
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 13 September 2022
Sec. Evolutionary and Population Genetics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.995783
 Zheng Ren1†
Zheng Ren1† Meiqing Yang1,2†
Meiqing Yang1,2† Xiaoye Jin1
Xiaoye Jin1 Qiyan Wang1
Qiyan Wang1 Yubo Liu1
Yubo Liu1 Hongling Zhang1
Hongling Zhang1 Jingyan Ji1
Jingyan Ji1 Chuan-Chao Wang2,3,4,5*
Chuan-Chao Wang2,3,4,5* Jiang Huang1*
Jiang Huang1*The genome-wide characteristics and admixture history of the Tai-Kadai-speaking populations are essential for understanding the population genetic diversity in southern China. We genotyped about 700,000 single nucleotide polymorphisms (SNPs) of 239 individuals from six Tai-Kadai-speaking populations residing in the mountainous Guizhou Province of southwestern China. We merged the genome-wide data with available populations and ancients in East and Southeast Asia to infer Tai-Kadai-speaking populations’ admixture history and genetic structure. We observed a genetic substructure within the studied six populations in the PCA, ADMIXTURE, ChromoPainter, GLOBETROTTER, f-statistics, and qpWave analysis. The Dong, Zhuang, and Bouyei people had a strong genetic affinity with other Tai-Kadai-speaking and Austronesian groups in the surrounding area. However, Gelao showed an affinity to Sino-Tibetan groups, and Mulao people were genetically close to Hmong-Mien populations. qpAdm further illuminated that Gelao and Dong_Tongren composited more Han-related ancestry than Dong, Zhuang, Bouyei, and Mulao people. Meanwhile, we observed high frequencies of Y-chromosome haplogroup O in studied Tai-Kadai-speaking groups except for Gelao people with a high haplogroup N frequency. From the maternal side, haplogroup M7 was frequent in studied populations except for Tongren Dong, who had a high frequency of haplogroup B5. Our newly reported data are helpful for further exploring population dynamics in southern China.
With the advent of genome-wide array genotyping and next-generation sequencing, the genetic history of East Asian populations has been finely reconstructed in the past decade. Previous studies have shown that after the separation of the northern and southern East Asian populations, the southern East Asian populations were further divided geographically into two genetic lineages corresponding to inland and coastal regions (Huang et al., 2020; Yang et al., 2020). Northern China was suggested to be the origin center of modern Sino-Tibetan languages and populations. While southern China was the homeland of multiple ethnic groups, including the ancestral groups of contemporary Hmong- Mien-, Tai- Kadai-, Austroasiatic-, and Austronesian-speaking populations. The high degrees of linguistic, cultural, and ethnic diversity in southern China make the region a focus for studying the historical evolution of these populations (Ning et al., 2020; Wang C. C. et al., 2021). The Tai-Kadai-speaking people were one of the main ethnic groups in southern East Asia. The short tandem repeat (STR), insertion-deletion polymorphism (Indel), and uniparental haplogroups have been used to explore the origin, genetic structure, and admixture history of this population (Deng et al., 2013; Ji et al., 2017; Guo et al., 2019; He et al., 2019; Luo et al., 2019; Ren et al., 2019; Xu et al., 2019; Sun et al., 2021). However, the population history of Tai-Kadai groups is far from clear due to the very limited resolution of the STR, Indel, and uniparental markers.
The ancient genomics has reconstructed the deep demographic history of East Asia and discovered a possible ancestor of southern Chinese related to the farmers in the Yangtze River Basin during the Neolithic period, which has made an important contribution to the genetic formation of the Austronesian-and Tai-Kadai-speaking populations (Wang C. C. et al., 2021). A recent study of the ancient genome in southern China found that the ancestors of the Tai-Kadai-speaking populations can be traced back to the BaBanQinCen people 1,500 years ago (Wang T. et al., 2021). The genetic profile of the Hainan Li from the southernmost part of China was less affected by Neolithic agricultural expansion or historical migration from northern China (He et al., 2020). Since the multiple southward expansion and migration of rice farming populations, the formation of the modern Southeast Asian people was greatly influenced by the populations from southern China (Lipson et al., 2018; Bin et al., 2021; Liu et al., 2021; Wang T. et al., 2021; Chen et al., 2022). Previous studies found a close genetic relationship between the Tai-Kadai-and Austronesian-speaking populations in Thailand by mitochondrial DNA (Kutanan et al., 2018), which further indicated a close relationship between the proto-Tai-Kadai and Austronesian-speaking people (Kutanan et al., 2021). According to archeology, the Tai-Kadai speakers were the indigenous people in southern China. However, the Tai-Kadai ethnic group is adjacent to the Hmong- Mien-, Austroasiatic-and Austronesian-speaking populations, and its origin and relationship are difficult to determine. The previous studies of the Tai-Kadai-speaking populations with high-density SNPs are mainly based on scattered geographical locations and a small number of samples. Given the limited exploration of genetic structure and admixture history for the Tai-Kadai group via STR, SNP, and Indel, we here collected the genome-wide data of the Zhuang, Dong, Bouyei, Mulao, Gelao, and other ethnic groups in different areas from Guizhou of southwest China and further clarified the demographic history of the inland Tai-Kadai-speaking population. The linguistic information for the samples was listed in Supplementary Table 1. Among the studied groups, Bouyei and Zhuang are the Tai branch, Dong belongs to the Kam-Sui branch, and Gelao and Majiang Mulao belong to the Kra branch.
We collected saliva samples from 239 individuals from the Tai-Kadai-speaking populations in Guizhou, southwest China. There were 50 individuals of Congjiang Zhuang in southeastern Guizhou (Zhuang_Congjiang), 51 individuals of Guanling Bouyei (Bouyei_Guanling), 22 individuals of Majiang Mulao in southeastern Guizhou (Mulao_Majiang, 木佬), 48 individuals of Liping Dong in southeastern Guizhou (Dong_Liping), 20 individuals of Dong in Dong_Tongren, and 48 individuals of Wuchuan Gelao in Zunyi (Gelao_Wuchuan). The geographical locations of the above six populations are shown in Supplementary Figure 1. Participants whose parents and grandparents are indigenous people reside in Guizhou for at least three generations and should have no consanguineous marriage with other groups. The Medical Ethics Committee of Guizhou Medical University approved the study (2019 Ethics Approval Document No. 74). We followed the recommendations provided by the revised Helsinki Declaration of 2000. All the participants signed written informed consent before participating in the study.
We further calculated the kinship coefficient via the KING software (Yang et al., 2011) using options “− kingship−degree 3.” We used PLINK v1.9 (Purcell et al., 2007) with the option “−missing” to calculate the SNP calling rate for each individual and “−remove” to exclude the individuals with the lowest SNP calling rate. We also excluded individuals with up to third-degree kinship with other collected samples (kinship coefficient > 0.125). A total of 222 unrelated samples were retained, including 43 individuals of Gelao_Wuchuan, 27 individuals of Zhuang_Congjiang, 45 individuals of Bouyei_Guanling, 28 individuals of Mulao_Majiang, 46 individuals of Dong_Liping, and 18 individuals of Dong_Wanshn.
Genomic DNA was extracted by QIAamp DNA Blood Mini kit (QIAGEN, Hilden, Germany) and quantified by the Nanodrop-2000 spectrophotometer (Thermo Scientific, Wilmington, DE, United States). We genotyped the genome-wide SNPs using the Infinium Global Screening Array, which included a total of 699,537 SNPs from the autosome, Y-chromosome, and mitochondrial DNA.
The genome-wide SNP data of the Tai-Kadai-speaking populations were merged with the reference dataset, including 1240K and Human Origin (HO) included in the Allen Ancient DNA Resource (AADR)1 (Patterson et al., 2012; Lipson et al., 2018; McColl et al., 2018; Liu D. et al., 2020; Ning et al., 2020; Yang et al., 2020; Wang et al., 2022). We generated two combined datasets covering 187,644 (1240K dataset) and 71,810 (HO-dataset) SNPs. The 1240K dataset has more SNPs than the HO dataset, but the HO dataset included more present-day reference populations than the 1240K, such as Maonan, Tibetan, Dong_Guizhou, Han, and others from China.
