- 1Molecular Ecology and Evolution Laboratory, Department of Biological Science, Sangji University, Wonju, Republic of Korea
- 2Korea National Park Research Institute, Korea National Park Service, Wonju, Republic of Korea
- 3Korean Entomological Institute, Korea University, Seoul, Republic of Korea
- 4National Institute of Ecology, Seocheon, Republic of Korea
Population or habitat connectivity is a key component in maintaining species and community-level regional biodiversity as well as intraspecific genetic diversity. Ongoing human activities cause habitat destruction and fragmentation, which exacerbate the connectivity due to restricted animal movements across local habitats, eventually resulting in the loss of biodiversity. The Baekdudaegan Mountain Range (BMR) on the Korean Peninsula represents “biodiversity hotspots” and eight of the 22 Korean national parks are located within the BMR. Given the striped field mouse (Apodemus agrarius) is the most common and ecologically important small mammals in these protected areas, the population genetic assessment of this species will allow for identifying “genetic diversity hotspots” and also “genetic barriers” that may hinder gene flow, and will therefore inform on effective conservation and management efforts for the national park habitats. We collected samples from hair, tail, or buccal swabs for 252 A. agrarius individuals in 2015 and 2019. By using mitochondrial DNA cytochrome b (cyt b) sequences and nine microsatellite loci, we determined levels of genetic diversity, genetic differentiation, and gene flow among eight national park populations of A. agrarius along the BMR. We found high levels of genetic diversity but the occurrences of inbreeding for all the nine samples analyzed. Our results also indicated that there was detectable temporal genetic variation between the 2015 and 2019 populations in the Jirisan National Park, which is probably due to a short-term decline in genetic diversity caused by reduced population sizes. We also found a well-admixed shared gene pool among the national park populations. However, a significant positive correlation between geographic and genetic distances was detected only in mtDNA but not microsatellites, which might be attributed to different dispersal patterns between sexes. There was a genetic barrier to animal movements around the Woraksan National Park areas. The poor habitat connectivity surrounding these areas can be improved by establishing an ecological corridor. Our findings of the presence of genetic barriers in some protected areas provide insights into the conservation and management efforts to improve the population or habitat connectivity among the national parks.
Introduction
Population connectivity or habitat connectivity, the integration of sub-populations of organisms into a functional unit (sensu Merriam, 1984) is a key component in maintaining species and community-level biodiversity and also ensuring genetic diversity, adaptive potential, and long term persistence of wild populations (Horskins et al., 2006). Knowledge of degrees of connectivity informs decisions regarding conservation and management plans to facilitate the movements of animals, particularly for fragmented populations or habitats (Epps et al., 2018; Thapa et al., 2018; Jang et al., 2021). Recently, habitat connectivity has become exacerbated in terrestrial ecosystems, primarily due to severe habitat destruction and fragmentation, resulting from ongoing human activities such as road and railway constructions, which ultimately leads to the declines in all levels of biodiversity (Pereira et al., 2010; Rands et al., 2010; Almasieh et al., 2022; Mohammadi et al., 2022; Rezaei et al., 2022). Habitat fragmentation has adverse effects on dispersal of organisms, leading to the reduction in gene flow among populations, which depletes within-population genetic diversity, causing escalating inbreeding and genetic drift effects (Khimoun et al., 2016). The subsequent reduced adaptability may cause the local extinction (Keyghobadi, 2007) as genetic diversity is associated with ecological integrity or ecological performance of a given population (Freeland et al., 2012). Gene flow by dispersal among populations helps to maintain within-population genetic diversity, which is critical for population persistence following environmental changes (Aitken et al., 2008; Boulding, 2008). Therefore, population connectivity may also play a crucial role in facilitating the population and community recovery after disturbances (Allendorf et al., 2013; Binks et al., 2019).
To determine a priority and establish an appropriate strategy for conservation planning of ecosystems and habitats, especially protected areas such as national parks, molecular-based assessment of levels of genetic diversity, genetic differentiation, and gene flow among populations as well as patterns of local adaptation to different environments is essential (Manel et al., 2003; Wultsch et al., 2016; Matocq et al., 2017; Hanson et al., 2021). While there are a handful of studies that have conducted population genetic analysis to develop habitat conservation and management plans, such as the establishment of ecological corridor between poorly connected areas (Streatfeild, 2009; Ballesteros-Mejia et al., 2020), little attention has been paid to the genetic diversity and population connectivity among protected reserves for the conservation and management efforts in general (e.g., Thapa et al., 2018; Jang et al., 2021).
The Baekdudaegan Mountain Range (BMR), located on the Korean Peninsula, functions as a key ecological axis with a “biodiversity hotspot” as an astonishingly high number of endemic (and endangered) wild animals and plants inhabits there (Choi, 2004; Chung et al., 2018). In the BMR, there are 26% of the total bird species occurring in Korea, 29% of mammal species, 60% of amphibians and reptiles, which represent the highest terrestrial biodiversity ever in the Korean Peninsula (Shin et al., 2016; Chung et al., 2018). Eight of the 22 national parks are located within the BMR. Forty-five percentage of Korean indigenous and 65% of Korean endangered species reside there (Kang and Jeong, 2017). Although some ecological studies have performed to investigate a biota within the BMR on birds (Lee, 2003), and vascular plants (Lim et al., 2004), population genetic studies for the purpose of conservation and management for the protected areas and mammalian species remain understudied (Byeon et al., 2018; Jang et al., 2021). The population genetic investigation of ecologically important and widespread animals will shed insights into designing conservation and management plans for wildlife and habitats, particularly national park areas.
Small mammals play ecologically important roles in terrestrial ecosystems as insect population regulators, pollinators, seed dispersal mediators, prey for carnivores, and also forest regeneration facilitators (Jones and Safi, 2011). In particular, rodents, one of the small mammals, serve as a keystone species and an ecosystem engineer in terrestrial forest ecosystems. Stephens’s kangaroo rats (Dipodomys stephensi), which are endemic to the Southern California, are found to regulate the abundance of the plants (genus Erodium) and thereby the whole plant community (Brock and Kelt, 2004). Burrows of prairie dogs (Cynomys gunnisoni and Cynomys ludovicianus) and banner-tailed kangaroo rats (Dipodomys spectabilis) can provide habitats for a diverse array of arthropods and therefore they facilitate arthropod abundances and species richness (Davidson and Lightfoot, 2007). Because small mammals have short lifespans, lessened generation time, and high turn-over rates may lead to a change in the genetic structure of populations over a relatively short time period (Merritt, 2010). In addition, most small social mammals show the tendency for sex differences in philopatry and dispersal, e.g., female philopatry with male-biased dispersal (Clutton-Brock and Lukas, 2012; García-Navas et al., 2016).
The most common small mammal in Korea is the striped field mouse (Apodemus agrarius), which is a socially monogamous rodent species widely distributed across farmlands and/or forests in Eurasia including eastern Asia (Johnson, 2001; Sakka et al., 2010). In the Korean Peninsula, the presence of four subspecies of A. agrarius (northern parts: A. a. manchuricus; coastal lowlands of the southernmost parts: A. a. pallescens; major central parts: A. a. coreae; Jeju Island: A. a. chejuensis) was initially suggested. However, A. a. pallescens and A. a. coreae were reported as a synonym and thus only two subspecies, A. a. coreae and A. a. chejuensis, probably exist (Koh and Yoo, 1992; Koh et al., 2000). Apodemus agrarius is the most widely distributed but is known to be susceptible to environmental changes such as ongoing climate changes (Serizawa et al., 2002; Liu et al., 2004; Zając et al., 2019). Previous studies of A. agrarius observed no significant genetic differences between northeastern and southwestern regions of the mainland of Korea, suggesting that A. agrarius populations are well connected along the BMR (Kim and Park, 2015). Also, Jo et al. (2018) found that genetic differences exist according to their habitat environments, and also that the areas with many anthropogenic influences act as a genetic barrier. Although only a handful of ecological and genetic studies on small mammals and rodents have been conducted in Korea, population genetic based assessments for the habitat connectivity in national park areas within the BMR, the core ecological axis of the Korean Peninsula, remain overlooked. The genetic structure assessment of A. agrarius inhabiting the national parks will allow for identifying “genetic diversity hotspots” and also “genetic barriers” that hinder gene flow and will provide crucial information for designing effective conservation and management plans for A. agrarius populations as well as the national park habitats.
In this study, by performing phylogeographic and population genetic analyses using mitochondrial DNA (mtDNA) cytochrome b (cyt b) sequences and nine microsatellite loci, we assessed the genetic diversity, population differentiation, and gene flow among the eight national park populations along the BMR to determine the level of population connectivity among these protected habitats. We further tested for temporal stability in the genetic structure by comparing genetic diversity and genetic composition of the Jirisan National Park population between 2015 and 2019. We hypothesized that A. agrarius populations show high dispersal ability but female philopatry, as suggested in previous studies (Clutton-Brock and Lukas, 2012; García-Navas et al., 2016; Jo et al., 2017). Therefore, we predicted that there would be elevated levels of population connectivity via high gene flow occurring across the BMR populations and also that maternally inherited mtDNA marker would exhibit more geographic structure than biparental microsatellites, due to female’s home-site fidelity with male dispersal.
Materials and methods
Study sites and sample collection
Eight national parks within the BMR house legally protected species (including CITES and endangered species) of mammals, including order Carnivora (leopard cat Prionailurus bengalensis, Eurasian otter Lutra lutra, yellow-throated marten Martes flavigula, and least weasel Mustela nivalis), order Artiodactyla (long-tailed goral Naemorhedus caudatus), and order Rodentia (Siberian flying squirrel Pteromys volans). Also several bird species, including Japanese sparrowhawk Accipiter gularis, Chinese sparrowhawk Accipiter soloensis, Eurasian sparrowhawk Accipiter nisus, long-billed plover Charadrius placidus, Eurasian eagle-owl Bubo bubo, black woodpecker Dryocopus martius, Mandarin duck Aix galericulata, common kestrel Falco tinnunculus, common buzzard Buteo buteo, and Eurasian hobby Falco Subbuteo occur in the protected areas (Kwon, 2009; Kim, 2010).
In 2015 and 2019, a total of 252 A. agrarius individuals [mtDNA (N = 221), microsatellites (N = 179)] were collected from eight national parks within the BMR [Seoraksan (SA), Odaesan (OD), Taebaeksan (TB), Sobaeksan (SB), Woraksan (WA), Songnisan (SN), Deogyusan (DY), and Jirisan (JR)] in South Korea (Table 1; Figure 1). The populations from SA, WA, and JR were sampled twice both in 2015 and in 2019 to test for the temporal stability in the genetic structure of A. agrarius. However, only the JR population was possible to be formally assessed, as sample sizes for the other populations [SA (N = 2 for the 2019 sample) and WA (N = 6 for the 2019 sample)] were insufficient (Table 1). At least more than 10 samples were attempted to be collected for every population, but we could only sample two to six individuals for three populations (SA_19, WA_19, and DY_19). To obtain A. agrarius samples, we set the Sherman traps at diverse habitat environments, including forest, grassland, and agricultural land. Hair, oral swab, or tail tissue samples were collected after captured using the traps and all the specimens were fixed in 99% ethanol and stored at −20°C. The mice were released immediately back to the original sites after taking samples. The tail tissue samples were only obtained when the already dead carcasses were found. Apodemus agrarius has been classified as “Least Concern (LC)” on the IUCN Red List (Kaneko et al., 2016). The capture and use of the study animals for this study were approved by Korea National Park Research Institute and National Institute of Ecology, Ministry of Environment in Korea. Sample collections were followed by guidelines (the Animal Protection Act) of Department of Animal Protection and Welfare, Animal and Plant Quarantine Agency (APQA) in Korea. We mainly took hair or oral swab samples to minimize unnecessary inhumane procedures.
