- 1Key Laboratory of Ecological Restoration and Conservation of Coastal Wetlands in Universities of Shandong, The Institute for Advanced Study of Coastal Ecology, Ludong University, Yantai, China
- 2Key Laboratory of Pollution Ecology and Environment Engineering, Institute of Applied Ecology, Chinese Academy of Sciences, Shenyang, China
- 3Dongying Academy of Agricultural Sciences, Dongying, China
Tidal creeks have attracted considerable attention in estuary wetland conservation and restoration with diverse micro-habitats and high hydrological connectivity. Bacterial communities act effectively as invisible engines to regulate nutrient element biogeochemical processes. However, few studies have unveiled the bacterial community structures and diversities of micro-habitats soils on the tidal creek section. Our study selected three sections cross a tidal creek with obviously belt-like habitats “pluff mudflat – bare mudflat – Tamarix chinensis community – T. chinensis-Suaeda salsa community– S. salsa community” in the Yellow River estuarine wetland. Based on soil samples, we dissected and untangled the bacterial community structures and special bacterial taxa of different habitats on the tidal creek section. The results showed that bacterial community structures and dominant bacterial taxa were significantly different in the five habitats. The bacterial community diversities significantly decreased with distance away from tidal creeks, as well as the dominant bacteria Flavobacteriia and δ-Proteobacteria, but in reverse to Bacteroidetes and Gemmatimonadetes. Moreover, the important biomarkers sulfate-reducing bacteria and photosynthetic bacteria were different distributions within the five habitats, which were closely associated with the sulfur and carbon cycles. We found that the bacterial communities were heterogeneous in different micro-habitats on the tidal creek section, which was related to soil salinity, moisture, and nutrients as well as tidal action. The study would provide fundamental insights into understanding the ecological functions of bacterial diversities and biogeochemical processes influenced by tidal creeks.
Introduction
Estuary wetlands, between ocean and terrestrial ecosystems, play important ecological functions in climate change regulation, biodiversity preservation, and environmental remediation (Fennessy, 2014; Murray et al., 2019; Xiao et al., 2019). In the front estuary wetlands by field investigation and aerial photography, there are a variety of small and different micro-habitats around tidal creeks, which compose the tidal creek salt marshes. Tidal creek salt marshes are active and critical to biogeochemical processes and biological diversities with high hydrological connectivity (Mallin and Lewitus, 2004; Vandenbruwaene et al., 2012; Cui et al., 2016). Along with the frequent and long-time tidal alternation, the tidal creek salt marshes comprise many different micro-habitats and have orderly occurred in habitat successions with unvegetated mudflat initiation, vegetation colonization, organic and inorganic sediment deposition, and upper marsh elevation (Wilson et al., 2014; Chirol et al., 2018; Wu et al., 2020). For example, the micro-habitats are bare lands and/or the Spartina alterniflora community near tidal creeks due to long-time flooding and in the upper lands away from tidal reeks colonized by more plant communities like Suaeda glauca, Salicornia europaea, and Phragmites communis (Kim, 2018; Liu et al., 2020). It should be notable that the diverse micro-habitats on the tidal creek section are to advantage to maintain the eco-functions, including biogeochemical processes, matter exchanges, environmental resilience, and so on (Arriola and Cable, 2017; Tan et al., 2020; Trifunovic et al., 2020; Wu et al., 2020).
It is extensively focused on that bacterial communities that are indispensable to understanding and untangling the estuary wetland eco-functions, because engines act as a driver for soil carbon and nitrogen turnover processes, environment resistance, resilience, and so on (Huang et al., 2020; Zou et al., 2020; Coban et al., 2022). Previous studies have demonstrated that bacterial communities are greatly affected by many environmental factors, such as pH, organic matter, nutrient availability, soil salinity, and moisture (Fierer, 2017). In coastal wetland, soil salinity and nutrients were regarded as the important environmental filters (An et al., 2019). As well, soil bacterial community structures and functions, like sulfate-reducing and nitrifying bacteria, were significantly different, ranging from oligohaline to hypersaline habitats (Li et al., 2019; Zhang et al., 2020). Thus, it is important to unveil the relationships between bacterial communities and habitats in tidal creek salt marshes to better understand eco-functions of estuary wetlands.
The Yellow River estuary wetland is focused by previous studies on micro-habitat types, biogeochemical processes, and eco-functions of wetlands (Zhao et al., 2010; Yu et al., 2018; Gong et al., 2021). Several recent studies have found that bacteria communities have existed in great variations, which were relevant to geographic patterns, season variations, plant types, and soil salinities (Lv et al., 2016; Zhang et al., 2017; Zhang et al., 2020; Li et al., 2021). However, it is poorly to announce bacterial community structures in the micro-habitat soils on the tidal creek section, which would limit our understanding of the biogeochemical processes and eco-functions in estuary wetland ecosystem. Herein, the present study has selected a tidal creek to investigate the bacterial community structures and unveil the keystone bacteria in response to the soil biogeochemical processes of different micro-habitats. Our hypotheses are that (1) dominant bacteria and bacterial biomarkers should be different across the five micro-habitats on the section of the tidal creek, (2) the differences may be related to the effects of tidal action and soil properties together.
