- 1Laboratoire d'Ethologie Expérimentale et Comparée UR 4443, Université Sorbonne Paris Nord, Villetaneuse, France
- 2Max Planck Institute for Chemical Ecology, Jena, Germany
- 3Evolution, Genomes, Behaviour and Ecology Laboratory, Université Paris-Saclay, CNRS, IRD, Gif-sur-Yvette, France
- 4Institut Universitaire de France, Paris, France
Agents that cause disease alter the cell metabolism of their hosts. Cells with an altered metabolism produce particular profiles of biomolecules, which are different from those of healthy cells. Such differences may be detected by olfaction. Historically, physicians used olfactory cues to diagnose sickness by smelling the breath or the urine of patients. However, other species have been shown to possess excellent olfactory abilities. Dogs, for instance, have been frequently used as biodetectors of human diseases, including cancer, viral and bacterial infections. Other mammalian species, such as rats, have been trained to perform similar tasks, but their disease detection abilities remain poorly explored. Here, we focus on the overlooked potential of invertebrate species and we review the current literature on olfactory detection of diseases by these animals. We discuss the possible advantages of exploring further the abilities of invertebrates as detection tools for human disease.
1. Introduction
Early detection of diseases is very often critical. Typically, the earlier a condition is diagnosed, the better are the chances of recovery. Diseases are characterised by an altered cell metabolism that can cause malfunction or loss of function in an organ with repercussions on the whole organism. An altered metabolism will produce specific patterns of volatile organic compounds (VOCs), which are different from those produced by a healthy organism and which can then be used as biomarkers for detecting the condition. Some pathogens, such as bacteria, also produce specific VOC patterns (Ratiu et al., 2019). In the laboratory, detection and identification of these VOCs can be performed using high-end chemical analysis. The most common technique includes the use of Solid-Phase Micro-Extraction (SPME) and Gas-Chromatography coupled with Mass Spectrometry (GC–MS) (Filipiak and Bojko, 2019). SPME fibres are composed of polymers that have strong affinity for specific compounds, allowing them to be adsorbed (binding process) by the fibre. The fibre is typically exposed to the empty space above the studied sample placed in a closed container (head space), allowing VOCs to bind to the fibre. The VOCs are later desorbed in a GC–MS. In the GC part, compounds will migrate through a column and will be separated according to their volatility or affinity with the stationary phase within the column. The identification of the compounds is made thanks to the MS, where the compounds are fragmented by ionisation, producing a unique pattern (mass spectrum) for each chemical compound. These analytical methods are very effective, but expensive and require well trained specialists.
Specific VOC patterns can also be perceived by natural detectors through olfaction. Since antiquity, the human nose has been used as a diagnostic tool in the practise of medicine to detect diseases such as diabetes (via the fruity smell of ketones in the breath) or diphtheria (putrid odour) (Bijland et al., 2013). Some individuals, so called “super smellers,” can detect volatiles emitted by human patients with Parkinson disease (Trivedi et al., 2019). Other species that also have excellent olfactory abilities may be trained to detect and identify suspect samples using standard olfactory conditioning protocols. Through training, some species can thus learn to indicate to the experimenter which samples contain the VOCs of interest by way of behavioural changes, whereas for other species the presence of a healthy or sick sample will be revealed by observing physiological modifications in the tested animal.
The literature on the use of non-human animals as biodetectors has been extensively reviewed, particularly for vertebrates (Oh et al., 2014; Pirrone and Albertini, 2017; Jendrny et al., 2021; Juge et al., 2022). Here, we focus on the overlooked potential of invertebrates as olfactory biodetectors. We therefore do not review the literature on vertebrates but we only briefly mention dogs and some other mammals, as case studies for comparison with invertebrates.
2. Use of animal olfactory detection abilities
2.1. Dogs
Currently, the most broadly used species for VOC detection is the dog (Canis lupus familiaris) (Guest and Otto, 2020). Dogs have been domesticated for millennia and were first trained for hunting and protection. Now, canine olfactory detection abilities are used to detect drugs (Francis et al., 2019), explosives (Furton and Myers, 2001), or disease in livestock (Welch, 1990). These abilities rely on dogs’ sharp sense of smell and on the fact that they can be conditioned to respond to specific samples via associative learning. During the conditioning, a stimulus (e.g., a typical drug odour) is associated with a reward (e.g., a treat or a toy). The individual associates the stimulus and the reward and will then look for the learned odour if it is motivated for the reward. Dogs are efficient biodetectors, and a success rate above 90% (percentage of correct responses) is often required before allowing the dog to operate in the field (Welch, 1990; Furton and Myers, 2001; Francis et al., 2019). The downside of this high threshold is that several months to years of constant individual training are needed to reach it, which can incur high costs for training and maintenance of these dogs. In addition, during this long training time, dogs form a strong attachment to their handler and cannot be optimally operated by others. Not only the dogs need to be trained, but to properly train dogs, handlers also require intensive training, involving additional time and costs.
2.2. Other mammals
Dogs are not the sole species used for olfactory detection tasks. Elephants (Loxodonta africana) can also detect the volatiles emitted by explosives (Miller et al., 2015). They can be quickly trained in controlled conditions (only 24 conditioning trials), and become efficient with a success rate around 90%. Indeed, these pachyderms have a strong sense of smell, with a high number of olfactory receptors (around 2000 – for comparison, dogs have around 800 receptors and mice around 1,200, Miller et al., 2015). However, as the authors of this study stated, they do not advocate using elephants directly in the field (due to the explosive risk linked to the size and weight of elephants), but these animals may be useful for screening samples brought to them. To solve this mine triggering problem, smaller species, like the African giant pouched rat, Cricetomys gambianus, have been considered (Edwards et al., 2015). Rats have the advantage of being too small to trigger mines, and as they do not form an emotional link with humans, different handlers can optimally use them. Rats have been shown to clear a minefield faster than humans, and with a 100% detection rate: no explosives could be found afterwards by humans using metal detectors. Rats are also cheap to train and to maintain. A non-governmental agency, APOPO, uses these trained rats to clear and rehabilitate fields in Mozambique (Edwards et al., 2015).
2.3. Invertebrates
In search of species that are smaller, cheaper and easier to train, researchers have tested the ability of invertebrates, and particularly of insects to detect explosives. Indeed, many insects can fly, are easy and cheap to maintain, and hundreds of individuals can be rapidly trained to detect a specific set of volatile compounds (Giurfa and Sandoz, 2012).
