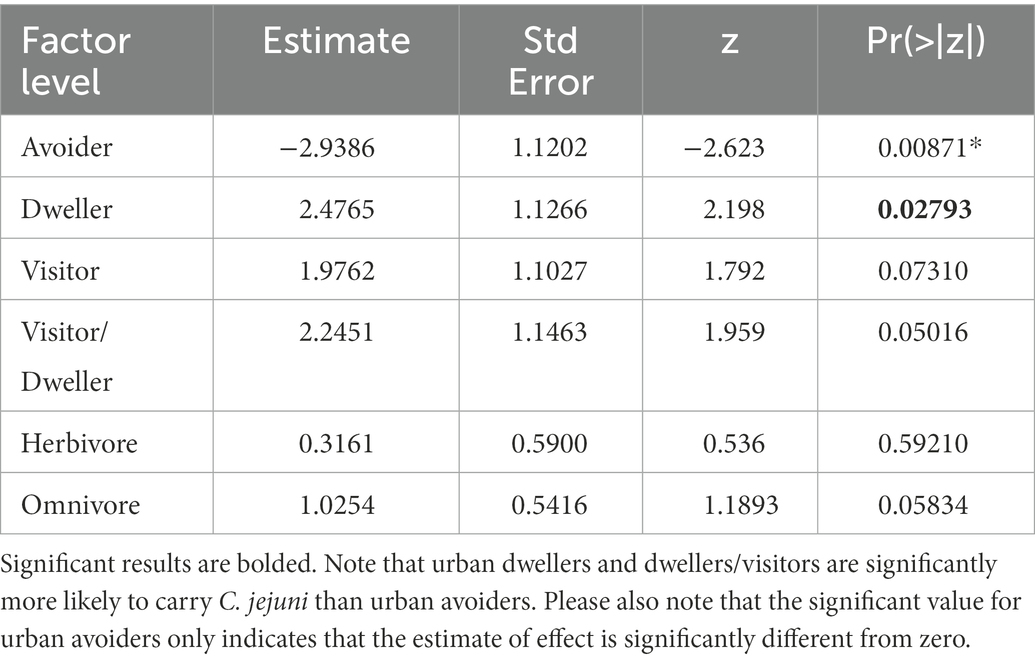
C. jejuni presence was best predicted by urban association and trophic level (ΔAICc value of 1.17, Table 3). There was not a significant interactive effect, as the model with an interactive term performed worse than a null model (carriage predicted by 1). Based on the model containing urban association and trophic level, urban dwellers were significantly more likely to be positive for C. jejuni than urban avoiders (p value = 0.0279, Table 4). While trophic level is included in the model with the lowest AICc value and omnivores seem to differ from carnivores in their likelihood of carrying C. jejuni, this effect was not quite to the level of statistical significance (p = 0.058, Table 4). This may be due to the relatively low sample size used in the analysis.
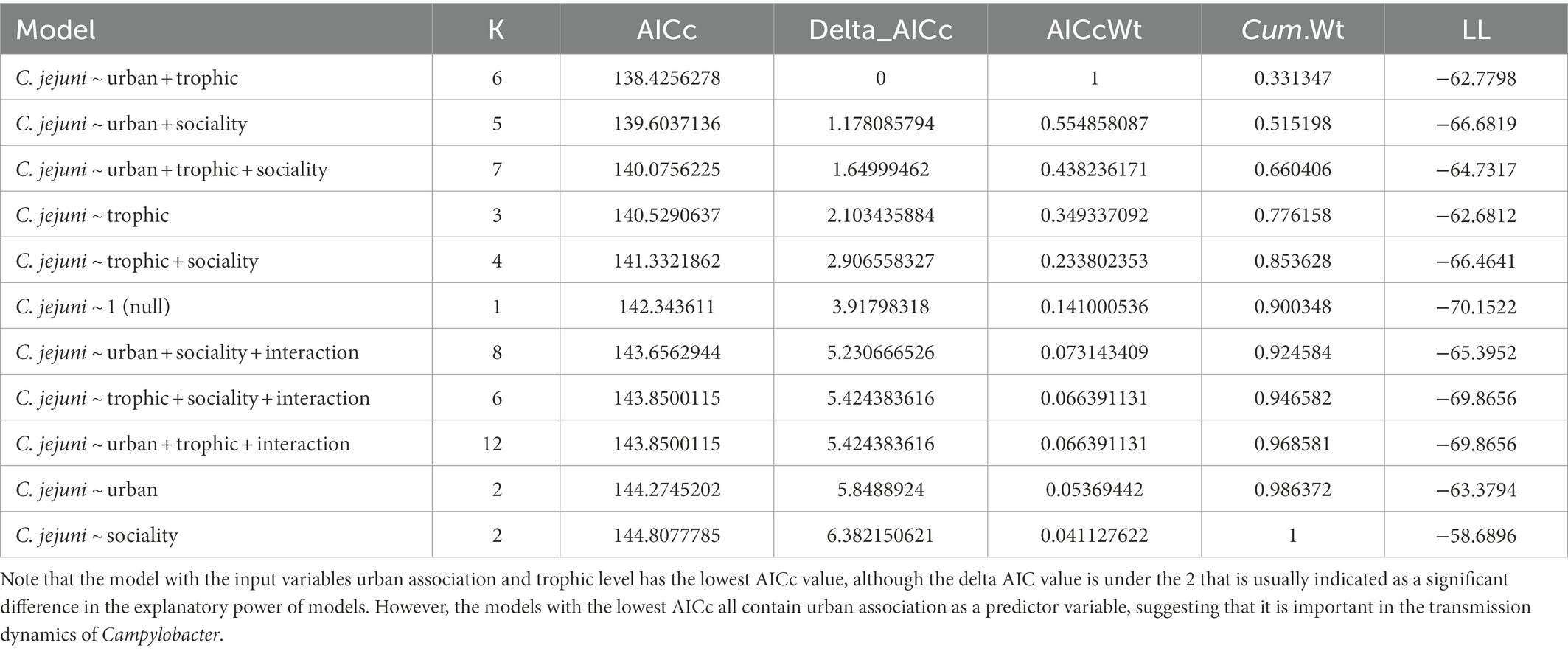
Table 3. AICc values for logistic regression models with C. jejuni presence/absence as the output variable.
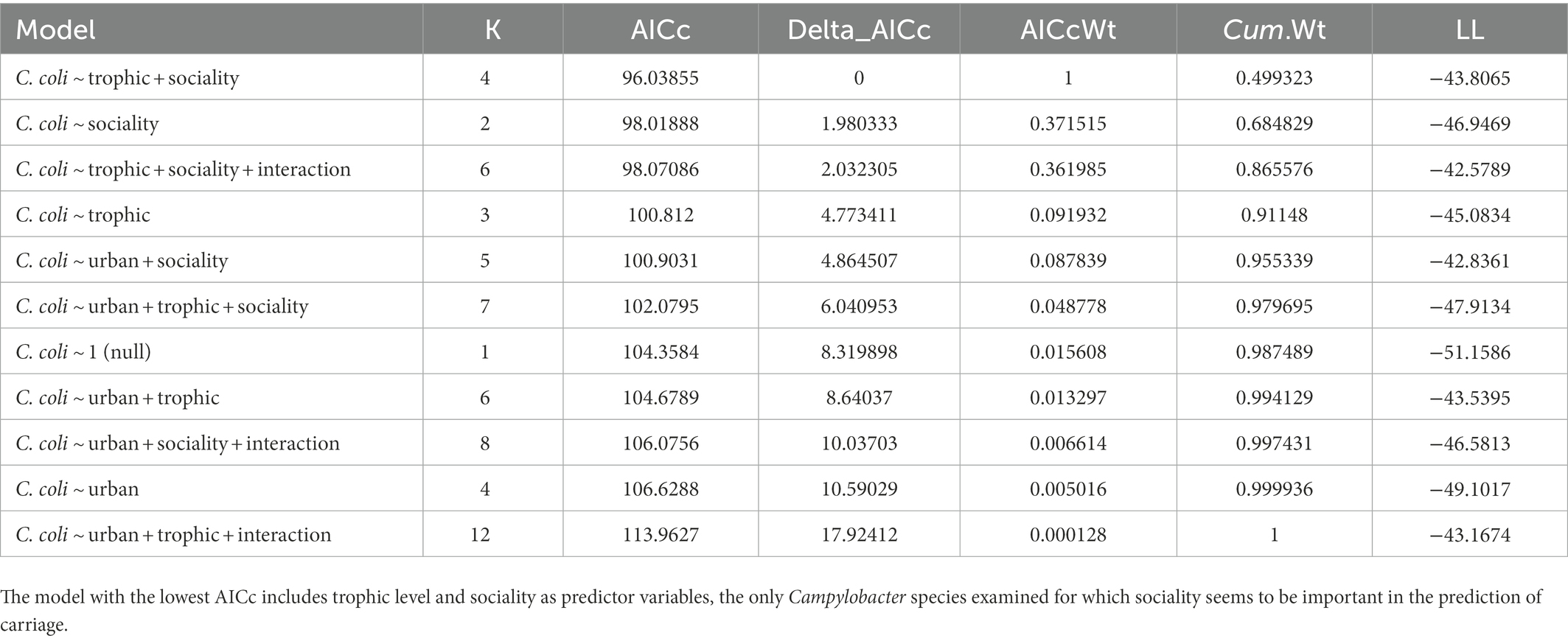
C. coli carriage was highly correlated with both sociality and trophic level in this analysis, as the model with the AICc included both variables (Table 5). Omnivores were significantly more likely than carnivores to carry the pathogen (p = 0.0282), with herbivores showing no significant difference from carnivores (p = 0.0977). Solitary species were less likely than social species to carry C. coli in our analysis (p = 0.0113, Table 6). Much like our analysis of C. jejuni, the interaction between these two variables does not appear to be significant in prediction of carriage.