Based on the HO dataset, we carried out PCA via the smartpca program of the EIGENSOFT v.6.1.4 (Patterson et al., 2006) package with default parameters and lsqproject: “YES.” We used the PLINK v.1.9 (Purcell et al., 2007) with the additional parameters “−indep-200 25 0.4” (Chang et al., 2015) to remove SNPs in strong linkage disequilibrium. Model-based clustering analysis was performed via ADMIXTURE v.1.3.0 (Alexander et al., 2009) based on the HO dataset with default parameters running 15 replicates from K = 2 to 8.
Based on the 1240K dataset, we conducted a series of f3/f4-statistics using the qp3Pop and qpDstat programs of ADMIXTOOLS (Patterson et al., 2012) with default parameters and estimated standard errors by the default blocked jackknife approach. We used the qp3Pop program to calculate the outgroup-f3-statistics in the form of f3(Population X, Tai-Kadai; Outgroup) using the default parameters to evaluate the shared genetic drift between Population X and Tai-Kadai-speaking populations since their separation from the Outgroup population. We also used the qp3pop to carry out the admixture-f3-statistics in the form of f3 (Tai-Kadai; Source 1, Source 2) to explore the admixture signals in the studied Tai-Kadai populations with different East and Southeast Asian ancestral source candidates, in which the Z-score < −3 denoted that Source 1 or Source 2 are related to the source populations that formed the Tai-Kadai groups. We computed the f4 (X, Outgroup; Y, Z) to formally test whether Population X shares more alleles with Y or Z, which could help determine the direction of gene flow.
We used qpWave and qpAdm (Haak et al., 2015) as implemented in the ADMIXTOOLS package to detect the minimum number of ancestral populations and quantitatively estimate corresponding admixture proportions based on the 1240K dataset. We also conducted a pairwise qpWave analysis to see whether the studied populations were genetic heterogeneous or homogeneous.
We used ALDER (Loh et al., 2013) to estimate the time of admixtures by assuming a generation time of 28 years based on the 1240K dataset. The newly studied groups were regarded as the “admixpop” (admixed population), and the remaining present-day East and Southeast Asian populations were used as the “refpops” (reference populations).
We ran TreeMix v1.13 (Pickrell and Pritchard, 2012) to infer the patterns of population splits and admixtures between our target populations and multiple ancestral populations. We used SHAPEIT v2 (Browning and Browning, 2011) to phase the genome-wide data and then used ChromoPainter v2 and FINESTRUCTURE v2 (Hellenthal et al., 2014) to explore the coincidence matrix based on the 1240K dataset. The haplotype sharing method is detecting the signal of recent genetic contact or common ancestor to combine continuous markers of linkage disequilibrium into haplotypes, improving the ability to dissect the fine-scale population structure and giving new insights into the demographic history.
Based on the Illumina array, we genotyped the lineage-informative SNPs (LISNPs) in mitochondrial DNA and Y-chromosome using an in-house script following the recommendations of the International Society of Genetic Genealogy2 and mtDNA tree Build 173 (van Oven and Kayser, 2009).
We have generated genome-wide data on approximately 700,000 SNPs for 222 Tai-Kadai-speaking individuals in Southwest China Guizhou Province. We merged our data with modern and ancient published populations. We first carried out a PC1-PC2 and PC1-PC3 to understand the general patterns of relatedness between the Tai-Kadai-speaking populations in Guizhou and the ancient and modern groups of East and Southeast Asia (Figure 1). The studied Tai-Kadai-speaking populations in Guizhou are marked in red. In the PC1-PC2 dimension, the Gelao_Wuchuan samples clustered with modern Han Chinese and the ancient millet farming populations in the Middle and Late Neolithic Yellow River Basin, indicating the Gelao_Wuchuan are genetically close to the Sino-Tibetan populations. The Dong_Tongren clustered with Guizhou Mongolians, Chuanqing, and She people in Southern China. The Dong_Liping and Bouyei_Guanling groups clustered with the present-day Tai-Kadai-speaking populations, ancient historical groups in Southern China, for example, BaBanQinCen, Layi, LaCen, Shenxian, Xitoucun, the Iron Age ancient samples in Thailand, and the Neolithic and Bronze Ages ancients in Vietnam. The Mulao_Majiang clustered with the Miao_Leishan in Guizhou, and PaThen in Vietnam, implying the population has the closest genetic relationship with the Hmong-Mien-speaking populations. The Zhuang_Congjiang are close to Tai-Kadai-speaking populations but quite scattered in the plot, showing the possible genetic influence from surrounding various populations. In the PC1–PC3 dimension, the clusters of Zhuang_Congjiang and Mulao_Majiang were consistent with clustering pattern of PC1-PC2, and other studied groups clustered with Tai-Kadai-and Sino-Tibetan-speaking populations.
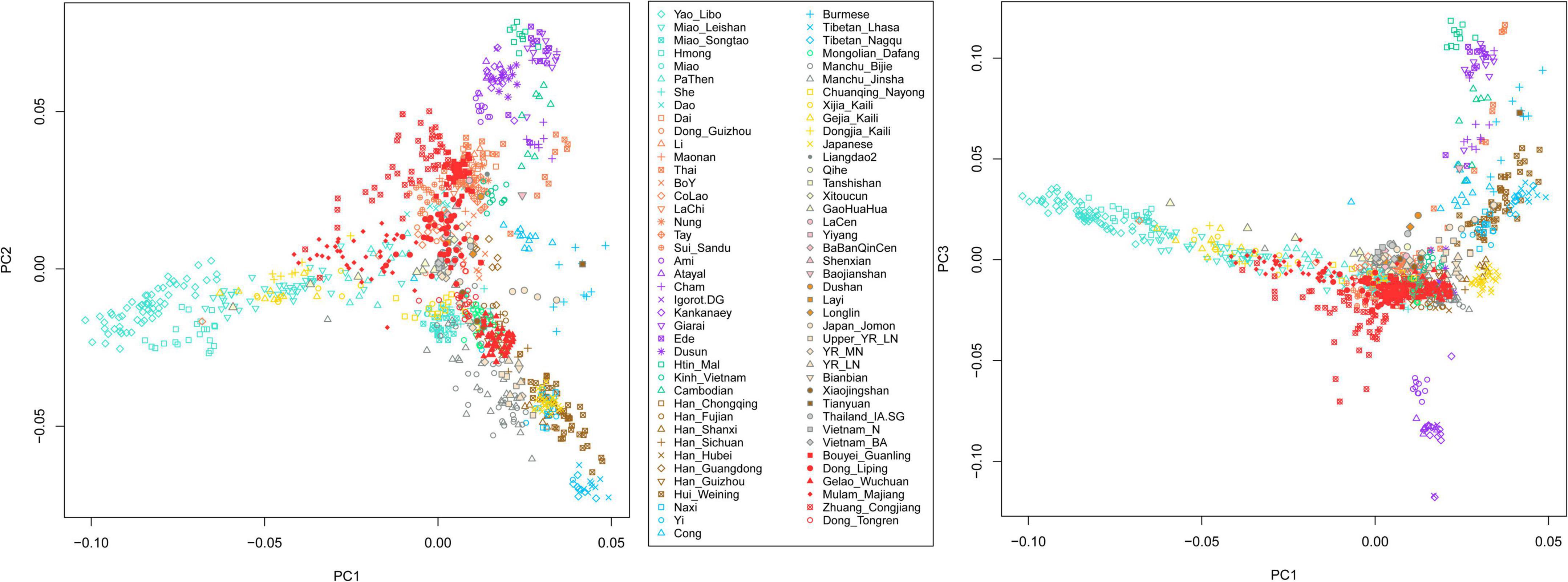
Figure 1. Principal component analysis between the Tai-Kadai-speaking group in Guizhou and other references.