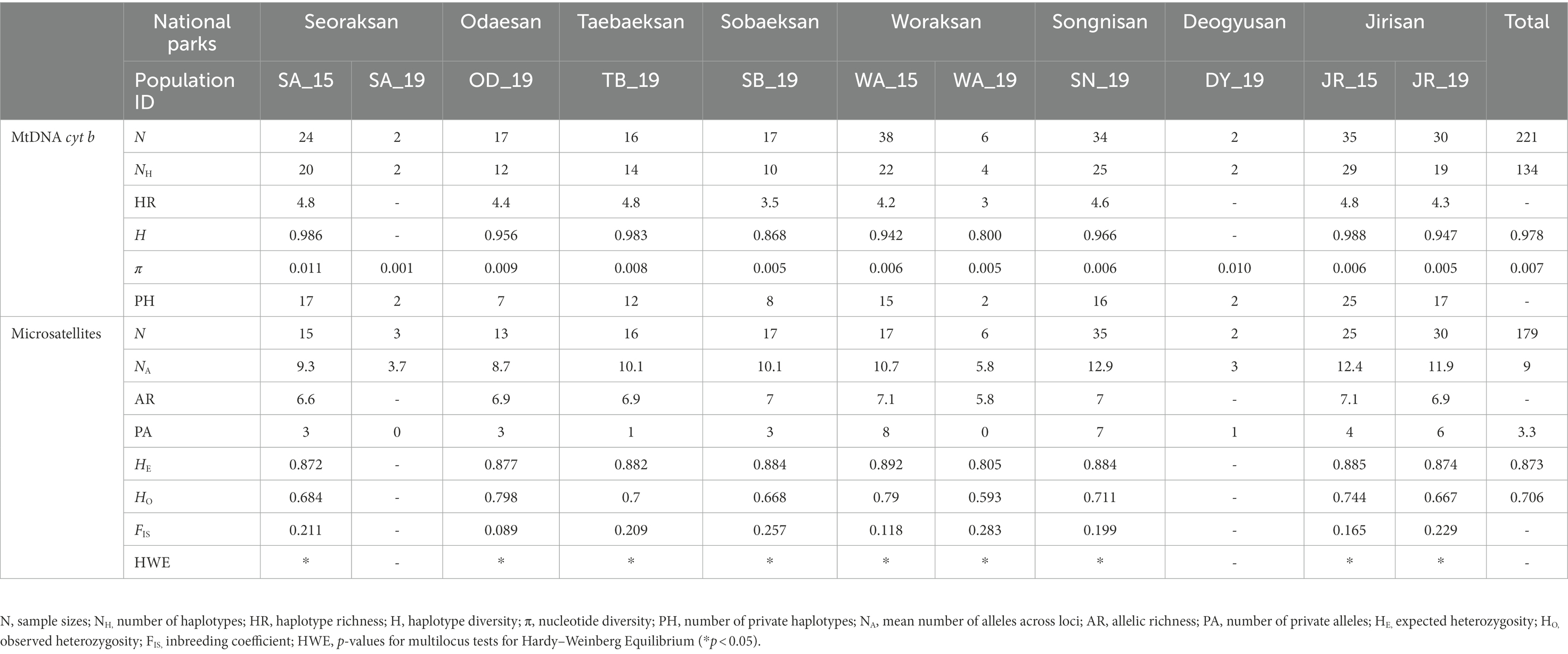
Table 1. Summary of genetic diversity statistics in 11 populations (including three temporal samples) of Apodemus agrarius in eight national parks within the Baekdudaegan Mountain Range (BMR) in South Korea at both mtDNA cyt b and nine microsatellite loci. The numbers 15 and 19 for population ID indicate sampling years of 2015 and 2019, respectively.
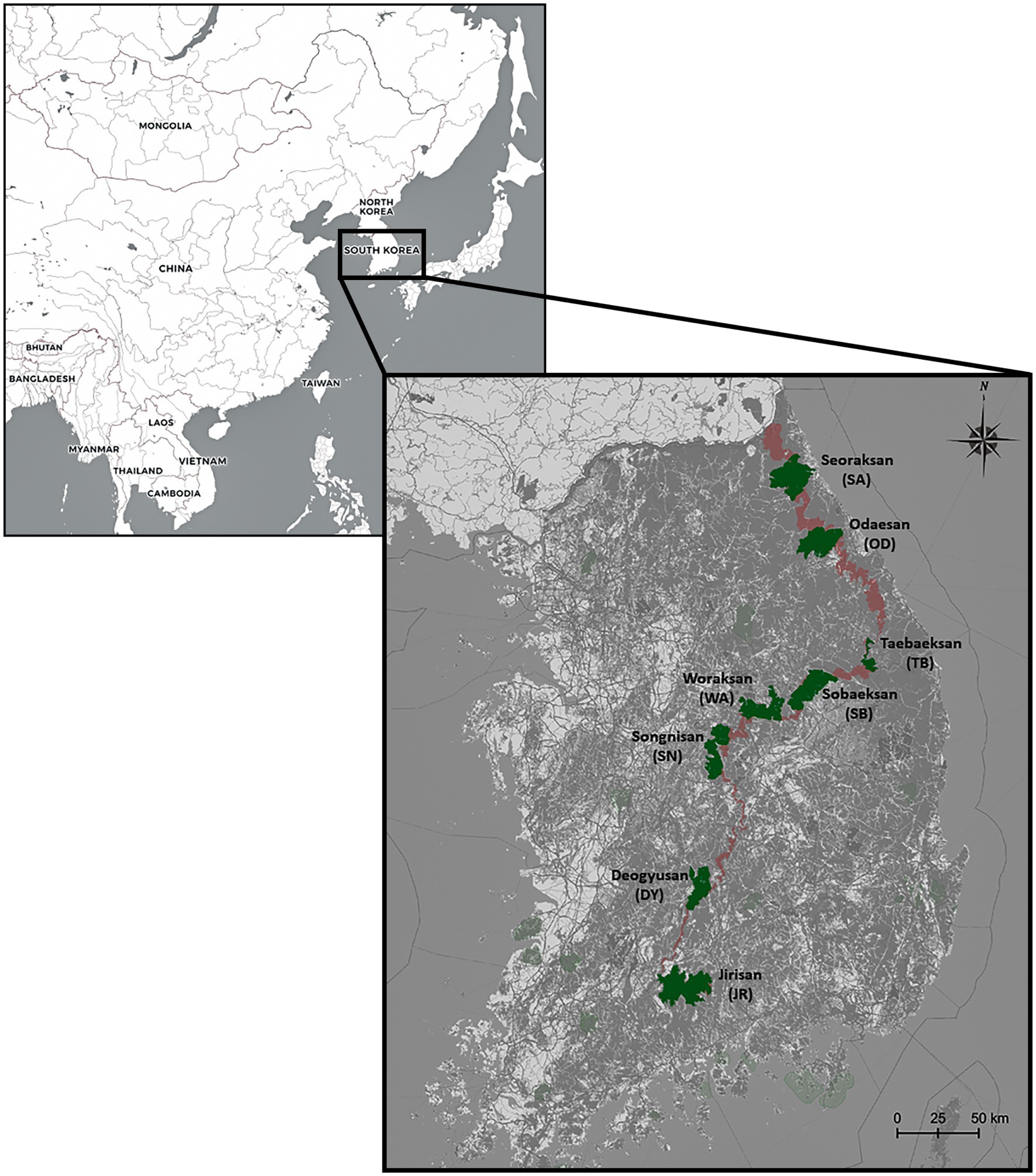
Figure 1. Sampling sites of the eight national park populations of Apodemus agrarius in South Korea. The range in dark red displays the Baekdudaegan Mountain Range (BMR) and areas in green denote the eight national parks examined in this study. The geographic map was generated by Quantum Geographical Information System (QGIS) software v3.10.
Sequencing of mitochondrial DNA cytochrome b gene
Genomic DNA was extracted using a DNeasy Blood and Tissue Kit (Qiagen, Germany), according to its protocol guidelines. We used published primers, L14724 and H15915 (Irwin et al., 1991) and also our newly developed A. agrarius specific primers for amplifying mtDNA cyt b gene in this study (FW: 5′-ACTTTGGCTCCCTCCTAGGT-3′; RV: 5′-GTTGTCCGCCGATTCAGGTA-3′). Polymerase chain reaction (PCR) amplification was carried out using a 2070 thermal cycler (Applied Biosystems, United States) in a total reaction volume of 15 μL containing 10× Dream Taq Green buffer (Thermo Fisher Scientific, United States), 2.0 mM dNTPs (Bio Basic Inc., Canada), 10 pmol each primer, 0.2 U of Taq polymerase (Thermo Fisher Scientific), and 4–30 ng/μL of DNA template. The following thermal cycling conditions were applied for every PCR: initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s and extension at 72°C for 1 min, and final extension at 72°C for 5 min. PCR products were checked in 2% agarose gels stained with Redsafe (iNtRON Biotechnology, Korea), purified enzymatically using Exonuclease I and Shrimp Alkaline Phosphatase (New England BioLabs, United States), and then sequenced in the reverse direction using an ABI 3730xl automated DNA sequencer (Applied Biosystems). The obtained cyt b sequences were edited using CHROMAS v2.4 computer software and aligned using BIOEDIT v7.2.5 (Hall et al., 2011) and finally verified manually.
Microsatellite genotyping
Nine polymorphic microsatellite loci were amplified with published primers, SFM3, SFM9, SFM14, CAA2A, GATAE10A, GACAB3A, GCATD7S, GTTC4A, and MSAA5 for genotyping (Makova et al., 1998; Ohnishi et al., 1998; Wu et al., 2008). Each of the forward primers was labeled with a fluorescent dye (FAM, HEX, or TAMRA) and PCR amplifications were conducted as the same as described for the mtDNA cyt b gene. PCR reactions were carried out using appropriate conditions for each primer pair: 95°C for 5 min, followed by 35 cycles at 95°C for 1 min, 56°C–62°C for 30 s and 72°C for 45 s, and final extension was conducted at 72°C for 20 min. The PCR products were verified, and then were electrophoresed on an ABI 3730xl automated DNA sequencer (Applied Biosystems). Sizes of amplified microsatellite fragments were determined with the ROX 500 bp size standard (ABI) using GENEMAPPER software v5.0 (Applied Biosystems). All alleles were scored manually and checked for an error using Microsatellite Toolkit in MS Excel (Park, 2000).
Population genetic analyses
Genetic diversity
Number of haplotypes (N), haplotype diversity (H), and nucleotide diversity (π) for the mtDNA cyt b were estimated for each national park population and the entire pooled sample of A. agrarius using ARLEQUIN v3.5 (Excoffier and Lischer, 2010). To calculate haplotype richness (HR), the rarefaction method was applied using CONTRIB v1.02 (Petit et al., 1998), which corrected for unbalanced sample sizes across the striped field mouse populations. Two populations (SA_19 and DY_19), which were sampled in 2019, were excluded from this and subsequent analyses, with the exception of mtDNA haplotype network and microsatellite STRUCTURE analyses as these populations had insufficient sample sizes (N < 5). We analyzed the WA and JR populations in 2015 and 2019 separately to compare the genetic diversity between the years. To construct a haplotype network, HAPSTAR v0.7 (Teacher and Griffiths, 2011) was used to infer the phylogenetic relationships among the haplotypes identified.
