
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 23 September 2022
Sec. Conservation and Restoration Ecology
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.976448
This article is part of the Research TopicBiodiversity Conservation and Ecological Function Restoration in Freshwater EcosystemsView all 16 articles
The red swamp crayfish, Procambarus clarkii, is a globally invasive species and has caused huge damage to aquaculture, biodiversity, and ecology worldwide. Antenna-expressed receptors are important for P. clarkii to detect chemosensory cues for mate attraction. In this study, we tested the behavior of male P. clarkii to the conditioned water from female P. clarkii during the mating and non-mating periods, and performed RNA sequencing to investigate the chemosensory-related genes of the antenna of male P. clarkii. The results of the behavioral assay have shown that for the female-conditioned water, male P. clarkii within the mating period can be significantly attracted, but not during the non-mating period. This suggested that the expressions of chemosensory-related genes in the antenna of male P. clarkii may change significantly with mating seasonal variation. Antenna transcriptomes found that a total of 59,218 unigenes with an average length of 1,056.41 bp, and 4,889 differentially expressed unigenes (DEGs), among which 2,128 were upregulated, while 2,761 were downregulated were obtained. A total of 12 upregulated and nine downregulated DEGs were associated with chemical reception, including four ionotropic receptors (IRs) or ionotropic glutamate receptors (iGluRs), eight G-protein-coupled receptors (GPCRs), five transient receptor potential channels (TRP channels), one sodium–calcium exchanger, one isomerase, and two uncharacterized proteins (chemosensory proteins-like, CSPs). CSPs were preliminarily classified as pheromone receptors in the antenna of male P. clarkii. Furthermore, the calcium transduction-related pathways may play an important role in the sex pheromone reception of the male P. clarkii’s antenna. The results of quantitative real-time reverse transcriptase PCR (RT-qPCR) showed that the trends of expression of eight selected unigenes were consistent with RNA-Seq results. Our results provide more comprehensive data for chemical communication mechanisms after P. clarkii enter the mating period and eventually would develop better control strategies in further.
Chemoreception is the dominant sensory modality for most crustaceans due to the complex aquatic environment (Harzsch and Krieger, 2018), which is the base of crustaceans for their daily activities, such as searching for food, reproduction, and communication (Blinova and Cherkashin, 2012). Previous studies suggest that the antenna is one of the predominant chemosensory organs of most crustaceans (Harzsch et al., 2011; Waldrop, 2013; Urbschat and Scholtz, 2019). Crustaceans receive chemical signals through several parallel channels, which can be summarized into two modes of “olfaction” and “distributed chemoreception” (Schmidt and Mellon, 2011). “Olfaction” is mediated by unimodal olfactory sensilla called “aesthetasc,” which are only present on the first antenna of the crustaceans (Harzsch and Krieger, 2018). Aesthetascs are innervated by olfactory sensory neurons (OSNs) (Derby et al., 2016). “Distributed chemoreception” is mediated by various bimodal sensillae in the first antenna (Solari et al., 2017), mouthparts (Garm et al., 2003; Garm et al., 2004), chelipeds, and walking appendages (Altner et al., 1983; Schmidt and Gnatzy, 1984). It is innervated by both chemoreceptor neurons (CRNs) and mechanoreceptor neurons (MRNs). However, there is a gap in the data for chemoreceptors of crustaceans. So far, the predicted chemosensory-related proteins in crustaceans include ionotropic receptors (IRs), ionotropic glutamate receptors (iGluRs), G-protein-coupled receptors (GPCRs), transient receptor potential channel (TRP channel), chemosensory proteins (CSPs), and gustatory receptor (GRs) (Eyun et al., 2017; Kozma et al., 2020a,b). Identification of candidate chemosensory-related genes helps to better understand the molecular basis of chemoreception in pests and eventually develop better control strategies (Wang et al., 2014; Gonzalez et al., 2021).
The red swamp crayfish, Procambarus clarkii (Girard, 1852), has its origins in North-eastern Mexico and South America (Shen et al., 2014), and was introduced to China from Japan during the 1930s (Shen et al., 2020). At present, it is an economically important animal (Liu et al., 2021). However, invasive crayfish P. clarkii into the natural ecosystems of China has caused huge losses to agriculture, biodiversity, and aquaculture. Moreover, P. clarkii is a carrier of the white spot syndrome virus (WSSV) and parasites, whereby it may cause infection and death of other commercial shrimps in case of spread without effective control (Zhu et al., 2009). Therefore, effectively and specifically controlling P. clarkii requires more attention. In recent years, control of invasive species using sex pheromone has been adopted for other species of insects (Gherardi et al., 2011; Keller-Costa et al., 2014; Johnson et al., 2015). Previous studies confirm that males can detect sex pheromones through their chemosensory organ uniaxially, and then search for the signal source or perform courtship behaviors (Oyama et al., 2020). In addition, the suitable reproduction temperatures of P. clarkii are ranging from 21 to 25°C, and it reduced mating behavior below 15°C (Jin et al., 2019; Dong et al., 2020). The male P. clarkii recognizes female mating receptivity by detecting the urinary components of females (Kubec et al., 2019). However, little is known about the chemosensory mechanism of action of these essential chemicals (such as sex pheromone components) in male P. clarkii.
Like other crustaceans, the P. clarkii’s antenna is a critical chemosensory organ that perceives and locates chemical signals or pheromones released by their mates (Breithaupt, 2011). Although the candidate chemosensory genes of the lateral flagellum of the antennule in P. clarkii were reported (Kozma et al., 2020a), the chemosensory genes in the antenna are still unknown. Therefore, to further understand the chemical communication mechanism through chemosensation in male P. clarkii, we performed the behavioral assays of male P. clarkii on the crude conditioned water from female P. clarkii (in the mating period, MP) during the mating (MP) and non-mating period (NMP), and collected the antenna of the male P. clarkii in the MP as the experimental group and the NMP as the control group. Subsequently, next-generation sequencing (NGS) and assembly were used to obtain a male antenna transcriptome of P. clarkii in MP vs. NMP. Our findings may help better understand the mechanisms of chemosensory responses in P. clarkii, and provide a solid foundation for further studies on the relationship between chemosensory-related genes and semiochemicals.
Procambarus clarkii (400 males and 20 females) was purchased from Guilin city, Guangxi province, China in August 2020. The average weight of the mature P. clarkii was 16.22 ± 1.82 g. The length of the antenna was longer than 5 cm. A total of 20 P. clarkii were cultured in one water tank and fed two times a day on artificial food throughout the experimental period (Li et al., 2012). A total of 20 mature female P. clarkii were fasted for 48 h and placed in a water tank with 1 L of ultrapure water (for crude conditioned water collections) for further cultivation for 24 h. The feeding temperature of female P. clarkii was between 23 and 25°C. The conditioned water of female P. clarkii was centrifuged for 20 min (8000 rpm, 4°C), and removed the insoluble substances to obtain the crude conditioned water (collected in the mating period, for behavior assay). The crude conditioned water samples were stored at −80°C until further behavior assay experiment.
All male P. clarkii had no contact with female crayfish or pheromones during the feeding process. We randomly divided male P. clarkii into two following groups: 200 individuals were fed until January 2021 (NMP, water temperature <15°C) and another 200 individuals were fed until April 2021 (the initial stage of the MP, water temperatures >22°C).
