- 1Integrative Biology Laboratory, College of Life Sciences, Nanjing Normal University, Nanjing, China
- 2Euler Genomics, Beijing, China
- 3School of Biology and Environmental Science, Queensland University of Technology, Brisbane, QLD, Australia
Vision plays an essential role in the life of many animals. While most mammals are night-active (nocturnal), many have adapted to novel light environments. This includes diurnal (day-active) and crepuscular (twilight-active) species. Here, we used integrative approaches to investigate the molecular evolution of 112 vision-related genes across 19 genomes representing most marsupial orders. We found that four genes (GUCA1B, GUCY2F, RGR, and SWS2) involved in retinal phototransduction likely became functionally redundant in the ancestor of marsupials, a group of largely obligate nocturnal mammals. We also show evidence of rapid evolution and positive selection of bright-light vision genes in the common ancestor of Macropus (kangaroos, wallaroos, and wallabies). Macropus-specific amino acid substitutions in opsin genes (LWS and SWS1), in particular, may be an adaptation for crepuscular vision in this genus via opsin spectral sensitivity tuning. Our study set the stage for functional genetics studies and provides a stepping stone to future research efforts that fully capture the visual repertoire of marsupials.
Introduction
Extant mammals include eutherians (placental mammals), metatherians (marsupials), and monotremes (egg-laying mammals). Mammals occupy a variety of light niches. In order to adapt to different light environments, they have activity patterns classified as nocturnal (night-active), diurnal (day-active), crepuscular (twilight-active), or non-circadian (Smale et al., 2003).
Approximately, two-thirds of mammals (including their ancestors) are nocturnal. These species inhabit poor light environments underwater (e.g., aquatic cetaceans), underground (e.g., mole rats), or in caves (e.g., bats) (Heesy and Hall, 2010; Bennie et al., 2014). Examples of diurnal species (∼20% of mammals) include primates and squirrels. It follows that vision impacts animal fitness by affecting survivorship through mating, foraging, and predator avoidance behaviors.
Visual perception in mammals is initiated by phototransduction, which converts light into an electrical signal in photoreceptor cells by signal transduction (Lamb, 2013). Cones and rods are the photoreceptors of the retina. Cones mediate vision in daylight (photopic vision) and color vision, while rods mediate vision in dim light (scotopic vision). These photoreceptors contain light-absorbing visual pigments that consist of opsin proteins (cone pigment in cones and rhodopsin, RH1, in rods) and other transduction components. When activated by light, opsin proteins activate G protein transduction (Gt), which subsequently activates phosphodiesterase (PDE), decreasing cytoplasmic-free cGMP levels and resulting in closed cGMP-gated channels and membrane hyperpolarization (Fu and Yau, 2007; Figure 1).
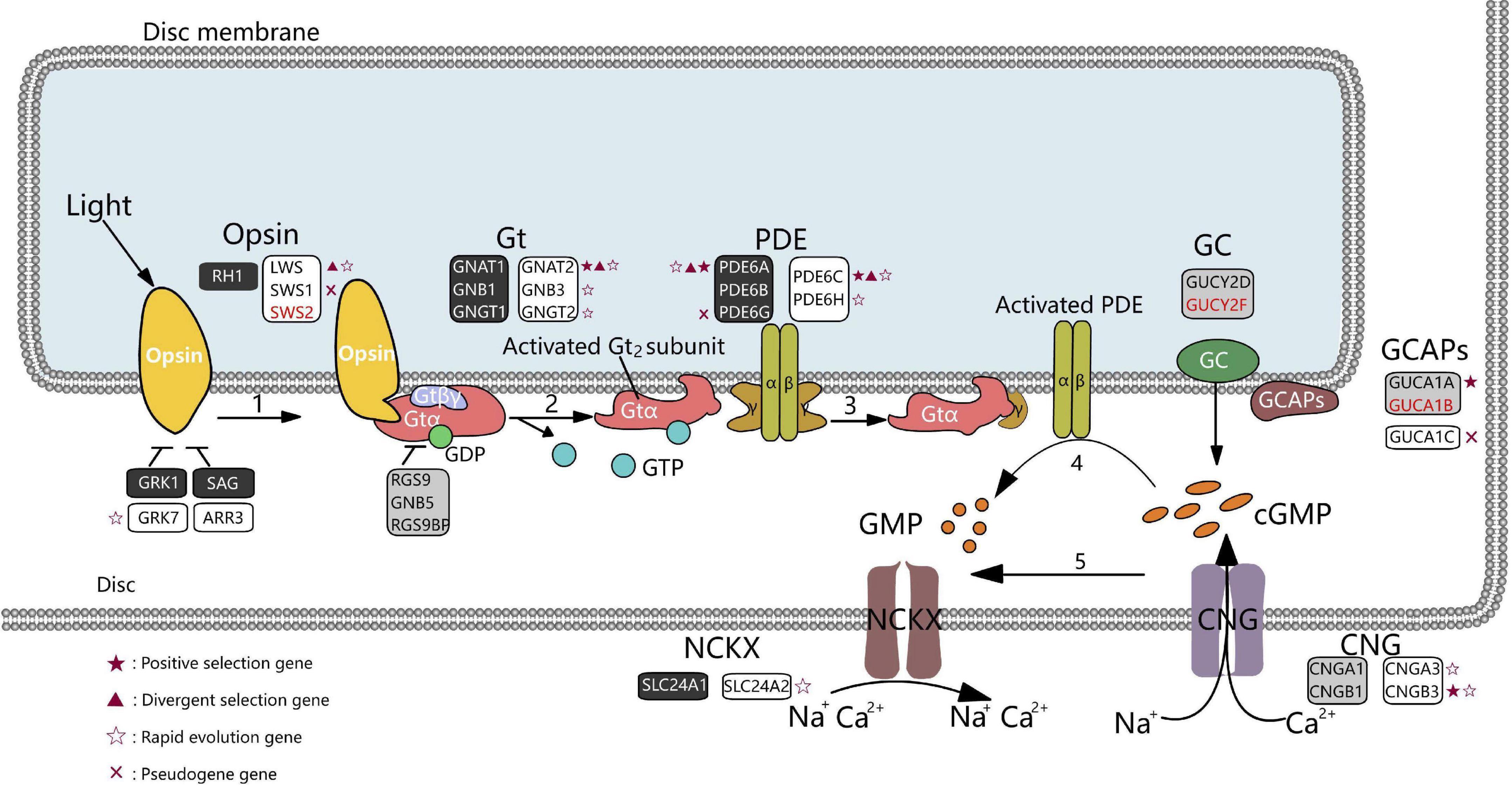
Figure 1. Diagram of phototransduction pathway genes in marsupials. Rectangles represent genes involved in the phototransduction pathway (black, rods; white, cones; gray, rods, and cones). Genes in red font could not be identified in any marsupial. Red crosses represent pseudogenes in at least one marsupial species. Genes under positive selection (branch-site model), divergent, or rapidly evolving in the crepuscular genus Macropus are shown as red stars, red triangles, and hollow stars, respectively. Gt denotes G protein transducing; PDE, phosphodiesterase; GC, guanylate cyclase; GCAPs, guanylate cyclase-activating proteins; CNG, cyclic nucleotide-gated ion channels; NCKX, Na+/Ca2+-K+ exchanger.
The visual system of mammals has undergone substantial anatomical and evolutionary modifications. For example, the eye of species that inhabit dim light conditions (e.g., cetaceans and microbats) are degraded, including decreased organisation of the retina, malformation of the lens, and subcutaneous positioning of the eye (Mass and Supin, 2007; Emerling and Springer, 2014; Emerling, 2018). Most studies have focused on the evolution of opsins and color vision in vertebrates. Cone opsins are classified into medium/long-wavelength sensitive (M/LWS) and short-wavelength sensitive (SWS) based on their peak absorption wavelength (λmax). Most eutherian mammals have two types of cone photoreceptors (SWS1 and M/LWS) and therefore possess dichromatic color vision. Notably, all Old World primates, apes, and humans have trichromatic color vision mediated by an opsin (M/LWS) gene duplication, which likely evolved for finding food in the forest (Dulai et al., 1999). Many visual genes have lost or diverged functions in certain mammals (Zhao et al., 2009). Previous studies demonstrated that cetaceans and the blind mole rat Nannospalax ehrenbergi lost the short-wavelength cone opsin OPN1SW (Peichl et al., 2001; David-Gray et al., 2002; Levenson and Dizon, 2003). Deep diving cetacean lineages (e.g., sperm whales and beaked whales), as well as some baleen whales, also lost the long-wavelength cone opsin OPN1LW, resulting in rod monochromatic vision (Peichl et al., 2001; Levenson and Dizon, 2003). The evolution of visual pigment tuning toward adaptation to dim light environments also involved amino acid mutations that modify spectral tuning and kinetics (Dungan and Chang, 2022). Meredith et al. (2013) reported that dim-light visual pigment rhodopsin (RH1) blue light-shifted at the base of Cetacea, ostensibly an adaptation to open-ocean environments. The rate of light-activated rhodopsin (meta II) decay in bats is slower compared to other mammals, indicating a bat-specific adaptation for vision in photic-limited conditions (Gutierrez et al., 2018). A recent study reported that parallel amino acid substitutions in the RH1 of deep-diving vertebrates affect retinal release and enable the visual systems of diving species to adjust quickly to changing light levels (Xia et al., 2021). In addition, multiple genes involved in cone phototransduction are pseudogenized in several whale lineages (Springer et al., 2016). Many genes associated with visual functions (e.g., RBP3, GUCY2F, ABCB5, RP1L1, CRB1, and ARR3) have inactivating mutations in subterranean mammals (Emerling, 2018; Zheng et al., 2022).
Marsupials are distributed between the Americas (Ameridelphia) and Australasia (Australidelphia) and diverged from a common ancestor approximately 80 million years ago (Deakin and O’Neill, 2020). This group of marsupials underwent considerable diversification, resulting in a range of activity patterns among extant species. They are largely nocturnal, but species with diurnal and crepuscular lifestyles exist. For instance, the Tasmanian devil (Sarcophilus harrisii) is a solitary and nocturnal carnivore predator that hunts between sunset and sunrise and spends most of the day in a den (Owen and Pemberton, 2005). Arboreal marsupials, such as the koala (Phascolarctos cinereus) and common brushtail possum (Trichosurus vulpecula), are usually nocturnal (Harper, 2005; Adam et al., 2021). Most species of the genus Macropus (including kangaroos, wallaroos, and wallabies) are nocturnal and crepuscular (later, we consider the evolution of the latter activity pattern) and spend the day in scrubs that they leave after dusk to feed in open grass plains (Inns, 1980; Kaleta and Chudzik, 2008). The crepuscular honey possum (Tarsipes rostratus) has low visual acuity (Arrese et al., 2002). Finally, the numbat (Myrmecobius fasciatus) is unique amongst marsupials, as they are diurnal and feed exclusively on termites (Cooper et al., 2003).