To investigate levels of microsatellite diversity in A. agrarius from eight national parks within the BMR, mean number of alleles per locus (NA), allelic richness (AR) corrected for unequal sample sizes, observed (HO) and expected (HE) heterozygosity, inbreeding coefficient (FIS), and Hardy–Weinberg equilibrium (HWE) were calculated using GENEPOP v4.0 (Rousset, 2008) and FSTAT v2.9.3.2 (Goudet, 2001). The 95% significance levels for exact test for HWE was corrected using a Bonferroni correction.
Spatio-temporal population structure
To assess the degree of spatial and temporal genetic differentiation among national park populations of A. agrarius, pair-wise FST estimates were statistically evaluated for both mtDNA cyt b and microsatellite genotypes using ARLEQUIN and GENEPOP. The 95% significance levels for pairwise population comparisons were also corrected using the Bonferroni correction. The population genetic structure was further analyzed using a Bayesian model-based clustering algorithm implemented in STRUCTURE v2.3.4 under a model of admixed ancestry among the stripe field mouse populations and correlated allele frequencies. STRUCTURE calculates a likelihood score when the data are forced into a given number of genetic clusters (K) = 1–11. We applied 10 iterations, with 100,000 burn-in steps followed by 1,000,000 Markov Chain Monte Carlo (MCMC) generations. The most likely number of genetic clusters (K value) was determined by using the delta K (ΔK) method implemented in the web-based tool STRUCTURE HARVERSTER website,1 based on the rate of change in the log probability of data between successive K values (Earl and VonHoldt, 2012). Also, STRUCTURE analyses were carried out to assess temporal variation between JR populations that were sampled between 2015 (JR_15) and 2019 (JR_19).
In addition, we tested for isolation by distance (IBD) among the six populations sampled in 2019 at both mtDNA cyt b and microsatellites using the Mantel test in GENALEX v6.502 (Peakall and Smouse, 2006). Populations sampled in 2015 were excluded from this analysis, as detectable temporal differentiation was found (see Results). The geographic surface distance in kilometers between two sampling sites was obtained from the website,2 based on the coordinate information (latitude/longitude) for each location.
We used BARRIER v2.2 (Manni et al., 2004) to investigate the presence of genetic barriers representing the largest genetic discontinuity (i.e., low connectivity) across the BMR populations, which used Voronoi tessellation and Delaunay triangulation to characterize the spatial relationships among the sites in GPS coordinates and applied the Monmonier’s maximum difference algorithm using the pairwise FST matrix generated in ARLEQUIN.
Continental-scale phylogenetic analyses
We investigated phylogenetic relationships across entire Eurasian populations of A. agrarius using previously determined 290 mtDNA cyt b sequences (Liu et al., 2004, 2018; Suzuki et al., 2008; Sakka et al., 2010; Oh et al., 2011, 2013; Yue et al., 2012; Koh et al., 2014; Kim and Park, 2015; Wang et al., 2015; Pereverzeva et al., 2017) (plus our determined 134 haplotype sequences; 802 bp; Supplementary Table S1). Apodemus chevrieri (HQ896683; Yue et al., 2012) and Apodemus peninsulae (HQ660074; Oh et al., 2011) were used as an outgroup. The phylogenetic tree was constructed with Neighbor Joining (NJ) method using as implemented in Mega v10.0.5 (Kumar et al., 2018) and the statistical support was estimated by 1,000 bootstrap replicates.
Results
Levels of genetic diversity in national park populations within the BMR
We found 134 haplotypes in the mtDNA cyt b (802 bp) segments among the 221 individuals from 11 national park populations of A. agrarius including three temporally replicated samples (Table 1; Supplementary Table S2). The analyses revealed that H ranged from 0.800 (WA_19) to 0.988 (JR_15), π from 0.001 (SA_19) to 0.011 (SA_15), and HR from 3.0 (WA_19) to 4.8 (SA_15, TB_19 and JR_15). GenBank accession numbers for the mtDNA haplotypes are ON569125-ON569256.
The identified 134 haplotypes of A. agrarius were connected by 1–38 mutational steps, which showed a “star-like” topology indicative of a relatively shallow evolutionary history (Figure 2A). H001, the most common haplotype, was found in eight populations (OD_19, TB_19, SB_19, WA_15, WA_19, SN_19, JR_15, and JR_19), with a frequency of 18.8% (30 out of 221 individuals), which is likely to be the most ancestral matrilineal lineage. The second most frequent haplotype (H002) occurred only in a frequency of 3.6% (eight individuals). The JR populations had a total of 46 haplotypes and the JR_15 and JR_19 samples had 29 and 19 haplotypes, respectively, and connected by 1–13 mutational steps (Figure 2B). There were only two haplotypes (H001 and H052) shared between JR_15 and JR_19 populations, suggesting that temporal variation might exist in mtDNA genetic composition of the JR populations of A. agrarius.
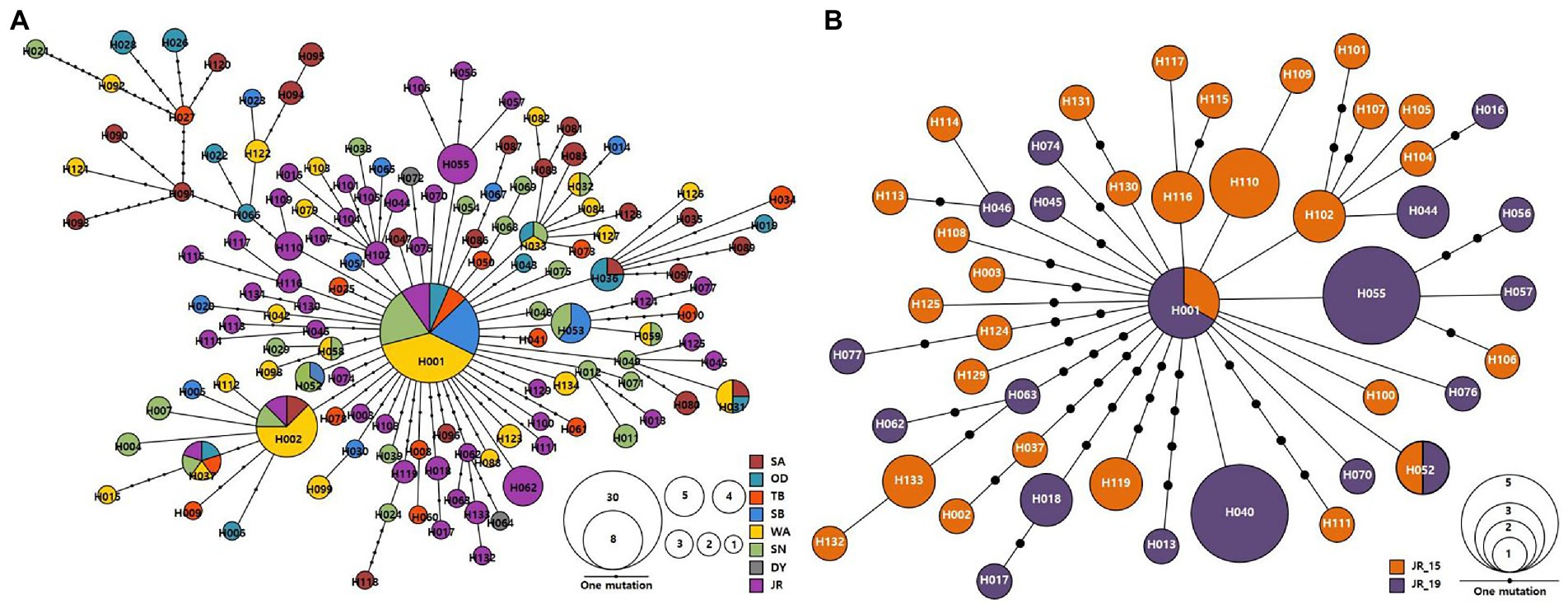
Figure 2. Haplotype network of the observed 134 haplotypes in Apodemus agrarius from entire pooled population (A) and 46 haplotypes from the Jirisan National Park population (JR_15 and JR_19) (B), based on mtDNA cyt b sequences (802 bp). Each line in the network represents a single mutational step between haplotypes, irrespective of its length. The size of the circle is proportional to individual numbers that belong to the respective haplotypes and small, filled circles denote intermediate haplotypes that are not present in our samples, but are necessary to connect all of the observed haplotypes to the network. Different colors indicate different national park populations. Population abbreviations as Table 1.
The NA per population was 9.0, ranging from 3.0 (DY_19) to 12.9 (SN_19). The AR values ranged from 5.8 (WA_19) to 7.1 (WA_15; mean = 6.8), and HO and HE each ranged from 0.593 (WA_19) to 0.798 (OD_19; mean = 0.706) and from 0.805 (WA_19) to 0.892 (WA_15; mean = 0.873), respectively. Most populations except populations of SA_19 and WA_19 had private alleles (PAs), and the highest number of PAs was found in the SN_19 (PAs = 7). The range of FIS values was from 0.089 (OD_19) to 0.283 (WA_19), suggesting moderately high levels of inbreeding occurring within the populations examined (Table 1).
Genetic structure and differentiation
STRUCTURE analyses based on the microsatellites revealed the most likely two genetic clusters (K = 2), but all populations showed similar distributions of individual genotypes, in which all individuals were assigned to approximately equal proportions of the inferred two genetic clusters (Figure 3A), probably because of very low levels of FST values [(overall mean FST = 0.011; Latch et al., 2006)]. The results suggest that populations in the BMR are comprised of a well admixed single gene pool in the national park populations and are impossible to be genetically differentiated. Pairwise FST estimates for mtDNA and microsatellites showed generally low levels of genetic differentiation, but some statistically significant genetic differentiation between populations, including JR_15 vs. JR_19 populations, although they were from the same national park (Table 2). When STRUCTURE analysis was performed for only two temporal samples of the JR population, approximately equal proportions of the two genetic groups exist (Figure 3B), although there was a significant difference in FST statistics (mtDNA cyt b: FST = 0.031; microsatellites: FST = 0.010). The magnitude of genetic differentiation at mtDNA (overall mean FST = 0.044) was found to be exactly four-fold higher than at microsatellites, which is what population genetic theory predicts, as effective population size (Ne) of mtDNA is one quarter of nuclear DNA due to its haploidy and unisexual inheritance (Hartl and Clark, 2007).