Honey bees’ learning abilities are well known. Bees can be trained in the controlled environment of a laboratory using the appetitive conditioning of the proboscis extension response (PER, Takeda, 1961; Bitterman et al., 1983). When the antennae of a hungry bee are touched with sucrose solution, the animal reflexively extends its proboscis to suck the sucrose. Neutral odours do not trigger this response in naive animals. However, if an odour is presented together with the sucrose solution, an association is formed and, after conditioning, the presentation of the odour alone triggers the PER. Using TNT vapour (a molecule present in ~90% of explosives) as stimulus, bees were able to react to concentrations as low as 11 ppt (Taylor-mccabe et al., 2008). Although the parameters of the conditioning protocol can be perfectly controlled within the laboratory, bringing the suspicious samples (in the case of explosives) to a building may be risky.
For this reason, bees were brought directly to the field to be trained to explosive VOCs. Sugar feeders were placed outside of a hive, with the VOCs of interest next to it, allowing bees to feed and associate odour and reward, as well as to bring this information to the hive and recruit naive nestmates within the hive (Rodacy et al., 2002). Once on a minefield, bees will fly and land near underground bombs (Bromenshenk et al., 2003). Since they are equipped with a tag, researchers are able to visualise hotspots of bees gathering around explosives on a map. Bees cannot trigger bombs, and can search for explosives in an area several kilometres wide with ease (although following the bees on this wide surface is not trivial). The disadvantages of such approaches are that bees will not explore the area at night and during bad weather. It is also unclear whether bees will screen the whole field or will focus on the strongest VOC sources and ignore others (Bromenshenk et al., 2003).
Combining the advantages of both techniques described above, the tobacco hornworm moth (Manduca sexta) was shown to be able to detect explosives VOCs in the field, after an initial appetitive conditioning with controlled parameters (e.g., number of training trials, stimulation time, inter-trial interval, King et al., 2004). In this study, the authors focused on moths’ appetitive response (similar to PER above) and used electrophysiology to measure muscular activity when the animals extend their proboscis. Trained moths extended their proboscis to the explosive odour, while naive moths did not. The difference between the behaviours of the two groups indicated the presence of explosives VOCs. Moths can be placed in a portable device, allowing researchers to use these insects directly in the field, which ensures that the whole field is scanned.
These different studies show that, similarly to mammals, insects can be trained individually (King et al., 2004; Taylor-mccabe et al., 2008) or in groups (Rodacy et al., 2002), and are able to detect VOCs both in the laboratory (Taylor-mccabe et al., 2008) and directly in the field (Bromenshenk et al., 2003; King et al., 2004). This adaptivity makes insects (and invertebrates in general), promising tools for the detection of VOCs of interest.
3. Olfaction and disease
3.1. Mammals
Animals’ sense of smell has not only been used for the detection of explosives or drugs. Recently, the possible detection of human diseases by dogs was investigated and this species was proven to be able to detect bacteria outbreaks in a hospital (Bomers et al., 2014), patients with epileptic seizure (Catala et al., 2019), or more recently patients infected by COVID-19 (Jendrny et al., 2020). However, one of the most studied diseases in the olfactory detection field is cancer, as it is the second most common cause of death in humans, with nearly 10 million people dying from the disease each year (WHO, 2020). Studies using dogs as cancer detectors have produced very diverse results. Since the first medical study reporting the ability of dogs to detect cancer (Williams and Pembroke, 1989), the first use of urine as sample (Willis et al., 2004), or the use of exhaled breath (McCulloch et al., 2006), dozens of studies have been performed, and some reported an excellent detection rate above 90% (Yoel et al., 2015; Junqueira et al., 2019; Thuleau et al., 2019), while others revealed a detection rate below 33% (Schallschmidt et al., 2015; Murarka et al., 2019, reviewed in Pirrone and Albertini, 2017). Such high variability can be explained by the different types and qualities of the samples used (blood, urine, cells, breath), the personality of the dogs, the abilities of the handlers and the training protocols. This last point is probably the most important, but most studies are elusive about the exact number of conditioning trials applied to the dogs, the inter-trial interval, the presentation order of the samples, and other critical details needed for proper replication.
Mice were also tested to detect the cancer state of conspecifics (Matsumura et al., 2010; Kokocinska-Kusiak et al., 2020; Gouzerh et al., 2022b) and humans (Sato et al., 2017). For instance, in choice tests in a Y-maze, 14 mice showed impressive abilities, by detecting the cancer state (pre or post operation) of patients with bladder cancer 100% of the time. Rodents can also detect tuberculosis, with a high efficiency (sensitivity from 70.8% to 86.6% and a specificity of 36.4% to 89.1% depending on the studies) (Fiebig et al., 2020).
3.2. The nematode Caenorhabditis elegans
The nematode Caernorhabditis elegans is about 1 mm long, has a nervous system including only about 300 neurons, but shows impressive olfactory abilities. Nematodes were tested for their ability to detect the odour of Mycobacterium tuberculosis, the bacteria responsible for tuberculosis (Neto et al., 2016). Previously identified biomarkers (Syhre and Chambers, 2008) found in the olfactory signature of the bacteria were used as stimuli (methyl nicotinate, methyl p-anisate, methyl phenylacetate, and o-phenylanisole). Nematodes were not conditioned to associate any odour with a reward, as they naturally express attraction or repulsion to different chemical compounds. Groups of nematodes were placed in the centre of a circular arena which was divided in four quadrants with an odorant in each quadrant to test for their olfactory preference. A control odour (known for being neither attractive nor repulsive) was placed in two opposite quadrants, whereas the tested compound was put in the two vacant quadrants. After 1 h, the number of nematodes present in each quadrant was counted, and a preference index was calculated to measure the attraction or repulsion induced by the tested odorant. For some of the tested compounds, nematodes were reacting to a concentration as low as 0.1 ng/μL and showed a linear reaction (the more concentrated, the more attractive/repulsive). Interestingly, for one of these compounds (methyl phenylacetate), nematodes showed repulsion at high concentration, and attraction when the concentration was low. Although compounds used here are single odorants and not a complex olfactory signature, the authors discussed the possibility of using automatic tracking to rapidly obtain a diagnosis of tuberculosis (Neto et al., 2016). Another study used the same circular arena apparatus to detect the presence of sepsis in urine of patients, and C. elegans nematodes were shown to be spontaneously attracted to the odour of sick patients (Tee et al., 2019).
This attraction/repulsion behaviour in nematodes was also used for testing cancer detection. Samples from different origins were used: from human cell lines and tissues (Hirotsu et al., 2015) to body fluids such as urine from humans (Hirotsu et al., 2015; Kusumoto et al., 2020; Asai et al., 2021; Lanza et al., 2021; Thompson et al., 2021), but also from mice (Ueda et al., 2019). These different studies did not focus on a single cancer type, but rather explored the abilities of the nematodes to detect several types of cancer, such as breast (e.g., Lanza et al., 2021), pancreatic (e.g., Asai et al., 2021), or prostate cancer (e.g., Thompson et al., 2021). Here again, researchers found that nematodes were attracted to sick samples, and could thus be used to diagnose cancer with a high efficiency. Over the 25 tests using urine from cancer patients (Hirotsu et al., 2015), nematodes were attracted to it 95% of the time (only one test was not successful). In this seminal study, the nematodes also showed repulsion to 95% of control samples (207/218), leading to an accuracy (calculated as the proportion of true positive and true negative results over the total number of tests) of 95% (in this case; ). Similar remarkable accuracy rates were also reported in recent studies testing more samples: 86% for breast cancer (Lanza et al., 2021), 73% for pancreatic cancer (Kobayashi et al., 2021) or 81% for prostate cancer (Thompson et al., 2021).