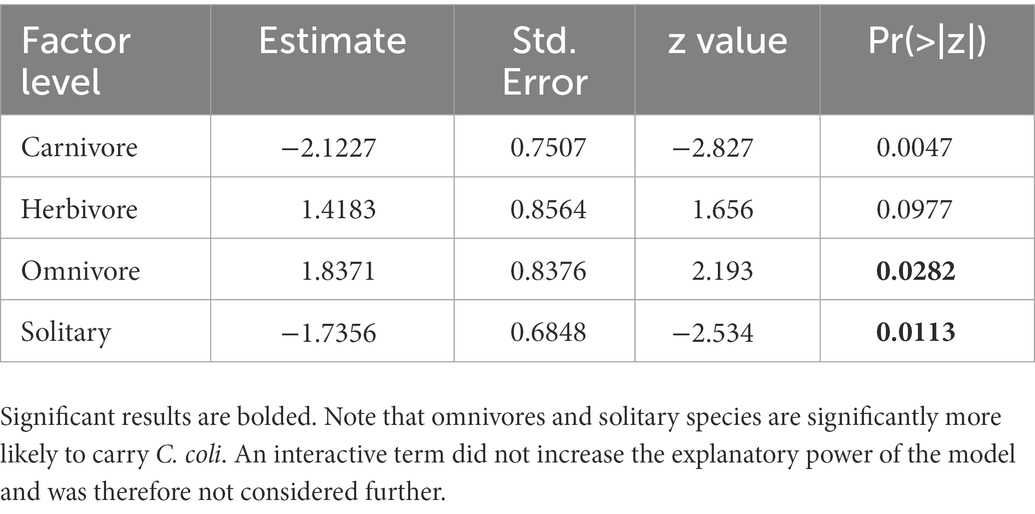
Table 6. Logistic regression output for the model with the highest explanatory power (C. coli ~ sociality + trophic).
Discussion
Across 65 reviewed publications meeting our criteria, infection detection was identified across a wide array and included 128 species with varying life traits and Campylobacter infection status. Our analysis indicates that life traits can have an important influence on the probability of Campylobacter carriage and transmission potential with dynamics in wildlife likely complex, involving interdependent interactions between host, pathogen, and the environment. Summary statistics by life history trait are provided in Table 7. Host factors driving infection in wildlife are multifaceted with life traits, such as sociality and trophic level appearing to influence disease transmission dynamics (Kriger and Hero, 2007; Johnson et al., 2012; VanderWaal et al., 2014). As an example, it has been demonstrated that parasite diversity is higher in bird species that utilize a wider variety of food sources (Gutiérrez et al., 2019). Host space use has been identified as an important factor influencing pathogen transmission and persistence dynamics (VanderWaal and Ezenwa, 2016). Similarly, trophic level and diet can alter pathogen exposure dynamics, with previous studies identifying an association between pathogen prevalence and carnivores in some multi-host pathogen systems (Moore et al., 2010; Hollings et al., 2013) which has recently been proposed as a form of accumulation due to the diversity of pathogen exposure from prey (Malmberg et al., 2021). Carnivores and water-associated species were shown to have a higher prevalence of antibiotic-resistant E. coli in a previous study (Jobbins and Alexander, 2015), further reinforcing the importance of life history traits in the evaluation of infectious disease dynamics.
Life history traits
Trophic level
Diet can provide a critical influence on infectious disease exposure and transmission (Fischer et al., 2005) with results from our meta-analysis indicating that they may influence Campylobacter spp. transmission patterns in wildlife as well. Omnivores are more likely to carry Campylobacter than herbivores or carnivores (Figure 1), which may be related to differences in host foraging behavior or bacterial survival rates in different environments. Trophic level did not seem to be as important in the transmission of C. coli, although the best performing model in the prediction of C. jejuni carriage included this variable, suggesting that it may be important for this species or that there is an interaction between trophic level and urban association. A model including an interactive term, however, did not reveal a significant interaction between these variables and did not perform as well in model comparison as the model without the interactive term (Table 2).
Urbanization
Urbanization did not appear to predict carriage of the Campylobacter genus, based on our analysis. However, wildlife species that utilize the urban landscape (urban dwellers) were significantly more likely using logistic regression to carry C. jejuni than urban avoiders (Figure 2). C. jejuni is considered an important zoonotic human pathogen and is ubiquitous in domestic poultry. Urban dwellers are more likely than avoiders to ingest waste from humans and domestic animals infected with C. jejuni. This has been demonstrated in wild birds, with those feeding on refuse more likely to carry Campylobacter than those that forage on natural food sources (Kapperud and Rosef, 1983). Additionally, C. jejuni has been found in raccoons and civets inhabiting urban centers (Lee et al., 2011), rodents overwintering in human dwellings (Lõhmus and Albihn, 2013) and even in wildlife feces on playgrounds (French et al., 2009; Abdollahpour et al., 2015). However, the occurrence of C. jejuni in wildlife species is not exclusively dependent on transmission from human sources. Wild birds are also known carriers of C. jejuni, having their own distinct C. jejuni subtypes (Ramonaite et al., 2014), and may act as a reservoir for other wildlife species in these environments. Wild-bird associated C. jejuni is also commonly found in water sources (Mughini-Gras et al., 2016) and may be responsible for a small fraction of human campylobacterosis cases annually (Cody et al., 2015). In a recent study, Persian fallow deer (Dama mesopotamica) in a wildlife refuge were found to have a high prevalence of Campylobacter spp. (52.3%, 33/63) and C. jejuni (27.0%, 17/63), with exposure to water sources used by migratory birds thought to be the primary source of infection (Khoshbakht et al., 2015). Interestingly, C. coli carriage was not associated with how an organism utilizes the urban environment, with no difference found between urban avoiders and dwellers or visitors/dwellers. Campylobacter coli is primarily associated with swine production systems, and previous studies have identified an overall reduced risk of C. coli infection in humans who live in urban areas (Roux et al., 2013) consistent with the limited number of swine expected in these environments.
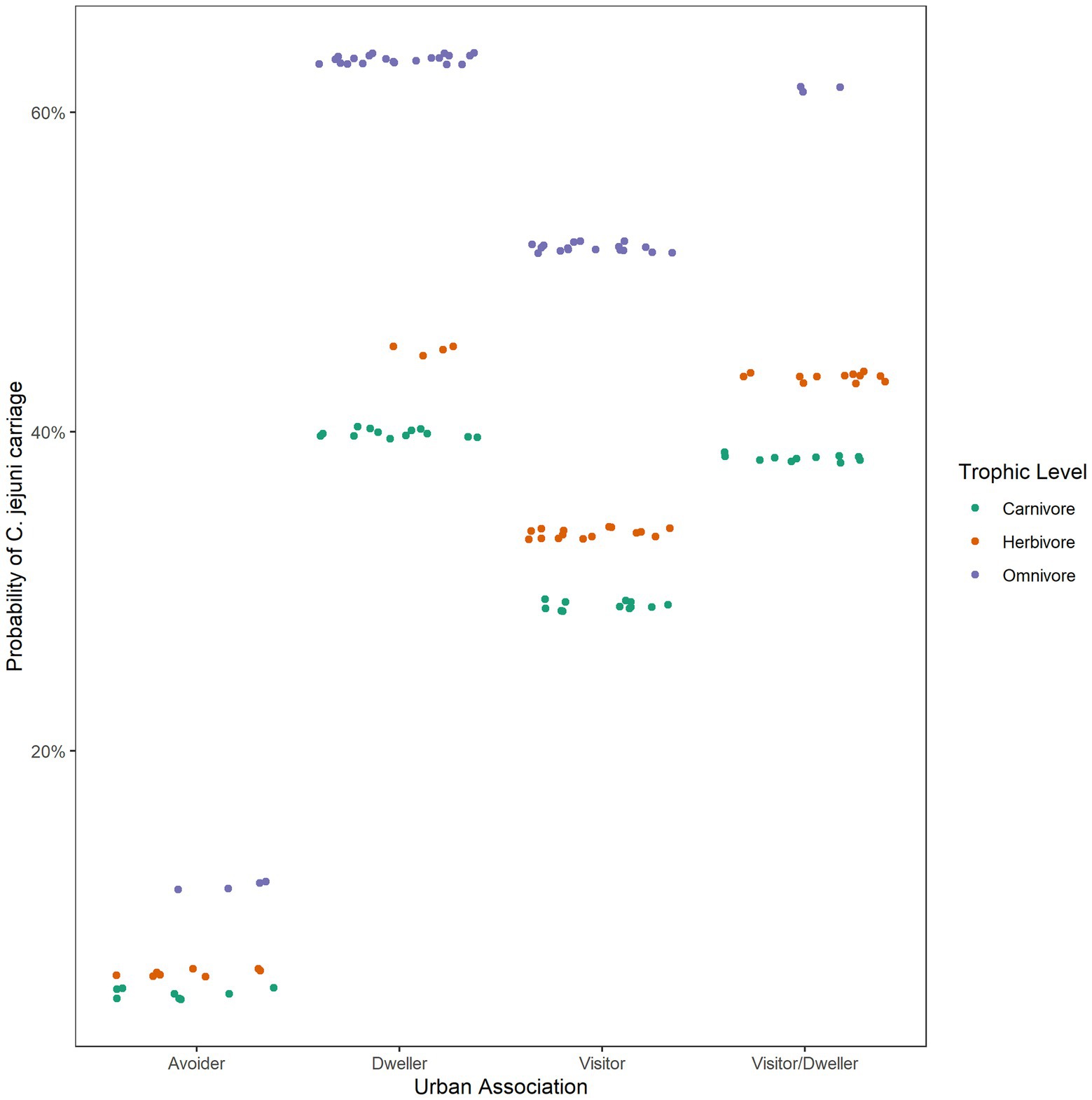
Figure 2. Probability of C. jejuni carriage at different trophic levels and degrees of urban association.