Materials and Methods
Soil Sampling and Soil Physicochemical Properties Analysis
We selected a tidal creek (37°46′−37°49′N, 119°5′−119°6′E), approximately 6,430-m length and 110–190-m width in the Yellow River estuarine wetland, located at Dongying City, Shandong Province, Eastern China (Figure 1A). Along the tidal creek, three sections, 200 m away from each other, were set, and each section was divided into five micro-habitats according to hydrologic conditions and the vegetation community, including pluff mudflat (PM), bare mudflat (BM), Tamarix chinensis community (TC), T. chinensis-Suaeda salsa community (TSC), S. salsa community (SC) (Figure 1B). In detail, PM was closest to the tidal creek with long-time flooding and no plant coverage. BM suffered medium-time flooding and uncovered vegetation, but more fiddler carb bioturbation. Compared with the lower micro-habitats of PM and BM, the upper micro-habitats of TC, TSC, and SC were short-time flooding by spring tidal current and colonized with two dominant halophytes of T. chinensis and S. salsa.
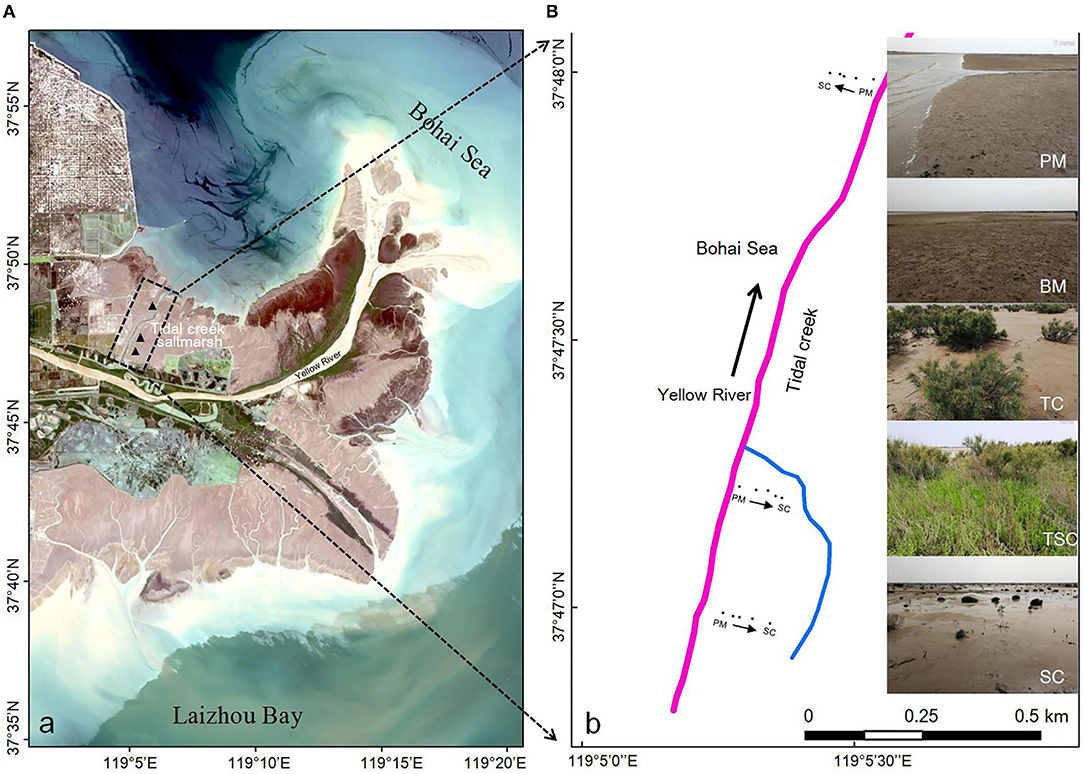
Figure 1. (A) Location of the micro-habitats around the tidal creek in the Yellow River estuary wetland. (B) Landforms of the micro-habitats around the tidal creek, namely PM (pluff mudflat), BM (bare mudflat), TC (Tamarix chinensis community), TSC (T. chinensis and Suaeda salsa community), and SC (S. salsa community).
The soil samples (2–10 cm) in every habitat were collected with three repetitions in July 2018. All of the soil subsamples were quickly sealed in sterile Eppendorf tubes and stored in liquid nitrogen for 16S rRNA analysis, and the others were air dried for soil physicochemical analyses in the laboratory.
Soil physicochemical properties consisting of soil moisture content (SMC), pH value, electrical conductivity (EC), soil organic carbon (SOC), total nitrogen (TN), and total phosphorus (TP) were measured using standard soil test methods as described by the agriculture protocols for China (Lu, 2000). In brief, SMC was determined by weight loss after drying 20 g of wet soil at 105°C for 24 h. Soil pH and EC were determined by a pH meter and an electronic probe (the soil-to-water ratio was 1:5). The SOC, TN, and TP were determined by the potassium dichromate heating oxidation-volumetric method, Kjeldahl nitrogen method, Mo-Sb anti spectrophotometric method, respectively.