The behavioral assays were performed using the Y-maze (self-made with glass) to determine the sensitivity of male P. clarkii to the crude conditioned water (Figure 1). There was a drain hole at the start arm of the Y-maze device to keep the water depth at 5 cm. Fifty male P. clarkii were tested for behavioral assays in NMP (in January) and MP (in April), respectively. The procedures of behavioral assays were performed as follows: First, 500 ml of ultrapure water was poured into Y-maze and regulated the water flow rate (20 ml/min) of infusion tubes to the same velocity by flow meters. Thereafter, cylindrical sponge blocks with stimulus (1 ml of crude conditioned water, experimental group) and ultrapure water (1 ml, control group) were placed in the end arm, respectively. Finally, the male P. clarkii was put into the start arm and observed the arm entry by P. clarkii. Each animal was tested for the crude conditioned water only once. At the end of each operation, the Y-maze would be emptied and wiped with alcohol on the surface to prevent leftover interference from the last operation. Each assay lasted 5 min. If no choice was made for more than 5 min, the male P. clarkii would be counted as unresponsive individuals, which would be excluded from the analysis. After every five P. clarkii were tested, the positions of the experimental group (crude conditioned water) and the control group (ultrapure water) were switched to prevent directional bias. The data were calculated using the Chi-squared test (Barnard et al., 2016) in SPSS v 25.0 at a significant level of P < 0.05.

Figure 1. The behavioral device: Y-maze. The height of Y-maze is all 5 cm. The Y-maze was symmetrical on both sides. 1: cylindrical sponge block, 2: flow meter, 3: water box, and 4: drain hole.
To reveal the difference in male P. clarkii responses to female-conditioned water, the antenna transcriptomics of male P. clarkii was performed in NMP and MP groups using RNA-Seq technology. When the behavior assay was done, the whole antenna with the hard cuticle of male P. clarkii (150 individuals in NMP and MP, respectively) was collected and transferred into sterile 15 ml centrifuge tubes without RNase and DNase (Corning, United States). In order to obtain enough RNA, 50 pairs of male antennae were combined into one sample. NMP and MP groups had three replicate samples, respectively. The samples of NMP and MP groups were frozen in liquid nitrogen and stored at −80°C until the extraction of total RNA. The library construction and sequencing of transcriptomes were performed by Major Technology Co. Ltd. (Shanghai, China). Total RNA extraction was performed using the TRIzol reagent (Invitrogen, Shanghai, China). Both concentration and purity were measured using a Nanodrop 2000 spectrophotometer (Invitrogen, MA, United States). The degradation of extracted RNA was detected on 1% agarose gels and RNA integrity number (RIN) was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies, United States).
mRNAs were enriched using magnetic beads with Oligo (dT) and randomly fragmented using fragmentation buffer. Under the action of reverse transcriptase, the fragmented mRNAs were used as templates for first-strand cDNA synthesis using random hexamer primers. Subsequently, second-strand cDNA was synthesized using DNA polymerase I and RNase H. We added an end-repair mix (including the end-repair enzyme mix and end-repair buffer) to patch the cohesive ends of the double-strand cDNA, followed by the addition of tail and sequencing adapters. Subsequently, cDNA was amplified by PCR. A total of six cDNA libraries were obtained after purifying the amplification products with AMPure XP beads. Thereafter, QuantiFluor dsDNA System and Quantus™ Fluorometer (Promega, Madison, WI, United States) were used to detect the concentration and inter size of the libraries, respectively. The effective concentration of the libraries was accurately quantified qPCR (quantitative polymerase chain reaction, effective concentration >2 nM) using the Qubit RNA Assay Kit in the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States). Finally, the constructed paired-end libraries were sequenced on the Illumina HiSeqXten/NovaSeq 6000 Sequencing Platform.
Using SeqPrep1 and Sickle,2 all clean reads from NMP and MP groups were obtained by filtering the sequencing adapters, poly-N, and the low-quality sequences (quality value, QV <20) from raw reads. The assembly of two sets of antenna was combined together and performed by using RNA-Seq de novo programs Trinity software (v2.8.5) (Liu et al., 2021). The completeness of the assembly was evaluated using the BUSCO software (v5.3.2). The assembled de novo unigenes were obtained, which were the encoding sequences. The redundancy was filtered using the Cd-hit-EST (Foquet et al., 2021). The Transrate (v1.0.3) was used to map reads to contigs and inspect the alignments (Marx et al., 2021).
The unigene sequences were compared using six databases, including the National Center for Biotechnology Information (NCBI) Non-Redundant Protein Sequence Database (Nr),3 Protein family (Pfam),4 Gene Ontology (GO),5 Swiss-Prot,6 Cluster of Orthologous Groups of proteins (COG),7 and Kyoto Encyclopedia of Genes and Genomes (KEGG).8 Functional annotation was processed using the Blast2GO software (Conesa et al., 2005).
The Bowtie software (Langmead et al., 2009) was used to compare clean reads with the unigene library and on combining with RSEM (Li and Dewey, 2011), the expression levels were estimated. The expression of unigenes was calculated using FPKM (fragments per kilobase per million) (Mortazavi et al., 2008) between six libraries. Then, the differentially expressed unigenes (DEGs) were analyzed using the DESeq2 software (v1.24.0) based on the false discovery rate (FDR) correction with Benjamini and Hochberg (BH) method (Varet et al., 2016). The resulting P-values were adjusted using the FDR. The DEGs were filtered using the threshold of FDR <0.05 and | log2 (fold change) | ≥1 (Ma et al., 2020). Subsequently, GO and KEGG pathway enrichment analyses were conducted based on Fisher’s precision probability test (BH method, P-adjust <0.05) using the Goatools software (Klopfenstein et al., 2018). All DEGs were mapped to terms in the KEGG and GO databases using the BLAST algorithm with an E-value of ≤1E-5, and the significantly (P < 0.05) enriched KEGG and GO terms in DEGs were identified compared with the transcriptome background. The data of pathways were analyzed on the online tool of Majorbio Cloud Platform (https://cloud.majorbio.com/page/tools/) (Ren et al., 2022).
To verify the results of our sequencing analyses, we selected eight sensory-related DEGs with high expressions in MP or NMP groups (Supplementary Table 1) from the male antenna of P. clarkii for quantitative real-time reverse transcriptase PCR (RT-qPCR) analysis. The total RNA was reversely transcribed into first-strand cDNA using the PrimeScript™ first-strand cDNA Synthesis Kit (TaKaRa, Shanghai, China), following which the newly synthesized cDNA was used as the template for RT-qPCR. Specific primers were designed using the Primer 5.0 software with a concentration of 10 μmol/L. The primer sequences of eight DEGs used for RT-qPCR are shown in Supplementary Table 1. The RT-qPCR was performed on an MA-6000 real-time fluorescence quantitative PCR instrument (Molarray, Jiangsu, China). The RT-qPCR reactions (20 μl) contained 1 μl of cDNA, 0.4 μl of each primer (0.2 μmol/L in this reaction system), 10 μl of 2 × SYBR real-time PCR premixture (dNTP and buffer), and added the RNase free sterilized ultrapure water. The reaction procedure was as follows: 95°C for 5 min, followed by 40 cycles with 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. To confirm reproducibility, the RT-qPCR reaction for each sample was performed in three technical replicates and three biological replicates based on the MP (>22°C) and NMP (<15°C) groups. The melt curves were used to evaluate the specificity of primers and amplification. All melt curves have a single peak, and PCR efficiency for each primer pair was ranging from 91 to 103%. The level of expression of selected genes was calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001), and the expression of selected genes was normalized against β-actin expression levels (Yu et al., 2022). Comparative analyses for each target gene among different samples were analyzed using the one-way ANOVA (SPSS, version 25.0).