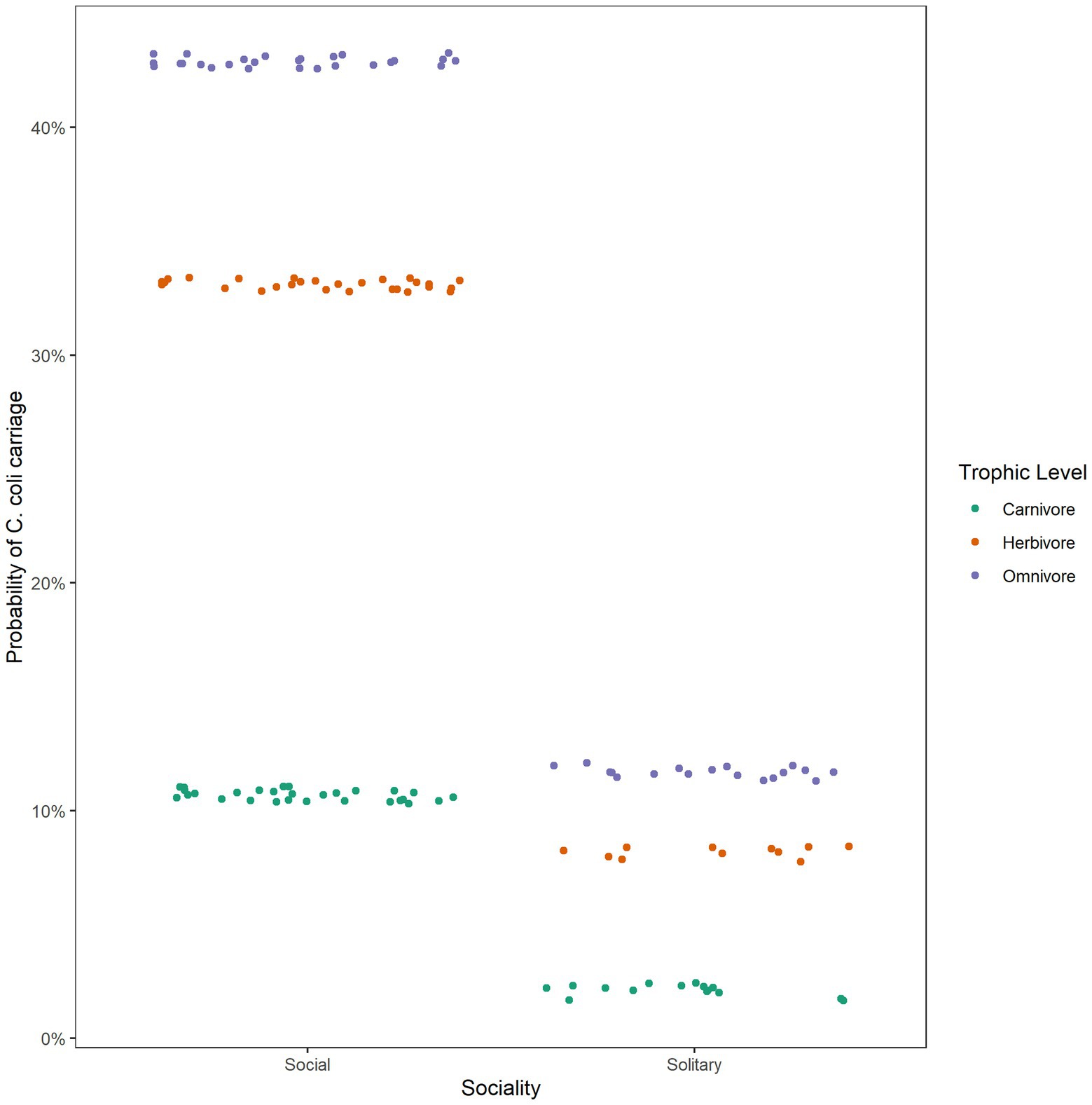
Sociality
Sociality did not appear to be important in the carriage of Campylobacter at the genus level or in the carriage of C. jejuni. Interestingly, from our analysis it has demonstrated importance in the carriage of C. coli, with social species more likely to carry this pathogen (Figure 3). A study of C. coli prevalence in feral pigs found a significant difference between males and females, with males more likely to carry the bacteria (Cummings et al., 2018). It was postulated that this was due to the social structure of pigs, as males are more likely to be solitary. Our results indicate that solitary species are less likely to carry C. coli, but there may exist heterogeneity across species, geographic areas, and a host of other factors. Different risk factors have been associated with C. coli and C. jejuni in human infections, including age, season and location (Gillespie et al., 2002; Roux et al., 2013), which implies that different risk factors could also be associated with C. coli in wildlife. Similar differences in transmission pathways may also explain why C. coli was more common in social than solitary species.
Public health implications
Consumption of wild game/bush meat
Overall, we found that almost 50% of wildlife species used as human food resources were positive for Campylobacter spp., with 31% positive for C. jejuni and 26% positive for C. coli, highlighting an important public health concern. This is a concern that is not entirely restricted to the developing world, however, with many developed countries having high prevalence of the pathogen group. For example, it was found that 9.1% of diarrheic patients in Alberta over a one-year period were positive for Campylobacter (Inglis et al., 2021). Additionally, some wildlife species consumed by humans contained antimicrobial resistant (AMR) strains of Campylobacter spp., which may have important implications to AMR transmission dynamics and public health (Sasaki et al., 2013).
Although there is an association between Campylobacter spp. presence and wildlife consumed by humans, there is not strong evidence from the literature implicating game meat as a significant public health threat. Campylobacter spp. were only rarely detected in moose (Alces alces; 6%) and deer (Odocoileus virginianus; 2%) carcasses in Finland (Sauvala et al., 2019), hares (Lepus timidus; 4.3%) in Norway (Rosef et al., 1983), and were not detected in any deer carcasses (Capreolus capreolus and Cervus elaphus) in Germany (Atanassova et al., 2008). Pyrenean chamois (Rupicapra pyrenaica) a game species harvested was also not found to harbor Campylobacter spp. (Espunyes et al., 2021) in a study conducted in Spain. Campylobacter was not detected in monkey, porcupine, duiker or river hog muscle meat (n = 104) in Gabon (Bachand et al., 2012). Results from wild boar studies have been more varied, with some detecting no or low Campylobacter spp. prevalence (Atanassova et al., 2008; Wacheck et al., 2009; Cummings et al., 2018), and others detecting high Campylobacter spp. prevalence (Jay-Russell et al., 2012; Díaz-Sánchez et al., 2013; Sasaki et al., 2013). It also may be that wild boar carry different Campylobacter spp. not detected by traditional culture methods. For example, a study in Spain found that 66% (188/287) of wild boar were positive for Campylobacter spp., but only one was positive for C. jejuni and no isolates were positive for C. coli or C. lari (Díaz-Sánchez et al., 2013). In contrast, another study in Spain identified C. lanienae as the dominant species in wild boar, making up 67.3% (34/49) of Campylobacter isolates (Navarro-Gonzalez et al., 2014), and found that it was significantly higher (p < 0.01) in wild boar than free-ranging cattle (Navarro-Gonzalez et al., 2014). Campylobacter lanienae has primarily been isolated from livestock and slaughterhouse workers, and its pathogenicity and role as a human gastrointestinal pathogen is not well understood (Guévremont et al., 2008; Oporto and Hurtado, 2011). Other species commonly isolated from wild boar include C. hyointestinalis (Sasaki et al., 2013), while C. coli and C. jejuni have been consistently less common (Jay-Russell et al., 2012; Cummings et al., 2018).
If there is a public health concern from wild game, it looks to be associated with cross-contamination during butchering and processing as in most food systems. For example, Stella et al. (2018) found that although only 16.7% (5/30) of wild boar carcass samples were positive for Campylobacter, 51.8% (29/56) of caecal contents were positive, and two carcass samples that were positive did not have positive caecal content. Further, processed wild game meat that was smoked in the Democratic Republic of Congo was contaminated with Campylobacter spp. in 11.1–26.6% of buffalo (Syncerus caffer), common duiker (Sylvicapra grimmia), and desert warthog (Phacochoerus aethiopicus) samples (Mpalang et al., 2013). Although the risk of infection from consuming wild game meat is lower than that from poultry, communities who rely heavily on this food source should exercise caution during butchering to reduce exposure.
Wildlife transmission to livestock and humans
As Campylobacter is primarily a foodborne pathogen, a pressing concern is the role of wildlife in pathogen transmission to domestic species commonly consumed by humans. There is conflicting evidence regarding the role of wildlife in Campylobacter transmission to livestock and poultry species. In one study from Spain, Campylobacter spp. from wild boar were deemed an unlikely reservoir for cattle because they each had their own dominant species: C. lanienae in boar and C. jejuni in cattle (Navarro-Gonzalez et al., 2014). In other studies, a low prevalence in wildlife coupled with minimal genetic similarity between wildlife and livestock genotypes suggests that transmission between wildlife and livestock is infrequent (French et al., 2005; Jensen et al., 2006; Meerburg et al., 2006; Sippy et al., 2012; Olkkola et al., 2021). When wildlife-specific strains were compared to generalist strains, there were genetic regions missing from wildlife-specific strains that may explain why they are unable to colonize a wide host range (Hepworth et al., 2011). Although Viswanathan et al. (2017) found most C. jejuni isolates phylogenetically grouped into distinct wildlife and livestock clusters, 31.25% (5/16) of dairy cattle isolates and 17.6% (3/17) of beef cattle isolates clustered with small mammal (raccoons, skunks and mice) isolates collected from farms, suggesting that transmission is occurring between wildlife and livestock in this landscape type. It may be that some wildlife species are more likely to be infected with livestock and human strains than others, as a study from wildlife and livestock in the United Kingdom found little genetic overlap between cattle and wild birds, but rabbit isolates were genetically similar to cattle isolates (Kwan et al., 2008).