Soil Bacterial DNA Extraction and PCR Amplification
Our study used the E.Z.N.A.® soil DNA Kit (Omega Bio-Tek, Norcross, GA, U.S.) to extract microbial DNA from soil samples. The final DNA concentration and purification were determined by a NanoDrop 2000 UV-vis spectrophotometer, and DNA quality was checked by 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of 16S rRNA were selected to identify bacteria species (Yang et al., 2018), and amplified using the barcoded primers 338F/806R (5'-ACTCCTACGGGAGGCAGCAG-3'/5'-GGACTACHVGGGTWTCTAAT-3') from the qualified DNA templates. Briefly, the PCR reactions were amplified using the following program: 3 min of initial denaturation at 95°C, followed by 27 cycles of denaturation at 95°C for 30 s, primer annealing at 55°C for 30 s, elongation at 72°C for 45 s, and a final extension of 10 min at 72°C. The resulted PCR products were extracted from a 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit. All of the purified PCR products were quantified using QuantiFluor™-ST (Promega, USA).
Illumina MiSeq16S rRNA High-Throughput Sequencing and Data Processing
Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Raw fastq files were quality-filtered by Trimmomatic and merged by FLASH with the following criteria: (1) The reads were truncated at any site receiving an average quality score <20 over a 50-bp sliding window. (2) Sequences of overlap being longer than 10 bp were merged according to their overlap with mismatch no more than 2 bp. (3) Sequences of each sample were separated according to barcodes (exactly matching) and Primers (allowing 2 nucleotide mismatching), and reads containing ambiguous bases were removed.
Operational taxonomic units (OTUs) were clustered with a 97% identity cutoff using UPARSE (version7.0.1090, http://drive5.com/uparse/), with a novel “greedy” algorithm that performs chimera filtering and OTU clustering simultaneously. The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier algorithm (https://sourceforge.net/projects/rdp-classifier/) against the Silva project's version 138 release (https://www.arb-silva.de/) using a confidence threshold of 70%. Mothur1.30.2 and Qiime 1.9.1 have calculated the bacterial α-diversity and β-diversity, respectively.
Statistical Analysis
The differences in soil physicochemical properties, bacterial α-diversity, and species were estimated by one-way ANOVA analysis in SPSS 22. Bacterial β-diversity analysis was displayed with principal coordinate analysis (PCoA) based on the weighted normalized unifrac (WNU) metric. Heatmap analysis and redundancy analysis (RDA) were used to find the differential OTUs, and the relationships between bacteria and soil physicochemical properties in the five habitats, respectively. All biomarkers of the five bacterial communities have been identified by linear discriminant analysis (LDA) effect size (LDA > 3, p <0.05) on the Galaxy (https://huttenhower.sph.harvard.edu/galaxy/root) to mine special biomarkers and explain the differences in different bacterial communities (Segata et al., 2011).
Results
Soil Physicochemical Properties
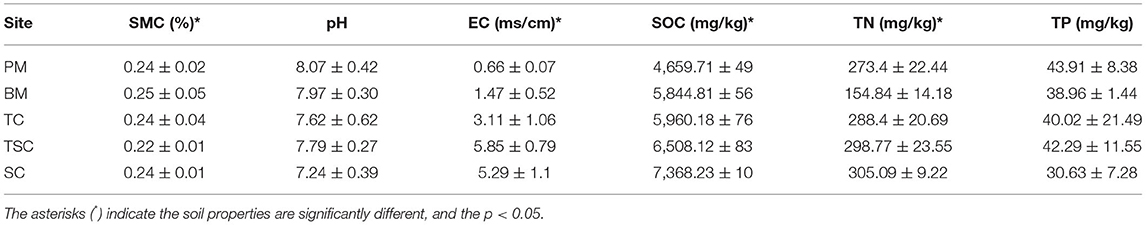
As shown in Table 1, the values of SOC, TN, EC, and SMC were significantly different among micro-habitats (p < 0.05). In detail, the SOC content was highest in SC and followed in TSC, TC, BM, and PM, with means of 7,368.23 ± 10, 6,508.12 ± 83, 5,960.18 ± 76, 5,844.81 ± 56, 4,659.71 ± 49 mg·kg−1, respectively. The TN contents in upper micro-habitats (TC, TSC, and SC) were higher than in lower micro-habitats (PM and BM), with the range from 298.77 to 328.42 mg·kg−1 vs. 164.84 to 240.07 mg·kg−1. As well, the soil EC in vegetation micro-habitats was higher than in the two bare mudflats, with a range from 3.11 to 5.85 ms·cm−1 vs. 66 to 1.47 ms·cm−1. The SMC was 34 ± 0.02% in PM, which was highest among all the micro-habitats. However, there were no significant differences in pH values (p = 0.208) and TP contents (p = 0.38) among the five micro-habitats.