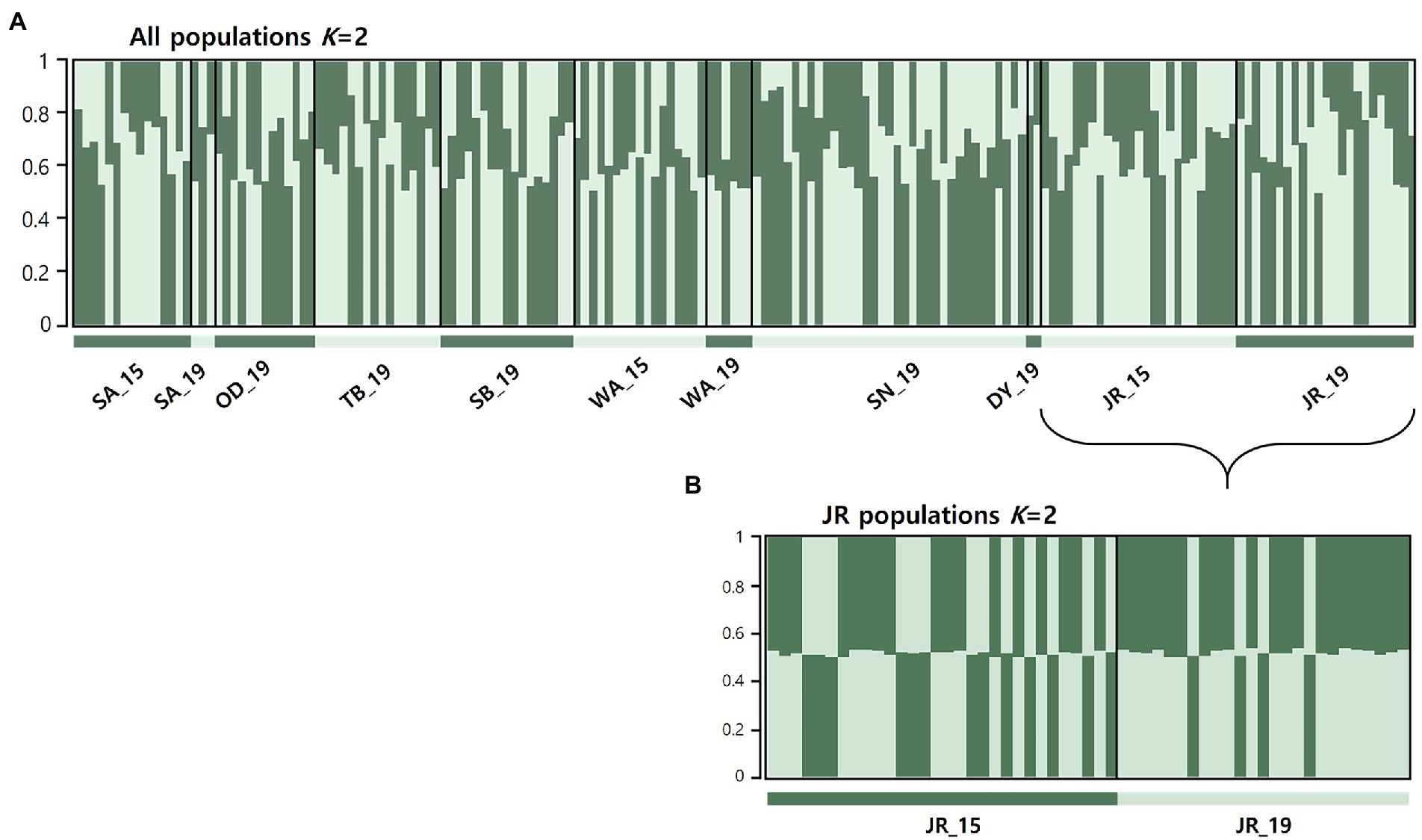
Figure 3. Results of genetic structure using a Bayesian population assignment test with STRUCTURE for the national park populations of Apodemus agrarius, based on nine microsatellite loci. Each individual is represented along the x-axis, and the y-axis denotes the probability of that individual belonging to each of the genetic clusters. (A) Analyses of population structure for the all 11 populations combined. (B) Results of the Jirisan National Park population assignment test with STRUCTURE assuming K = 2.
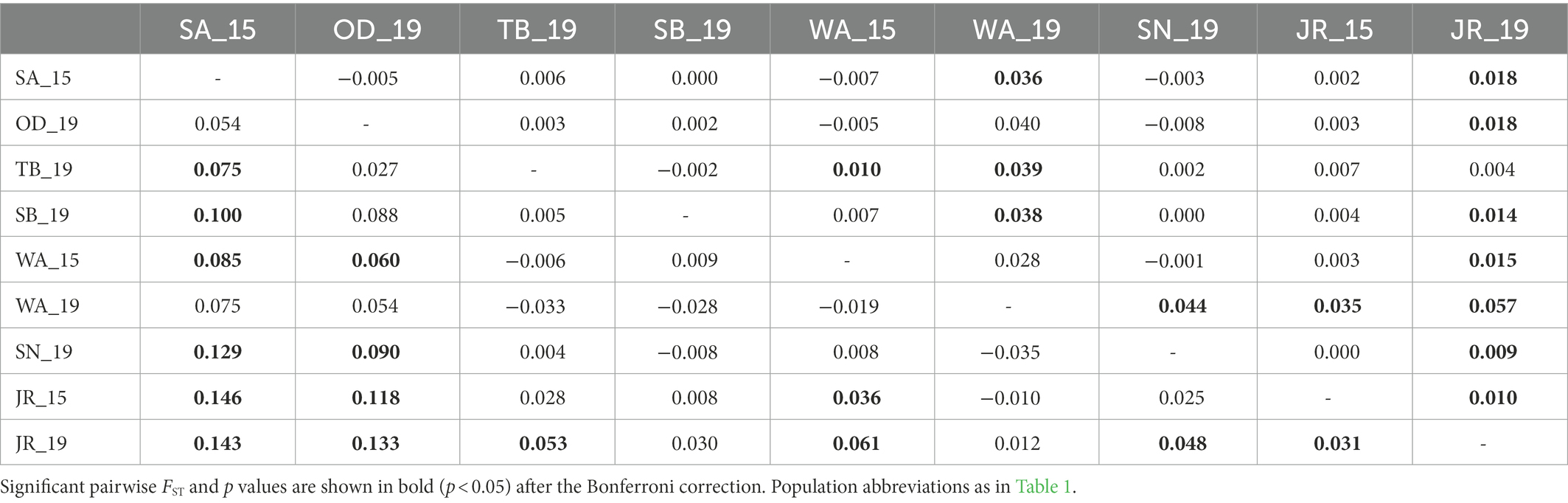
Table 2. Pairwise genetic differentiation (FST) based on mtDNA cyt b sequences (below diagonal) and nine microsatellite loci genotypes (above diagonal) for nine national park populations of Apodemus agrarius within the Baekdudaegan Mountain Range (BMR).
The Mantel test of six populations sampled in 2019 showed a significant positive correlation between geographic (km) and genetic (FST values) distances only in mtDNA but not in microsatellites (mtDNA: r = 0.76, p = 0.04; microsatellites: r = 0.07, p = 0.57; Figure 4), which may represent sex-biased gene flow (Jang et al., 2021). The first barrier identified by Monmonier’s maximum difference algorithm is located between Sobaeksan (SB) and Woraksan (WA) National Parks, and the second barrier existed between Woraksan (WA) and Songnisan (SN) National Parks, as shown in both mtDNA and microsatellites (Figure 5). These results suggest that national parks located at halfway points along the BMR in South Korea, where the geographic orientation of mountain ranges changes, may act as a major physical barrier to the movements of the small rodents.
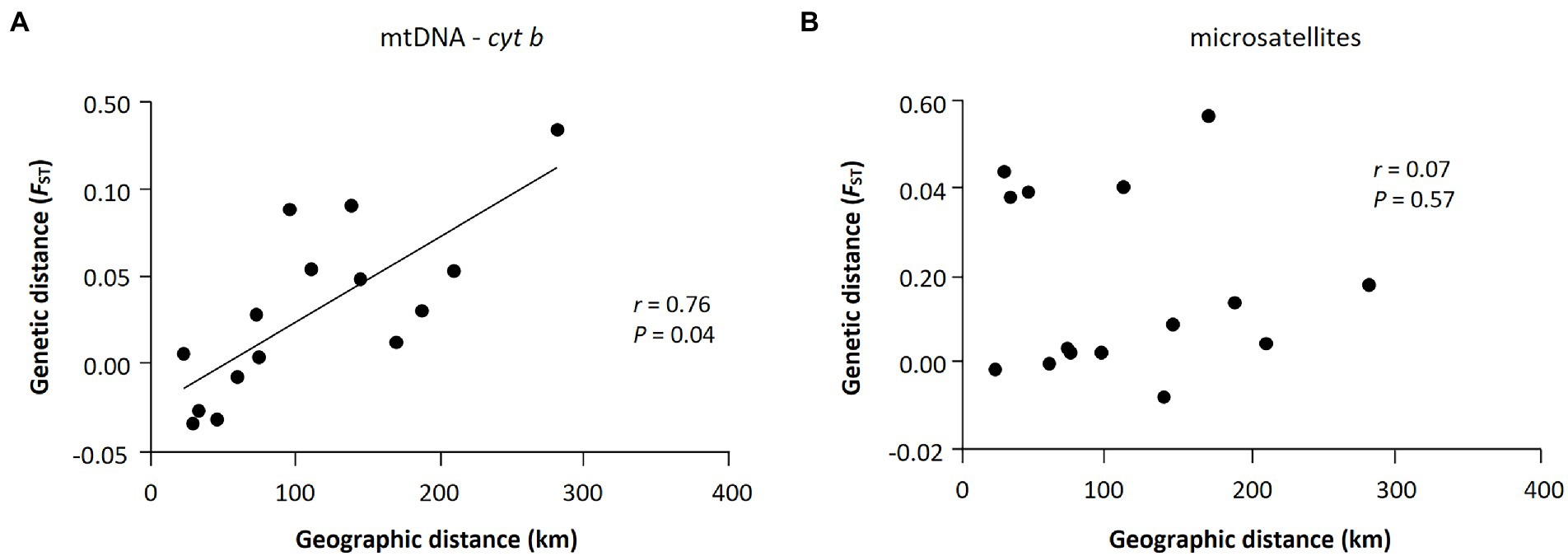
Figure 4. Results of isolations-by-distance (IBD) analysis with the Mantel tests for Apodemus agrarius in national parks within the Baekdudaegan Mountain Range (BMR). (A) and (B) are the results of mtDNA and microsatellites, respectively. The relationship between genetic (FST) and geographic distances was statistically significant only for the mtDNA data set (r = 0.76, p = 0.04).