In contrast to eutherian mammals, the molecular evolution of marsupial vision remains largely unexplored. Studies on marsupial vision have been limited to very few species and have focused on opsin proteins and color vision. For instance, two classes of cone opsin (i.e., SWS1 and M/LWS) were found in the gray short-tailed opossum (Monodelphis domestica), the big-eared opossum (Didelphis aurita), and tammar wallaby (Macropus eugenii), suggesting that marsupials generally have dichromatic color vision, which is consistent with color discrimination test data (Hemmi, 1999; Deeb et al., 2003; Hunt et al., 2009). It has been reported that some marsupials are trichromatic, but the genetic or cytological basis remains unresolved. Potentially trichromatic species include the fat-tailed dunnart (Sminthopsis crassicaudata), honey possum, quokka (Setonix brachyurus), and quenda (Isoodon obesulus); however, there is no genetic evidence of a third cone pigment gene (Cowing et al., 2008; Ebeling et al., 2010; Upton et al., 2021).
In this study, we performed a comparative evolutionary analysis of vision-related genes of 19 marsupials. We found that gene loss in the marsupial ancestor might be related to the nocturnal activity of this group of mammals. We also report signals of positive selection and specific amino acid changes in the visual phototransduction genes of the crepuscular genus Macropus, suggesting adaptations for a twilight environment. In summary, our work provides new molecular insights into vision adaptations to different photic niches in marsupials.
Materials and methods
Species coverage
To study the evolution of vision-related genes in marsupials, the genome assemblies of 19 marsupials were downloaded from NCBI (Kitts et al., 2016) or DNA Zoo (Dudchenko et al., 2017, 2018). The three species from the Americas (Ameridelphia) were agile gracile mouse opossum (Gracilinanus agilis; NCBI AgileGrace; Tian et al., 2021), Virginia opossum (Didelphis virginiana; DNA Zoo dv-2k; Dudchenko et al., 2018), and gray short-tailed opossum (Monodelphis domestica; DNA Zoo MonDom5_HiC; Mikkelsen et al., 2007). Sixteen species from Australasia (Australidelphia) were interrogated: Diprotodontia: tammar wallaby (Macropus eugenii; NCBI Meug_1.1; Renfree et al., 2011), red kangaroo (Macropus rufus; DNA Zoo mr-2k), eastern gray kangaroo (Macropus giganteus; DNA Zoo mg-2k), and western gray kangaroo (Macropus fuliginosus; DNA Zoo mf-2k); Phalangeridae: common brushtail possum (Trichosurus vulpecula; NCBI mTriVul1.pri) and ground cuscus (Phalanger gymnotis; DNA Zoo pg-2k); Petauridae: Leadbeater’s possum (Gymnobelideus leadbeateri; DNA Zoo LBP_v1_HiC); Phascolarctidae: koala (Phascolarctos cinereus; NCBI phaCin_unsw_v4.1; Johnson et al., 2018) and common wombat (Vombatus ursinus; DNA Zoo vu-2k; Dudchenko et al., 2018); Dasyuridae: Tasmanian devil (Sarcophilus harrisii; NCBI mSarHar1.11; Miller et al., 2011; Murchison et al., 2012), thylacine (Thylacinus cynocephalus; NCBI UniMelb_ThyCyn2.0_hybrid_assembly; Feigin et al., 2018), yellow-footed antechinus (Antechinus flavipes; NCBI AdamAnt; Tian et al., 2022), brown antechinus (Antechinus stuartii; NCBI USYD_AStu_M; Brandies et al., 2020), as well as gapfilled nuclear genome assemblies of black-tailed dusky antechinus (Antechinus arktos), silver-headed antechinus (Antechinus argentus), and black-tailed dasyure (Murexia melanurus) generated by mapping to the A. flavipes assembly AdamAnt (Seim, 2020; Tian et al., 2022). We employed platypus (Ornithorhynchus anatinus; NCBI mOrnAna1.pri.v4; Warren et al., 2008; Zhou et al., 2021) as the outgroup to the marsupials.
Information retrieval
Human vision-related genes were collected from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2017) and the literature (Schott et al., 2019; Espíndola-Hernández et al., 2020). A total of 112 genes were obtained, including visual and non-visual opsins, vision genes, visual phototransduction, photoreceptor development, optic nerve development, nerve conduction, and ocular structure development genes (Figure 2A and Supplementary Table 1). Vision-related genes of other species were identified by BLASTn v2.13.0 (Johnson et al., 2008) search (E-value cutoff 1 × 10–5) on a local instance of sequenceserver v1.0.14 (Priyam et al., 2019) using gray short-tailed opossum genes as queries. Protein-coding sequence (CDS) of the gray short-tailed opossum was downloaded from the NCBI. For each gene, the CDS was used to conduct codon-based multiple sequence alignment using PRANK v170427, which implements a phylogeny-aware alignment algorithm to distinguish alignment gaps caused by insertions or deletions (Löytynoja and Goldman, 2010). Gaps and ambiguous bases were removed by Gblocks v0.91b (Castresana, 2000) to obtain reliable CDS alignments. We removed gene alignments with lengths less than 150 bp. Genes with disruptive mutations (e.g., frameshift insertions and deletions, premature stop codons, and splice site mutations at intron/exon boundaries) were verified by interrogating raw sequence reads at NCBI’s Sequence Read Archive (SRA) database using the method described in Jebb and Hiller (2018).
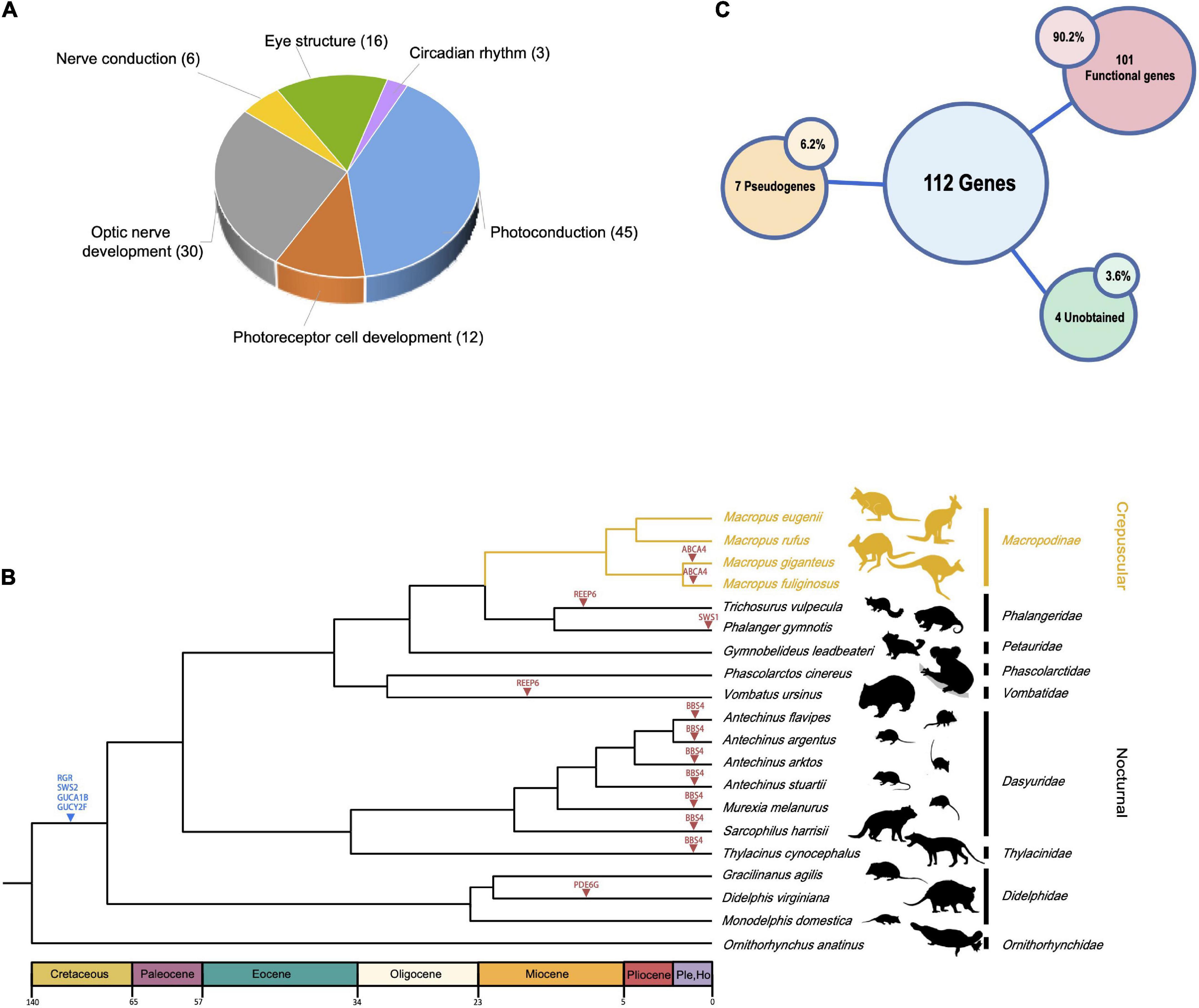
Figure 2. Overview of vision-related genes examined and species tree of marsupials. (A) Pie diagram showing broad functional categories of the 112 vision-related genes investigated. (B) Phylogenetic relationships of 19 marsupial species. The crepuscular Macropus lineage is shown in yellow. Four genes lost in the ancestor of marsupials are indicated by a blue triangle, pseudogenes found in one or more lineages as a red triangle. Silhouettes of mammalian species are reproduced from http://phylopic.org under a Public Domain or Creative Commons license. (C) Overview of the number of genes in (A) that are lost in all marsupials (“Unobtained”), pseudogenes (in at least one marsupial), and genes intact in all marsupials (“Functional genes”).