Overview of 16S rRNA High-Throughput Sequencing and Bacterial OTU Diversity
The study has got 702,472 high-quality amplicons with an average length of 440 bp in 15 soil samples, and 99.81% of them were even distributed into the ranges from 401 to 460 bp in length (Figure 2A). After picking out the chimeras and single sequences, it generalized 534,284 valid reads and clustered into a total of 4,657 valid OTUs in the five micro-habitats (Table 2). With Good's coverage scores ranging from 98.42 to 99.41% as a result, there were most OTUs in BM (2,155), and followed by TC (1,947), PM (1,915), SC (1,384), and TSC (1,048). The bacterial α-diversity analysis showed that community richness estimators ACE, Chao1, and diversity index Shannon were significantly different in the five micro-habitat soils (all p < 0.01), except for Simpson index (p = 0.107). ACE, Chao1, and Shannon index of PM, BM, and TC were significantly higher than that of the TSC and SC. With the increase of sample sequences, the rarefaction curves of bacterial OTUs and Shannon-Wiener index have approached the plateaus (Figures 2B,C), which indicated that our subsampling analysis can reflected the bacteria diversity in each sample. Furthermore, the bacterial β-diversity explained the OTUs separation with PCoA (R2 = 0.558, p = 0.001), which showed a significant variance among the five micro-habitats. The first axis PC1 and second axis PC2 explained 55.16% of the total variance with PCoA (Figure 2D). Together, the bacterial α- and β-diversity implied that bacterial community structures were different between the lower micro-habitats vs. the upper micro-habitats.
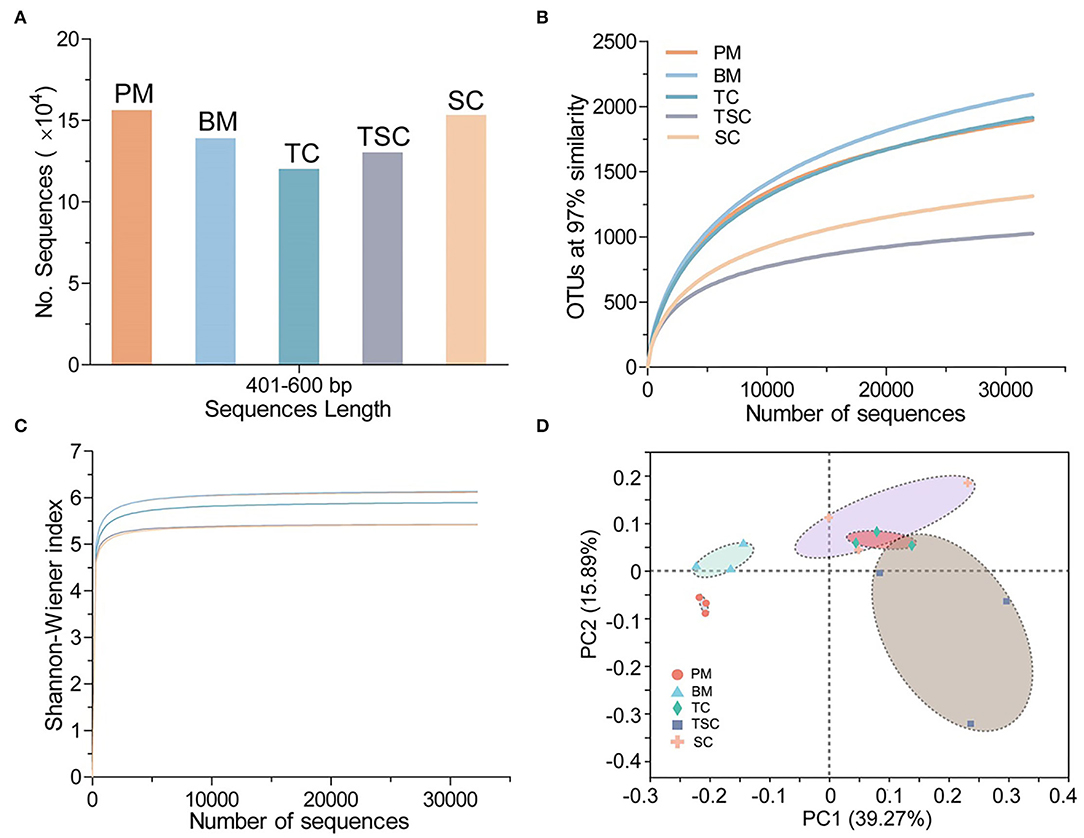
Figure 2. (A) 16S rRNA V3-V4 amplicons ranged from 401 to 460 bp in the five habitats. (B) Rarefaction curves of 97% similar OTUs. (C) Shannon-Wiener curves in the five habitats. (D) β-diversity of the bacterial community in the five habitats.