Since no conditioning is involved in the nematodes’ choices, it is unclear why they show attraction/repulsion to particular compounds. Also, as these studies showed, different dilutions of the same samples could elicit opposite behaviours (attraction or repulsion) in the same species, which should be carefully documented. Nevertheless, the quest for disease biomarkers could help find the best animal species with higher natural abilities to detect compounds of interest. The production of mutants eliciting a behaviour only in the presence of specific biomarkers is also a possibility. The VOCs related to diseases with a single origin (e.g., tuberculosis originating from a bacteria) were already described and tested (Neto et al., 2016), however, for more complex diseases with multiple possible origins (e.g., cancer), standardised studies, using the same experimental protocols, are needed (Gouzerh et al., 2022a) to identify the specific VOCs that could be used during choice tests. Nevertheless, the use of animals naturally expressing specific behaviours toward healthy/sick samples should be further explored, as this could lead to rapid detection assays, avoiding a disadvantage of using conditioned individuals: the conditioning time (Figure 1). It is also important to note that this method is available for the public in Japan for a cost of ~100$ (di Luccio et al., 2022).
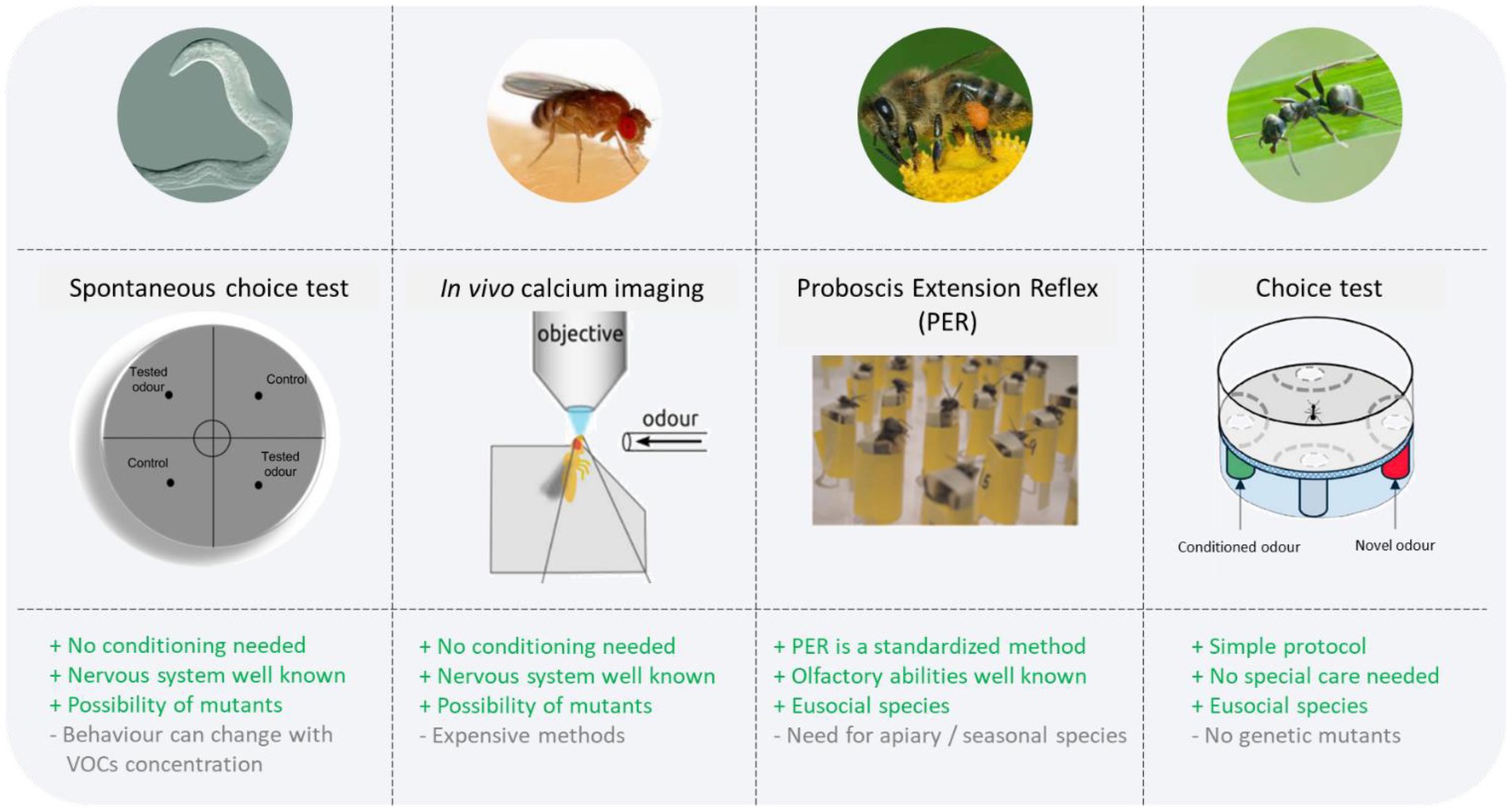
Figure 1. Advantages and constraints of the invertebrate species for disease detection. The four species already tested – the nematode Caenorhabditis elegans, the fly Drosophila melanogaster, the bee Apis mellifera, and the ant Formica fusca – are presented together with the method used, as well as with the advantages and current constraints of these species for disease detection. Pictures for the methods are adapted from Giurfa and Sandoz (2012), Strauch et al. (2014), Thompson et al. (2021), Piqueret et al. (2022a).
3.3. The fruit fly: Drosophila melanogaster
Amongst insects, the fruit fly Drosophila melanogaster is one of the most studied species, and its ability to detect cancer samples has also been investigated (Strauch et al., 2014). Similar to the studies using nematodes, individuals were not trained to specific cancer VOCs. Instead, researchers used in vivo calcium imaging to record neural activity patterns (from olfactory receptor neurons) on the fly antenna. They analysed the antenna responses to the VOCs emanating from cancer cell line samples and cancer-free cell lines. The authors found that the VOCs of different cell lines produce different and specific neuronal activation patterns. Using multidimensional analysis, the samples could later be classified as cancerous or healthy based on the evoked activation patterns (Strauch et al., 2014).