To understand the processes shaping trait evolution along the branches of a phylogenetic tree, we reconstructed the ancestral vision states. Information on activity patterns of marsupials was obtained from Animal Diversity Web1 and literature searches (Pemberton, 1990; Cooper et al., 1999; Stokes et al., 2004). The working trees used for evolutionary analyses were retrieved using TimeTree (Kumar et al., 2017).
Molecular evolution analyses
Several different codon-based likelihood methods, implemented in the CodeML programme of PAML v4.7 (Yang, 2007), were used to explore the strength and form of selection acting on vision-related genes. Briefly, the ratio of non-synonymous (dN)/synonymous (dS) substitution rates (ω = dN/dS) was estimated. We conducted analyses on a dataset that included 20 eutherian mammals and the platypus (Supplementary Figure 1). We also built a marsupial-only dataset to investigate whether there are distinct evolutionary trajectories between marsupials with different activity patterns (e.g., nocturnal vs. crepuscular) (Figure 2B). The free-ratios model (model = 1) was used to estimate the evolutionary rate along each lineage. The mean dN/dS ratios for each lineage were calculated for all genes. The branch-site model was used to test for episodes of positive selection along particular foreground branches in an otherwise conservatively evolving background (Zhang et al., 2005). Compared with the null model Ma0 (dN/dS = 1), the modified model A (Ma) allows a codon site class with dN/dS > 1 along specified branches of the phylogeny (foreground branches) as an indicator of positive selection within the specific lineages (Zhang et al., 2005). The Clade model C was performed to evaluate divergent selective pressures. This model is designed to detect sites that vary in strength and form of selection among clades (Bielawski and Yang, 2004). We utilised the branch-site model and the clade model C with branches along the last common ancestor of marsupials in the mammalian dataset and crepuscular Macropus species in the marsupial dataset set as the foreground separately. We identified sites under positive selection using Bayes’ Empirical Bayes (BEB) in PAML (Yang et al., 2005). The significance of the difference between the non-nested and nested models was evaluated using a likelihood ratio test (LRT). The Benjamini–Hochberg method was applied to correct P-values for multiple testing (cutoff set at 0.05). As a complementary to PAML models, we employed the aBSREL model in HYPHY to estimate the dN/dS ratio on each branch of the phylogeny without any a priori input (Kosakovsky Pond et al., 2020).
Disruptive mutations in genes were detected as relaxed selection by PAML branch models (Yang, 2007)2. Purifying selection was detected by comparing models A (all branches have a single ω value) and B (ω = 1 in all branches). Selective pressure relaxation on pseudogenized genes was assessed by comparing model A and model C (pseudogenized branches had a ω2, while all others had ω1). Model C was also compared to Model D (fixed ω2 = 1 for pseudogenized branches) to investigate whether the selective pressure is completely removed in pseudogenized branches. We also performed model E, where ω is allowed to vary among branches. RELAX uses a descriptive model to infer a relaxation parameter k for every gene in every species (k > 1 indicates intensified selection, i.e., positive or purifying selection, k < 1 suggests relaxed selection) (Wertheim et al., 2015).
A further approach, described by Sharma et al. (2018), was used to estimate when pseudogenes were inactivated in marsupial lineages. In the formula, K = KsTs/T + KnTn/T, K = Ka/Ks, Kn = 1, and T is the time since the species split from the last common ancestor. We estimated a lower and upper bound for Tn (how long pseudogenes evolved neutrally) as T (K−Ks)/(1 − Ks).
Specific amino acid substitutions and functional effect prediction in crepuscular marsupials
Amino acid substitutions have been associated with changes in protein function (Yadava et al., 2002). The segregating site extraction module implemented in FasParser v2.10.0 (Sun, 2018) was used to identify species-specific amino acid mutations in crepuscular marsupials, that is, residues shared in all crepuscular species and different from other marsupials. Potential functional effects of these substitutions were predicted by PolyPhen-2 (Adzhubei et al., 2010), PROVEAN (Choi and Chan, 2015), and SIFT (Sim et al., 2012). PolyPhen-2 predicts the possible impact of amino acid substitutions on the stability and function of human proteins using structural and comparative evolutionary considerations (Adzhubei et al., 2010). PROVEAN generates predictions not only for single amino acid substitutions but also for multiple amino acid substitutions, insertions, and deletions using an alignment-based score approach (Choi and Chan, 2015). SIFT is an algorithm that predicts the potential impact of amino acid substitutions or indels on protein function (Sim et al., 2012). We used the protein–protein interactions database STRING v11.53 to explore the networks of genes with specific non-synonymous changes.
Results
Pseudogenes in marsupial vision-related genes
Among our list of 112 vision-related genes present in humans (Supplementary Table 1), we failed to obtain four genes (Figure 2C, GUCA1B, GUCY2F, RGR, and SWS2) in marsupials, suggesting that these genes were lost in the marsupial ancestor. In addition, we identified several independent marsupial pseudogenization events (Supplementary Figure 2). ABCA4 is pseudogenized in the common ancestor of two out of the four Macropus species examined (the Western gray kangaroo, Macropus fuliginosus, and the eastern gray kangaroo, Macropus giganteus). The same change (a premature stop codon at exon 13) was found in both genomes. We also found two genes (PDE6G and GUCA1C) that were inactivated via premature stop codons in Didelphis virginiana. Two genes (REEP6 and EYS) with premature stop codons were found in Vombatus ursinus. A pseudogenized REEP6 was also found in Trichosurus vulpecula (one premature stop codon and a 2-bp deletion). A premature stop codon was found in Phalanger gymnotis SWS1. Finally, BBS4 has a premature stop codon and frameshift in all Dasyuromorphia species, including six of the family Dasyuridae and one of the family Thylacinidae.
All the above genes showed strong purifying selection in marsupials (Supplementary Table 2). The selective purifying pressure is markedly increased in branches with a pseudogenized ABCA4 (ω2 = 0.613, ω1 = 0.140) and REEP6 (Vombatus ursinus: ω2 = 0.634, ω1 = 0.127; Trichosurus vulpecula: ω2 = 0.739, ω1 = 0.147), suggesting that the selective pressure on these genes was relaxed in marsupial lineages (Supplementary Table 2). Only Model C that allowed Vombatus ursinus lineage with pseudogenized REEP6 with ω2 = 0.634 better fit Model D, where pseudogenized branches were fixed at ω2 = 1. This indicates that the selective pressure on REEP6 completely relaxed in the common ancestor of Vombatus ursinus, as further corroborated by the results of RELAX when Vombatus ursinus with inactivating mutations were used as a test branch (k = 0.15, p < 0.0001). We estimate that Vombatus ursinus REEP6 was inactivated 18.0–23.2 million years ago (Mya) (Supplementary Table 3) after this species emerged (31–40 Mya).
Molecular evolution of marsupial vision-related genes
To explore variation in selective pressure among marsupials, we used codon-based models on two datasets: mammals (20 eutherians, 19 marsupials, and 1 monotreme) and marsupial-only (plus monotreme as an outgroup). To test the possible occurrence of nocturnality in ancestral marsupials, we analysed the adaptive evolution of vision genes along the ancestral branch of all marsupials in the mammalian dataset. Positive selection was detected in three genes (RRH, RDH10, and CNGA1) when the ancestral lineage of marsupials was set as the foreground (Supplementary Table 4). In addition to positive selection, the clade model C analyses showed evidence for a burst of selection occurring along the lineages leading to the diversification of the major clades. We identified eight genes (GNB5, GRK1, RCVRN, SLC24A2, RHO, PDE6A, GUCY2D, and CNGA3) with significant improvement over the null model when allowing for divergent selection pressure between marsupials and outgroups (Supplementary Table 5).
In the marsupials-only dataset, we found evidence for selection along the last common ancestor (LCA) of the crepuscular genus Macropus (includes kangaroos, wallaroos, and wallabies). The lineage-specific dN/dS ratios obtained by the free-ratio model of each gene across each branch showed that Macropus lineages had significantly higher dN/dS ratios of cone phototransduction pathway genes (i.e., bright-light vision genes: ARR3, CNGA3, CNGB3, GNAT2, GNB3, GNGT2, GRK7, LWS, PDE6C, PDE6H, and SLC24A2) than their paired control nocturnal marsupial species (P < 0.048, Wilcoxon test) (Figure 3A). Moreover, we also found that all nocturnal marsupial lineages exhibited significantly elevated dN/dS ratios of rod phototransduction pathway genes (i.e., dim-light vision genes: CNGA1, CNGB1, GNAT1, GNB1, PDE6A, PDE6B, RHO, SAG, and SLC24A1) than crepuscular marsupials (in our dataset, genus Macropus) (P < 0.0059) (Figure 3B). The Clade model analyses also revealed that 13 genes (COL11A1, RP1, GNAT2, PDE6A, ATP8A2, PDE5A, LWS, PDE6C, CRYBA1, SEMA3A, SLITRK6, CACNB4, and RDH8) showed significantly higher elevated dN/dS ratios in crepuscular Macropus species than nocturnal marsupial species (Figure 3C and Supplementary Table 6). These results suggest that crepuscular Macropus species show more rapid vision gene evolution than nocturnal marsupials.