The results of behavioral assays have shown that the conditioned water from female P. clarkii can significantly attract the male individuals during the MP (χ2 = 4.12, P = 0.04), but cannot significantly attract the males in NMP (χ2 = 0.31, P = 0.58) (Figure 2 and Supplementary Table 2).
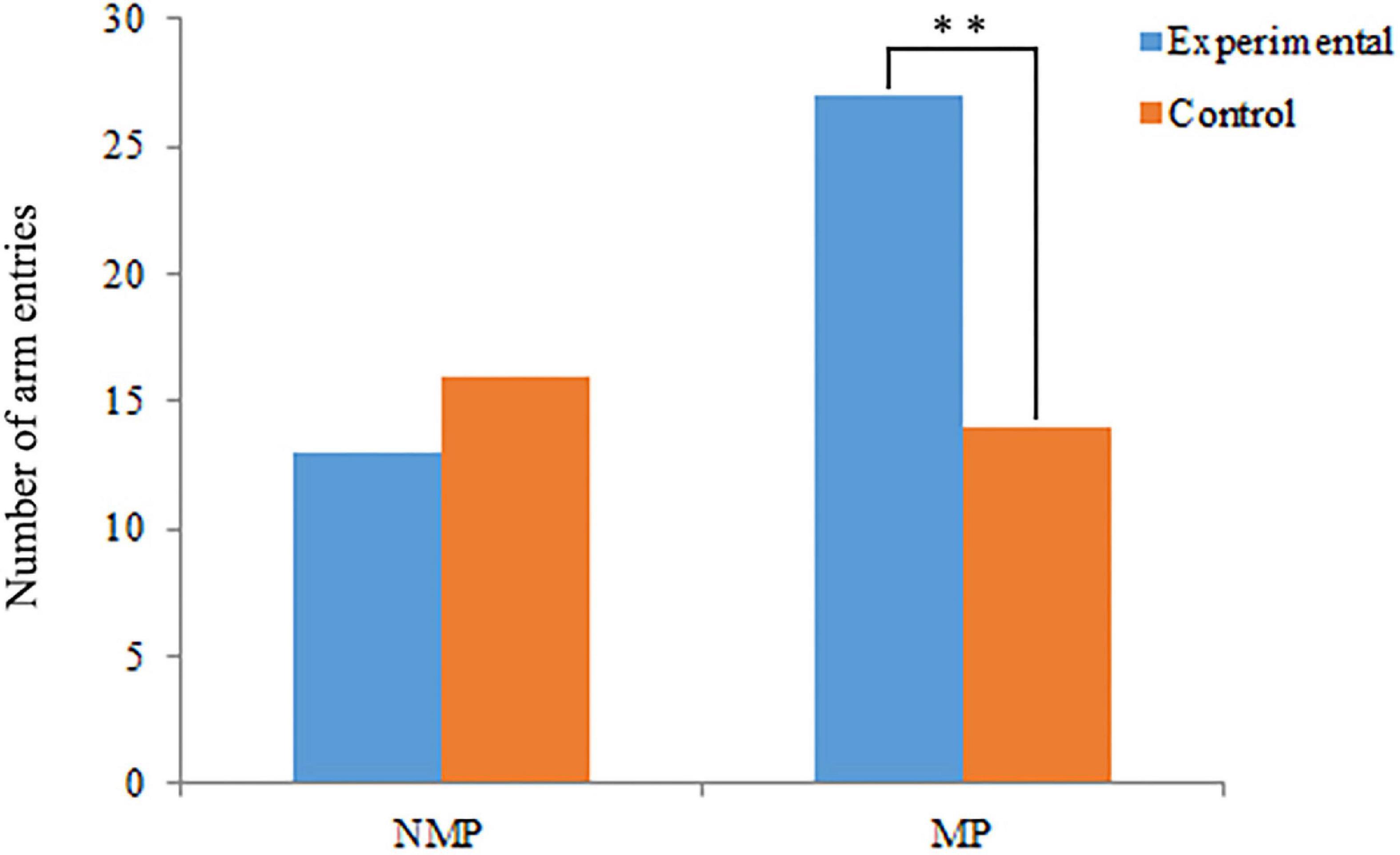
Figure 2. Behavioral assays of male Procambarus clarkii during the non-mating period (NMP) and mating period (MP). *Represents the significant differences between experimental and control groups (P < 0.05, n = 50). The numbers of no choice were excluded from the statistical analysis.
A total of 258.48 million raw reads and 252.98 million clean reads were obtained from six libraries (Supplementary Table 3). The Q20 and Q30 were greater than 97 and 92%, respectively. The GC contents were ranging from 44.38 to 49.05% (Supplementary Table 3). Moreover, all clean reads from NMP and MP groups were combined to de novo assemble the unigenes (Table 1 and Supplementary Figure 1). The BUSCO score showed that the transcriptome was with high completeness (single-copy: 91.8%; duplicated: 4.2%). The raw data files have been uploaded to the NCBI sequence read archive (BioProject accession number: PRJNA839237).
To investigate information on gene functions more comprehensively, a total of 57,812 unigenes were annotated using six databases including Nr, Pfam, Swiss-Prot, COG, GO, and KEGG. The results of annotation of the unigenes are as follows: 18,388 (31.81%) unigenes were annotated in Nr; 14,039 (24.28%) in Pfam; 11,986 (20.73%) in Swiss-Prot; 14,479 (25.04%) in COG; 14,129 (24.44%) in GO; and 9,176 (15.87%) in KEGG (Supplementary Figure 3 and Supplementary Table 4). The annotations of unigenes to different databases are shown in Supplementary Figure 4. Moreover, a total of 111 unigenes were annotated to chemosensory-related functions, including IRs or iGluRs (43 unigenes annotated), TRP channels (30), CSPs-like (2), GPCRs (17), GR (1), and other olfactory-related proteins (18) (Supplementary Excel Table 1).
Based on the DESeq2 software, a total of 4,889 DEGs with the FDR <0.05 and | log2 (fold change) | ≥1 were identified between MP and NMP groups, including 2,128 upregulated and 2,761 downregulated unigenes (Supplementary Figure 4). The fold change (FC) and annotations of all DEGs are shown in Supplementary Excel Table 2. Furthermore, the annotations from the GO analysis of DEGs were classified into 44 GO categories, including BP (18), CC (14), and MF (12). The top three GO terms in CC were cell part (GO:0044464), membrane part (GO:0044425), and protein-containing complex (GO:0032991); in BP were, cellular process (GO:0009987), metabolic process (GO:0008152), and biological regulation (GO:0065007); in MF were, binding (GO:0005488), catalytic activity (GO:0003824), and structural molecule activity (GO:0005198) (Supplementary Figure 5A). Moreover, the KEGG enrichments have shown that the “Ribosome (P-value = 5.067E-31, 156 upregulated and 39 downregulated DEGs)” is the most enriched pathway, followed by “Amoebiasis” and “Complement and coagulation cascades” (Supplementary Figure 5B).