Additional studies have identified identical Campylobacter spp. clones from rodents, sparrows, flies and livestock (Adhikari et al., 2004), and high genetic similarity between Campylobacter spp. isolated from wild birds, mice and poultry (Hiett et al., 2002). This genetic similarity across isolates can create increased transmission risk to poultry flocks, as an environment contaminated with feces from other animals is a risk factor for poultry infection with Campylobacter spp. (Ellis-Iversen et al., 2012). An increased transmission risk to poultry flocks may also increase risk to humans, as demonstrated by a study in England which found significant overlap of Campylobacter sequence types between animals and humans (Wilson et al., 2008). It is also worth noting that Campylobacter has been demonstrated to form distinct genetic clusters that differ in pathogenicity (Carvalho et al., 2001). If such clusters are found circulating in animal species, they may represent a heightened risk to associated humans.
It is unclear whether wildlife may acquire Campylobacter spp. infections from livestock and/or the environment. In a study of wild rodents across chicken and pig farms, Campylobacter species in rodents varied between farm type, with C. jejuni more common on poultry farms and C. coli more common on pig farms (Backhans et al., 2013). A survey of omnivorous mesopredators in Canada (skunks, raccoons, and opossums) associated with swine farms revealed the presence of C. jejuni and C. upsaliensis, but not C. coli which is typically associated with swine (Bondo et al., 2019). Further analysis of mammalian wildlife and livestock species that incorporates both behavior and phylogenetic analysis of Campylobacter spp. is necessary and has demonstrated value in wild bird studies (Taff et al., 2016). It is difficult to make broad conclusions on the role of wildlife in agricultural Campylobacter transmission dynamics as most studies have been limited to the United States and Europe. Food production systems are increasinly globalized, so it will be important to understand the role of the various wildlife species across landscapes (Van Nierop et al., 2005; Suzuki and Yamamoto, 2009; Adzitey et al., 2012; Perdoncini et al., 2015).
Limitations and knowledge gaps
This study provides a broad overview of Campylobacter spp. infection in free-ranging mammalian wildlife. Statistical analyses in this study were based on presence/absence and did not include Campylobacter prevalence data, removing some of the complexity of the original datasets. Prevalence often varied greatly between species and studies, ranging between 1 to 87.5%. These differences in prevalence likely create differential transmission risks, a potential area of future research. Culture and detection methods also varied greatly between the studies which can have a significant impact on the prevalence, species and genotypes isolated (Williams et al., 2012; Hetman et al., 2020). Detection methods often are biased toward C. jejuni and C. coli isolation which can skew the representation of Campylobacter species present (Lastovica and le Roux, 2000). Importantly, there have been relatively few studies about Campylobacter upsaliensis, especially in free-ranging wild mammals. This species is suspected to be an emerging human pathogen and has been isolated from domestic animals such as dogs with some frequency (Bourke et al., 1998; Man, 2011; Couturier et al., 2012). The age and storage of feces also impacts Campylobacter isolation, with fresh fecal samples significantly more likely to likely to be positive for Campylobacter spp. than older feces using culture methods (Abdollahpour et al., 2015). In a study performed using sheep feces sampled and placed back into the environment (Jackson et al., 2009), the authors found that Campylobacter died rapidly. Another phenomenon in Campylobacter spp. that complicates isolation is the progression to a viable but non-culturable (VBNC) state under harsh conditions (Moriarty et al., 2011). This may be particularly relevant for Campylobacter isolation from wildlife samples, especially if samples are taken from remote sites. Culture methods have also been found to be inferior to immunoassay and molecular methods for sensitivity and specificity of Campylobacter detection (Buss et al., 2019).
Pathogen prevalence and transmission have been well studied in European and North American wildlife and bird species, but relatively few studies on Campylobacter prevalence and transmission in wildlife have occurred in developing countries where Campylobacter burden is highest (Troeger et al., 2017), transmission dynamics may differ (Platts-Mills and Kosek, 2014), and limited resources may increase the difficulty of investigations. There is also a lack of phylogenetic information on Campylobacter spp. in mammalian wildlife. Campylobacter spp. possess the ability to evolve rapidly (Wilson et al., 2009) which suggests that the relationship between wildlife-associated Campylobacter and human-associated Campylobacter may be obscured. Most papers that incorporate genetic or phylogenetic information rely on MLST techniques which have been the gold-standard for defining clonal groups but have been shown to be inferior at establishing epidemiological dynamics (Llarena et al., 2017). Only two recent papers incorporate WGS on free-ranging wildlife and both papers found human-associated Campylobacter genotypes in their respective wildlife species (Baily et al., 2015; Hazeleger et al., 2018). Future research should incorporate WGS methods into targeted studies of species at the human-wildlife interface that are most likely to contribute to Campylobacter transmission and human disease.
Conclusion
Multi-host pathogens, such as Campylobacter, have complex interdependent transmission dynamics that can be altered by landscape changes and host life traits. Our understanding of Campylobacter spp. in mammals at the human-wildlife interface is limited, but it is important to understand these dynamics to effectively model and predict transmission risk and public and animal health needs. As large-scale surveillance of Campylobacter is economically unfeasible, this study may optimize efforts by identifying the wildlife species most likely to contribute to Campylobacter infections in humans. In this study, we found that life traits significantly influenced the presence of Campylobacter spp. in free-ranging wildlife. Most importantly, urban association is strongly correlated with C. jejuni presence (Figure 4) and trophic level was important across all pathogens, with omnivores more likely to carry Campylobacter spp. Additionally, sociality is strongly correlated with C. coli presence. These results demonstrate that life traits must be leveraged as a powerful tool for identifying points of increased zoonotic spillover risk across the landscape. Campylobacter is a rapidly evolving pathogen at the human-animal interface; therefore, targeted monitoring of urban-associated species will be important for understanding transmission dynamics and protecting public health.
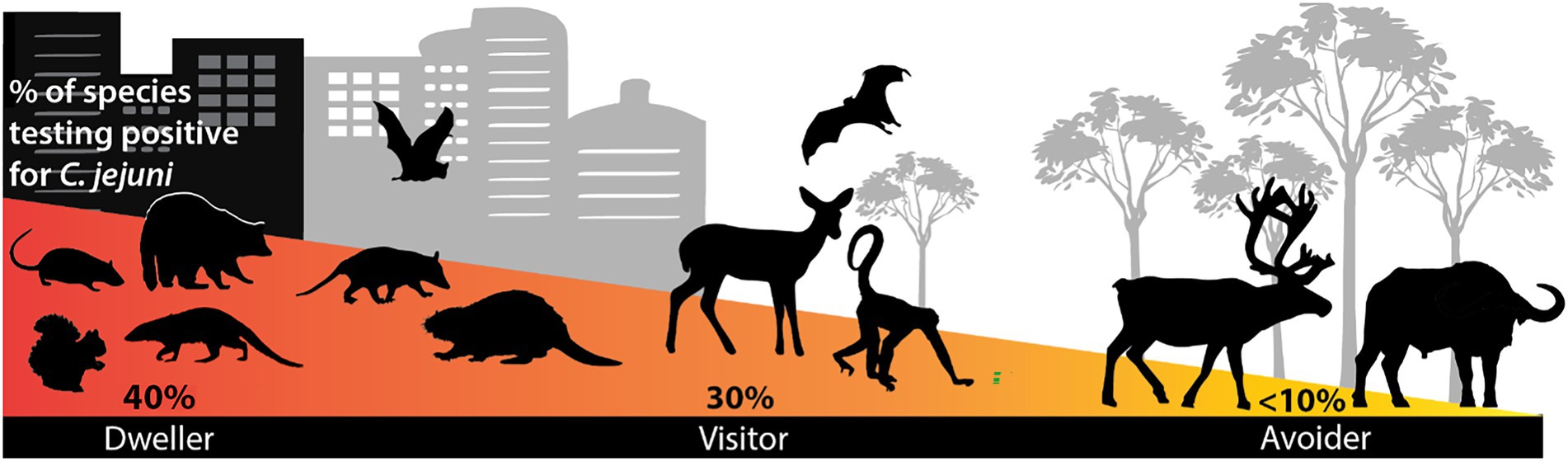
Figure 4. Difference in C. jejuni detection among wild mammalian species based on urban behavior. Urban dwellers had a higher probability of carrying C. jejuni than urban avoiders.
Author’s note
As demonstrated with the recent emergence of SARS-CoV-2 and the ensuing global pandemic, our ability to predict pathogen emergence remains extremely limited, influenced strongly by the complexity of interdependent factors that drive these processes. Using published literature and Campylobacter spp. infection detection in wild mammalian species, we identify the potential usefulness of key life traits that may predict a heightened risk of infection with this multi-host pathogen. Across host-pathogen systems, this approach may provide a useful framework for identifying host factors that influence pathogen transmission potential.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
KA: conceptualization, funding acquisition, project administration, and resources. MB, SM, and KA: data curation, methodology, software, visualization, and validation. MB and SM: formal analysis and Investigation. KA and MP: supervision. KA, MB, SM, and MP: writing – original draft preparation. KA, MB, and MP: writing – reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
Funding for this research was provided in part by the National Science Foundation (CNH2-L: Human waste and its role in creating at an integrated socioenvironmental system at an urban-wilderness continuum). Funding for MP was provided in part by the Virginia Agricultural Experiment Station and the Hatch Program of the National Institute of Food and Agriculture, U.S. Department of Agriculture. Funding for MB was provided by the National Institutes of Health, award #5T32OD028239–02.