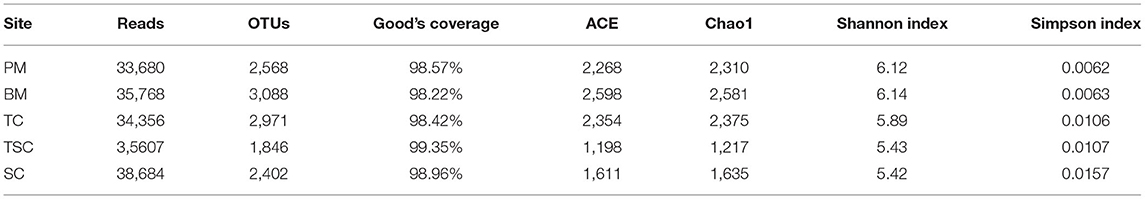
Table 2. The bacterial communities in valid sequence reads, 97% OTUs, Good's Coverage, and bacterial α-diversity indices.
Bacterial Community Structures in Different Micro-Habitats
At the bacterial OTU level, there were 573 shared bacterial OTUs in five micro-habitats, accounting for 22.31% (PM), 18.56% (BM), 19.29% (TC), 31.04% (TSC), and 23.86% (SC), respectively (Figure 3A). On the contrary, 170 OTUs were individual in PM (6.62%), BM (5.05%), TC (4.91%), TSC (11.38%), and SC (8.03%), respectively. Moreover, 67 bacterial classes were shared and accounted for 59.82% (PM), 56.78% (BM), 55.83% (TC), 79.76% (TSC), and 66.34% (SC) (Figure 3B). However, only a few bacteria were uniquely or commonly observed in the other combinations with 1 to 16 overlapped classes, even no shared, which implied that bacterial taxa were relatively stable at the class level vs. at the OTU level across the five micro-habitats.
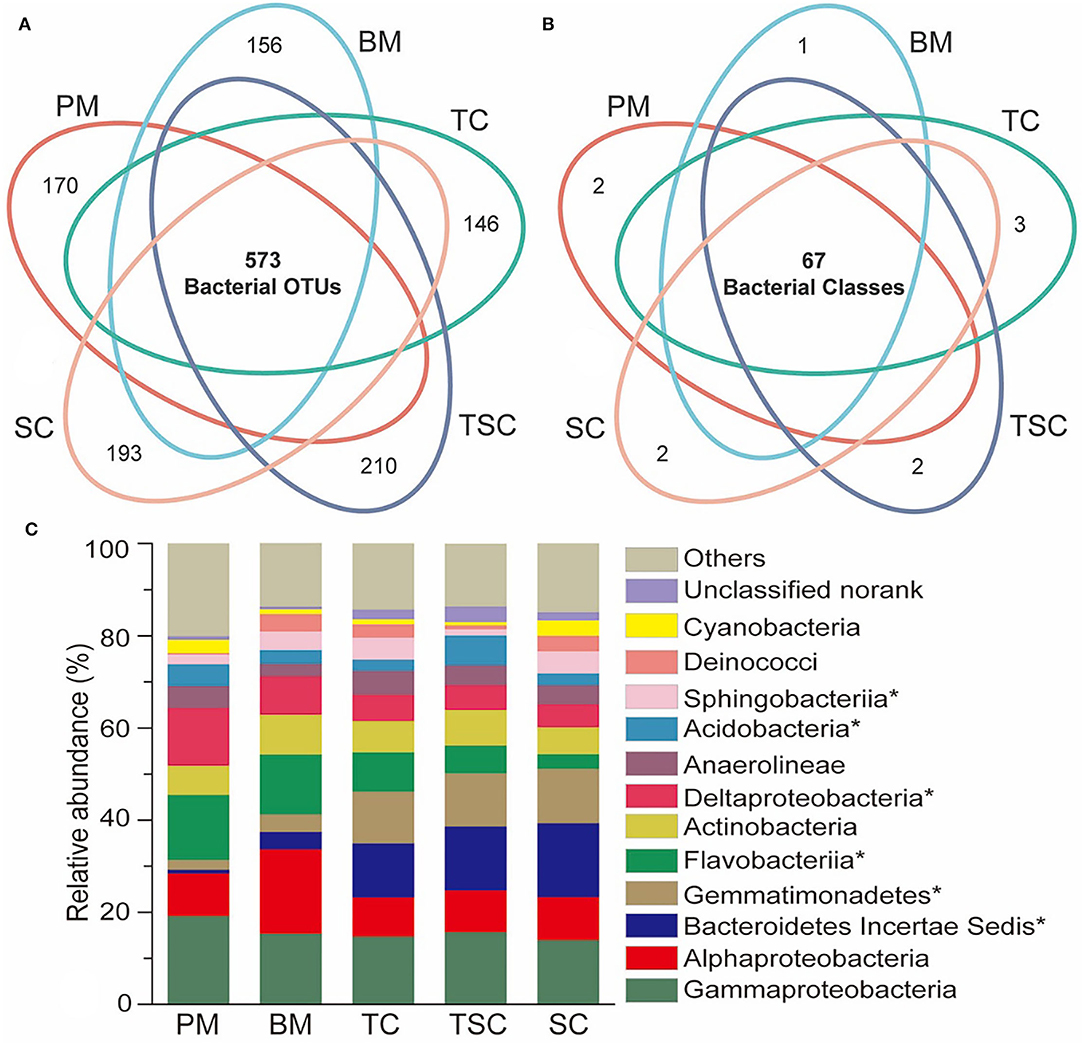
Figure 3. Venn diagram of bacterial OTUs (A) and classes (B) among the five habitats. (C) Dominant bacteria in the five habitats at bacterial class level, and the asterisks (*) indicate the significantly different bacteria (p < 0.05).