The technique used in this study is an expensive one that requires highly trained personnel to be operated (Figure 1), and as the authors stated, this study was a proof of concept. However, they propose to integrate odorant receptors to artificial systems (i.e., electronic noses) to improve the sensitivity of the existing systems. Fruit flies are also studied for their learning abilities and many neurogenetic tools are available in this insect, so that learning assays involving cancer odours are possible, as would be the use of specific mutants detecting particular VOCs.
3.4. The honey bee Apis mellifera
Honey bees (Apis mellifera) are a prominent insect model for the study of olfactory perception learning and memory (Menzel and Giurfa, 2001; Sandoz, 2011; Paoli and Galizia, 2021). Compared to the plethora of olfactory learning studies in this insect, very few studies were dedicated to disease detection. Using PER conditioning, the researchers tested whether lung cancer cell line odours could be associated with a reward (Schallschmidt et al., 2015). Bees underwent 10 training trials followed by two memory tests. At the end of the training, bees did not discriminate cancer cells from the control (culture medium alone). However, in this study the number of trained bees was low (n = 14) compared to the majority of associative learning studies in this model, which typically test at least 30 individuals for each condition (Giurfa and Sandoz, 2012). Moreover, in the same study (Schallschmidt et al., 2015), the researchers trained dogs using the same cancer samples, and the dogs were also unable to differentiate these cancer samples from the control. This result is intriguing, since as we reported above dogs are known to detect cancer biomarkers (review in Pirrone and Albertini, 2017). Therefore, the poor performance shown by bees in this study cannot be taken as an inability of this species to be used as a biodetector for cancer. It could be explained by possible problems with sample origin or sample preparation. Further studies on bee cancer detection abilities thus need to be conducted.
Bees were successfully trained to detect the odour of tuberculosis (Suckling and Sagar, 2011) (see also the detection abilities of nematodes; Neto et al., 2016). Three chemical biomarkers emitted by this pathogen were previously identified, and used as odour stimuli. During PER conditioning, different groups of bees were trained to a single concentration of one of these odorants, and were then tested using different concentrations of the trained odorant with a dilution factor going from 10−6 to 101. For two of the compounds tested, bees started responding when the concentration was above 10 pg., which is equivalent to a magnitude of 10−4 (Suckling and Sagar, 2011). This fits with honey bees’ well known olfactory detection abilities (Sandoz, 2011; Paoli and Galizia, 2021). In this study, bees were trained using single biomarker compounds. Future studies should use samples of biological origin to assess bees’ detection abilities in a more realistic situation, e.g., by using human samples.
Recently, honeybees were trained to detect the odour of the virus SARS-Cov-2 in infected mink (Neovison vison) samples (Kontos et al., 2022). Throat swabs were collected from healthy and infected individuals, and placed into culture medium (DMEM). Filter paper strips were then soaked with these solutions and used as odour stimuli. Using the PER method, bees were trained to associate the infected sample with a reward (sugar solution), and the healthy sample with a punishment (quinine solution). After 18 conditioning trials (nine rewarded, nine punished), bees were able to discriminate the infected samples from healthy ones. Researchers estimate that 10 bees would be enough to obtain a success score above 90%, which is equivalent to the available antigen tests.
One of the disadvantages of using honey bees is the need for an apiary and experienced beekeepers to successfully rear them. On the other hand, the advantage of using bees is that their olfactory learning abilities are well documented, and comparisons between studies are facilitated due to the use of a well described and broadly used protocol, PER conditioning (Bitterman et al., 1983; Giurfa and Sandoz, 2012; Figure 1). Multiplication of researches using bees with this standardised protocol should facilitate cross-validation and replication of results from different teams, thus adding more weight to the published results.
3.5. The ant Formica fusca
Like honeybees, ants are social insects that live in groups and rely mostly on olfaction for communication. In recent years, ants have been increasingly used in olfactory learning studies (Guerrieri and d’Ettorre, 2010; Perez et al., 2015; Piqueret et al., 2019; Czaczkes and Kumar, 2020; Wagner et al., 2022). Using a conditioning protocol, the detection abilities of Formica fusca ants were tested using cancer cell lines as the odour source (Piqueret et al., 2022a). Human cancer cells were cultivated in cultured medium (DMEM) and, after 3 days, the supernatant was used as olfactory stimulus. Individual ants were trained to associate this odour with a sugar solution in a circular arena. After only three conditioning trials, the preference of the ants was tested between the learnt odour and a novel odour, in the absence of any reward. Ants were challenged with various discrimination tasks: first, they had to differentiate between a cell line and the culture medium alone; second, they had to discriminate a cancer cell line from a healthy cell line; and finally, ants had to choose between a first cancer cell line and a second cancer cell line originating from the same tissue (human breast). In all cases, ants were able to discriminate between the conditioned samples and the novel samples. The VOCs profile produced by a single cell line is composed of dozens of single compounds (Piqueret et al., 2022a), but it is less complex than the odour of a whole organism composed of a myriad of different cell types. Even when using complex body odours, Formica ants demonstrated their abilities to discriminate cancer-free individuals from sick ones (Piqueret et al., 2022b). Human tumours were grafted on mice (Patient derived xenograft – PDX; Dobrolecki et al., 2016), and their urine served as odour stimuli. Half of the ants had to associate the odour of tumour-bearing mice with a sugar solution, while the other half learned that the odour of tumour-free mice was associated with the reward. After three conditioning trials using urine from three different mice, the preference of ants was tested using urine from novel mice (to control if ants learned the individual odours or the tumour odour itself). Ants showed a clear preference for the odour they were conditioned to, indicating that they could differentiate between urine odours from tumour-bearing and tumour-free mice. Together, these studies open the possibility of using ants in a non-invasive cancer detection protocol.
The use of ants has a number of advantages. The protocol used for the detection of cancer-related odours is relatively simple (Piqueret et al., 2019) and the conditioning and preference tests can be performed within 1 h. The species used (F. fusca) form long-lasting memories, and can be tested up to nine times for its olfactory preferences without requiring novel training. Recently, the first CRISPR/Cas-9 olfactory mutant lineages were established (Friedman et al., 2017; Trible et al., 2017) using another ant species (Ooceranea biroi) that have the particularity of being clonal. CRISPR/Cas-9 mutants were also developed in Harpegnatos saltator (Yan et al., 2017), Solenopsis invicta (Chiu et al., 2020), and Lasius niger (Konu et al., 2022). Similarly, honeybees (that can be used to detect disease – see previous point) had their own CRISPR/Cas-9 mutants developed as well (Roth et al., 2019; Deǧirmenci et al., 2020; Chen et al., 2021).
Combined with the proper genes, it would be in principle possible to produce entire colonies (of genetically identical individuals) expressing a natural attraction (or repulsion) behaviour to disease-related VOCs, cutting down the conditioning time. Lastly, a strong advantage of using ants is that they can be kept in the laboratory in simple artificial nests and do not require special care, allowing to consider their use by non-specialists (Figure 1).