As shown in Table 2, 21 identified DEGs were related to chemosensory functions. On comparing the MP and NMP groups, nine DEGs were found to be downregulated, while 13 were upregulated, including four IRs or iGluRs, eight G-protein-coupled receptors, five TRP channels, one sodium–calcium exchanger, one isomerase, and two uncharacterized protein (CSP-like). The sequences of these DEGs are shown in Supplementary Excel Table 3. Among these 21 DEGs, the DEG related to sodium–calcium exchanger (solute carrier family, NCX) was involved in the olfactory transduction and calcium signaling pathway (Figure 3).
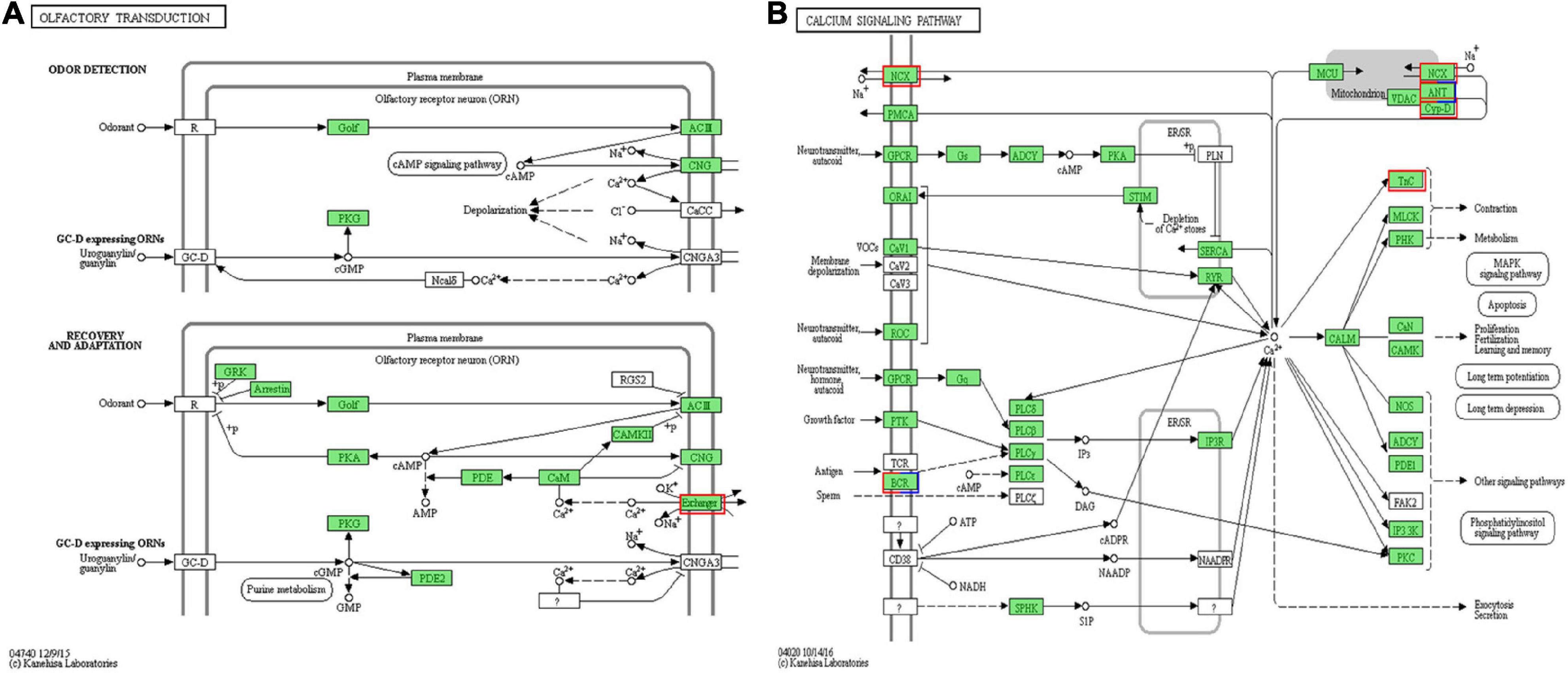
Figure 3. The calcium-related differentially expressed unigenes (DEGs) were enriched in (A) the olfactory transduction pathway and (B) the calcium signaling pathway. The red box represents the upregulated DEGs in mating period (MP) groups. The blue box represents the downregulated DEGs in MP groups, and the box with both red and blue represents that there are both up and downregulated DEGs in MP groups. The green box represents the genes annotated in the pathway without significant difference (Image credits: https://www.kegg.jp/pathway/map=map04740&keyword=olfactory%20transduction and https://www.kegg.jp/pathway/map=map04020&keyword=calcium).
To verify the accuracy of the RNA-Seq data, eight sensory-involved genes were selected from among the DEGs and validated by RT-qPCR analysis. The expression levels of six DEGs were significantly upregulated (GPCR moody, TRIM 32, CSP-like, ankyrin-1, NCX 1, and SSTR 2) in MP groups and two (ATP 1A and STK 26-like) were significantly downregulated in the MP groups. The results of RT-qPCR were consistent with those of RNA-Seq, indicating the reliability of the RNA-Seq data (Figure 4, Supplementary Table 5, and Supplementary Excel Table 4).
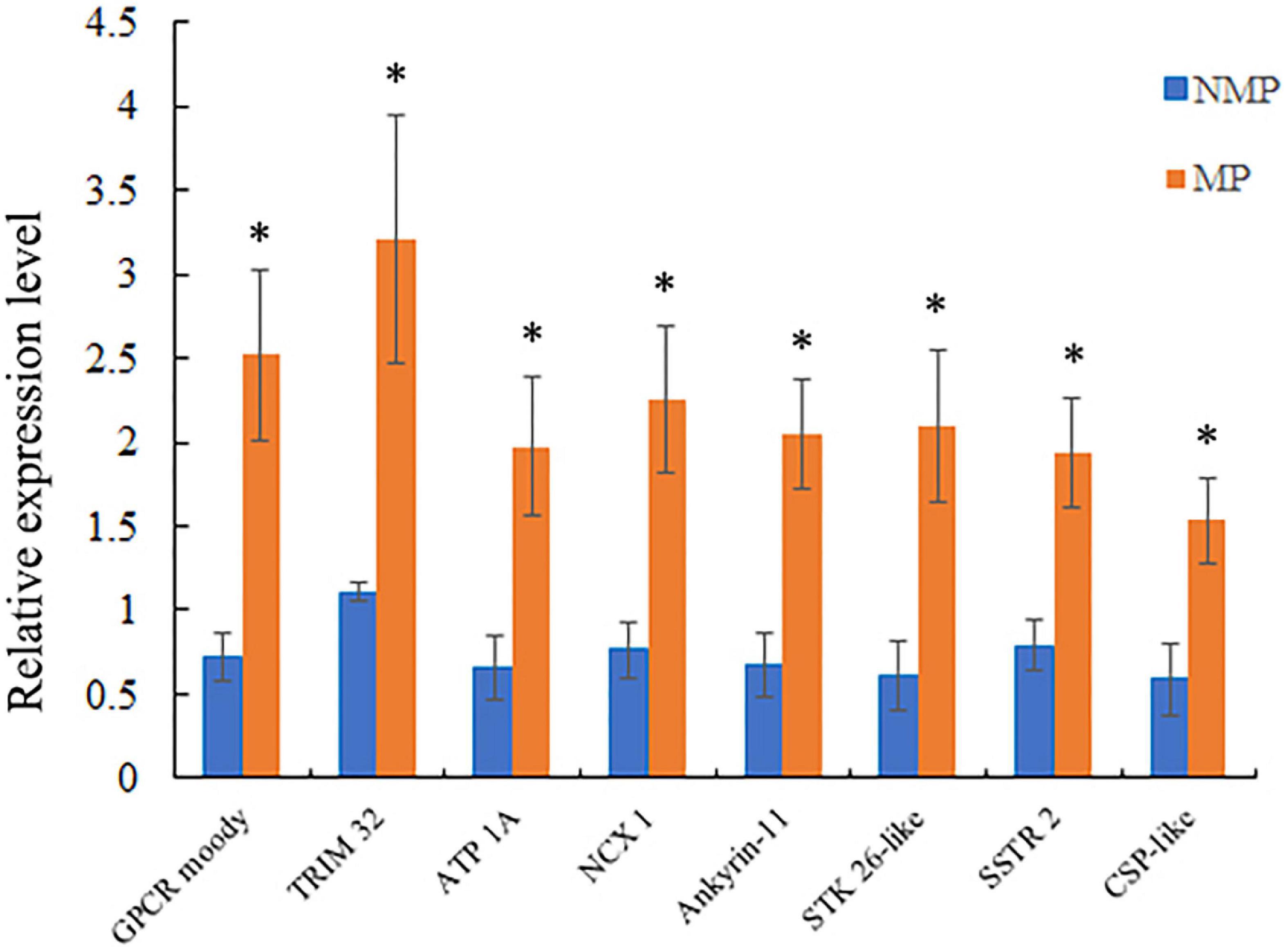
Figure 4. Relative levels of selected differentially expressed unigene (DEG) expression by RT-qPCR. Levels were measured using the 2–ΔΔCT method; β-actin was the internal normalization control. Data were shown as mean ± SD, n = 3; *P < 0.05.