We conducted an ADMIXTURE clustering analysis using the HO dataset to dissect the genetic composition of our studied populations. We provided an ADMIXTURE analysis with K from 2 to 6 in Supplementary Figure 2. We observed the lowest cross-validation error was 0.5204 at K = 4. At K = 4, we observed four ancestry components in the Tai-Kadai-speaking populations in Guizhou Province (Figure 2): the blue component mainly found in the Hmong-Mien-speaking populations; the pink component enriched in the Austronesian-speaking populations in Southeast Asia; the purple component primarily present in the ancient samples of Northern East Asia and modern Sino-Tibetans; the yellow component mainly detected in present-day Austroasiatic-speaking populations and the ancient samples in southern China and Southeast Asia. We observed a genetic substructure in our newly genotyped Tai-Kadai groups. The Mulao_Majiang and Zhuang_Congjiang are genetically similar, showing an affinity with Hmong-Mien speaking populations. The Bouyei_Guanling and Dong_Liping clustered together with other published Tai-Kadai groups such as Li, Maonan, Sui, and Dong with increasing amounts of Austronesian-related components. The Gelao_Wuchuan and Dong_Tongren clustered with southern Han Chinese with amounts of Sino-Tibetan-related components.
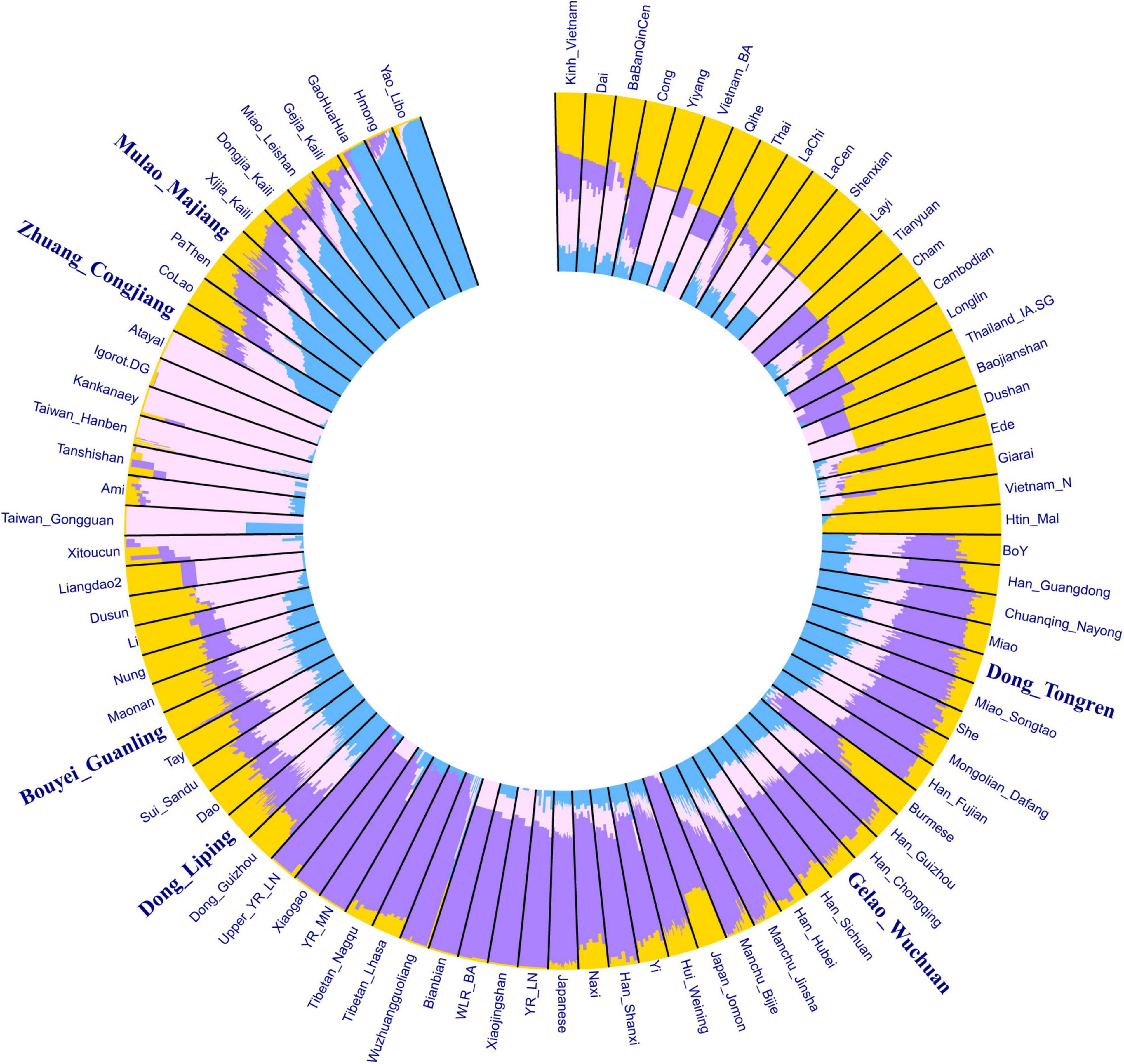
Figure 2. HO-SNP-based ADMIXTURE analysis showed the admixture composition of the Tai-Kadai-speaking in Guizhou and other references (K = 4).
We applied the haplotype sharing method to dissect a fine-scale population structure using the 1240K dataset. We observed a genetic substructure in studied Tai-Kadai groups based on the pairwise coincidence matrix (Figure 3). We found the Gelao_Wuchuan mainly clustered with Guizhou local Han Chinese, while the Dong_Tongren and Dong_Liping clustered together with Guizhou Dong, Guizhou Mongolian and Sui people. The Mulao_Majiang and Bouyei_Guanling mainly clustered closely together to Tai-Kadai speaking Maonan people. The Zhuang_Congjiang clustered together with Hmong-Mien speaking groups in Guizhou.
To explore the relationship between the studied populations and other East and Southeast Asians, we measured the shared genetic drift and possible gene flow via f3-and f4- statistics. The outgroup-f3 statistics were carried out in the form of f3 (X, Tai-Kadai; Outgroup), in which Population X represents the ancient and modern populations in East and Southeast Asia. We showed the results of outgroup-f3 statistics in Figure 4.