Acknowledgments
The authors would like to thank Claire Sanderson for her assistance in the creation of figures and Stephen Warre for his feedback on statistical analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1070519/full#supplementary-material
References
Abdollahpour, N., Zendehbad, B., Alipour, A., and Khayatzadeh, J. (2015). Wild-bird feces as a source of Campylobacter jejuni infection in children's playgrounds in Iran. Food Control 50, 378–381. doi: 10.1016/j.foodcont.2014.09.007
Abraham, S., Sahibzada, S., Hewson, K., Laird, T., Abraham, R., Pavic, A., et al. (2020). Emergence of fluoroquinolone-resistant Campylobacter jejuni and Campylobacter coli among Australian chickens in the absence of fluoroquinolone use. Appl. Environ. Microbiol. 86, E02765–E02719. doi: 10.1128/AEM.02765-19
Adhikari, B., Connolly, J. H., Madie, P., and Davies, P. R. (2004). Prevalence and clonal diversity of campylobacter Jejuni from dairy farms and urban sources. N. Z. Vet. J. 52, 378–383. doi: 10.1080/00480169.2004.36455
Adzitey, F., Rusul, G., Huda, N., Cogan, T., and Corry, J. (2012). Prevalence, antibiotic resistance and rapd typing of campylobacter species isolated from ducks, their rearing and processing environments in Penang, Malaysia. Int. J. Food Microbiol. 154, 197–205. doi: 10.1016/j.ijfoodmicro.2012.01.006
Aho, K., Derryberry, D., and Peterson, T. (2014). Model selection for ecologists: the worldviews of Aic and Bic. Ecology 95, 631–636. doi: 10.1890/13-1452.1
Alexander, K. A., Mcnutt, J., Briggs, M., Standers, P., Funston, P., Hemson, G., et al. (2010). Multi-host pathogens and carnivore management in Southern Africa. Comp. Immunol. Microbiol. Infect. Dis. 33, 249–265. doi: 10.1016/j.cimid.2008.10.005
Arsenault, J., Berke, O., Michel, P., Ravel, A., and Gosselin, P. (2012). Environmental and demographic risk factors for Campylobacteriosis: do various geographical scales tell the same story? BMC Infect. Dis. 12:318. doi: 10.1186/1471-2334-12-318
Atanassova, V., Apelt, J., Reich, F., and Klein, G. (2008). Microbiological quality of freshly shot game in Germany. Meat Sci. 78, 414–419. doi: 10.1016/j.meatsci.2007.07.004
Audet, J.-N., Ducatez, S., and Lefebvre, L. (2015). The town bird and the country bird: problem solving and Immunocompetence vary with urbanization. Behav. Ecol. 27, 637–644. doi: 10.1093/beheco/arv201
Bachand, N., Ravel, A., Onanga, R., Arsenault, J., and Gonzalez, J.-P. (2012). Public health significance of zoonotic bacterial pathogens from bushmeat sold in Urban Markets of Gabon, Central Africa, bioone. J. Wildl. Dis. 48, 785–789. doi: 10.7589/0090-3558-48.3.785
Backhans, A., Jacobson, M., Hansson, I., Lebbad, M., Lambertz, S. T., Gammelgård, E., et al. (2013). Occurrence of pathogens in wild rodents caught on Swedish pig and chicken farms. Epidemiol. Infect. 141, 1885–1891. doi: 10.1017/S0950268812002609
Baily, J. L., Méric, G., Bayliss, S., Foster, G., Moss, S. E., Watson, E., et al. (2015). Evidence of land-sea transfer of the zoonotic pathogen campylobacter to a wildlife marine sentinel species. Mol. Ecol. 24, 208–221. doi: 10.1111/mec.13001
Bedrick, E. J., and Tsai, C.-L. (1994). Model selection for multivariate regression in small samples. Biometrics 50, 226–231. doi: 10.2307/2533213
Bondo, K. J., Pearl, D. L., Janecko, N., Reid-Smith, R. J., Parmley, E. J., Weese, J. S., et al. (2019). Salmonella, campylobacter, clostridium difficile, and anti-microbial resistant Escherichia coli in the faeces of sympatric meso-mammals in southern Ontario, Canada. Zoonoses Public Health 66, 406–416. doi: 10.1111/zph.12576
Bourke, B., Chan, V. L., and Sherman, P. (1998). Campylobacter upsaliensis: waiting in the wings. Clin. Microbiol. Rev. 11, 440–449. doi: 10.1128/CMR.11.3.440
Bradley, C. A., and Altizer, S. (2007). Urbanization and the ecology of wildlife diseases. Trends Ecol. Evol. 22, 95–102. doi: 10.1016/j.tree.2006.11.001
Brearley, G., Rhodes, J., Bradley, A., Baxter, G., Seabrook, L., Lunney, D., et al. (2013). Wildlife disease prevalence in human-modified landscapes. Biol. Rev. 88, 427–442. doi: 10.1111/brv.12009
Brunton, E. A., Clemente, C. J., and Burnett, S. E. (2020). Not all urban landscapes are the same: interactions between urban land use and stress in a large herbivorous mammal. Ecol. Appl. 30:E02055. doi: 10.1002/eap.2055
Buss, J. E., Cresse, M., Doyle, S., Buchan, B. W., Craft, D. W., and Young, S. (2019). Campylobacter culture fails to correctly detect campylobacter in 30% of positive patient stool specimens compared to non-cultural methods. Eur. J. Clin. Microbiol. Infect. Dis. 38, 1087–1093. doi: 10.1007/s10096-019-03499-x
Carvalho, A. C., Ruiz-Palacios, G. M., Ramos-Cervantes, P., Cervantes, L. E., Jiang, X., and Pickering, L. K. (2001). Molecular characterization of invasive and noninvasive Campylobacter jejuni and Campylobacter coli isolates. J. Clin. Microbiol. 39, 1353–1359. doi: 10.1128/JCM.39.4.1353-1359.2001
Chaban, B., Ngeleka, M., and Hill, J. E. (2010). Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals. BMC Microbiol. 10:73. doi: 10.1186/1471-2180-10-73
Cody, A. J., Mccarthy, N. D., Bray, J. E., Wimalarathna, H. M. L., Colles, F. M., Jansen Van Rensburg, M. J., et al. (2015). Wild bird-associated Campylobacter jejuni isolates are a consistent source of human disease, in Oxfordshire, United Kingdom. Environ. Microbiol. Rep. 7, 782–788. doi: 10.1111/1758-2229.12314
Couturier, B. A., Couturier, M. R., Kalp, K. J., and Fisher, M. A. (2013). Detection of non-jejuni and-coli campylobacter species from stool specimens with an immunochromatographic antigen detection assay. J. Clin. Microbiol. 51, 1935–1937. doi: 10.1128/JCM.03208-12
Couturier, B. A., Hale, D. C., and Couturier, M. R. (2012). Association of Campylobacter upsaliensis with persistent bloody diarrhea. J. Clin. Microbiol. 50, 3792–3794. doi: 10.1128/JCM.01807-12
Cummings, K. J., Rodriguez-Rivera, L. D., Mcneely, I., Suchodolski, J. S., Mesenbrink, B. T., Leland, B. R., et al. (2018). Fecal shedding of Campylobacter jejuni and Campylobacter coli among feral pigs in Texas. Zoonoses Public Health 65, 215–217. doi: 10.1111/zph.12390
Devleesschauwer, B., Bouwknegt, M., Mangen, M.-J. J., and Havelaar, A. H. 2017. Health and economic burden of campylobacter. Campylobacter. Elsevier.
Díaz-Sánchez, S., Sánchez, S., Herrera-León, S., Porrero, C., Blanco, J., Dahbi, G., et al. (2013). Prevalence of Shiga toxin-producing Escherichia coli, Salmonella spp. and Campylobacter spp. in large game animals intended for consumption: relationship with management practices and livestock influence. Vet. Microbiol. 163, 274–281. doi: 10.1016/j.vetmic.2012.12.026
Du, J., Luo, J., Huang, J., Wang, C., Li, M., Wang, B., et al. (2019). Emergence of genetic diversity and multi-drug resistant Campylobacter jejuni from wild birds in Beijing, China. Front. Microbiol. 10:2433. doi: 10.3389/fmicb.2019.02433
Ellis-Iversen, J., Ridley, A., Morris, V., Sowa, A., Harris, J., Atterbury, R., et al. (2012). Persistent environmental reservoirs on farms as risk factors for campylobacter in commercial poultry. Epidemiol. Infect. 140, 916–924. doi: 10.1017/S095026881100118X
Espunyes, J., Cabezon, O., Dias-Alves, A., Miralles, P., Ayats, T., and Cerda-Cuellar, M. (2021). Assessing the role of livestock and sympatric wild ruminants in spreading antimicrobial resistant Campylobacter and Salmonella in alpine ecosystems. BMC Vet. Res. 17:79. doi: 10.1186/s12917-021-02784-2
Firke, S. 2021. Simple tools for examining and cleaning dirty data [R Package Janitor Version 2.1.0].
Fischer, C., Reperant, L., Weber, J., Hegglin, D., and Deplazes, P. (2005). Echinococcus multilocularis infections of rural, residential and urban foxes (Vulpes Vulpes) in the Canton of Geneva, Switzerland. Parasite 12, 339–346. doi: 10.1051/parasite/2005124339
Fox, J., Weisberg, S., Adler, D., Bates, D., Baud-Bovy, G., Ellison, S., Firth, D., Friendly, M., Gorjanc, G., and Graves, S. 2012. Package ‘Car’. Vienna: R Foundation For Statistical Computing, 16.