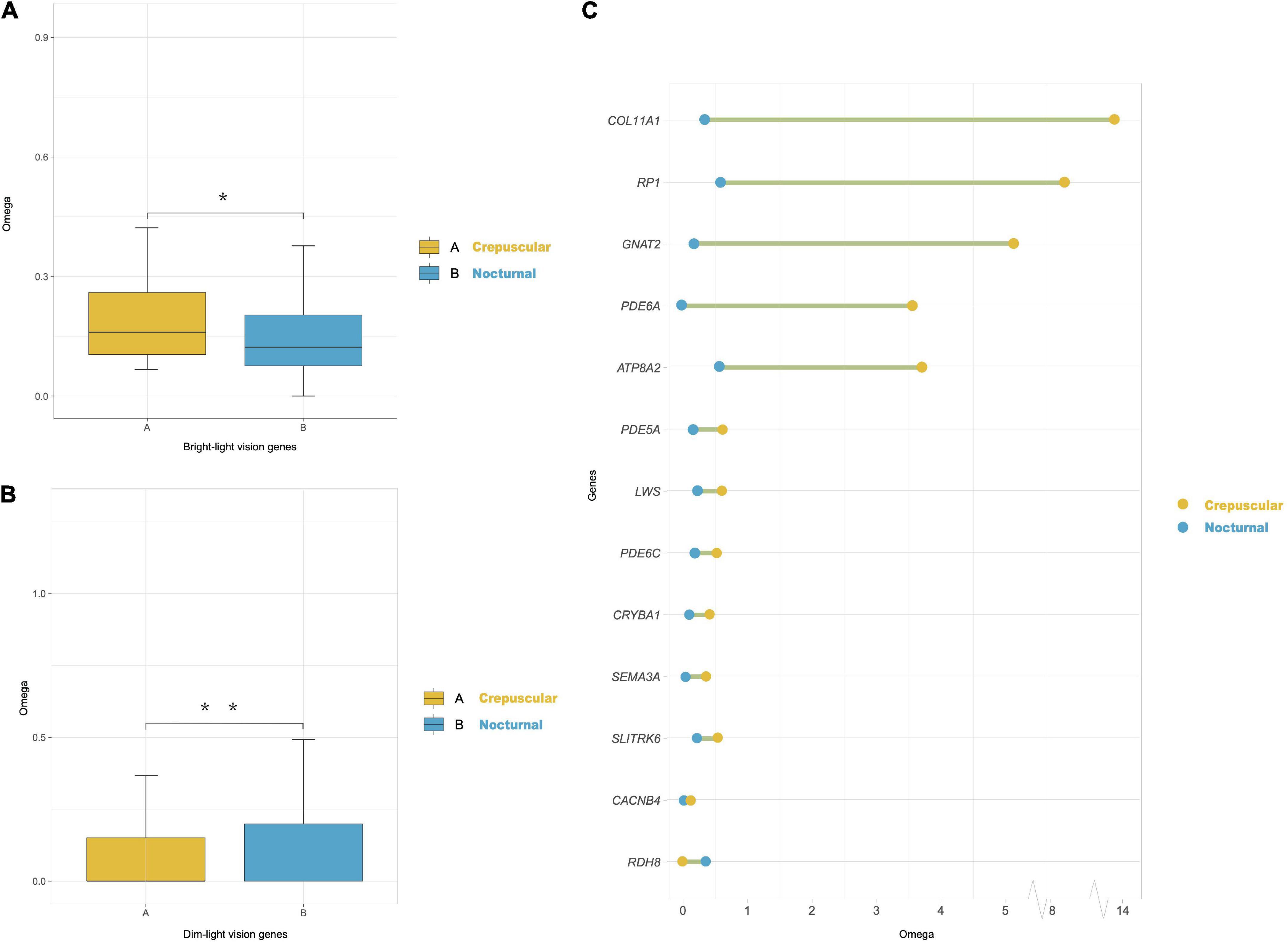
Figure 3. Divergently selected genes in marsupials with different activity patterns. (A) Omega (dN/dS) ratios of cone phototransduction pathway genes (i.e., bright-light vision genes) of crepuscular and nocturnal marsupial lineages obtained using the PAML free-ratios model. We compared each pair of groups by the Wilcoxon test (*P < 0.05). (B) Omega (dN/dS) ratios of rod phototransduction pathway genes (i.e., dim-light vision genes) of crepuscular and nocturnal marsupials. We compared each pair of groups by the Wilcoxon test (**P < 0.01). (C) Overview of a shift in selection pressure on vision genes between crepuscular and nocturnal marsupials using PAML clade model C. Blue dots indicate the omega ratio of background nocturnal species, and yellow dots represent the omega ratio of crepuscular Macropus species. A green line between the two dots indicates a significant P-value (LRT, P < 0.05).
We employed the branch-site model to detect positively selected genes (PSGs) in marsupial branches. Three different likelihood ratio tests were performed on the transition lineage from a nocturnal to a crepuscular lifestyle (i.e., the ancestral branch of Macropus) and all Macropus terminal branches (Figure 2). RDH8 was positively selected along the transition lineage, and eight PSGs (COL11A1, PDE6D, CNGB3, GNAT2, PDE6A, PED6C, RPGR, and GUCA1A) were found along Macropus (Supplementary Table 7). We conducted a complementary analysis using aBSREL, yielding five PSGs (ATP8A2, CNGA1, RDH8, RHO, and SEMA3A) along the transition lineage and three PSGs (RHO, RPGR, and CNGA1) in Macropus species (Supplementary Table 8).
Widespread species-specific amino acid substitutions in the crepuscular genus Macropus
We found 170 specific non-synonymous changes in 50 proteins in all Macropus lineages (Supplementary Table 9). Seven genes (COL11A1, GUCA1A, PDE6A, PDE6C, RPGR, DMD, and TMEM126A) possess both specific amino acid residues and a positive selection signal. Twenty-five amino acid substitutions in 15 proteins were predicted to be probably deleterious by PROVEAN, and 44 sites in 29 proteins were also predicted to be probably damaging (score ≥ 0.909) or possibly damaging (0.447 ≤ score < 0.909) by PolyPhen-2 (Supplementary Table 9). In addition, 13 specific substitutions in nine Macropus proteins were predicted to have damaging effects using SIFT (score < 0.05) (Supplementary Table 9). Notably, 17 substitutions in 13 genes (ARR3, CACNA2D4, CCDC66, CLN5, DMD, GRK7, LAMC3, NTRK2, OPA1, OPTC, PDE6A, RPGR, and TMEM126A) were identified by at least two methods, and seven sites in four proteins (LAMC3: Y741H, L620F, N473D, and D27N; OPTC: E67V; RPGR: E210G; TMEM126A: F91L) were predicted to be damaging by three methods. STRING analysis revealed that these proteins interact with a relatively high degree of connectivity (P < 1 × 10–16, Figure 4A and Supplementary Table 10) and show enrichment for gene ontology (GO) terms, such as “sensory perception of light stimulus,” “visual perception,” “opsin binding,” “photoreceptor outer segment,” and “photoreceptor disc membrane” (Figure 4B and Supplementary Table 10).
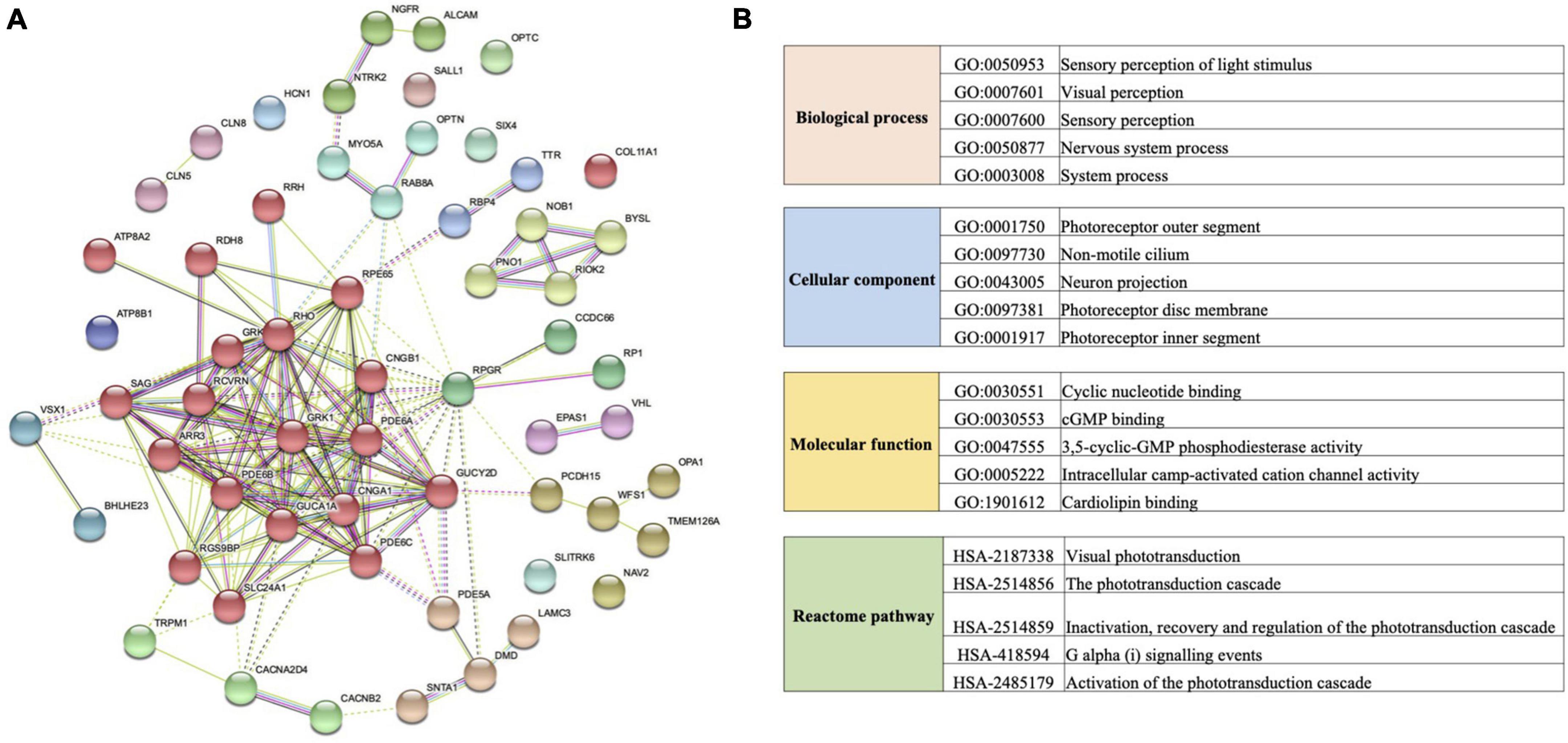
Figure 4. Protein interaction network and functional enrichment of vision genes with specific amino acid replacement in the crepuscular genus Macropus. (A) Protein–protein interaction network generated using the STRING database. Lines between each node indicate inferred or experimentally demonstrated biological associations. (B) Gene ontology (GO) term and Reactome pathway enrichments of genes examined in (A).