Numerous behavioral studies indicate that the male P. clarkii have “unidirectional receptors” for the sex pheromones released by females during the MP (Monteclaro et al., 2010; Peddio et al., 2019; Oyama et al., 2020). Male individuals can recognize different phases of females by detecting the pheromones and then performing courtship behaviors after checking the sex pheromone levels (Kubec et al., 2019). However, previous research suggests that the frequencies of copulation and oviposition in P. clarkii are significantly reduced when the temperature goes below 15°C (Egly et al., 2019). In this study, we found that male P. clarkii were attracted to female-conditioned water in MP but not in NMP, which was consistent with previous studies. We speculated that the expression of chemosensory genes in antenna may be altered due to different temperatures and periods. Therefore, we identified the chemosensory genes and pathways in the antenna of P. clarkii between MP vs. NMP by RNA-Seq for the first time. However, the information on chemosensory genes was as sparse as expected, presumably since genetic annotation of species closely related to P. clarkii is scarce in public genetic databases; in general, the genetic information on chemoreception in crustaceans is lacking. This study is a necessary supplement to the genetic information library for P. clarkii.
Procambarus clarkii is one of the main model animals in the study of chemosensory receptors in crustaceans. The previous study had found several types of chemoreceptors in the antennule and legs of P. clarkii, including the IRs, iGluRs, TRP channels, and GRs (Kozma et al., 2020a). In our study, we found these four types of chemosensory receptors in the antenna of P. clarkii as well, indicating that these chemosensory receptors are not organ-specific, but expressed generally in a variety of sensory organs of P. clarkii. Furthermore, we found only one GR in the antenna of P. clarkii, which was consistent with the transcriptomic results of the antennule and legs in P. clarkii (Kozma et al., 2020a). This suggested that GRs remain low in numbers in P. clarkii and other decapod crustaceans. However, compared with the transcriptomic results of antennule and legs in P. clarkii (Kozma et al., 2020a), we found that the numbers of TRP channels were more in the antenna than in the two organs (antennule and leg), which we suggested that TRP channels might mediate more chemosensory processes in the antenna of P. clarkii. Overall, a lot of research on the chemosensory genes and receptors of crustaceans is still in the recognition stage, and there are still numerous receptor functions that have not been studied and reported yet and need more studies to investigate.
Notably, the DEGs related to the TRP channels were typically upregulated. TRP channels are homotetramers or heterotetramers with six transmembrane segments. There are eight subfamilies of TRP channels, including TRPC, TRPA, TRPP, TRPN, TRPV, TRPM, TRPM, and TRPT (Venkatachalam and Montell, 2007; Venkatachalam et al., 2014; Van den Eynde et al., 2021). In this study, six types of TRP channels (TRPA1, Pyrexia TRPA, TRPM2, TRPM3, TRPC4, and TRPV5) were identified. TRP channels participate in many sensory processes, such as vision, olfactory, audition, and temperature sensation, which affect the behaviors of creatures profoundly (Fowler and Montell, 2013). As the climate becomes warmer, more movements are generated by benthos and crayfish (Larson and Magoulick, 2011; Johnson et al., 2014; Wittwer et al., 2018). We speculated that P. clarkii could enhance self-abilities for various senses instinctively for environmental adaptation, which include the sex pheromone receptions as well.
Many DEGs have been annotated to uncharacterized proteins in our study, of which two DEGs were annotated to CSPs in other species via the Pfam database. The expressions of the two CSPs-like were significantly higher in the MP group than in the NMP group. In addition, the expressions of CSPs were also much higher relative to other unigenes. Previous studies supported that CSPs were sensory-related proteins with pheromone binding function in insects (Bohbot et al., 1998; Jacquin-Joly et al., 2001). Unlike insect-specific proteins (OBPs), CSPs were found in crustaceans such as Daphnia carinata and were expressed in ovaries, thoracic limbs, rectum, and second antenna in both sexual and asexual females (Li et al., 2016). This suggested that the CSPs might respond to environmental signals and control the reproductive switch from sexual to asexual reproduction in D. carinata (Li et al., 2016). Generally, owing to the lack of OBPs, crustaceans only have 0–2 CSPs (Derby et al., 2016). Our results were consistent. Even though CSPs were broadly reported in insects, crustaceans, and myriapods (Zhou et al., 2006; Chipman et al., 2014; Pelosi et al., 2014), their functions in crustaceans remained unknown. Based on our results, we speculated that CSPs might be one of the dominant signaling receptors of pheromone binding in male P. clarkii. More attention to the expressions and changes in CSPs is needed in further studies. Moreover, the functions of lots of DEGs in the antenna of P. clarkii have not been annotated in the six databases, and have been classified as uncharacterized proteins, indicating that the information on pheromone-related receptors in P. clarkii is very scarce, which needs a lot of research to investigate. We suggested that there are still many candidate sex pheromone receptors in P. clarkii that have not been discovered and reported.
The ribosome is the molecular machine for biological protein synthesis (Lv et al., 2017). A previous study has reported that the inactivation of ribosomes can disrupt chemoreception in Caenorhabditis elegans (McConnell et al., 2015). Researchers suggested that the ribosomes affect the process of translation, and further affect the biological synthesis of chemosensation (McConnell et al., 2015). In this study, the ribosome was the most enriched pathway, of which most DEGs related to ribosome were upregulated. In Apis Mellifera ligustica, ribosomes were considered to play an important role in olfactory learning behavior, and the learning ability can be significantly reduced after being irritated by insecticide (Hou et al., 2021). Despite the interactive relationship between ribosomes and chemosensory processes in crustaceans remaining unknown, our results can still indicate that biological protein synthesis is more active after the male P. clarkii enters the mating period.