The dominant bacterial classes of the five habitats were γ-Proteobacteria (15.76%), α-Proteobacteria (10.89%), Bacteroidetes IncertaeSedis (9.04%), Gemmatimonadetes (8.3%), Flavobacteriia (8.94%), Actinobacteria (7.31%), δ-Proteobacteria (7.22%), Anaerolineae (4.2%), Acidobacteria (3.8%), Sphingobacteriia (3.41%), and so on (Figure 3C). Of them, Bacteroidetes IncertaeSedis and Gemmatimonadetesin upper micro-habitats (SC, TSC, and TC) were obviously higher than them in lower micro-habitats (PM and BM), with 16.04–0.8% (p = 0.02) and 11.74–2.09% (p = 0.02), while Flavobacteriia and δ-Proteobacteria were decreased from PM to SC with 14.16–3.25% (p = 0.04) and 12.58–4.96% (p = 0.02). In addition, Acidobacteria (p = 0.03) and Sphingobacteriia (p = 0.04) were significantly different in the five micro-habitats.
The top 50 bacterial OTUs were selected to compare the OTU abundances and taxa to characterize the variations of bacterial community structures (Figure 4). Fourteen OTUs were significantly enriched in PM, BM, and TC (p < 0.05), which belonged to eight bacterial families of JTB255, Flavobacteriaceae, Unknown γ-Proteobacteria, Sva1033, Rhodobacteraceae, Rhodobiaceae, Rhodospirillaceae, and OM1 clade. Meanwhile, about 20 OTUs were abundant in TC, TSC, and SC, widely mapped into Flavobacteriaceae, Rhodobacteraceae, Trueperaceae, unclassified Chloroflexi, OM1 clade, etc.
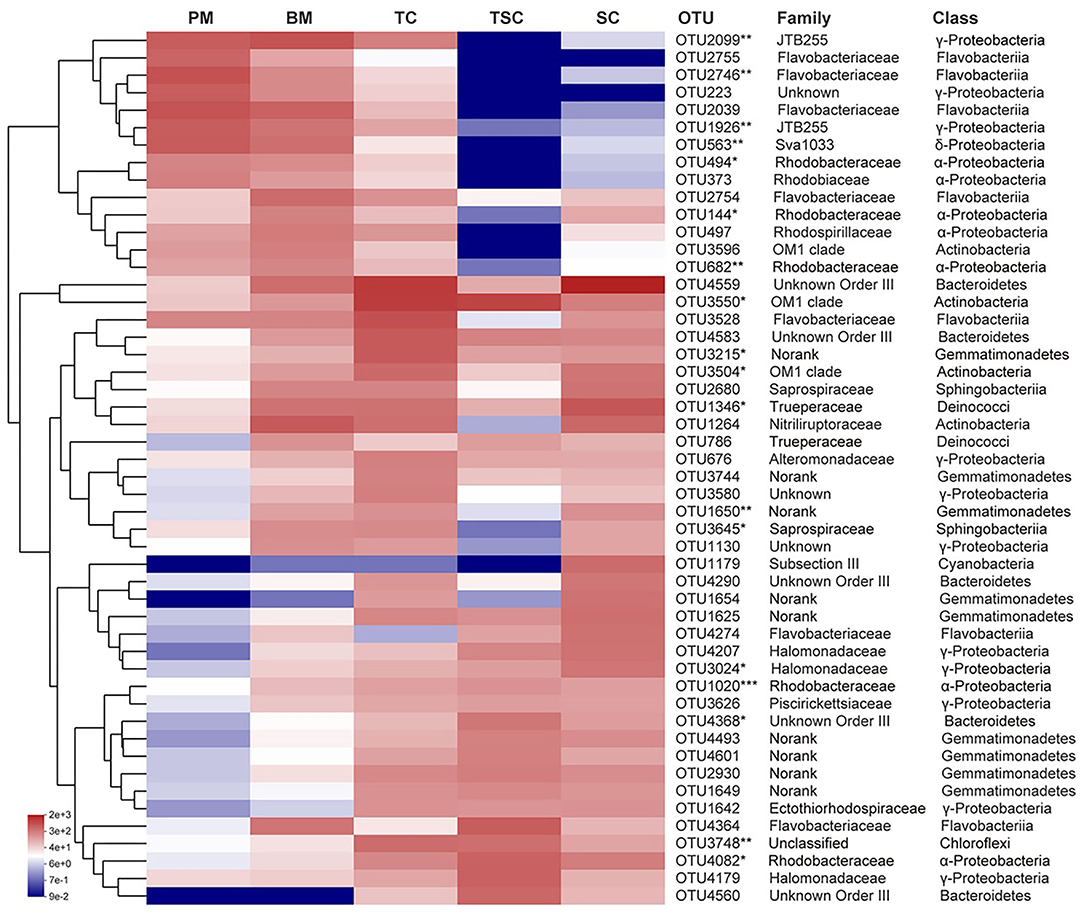
Figure 4. The relative abundances of top 50 OTUs at the family and class levels in the five habitats. The symbol (*) indicates the significantly different bacteria. The * symbol indicates the value of p < 0.05, the ** symbol indicates the value of p < 0.01, and the *** symbol indicates the value of p < 0.001.