Discussion
Vision gene loss may be a marsupial dim-light adaptation
Eighty years ago, Walls proposed the concept of a “nocturnal bottleneck” in placental mammals, where these species evolved a nocturnal lifestyle to avoid daytime activity during the dinosaur era (Walls, 1942). This hypothesis is supported by several lines of evidence, from early paleontological records to more recent comparative genomics and phylogenetic studies (Heesy and Hall, 2010; Borges et al., 2018). For example, when placental mammals adopted nocturnality, they lost some vision-related genes, such as the visual pigmentation genes RH2 and SWS2, manifesting as dichromacy in most mammals (Gerkema et al., 2013). Regressive evolution is widespread among the visual systems of species that have invaded dim-light niches, including caves (Policarpo et al., 2021), deep oceans (Meredith et al., 2013), nocturnal environments (Wu et al., 2016), and subterranean habitats (Kim et al., 2011; Emerling and Springer, 2014). It is generally appreciated that marsupials have adapted to dim-light conditions. However, studies have largely overlooked marsupials despite the wide variety of species and ecological niches in this group of mammals.
Retinal photoreceptor cells adjust their sensitivity to allow photons to be transduced over a wide range of light intensities. One mechanism of sensitivity adjustments is the Ca2+ regulation of guanylate cyclase (GC) by GC-activating proteins (GUCA1A and GUCA1B) (Figure 1). In the present study, we found that two genes (GUCA1B and GUCY2F) required for normal photoreception were lost in the marsupial ancestor. Mouse knockout studies have shown that GUCA1A and GUCA1B paralogs are capable of facilitating vision, and GUCA1A restores the recovery of photoreceptor responses in the absence of GUCA1B (Howes et al., 2002; Pennesi et al., 2003). GUCY2F (GC2) is a retina-specific gene. No human retinal disease is linked to a GUCY2F defect, and knockout mice show normal electroretinographic responses (Baehr et al., 2007). These results hint that GUCA1B and GUCY2F loss in marsupials has a limited visual fitness consequence. RGR (retinal G-protein receptor) was also lost in the ancestor of marsupials. RGR is a non-rod or non-cone opsin localised to the membranes of retinal pigment epithelium and Müller cells (Kumbalasiri and Provencio, 2005). A study in Rgr-null mice demonstrated that RGR is involved in generating 11-cis-retinaldehyde and functions in the classical retinoid (visual) cycle (Chen et al., 2001). Rgr-null mice show normal morphological development and no apparent retinal degeneration in adults (Chen et al., 2001). Thus, similar to the above finding, loss of marsupial RGR is likely tolerated.
Several other visual genes (e.g., REEP6 and BBS4) are inactivated in one or more marsupial species. Receptor expression enhancing protein 6 (REEP6) belongs to the REEP family of proteins and has been implicated in shaping tubular organelles, such as the endoplasmic reticulum (ER) and Golgi. Reep6 knockout mice exhibit progressive retinal degeneration from disrupted ER homeostasis and protein trafficking (Agrawal et al., 2017). This gene evolved under relaxed selection in Vombatus ursinus and Trichosurus vulpecula. Both are burrowing species: Vombatus ursinus spends long periods resting in deep, thermally favorable burrows (Evans, 2008), while Trichosurus vulpecula rests in hollow-bearing trees (Cawthen and Munks, 2011). REEP6 loss might be an adaptation for the dim-light habitat of these species. Furthermore, we found that BBS4 is inactivated in all species of the carnivorous Australian order Dasyuromorphia. Knockdown of Bbs4 in mice is associated with photoreceptor cell damage and retinal degeneration (Swiderski et al., 2007).
Interestingly, ATP Binding Cassette Subfamily A Member 4 (ABCA4) is pseudogenized in both species of gray kangaroos (eastern gray kangaroo, Macropus giganteus, and western gray kangaroo, M. fuliginosus). For example, the eastern gray kangaroo actively forages during both daylight and twilight hours (Clarke et al., 1995), different from the red kangaroo (M. rufus) and tammar wallaby (M. eugenii) also examined here. Although the loss of ABCA4 in eutherian mammals (trichromatic humans and dichromatic mice alike) is associated with progressive vision loss (Al-Khuzaei et al., 2021), we speculate that its pseudogenization facilitated the evolution of diurnal foraging in a marsupial genus that is ostensibly crepuscular.
Considering that gene losses or pseudogenization may occur as an adaptation or because the gene function becomes obsolete and the gene sequence drifts (Albalat and Cañestro, 2016), we hypothesise that the above vision genes are functionally redundant in marsupials, possibly owing to a decreased demand to quickly regenerate photoreceptors in a nocturnal lifestyle.
Molecular evidence of a shift in selection pressure of Macropus vision genes
A previous study on deep-sea pearlside fishes reported that their retina is composed almost exclusively of transmuted cone photoreceptors, implying an adaptation to twilight light conditions (de Busserolles et al., 2017). Even though Macropus species, such as the tammar wallaby, are crepuscular animals, they are active during the day to varying degrees (Hemmi and Mark, 1998) and see discussion on ABCA4 above). Therefore, we hypothesised that cone phototransduction genes of crepuscular Macropus experienced shifts in selection pressure. We found evidence for a burst of accelerated, positive, and divergent selection on the branch leading to Macropus species and the transition branch representing a nocturnal to crepuscular activity pattern (Figure 5A), suggesting that there has been some vision modifications along this branch since its emergence. Most genes with selection shifts are bright-light genes and have roles in photoconduction (e.g., CNGA1, CNGB3, GNAT2, GUCA1A, PDE6A, PDE6C, PDE6D, RDH8, and RHO) or are photoreceptors (e.g., COL11A1 and RPGR). Intriguingly, several models revealed strong signals of selection on PDE6C and GNAT2 along the ancestral branch of Macropus. We also found two radical amino acid substitutions in Macropus PDE6C (Figure 5B). PDE6C encodes the cone α subunit of the cyclic guanosine monophosphate (cGMP) phosphodiesterase, which converts cGMP to 5-GMP and plays an essential role in cone phototransduction (Thiadens et al., 2009). At the level of the phototransduction cascade, the Gα subunit of transducin is critical for signal transduction (Chabre et al., 1988). GNAT2 encodes cone-specific G-protein transducin alpha subunit, and loss of Gnat2 expression in mice abolished cone phototransduction (Ronning et al., 2018). In addition, we found one positively selected and accelerated cone-expressed gene, CNGB3. This gene encodes the β subunit of the cyclic nucleotide-gated channels in cone photoreceptors (Wu et al., 2016). The strong positive selection of this gene may enhance the photoresponse and visual acuity of Macropus species. Taken together, these genes may serve to optimise the vision of Macropus for a twilight environment.
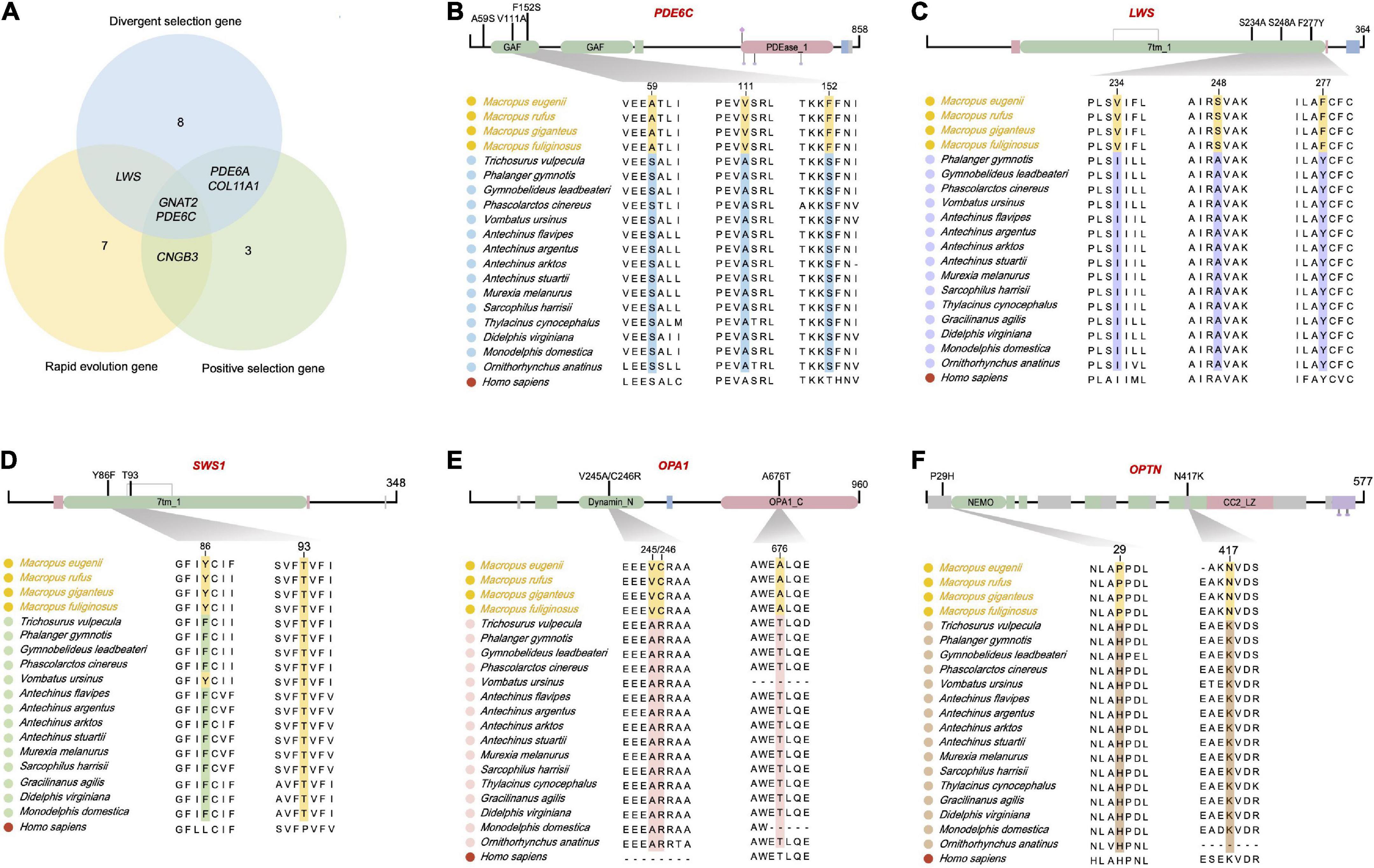
Figure 5. Five retinal phototransduction proteins have fixed amino acid substitutions in the crepuscular genus Macropus. (A) Venn graph of positively selected genes, rapidly evolving genes, and divergently selected genes in genus Macropus. (B–F) Proteins with fixed amino acid substitutions in Macropus. PDE6C (phosphodiesterase 6C, cGMP-specific, cone, and alpha prime), LWS (long wavelength-sensitive opsin), SWS1 (short wavelength-sensitive opsin 1), OPA1 (OPA1 mitochondrial dynamin-like GTPase), and OPTN (optineurin). Top, overview of amino acid change locations (functional domains were predicted using Pfam). Bottom, Multiple sequence alignments. The human sequence was used as the alignment reference.