3.6. What comes next?
In this review, we discussed the potential of invertebrates for human disease detection using their ability to detect and learn the VOCs emitted by pathogens (Table 1). Invertebrates have the advantage of being small, easy to maintain and operate, as well as to be available in large numbers. Until now, only a few species have been tested for their olfactory abilities for disease detection. However, the species that have been tested were chosen for several reasons (Figure 1).
On the one hand, honey bees and ants are social insects and live in colonies that can be composed of thousands of insects. Apart from their use of olfactory cues while foraging for food, communication between nestmates is critical, and individuals can share learned information socially with other nestmates. During trophallaxis, two (or more) individuals exchange food, but also information about odorant food cues (Farina et al., 2005; Provecho and Josens, 2009). Bees can also exchange information based on the antennal movement of conspecifics (Cholé et al., 2019). Although the time required to condition individuals is relatively short (minutes to hours), it may also be possible to condition only a small number of individuals per colony that will later go on to share the information with naive nestmates, as social insects possess impressive social learning abilities (Chittka and Rossi, 2022). Another possibility would be letting a whole colony condition itself, by presenting the VOCs of interest next to a reward outside the nest (Rodacy et al., 2002). Each member of the colony would then already be conditioned (or at least non-naive) to a specific disease/biomarker, dramatically reducing the individual training time.
On the other hand, fruit flies and nematodes are well established models for genetic manipulation, and mutants can easily be produced (note that this was also achieved in ants and bees; Friedman et al., 2017; Kohno and Kubo, 2019). Mutants with specific disease-related olfactory receptors could therefore be developed to reach higher detection thresholds, or to detect new odorants and the associated disease (Neto et al., 2016). Mutants expressing a particular behaviour (e.g., repulsion/attraction) only in the presence of the VOCs from a disease would greatly simplify the diagnosis. This could be performed by non-specialised personnel, or via computer assisted tracking.
Electronic biosensors are also being developed for disease detection. Electronic noses apply different techniques based on high-end polymers or metal-based sensors. VOCs that are present in the environment will bind to sensors if they have the according affinity. This binding process will produce a specific signal, as different VOCs have different affinities with a given sensor (Behera et al., 2019). E-noses can have very low detection thresholds, but current sensors are rather unspecific and a range of volatiles can bind with each sensor. Efficient systems thus currently combine an array of different sensors (Frenois et al., 2014; Claverie, 2022) including for example metal based and fluorescent detectors (absorption of a compound change the fluorescence of the sensor) in order to detect more specific compounds. Insect olfaction may revolutionise such systems, by integrating insect olfactory receptors as biosensors, ensuring a highly specific binding process with particular VOCs (Bohbot and Vernick, 2020; Saha et al., 2020; Claverie, 2022; Farnum et al., 2022). The development of neurogenetic tools to identify and precisely characterise olfactory coding and processing in the insect brain (Carcaud et al., 2022) could help design solutions for silencing specific neurons or implementing them in biosensors.
The use of olfactory detection abilities of invertebrates is at an early stage with extremely few species/diseases tested so far. Nevertheless, invertebrates show a significant potential when it comes to implementation, replication, rapidity, and efficiency of disease detection. Invertebrates account for up to 97% of all animal species, and thus represent a mostly unexplored and thriving pool of millions of species with potentially impressive detection abilities. They should receive more attention from scientists working on disease detection. Invertebrates have the advantages of being cheap and easy to maintain, and easy to train under standardised procedures in controlled laboratory conditions. Some species do not need to be trained and can express natural behaviour toward specific samples, while others can present interesting mutations (e.g., on olfactory receptors used for disease detection), and finally some are eusocial species, able to transmit learned information to conspecifics (Figure 1). We hope that this short review will prompt further efforts to make use of the rich diversity of species and behavioural and neurophysiological protocols available in invertebrates, to develop efficient new assays for the early detection of deadly diseases.
Author contributions
Pd’E had the idea of the manuscript. BP, J-CS, and Pd’E discussed the topic. BP wrote the manuscript with inputs from J-CS and Pd’E. All the authors approved the final version.
Funding
Pd’E funded BP with a grant from the Institut Universitaire de France (IUF).
Acknowledgments
We would like to thank Fatima Mechta-Grigoriou for allowing us to start exploring the stimulating field of disease detection by animals. We also thank Erika Dawson for improving the first draft of the manuscript with helpful comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Asai, A., Konno, M., Ozaki, M., Kawamoto, K., Chijimatsu, R., Kondo, N., et al. (2021). Scent test using Caenorhabditis elegans to screen for early-stage pancreatic cancer. Oncotarget 12, 1687–1696. doi: 10.18632/oncotarget.28035
Behera, B., Joshi, R., Anil Vishnu, G. K., Bhalerao, S., and Pandya, H. J. (2019). Electronic nose: a non-invasive technology for breath analysis of diabetes and lung cancer patients. J. Breath Res. 13:024001. doi: 10.1088/1752-7163/aafc77
Bijland, L. R., Bomers, M. K., and Smulders, Y. M. (2013). Smelling the diagnosis A review on the use of scent in diagnosing disease. Neth. J. Med. 71, 300–307.
Bitterman, M. E., Menzel, R., Fietz, A., and Schäfer, S. (1983). Classical conditioning of proboscis extension in honeybees (Apis mellifera). J. Comp. Psychol. 97, 107–119. doi: 10.1037/0735-7036.97.2.107
Bohbot, J. D., and Vernick, S. (2020). The emergence of insect odorant receptor-based biosensors. Biosensors 10, 1–22. doi: 10.3390/bios10030026
Bomers, M. K., van Agtmael, M. A., Luik, H., Vandenbroucke-Grauls, C. M. J. E., and Smulders, Y. M. (2014). A detection dog to identify patients with Clostridium difficile infection during a hospital outbreak. J. Infect. 69, 456–461. doi: 10.1016/j.jinf.2014.05.017
Bromenshenk, J., Henderson, C., Seccomb, R., Rice, S., Etter, R., Jerry Bromenshenk, A., et al. (2003). Can honey bees assist in area reduction and landmine detection? James J. Conv. Weapons Destr. 7, 24–27. doi: 10.1017/CBO9781107415324.004
Carcaud, J., Otte, M., Grünewald, B., Haase, A., Sandoz, J.-C., and Beye, M. (2022). Multisite imaging of neural activity using a genetically encoded calcium sensor in the honey bee Apis mellifera. bio Rxiv. doi: 10.1101/2022.04.22.489138
Catala, A., Grandgeorge, M., Schaff, J. L., Cousillas, H., Hausberger, M., and Cattet, J. (2019). Dogs demonstrate the existence of an epileptic seizure odour in humans. Sci. Rep. 9, 4103–4107. doi: 10.1038/s41598-019-40721-4
Chen, Z., Traniello, I. M., Rana, S., Cash-Ahmed, A. C., Sankey, A. L., Yang, C., et al. (2021). Neurodevelopmental and transcriptomic effects of CRISPR/Cas9-induced somatic orco mutation in honey bees. J. Neurogenet. 35, 320–332. doi: 10.1080/01677063.2021.1887173
Chittka, L., and Rossi, N. (2022). Social cognition in insects. Trends Cogn. Sci. 26, 578–592. doi: 10.1016/j.tics.2022.04.001
Chiu, Y. K., Hsu, J. C., Chang, T., Huang, Y. C., and Wang, J. (2020). Mutagenesis mediated by CRISPR/Cas9 in the red imported fire ant, Solenopsis invicta. Insectes Soc. 67, 317–326. doi: 10.1007/s00040-020-00755-8
Cholé, H., Carcaud, J., Mazeau, H., Famié, S., Arnold, G., and Sandoz, J. C. (2019). Social contact acts as appetitive reinforcement and supports associative learning in honeybees. Curr. Biol. 29, 1407–1413.e3. doi: 10.1016/j.cub.2019.03.025
Claverie, N. (2022). Olfaction active bioinspirée de vapeurs pour la détection de substances spécifiques. Thèse de doctorat. Tours: Université de Tours.