Bacterial Biomarkers in Different Micro-Habitats
A total of 129 bacterial biomarkers were identified in the five micro-habitats, with 63 biomarkers (12 bacterial classes) in PM, 16 (5 classes) in BM, 9 (4 classes) in TC, 33 (2 phyla and 8 classes) in TSC, and 8 (2 classes) in SC, respectively (Supplementary Table S1). The bacterial communities of PM and TSC had more biomarkers, which mainly clustered into the classes of α-, δ-, γ-Proteobacteria, Flavobacteriia, Acidobacteria, Actinobacteria, etc. At the bacterial genus level, these biomarkers belonged to 60 genera, and most enriched in PM with 29 bacterial genera, followed by in BM (12), TSC (10), TC (5), and SC (4) (Figure 5). For example, the result found that 10 genera (Desulfococcus, Desulfosarcina, the Sva0081 sediment group, no-rank Desulfobulbaceae, unclassified Desulfobulbaceae, Desulfobulbus, Syntrophobacteraceae, Desulfuromonadales, unclassified Desulfuromonadales, and Geothermobacter) of sulfate-reducing bacteria (SRB) were the biomarkers in PM and BM. In addition, aerobic anoxygenic phototrophic bacteria (AAPB) were also significantly different and diverse in the five micro-habitats with 14 bacterial genera, such as Rhodobacteraceae, Rhodospirillaceae, and Roseovarius. It would be noted that several AAPBs were abundant in Rhodobacteraceae (5.42%), Rhodospirillaceae (1.68%), Ectothiorhodospiraceae (1.05%), Subsection III (0.56%), Rhodobiaceae (0.48%).
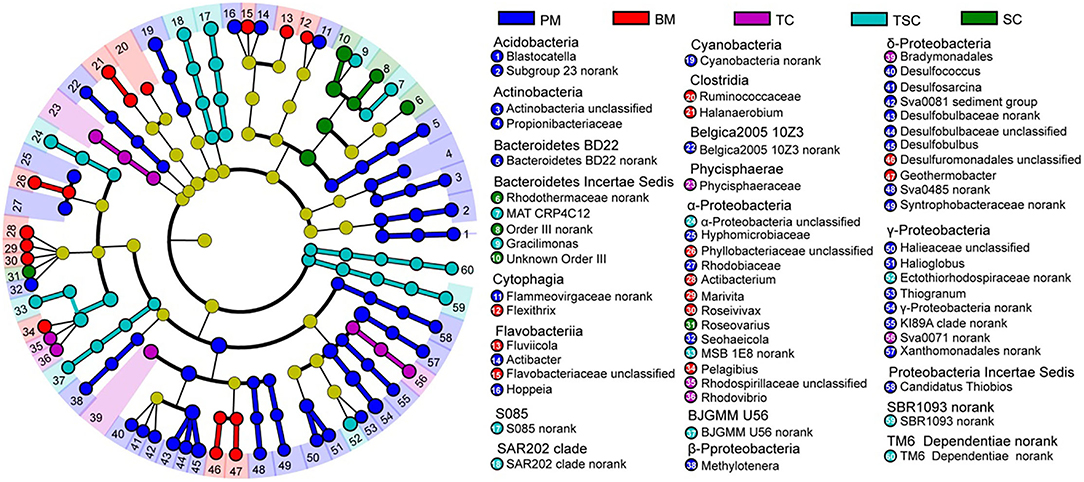
Figure 5. Linear discriminant analysis effect size (LefSe) of bacterial biomarkers in the five habitats.
The Relationship Between Bacterial Community Structures and Soil Physicochemical Properties
At the bacterial class level, the first axis RDA1 and second axis RDA2 have explained 47.56% variances between bacterial community structures and soil physicochemical properties (Figure 6A). The RDA result showed that bacterial community structures of TC, TSC, and SC were positively related to SOC, TN, and EC but negatively related to SMC, pH, and TP, that of PM and BM in reverse. Furthermore, our study selected the top 20 bacterial classes and 50 bacterial families to unveil the relationship with soil properties. In detail, heatmap analysis showed that the classes Bacteroidetes, Phycisphaerae, Gemmatimonadetes, and unclassified Chloroflexi were positively related to SOC, TN, and EC, in contrast to α-, δ-Proteobacteria, and Flavobacteriia (Figure 6B). Moreover, the bacterial classes (e.g., Flavobacteriia, Verrucomicrobiae, Phycisphaerae) were significantly affected by SMC and pH. At the family level, the result showed that 26 bacteria were closely related to SOC, TN, EC, and SMC (Figure 6C). The majority of them were the dominant class α-, δ-, γ-Proteobacteria, Gemmatimonadetes, Bacteroidetes, and Flavobacteriia, suggesting that the soil properties of SOC, TN, EC, and SMC were the important environmental factors to alter the bacterial community structures in the five micro-habitats.