Frasao, B. D. S., Marin, V. A., and Conte-Junior, C. A. (2017). Molecular detection, typing, and quantification of Campylobacter Spp. in foods of animal origin. Compr. Rev. Food Sci. Food Saf. 16, 721–734. doi: 10.1111/1541-4337.12274
French, N., Barrigas, M., Brown, P., Ribiero, P., Williams, N., Leatherbarrow, H., et al. (2005). Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7, 1116–1126. doi: 10.1111/j.1462-2920.2005.00782.x
French, N. P., Midwinter, A., Holland, B., Collins-Emerson, J., Pattison, R., Colles, F., et al. (2009). Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children's playgrounds. Appl. Environ. Microbiol. 75, 779–783. doi: 10.1128/AEM.01979-08
Gillespie, I. A., O’brien, S. J., Frost, J. A., Adak, G. K., Horby, P., Swan, A. V., et al. (2002). A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8, 937–942. doi: 10.3201/eid0809.010817
Guévremont, E., Normand, V., Lamoureux, L., and Côté, C. (2008). Genetic detection of campylobacter lanienae in fecal matter and stored manure from swine and dairy cattle. Foodborne Pathog. Dis. 5, 361–364. doi: 10.1089/fpd.2007.0054
Gusenbauer, M., and Haddaway, N. R. (2020). Which academic search systems are suitable for systematic reviews or meta-analyses? Evaluating retrieval qualities of Google Scholar, Pubmed, and 26 other resources. Res. Synth. Methods 11, 181–217. doi: 10.1002/jrsm.1378
Gutiérrez, J. S., Piersma, T., and Thieltges, D. W. (2019). Micro-and macroparasite species richness in birds: the role of host life history and ecology. J. Anim. Ecol. 88, 1226–1239. doi: 10.1111/1365-2656.12998
Haddaway, N. R., Collins, A. M., Coughlin, D., and Kirk, S. (2015). The role of Google Scholar in evidence reviews and its applicability to grey literature searching. PLoS One 10:E0138237. doi: 10.1371/journal.pone.0138237
Hassell, J. M., Begon, M., Ward, M. J., and Fèvre, E. M. (2017). Urbanization and disease emergence: dynamics at the wildlife–livestock–human Interface. Trends Ecol. Evol. 32, 55–67. doi: 10.1016/j.tree.2016.09.012
Hazeleger, W. C., Jacobs-Reitsma, W. F., Lina, P. H. C., De Boer, A. G., Bosch, T., Van Hoek, A. H. A. M., et al. (2018). Wild, insectivorous bats might be carriers of Campylobacter spp. PLoS One 13, E0190647–E0190647. doi: 10.1371/journal.pone.0190647
He, Y., Yuan, Q., Mathieu, J., Stadler, L., Senehi, N., Sun, R., et al. (2020). Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. NPJ Clean Water 3:4. doi: 10.1038/s41545-020-0051-0
Hepworth, P. J., Ashelford, K. E., Hinds, J., Gould, K. A., Witney, A. A., Williams, N. J., et al. (2011). Genomic variations define divergence of water/wildlife-associated Campylobacter jejuni niche specialists from common clonal complexes. Environ. Microbiol. 13, 1549–1560. doi: 10.1111/j.1462-2920.2011.02461.x
Hetman, B. M., Mutschall, S. K., Carrillo, C. D., Thomas, J. E., Gannon, V. P., Inglis, G. D., et al. (2020). “These Aren’t the strains you’re looking for”: recovery bias of common Campylobacter jejuni subtypes in mixed cultures. Front. Microbiol. 11:541. doi: 10.3389/fmicb.2020.00541
Hiett, K., Stern, N., Fedorka-Cray, P., Cox, N., Musgrove, M., and Ladely, S. (2002). Molecular subtype analyses of Campylobacter spp. from Arkansas and California poultry operations. Appl. Environ. Microbiol. 68, 6220–6236. doi: 10.1128/AEM.68.12.6220-6236.2002
Hollings, T., Jones, M., Mooney, N., and Mccallum, H. (2013). Wildlife disease ecology in changing landscapes: mesopredator release and toxoplasmosis. Int. J. Parasitol. Parasites Wildl. 2, 110–118. doi: 10.1016/j.ijppaw.2013.02.002
Iglesias-Carrasco, M., Aich, U., Jennions, M. D., and Head, M. L. (2020). Stress in the city: meta-analysis indicates no overall evidence for stress in urban vertebrates. Proc. Biol. Sci. 287:20201754.
Inglis, G., Kalischuk, L., and Busz, H. (2004). Chronic shedding of campylobacter species in beef cattle. J. Appl. Microbiol. 97, 410–420. doi: 10.1111/j.1365-2672.2004.02313.x
Inglis, G. D., Teixeira, J. S., and Boras, V. F. (2021). Comparative prevalence and diversity of Campylobacter jejuni strains in water and human beings over a 1-year period in southwestern Alberta, Canada. Can. J. Microbiol. 67, 851–863. doi: 10.1139/cjm-2021-0158
Iucn. 2019. The Iucn Red List Of Threatened Species [Online]. Available at: https://www.iucnredlist.org [Accessed April 18, 2019].
Jackson, D. N., Davis, B., Tirado, S. M., Duggal, M., Van Frankenhuyzen, J. K., Deaville, D., et al. (2009). Survival mechanisms and culturability of Campylobacter jejuni under stress conditions. Antonie Van Leeuwenhoek 96, 377–394. doi: 10.1007/s10482-009-9378-8
Jaing, C., Thissen, J. B., Gardner, S., Mcloughlin, K., Slezak, T., Bossart, G. D., et al. (2015). Pathogen surveillance in wild bottlenose dolphins tursiops truncatus. Dis. Aquat. Org. 116, 83–91. doi: 10.3354/dao02917
Jay-Russell, M. T., Bates, A., Harden, L., Miller, W. G., and Mandrell, R. E. (2012). Isolation of campylobacter from feral swine (sus Scrofa) on the ranch associated with the 2006 Escherichia coli O157:H7 spinach outbreak investigation in California. Zoonoses Public Health 59, 314–319. doi: 10.1111/j.1863-2378.2012.01465.x
Jensen, A. N., Dalsgaard, A., Baggesen, D. L., and Nielsen, E. (2006). The occurrence and characterization of Campylobacter jejuni and C. coli in organic pigs and their outdoor environment. Vet. Microbiol. 116, 96–105. doi: 10.1016/j.vetmic.2006.03.006
Jobbins, S. E., and Alexander, K. A. (2015). From whence they came—antibiotic-resistant Escherichia coli in African wildlife. J. Wildl. Dis. 51, 811–820. doi: 10.7589/2014-11-257
Johnson, P. T. J., Rohr, J. R., Hoverman, J. T., Kellermanns, E., Bowerman, J., and Lunde, K. B. (2012). Living fast and dying of infection: host life history drives interspecific variation in infection and disease risk. Ecol. Lett. 15, 235–242. doi: 10.1111/j.1461-0248.2011.01730.x
Jones, K. E., Bielby, J., Cardillo, M., Fritz, S. A., O'dell, J., Orme, C. D. L., et al. (2009). Pantheria: a species-level database of life history, ecology, and geography of extant and recently extinct mammals: ecological archives E090-184. Ecology 90, 2648–2648. doi: 10.1890/08-1494.1
Kalashnikova, V., Dzhikidze, E., Stasilevich, Z., and Chikobava, M. (2002). Detection of Campylobacter jejuni in healthy monkeys and monkeys with enteric infections by Pcr. Bull. Exp. Biol. Med. 134, 299–300. doi: 10.1023/A:1021528122942
Kapperud, G., Espeland, G., Wahl, E., Walde, A., Herikstad, H., Gustavsen, S., et al. (2003). Factors associated with increased and decreased risk of campylobacter infection: a prospective case-control study in Norway. Am. J. Epidemiol. 158, 234–242. doi: 10.1093/aje/kwg139
Kapperud, G., And & Rosef, O. 1983. Avian wildlife reservoir of Campylobacter fetus Subsp. Jejuni, Yersinia Spp., and Salmonella Spp. in Norway. Appl. Environ. Microbiol., 45, 375–380, doi: 10.1128/aem.45.2.375-380.1983
Khoshbakht, R., Tabatabaei, M., Shirzad Aski, H., and Shayegh, H. (2015). Distribution of salmonella, Arcobacter, and thermophilic Campylobacter Spp. among Persian fallow deer (Dama Mesopotamica) population in Dasht-E-Arzhan wildlife refuge, southern Iran. Comp. Clin. Pathol. 24, 777–781. doi: 10.1007/s00580-014-1981-z
Kirk, M. D., Pires, S. M., Black, R. E., Caipo, M., Crump, J. A., Devleesschauwer, B., et al. (2015). World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 12:E1001921. doi: 10.1371/journal.pmed.1001921
Kriger, K. M., and Hero, J. M. (2007). The chytrid fungus batrachochytrium dendrobatidis is non-randomly distributed across amphibian breeding habitats. Divers. Distrib. 13, 781–788. doi: 10.1111/j.1472-4642.2007.00394.x
Kwan, P. S. L., Barrigas, M., Bolton, F. J., French, N. P., Gowland, P., Kemp, R., et al. (2008). Molecular epidemiology of Campylobacter jejuni populations in dairy cattle, wildlife, and the environment in a farmland area. Appl. Environ. Microbiol. 74, 5130–5138. doi: 10.1128/AEM.02198-07
Lastovica, A. J., and Le Roux, E. (2000). Efficient isolation of Campylobacteria from stools. J. Clin. Microbiol. 38, 2798–2799. doi: 10.1128/JCM.38.7.2798-2799.2000