Ca2+ is the regulator of almost all eukaryotic signaling cascades, and Ca2+-entry is the first step to converting chemical signals into electrical ones (Pyrski et al., 2007). The previous study has shown that the concentration ratio of intracellular Ca2+ and cyclic nucleotide levels can mediate the ion channels synergistically (Stengl, 2010). After the sex pheromone binds to the receptor, the latter can activate the type III adenylyl cyclase via a GTP-binding protein, resulting in raising the cAMP level (Castillo et al., 2007). High concentrations of cAMP can open the cyclic nucleotide-gated channels, leading to the Na+ and Ca2+ flow into the cell, further leading to depolarization (Firestein and Zufall, 1994). The sodium carrier family (NCX and sodium–calcium exchanger) is one of the membrane transporters, which has been considered the main mechanism for Ca2+ efflux in crustaceans (Flik et al., 1994). The NCX has a fundamental role in controlling the intracellular concentrations of Na+ and Ca2+ (Canitano et al., 2002), which is the efficient Ca2+ extrusion system in the plasma membrane. In this study, we further found that the DEG related to NCX and sodium–calcium exchanger was significantly upregulated in MP groups and was involved in olfactory transduction and calcium signaling pathways. In the calcium signaling pathway, the upregulation of NCX could further affect the calcium concentrations of mitochondria, which plays a central role in cell signalings (Jacobson and Duchen, 2004). The previous study had shown that the mitochondria acted as the calcium buffer and reacted with the signals via upregulating the tricarboxylic acid cycle (Jacobson and Duchen, 2004). The activations of mitochondria might be more active for signal transductions. Therefore, we suggested that the calcium signaling pathway might be involved in sex pheromone signal transductions and the changes in antenna potential to sex pheromone signals were more sensitive when P. clarkii entered the mating period.
In this study, we found the sensitivity of male P. clarkii to conditioned water was different in the mating and non-mating period, and they were more sensitive in the mating period. Furthermore, a total of 12 upregulated and nine downregulated DEGs were associated with chemosensory-related functions, of which two related to CSPs were remarkably upregulated after P. clarkii entered MP. Based on the levels of CSP-related DEG expression, we suggested that CSP might be the key receptor for chemical signals or sex pheromone reception in the antenna of P. clarkii. In addition, the calcium-related DEG and signaling pathway might be one of the dominant factors in the differences in chemosensory sensitivity of male P. clarkii between the mating and non-mating periods. The results presented herein will be fundamental for future functional studies on chemosensory genes in P. clarkii. The findings are expected to clarify the chemical communication mechanisms in P. clarkii and provide new targets for invasion management in future.
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: National Center for Biotechnology Information (NCBI) BioProject, https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA839237.
ZZ, JH, and ZW designed the study. ZZ, HW, DL, and WZ performed the experiments and data analysis. ZZ drafted the manuscript. ZZ, HW, LM, JH, and ZW discussed the study design and data. JH and ZW revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (31960280), Science and Technology Base and Talent Special Project of Guangxi Province (AD20159042), Guangxi Natural Science Foundation of China (No. 2018GXNSFAA294024), Guangxi Universities Young and Middle-aged Teachers Scientific Research Basic Ability Improvement Project (2019KY0055), and Key Research Program of Guangxi Normal University (2017ZD009).
We are grateful to the lab members (Tingting Wei, Ronghui Zhao, and Xiufang Nong) for their help in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.976448/full#supplementary-material
Altner, I., Hatt, H., and Altner, H. (1983). Structural properties of bimodal chemo- and mechanosensitive setae on the pereiopod chelae of the crayfish, Austropotamobius torrentium. Cell Tissue Res. 228, 357–374. doi: 10.1007/BF00204885
Barnard, S., Matthews, L., Messori, S., Podaliri-Vulpiani, M., and Ferri, N. (2016). Laterality as an indicator of emotional stress in ewes and lambs during a separation test. Anim. Cogn. 19, 207–214. doi: 10.1007/s10071-015-0928-3
Blinova, N. K., and Cherkashin, S. A. (2012). The olfactory system of crustaceans as a model for ecologo-toxicological studies. J. Evol. Biochem. Physiol. 48, 155–165. doi: 10.1134/s0022093012020053
Bohbot, J., Sobrio, F., Lucas, P., and Nagnan-Le Meillour, P. (1998). Functional characterization of a new class of odorant-binding proteins in the moth Mamestra brassicae. Biochem. Biophys. Res. Commun. 253, 489–494. doi: 10.1006/bbrc.1998.9806
Breithaupt, T. (2011). “Chemical communication in crayfish,” in Chemical communication in crustaceans, eds T. Breithaupt and M. Thiel (New York, NY: Springer), 257–276.
Canitano, A., Papa, M., Boscia, F., Castaldo, P., Sellitti, S., Taglialatela, M., et al. (2002). Brain distribution of the Na+/Ca2+ exchanger-encoding genes NCX1, NCX2, and NCX3 and their related proteins in the central nervous system. Ann. N. Y. Acad. Sci. 976, 394–404. doi: 10.1111/j.1749-6632.2002.tb04766.x
Castillo, K., Delgado, R., and Bacigalupo, J. (2007). Plasma membrane Ca(2+)-ATPase in the cilia of olfactory receptor neurons: Possible role in Ca(2+) clearance. Eur. J. Neurosci. 26, 2524–2531. doi: 10.1111/j.1460-9568.2007.05863.x
Chipman, A. D., Ferrier, D. E. K., Brena, C., Qu, J., Hughes, D. S. T., Schroeder, R., et al. (2014). The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12:e1002005. doi: 10.1371/journal.pbio.1002005
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Derby, C. D., Kozma, M. T., Senatore, A., and Schmidt, M. (2016). Molecular mechanisms of reception and perireception in crustacean chemoreception: A comparative review. Chem. Sens. 41, 381–398. doi: 10.1093/chemse/bjw057
Dong, Y. F., Li, J. T., Zhang, X. X., Xian, J. A., and Wang, D. M. (2020). A review: Research advance on seeding breeding of red swamp crayfish Procambarus clarkii (in chinese). Chin. J. Fish. 33, 68–74.