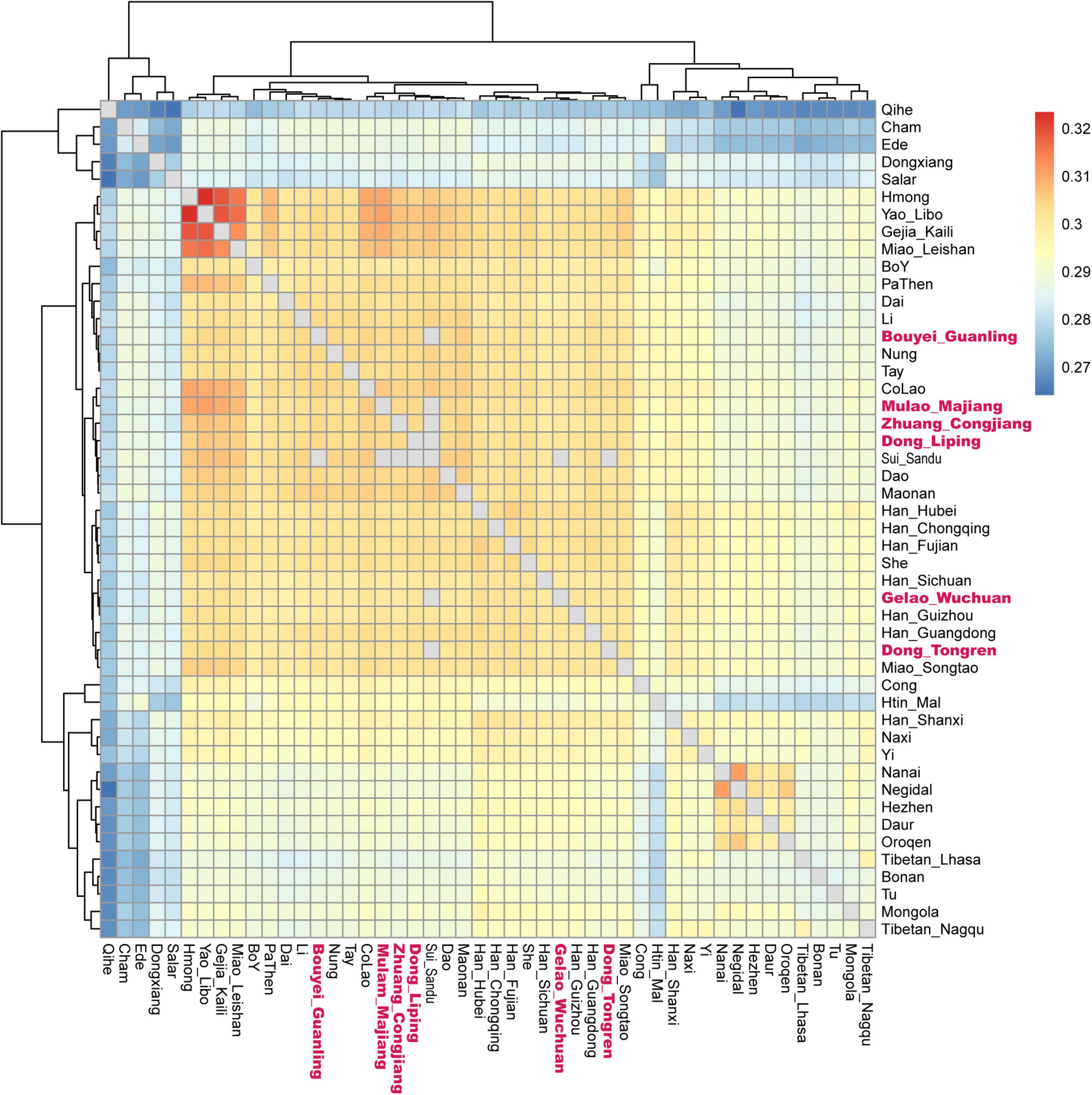
Figure 4. Genetic relation between Guizhou Tai-Kadai-speaking population and reference populations based on outgroup-f3 statistics.
The Bouyei_Guanling shared more genetic drift with the Austronesian speaking Ami from the southeast coast of China (f3 = 0.3186), the Hmong-Mien speaking Dongjia, Gejia, and Yao_Libo from Guizhou (f3 = 0.3176, 0.3165, and 0.3164, respectively) and the ancient BaBanQinCen samples from Guangxi 1,500 years ago (f3 = 0.3164). The Dong_Liping shared more genetic drift with the Hmong-Mien speaking Yao_Libo, Gejia, Miao_Leishan, and Dongjia from Guizhou (f3 = 0.3191, 0.3181, 0.3180, and 0.3180, respectively) and Ami (f3 = 0.3183). The Dong_Tongren shared more genetic drift with Hmong-Mien speaking Yao_Libo, Gejia, Miao, Dongjia, and Xijia (f3 = 0.3163, 0.3160, 0.3155, 0.3155, and 0.3151, respectively), Ami (f3 = 0.3157), and southern Han (f3 = 0.3148). The Gelao_Wuchuan shared more genetic drift with the southern Han (f3 = 0.3147), She (f3 = 0.3144), and Ami (f3 = 0.3144). The Zhuang_Congjiang shared more genetic drift with the Hmong-Mien speaking Yao_Libo, Gejia, Dongjia, and Miao_Leishan (f3 = 0.3200, 0.3188, 0.3183, and 0.3181, respectively) and Ami (f3 = 0.3180). The Mulao_Majiang shared more genetic drift with Hmong-Mien speaking Yao_Libo and Gejia (f3 = 0.3238 and 0.3218, respectively), the ancient GaoHuaHua from Guangxi (f3 = 0.3211), and Hmong-Mien speaking Dongjia and Miao_Leishan (f3 = 0.3209 and 0.3207, respectively). From outgroup-f3 statistics, we observed a close genetic relationship between the Tai-Kadai-speaking and the local Hmong-Mien-speaking populations in Guizhou. Mulao_Majiang was clustered with CoLao (Gelao from Vietnam), both influenced much from the Hmong-Mien populations, might also indicated the affinity of Majiang Mulao to the Kra branch. We also found that Gelao_Wuchuan and Dong_Tongren shared more genetic drift with the southern Han Chinese.
To further explore the influence of gene flow from surrounding populations, we performed the f4 statistics in the form of f4 (X, Outgroup; Y, Z). The results are shown in Supplementary Table 2. Dai, Ami, Yao, and Southern Han were regarded as the representative populations of the Tai-Kadai, Austronesian, Hmong-Mien-speaking populations, and Han Chinese in Southern China, respectively. We found Zhuang_Congjiang, Dong_Liping, and Mulao_Majiang shared more alleles with the Hmong-Mien-speaking Yao people than with other East Asians, including Dai, Ami, and Southern Han, as shown in the Z-score > 3 in the f4 (Zhuang/Dong/Mulao, Mbuti; Yao_Libo, Z). While the other two Tai-Kadai-speaking populations, Dong_Tongren and Gelao_Wuchuan, shared more alleles with the Yao, Han, and Ami than with Dai, which means that these two populations are less affected by the Tai-Kadai-speaking populations.
We next used the pairwise f4(X, Outgroup; Tai-Kadai1, Tai-Kadai2) to investigate whether there are genetic differences among Tai-Kadai-speaking populations in Guizhou and showed the results in Supplementary Table 3.
In the f4(X, Mbuti; Bouyei_Guanling, other Tai-Kadai groups), we found Sino-Tibetan related populations, including Han Chinese and ancient Yellow River farmers, shared more alleles with Dong_Liping, Dong_Tongren, Gelao_Wuchuan, Mulao_Majiang, and Sui_Sandu than with Bouyei_Guanling. While Austronesian speaking Ami and Tai-Kadai speaking Dai shared more alleles with Bouyei_Guanling than with Mulao_Majiang, Gelao_Wuchuan, and Dong_Tongren. We also found Hmong-Mien speaking groups shared more alleles with Dong_Liping, Mulao_Majiang, Sui_Sandu, and Zhuang_Congjiang than with Bouyei_Guanling, but Hmong-Mien groups shared more alleles with Bouyei_Guanling than with Gelao_Wuchuan.
In the f4(X, Mbuti; Dong_Liping, other Tai-Kadai groups), we found Austronesian, Hmong-Mien, and Tai-Kadai speaking populations shared more alleles with Dong_Liping than with Dong_Tongren and Gelao_Wuchuan. While Sino-Tibetan-related populations, including Han Chinese and ancient Yellow River farmers, shared more alleles with Dong_Liping than with Zhuang_Congjiang, Sui_Sandu, and Mulao_Majiang, but shared fewer alleles than with Gelao_Wuchuan. We also detected that Hmong-Mien groups shared more alleles with Mulao_Majiang than with Dong_Liping.
In the f4(X, Mbuti; Gelao_Wuchuan, other Tai-Kadai groups), we found Sino-Tibetan related populations, including Han Chinese and ancient Yellow River farmers, shared more alleles with Gelao_Wuchuan than with Dong_Tongren, Mulao_Majiang, Sui_Sandu, and Zhuang_Congjiang, but Austronesian, Hmong-Mien, and Tai-Kadai speaking populations shared more alleles with Dong_Tongren, Mulao_Majiang, Sui_Sandu, and Zhuang_Congjiang than with Gelao_Wuchuan.
In the f4(X, Mbuti; Mulao_Majiang, other Tai-Kadai groups), we found Hmong-Mien speaking populations shared more alleles with Mulao_Majiang than with Dong_Tongren, Sui_Sandu, and Zhuang_Congjiang. Austronesian and Tai-Kadai speaking populations shared more alleles with Sui_Sandu than with Mulao_Majiang. Sino-Tibetan-related populations, including Han Chinese and ancient Yellow River farmers, shared more alleles with Dong_Tongren than Mulao_Majiang, but Tai-Kadai groups shared more alleles with Mulao_Majiang than with Dong_Tongren.