Czaczkes, T. J., and Kumar, P. (2020). Very rapid multi-odour discrimination learning in the ant Lasius niger. Insect. Soc. 67, 541–545. doi: 10.1007/s00040-020-00787-0
Deǧirmenci, L., Geiger, D., Rogé Ferreira, F. L., Keller, A., Krischke, B., Beye, M., et al. (2020). CRISPR/Cas 9-mediated mutations as a new tool for studying taste in honeybees. Chem. Senses 45, 655–666. doi: 10.1093/chemse/bjaa063
di Luccio, E., Morishita, M., and Hirotsu, T. (2022). C. elegans as a powerful tool for cancer screening. Biomedicine 10:2371. doi: 10.3390/biomedicines10102371
Dobrolecki, L. E., Airhart, S. D., Alferez, D. G., Aparicio, S., Behbod, F., Bentires-Alj, M., et al. (2016). Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 35, 547–573. doi: 10.1007/s10555-016-9653-x
Edwards, T. L., Cox, C., Weetjens, B., Tewelde, T., and Poling, A. (2015). Giant African pouched rats (Cricetomys gambianus) that work on tilled soil accurately detect land mines. J. Appl. Behav. Anal. 48, 696–700. doi: 10.1002/jaba.214
Farina, W. M., Grüater, C., and Díaz, P. C. (2005). Social learning of floral odours inside the honeybee hive. Proc. R. Soc. B Biol. Sci. 272, 1923–1928. doi: 10.1098/rspb.2005.3172
Farnum, A., Parnas, M., Hoque Apu, E., Cox, E., Lefevre, N., Contag, C. H., et al. (2022). Harnessing insect olfactory neural circuits for detecting and discriminating human cancers. Biosens. Bioelectron. 219:114814. doi: 10.1016/j.bios.2022.114814
Fiebig, L., Beyene, N., Burny, R., Fast, C. D., Cox, C., and Mgode, G. F. (2020). From pests to tests: Training rats to diagnose tuberculosis. Eur. Respir. J. 55, 4–8. doi: 10.1183/13993003.02243-2019
Filipiak, W., and Bojko, B. (2019). SPME in clinical, pharmaceutical, and biotechnological research – How far are we from daily practice? Trends Anal. Chem. 115, 203–213. doi: 10.1016/j.trac.2019.02.029
Francis, V. S., Holness, H. K., and Furton, K. G. (2019). The ability of narcotic detection canines to detect illegal synthetic cathinones (bath salts). Front. Vet. Sci. 6, 1–6. doi: 10.3389/fvets.2019.00098
Frenois, C., Barthet, C., Pereira, F., Minot, B., Veignal, F., Besnard, S., et al. (2014). Detection of vapour explosives by a multi-sensor prototype-performance evaluation under laboratory and real conditions. In Proceedings of the 2014 IEEE Sensors, Valencia, Spain, 2–5 November 2014. 1057–1060.
Friedman, D. A., Gordon, D. M., and Luo, L. (2017). The MutAnts are here. Cells 170, 601–602. doi: 10.1016/j.cell.2017.07.046
Furton, K. G., and Myers, L. J. (2001). The scientific foundation and efficacy of the use of canines as chemical detectors for explosives. Talanta 54, 487–500. doi: 10.1016/S0039-9140(00)00546-4
Giurfa, M., and Sandoz, J.-C. (2012). Invertebrate learning and memory: fifty years of olfactory conditioning of the proboscis extension response in honeybees. Learn. Mem. 19, 54–66. doi: 10.1101/lm.024711.111
Gouzerh, F., Bessière, J.-M., Ujvari, B., Thomas, F., Dujon, A. M., and Dormont, L. (2022a). Odors and cancer: current status and future directions. Biochim. Biophys. Acta Rev. Cancer 1877:188644. doi: 10.1016/j.bbcan.2021.188644
Gouzerh, F., Buatois, B., Hervé, M. R., Mancini, M., Maraver, A., Dormont, L., et al. (2022b). Odours of cancerous mouse congeners: detection and attractiveness. Biol. Open 11:bio059208. doi: 10.1242/bio.059208
Guerrieri, F. J., and d’Ettorre, P. (2010). Associative learning in ants: conditioning of the maxilla-labium extension response in Camponotus aethiops. J. Insect Physiol. 56, 88–92. doi: 10.1016/j.jinsphys.2009.09.007
Guest, C., and Otto, C. M. (2020). Editorial: canine olfactory detection. Front. Vet. Sci. 7, 1–3. doi: 10.3389/fvets.2020.00100
Hirotsu, T., Sonoda, H., Uozumi, T., Shinden, Y., Mimori, K., Maehara, Y., et al. (2015). A highly accurate inclusive cancer screening test using Caenorhabditis elegans scent detection. PLoS One 10:e0118699. doi: 10.1371/journal.pone.0118699
Jendrny, P., Schulz, C., Twele, F., Meller, S., Von Köckritz-Blickwede, M., Osterhaus, A. D. M. E., et al. (2020). Scent dog identification of samples from COVID-19 patients – A pilot study. BMC Infect. Dis. 20, 1–7. doi: 10.1186/s12879-020-05281-3
Jendrny, P., Twele, F., Meller, S., Osterhaus, A. D. M. E., Schalke, E., and Volk, H. A. (2021). Canine olfactory detection and its relevance to medical detection. BMC Infect. Dis. 21, 838–815. doi: 10.1186/s12879-021-06523-8
Juge, A. E., Foster, M. F., and Daigle, C. L. (2022). Canine olfaction as a disease detection technology: a systematic review. Appl. Anim. Behav. Sci. 253:105664. doi: 10.1016/j.applanim.2022.105664
Junqueira, H., Quinn, T. A., Biringer, R., Hussein, M., Smeriglio, C., Barrueto, L., et al. (2019). Accuracy of canine scent detection of non–small cell lung cancer in blood serum. J. Am. Osteopath. Assoc. 119, 413–418. doi: 10.7556/jaoa.2019.077
King, T. L., Horine, F. M., Daly, K. C., and Smith, B. H. (2004). Explosives detection with hard-wired moths. IEEE Trans. Instrum. Meas. 53, 1113–1118. doi: 10.1109/TIM.2004.831455
Kobayashi, M., Fujita, A., Ogawa, T., Tanisaka, Y., Mizuide, M., Kondo, N., et al. (2021). Caenorhabditis elegans as a diagnostic aid for pancreatic cancer. Pancreas 50, 673–678. doi: 10.1097/MPA.0000000000001814