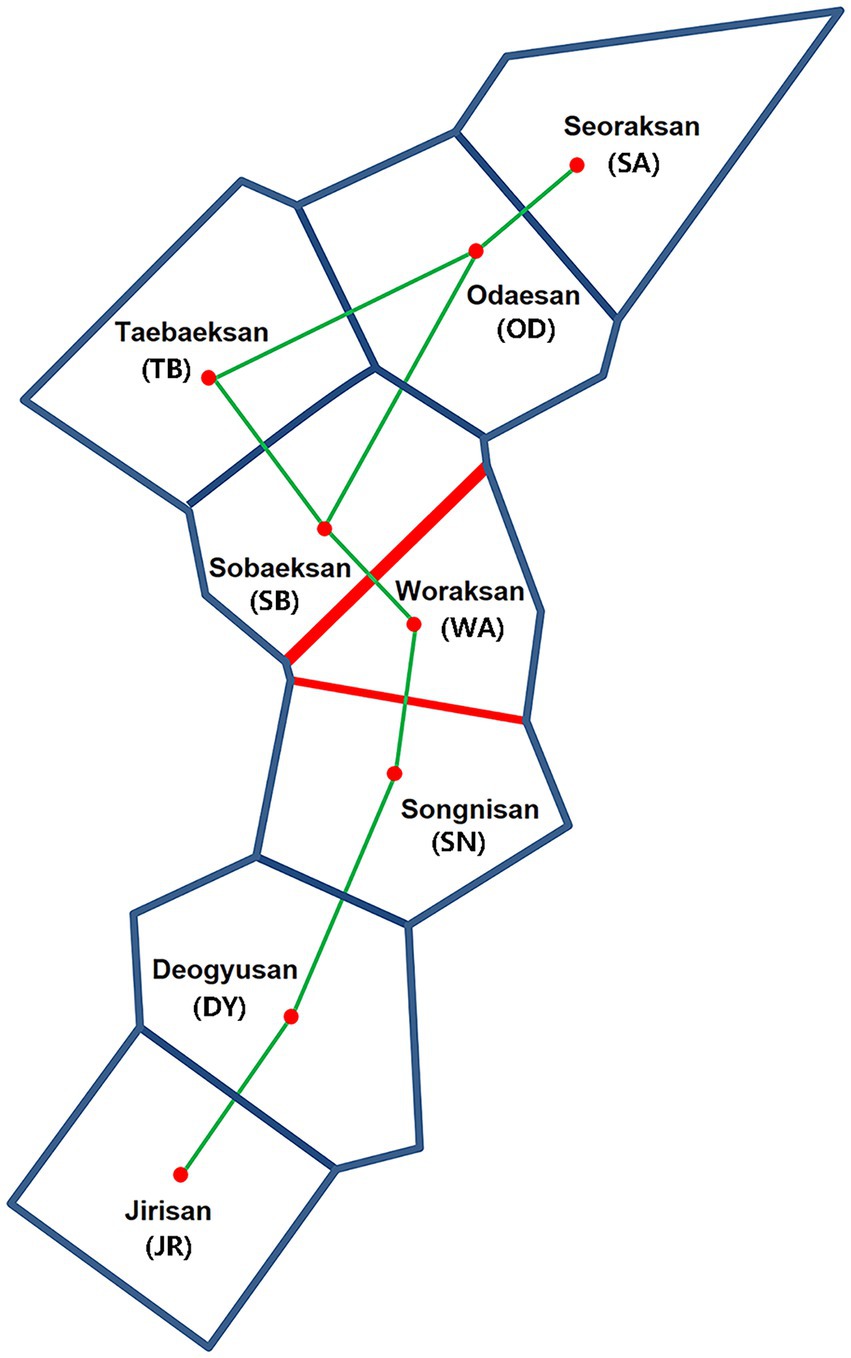
Figure 5. Genetic barriers (shown by red lines) of Apodemus agrarius populations ranked in an order of impermeability (indicated by line thickness) using Delaunay triangulation (connection of each national park; green lines) and Voronoi tessellation (GPS information for each national park; blue lines) as implemented in BARRIER, based on FST values of mtDNA and microsatellites.
Phylogenetic relationships among Eurasian Apodemus agrarius populations
The large-scale mtDNA phylogeny of A. agrarius populations inhabiting Eurasian regions, including the mainland of South Korea, Jeju Island (the southernmost region of South Korea), China, Taiwan, Kazakhstan, Russia, Germany, Italy, and Hungary revealed that three major clades have evolved and Taiwanese clades (Clade 3) were the most ancestral (Supplementary Figure S1). Asian and European populations including our samples (the mainland of South Korea) formed a monophyletic group (Clade 1). Apodemus chejuensis occurring in Jeju Island formed a separate clade (Clade 2).
Discussion
Genetic diversity and inbreeding
We found that while A. agrarius populations inhabiting eight national parks in South Korea have high genetic diversity, they show a genetic signal of non-random mating (based on microsatellites), which strongly suggests the occurrences of inbreeding, as HO is significantly lower than HE for every population analyzed (Table 1). A previous study (Jo et al., 2017) showed that FIS values for 13 populations of A. agrarius in South Korea ranged from 0.023 to 0.280, which are similar levels as what we observed (FIS = 0.089–0.283), supporting the “inbreeding prevalence” hypothesis. Many mammalian species are known to have evolved behaviors that lower the incidences of inbreeding as it may be selected against due to its adverse effects on the fitness (Blouin and Blouin, 1988). However, rodents are generally philopatric, particularly females, despite their large home range for small body size. Therefore, inbreeding can often occur because they do not move away far from their habitat range (Dewsbury, 1988). The observed significant results of IBD at only mtDNA, but not microsatellites may support the hypothesis of female philopatry and male-mediated dispersal in the striped field mouse (Jang et al., 2021). A paradoxical hypothesis would be that the striped field mouse populations may be selected for inbreeding. Some recent studies of mammalian species have suggested that several species may not discriminate against inbreeding and actually seek for inbreeding, particularly when the reproductive cost of inbreeding is low (Szulkin et al., 2009; Rioux-Paquette et al., 2010; Olson et al., 2012). Although the ultimate ecological and evolutionary explanations as to the mechanisms of inbreeding are uncertain, the significant IBD patterns only found in mtDNA would provide genetic evidence supporting the notion that especially female A. agrarius has a higher homing fidelity and therefore more tends to be sedentary (García-Navas et al., 2016; Jang et al., 2021). However, additional analysis using more microsatellite markers would be required to substantiate the findings, as larger number of microsatellite markers (e.g., N > 12) could provide more accurate results, although the number of markers needed may differ among species (Wang et al., 2021).
Population connectivity and female philopatry with male dispersal
The results of our genetic structure analyses at both mtDNA and microsatellites show overall low inter-population genetic differentiation, suggesting their shared a well-admixed gene pool, supported by low FST values of pairwise comparisons and STRUCTURE analyses. Weak genetic differentiation suggests high gene flow taking place among populations (i.e., high connectivity) due to their large dispersal capacity (Jo et al., 2017). However, we should be cautious about the conclusion of the lack of genetic structure, given significant IBD patterns in mtDNA and also the presence of genetic barriers. Although some studies showed that there is a significant relationship between genetic and geographic distances at microsatellites (Jo et al., 2017, 2018), our results revealed the IBD present only in mtDNA, but not in microsatellites. This pattern can be explained by the fact that since Ne of mtDNA is one-quarter of the nuclear genome, population differentiation/divergence time is theoretically predicted to progress four times faster (Moore, 1995). However, a more plausible hypothesis would be that different behavioral patterns between sexes of A. agrarius (i.e., female homing and male-biased dispersal) give rise to the contrasting patterns of IBD between two different marker sets. Most small mammals (e.g., Microtus oeconomus, Mus spicilegus, Eptesicus fuscus, Ctenomys australis, and Rhinolophus ferrumequinum) have male-biased long-distance dispersal selected for inbreeding avoidance (Gundersen and Andreassen, 1998; Poteaux et al., 2008; Mora et al., 2010; Jang et al., 2021; Richardson et al., 2021). The sex differences in dispersal in A. agrarius can therefore likely contribute to the observed patterns of IBD between two marker sets with contrasting modes of inheritance.
Our results of genetic structure and differentiation suggest relatively high gene flow and high connectivity among A. agrarius populations in eight national parks within the BMR, although genetic isolation is also present in some national park areas. In particular, there is a genetic barrier to the dispersal of animals centered around the Woraksan National Park (Figure 5). The previous findings that the Woraksan population of another mammalian species, the great horseshoe bat Rhinolophus ferrumequinum serves as a major “sink” as well as its surrounding areas being as a major genetic barrier support the notion that animal movements in this region would be somehow problematic (Jang et al., 2021). One plausible explanation would be that a provincial highway line 597, which is notorious for a very high risk of roadkill occurrences, traverses the Woraksan National Park and this national road may act as a genetic barrier (Song et al., 2009). Roads can also cause noise pollution by traffic and it has been documented that the main source of noise in the Woraksan comes from roads, which can be stressful for wildlife animals (Korea National Park Research Institute, 2021). Noise can act as a stressor causing physiological and behavioral impairments in wildlife animals (e.g., memory and hearing impairments, depression), which may have led to the isolation of the Woraksan population from nearby populations (Ravindran et al., 2005; Haider et al., 2012; Barzegar et al., 2015). In addition, campsites are increasing now in the Woraksan National Park, which renders large human interference and also grassland, the main habitat of A. agrarius, is steadily declining (Korea National Park Research Institute, 2021). In general, road and rail construction is the main cause of habitat fragmentation, which has severe negative impacts on organism’s movements. In a previous study of investigating landscape elements to pinpoint habitat fragmentation effects on four rodent species (Clethrionomys glareolus, Apodemus flavicollis, Apodemus sylvaticus, and A. agrarius) in Poland, different species responded differently to spatial characteristics because of species-specific behaviors, mobility, and habitat preferences (Kozakiewicz et al., 1999). With respect to the habitat preferences, the impact of habitat fragmentation may be species specific as well, and it can affect demographic characteristics of rodent species in different ways, such as increased mortality due to road kills, modification of behavior, and/or changes in environmental factors (Forman and Deblinger, 2000; Almasieh et al., 2022; Rhim et al., 2003). A study evaluating the role of road edges for wood mice, A. sylvaticus showed that while roads themselves serve as barriers to movement of this species by reducing overall connectivity, road verges may play a role in promoting short-distance movement, suggesting that road edges may be important for improving “small-scale connectivity” in small mammals in Mediterranean woodland (Galantinho et al., 2020). A previous study that calculated minimum-cost path modeling to conserve key habitats and connectivity for large mammals Hyaena hyaena found that roads represent a high risk and therefore proposed a management considering conservation of corridors and reduction of roadkills (Almasieh et al., 2022). For the effective habitat management and conservation, we need to improve the connectivity surrounding the Woraksan National Park by establishing an ecological corridor between poorly connected areas or preserving the landscaping at the edges of roads. As the connectivity of small mammals increases, it would be expected that the entire ecosystem of the BMR protected habitats might more stably sustain.
Temporal genetic structure in the Jirisan National Park population
The results of our temporal analysis indicate that the 2015 and 2019 Jirisan National Park populations considerably differed genetically. A plausible hypothesis would be that environmental changes such as ongoing climate change cause temporal variation in demographic characteristics (e.g., abundances) of the population, which will eventually lead to detectable temporal genetic structure caused by a reduction in genetic diversity. It is well-documented that ongoing climate change has drastic effects on local biodiversity, geographic distribution, life-history evolution, and population sizes (Martinez et al., 2018; Hidasi-Neto et al., 2019; Ruaro et al., 2019). In South Korea, average annual temperature has risen at a rate of 0.23°C decade−1 over the past 50 years during 1954 to 2000s with much faster warming trends since 1980s (Jung et al., 2002). Annual precipitation has also increased sharply during the last several decades, while this rise is primarily due to the increase in frequency and intensity of heavy precipitation during summer (albeit a decrease in spring and winter; Jung et al., 2002, 2011). We postulated that these regional climate changes might have negatively affected A. agrarius abundances, which causes a decrease in Ne and genetic diversity, resulting in the observed temporal structure. This hypothesis would be conceivable, given the mice could not be observed at all at a particular site (the Daeseong area) in Jirisan National Park in 2020, although they had been observed there during 2006 to 2010, based on the 5th Natural Environment Survey (National Institute of Ecology, 2021). The climate change effects would be prevalent across all the national parks (Kim and Kim, 2016). The striped field mice are known to prefer wet habitat environments (Herzig-Straschil et al., 2003). However, during our study period, the Jirisan National Park region encountered a severe drought. According to the Korea Meteorological Administration,3 the average temperature in the Jirisan National Park area gradually increased from 12.8°C in (2014), 13.0°C (2015), 13.6°C (2016), 12.9°C (2017), 12.9°C (2018), and 13.4°C (2019), and annual average precipitation was 1,368 mm (2014), 1,084 mm (2015), 1,349 mm (2016), 746 mm (2017), 1,567 mm (2018), and 1,522 mm (2019) with a severe drought in 2015 and 2017. The catastrophic environmental events and climate change may cause a decline in the population sizes of A. agrarius in recent years, resulting in the temporal genetic structure as an outcome of genetic drift effects. The lower genetic diversity of the Jirisan population sampled in 2019 than in 2015 supports this hypothesis (Table 1). Moreover, our median estimates of linkage disequilibrium (LD)-based contemporary Ne for the Jirisan population are infinity of 246.9~∞ (95% CI) and 100.4 of 48.3~2105.1 for JR_15 and JR_19, respectively, further supporting the hypothesis. Alternative hypothesis would be a simply sampling artifact (e.g., insufficient sample size) that could explain the reason why we observed some temporal variation over 4 years in the gene pool of the populations.