Retinal opsin photopigments initiate mammalian vision when stimulated by light (Figure 1). It is interesting to note that LWS, a long-wavelength sensitive opsin, was positively selected in the Macropus ancestor. In addition, we found two radical amino acid replacements (sites 248 and 277) in the LWS cytoplasmic domain (Figure 5C). Among these, the Y277F substitution is associated with a short-wavelength shift in mammals (Yokoyama and Radlwimmer, 2001; Davies et al., 2012). Twilight is primarily characterized by relative enrichment of shorter wavelength light (Roenneberg and Foster, 1997). We propose that Macropus LWS, with positively selected radical amino acid changes, may increase photon absorption in a twilight environment. Short wavelength-sensitive opsin 1 (SWS1) is also intriguing. It has been suggested that mammal SWS1 amino acid residue changes are associated with photic niche adaptation by spectral tuning (Emerling et al., 2015). SWS1 of different species has a peak absorption wavelength (λmax) that range from ultraviolet-sensitive (UVS) to violet-sensitive (VS) (Hunt and Peichl, 2014). We found that all nocturnal marsupial SWS1 proteins harbor F86 and T93 substitutions (Figure 5D), suggesting that these species may have UVS pigments. In contrast, all Macropus species and Vombatus ursinus are more likely to have VS pigment due to their SWS1 possessing Y86 and T93 (Figure 5D). In agreement with a previous hypothesis on mammalian SWS1 evolution (Emerling et al., 2015), our finding supports a scenario where nocturnal marsupials have UVS pigments to facilitate the detection of a broader spectrum of light while crepuscular Macropus species evolved reduced UV lens transmittance to limit the retina exposure to damaging UV light and improve visual acuity under twilight conditions.
Finally, several genes (e.g., OPA1 and OPTN) with Macropus-specific amino acid sites are associated with the development and integrity of the retina. OPA1 (OPA1 mitochondrial dynamin-like GTPase) encodes a dynamin-related mitochondrial protein essential for retinal ganglion cell synaptic architecture and connectivity (Williams et al., 2012). OPA1 deficiency has been associated with increased autophagy in retinal ganglion cells in a murine model of dominant optic atrophy (White et al., 2009). Three radical amino acid substitutions (residues 245, 246, and 676) were identified in Macropus OPA1. One was predicted to be deleterious using PROVEAN and Polyphen-2 (Figure 5E and Supplementary Table 9, T676A). OPTN (optineurin) has two amino acid substitutions in Macropus (Figure 5F). Optineurin is an autophagy receptor (Sarfarazi and Rezaie, 2003). Considering that the retina of Macropus species, such as tammar wallaby, shows a high ganglion and cone cell density (Hemmi and GRUeNERT, 1999; Wimborne et al., 1999), the OPA1 and OPTN amino acid changes may confer neuroprotection or improve visual acuity.
Conclusion
This study provides new insights into the molecular evolution of marsupial vision. While a limited number of marsupials have been genetically sequenced to date, the genomes of many species are forthcoming (reviewed in Deakin and O’Neill, 2020) and should enable a photic niche survey across the diverse marsupial taxonomy. Further research is also required to determine whether the gene changes identified have a functional significance. Our work provides a gene set that can now be tested in various animal models, including marsupials (Kiyonari et al., 2021).
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author. The dataset presented in this study can be found at https://doi.org/10.6084/m9.figshare.20357475.
Author contributions
IS conceived the study and assisted with manuscript revision. RT, HG, ZJ, FZ, and JZ participated in the data analysis. RT and HG interpreted the data. RT wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (Grant No. 31900310 to RT), the Jiangsu Specially-Appointed Professors Programme (to IS), Jiangsu Province’s Innovation Programme (JSSCTD202142) and the Priority Academic Programme Development of Jiangsu Higher Education Institutions (PAPD) to RT and IS, and the Postgraduate Research & Practice Innovation Programme of Jiangsu Province. The funding bodies for this study had no role in the design of the study, collection of data, data analysis and interpretation, or in writing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organisations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.982073/full#supplementary-material
Footnotes
References
Adam, D., Johnston, S., Beard, L., Nicolson, V., Lisle, A., Gaughan, J., et al. (2021). Temporal effect of feeding on the body temperature and behaviour of captive koalas (Phascolarctos cinereus). Aus. Mammal. 44, 16–23. doi: 10.1071/AM20024
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P., et al. (2010). A method and server for predicting damaging missense mutations. Nat. Methods. 7, 248–249. doi: 10.1038/nmeth0410-248
Agrawal, S. A., Burgoyne, T., Eblimit, A., Bellingham, J., Parfitt, D. A., Lane, A., et al. (2017). REEP6 deficiency leads to retinal degeneration through disruption of ER homeostasis and protein trafficking. Hum. Mol. Genet. 26, 2667–2677. doi: 10.1093/hmg/ddx149
Albalat, R., and Cañestro, C. (2016). Evolution by gene loss. Nat. Rev. Genet. 17, 379–391. doi: 10.1038/nrg.2016.39
Al-Khuzaei, S., Broadgate, S., Foster, C. R., Shah, M., Yu, J., Downes, S. M., et al. (2021). An overview of the genetics of ABCA4 retinopathies, an evolving story. Genes 12, 1241. doi: 10.3390/genes12081241
Arrese, C., Archer, M., and Beazley, L. (2002). Visual capabilities in a crepuscular marsupial, the honey possum (Tarsipes rostratus): A visual approach to ecology. J. Zool. 256, 151–158. doi: 10.1017/S0952836902000183
Baehr, W., Karan, S., Maeda, T., Luo, D.-G., Li, S., Bronson, J. D., et al. (2007). The function of guanylate cyclase 1 and guanylate cyclase 2 in rod and cone photoreceptors. J. Biol. Chem. 282, 8837–8847. doi: 10.1074/jbc.M610369200
Bennie, J. J., Duffy, J. P., Inger, R., and Gaston, K. J. (2014). Biogeography of time partitioning in mammals. Proc. Natl. Acad. Sci. U.S.A. 111, 13727–13732. doi: 10.1073/pnas.1216063110
Bielawski, J. P., and Yang, Z. (2004). A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol Evol. 59, 121–132. doi: 10.1007/s00239-004-2597-8
Borges, R., Johnson, W. E., O’Brien, S. J., Gomes, C., Heesy, C. P., and Antunes, A. (2018). Adaptive genomic evolution of opsins reveals that early mammals flourished in nocturnal environments. BMC Genomics 19:121. doi: 10.1186/s12864-017-4417-8
Brandies, P. A., Tang, S., Johnson, R. S. P., Hogg, C. J., and Belov, K. (2020). The first Antechinus reference genome provides a resource for investigating the genetic basis of semelparity and age-related neuropathologies. Gigabyte 1:7. doi: 10.46471/gigabyte.7
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Cawthen, L., and Munks, S. (2011). The use of hollow-bearing trees retained in multi-aged regenerating production forest by the Tasmanian common brushtail possum (Trichosurus vulpecula fuliginosus). Wildlife Res. 38, 687–695. doi: 10.1071/WR10125
Chabre, M., Bigay, J., Bruckert, F., Bornancin, F., Deterre, P., Pfister, C., et al. (1988). Visual Signal Transduction; The Cycle of Transducin Shuttling between Rhodopsin and cGMP Phosphodiesterase. Cold Spring Harb. Symp. Quant. Biol. 53, 313–324. doi: 10.1101/SQB.1988.053.01.038
Chen, P., Hao, W., Rife, L., Wang, X. P., Shen, D., Chen, J., et al. (2001). A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat. Genet. 28, 256–260. doi: 10.1038/90089
Choi, Y., and Chan, A. P. (2015). PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747. doi: 10.1093/bioinformatics/btv195
Clarke, J. L., Jones, M. E., and Jarman, P. J. (1995). Diurnal and nocturnal grouping and foraging behaviors of free-ranging eastern grey kangaroos. Aust. J. Zool. 43, 519–529. doi: 10.1071/ZO9950519
Cooper, C., Withers, P., and Bradshaw, S. (2003). Field metabolic rate and water turnover of the numbat (Myrmecobius fasciatus). J. Comp. Physiol. B. 173, 687–693. doi: 10.1007/s00360-003-0380-6
Cooper, D. W., Ord, T. J., and Evans, C. S. (1999). Nocturnal behaviour of the parma wallaby, Macropus parma (Marsupialia: Macropodoidea). Aust. J. Zool. 47, 155–167. doi: 10.1071/ZO98047
Cowing, J. A., Arrese, C. A., Davies, W. L., Beazley, L. D., and Hunt, D. M. (2008). Cone visual pigments in two marsupial species: The fat-tailed dunnart (Sminthopsis crassicaudata) and the honey possum (Tarsipes rostratus). Proc. Royal Soc. B 275, 1491–1499. doi: 10.1098/rspb.2008.0248
David-Gray, Z., Bellingham, J., Munoz, M., Avivi, A., Nevo, E., and Foster, R. (2002). Adaptive loss of ultraviolet-sensitive/violet-sensitive (UVS/VS) cone opsin in the blind mole rat (Spalax ehrenbergi). Eur. J. Neurosci. 16, 1186–1194. doi: 10.1046/j.1460-9568.2002.02161.x
Davies, W. I., Wilkie, S. E., Cowing, J. A., Hankins, M. W., and Hunt, D. M. (2012). Anion sensitivity and spectral tuning of middle-and long-wavelength-sensitive (MWS/LWS) visual pigments. Cell. Mol. Life Sci. 69, 2455–2464. doi: 10.1007/s00018-012-0934-4