Egly, R. M., Annis, G. M., Chadderton, W. L., Peters, J. A., and Larson, E. R. (2019). Predicting the potential distribution of the non-native red swamp crayfish Procambarus clarkii in the Laurentian Great Lakes. J. Great Lakes Res. 45, 150–159. doi: 10.1016/j.jglr.2018.11.007
Eyun, S. I., Soh, H. Y., Posavi, M., Munro, J. B., Hughes, D. S. T., Murali, S. C., et al. (2017). Evolutionary history of chemosensory-related gene families across the Arthropoda. Mol. Biol. Evol. 34, 1838–1862. doi: 10.1093/molbev/msx147
Firestein, S., and Zufall, F. (1994). The cyclic nucleotide gated channel of olfactory receptor neurons. Semin. Cell Biol. 5, 39–46. doi: 10.1006/scel.1994.1006
Flik, G., Verbost, P. M., Atsma, W., and Lucu, C. (1994). Calcium transport in gill plasma membranes of the crab Carcinus maenas: Evidence for carriers driven by ATP and a Na+ gradient. J. Exp. Biol. 195, 109–122. doi: 10.1242/jeb.195.1.109
Foquet, B., Castellanos, A. A., and Song, H. (2021). Comparative analysis of phenotypic plasticity sheds light on the evolution and molecular underpinnings of locust phase polyphenism. Sci. Rep. 11:11925. doi: 10.1038/s41598-021-91317-w
Fowler, M. A., and Montell, C. (2013). Drosophila TRP channels and animal behavior. Life Sci. 92, 394–403. doi: 10.1016/j.lfs.2012.07.029
Garm, A., Derby, C. D., and Hoeg, J. T. (2004). Mechanosensory neurons with bend- and osmo-sensitivity in mouthpart setae from the spiny lobster Panulirus argus. Biol. Bull. 207, 195–208. doi: 10.2307/1543208
Garm, A., Hallberg, E., and Hoeg, J. T. (2003). Role of maxilla 2 and its setae during feeding in the shrimp Palaemon adspersus (Crustacea: Decapoda). Biol. Bull. 204, 126–137. doi: 10.2307/1543548
Gherardi, F., Aquiloni, L., Dieguez-Uribeondo, J., and Tricarico, E. (2011). Managing invasive crayfish: Is there a hope? Aquat. Sci. 73, 185–200. doi: 10.1007/s00027-011-0181-z
Gonzalez, F., Johny, J., Walker, W. B. III, Guan, Q., Mfarrej, S., Jakse, J., et al. (2021). Antennal transcriptome sequencing and identification of candidate chemoreceptor proteins from an invasive pest, the American palm weevil, Rhynchophorus palmarum. Sci. Rep. 11:8334. doi: 10.1038/s41598-021-87348-y
Harzsch, S., and Krieger, J. (2018). Crustacean olfactory systems: A comparative review and a crustacean perspective on olfaction in insects. Progr. Neurobiol. 161, 23–60. doi: 10.1016/j.pneurobio.2017.11.005
Harzsch, S., Rieger, V., Krieger, J., Seefluth, F., Strausfeld, N. J., and Hansson, B. S. (2011). Transition from marine to terrestrial ecologies: Changes in olfactory and tritocerebral neuropils in land-living isopods. Arthropod Struct. Dev. 40, 244–257. doi: 10.1016/j.asd.2011.03.002
Hou, M.-S., Qiu, Y.-M., Zhao, B.-A., Yu, T.-T., Liang, L.-Q., Su, S.-K., et al. (2021). Effects of a sublethal dose of imidacloprid on the olfactory learning behavior of Apis mellifera ligustica workers and an analysis of their brain transcriptomes. Acta Entomol. Sin. 64, 817–827. doi: 10.16380/j.kcxb.2021.07.006
Jacobson, J., and Duchen, M. (2004). Interplay between mitochondria and cellular calcium signalling. Mol. Cell. Biochem. 256-257, 209–218. doi: 10.1023/B:MCBI.0000009869.29827.df
Jacquin-Joly, E., Vogt, R. G., Francois, M. C., and Nagnan-Le Meillour, P. (2001). Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. Sens. 26, 833–844. doi: 10.1093/chemse/26.7.833
Jin, S., Jacquin, L., Huang, F., Xiong, M., Li, R., Lek, S., et al. (2019). Optimizing reproductive performance and embryonic development of red swamp crayfish Procambarus clarkii by manipulating water temperature. Aquaculture 510, 32–42. doi: 10.1016/j.aquaculture.2019.04.066
Johnson, M. F., Rice, S. P., and Reid, I. (2014). The activity of signal crayfish (Pacifastacus leniusculus) in relation to thermal and hydraulic dynamics of an alluvial stream, UK. Hydrobiologia 724, 41–54. doi: 10.1007/s10750-013-1708-1
Johnson, N. S., Tix, J. A., Hlina, B. L., Wagner, C. M., Siefkes, M. J., Wang, H., et al. (2015). A sea lamprey (Petromyzon marinus) sex pheromone mixture increases trap catch relative to a single synthesized component in specific environments. J. Chem. Ecol. 41, 311–321. doi: 10.1007/s10886-015-0561-2
Keller-Costa, T., Hubbard, P. C., Paetz, C., Nakamura, Y., da Silva, J. P., Rato, A., et al. (2014). Identity of a tilapia pheromone released by dominant males that primes females for reproduction. Curr. Biol. 24, 2130–2135. doi: 10.1016/j.cub.2014.07.049
Klopfenstein, D. V., Zhang, L., Pedersen, B. S., Ramirez, F., Vesztrocy, A. W., Naldi, A., et al. (2018). GOATOOLS: A python library for gene ontology analyses. Sci. Rep. 8:10872. doi: 10.1038/s41598-018-28948-z
Kozma, M. T., Hanh, N.-V., Wong, Y. Y., Shukla, N. S., Pawar, S. D., Senatore, A., et al. (2020a). Comparison of transcriptomes from two chemosensory organs in four decapod crustaceans reveals hundreds of candidate chemoreceptor proteins. PLoS One 15:e0230266. doi: 10.1371/journal.pone.0230266
Kozma, M. T., Ngo-Vu, H., Rump, M. T., Bobkov, Y. V., Ache, B. W., and Derby, C. D. (2020b). Single cell transcriptomes reveal expression patterns of chemoreceptor genes in olfactory sensory neurons of the Caribbean spiny lobster, Panulirus argus. BMC Genom. 21:649. doi: 10.1186/s12864-020-07034-7
Kubec, J., Kouba, A., and Buric, M. (2019). Communication, behaviour, and decision making in crayfish: A review. Zool. Anzeig. 278, 28–37. doi: 10.1016/j.jcz.2018.10.009
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Larson, E. R., and Magoulick, D. D. (2011). Life-history notes on Cambarus hubbsi creaser (hubbs crayfish) from the South Fork Spring River, Arkansas. Southeast. Nat. 10, 121–132. doi: 10.1656/058.010.0110
Li, B., and Dewey, C. N. (2011). RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, H., Yang, Y., Xu, G., Wu, D., Lv, W., Jiang, Q., et al. (2016). Cloning, expression and localization of DacaCSP2 and DacaCSP3 during different reproductive stages in Daphnia carinata. Gene 582, 59–68. doi: 10.1016/j.gene.2016.01.048
Li, Y., Deng, W., Yang, K., and Wang, W. (2012). The expression of prophenoloxidase mRNA in red swamp crayfish, Procambarus clarkii, when it was challenged. Genomics 99, 355–360. doi: 10.1016/j.ygeno.2012.04.001
Liu, Q.-N., Tang, Y.-Y., Li, Y.-T., Zha, X.-H., Yang, T.-T., Zhang, D.-Z., et al. (2021). Differentially expressed genes in hemocytes of red swamp crayfish Procambarus clarkii following lipopolysaccharide challenge. Aquaculture 533:735943. doi: 10.1016/j.aquaculture.2020.735943