Evolutionary history of Eurasian populations of Apodemus agrarius
The mtDNA based phylogenetic tree shows that Eurasian A. agrarius populations have diverged into three well-separated lineages, Clade 1 representing the mainland of South Korea, Russia, China, Kazakhstan, and Europe, Clade 2 representing Jeju Island and Clade 3 the Taiwan population, which is the most ancestral lineage. These phylogenetic patterns are consistent with previous studies (Kim and Park, 2015; Latinne et al., 2020; Kozyra et al., 2021) that found that geographically distinct clades have evolved in A. agrarius throughout its entire geographical range. Apodemus agrarius seems to share the recently evolved common lineage probably because this species has colonized and expanded its distributional range rapidly throughout Central Asia and Europe during the last interglacial periods, which would have been facilitated by its ecological characteristics such as high ecological plasticity and synanthropic habits of the striped field mouse (Latinne et al., 2020).
Also, our haplotype network analysis for the BMR populations forms a star-like structure, which means that A. agrarius population has occurred a rapid differentiation during a short period of time (Slatkin and Hudson, 1991). As suggested in previous studies, due to the influence of the LGM (last glacial maximum) about 20,000 to 30,000 years ago, A. agrarius population experienced a bottleneck at the BMR during the Ice Age, which is known as a refuge on the Korean Peninsula and it is estimated that the populations rapidly increased for a short period of time after the retreat of the glacier (Suzuki et al., 2008; Sakka et al., 2010; Kim and Park, 2015). Also, considering the overall low genetic distances among the haplotypes, it would be conceivable that geographic/historic isolation following population expansion after the LGM may have led to the shallow phylogeographic structure (Hewitt, 2000).
Implications for conservation and management
Small mammals might be susceptible to deteriorating population connectivity caused by habitat destruction and fragmentation (Streatfeild, 2009). Small social mammals may be even more vulnerable to the low habitat connectivity as they often show different behavioral patterns between sexes (female home-site fidelity and male-mediated dispersal; García-Navas et al., 2016; Jang et al., 2021). Because the connectivity largely affects dispersal of not only mice, but also other mammalian species across the national park habitats, large efforts need to be made to improve the connectivity for the purpose of sustaining regional biodiversity (Kahilainen et al., 2014; Almasieh et al., 2022). Particular attention should be paid to restoring possible “genetic barrier” located in the Woraksan National Park and continuous monitoring and priority on development of conservation and management program, such as prioritization or construction of ecological corridor would be required (Thapa et al., 2018; Rezaei et al., 2022). In order to improve the connectivity, it is necessary to identify physical barriers (e.g., roads) to animal movements that cause population disconnection or isolation (Almasieh et al., 2022). For this, we need to conduct ecological and genetic surveys at a smaller geographic scale surrounding the Workasan National Park areas and pinpoint the exact geographical points where the genetic barrier really occurs. Moreover, we suggest that Seoraksan (SA) and Songnisan (SN) National Parks should be given high priority for conservation and management by considering them as “genetic diversity hotspots” for the protection of A. agrarius’s diversity and endemism, as they have relatively higher haplotype/allelic richness and more unique haplotypes/alleles. Studying other coexisting rodents with a similar dispersal range and habitat utilization in the national parks, such as Apodemus peninsulae and Myodes rufocanus will help to better understand the habitat connectivity and landscape genetic structure in these protected areas. Continuous ecological as well as genetic monitoring will be important for identifying how the biodiversity hotspot regions in Korea will sustain over the long term under ongoing climate changes.
Overall, our study shows that national park populations of the most common rodent species, A. agrarius harbor extraordinarily high levels of genetic diversity, despite the occurrences of inbreeding, which is partly due to female’s home-site fidelity. Unexpectedly, there is temporal genetic variation detected in the Jirisan National Park population over 4 years, which is likely due to the declines in population sizes caused by environmental changes. While A. agrarius populations in the BMR represents relatively high gene flow and high connectivity, genetic barriers also exist in some areas. The discovery of these genetic barriers advocates a need for a variety of effective habitat management, e.g., the prioritization and construction of ecological corridors, to improve the connectivity surrounding the Woraksan National Park areas. Our study highlights the need for population or habitat connectivity managements of small mammals in protected areas in South Korea and provide insights into habitat managements for the terrestrial biodiversity hotspots.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession numbers can be found at: NCBI; ON569125-ON569256.
Ethics statement
The capture and use of the study animals for this study were approved by Korea National Park Research Institute and National Institute of Ecology, Ministry of Environment in Korea. Sample collections were followed by guidelines (the Animal Protection Act) of Department of Animal Protection and Welfare, Animal and Plant Quarantine Agency (APQA) in Korea. We mainly took hair or oral swab samples to minimize unnecessary inhumane procedures. However, no research permits were required for the collection of mouse samples as this species is not listed as endangered in Korea.
Author contributions
HRK, HHM, B-JK, and HJL conceived and designed the study. HRK, JYK, and B-JK performed the field collection. YRK, JHK, and HJL conducted the molecular experiments and data analyses. HJL coordinated the research project. YRK, JHK, B-JK, and HJL wrote an original draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by a grant from the Korea National Park Research Institute (project number: NPRI 2019-11) and Korea National Park Service in South Korea. This work was supported in part by the National Institute of Ecology, funded by Korea Ministry of Environment (MOE) (No. NIE-B-2023-12), and by Korea Environment Industry and Technology Institute through development of modelling for forecasting expansion and risk assessment of alien species, funded by Korea MOE (No. 20180022700013).
Acknowledgments
We thank members of the Korea National Park Research Institute for organizing to collect the striped field mouse samples in the field. We are grateful to Song Yi Baek for microsatellite genotyping on the 2015 samples and to Ji Eun Jang for helping to prepare for the reports of the research project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1038058/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Phylogenetic relationships based on mtDNA cyt b gene of Apodemus agrarius from Asia and Europe, including South Korea (the mainland, Jeju Island), Russia, China, Kazakhstan, Europe (Germany, Italy, Hungary) and Taiwan. Numbers at the branches represent the bootstrap support values for neighbor joining (NJ) tree.
Footnotes
1. ^http://taylor0.biology.ucla.edu/structureHarvester/
2. ^http://www.movable-type.co.uk/scripts/latlong-nomodule.html
References
Aitken, S. N., Yeaman, S., Holliday, J. A., Wang, T., and Curtis-McLane, S. (2008). Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 1, 95–111. doi: 10.1111/j.1752-4571.2007.00013.x
Allendorf, F., Luikart, G., and Aitken, S. (2013). Conservation and the genetics of populations. Oxford: Wiley-Blackwell.
Almasieh, K., Mohammadi, A., and Alvandi, R. (2022). Identifying core habitats and corridors of a near threatened carnivore, striped hyaena (Hyaena hyaena) in southwestern Iran. Sci. Rep. 12:3425. doi: 10.1016/j.scitotenv.2022.155753
Ballesteros-Mejia, L., Lima, J. S., and Collevatti, R. G. (2020). Spatially-explicit analyses reveal the distribution of genetic diversity and plant conservation status in Cerrado biome. Biodivers. Conserv. 29, 1537–1554. doi: 10.1007/s10531-018-1588-9
Barzegar, M., Sajjadi, F. S., Talaei, S. A., Hamidi, G., and Salami, M. (2015). Prenatal exposure to noise stress: anxiety, impaired spatial memory, and deteriorated hippocampal plasticity in postnatal life. Hippocampus 25, 187–196. doi: 10.1002/hipo.22363
Binks, R. M., Byrne, M., McMahon, K., Pitt, G., Murray, K., and Evans, R. D. (2019). Habitat discontinuities form strong barriers to gene flow among mangrove populations, despite the capacity for long-distance dispersal. Divers. Distrib. 25, 298–309. doi: 10.1111/ddi.12851
Blouin, S. F., and Blouin, M. (1988). Inbreeding avoidance behaviors. Trends Ecol. Evol. 3, 230–233. doi: 10.1016/0169-5347(88)90164-4
Boulding, E. G. (2008). “Genetic diversity, adaptive potential, and population viability in changing environments” in Conservation biology: Evolution in action. eds. S. Carroll and C. Fox (Oxford: Oxford University Press), 199–219.
Brock, R. E., and Kelt, D. A. (2004). Keystone effects of the endangered stephens' kangaroo rat (Dipodomys stephensi). Biol. Conserv. 116, 131–139. doi: 10.1016/S0006-3207(03)00184-8
Byeon, S., Jang, J., Choi, H., Kim, H., and Lee, H. (2018). Genetic diversity and phylogenetic relationships of the greater horseshoe bat, Rhinolophus ferrumequinum, from national parks of Korea including the Baekdudaegan mountain range. J. Natl. Park. Res. 9, 336–342.
Choi, Y.-K. (2004). “Baekdudaegan, the central axis of the Korean peninsular: the path toward management strategies regarding to its concepts” in Ecological issues in a changing world. eds. S. Hong, et al. (Dordrecht: Springer), 355–383.