In the f4(X, Mbuti; Zhuang_Congjiang, other Tai-Kadai groups), we found Austronesian, Hmong-Mien, and Tai-Kadai speaking populations shared more alleles with Zhuang_Congjiang than with Dong_Tongren, but Sino-Tibetan related populations including Han Chinese and ancient Yellow River farmers shared fewer alleles with Zhuang_Congjiang than with Dong_Tongren. Austronesian and Tai-Kadai speaking populations shared fewer alleles with Zhuang_Congjiang than with Sui_Sandu.
In the f4(X, Mbuti; Dong_Tongren, Sui_Sandu), we found Sino-Tibetan related populations, including Han Chinese and ancient Yellow River farmers, shared more alleles with Dong_Tongren than with Sui_Sandu, but Austronesian, Hmong-Mien, and Tai-Kadai related ancient, and present-day populations shared more alleles with Sui_Sandu than with Dong_Tongren.
We next applied qpWave analysis to pairwisely determine if the two populations are consistent in deriving from a single source. We used Mbuti, Yana_UP, Ust_Ishim, Kolyma_M, Tianyuan, Liangdao2, AR_EN, Longlin, Papuan, and GaoHuaHua as outgroups. We found a significant genetic difference between any pairs of the studied populations (Supplementary Table 4, p_rank0 < 0.05).
The above f4 statistics and qpWave analysis showed a genetic substructure among the Tai-Kadai-speaking populations in Guizhou. There are genetic differences among the same ethnic groups in different regions.
We ran TreeMix analysis to infer the pattern of population splits and admixtures based on genome-wide allele frequency data and showed the results in Figure 5. We observed the Dong in southeastern Guizhou, Mulao, Hmong-Mien-speaking populations, Zhuang_Congjiang, Bouyei_Guanling, and other Tai-Kadai-speaking populations clustered together into a branch. While Gelao_Wuchuan and Dong_Tongren clustered with Sino-Tibetan groups as another branch. We detected gene flows from Hmong-Mien groups to Dong_Tongren, to Dong_Liping, and also to Sui and Dai people.
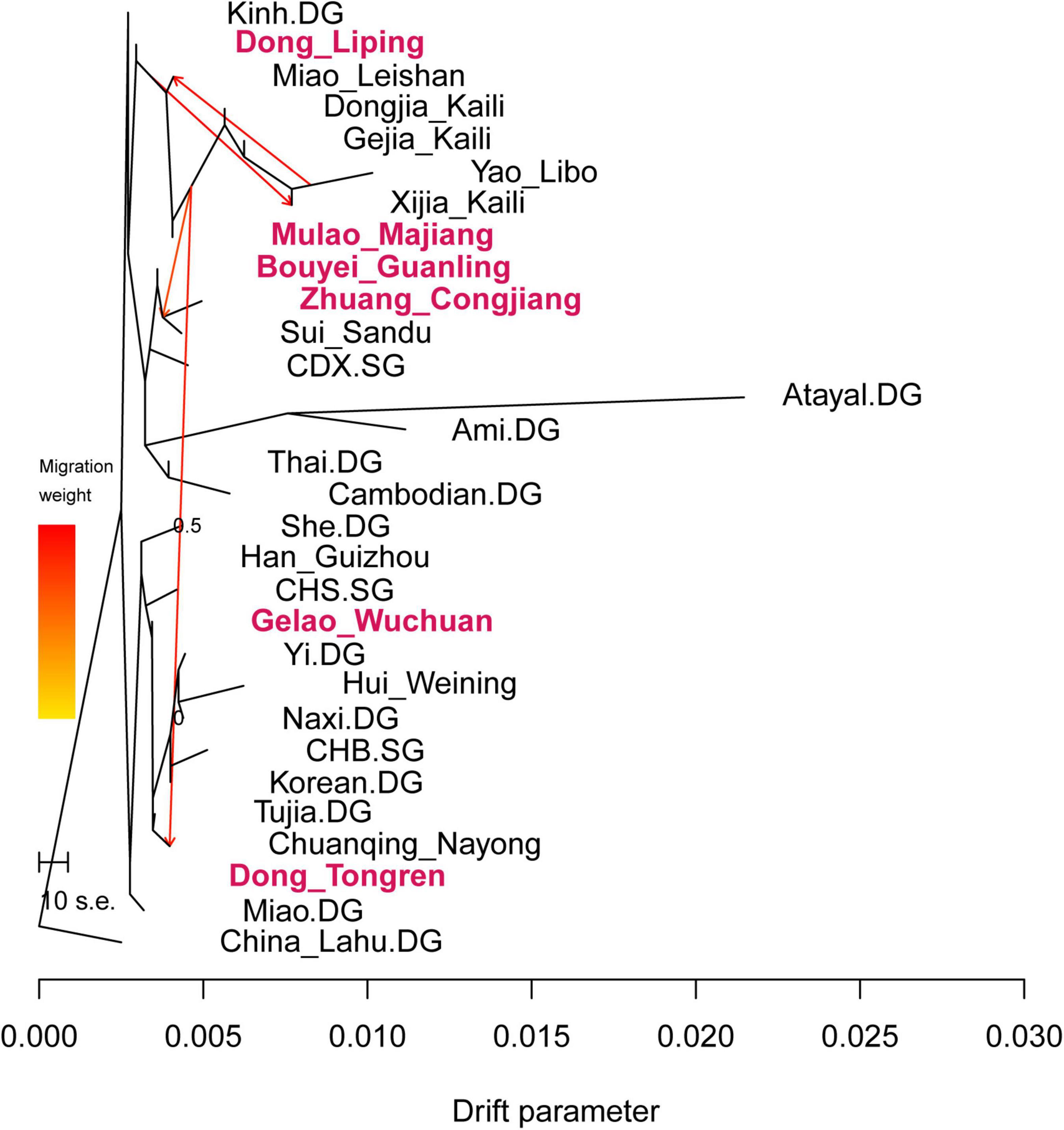
Figure 5. Genetic splits and admixture pattern of Guizhou Tai-Kadai-speaking and reference populations.
We used all possible reference populations as genetic sources (Source 1 and Source 2) to detect the possible admixture signals in each studied Guizhou Tai-Kadai-speaking population via admixture-f3-statistics in the form of f3(Tai-Kadai; Source 1, Source 2). We showed the results in Supplementary Table 5. We observed the pairs of a Hmong-Mien group with Yellow River farmers or Han Chinese could generate the most significant negative f3 values for Gelao_Wuchuan, Dong_Liping, Mulao_Majiang, and Dong_Tongren.
We further used qpAdm analysis to calculate the admixture proportion of the Guizhou Tai-Kadai-speaking populations. The Neolithic millet farming population (YR_MN) in the Yellow River Basin was used to represent the ancestors of Sino-Tibetans, and the Atayal was used to represent the lineage of the southern East Asians. As shown in Figure 6, the proportions of Yellow River farmer-related ancestry in Bouyei_Guanling, Zhuang_Congjiang, Dong_Liping, Mulao_Majiang, Dong_Tongren, and Gelao_Wuchuan are 24.7, 30.3, 41, 47.1, 56.3, and 70.1%, respectively, and the rest of the ancestry was from Atayal related southern East Asians. The Gelao_Wuchuan and Dong_Tongren contained a large proportion of the ancestry related to Yellow River farmers, which is close to the proportion in modern Han populations (62.4–81.4%). However, the proportions of Yellow River farmer-related ancestry in other Tai-Kadai-speaking populations are all less than 50%, confirming a genetic substructure in the Tai-Kadai populations.