Laver, P. N., Ganswindt, A., Ganswindt, S. B., and Alexander, K. A. (2012). Non-invasive monitoring of glucocorticoid metabolites in banded mongooses (Mungos Mungo) in response to physiological and biological challenges. Gen. Comp. Endocrinol. 179, 178–183. doi: 10.1016/j.ygcen.2012.08.011
Lawton, S. J., Weis, A. M., Byrne, B. A., Fritz, H., Taff, C. C., Townsend, A. K., et al. (2018). Comparative analysis of campylobacter isolates from wild birds and chickens using Maldi-Tof Ms, biochemical testing, and DNA sequencing. J. Vet. Diagn. Investig. 30, 354–361. doi: 10.1177/1040638718762562
Lee, K., Iwata, T., Nakadai, A., Kato, T., Hayama, S., Taniguchi, T., et al. (2011). Prevalence of Salmonella, Yersinia and Campylobacter Spp. in feral raccoons (Procyon Lotor) and masked palm civets (Paguma Larvata) in Japan. Zoonoses Public Health 58, 424–431. doi: 10.1111/j.1863-2378.2010.01384.x
Levesque, S., Fournier, E., Carrier, N., Frost, E., Arbeit, R. D., and Michaud, S. (2013). Campylobacteriosis in urban versus rural areas: a case-case study integrated with molecular typing to validate risk factors and to attribute sources of infection. PLoS One 8:E83731. doi: 10.1371/journal.pone.0083731
Llarena, A.-K., Taboada, E., and Rossi, M. (2017). Whole-genome sequencing in epidemiology of Campylobacter jejuni infections. J. Clin. Microbiol. 55, 1269–1275. doi: 10.1128/JCM.00017-17
Lõhmus, M., and Albihn, A. (2013). Gastrointestinal pathogens in rodents overwintering in human facilities around Uppsala, Sweden. J. Wildl. Dis. 49, 747–749. doi: 10.7589/2013-02-028
Lopes, B. S., Strachan, N. J. C., Ramjee, M., Thomson, A., Macrae, M., Shaw, S., et al. (2019). Nationwide stepwise emergence and evolution of multidrug-resistant Campylobacter jejuni sequence type 5136, United Kingdom. Emerg. Infect. Dis. 25, 1320–1329. doi: 10.3201/eid2507.181572
Luangtongkum, T., Jeon, B., Han, J., Plummer, P., Logue, C. M., and Zhang, Q. (2009). Antibiotic resistance in campylobacter: emergence, transmission and persistence. Future Microbiol. 4, 189–200. doi: 10.2217/17460913.4.2.189
Luo, N., Pereira, S., Sahin, O., Lin, J., Huang, S., Michel, L., et al. (2005). Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. U. S. A. 102, 541–546. doi: 10.1073/pnas.0408966102
Lyons, J., Mastromonaco, G., Edwards, D. B., and Schulte-Hostedde, A. I. (2017). Fat and happy in the city: eastern chipmunks in urban environments. Behav. Ecol. 28, 1464–1471. doi: 10.1093/beheco/arx109
Macdonald, E., White, R., Mexia, R., Bruun, T., Kapperud, G., Lange, H., et al. (2015). Risk factors for sporadic domestically acquired campylobacter infections in Norway 2010-2011: a National Prospective Case-Control Study. PLoS One 10:E0139636. doi: 10.1371/journal.pone.0139636
Malmberg, J. L., White, L. A., and Vandewoude, S. (2021). Bioaccumulation of pathogen exposure in top predators. Trends Ecol. Evol. 36, 411–420. doi: 10.1016/j.tree.2021.01.008
Man, S. M. (2011). The clinical importance of emerging campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 8, 669–685. doi: 10.1038/nrgastro.2011.191
Meerburg, B., Jacobs-Reitsma, W., Wagenaar, J., and Kijlstra, A. (2006). Presence of Salmonella and Campylobacter spp. in wild small mammals on organic farms. Appl. Environ. Microbiol. 72, 960–962. doi: 10.1128/AEM.72.1.960-962.2006
Melnyk, A. H., Wong, A., and Kassen, R. (2015). The fitness costs of antibiotic resistance mutations. Evol. Appl. 8, 273–283. doi: 10.1111/eva.12196
Mildenstein, T., Tanshi, I., and Racey, P. A. 2016. Exploitation of bats for Bushmeat and medicine. Bats in the anthropocene: Conservation of bats in a changing world. Springer, Cham.
Molina-Lopez, R., Valverdú, N., Martin, M., Mateu, E., Obon, E., Cerdà-Cuéllar, M., et al. (2011). Wild raptors as carriers of antimicrobial-resistant salmonella and Campylobacter strains. Vet. Rec. 168:565. doi: 10.1136/vr.c7123
Moore, S. M., Borer, E. T., and Hosseini, P. R. (2010). Predators indirectly control vector-borne disease: linking predator–prey and host–pathogen models. J. R. Soc. Interface 7, 161–176. doi: 10.1098/rsif.2009.0131
Moriarty, E. M., Mackenzie, M. L., Karki, N., and Sinton, L. W. (2011). Survival of Escherichia coli, Enterococci, and Campylobacter spp. in sheep feces on pastures. Appl. Environ. Microbiol. 77, 1797–1803. doi: 10.1128/AEM.01329-10
Mpalang, R. K. A., Mpalang, M. K. A., Kaut, C. M., Boreux, R., Melin, P., Bitiang, F., et al. (2013). Bacteriological assessment of smoked game meat in Lubumbashi, Drc. Biotechnol. Agron. Soc. Environ. 17, 441–449.
Mughini Gras, L., Smid, J. H., Wagenaar, J. A., Koene, M. G., Havelaar, A. H., Friesema, I. H., et al. (2013). Increased risk for Campylobacter jejuni and C. coli infection of pet origin in dog owners and evidence for genetic association between strains causing infection in humans and their pets. Epidemiol. Infect. 141, 2526–2535. doi: 10.1017/S0950268813000356
Mughini-Gras, L., Penny, C., Ragimbeau, C., Schets, F. M., Blaak, H., Duim, B., et al. (2016). Quantifying potential sources of surface water contamination with Campylobacter jejuni and Campylobacter coli. Water Res. 101, 36–45. doi: 10.1016/j.watres.2016.05.069
Myers, P., Espinosa, R., Parr, C., Jones, T., Hammond, G., and Dewey, T. 2019. The Animal Diversity Web [Online]. Online. Available at: https://animaldiversity.org/ (Accessed April 8, 2019).
Navarro-Gonzalez, N., Ugarte-Ruiz, M., Porrero, M. C., Zamora, L., Mentaberre, G., Serrano, E., et al. (2014). Campylobacter shared between free-ranging cattle and sympatric wild ungulates in a natural environment (ne Spain). EcoHealth 11, 333–342. doi: 10.1007/s10393-014-0921-3
Neiderud, C.-J. (2015). How urbanization affects the epidemiology of emerging infectious diseases. Infect. Ecol. Epidemiol. 5:27060. doi: 10.3402/iee.v5.27060
Newell, D. G., Mughini-Gras, L., Kalupahana, R. S., and Wagenaar, J. A. (2017). Campylobacter epidemiology—sources and routes of transmission for human infection. Campylobacter 2017, 85–110. doi: 10.1016/B978-0-12-803623-5.00005-8
Ngotho, M., Ngure, R., Kamau, D., Kagira, J., Gichuki, C., Farah, I., et al. (2006). A fatal outbreak of Campylobacter jejuni enteritis in a colony of vervet monkeys in Kenya. Scand. J. Lab. Anim. Sci. 33, 205–210. doi: 10.23675/sjlas.v33i4.109
Olkkola, S., Rossi, M., Jaakkonen, A., Simola, M., Tikkanen, J., Hakkinen, M., et al. (2021). Host-dependent clustering of campylobacter strains from small mammals in Finland. Front. Microbiol. 11:3520. doi: 10.3389/fmicb.2020.621490
Oporto, B., and Hurtado, A. (2011). Emerging thermotolerant Campylobacter species in healthy ruminants and swine. Foodborne Pathog. Dis. 8, 807–813. doi: 10.1089/fpd.2010.0803
Osbjer, K., Boqvist, S., Sokerya, S., Chheng, K., San, S., Davun, H., et al. (2016). Risk factors associated with campylobacter detected by PCR in humans and animals in rural Cambodia. Epidemiol. Infect. 144, 2979–2988. doi: 10.1017/S095026881600114X
Page, M. J., Mckenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The Prisma 2020 statement: an updated guideline for reporting systematic reviews. Systematic Reviews 10, 1–11. doi: 10.1186/s13643-021-01626-4
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Perdoncini, G., Sierra-Arguello, Y. M., Lima, L. M., Trindade, M. M., Gomes, M. J. P., Santos, L. R. D., et al. (2015). Occurrence of Campylobacter jejuni and C. coli on broiler carcasses after chilling in southern Brazil. Pesqui. Vet. Bras. 35, 349–352. doi: 10.1590/S0100-736X2015000400006
Platts-Mills, J. A., and Kosek, M. (2014). Update on the burden of campylobacter in developing countries. Curr. Opin. Infect. Dis. 27, 444–450. doi: 10.1097/QCO.0000000000000091
Potter, R. C., Kaneene, J. B., and Hall, W. N. (2003). Risk factors for sporadic Campylobacter jejuni infections in rural Michigan: a prospective case-control study. Am. J. Public Health 93, 2118–2123. doi: 10.2105/AJPH.93.12.2118
R Core Development Team 2022. R: A Language And Environment For Statistical Computing. R Foundation For Statistical Computing. Vienna, Austria.