Chung, M. Y., Son, S., Suh, G. U., Herrando-Moraira, S., Lee, C. H., López-Pujol, J., et al. (2018). The Korean Baekdudaegan Mountains: a glacial refugium and a biodiversity hotspot that needs to be conserved. Front. Genet. 9:489. doi: 10.3389/fgene.2018.00489
Clutton-Brock, T. H., and Lukas, D. (2012). The evolution of social philopatry and dispersal in female mammals. Mol. Ecol. 21, 472–492. doi: 10.1111/j.1365-294X.2011.05232.x
Davidson, A. D., and Lightfoot, D. C. (2007). Interactive effects of keystone rodents on the structure of desert grassland arthropod communities. Ecography 30, 515–525. doi: 10.1111/j.0906-7590.2007.05032.x
Dewsbury, D. A. (1988). Kin discrimination and reproductive behavior in muroid rodents. Behav. Genet. 18, 525–536. doi: 10.1007/BF01065519
Earl, D. A., and VonHoldt, B. M. (2012). Structure harvester: a website and program for visualizing structure output and implementing the evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Epps, C. W., Crowhurst, R. S., and Nickerson, B. S. (2018). Assessing changes in functional connectivity in a desert bighorn sheep metapopulation after two generations. Mol. Ecol. 27, 2334–2346. doi: 10.1111/mec.14586
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Forman, R. T., and Deblinger, R. D. (2000). The ecological road-effect zone of a Massachusetts (USA) suburban highway. Conserv. Biol. 14, 36–46. doi: 10.1046/j.1523-1739.2000.99088.x
Freeland, J. R., Biss, P., and Silvertown, J. (2012). Contrasting patterns of pollen and seed flow influence the spatial genetic structure of sweet vernal grass (Anthoxanthum odoratum) populations. J. Hered. 103, 28–35. doi: 10.1093/jhered/esr111
Galantinho, A., Herrera, J. M., Eufrázio, S., Silva, C., Carvalho, F., Alpizar-Jara, R., et al. (2020). Road verges provide connectivity for small mammals: a case study with wood mice (Apodemus sylvaticus) in an agro-silvo pastoral system. J. Environ. Manage. 258:110033. doi: 10.1016/j.jenvman.2019.110033
García-Navas, V., Bonnet, T., Waldvogel, D., Camenisch, D., and Postma, E. (2016). Consequences of natal philopatry for reproductive success and mate choice in an alpine rodent. Behav. Ecol. 27, 1158–1166. doi: 10.1093/beheco/arw031
Goudet, J. (2001). Fstat, a program to estimate and test gene diversities and fixation indices, version 2.9.3. Available at: http://www2.unil.ch/popgen/softwares/fstat.htm
Gundersen, G., and Andreassen, H. P. (1998). Causes and consequences of natal dispersal in root voles, Microtus oeconomus. Anim. Behav. 56, 1355–1366. doi: 10.1006/anbe.1998.0911
Haider, S., Naqvi, F., Batool, Z., Tabassum, S., Perveen, T., Saleem, S., et al. (2012). Decreased hippocampal 5-HT and DA levels following sub-chronic exposure to noise stress: impairment in both spatial and recognition memory in male rats. Sci. Pharm. 80, 1001–1011. doi: 10.3797/scipharm.1207-15
Hall, T., Biosciences, I., and Carlsbad, C. (2011). Bioedit: an important software for molecular biology. GERF Bull. Biosci. 2, 60–61.
Hanson, J. O., Veríssimo, A., Velo-Antón, G., Marques, A., Camacho-Sanchez, M., Martínez-Solano, Í., et al. (2021). Evaluating surrogates of genetic diversity for conservation planning. Conserv. Biol. 35, 634–642. doi: 10.1111/cobi.13602
Hartl, D. L., and Clark, A. G. (2007). Principles of population genetics. 4th. Sinauer Associates. Sunderland, MA.
Herzig-Straschil, B., Bihari, Z., and Spitzenberger, F. (2003). Recent changes in the distribution of the field mouse (Apodemus agrarius) in the western part of the Carpathian basin. Ann. Naturhist. Mus. Wien B. 105, 421–428.
Hewitt, G. (2000). The genetic legacy of the quaternary ice ages. Nature 405, 907–913. doi: 10.1038/35016000
Hidasi-Neto, J., Joner, D. C., Resende, F., de Macedo Monteiro, L., Faleiro, F. V., Loyola, R. D., et al. (2019). Climate change will drive mammal species loss and biotic homogenization in the Cerrado biodiversity hotspot. Perspect. Ecol. Conserv. 17, 57–63. doi: 10.1016/j.pecon.2019.02.001
Horskins, K., Mather, P. B., and Wilson, J. C. (2006). Corridors and connectivity: when use and function do not equate. Landsc. Ecol. 21, 641–655. doi: 10.1007/s10980-005-5203-6
Irwin, D. M., Kocher, T. D., and Wilson, A. C. (1991). Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 32, 128–144. doi: 10.1007/BF02515385
Jang, J. E., Byeon, S. Y., Kim, H. R., Kim, J. Y., Myeong, H. H., and Lee, H. J. (2021). Genetic evidence for sex-biased dispersal and cryptic diversity in the greater horseshoe bat. Rhinolophus ferrumequinum. Biodivers. Conserv. 30, 847–864. doi: 10.1007/s10531-021-02120-y
Jo, Y.-S., Kim, H.-N., Baccus, J. T., and Jung, J. (2017). Genetic differentiation of the Korean striped field mouse, Apodemus agrarius (Muridae, Rodentia), based on microsatellite polymorphism. Mammalia 81, 297–307. doi: 10.1515/mammalia-2015-0152
Jo, Y.-S., Lee, S.-R., Baccus, J. T., Jung, J., and Forstner, M. R. (2018). Environmental factors affecting population level genetic divergence of the striped field mouse (Apodemus agrarius) in South Korea. Ecol. Res. 33, 989–999. doi: 10.1007/s11284-018-1613-1
Johnson, K. (2001). Hantaviruses: history and overview. Hantaviruses 256, 1–14. doi: 10.1007/978-3-642-56753-7_1
Jones, K. E., and Safi, K. (2011). Ecology and evolution of mammalian biodiversity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2451–2461. doi: 10.1098/rstb.2011.0090
Jung, I.-W., Bae, D.-H., and Kim, G. (2011). Recent trends of mean and extreme precipitation in Korea. Int. J. Climatol. 31, 359–370. doi: 10.1002/joc.2068
Jung, H. S., Choi, Y., Oh, J. H., and Lim, G. H. (2002). Recent trends in temperature and precipitation over South Korea. Int. J. Climatol. 22, 1327–1337. doi: 10.1002/joc.797
Kahilainen, A., Puurtinen, M., and Kotiaho, J. S. (2014). Conservation implications of species–genetic diversity correlations. Glob. Ecol. Conserv. 2, 315–323. doi: 10.1016/j.gecco.2014.10.013
Kaneko, Y., Kryštufek, B., Zagarondnyuk, I., Vohralík, V., Batsaikhan, N., Avirmed, D., et al. (2016). Apodemus agrarius (errata version published in 2017). The IUCN red list of threatened species 2016: e.T1888A115057408. Available at: https://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T1888A22422191.en (Accessed December 05, 2022).
Kang, S., and Jeong, J. (2017). A report of Korea national park species list (2016). J. Natl. Park. Res. 8, 83–88.
Keyghobadi, N. (2007). The genetic implications of habitat fragmentation for animals. Can. J. Zool. 85, 1049–1064. doi: 10.1139/Z07-095
Khimoun, A., Eraud, C., Ollivier, A., Arnoux, E., Rocheteau, V., Bely, M., et al. (2016). Habitat specialization predicts genetic response to fragmentation in tropical birds. Mol. Ecol. 25, 3831–3844. doi: 10.1111/mec.13733
Kim, E.-K. (2010). Terrestrial mammalia of the Korea national parks in Baekdudaegan. J. Natl. Park. Res. 1, 104–107.
Kim, C.-C., and Kim, T.-G. (2016). Evaluation on climate change vulnerability of Korea national parks. Korean. J. Ecol. Environ 49, 42–50. doi: 10.11614/KSL.2016.49.1.042
Kim, H. R., and Park, Y. C. (2015). Genetic diversity and genetic structure of the striped field mouse Apodemus agrarius coreae (Muridae, Rodentia) in Korea. Gene 572, 292–297. doi: 10.1016/j.gene.2015.08.014
Koh, H. S., Lee, W.-J., and Kocher, T. D. (2000). The genetic relationships of two subspecies of striped field mice, Apodemus agrarius coreae and Apodemus agrarius chejuensis. Heredity 85, 30–36. doi: 10.1046/j.1365-2540.2000.00723.x
Koh, H. S., Shaner, P. J., Csorba, G., Wang, Y. J., Jang, K. H., and Lee, J. H. (2014). Comparative genetics of Apodemus agrarius (Rodentia: Mammalia) from insular and continental Eurasian populations: cytochrome b sequence analyses. Acta Zool. Acad. Sci. Hung. 60, 73–84.
Koh, H.-S., and Yoo, B.-S. (1992). Variation of mitochondrial DNA in two subspecies of striped field mice, Apodemus agrarius coreae and Apodemus agrarius chejuensis, from Korea. Korean J. Zool. 35, 332–338.
Korea National Park Research Institute (2021). 2020 Woraksan Gungnip Gongwon gongwon jawon josa. South Korea: Korea National Park Research Institute.
Kozakiewicz, M., Gortat, T., Kozakiewicz, A., and Barkowska, M. (1999). Effects of habitat fragmentation on four rodent species in a polish farm landscape. Landsc. Ecol. 14, 391–400. doi: 10.1023/A:1008070610187
Kozyra, K., Zając, T. M., Ansorge, H., Wierzbicki, H., Moska, M., Stanko, M., et al. (2021). Late Pleistocene expansion of small murid rodents across the palearctic in relation to the past environmental changes. Genes 12:642. doi: 10.3390/genes12050642
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwon, Y.-S. (2009). Comparison of birds distribution between national park and province park – Sobaeksan national Park and Taebaeksan Province Park. J. Natl. Park. Res. 1, 45–52.
Latch, E. K., Dharmarajan, G., Glaubitz, J. C., and Rhodes, O. E. (2006). Relative performance of bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv. Genet. 7, 295–302. doi: 10.1007/s10592-005-9098-1
Latinne, A., Navascués, M., Pavlenko, M., Kartavtseva, I., Ulrich, R. G., Tiouchichine, M.-L., et al. (2020). Phylogeography of the striped field mouse, Apodemus agrarius (Rodentia: Muridae), throughout its distribution range in the Palaearctic region. Mamm. Biol. 100, 19–31. doi: 10.1007/s42991-019-00001-0
Lee, D. P. (2003). Birds in the Baekdudaegan from Jirisan to Deokyusan: a review. Korean J. Environ. Ecol. 16, 487–497.
Lim, D.-O., Kim, Y.-S., Park, Y.-K., Ryu, Y.-M., and Koh, M.-H. (2004). Vascular plants of mt. Deog-yu area in the Baekdudaegan. Korean J. Environ. Ecol. 18, 107–123.
Liu, S. Y., He, K., Chen, S. D., Jin, W., Murphy, R. W., Tang, M. K., et al. (2018). How many species of Apodemus and Rattus occur in China? A survey based on mitochondrial cyt b and morphological analyses. Zool. Res. 39, 309–320. doi: 10.24272/j.issn.2095-8137.2018.053
Liu, X., Wei, F., Li, M., Jiang, X., Feng, Z., and Hu, J. (2004). Molecular phylogeny and taxonomy of wood mice (genus Apodemus Kaup, 1829) based on complete mtDNA cytochrome b sequences, with emphasis on Chinese species. Mol. Phylogenet. Evol. 33, 1–15. doi: 10.1016/j.ympev.2004.05.011
Makova, K., Patton, J., Krysanov, E. Y., Chesser, R., and Baker, R. (1998). Microsatellite markers in wood mouse and striped field mouse (genus Apodemus). Mol. Ecol. 7, 247–249. doi: 10.1111/j.1365-294x.1998.00315.x
Manel, S., Schwartz, M. K., Luikart, G., and Taberlet, P. (2003). Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18, 189–197. doi: 10.1016/S0169-5347(03)00008-9
Manni, F., Guérard, E., and Heyer, E. (2004). Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using monmonier's algorithm. Hum. Biol. 76, 173–190. doi: 10.1353/hub.2004.0034
Martinez, A. S., Willoughby, J. R., and Christie, M. R. (2018). Genetic diversity in fishes is influenced by habitat type and life-history variation. Ecol. Evol. 8, 12022–12031. doi: 10.1002/ece3.4661
Matocq, M., Kelly, P., Rippert, J., and Phillips, S. (2017). Population genetic structure of the riparian brush rabbit (Sylvilagus bachmani riparius): using multiple marker systems to gain insight into historic and ongoing genetic connectivity. University of Nevada, Reno and California State University, Stanislaus.