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, J., Zhang, L., Liu, P., and Li, J. (2017). Transcriptomic variation of eyestalk reveals the genes and biological processes associated with molting in Portunus trituberculatus. PLoS One 12:e0175315. doi: 10.1371/journal.pone.0175315
Ma, X. H., Ren, H. J., Peng, R. Y., Li, Y., and Ming, L. (2020). Comparative expression profiles of host circulating miRNAs in response to Trichinella spiralis infection. Vet. Res. 51:39. doi: 10.1186/s13567-020-00758-0
Marx, H. E., Jorgensen, S. A., Wisely, E., Li, Z., Dlugosch, K. M., and Barker, M. S. (2021). Pilot RNA-seq data from 24 species of vascular plants at Harvard Forest. Appl. Plant Sci. 9:e11409. doi: 10.1002/aps3.11409
McConnell, M.-T., Lisgarten, D. R., Byrne, L. J., Harvey, S. C., and Bertolo, E. (2015). Winter aconite (Eranthis hyemalis) lectin as a cytotoxic effector in the lifecycle of Caenorhabditis elegans. PeerJ 3:e1206. doi: 10.7717/peerj.1206
Monteclaro, H. M., Anraku, K., and Matsuoka, T. (2010). Response properties of crayfish antennules to hydrodynamic stimuli: Functional differences in the lateral and medial flagella. J. Exp. Biol. 213, 3683–3691. doi: 10.1242/jeb.046011
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Oyama, T., Momohara, Y., Yano, H., Kamio, M., Fujiyama, N., and Nagayama, T. (2020). Sex recognition and agonistic strategies of male and female crayfish. Behaviour 157, 575–596. doi: 10.1163/1568539X-bja10014
Peddio, S., Sollai, G., Podda, C., Frau, G., Palmas, F., Sabatini, A., et al. (2019). The success in the short-distance communication for mating does not depend on chemical signals in the crustacean decapod Procambarus clarkii (Girard, 1852). Adv. Oceanogr. Limnol. 10, 67–73. doi: 10.4081/aiol.2019.8617
Pelosi, P., Iovinella, I., Felicioli, A., and Dani, F. R. (2014). Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 5:320. doi: 10.3389/fphys.2014.00320
Pyrski, M., Koo, J. H., Polumuri, S. K., Ruknudin, A. M., Margolis, J. W., Schulze, D. H., et al. (2007). Sodium/calcium exchanger expression in the mouse and rat olfactory systems. J. Comp. Neurol. 501, 944–958. doi: 10.1002/cne.21290
Ren, Y., Yu, G., Shi, C., Liu, L., Guo, Q., Han, C., et al. (2022). Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 1:e12. doi: 10.1002/imt2.12
Schmidt, M., and Gnatzy, W. (1984). Are the funnel-canal organs the ‘campaniform sensilla’ of the shore crab, Carcinus maenas (Decapoda, Crustacea)? II. Ultrastructure. Cell Tissue Res. 237, 81–93. doi: 10.1007/BF00229202
Schmidt, M., and Mellon, D. Jr. (2011). “Neuronal processing of chemical information in crustaceans,” in Chemical communication in crustaceans, eds T. Breithaupt and M. Thiel (New York, NY: Springer), 123–147.
Shen, G., Zhang, X., Gong, J., Wang, Y., Huang, P., Shui, Y., et al. (2020). Transcriptomic analysis of Procambarus clarkii affected by “Black May” disease. Sci. Rep. 10:21225. doi: 10.1038/s41598-020-78191-8
Shen, H., Hu, Y., Ma, Y., Zhou, X., Xu, Z., Shui, Y., et al. (2014). In-depth transcriptome analysis of the red swamp crayfish Procambarus clarkii. PLos One 9:e110548. doi: 10.1371/journal.pone.0110548
Solari, P., Sollai, G., Masala, C., Loy, F., Palmas, F., Sabatini, A., et al. (2017). Antennular morphology and contribution of aesthetascs in the detection of food-related compounds in the shrimp Palaemon adspersus Rathke, 1837 (Decapoda: Palaemonidae). Biol. Bull. 232, 110–122. doi: 10.1086/692696
Stengl, M. (2010). Pheromone transduction in moths. Front. Cell. Neurosci. 4:133. doi: 10.3389/fncel.2010.00133
Urbschat, N., and Scholtz, G. (2019). Comparative analysis of the antennae of three amphipod species with different lifestyles. Arthropod Struct. Dev. 53:100886. doi: 10.1016/j.asd.2019.100886
Van den Eynde, C., Vriens, J., and De Clercq, K. (2021). Transient receptor potential channel regulation by growth factors. Biochim. Biophys. Acta Mol. Cell Res. 1868:118950. doi: 10.1016/j.bbamcr.2021.118950
Varet, H., Brillet-Gueguen, L., Coppee, J.-Y., and Dillies, M.-A. (2016). SARTools: A DESeq2-and EdgeR-Based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS One 11:e0157022. doi: 10.1371/journal.pone.0157022
Venkatachalam, K., and Montell, C. (2007). TRP channels. Annu. Rev. Biochem. 76, 387–417. doi: 10.1146/annurev.biochem.75.103004.142819
Venkatachalam, K., Luo, J., and Montell, C. (2014). “Evolutionarily conserved, multitasking TRP channels: Lessons from worms and flies,” in Mammalian transient receptor potential, eds B. Nilius and V. Flockerzi (Cham: Springer), 937–962. doi: 10.1007/978-3-319-05161-1_9
Waldrop, L. D. (2013). Ontogenetic scaling of the olfactory antennae and flicking behavior of the shore crab, Hemigrapsus oregonensis. Chem. Sens. 38, 541–550. doi: 10.1093/chemse/bjt024
Wang, J., Li, D.-Z., Min, S.-F., Mi, F., Zhou, S.-S., and Wang, M.-Q. (2014). Analysis of chemosensory gene families in the beetle Monochamus alternatus and its parasitoid Dastarcus helophoroides. Comp. Biochem. Physiol. Part D Genomics Proteomics 11, 1–8. doi: 10.1016/j.cbd.2014.05.001
Wittwer, C., Stoll, S., Strand, D., Vradalstad, T., Nowak, C., and Thines, M. (2018). eDNA-based crayfish plague monitoring is superior to conventional trap-based assessments in year-round detection probability. Hydrobiologia 807, 87–97. doi: 10.1007/s10750-017-3408-8
Yu, D., Zhai, Y., He, P., and Jia, R. (2022). Comprehensive transcriptomic and metabolomic analysis of the Litopenaeus vannamei hepatopancreas after WSSV challenge. Front. Immunol. 13:826794. doi: 10.3389/fimmu.2022.826794
Zhou, J.-J., Kan, Y., Antoniw, J., Pickett, J. A., and Field, L. M. (2006). Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chem. Sens. 31, 453–465. doi: 10.1093/chemse/bjj050
Keywords: Procambarus clarkii, antenna, transcriptome, RNA-Seq, chemosensory, chemical communication
Citation: Zhou Z, Mo L, Li D, Zeng W, Wu H, Wu Z and Huang J (2022) Comparative transcriptomics analyses of chemosensory genes of antenna in male red swamp crayfish Procambarus clarkii. Front. Ecol. Evol. 10:976448. doi: 10.3389/fevo.2022.976448
Received: 23 June 2022; Accepted: 22 August 2022;
Published: 23 September 2022.
Edited by:
Naicheng Wu, Ningbo University, ChinaReviewed by:
Scott Francis Cummins, University of the Sunshine Coast, AustraliaCopyright © 2022 Zhou, Mo, Li, Zeng, Wu, Wu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinlong Huang, amxfaHVhbmdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.