Kohno, H., and Kubo, T. (2019). Genetics in the honey bee: achievements and molecular and neural mechanisms underlying. Insects 10:348. doi: 10.3390/insects10100348
Kokocinska-Kusiak, A., Matalińska, J., Sacharczuk, M., Sobczyńska, M., Góral-Radziszewska, K., Wileńska, B., et al. (2020). Can mice be trained to discriminate urine odor of conspecifics with melanoma before clinical symptoms appear? J. Vet. Behav. 39, 64–76. doi: 10.1016/j.jveb.2020.04.004
Kontos, E., Samimi, A., Hakze-van der Honing, R. W., Priem, J., Avarguès-Weber, A., Haverkamp, A., et al. (2022). Bees can be trained to identify SARS-CoV-2 infected samples. Biol. Open 11. doi: 10.1242/bio.059111
Konu, M., Kulmuni, J., and Viljakainen, L. (2022). Genetic modification of the ant Lasius niger using CRISPR-Cas9 technology. Insect Mol. Biol., 1–15. doi: 10.1111/imb.12809
Kusumoto, H., Tashiro, K., Shimaoka, S., Tsukasa, K., Baba, Y., Furukawa, S., et al. (2020). Efficiency of gastrointestinal cancer detection by nematode-NOSE (N-NOSE). In Vivo 34, 73–80. doi: 10.21873/invivo.11747
Lanza, E., Di Rocco, M., Schwartz, S., Caprini, D., Milanetti, E., Ferrarese, G., et al. (2021). C. elegans-based chemosensation strategy for the early detection of cancer metabolites in urine samples. Sci. Rep. 11, 17133–17116. doi: 10.1038/s41598-021-96613-z
Matsumura, K., Opiekun, M., Oka, H., Vachani, A., Albelda, S. M., Yamazaki, K., et al. (2010). Urinary volatile compounds as biomarkers for lung cancer: a proof of principle study using odor signatures in mouse models of lung cancer. PLoS One 5:e8819. doi: 10.1371/journal.pone.0008819
McCulloch, M., Jezierski, T., Broffman, M., Hubbard, A., Turner, K., and Janecki, T. (2006). Diagnostic accuracy of canine scent detection in early-and late-stage lung and breast cancers. Integr. Cancer Ther. 5, 30–39. doi: 10.1177/1534735405285096
Menzel, R., and Giurfa, M. (2001). Cognitive architecture of a mini-brain: the honeybee. Trends Cogn. Sci. 5, 62–71. doi: 10.1016/S1364-6613(00)01601-6
Miller, A. K., Hensman, M. C., Hensman, S., Schultz, K., Reid, P., Shore, M., et al. (2015). African elephants (Loxodonta africana) can detect TNT using olfaction: implications for biosensor application. Appl. Anim. Behav. Sci. 171, 177–183. doi: 10.1016/j.applanim.2015.08.003
Murarka, M., Vesley-Gross, Z. I., Essler, J. L., Smith, P. G., Hooda, J., Drapkin, R., et al. (2019). Testing ovarian cancer cell lines to train dogs to detect ovarian cancer from blood plasma: a pilot study. J. Vet. Behav. 32, 42–48. doi: 10.1016/j.jveb.2019.04.010
Neto, M. F., Nguyen, Q. H., Marsili, J., McFall, S. M., and Voisine, C. (2016). The nematode Caenorhabditis elegans displays a chemotaxis behavior to tuberculosis-specific odorants. J. Clin. Tuberc. Other Mycobact. Dis. 4, 44–49. doi: 10.1016/j.jctube.2016.06.001
Oh, Y., Lee, Y., Heath, J., and Kim, M. (2014). Applications of animal biosensors: a review. IEEE Sensors J. 15, 637–645. doi: 10.1109/JSEN.2014.2358261
Paoli, M., and Galizia, G. C. (2021). Olfactory coding in honeybees. Cell Tissue Res. 383, 35–58. doi: 10.1007/s00441-020-03385-5
Perez, M., Giurfa, M., and d’Ettorre, P. (2015). The scent of mixtures: rules of odour processing in ants. Sci. Rep. 5:8659. doi: 10.1038/srep08659
Piqueret, B., Bourachot, B., Leroy, C., Devienne, P., Mechta-Grigoriou, F., d’Ettorre, P., et al. (2022a). Ants detect cancer cells through volatile organic compounds. iScience 25:103959. doi: 10.1016/j.isci.2022.103959
Piqueret, B., Montaudon, É., Devienne, P., Leroy, C., Marangoni, E., Sandoz, J.-C., et al. (2022b). Ants act as olfactory bio-detectors of tumour in patient-derived xenograft mice. bio Rxiv. doi: 10.1101/2022.05.16.492058
Piqueret, B., Sandoz, J. C., and d’Ettorre, P. (2019). Ants learn fast and do not forget: associative olfactory learning, memory and extinction in Formica fusca. R. Soc. Open Sci. 6. doi: 10.1098/rsos.190778
Pirrone, F., and Albertini, M. (2017). Olfactory detection of cancer by trained sniffer dogs: A systematic review of the literature. J. Vet. Behav. Clin. Appl. Res. 19, 105–117. doi: 10.1016/j.jveb.2017.03.004
Provecho, Y., and Josens, R. (2009). Olfactory memory established during trophallaxis affects food search behaviour in ants. J. Exp. Biol. 212, 3221–3227. doi: 10.1242/jeb.033506
Ratiu, I. A., Bocos-Bintintan, V., Monedeiro, F., Milanowski, M., Ligor, T., and Buszewski, B. (2019). An optimistic vision of future: diagnosis of bacterial infections by sensing their associated volatile organic compounds. Crit. Rev. Anal. Chem. 50, 501–512. doi: 10.1080/10408347.2019.1663147
Rodacy, P., Bender, F. A., Bromenshenk, J., Henderson, C., and Bender, G. (2002). The training and deployment of honeybees to detect explosives and other agents of harm. Proc. SPIE 4742, 474–481. doi: 10.1117/12.479119
Roth, A., Vleurinck, C., Netschitailo, O., Bauer, V., Otte, M., Kaftanoglu, O., et al. (2019). A genetic switch for worker nutritionmediated traits in honeybees. PLoS Biol. 17, e3000171–e3000118. doi: 10.1371/journal.pbio.3000171