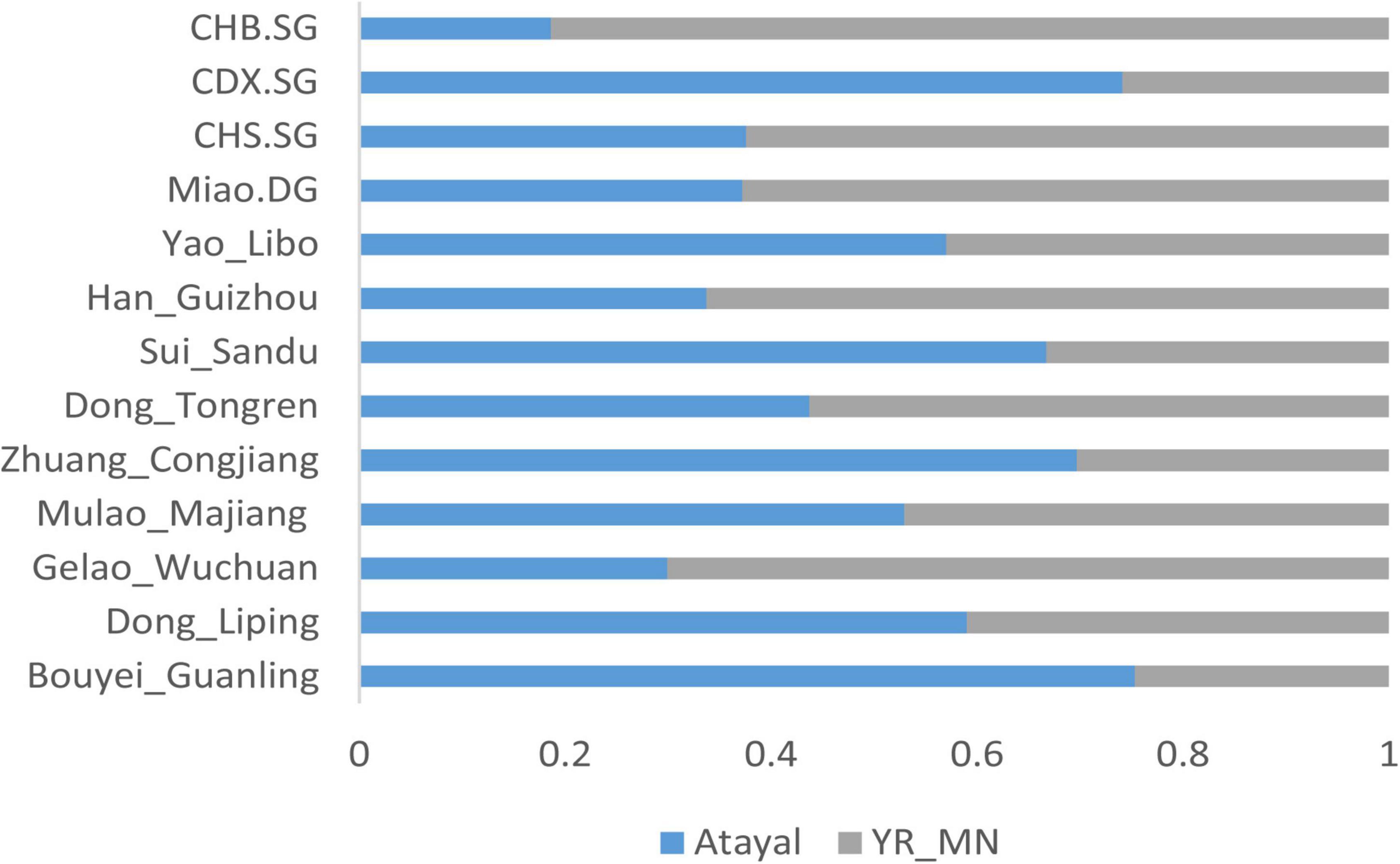
Figure 6. Two-way admixture models showed the admixture landscape of the Guizhou Tai-Kadai-speaking population.
We investigated the approximate admixture date based on the decay of admixture-induced linkage disequilibrium (Supplementary Table 6). We observed admixture signals between Tai-Kadai and Sino-Tibetan, Hmong-Mien or Austroasiatic groups. The genetic influences from Tibeto-Burman, Tujia, Naxi, and Yi groups to Tai-Kadai-speakers were suggested to have happened before 1,000 years. We also detected admixture events between Bouyei_Guanling, Dong_Liping, Mulao_Majiang and Han Chinese ∼200∼3,000 years ago. All studied Tai-Kadai-speaking populations are affected by the gene flow of Hmong-Mien-speaking populations in different periods. For example, Bouyei and Gelao groups were affected by Hmong-Mien populations at an earlier time (about 1,000∼6,200 years) than other groups. In the recent hundred years, there was an admixture event between Zhuang_Congjiang and Austroasiatic speaking Kinh.
The patrilineal Y-chromosome haplogroups of each ethnic group are diverse. As shown in Supplementary Table 7, we observed high frequencies of the haplogroup O in Guizhou Tai-Kadai populations except for the Gelao_Wuchuan. The haplogroup O1a1a-P203.2 has the highest frequency in Bouyei (94.12%), haplogroup O2a2a-M188 has the highest frequency in Zhuang (72.41%), haplogroup O1a1a-P203.2 has the highest frequency in Mulao (97.12%), haplogroup O1a1a-P203.2, O1a2-M103, and O2a1a-F3143 are the most frequent lineage in Dong_Liping (15.38%), haplogroup D1a1a-F3306, O1b1a-M1470, and O2a2a-M188 have the highest frequency in Dong_Tongren (20%). In comparison, N1b1b-Y23802 is the most frequent haplogroup in Gelao_Wuchuan (57.58%). The haplogroups with the highest frequencies among the Bouyei, Mulao, and Dong_Liping are also commonly found in other Tai-Kadai-speaking populations. The haplogroups of the Gelao_Wuchuan are mainly from the north of East Asia.
From the maternal mitochondrial DNA side, we observed the high frequency of haplogroup M7 in all the populations except the Dong_Tongren. The highest frequencies of M7 were found in Bouyei_Guanling and Mulao_Majiang, which were 26.67 and 25.64%, respectively. The frequencies of haplogroup M7 and C7 in Gelao_Wuchuan were the highest; both are 12.24%. Haplogroup D4, F1, M7, and B4 appeared at high frequencies in Zhuang_Congjiang, while B5, B4, M7, and F1 appeared at high frequencies in Dong_Liping. Haplogroup B5 occurred at the highest frequency of 20% in Dong_Tongren. The matrilineal haplogroups of the Guizhou Tai-Kadai-speaking populations in our study are dominated by the specific lineages in Southern East Asia.
Southern China was suggested as the birthplace of the Tai-Kadai-speaking populations. The rice farmers in Southern China had continuously migrated southward to Southeast Asia, such as Thailand and Vietnam, since the Middle Neolithic (Lipson et al., 2018; Kutanan et al., 2019). In addition, the southward migration of the millet farming populations and Han Chinese from the Yellow River further contributed to the abundant genetic diversity in southern China (Chi and Hung, 2015). From the end of the Warring States period to the Qin and Han Dynasties, the ancestors of the Tai-Kadai-speaking populations were called “Baiyue”. During the Han Dynasty, the military-civilian of the Central Plain entered the Baiyue area, and contacted and communicated with various southern tribes. The present-day Tai-Kadai-speaking people living in Guizhou include a variety of ethnic groups, such as Dong, Bouyei, Sui, Gelao, Zhuang, Mulao, Maonan, and so on.
Previous studies have also observed extensive genetic admixture between the Han Chinese and southern aborigines (Wen et al., 2004; He et al., 2020). The results of autosomal, X, and Y chromosomal STR showed a close genetic relationship between the Guizhou Tai-Kadai-speaking people and neighboring Tai-Kadai-and Hmong-Mien-speaking populations. For example, the Guizhou Dong is closely related to the Guizhou Bouyei and Han (Zhang, 2015; Feng et al., 2020; Liu Y. et al., 2020; Yang et al., 2021). However, the genetic diversity and population history of the Tai-Kadai populations are far from clear due to the limited sampling and the low resolution of genetic markers.