de Busserolles, F., Cortesi, F., Helvik, J. V., Davies, W. I., Templin, R. M., Sullivan, R. K., et al. (2017). Pushing the limits of photoreception in twilight conditions: The rod-like cone retina of the deep-sea pearlsides. Sci. Adv. 3:eaao4709. doi: 10.1126/sciadv.aao4709
Deakin, J. E., and O’Neill, R. J. (2020). Evolution of marsupial genomes. Annu. Rev. Anim. Biosci. 8, 25–45. doi: 10.1146/annurev-animal-021419-083555
Deeb, S. S., Wakefield, M. J., Tada, T., Marotte, L., Yokoyama, S., and Marshall Graves, J. A. (2003). The cone visual pigments of an Australian marsupial, the tammar wallaby (Macropus eugenii): Sequence, spectral tuning, and evolution. Mol. Biol. Evol. 20, 1642–1649. doi: 10.1093/molbev/msg181
Dudchenko, O., Batra, S. S., Omer, A. D., Nyquist, S. K., Hoeger, M., Durand, N. C., et al. (2017). De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95. doi: 10.1126/science.aal3327
Dudchenko, O., Shamim, M. S., Batra, S. S., Durand, N. C., Musial, N. T., Mostofa, R., et al. (2018). The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. BioRxiv [Preorint]. doi: 10.1101/254797
Dulai, K. S., von Dornum, M., Mollon, J. D., and Hunt, D. M. (1999). The evolution of trichromatic color vision by opsin gene duplication in New World and Old World primates. Genome Res. 9, 629–638. doi: 10.1101/gr.9.7.629
Dungan, S. Z., and Chang, B. S. (2022). Ancient whale rhodopsin reconstructs dim-light vision over a major evolutionary transition: Implications for ancestral diving behavior. Proc. Natl. Acad. Sci. U.S.A. 119:e2118145119. doi: 10.1073/pnas.2118145119
Ebeling, W., Natoli, R. C., and Hemmi, J. M. (2010). Diversity of color vision: Not all Australian marsupials are trichromatic. PLoS One 5:e14231. doi: 10.1371/journal.pone.0014231
Emerling, C. A. (2018). Regressed but not gone: Patterns of vision gene loss and retention in subterranean mammals. Integr. Comp. Biol. 58, 441–451. doi: 10.1093/icb/icy004
Emerling, C. A., Huynh, H. T., Nguyen, M. A., Meredith, R. W., and Springer, M. S. (2015). Spectral shifts of mammalian ultraviolet-sensitive pigments (short wavelength-sensitive opsin 1) are associated with eye length and photic niche evolution. Proc. Royal Soc. B. 282:20151817. doi: 10.1098/rspb.2015.1817
Emerling, C. A., and Springer, M. S. (2014). Eyes underground: Regression of visual protein networks in subterranean mammals. Mol. Phylogenet. Evol. 78, 260–270. doi: 10.1016/j.ympev.2014.05.016
Espíndola-Hernández, P., Mueller, J. C., Carrete, M., Boerno, S., and Kempenaers, B. (2020). Genomic evidence for sensorial adaptations to a nocturnal predatory lifestyle in owls. Genome Biol. Evol. 12, 1895–1908. doi: 10.1093/gbe/evaa166
Evans, M. C. (2008). Home range, burrow-use and activity patterns in common wombats (Vombatus ursinus). Wildlife Res. 35, 455–462. doi: 10.1071/WR07067
Feigin, C. Y., Newton, A. H., Doronina, L., Schmitz, J., Hipsley, C. A., Mitchell, K. J., et al. (2018). Genome of the Tasmanian tiger provides insights into the evolution and demography of an extinct marsupial carnivore. Nat. Ecol. Evol. 2, 182–192. doi: 10.1038/s41559-017-0417-y
Fu, Y., and Yau, K.-W. (2007). Phototransduction in mouse rods and cones. Pflug. Arch. Eur. J. Phy. 454, 805–819. doi: 10.1007/s00424-006-0194-y
Gerkema, M. P., Davies, W. I., Foster, R. G., Menaker, M., and Hut, R. A. (2013). The nocturnal bottleneck and the evolution of activity patterns in mammals. Proc. Royal Soc. B 280:20130508. doi: 10.1098/rspb.2013.0508
Gutierrez, E. D. A., Castiglione, G. M., Morrow, J. M., Schott, R. K., Loureiro, L. O., Lim, B. K., et al. (2018). Functional shifts in bat dim-light visual pigment are associated with differing echolocation abilities and reveal molecular adaptation to photic-limited environments. Mol. Biol. Evol. 35, 2422–2434. doi: 10.1093/molbev/msy140
Harper, M. J. (2005). Home range and den use of common brushtail possums (Trichosurus vulpecula) in urban forest remnants. Wildlife Res. 32, 681–687. doi: 10.1071/WR04072
Heesy, C. P., and Hall, M. I. (2010). The nocturnal bottleneck and the evolution of mammalian vision. Brain Behav. Evol. 75, 195–203. doi: 10.1159/000314278
Hemmi, J. (1999). Dichromatic colour vision in an Australian marsupial, the tammar wallaby. J. Comp. Physiol. A. 185, 509–515. doi: 10.1007/s003590050411
Hemmi, J., and Mark, R. (1998). Visual acuity, contrast sensitivity and retinal magnification in a marsupial, the tammar wallaby (Macropus eugenii). J. Comp. Physiol. A. 183, 379–387. doi: 10.1007/s003590050264
Hemmi, J. M., and GRUeNERT, U. (1999). Distribution of photoreceptor types in the retina of a marsupial, the tammar wallaby (Macropus eugenii). Visual Neurosci. 16, 291–302. doi: 10.1017/S0952523899162102
Howes, K. A., Pennesi, M. E., Sokal, I., Church-Kopish, J., Schmidt, B., Margolis, D., et al. (2002). GCAP1 rescues rod photoreceptor response in GCAP1/GCAP2 knockout mice. EMBO J. 21, 1545–1554. doi: 10.1093/emboj/21.7.1545
Hunt, D. M., Chan, J., Carvalho, L. S., Hokoc, J. N., Ferguson, M. C., Arrese, C. A., et al. (2009). Cone visual pigments in two species of South American marsupials. Gene 433, 50–55. doi: 10.1016/j.gene.2008.12.006
Hunt, D. M., and Peichl, L. (2014). S cones: Evolution, retinal distribution, development, and spectral sensitivity. Visual Neurosci. 31, 115–138. doi: 10.1017/S0952523813000242
Inns, R. W. (1980). Ecology of the Kangaroo Island wallaby, Macropus eugenii (Desmarest), in Flinders Chase National Park, Kangaroo Island/by Robert W. Inns. Adelaide, SA: University of Adelaide.
Jebb, D., and Hiller, M. (2018). Recurrent loss of HMGCS2 shows that ketogenesis is not essential for the evolution of large mammalian brains. eLife 7:e38906. doi: 10.7554/eLife.38906
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: A better web interface. Nucleic Acids Res. 36:W5–W9. doi: 10.1093/nar/gkn201
Johnson, R. N., O’Meally, D., Chen, Z., Etherington, G. J., Ho, S. Y. W., Nash, W. J., et al. (2018). Adaptation and conservation insights from the koala genome. Nat. Genet. 50, 1102–1111. doi: 10.1038/s41588-018-0153-5
Kaleta, T., and Chudzik, P. (2008). The comparison of behaviour in two macropodid marsupials: Red kangaroo Macropus rufus and red-necked wallaby Macropus rufogriseus kept in adjacent enclosures at Warsaw ZOO. Anim. Sci. 45, 53–58.
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45:D353–D361. doi: 10.1093/nar/gkw1092
Kim, E. B., Fang, X., Fushan, A. A., Huang, Z., Lobanov, A. V., Han, L., et al. (2011). Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479, 223–227. doi: 10.1038/nature10533
Kitts, P. A., Church, D. M., Thibaud-Nissen, F., Choi, J., Hem, V., Sapojnikov, V., et al. (2016). Assembly: A resource for assembled genomes at NCBI. Nucleic Acids Res. 44:D73–D80. doi: 10.1093/nar/gkv1226
Kiyonari, H., Kaneko, M., Abe, T., Shiraishi, A., Yoshimi, R., Inoue, K. I., et al. (2021). Targeted gene disruption in a marsupial, Monodelphis domestica, by CRISPR/Cas9 genome editing. Curr. Biol. 31, 3956–3963.e3954. doi: 10.1016/j.cub.2021.06.056
Kosakovsky Pond, S. L., Poon, A. F., Velazquez, R., Weaver, S., Hepler, N. L., Murrell, B., et al. (2020). HyPhy 2.5—a customizable platform for evolutionary hypothesis testing using phylogenies. Mol. Biol. Evol. 37, 295–299. doi: 10.1093/molbev/msz197
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Kumbalasiri, T., and Provencio, I. (2005). Melanopsin and other novel mammalian opsins. Exp. Eye Res. 81, 368–375. doi: 10.1016/j.exer.2005.05.004
Lamb, T. D. (2013). Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119. doi: 10.1016/j.preteyeres.2013.06.001
Levenson, D. H., and Dizon, A. (2003). Genetic evidence for the ancestral loss of short-wavelength-sensitive cone pigments in mysticete and odontocete cetaceans. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 270, 673–679. doi: 10.1098/rspb.2002.2278
Löytynoja, A., and Goldman, N. (2010). webPRANK: A phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC bioinformatics 11:579. doi: 10.1186/1471-2105-11-579
Mass, A. M., and Supin, A. Y. (2007). Adaptive features of aquatic mammals’ eye. Anat. Rec. 290, 701–715. doi: 10.1002/ar.20529
Meredith, R. W., Gatesy, J., Emerling, C. A., York, V. M., and Springer, M. S. (2013). Rod monochromacy and the coevolution of cetacean retinal opsins. Plos Genet. 9:e1003432. doi: 10.1371/journal.pgen.1003432
Mikkelsen, T. S., Wakefield, M. J., Aken, B., Amemiya, C. T., Chang, J. L., Duke, S., et al. (2007). Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447, 167–177. doi: 10.1038/nature05805
Miller, W., Hayes, V. M., Ratan, A., Petersen, D. C., Wittekindt, N. E., Miller, J., et al. (2011). Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil). Proc. Natl. Acad. Sci. U.S.A. 108, 12348–12353. doi: 10.1073/pnas.1102838108
Murchison, E. P., Schulz-Trieglaff, O. B., Ning, Z., Alexandrov, L. B., Bauer, M. J., Fu, B., et al. (2012). Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 148, 780–791. doi: 10.1016/j.cell.2011.11.065
Owen, D., and Pemberton, D. (2005). Tasmanian Devil: A Unique And Threatened Animal. Crows Nest, SB: Allen & Unwin.