Saha, D., Mehta, D., Altan, E., Chandak, R., Traner, M., Lo, R., et al. (2020). Explosive sensing with insect-based biorobots. Biosens. Bioelectron. X 6:100050. doi: 10.1016/j.biosx.2020.100050
Sandoz, J. C. (2011). Behavioral and neurophysiological study of olfactory perception and learning in honeybees. Front. Syst. Neurosci. 5. doi: 10.3389/fnsys.2011.00098
Sato, T., Katsuoka, Y., Yoneda, K., Nonomura, M., Uchimoto, S., Kobayakawa, R., et al. (2017). Sniffer mice discriminate urine odours of patients with bladder cancer: a proof-of-principle study for non-invasive diagnosis of cancer-induced odours. Sci. Rep. 7:14628. doi: 10.1038/s41598-017-15355-z
Schallschmidt, K., Becker, R., Zwaka, H., Menzel, R., Johnen, D., Fischer-Tenhagen, C., et al. (2015). In vitro cultured lung cancer cells are not suitable for animal-based breath biomarker detection. J. Breath Res. 9. doi: 10.1088/1752-7155/9/2/027103
Strauch, M., Lüdke, A., Münch, D., Laudes, T., Galizia, C. G., Martinelli, E., et al. (2014). More than apples and oranges – detecting cancer with a fruit fly’s antenna. Sci. Rep. 4. doi: 10.1038/srep03576
Suckling, D. M., and Sagar, R. L. (2011). Honeybees Apis mellifera can detect the scent of mycobacterium tuberculosis. Tuberculosis 91, 327–328. doi: 10.1016/j.tube.2011.04.008
Syhre, M., and Chambers, S. T. (2008). The scent of mycobacterium tuberculosis. Tuberculosis 88, 317–323. doi: 10.1016/j.tube.2008.01.002
Takeda, K. (1961). Classical conditioned response in the honey bee. J. Insect Physiol. 6, 168–179. doi: 10.1016/0022-1910(61)90060-9
Taylor-mccabe, K. J., Wingo, R. M., and Haarmann, T. K. (2008). Honey bees (Apis mellifera) as explosives detectors: exploring proboscis extension reflex conditioned response to trinitrotolulene (TNT). Apidologie 836.
Tee, L. F., Tan, T. L., Neoh, H., and Jamal, R. (2019). Rapid detection of sepsis using CESDA: the Caenorhabditis elegans sepsis detection assay. J. Brazilian Soc. Trop. Med. 52:e20180300. doi: 10.1590/0037-8682-0300-2018
Thompson, M., Feria, N. S., Yoshioka, A., Tu, E., Civitci, F., Estes, S., et al. (2021). A Caenorhabditis elegans behavioral assay distinguishes early stage prostate cancer patient urine from controls. Biol. Open 10. doi: 10.1242/BIO.057398
Thuleau, A., Gilbert, C., Bauër, P., Alran, S., Fourchotte, V., Guillot, E., et al. (2019). A new transcutaneous method for breast cancer detection with dogs. Oncologia 96, 110–113. doi: 10.1159/000492895
Trible, W., Olivos-Cisneros, L., McKenzie, S. K., Saragosti, J., Chang, N. C., Matthews, B. J., et al. (2017). Orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cells 170, 727–735.e10. doi: 10.1016/j.cell.2017.07.001
Trivedi, D. K., Sinclair, E., Xu, Y., Sarkar, D., Walton-Doyle, C., Liscio, C., et al. (2019). Discovery of volatile biomarkers of Parkinson’s disease from sebum. ACS Cent. Sci. 5, 599–606. doi: 10.1021/acscentsci.8b00879
Ueda, Y., Kawamoto, K., Konno, M., Noguchi, K., Kaifuchi, S., Satoh, T., et al. (2019). Application of C. elegans cancer screening test for the detection of pancreatic tumor in genetically engineered mice. Oncotarget 10, 5412–5418. doi: 10.18632/oncotarget.27124
Wagner, T., Galante, H., and Czaczkes, T. J. (2022). A systematic examination of learning in the invasive ant Linepithema humile reveals very rapid development of short and long-term memories. bioRxiv. doi: 10.1101/2022.04.12.487867
Welch, J. B. (1990). A detector dog for screwworms (Diptera: Calliphoridae). J. Econ. Entomol. 83, 1932–1934. doi: 10.1093/jee/83.5.1932
WHO (2020). WHO report on cancer: Setting priorities, investing wisely and providing care for all. World Health Organization. Available at: https://www.who.int/publications-detail/who-report-on-cancer-setting-priorities-investing-wisely-and-providing-care-for-all (Accessed December 28, 2022).
Williams, H., and Pembroke, A. (1989). Sniffer dogs in the melanoma clinic? Lancet 1, 734–735. doi: 10.1016/s0140-6736(89)92257-5
Willis, C. M., Church, S. M., Guest, C. M., Cook, W. A., McCarthy, N., Bransbury, A. J., et al. (2004). Olfactory detection of human bladder cancer by dogs: proof of principle study. BMJ 329:712. doi: 10.1136/bmj.329.7468.712
Yan, H., Opachaloemphan, C., Mancini, G., Yang, H., Gallitto, M., Mlejnek, J., et al. (2017). An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cells 170, 736–747.e9. doi: 10.1016/j.cell.2017.06.051
Keywords: human diseases, biodetectors, insects, cancer, volatile organic compounds
Citation: Piqueret B, Sandoz J-C and d’Ettorre P (2023) The neglected potential of invertebrates in detecting disease via olfaction. Front. Ecol. Evol. 10:960757. doi: 10.3389/fevo.2022.960757
Edited by:
Magdalena Ruiz Rodriguez, University of Granada, SpainReviewed by:
Bruce A. Kimball, Monell Chemical Senses Center, United StatesClaire Martin, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2023 Piqueret, Sandoz and d’Ettorre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrizia d’Ettorre, ✉ ZC1ldHRvcnJlQHVuaXYtcGFyaXMxMy5mcg==
 Baptiste Piqueret
Baptiste Piqueret Jean-Christophe Sandoz
Jean-Christophe Sandoz Patrizia d’Ettorre
Patrizia d’Ettorre