Merriam, G. (1984). “Connectivity: a fundamental ecological characteristic of landscape pattern” in Proceedings of the first international seminar on methodology in landscape ecological research and planning. eds. J. Brandt and P. A. Agger (Rosskilde: Rosskilde University Centre)
Mohammadi, A., Almasieh, K., Nayeri, D., Adibi, M. A., and Wan, H. Y. (2022). Comparison of habitat suitability and connectivity modelling for three carnivores of conservation concern in an Iranian montane landscape. Landsc. Ecol. 37, 411–430. doi: 10.1007/s10980-021-01386-5
Moore, W. S. (1995). Inferring phylogenies from mtDNA variation: mitochondrial-gene trees versus nuclear-gene trees. Evolution 49, 718–726. doi: 10.1111/j.1558-5646.1995.tb02308.x
Mora, M. S., Mapelli, F. J., Gaggiotti, O. E., Kittlein, M. J., and Lessa, E. P. (2010). Dispersal and population structure at different spatial scales in the subterranean rodent Ctenomys australis. BMC Genet. 11, 1–14. doi: 10.1186/1471-2156-11-9
National Institute of Ecology. (2021). 5th Natural Environment Survey. National Institute of Ecology, Korea.
Oh, D. J., Kim, T. W., Chang, M. H., Han, S. H., Oh, H. S., and Kim, S. J. (2011). The mitochondrial genome of Apodemus peninsulae (Rodentia, Muridae). Mitochondrial DNA 22, 99–101. doi: 10.3109/19401736.2011.624612
Oh, D. J., Kim, T. W., Chang, M. H., Han, S. H., Oh, H. S., and Kim, S. J. (2013). Migration route estimation of the Jeju striped field mouse Apodemus agrarius chejuensis (Rodentia, Muridae). Mitochondrial DNA 24, 137–144. doi: 10.3109/19401736.2012.726619
Ohnishi, N., Ishibashi, Y., Saitoh, T., Abe, S., and Yoshida, M. (1998). Polymorphic microsatellite DNA markers in the Japanese wood mouse Apodemus argenteus. Mol. Ecol. 7, 1431–1432. doi: 10.1111/j.1365-294x.1998.00445.x
Olson, L., Blumstein, D., Pollinger, J., and Wayne, R. (2012). No evidence of inbreeding avoidance despite demonstrated survival costs in a polygynous rodent. Mol. Ecol. 21, 562–571. doi: 10.1111/j.1365-294X.2011.05389
Park, S. (2000). Microsatellite toolkit for ms excel 97 or 2000. Dublin, Ireland, Molecular Population Genetics Lab, Smurfit Institute of Genetics, Trinity College.
Peakall, R., and Smouse, P. E. (2006). Genalex 6: genetic analysis in excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Pereira, H. M., Leadley, P. W., Proença, V., Alkemade, R., Scharlemann, J. P., Fernandez-Manjarrés, J. F., et al. (2010). Scenarios for global biodiversity in the 21st century. Science 330, 1496–1501. doi: 10.1126/science.1196624
Pereverzeva, V. V., Primak, A. A., Pavlenko, M. V., Dokuchaev, N. E., and Evdokimova, A. A. (2017). Genetic features and the putative sources of formation of isolated populations of the striped field mouse Apodemus agrarius Pallas, 1771 in Magadan oblast. Russ. J. Biol. Invasions 8, 87–100. doi: 10.1134/S2075111717010106
Petit, R. J., El Mousadik, A., and Pons, O. (1998). Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12, 844–855. doi: 10.1046/j.1523-1739.1998.96489.x
Poteaux, C., Busquet, N., Gouat, P., Katona, K., and Baudoin, C. (2008). Socio-genetic structure of mound-building mice, Mus spicilegus, in autumn and early spring. Biol. J. Linn. Soc. Lond. 93, 689–699. doi: 10.1111/j.1095-8312.2007.00944.x
Rands, M. R., Adams, W. M., Bennun, L., Butchart, S. H., Clements, A., Coomes, D., et al. (2010). Biodiversity conservation: challenges beyond 2010. Science 329, 1298–1303. doi: 10.1126/science.1189138
Ravindran, R., Devi, R. S., Samson, J., and Senthilvelan, M. (2005). Noise-stress-induced brain neurotransmitter changes and the effect of Ocimum sanctum (Linn) treatment in albino rats. J. Pharmacol. Sci. 98, 354–360. doi: 10.1254/jphs.FP0050127
Rezaei, S., Mohammadi, A., Malakoutikhah, S., and Khosravi, R. (2022). Combining multiscale niche modeling, landscape connectivity, and gap analysis to prioritize habitats for conservation of striped hyaena (Hyaena hyaena). PLoS One 17:e0260807. doi: 10.1371/journal.pone.0260807
Rhim, S.-J., Lee, C.-B., Hur, W.-H., Park, Y.-S., Choi, S.-Y., Piao, R., et al. (2003). Influence of roads on small rodents population in fragmented forest areas, South Korea. J. For. Res. 14, 155–158. doi: 10.1007/BF02856784
Richardson, J. L., Michaelides, S., Combs, M., Djan, M., Bisch, L., Barrett, K., et al. (2021). Dispersal ability predicts spatial genetic structure in native mammals persisting across an urbanization gradient. Evol. Appl. 14, 163–177. doi: 10.1111/eva.13133
Rioux-Paquette, E., Festa-Bianchet, M., and Coltman, D. W. (2010). No inbreeding avoidance in an isolated population of bighorn sheep. Anim. Behav. 80, 865–871. doi: 10.1016/j.anbehav.2010.08.006
Rousset, F. (2008). Genepop’007: a complete re-implementation of the genepop software for windows and linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Ruaro, R., Conceição, E. O., Silva, J. C., Cafofo, E. G., Angulo-Valencia, M. A., Mantovano, T., et al. (2019). Climate change will decrease the range of a keystone fish species in La Plata River basin, South America. Hydrobiologia 836, 1–19. doi: 10.1007/s10750-019-3904-0
Sakka, H., Quere, J. P., Kartavtseva, I., Pavlenko, M., Chelomina, G., Atopkin, D., et al. (2010). Comparative phylogeography of four Apodemus species (Mammalia: Rodentia) in the Asian Far East: evidence of quaternary climatic changes in their genetic structure. Biol. J. Linn. Soc. Lond. 100, 797–821. doi: 10.1111/j.1095-8312.2010.01477.x
Serizawa, K., Suzuki, H., Iwasa, M. A., Tsuchiya, K., Pavlenko, M. V., Kartavtseva, I. V., et al. (2002). A spatial aspect on mitochondrial DNA genealogy in Apodemus peninsulae from East Asia. Biochem. Genet. 40, 149–161. doi: 10.1023/a:1015841424598
Shin, M.-H., Kim, J.-H., Kwon, J., Lim, J.-H., Choi, H. T., and Park, C. (2016). Comparison of survey methods and results for natural environment in Baekdudaegan mountain system. J. Korea Soc. Environ. Restor. Technol. 19, 1–18. doi: 10.13087/kosert.2016.19.2.1
Slatkin, M., and Hudson, R. R. (1991). Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129, 555–562. doi: 10.1093/genetics/129.2.555
Song, J.-Y., Kim, M.-S., Kim, I.-S., Kim, T.-H., Roh, I., Seo, S.-W., et al. (2009). Roadkill of amphibians in the Korea national park. Korean J. Environ. Ecol. 23, 187–193.
Streatfeild, C. A. (2009). The effects of habitat fragmentation on the demography and population genetic structure of Uromys caudimaculatus. PhD thesis, Queensland University of Technology.
Suzuki, H., Filippucci, M. G., Chelomina, G. N., Sato, J. J., Serizawa, K., and Nevo, E. (2008). A biogeographic view of Apodemus in Asia and Europe inferred from nuclear and mitochondrial gene sequences. Biochem. Genet. 46, 329–346. doi: 10.1007/s10528-008-9149-7
Szulkin, M., Zelazowski, P., Nicholson, G., and Sheldon, B. C. (2009). Inbreeding avoidance under different null models of random mating in the great tit. J. Anim. Ecol. 78, 778–788. doi: 10.1111/j.1365-2656.2009.01544.x
Teacher, A., and Griffiths, D. (2011). Hapstar: automated haplotype network layout and visualization. Mol. Ecol. Resour. 11, 151–153. doi: 10.1111/j.1755-0998.2010.02890.x
Thapa, K., Manandhar, S., Bista, M., Shakya, J., Sah, G., Dhakal, M., et al. (2018). Assessment of genetic diversity, population structure, and gene flow of tigers (Panthera tigris tigris) across Nepal's terai arc landscape. PLoS One 13:e0193495. doi: 10.1371/journal.pone.0193495
Wang, W., Lin, X. D., Guo, W. P., Zhou, R. H., Wang, M. R., Wang, C. Q., et al. (2015). Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology 474, 19–27. doi: 10.1016/j.virol.2014.10.017
Wang, H., Yang, B., Wang, H., and Xiao, H. (2021). Impact of different numbers of microsatellite markers on population genetic results using SLAF-seq data for rhododendron species. Sci. Rep. 11:8597. doi: 10.1038/s41598-021-87945-x
Wu, H., Zhan, X.-J., Yan, L., Liu, S.-Y., Li, M., Hu, J.-C., et al. (2008). Isolation and characterization of fourteen microsatellite loci for striped field mouse (Apodemus agrarius). Conserv. Genet. 9, 1691–1693. doi: 10.1007/s10592-008-9535-z
Wultsch, C., Caragiulo, A., Dias-Freedman, I., Quigley, H., Rabinowitz, S., and Amato, G. (2016). Genetic diversity and population structure of Mesoamerican jaguars (Panthera onca): implications for conservation and management. PLoS One 11:e0162377. doi: 10.1371/journal.pone.0162377
Yue, H., Fan, Z., Liu, S., Liu, Y., Song, Z., and Zhang, X. (2012). A mitogenome of the Chevrier's field mouse (Apodemus chevrieri) and genetic variations inferred from the cytochrome b gene. DNA Cell Biol. 31, 460–469. doi: 10.1089/dna.2011.1301
Keywords: biodiversity hotspot, connectivity, female philopatry, genetic diversity, habitat management, male-biased gene flow, national parks, non-random mating
Citation: Kim YR, Kim HR, Kim JY, Myeong HH, Kang JH, Kim B-J and Lee HJ (2023) Spatio-temporal genetic structure of the striped field mouse (Apodemus agrarius) populations inhabiting national parks in South Korea: Implications for conservation and management of protected areas. Front. Ecol. Evol. 11:1038058. doi: 10.3389/fevo.2023.1038058
Edited by:
Paolo Franchini, University of Tuscia, ItalyReviewed by:
Alireza Mohammadi, University of Jiroft, IranLaima Baltrunaite, Nature Research Centre, Lithuania
Copyright © 2023 Kim, Kim, Kim, Myeong, Kang, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyuk Je Lee, ✉ aHl1a2plbGVlQHNhbmdqaS5hYy5rcg==; bGhqazYyMkBnbWFpbC5jb20=
 Yu Rim Kim1
Yu Rim Kim1 Ji Hyoun Kang
Ji Hyoun Kang Hyuk Je Lee
Hyuk Je Lee