Peichl, L., Behrmann, G., and Kröger, R. H. (2001). For whales and seals the ocean is not blue: A visual pigment loss in marine mammals. Eur. J. Neurosci. 13, 1520–1528. doi: 10.1046/j.0953-816x.2001.01533.x
Pemberton, D. (1990). Social Organisation And Behaviour Of The Tasmanian Devil, Sarcophilus Harrisii, Ph.D Thesis. Hobart, TAS: University of Tasmania.
Pennesi, M. E., Howes, K. A., Baehr, W., and Wu, S. M. (2003). Guanylate cyclase-activating protein (GCAP) 1 rescues cone recovery kinetics in GCAP1/GCAP2 knockout mice. Proc. Natl. Acad. Sci. U.S.A. 100, 6783–6788. doi: 10.1073/pnas.1130102100
Policarpo, M., Fumey, J., Lafargeas, P., Naquin, D., Thermes, C., Naville, M., et al. (2021). Contrasting gene decay in subterranean vertebrates: Insights from cavefishes and fossorial mammals. Mol. Biol. Evol. 38, 589–605. doi: 10.1093/molbev/msaa249
Priyam, A., Woodcroft, B. J., Rai, V., Moghul, I., Munagala, A., Ter, F., et al. (2019). Sequenceserver: A Modern Graphical User Interface for Custom BLAST Databases. Mol. Biol. Evol. 36, 2922–2924. doi: 10.1093/molbev/msz185
Renfree, M. B., Papenfuss, A. T., Deakin, J. E., Lindsay, J., Heider, T., Belov, K., et al. (2011). Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 12:R81.
Roenneberg, T., and Foster, R. G. (1997). Twilight times: Light and the circadian system. Photochem. Photobiol. 66, 549–561. doi: 10.1111/j.1751-1097.1997.tb03188.x
Ronning, K. E., Allina, G. P., Miller, E. B., Zawadzki, R. J., Pugh, E. N. Jr., Herrmann, R., et al. (2018). Loss of cone function without degeneration in a novel Gnat2 knock-out mouse. Exp. Eye Res. 171, 111–118. doi: 10.1016/j.exer.2018.02.024
Sarfarazi, M., and Rezaie, T. (2003). Optineurin in primary open angle glaucoma. Ophthalmol. Clin. North. Am. 16, 529–541. doi: 10.1016/S0896-1549(03)00061-0
Schott, R. K., Bhattacharyya, N., and Chang, B. S. (2019). Evolutionary signatures of photoreceptor transmutation in geckos reveal potential adaptation and convergence with snakes. Evolution 73, 1958–1971. doi: 10.1111/evo.13810
Seim, I. (2020). Data from: Gapfilled nuclear genome assemblies of black- tailed dusky antechinus (Antechinus arktos), silver-headed antechinus (Antechinus argentus), and black-tailed dasyure (Murexia melanurus). Genève, CH: Zenodo
Sharma, V., Hecker, N., Roscito, J. G., Foerster, L., Langer, B. E., and Hiller, M. (2018). A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 9:1215. doi: 10.1038/s41467-018-03667-1
Sim, N.-L., Kumar, P., Hu, J., Henikoff, S., Schneider, G., and Ng, P. C. (2012). SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40:W452–W457. doi: 10.1093/nar/gks539
Smale, L., Lee, T., and Nunez, A. A. (2003). Mammalian diurnality: Some facts and gaps. J. Biol. Rhythm. 18, 356–366. doi: 10.1177/0748730403256651
Springer, M. S., Emerling, C. A., Fugate, N., Patel, R., Starrett, J., Morin, P. A., et al. (2016). Inactivation of cone-specific phototransduction genes in rod monochromatic cetaceans. Front. Ecol. Evol. 4:61. doi: 10.3389/fevo.2016.00061
Stokes, V. L., Pech, R. P., Banks, P. B., and Arthur, A. D. (2004). Foraging behaviour and habitat use by Antechinus flavipes and Sminthopsis murina (Marsupialia: Dasyuridae) in response to predation risk in eucalypt woodland. Biol. Conserv. 117, 331–342. doi: 10.1016/j.biocon.2003.12.012
Sun, Y. B. (2018). FasParser2: A graphical platform for batch manipulation of tremendous amount of sequence data. Bioinformatics 34, 2493–2495. doi: 10.1093/bioinformatics/bty126
Swiderski, R. E., Nishimura, D. Y., Mullins, R. F., Olvera, M. A., Ross, J. L., Huang, J., et al. (2007). Gene expression analysis of photoreceptor cell loss in bbs4-knockout mice reveals an early stress gene response and photoreceptor cell damage. Invest. Ophth. Vis. Sci. 48, 3329–3340. doi: 10.1167/iovs.06-1477
Thiadens, A. A., den Hollander, A. I., Roosing, S., Nabuurs, S. B., Zekveld-Vroon, R. C., Collin, R. W., et al. (2009). Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am. J. Hum. Genet. 85, 240–247. doi: 10.1016/j.ajhg.2009.06.016
Tian, R., Han, K., Geng, Y., Yang, C., Guo, H., Shi, C., et al. (2021). A Chromosome-Level Genome of the Agile Gracile Mouse Opossum (Gracilinanus agilis). Genome. Biol. Evol 13:evab162. doi: 10.1093/gbe/evab162
Tian, R., Han, K., Geng, Y., Yang, C., Shi, C., Thomas, P. B., et al. (2022). A chromosome-level genome of Antechinus flavipes provides a reference for an Australian marsupial genus with male death after mating. Mol. Ecol. Resour. 22, 740–754. doi: 10.1111/1755-0998.13501
Upton, B. A., Díaz, N. M., Gordon, S. A., Van Gelder, R. N., Buhr, E. D., and Lang, R. A. (2021). Evolutionary constraint on visual and nonvisual mammalian opsins. J. Biol. Rhythm. 36, 109–126. doi: 10.1177/0748730421999870
Walls, G. L. (1942). The Vertebrate Eye And Its Adaptive Radiation. Toowoomba City, QLD: Cranbrook Press. doi: 10.5962/bhl.title.7369
Warren, W. C., Hillier, L. W., Marshall Graves, J. A., Birney, E., Ponting, C. P., Grutzner, F., et al. (2008). Genome analysis of the platypus reveals unique signatures of evolution. Nature 453, 175–183. doi: 10.1038/nature06936
Wertheim, J. O., Murrell, B., Smith, M. D., Kosakovsky Pond, S. L., and Scheffler, K. (2015). RELAX: Detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 32, 820–832. doi: 10.1093/molbev/msu400
White, K. E., Davies, V. J., Hogan, V. E., Piechota, M. J., Nichols, P. P., Turnbull, D. M., et al. (2009). OPA1 deficiency associated with increased autophagy in retinal ganglion cells in a murine model of dominant optic atrophy. Invest. Ophth. Vis. Sci. 50, 2567–2571. doi: 10.1167/iovs.08-2913
Williams, P. A., Piechota, M., von Ruhland, C., Taylor, E., Morgan, J. E., and Votruba, M. (2012). Opa1 is essential for retinal ganglion cell synaptic architecture and connectivity. Brain 135, 493–505. doi: 10.1093/brain/awr330
Wimborne, B., Mark, R., and Ibbotson, M. (1999). Distribution of retinogeniculate cells in the tammar wallaby in relation to decussation at the optic chiasm. J. Comp. Neurol. 405, 128–140. doi: 10.1002/(SICI)1096-9861(19990301)405:1<128::AID-CNE9>3.0.CO;2-H
Wu, Y., Hadly, E. A., Teng, W., Hao, Y., Liang, W., Liu, Y., et al. (2016). Retinal transcriptome sequencing sheds light on the adaptation to nocturnal and diurnal lifestyles in raptors. Sci. Rep. 6:33578. doi: 10.1038/srep33578
Xia, Y., Cui, Y. M., Wang, A. S., Liu, F. N., Chi, H., Potter, J. H. T., et al. (2021). Convergent phenotypic evolution of rhodopsin for dim-light sensing across deep-diving vertebrates. Mol. Biol. Evol. 38, 5726–5734. doi: 10.1093/molbev/msab262
Yadava, N., Potluri, P., Smith, E. N., Bisevac, A., and Scheffler, I. E. (2002). Species-specific and mutant MWFE proteins: Their effect on the assembly of a functional mammalian mitochondrial complex I. J. Biol. Chem. 277, 21221–21230. doi: 10.1074/jbc.M202016200
Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z., Wong, W. S., and Nielsen, R. (2005). Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Yokoyama, S., and Radlwimmer, F. B. (2001). The molecular genetics and evolution of red and green color vision in vertebrates. Genetics 158, 1697–1710. doi: 10.1093/genetics/158.4.1697
Zhang, J., Nielsen, R., and Yang, Z. (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479. doi: 10.1093/molbev/msi237
Zhao, H., Rossiter, S. J., Teeling, E. C., Li, C., Cotton, J. A., and Zhang, S. (2009). The evolution of color vision in nocturnal mammals. Proc. Natl. Acad. Sci. U.S.A. 106, 8980–8985. doi: 10.1073/pnas.0813201106
Zheng, Z., Hua, R., Xu, G., Yang, H., and Shi, P. (2022). Gene losses may contribute to subterranean adaptations in naked mole-rat and blind mole-rat. BMC Biol. 20:44. doi: 10.1186/s12915-022-01243-0
Keywords: vision-related genes, marsupials, nocturnal vision, crepuscular vision, molecular evolution
Citation: Tian R, Guo H, Jin Z, Zhang F, Zhao J and Seim I (2022) Molecular evolution of vision-related genes may contribute to marsupial photic niche adaptations. Front. Ecol. Evol. 10:982073. doi: 10.3389/fevo.2022.982073
Received: 30 June 2022; Accepted: 03 August 2022;
Published: 26 August 2022.
Edited by:
Lin Zhang, Hubei University of Chinese Medicine, ChinaReviewed by:
Zhengfei Wang, Yancheng Teachers University, ChinaRobert William Meredith, Montclair State University, United States
Copyright © 2022 Tian, Guo, Jin, Zhang, Zhao and Seim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inge Seim, aW5nZUBzZWltbGFiLm9yZw==
†These authors have contributed equally to this work
 Ran Tian
Ran Tian Han Guo1†
Han Guo1† Inge Seim
Inge Seim