In this study, we synthetically analyzed the high-density SNP variation data of the Tai-Kadai-speaking populations in Guizhou. We combined it with the genome-wide data of both ancient and modern populations to explore the fine-scale genetic structure and admixture history. We detected Zhuang_Congjiang, Bouyei_Guanling, and Dong_Liping derived ancestry mainly from an ancestral lineage related to the present-day Tai-Kadai-speaking populations, which is consistent with their linguistic classification. This type of Tai-Kadai related ancestry can be traced back to at least 1,500 years ago represented by the ancient BaBanQinCen sample in Guangxi. Wang T. et al. (2021) also revealed that the Tai-Kadai and Austronesian populations shared a common ancestor who might be related to the Neolithic rice farmers in the Yangtze River basin. The current study found that Zhuang, Dong, and Bouyei people shared more alleles with Austronesian populations such as Ami and Atayal than Hmong-Mien, Austroasiatic-, and Austronesian-speaking populations, supporting the common origin of Tai-Kadai and Austronesian populations.
We also observed a genetic substructure in the Tai-Kadai populations of Guizhou, mainly caused by the different proportions of Yellow River farmer-related ancestry in different populations. Tai-Kadai populations in Guizhou also were largely affected by the gene flow from the Hmong-Mien and Sino-Tibetan related groups. For example, unlike other Tai-Kadai groups, Gelao_Wuchuan and Dong_Tongren are closer to Sino-Tibetan populations. The genetic substructure implied that the surrounding populations had played an important role in the genetic formation of the Tai-Kadai groups. According to the migration routes of Tai-Kadai speaking peoples, we speculated that Tai-Kadai peoples were continuously influenced by Han and Hmong-Mien speaking groups during the migration from Southeast to Southwest China.
The Majiang Mulao people were officially classified as Mulam, but they claimed to be Miao or Bouyei people. We found Majiang Mulao samples were genetically closer to the Hmong-Mien groups, while Mulam (仫佬) people were more closely related to Tai-Kadai-speakers (Wu, 1995), providing genetic evidence for understanding the relationship of Mulao and Mulam.
The Wuchuan Gelao in our study showed genetic affinity with Han Chinese groups, suggesting the possible influence from Han Chinese to Gelao. But we note that although Wuchuan is the county with largest official Gelao ethnic population, no Gelao-speaking people are left there now. Many Han people changed the ethnicity to Gelao in the recent 40 years. Therefore, some of the sampled Wùchuân Gelao people in our study may also be from the Han descendants.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://zenodo.org/record/6816509.
The studies involving human participants were reviewed and approved by Ethical Committee of Guizhou Medical University. The patients/participants provided their written informed consent to participate in this study.
C-CW and JH designed this study. ZR, MY, QW, YL, and JJ collected the samples and conducted the experiment. MY wrote the manuscript. XJ, HZ, and JJ analyzed the results. C-CW modified the manuscript. All authors reviewed and approved the submitted version of the manuscript.
This work was funded by Guizhou Scientific Support Project, Qian Science Support (2021) General 448, and National Natural Science Foundation (No. 82160324). Guizhou Scientific Support Project, Qian Science Support (2021) General 448, the Guizhou Province Education Department, Characteristic Region Project, Qian Education KY No. (2021)065, the Guizhou Province Engineering Technology Research Center Project [Qian High-Tech of Development and Reform Commission No. (2016)1345], the Guizhou Scientific Support Project [Qian Science Support (2019)2825], the Guizhou “Hundred” innovative talents project [Qian Science Talent Platform (2020)6012], the Guizhou Scientific Support Project [Qian Science Support (2020)4Y057], the Guizhou Science Project [Qian Science Foundation (2020)1Y353], the “Double First Class University Plan” key construction project of Xiamen University (the origin and evolution of East Asian populations and the spread of Chinese civilization, 0310/X2106027), Nanqiang Outstanding Young Talents Program of Xiamen University (X2123302), the Major Project of National Social Science Foundation of China granted to C-CW (21&ZD285), the Major Project of National Social Science Foundation of China (20&ZD248), the Major Project of National Social Science Foundation of China (2021MZD014), and the European Research Council (ERC) grant (ERC-2019-ADG-883700-TRAM).
S. Fang and Z. Xu from the Information and Network Center of Xiamen University are acknowledged their help with the high-performance computing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.995783/full#supplementary-material
Supplementary Figure 1 | The geographical location distribution of sample collection.
Supplementary Figure 2 | HO-SNP-based ADMIXTURE analysis showed the admixture composition of the Tai-Kadai-speaking in Guizhou and other references (K = 2 to 6).
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Bin, X., Wang, R., Huang, Y., Wei, R., Zhu, K., Yang, X., et al. (2021). Genomic Insight Into the Population Structure and Admixture History of Tai-Kadai-Speaking Sui People in Southwest China. Front. Genet. 12:735084. doi: 10.3389/fgene.2021.735084
Browning, B. L., and Browning, S. R. (2011). A fast, powerful method for detecting identity by descent. Am. J. Hum. Genet. 88, 173–182. doi: 10.1016/j.ajhg.2011.01.010
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4:7. doi: 10.1186/s13742-015-0047-8
Chen, J., He, G., Ren, Z., Wang, Q., Liu, Y., Zhang, H., et al. (2022). Fine-Scale Population Admixture Landscape of Tai-Kadai-Speaking Maonan in Southwest China Inferred From Genome-Wide SNP Data. Front. Genet. 13:815285. doi: 10.3389/fgene.2022.815285
Chi, Z., and Hung, H.-C. (2015). The emergence of agriculture in southern China. Antiquity 84, 11–25. doi: 10.1017/s0003598x00099737
Deng, Q. Y., Wang, C. C., Wang, X. Q., Wang, L. X., Wang, Z. Y., Wu, W. J., et al. (2013). Genetic affinity between the Kam-Sui speaking Chadong and Mulam people. J. Syst. Evol. 51, 263–270.