Ramonaite, S., Kudirkiene, E., Tamuleviciene, E., Leviniene, G., Malakauskas, A., Gölz, G., et al. (2014). Prevalence and genotypes of Campylobacter jejuni from urban environmental sources in comparison with clinical isolates from children. J. Med. Microbiol. 63, 1205–1213. doi: 10.1099/jmm.0.072892-0
Renteria-Solis, Z., Forster, C., Aue, A., Wittstatt, U., Wibbelt, G., and Konig, M. (2014). Canine distemper outbreak in raccoons suggests pathogen interspecies transmission amongst alien and native carnivores in urban areas from Germany. Vet. Microbiol. 174, 50–59. doi: 10.1016/j.vetmic.2014.08.034
Rosef, O., Gondrosen, B., Kapperud, G., and Underdal, B. (1983). Isolation and characterization of Campylobacter jejuni and Campylobacter coli from domestic and wild mammals in Norway. Appl. Environ. Microbiol. 46, 855–859. doi: 10.1128/aem.46.4.855-859.1983
Roux, F., Sproston, E., Rotariu, O., Macrae, M., Sheppard, S. K., Bessell, P., et al. (2013). Elucidating the aetiology of human Campylobacter coli infections. PLoS One 8:E64504. doi: 10.1371/journal.pone.0064504
Santini, L., González-Suárez, M., Russo, D., Gonzalez-Voyer, A., Von Hardenberg, A., and Ancillotto, L. (2019). One strategy does not fit all: determinants of urban adaptation in mammals. Ecol. Lett. 22, 365–376. doi: 10.1111/ele.13199
Sasaki, Y., Goshima, T., Tetsuya, M., Murakami, M., Haruna, M., Ito, K., et al. (2013). Prevalence and antimicrobial susceptibility of foodborne bacteria in wild boars (sus Scrofa) and wild deer (Cervus Nippon) in Japan. Foodborne Pathog. Dis. 10, 985–991. doi: 10.1089/fpd.2013.1548
Sauvala, M., Laaksonen, S., Laukkanen-Ninios, R., Jalava, K., Stephan, R., and Fredriksson-Ahomaa, M. (2019). Microbial contamination of moose (Alces Alces) and White-tailed deer (Odocoileus Virginianus) carcasses harvested by hunters. Food Microbiol. 78, 82–88. doi: 10.1016/j.fm.2018.09.011
Sheppard, S. K., Mccarthy, N. D., Falush, D., and Maiden, M. C. (2008). Convergence of campylobacter species: implications for bacterial evolution. Science 320, 237–239. doi: 10.1126/science.1155532
Sippy, R., Sandoval-Green, C. M., Sahin, O., Plummer, P., Fairbanks, W. S., Zhang, Q., et al. (2012). Occurrence and molecular analysis of Campylobacter in wildlife on livestock farms. Vet. Microbiol. 157, 369–375. doi: 10.1016/j.vetmic.2011.12.026
Stella, S., Tirloni, E., Castelli, E., Colombo, F., and Bernardi, C. (2018). Microbiological evaluation of carcasses of wild boar hunted in a hill area of northern Italy. J. Food Prot. 81, 1519–1525. doi: 10.4315/0362-028X.JFP-18-077
Strachan, N. J. C., Rotariu, O., Gormley, F. J., Ogden, I. D., Dallas, J. F., Forbes, K. J., et al. (2009). Attribution of campylobacter infections in Northeast Scotland to specific sources by use of multilocus sequence typing. J. Infect. Dis. 199, 1205–1208. doi: 10.1086/597417
Suzuki, H., and Yamamoto, S. (2009). Campylobacter contamination in retail poultry meats and by-products in the world: a literature survey. J. Vet. Med. Sci. 71, 255–261. doi: 10.1292/jvms.71.255
Taff, C. C., and Townsend, A. K. (2017). Campylobacter jejuni infection associated with relatively poor condition and low survival in a wild bird. J. Avian Biol. 48, 1071–1076. doi: 10.1111/jav.01282
Taff, C. C., Weis, A. M., Wheeler, S., Hinton, M. G., Weimer, B. C., Barker, C. M., et al. (2016). Influence of host ecology and behavior on Campylobacter jejuni prevalence and environmental contamination risk in a synanthropic wild bird species. Appl. Environ. Microbiol. 82, 4811–4820. doi: 10.1128/AEM.01456-16
Troeger, C., Forouzanfar, M., Rao, P. C., Khalil, I., Brown, A., Reiner, R. C. Jr., et al. (2017). Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the global burden of disease study 2015. Lancet Infect. Dis. 17, 909–948. doi: 10.1016/S1473-3099(17)30276-1
Usio, N. (2020). Spatiotemporal patterns of latrine-site use by small-clawed otters in a heterogeneous Rice field landscape. Mammal Study 45, 103–110. doi: 10.3106/ms2019-0031
Van Nierop, W., Duse, A., Marais, E., Aithma, N., Thothobolo, N., Kassel, M., et al. (2005). Contamination of chicken carcasses in Gauteng, South Africa, by Salmonella, Listeria Monocytogenes and Campylobacter. Int. J. Food Microbiol. 99, 1–6. doi: 10.1016/j.ijfoodmicro.2004.06.009
Vanderwaal, K. L., Atwill, E. R., Isbell, L. A., and Mccowan, B. (2014). Linking social and pathogen transmission networks using microbial genetics in giraffe (Giraffa Camelopardalis). J. Anim. Ecol. 83, 406–414. doi: 10.1111/1365-2656.12137
Vanderwaal, K. L., and Ezenwa, V. O. (2016). Heterogeneity in pathogen transmission: mechanisms and methodology. Funct. Ecol. 30, 1606–1622. doi: 10.1111/1365-2435.12645
Verble, K., Hallerman, E. M., and Alexander, K. A. (2021). Urban landscapes increase dispersal, gene flow, and pathogen transmission potential in banded mongoose (Mungos Mungo) in northern Botswana. Ecol. Evol. 11, 9227–9240. doi: 10.1002/ece3.7487
Viswanathan, M., Pearl, D. L., Taboada, E. N., Parmley, E. J., Mutschall, S. K., and Jardine, C. M. (2017). Cluster analysis of campylobacter Jejuni genotypes isolated from small and medium-sized mammalian wildlife and bovine livestock from Ontario farms. Zoonoses Public Health 64, 185–193. doi: 10.1111/zph.12294
Wacheck, S., Fredriksson-Ahomaa, M., König, M., Stolle, A., and Stephan, R. (2009). Wild boars as an important reservoir for foodborne pathogens. Foodborne Pathog. Dis. 7, 307–312. doi: 10.1089/fpd.2009.0367
Wagenaar, J. A., French, N. P., and Havelaar, A. H. (2013). Preventing campylobacter at the source: why is it so difficult? Clin. Infect. Dis. 57, 1600–1606. doi: 10.1093/cid/cit555
Waldenstrom, J., Axelsson-Olsson, D., Olsen, B., Hasselquist, D., Griekspoor, P., Jansson, L., et al. (2010). Campylobacter jejuni colonization in wild birds: results from an infection experiment. PLoS One 5:E9082. doi: 10.1371/journal.pone.0009082
Wei, B., Kang, M., and Jang, H.-K. (2019). Genetic characterization and epidemiological implications of campylobacter isolates from wild birds in South Korea. Transbound. Emerg. Dis. 66, 56–65. doi: 10.1111/tbed.12931
Weis, A. M., Storey, D. B., Taff, C. C., Townsend, A. K., Huang, B. C., Kong, N. T., et al. (2016). Genomic comparison of Campylobacter spp. and their potential for zoonotic transmission between birds, primates, and livestock. Appl. Environ. Microbiol. 82, 7165–7175. doi: 10.1128/AEM.01746-16
Wickham, H., Chang, W., and Wickham, M. H. 2016. Package ‘Ggplot2’. Create Elegant Data Visualisations Using The Grammar Of Graphics. Version, 2, 1–189.
Williams, L., Sait, L., Cogan, T., Jørgensen, F., Grogono-Thomas, R., and Humphrey, T. (2012). Enrichment culture can bias the isolation of campylobacter subtypes. Epidemiol. Infect. 140, 1227–1235. doi: 10.1017/S0950268811001877
Wilson, D. J., Gabriel, E., Leatherbarrow, A. J., Cheesbrough, J., Gee, S., Bolton, E., et al. (2008). Tracing the source of Campylobacteriosis. PLoS Genet. 4:E1000203. doi: 10.1371/journal.pgen.1000203
Wilson, D. J., Gabriel, E., Leatherbarrow, A. J., Cheesbrough, J., Gee, S., Bolton, E., et al. (2009). Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol. Biol. Evol. 26, 385–397. doi: 10.1093/molbev/msn264
Keywords: zoonotic, Campylobacter, life histories, spillover, wildlife, foodbome pathogen
Citation: Brooks MR, Medley S, Ponder M and Alexander KA (2023) Campylobacter in aquatic and terrestrial mammals is driven by life traits: A systematic review and meta-analysis. Front. Ecol. Evol. 11:1070519. doi: 10.3389/fevo.2023.1070519
Edited by:
Andreas Erich Zautner, University Hospital Magdeburg, GermanyReviewed by:
Ben Pascoe, University of Oxford, United KingdomFauzy Negiub Ali Nasher, University of London, United Kingdom
Hagen Frickmann, Bundeswehrkrankenhaus, Germany
Copyright © 2023 Brooks, Medley, Ponder and Alexander. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kathleen A. Alexander, ✉ a2F0aHlhbHhAdnQuZWR1
 Michael R. Brooks
Michael R. Brooks Sarah Medley2
Sarah Medley2 Monica Ponder
Monica Ponder Kathleen A. Alexander
Kathleen A. Alexander