Feng, R., Zhao, Y., Chen, S., Li, Q., Fu, Y., Zhao, L., et al. (2020). Genetic analysis of 50 Y-STR loci in Dong, Miao, Tujia, and Yao populations from Hunan. Int. J. Legal. Med. 134, 981–983. doi: 10.1007/s00414-019-02115-z
Guo, J., Ji, J., He, G., Ren, Z., Zhang, H., Wang, Q., et al. (2019). Genetic structure and forensic characterisation of 19 X-chromosomal STR loci in Guizhou Sui population. Ann. Hum. Biol. 46, 246–253. doi: 10.1080/03014460.2019.1623911
Haak, W., Lazaridis, I., Patterson, N., Rohland, N., Mallick, S., Llamas, B., et al. (2015). Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211. doi: 10.1038/nature14317
He, G., Ren, Z., Guo, J., Zhang, F., Zou, X., Zhang, H., et al. (2019). Population genetics, diversity and forensic characteristics of Tai-Kadai-speaking Bouyei revealed by insertion/deletions markers. Mol. Genet. Genom. 294, 1343–1357. doi: 10.1007/s00438-019-01584-6
He, G., Wang, Z., Guo, J., Wang, M., Zou, X., Tang, R., et al. (2020). Inferring the population history of Tai-Kadai-speaking people and southernmost Han Chinese on Hainan Island by genome-wide array genotyping. Eur. J. Hum. Genet. 28, 1111–1123. doi: 10.1038/s41431-020-0599-7
Hellenthal, G., Busby, G. B. J., Band, G., Wilson, J. F., Capelli, C., Falush, D., et al. (2014). A genetic atlas of human admixture history. Science 343, 747–751. doi: 10.1126/science.1243518
Huang, X., Xia, Z.-Y., Bin, X., He, G., Guo, J., Lin, C., et al. (2020). Genomic Insights into the Demographic History of Southern Chinese. Front. Ecol. Evol. 10:853391. doi: 10.3389/fevo.2022.853391
Ji, J., Ren, Z., Zhang, H., Wang, Q., Wang, J., Kong, Z., et al. (2017). Genetic profile of 23 Y chromosomal STR loci in Guizhou Shui population, southwest China. Forensic Sci. Int. Genet. 28:e16–e17. doi: 10.1016/j.fsigen.2017.01.010
Kutanan, W., Kampuansai, J., Brunelli, A., Ghirotto, S., Pittayaporn, P., Ruangchai, S., et al. (2018). New insights from Thailand into the maternal genetic history of Mainland Southeast Asia. Eur. J. Hum. Genet. 26, 898–911. doi: 10.1038/s41431-018-0113-7
Kutanan, W., Kampuansai, J., Srikummool, M., Brunelli, A., Ghirotto, S., Arias, L., et al. (2019). Contrasting Paternal and Maternal Genetic Histories of Thai and Lao Populations. Mol. Biol. Evol. 36, 1490–1506. doi: 10.1093/molbev/msz083
Kutanan, W., Liu, D., Kampuansai, J., Srikummool, M., Srithawong, S., Shoocongdej, R., et al. (2021). Reconstructing the Human Genetic History of Mainland Southeast Asia: Insights from Genome-Wide Data from Thailand and Laos. Mol. Biol. Evol. 38, 3459–3477. doi: 10.1093/molbev/msab124
Lipson, M., Cheronet, O., Mallick, S., Rohland, N., Oxenham, M., Pietrusewsky, M., et al. (2018). Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science 361, 92–95. doi: 10.1126/science.aat3188
Liu, D., Duong, N. T., Ton, N. D., Van Phong, N., Pakendorf, B., Van Hai, N., et al. (2020). Extensive Ethnolinguistic Diversity in Vietnam Reflects Multiple Sources of Genetic Diversity. Mol. Biol. Evol. 37, 2503–2519. doi: 10.1093/molbev/msaa099
Liu, Y., Zhang, H., He, G., Ren, Z., Zhang, H., Wang, Q., et al. (2020). Forensic Features and Population Genetic Structure of Dong, Yi, Han, and Chuanqing Human Populations in Southwest China Inferred From Insertion/Deletion Markers. Front. Genet. 11:360. doi: 10.3389/fgene.2020.00360
Liu, Y., Xie, J., Wang, M., Liu, C., Zhu, J., Zou, X., et al. (2021). Genomic Insights Into the Population History and Biological Adaptation of Southwestern Chinese Hmong-Mien People. Front. Genet. 12:815160. doi: 10.3389/fgene.2021.815160
Loh, P. R., Lipson, M., Patterson, N., Moorjani, P., Pickrell, J. K., Reich, D., et al. (2013). Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193, 1233–1254. doi: 10.1534/genetics.112.147330
Luo, Y., Wu, Y., Qian, E., Wang, Q., Wang, Q., Zhang, H., et al. (2019). Population genetic analysis of 36 Y-chromosomal STRs yields comprehensive insights into the forensic features and phylogenetic relationship of Chinese Tai-Kadai-speaking Bouyei. PLoS One 14:e0224601. doi: 10.1371/journal.pone.0224601
McColl, H., Racimo, F., Vinner, L., Demeter, F., Gakuhari, T., Moreno-Mayar, J. V., et al. (2018). The prehistoric peopling of Southeast Asia. Science 361, 88–92. doi: 10.1126/science.aat3628
Ning, C., Li, T., Wang, K., Zhang, F., Li, T., Wu, X., et al. (2020). Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat. Commun. 11:2700. doi: 10.1038/s41467-020-16557-2
Patterson, N., Moorjani, P., Luo, Y., Mallick, S., Rohland, N., Zhan, Y., et al. (2012). Ancient admixture in human history. Genetics 192, 1065–1093. doi: 10.1534/genetics.112.145037
Patterson, N., Price, A. L., and Reich, D. (2006). Population structure and eigenanalysis. PLoS Genet. 2:e190. doi: 10.1371/journal.pgen.0020190
Pickrell, J. K., and Pritchard, J. K. (2012). Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. doi: 10.1371/journal.pgen.1002967
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Ren, Z., Guo, J., He, G., Zhang, H., Zou, X., Zhang, H., et al. (2019). Forensic genetic polymorphisms and population structure of the Guizhou Bouyei people based on 19 X-STR loci. Ann. Hum. Biol. 46, 574–580. doi: 10.1080/03014460.2019.1697362
Sun, J., Li, Y. X., Ma, P. C., Yan, S., Cheng, H. Z., Fan, Z. Q., et al. (2021). Shared paternal ancestry of Han, Tai-Kadai-speaking, and Austronesian-speaking populations as revealed by the high resolution phylogeny of O1a-M119 and distribution of its sub-lineages within China. Am. J. Phys. Anthropol. 174, 686–700. doi: 10.1002/ajpa.24240
van Oven, M., and Kayser, M. (2009). Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30:E386–E394. doi: 10.1002/humu.20921
Wang, C. C., Yeh, H. Y., Popov, A. N., Zhang, H. Q., Matsumura, H., Sirak, K., et al. (2021). Genomic insights into the formation of human populations in East Asia. Nature 591, 413–419. doi: 10.1038/s41586-021-03336-2
Wang, T., Wang, W., Xie, G., Li, Z., Fan, X., Yang, Q., et al. (2021). Human population history at the crossroads of East and Southeast Asia since 11,000 years ago. Cell 184, 3829–3841.e21. doi: 10.1016/j.cell.2021.05.018
Wang, M., Du, W., Tang, R., Liu, Y., Zou, X., Yuan, D., et al. (2022). Genomic history and forensic characteristics of Sherpa highlanders on the Tibetan Plateau inferred from high-resolution InDel panel and genome-wide SNPs. Forensic Sci. Int. Genet. 56:102633. doi: 10.1016/j.fsigen.2021.102633
Wen, B., Li, H., Lu, D., Song, X., Zhang, F., He, Y., et al. (2004). Genetic evidence supports demic diffusion of Han culture. Nature 431, 302–305. doi: 10.1038/nature02878
Wu, G. (1995). “Mulam” is Not “Mulao”− Rethinking the Name and Origin of Mulam Ethnic Group. Ethnic Stud. Guangxi 1995, 2.82–88.
Xu, B., Guo, J., Huang, Y., Chen, X., Deng, X., and Wang, C. C. (2019). The paternal genetic structure of Jingpo and Dai in southwest China. Ann. Hum. Biol. 46, 279–283. doi: 10.1080/03014460.2019.1624821
Yang, J., Lee, S. H., Goddard, M. E., and Visscher, P. M. (2011). GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. doi: 10.1016/j.ajhg.2010.11.011
Yang, M., Jin, X., Ren, Z., Wang, Q., Zhang, H., Zhang, H., et al. (2021). X-chromosomal STRs for genetic composition analysis of Guizhou Dong group and its phylogenetic relationships with other reference populations. Ann. Hum. Biol. 48, 621–626. doi: 10.1080/03014460.2021.2008001
Yang, M. A., Fan, X., Sun, B., Chen, C., Lang, J., Ko, Y. C., et al. (2020). Ancient DNA indicates human population shifts and admixture in northern and southern China. Science 369, 282–288. doi: 10.1126/science.aba0909
Keywords: Tai-Kadai, genome-wide data, genetic profile, SNP, demographic history
Citation: Ren Z, Yang M, Jin X, Wang Q, Liu Y, Zhang H, Ji J, Wang C-C and Huang J (2022) Genetic substructure of Guizhou Tai-Kadai-speaking people inferred from genome-wide single nucleotide polymorphisms data. Front. Ecol. Evol. 10:995783. doi: 10.3389/fevo.2022.995783
Received: 16 July 2022; Accepted: 26 August 2022;
Published: 13 September 2022.
Edited by:
Guanglin He, Sichuan University, ChinaReviewed by:
Jatupol Kampuansai, Chiang Mai University, ThailandCopyright © 2022 Ren, Yang, Jin, Wang, Liu, Zhang, Ji, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiang Huang, bW1tX2hqQDEyNi5jb20=; Chuan-Chao Wang, d2FuZ0B4bXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.