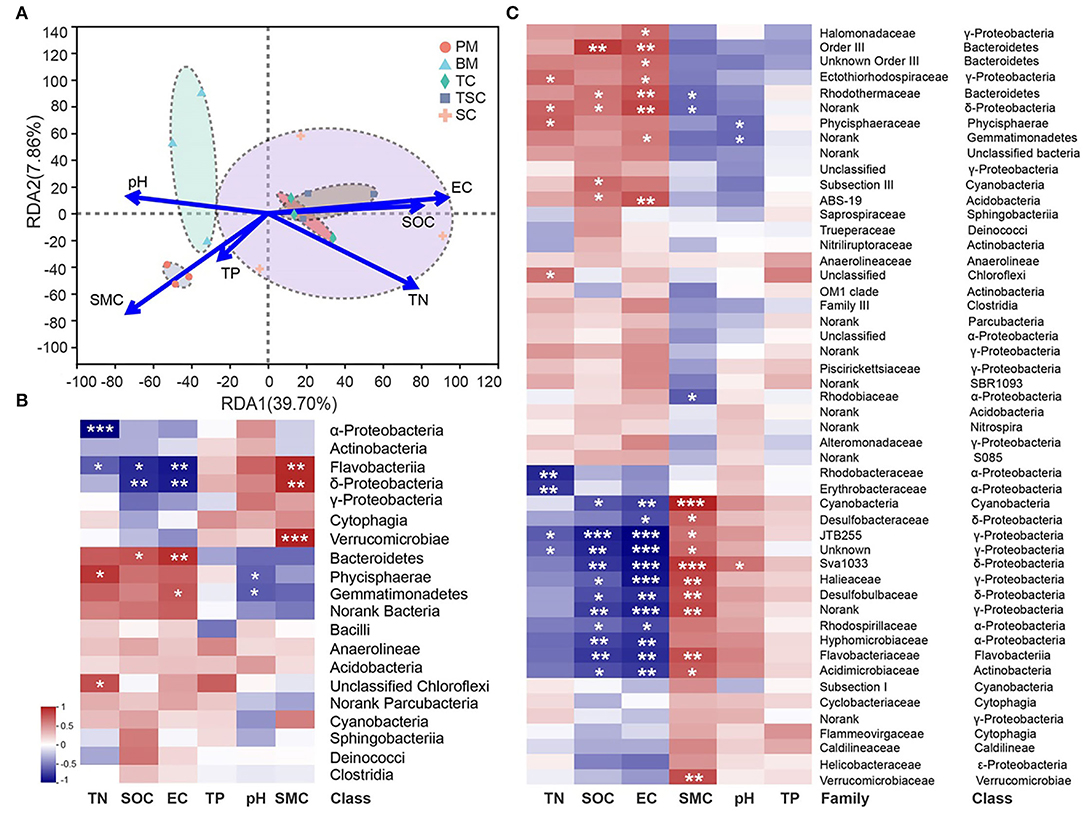
Figure 6. (A) Redundancy analysis (RDA) between soil properties and bacterial community structures at class level. The correlation heatmaps of soil properties and dominant bacteria at class (B) and family (C) level. The symbol (*) indicates the significantly different bacteria. The * symbol indicates the value of p < 0.05, the ** symbol indicates the value of p < 0.01, and the *** symbol indicates the value of p < 0.001.
Discussion
Tidal creek salt marshes have characterized as frequently tidal flooding and strongly hydrological connectivity, and shape active and uneven ecosystems with several small and diverse micro-habitats (Mooraki et al., 2009; Cabezas et al., 2017; Wang et al., 2021). Some studies have shown that these micro-habitats were important to maintain health estuary wetland eco-functions, such as carbon capture and storage, wetland restoration and buffering disturbances (Wu et al., 2020; Glaser et al., 2021). In field investigation, there were five micro-habitats of pluff mudflat, bare mudflat, T. chinensis, T. chinensis-S. salsa, and S. salsa around the tidal creek. This may be due to that tidal creek greatly altered microtopography, soil seed colonization, and vegetation succession via tidal fluctuation in the estuary wetland (Chang et al., 2010; Zhu et al., 2021). These micro-habitats around tidal creeks generated significantly different soil properties of soil moisture, salinity, and nutrients (Table 1). Because of near tidal creeks and frequent tidal flooding, SMC of the lower micro-habitats (pluff and bare mudflats) was higher. However, the upper micro-habitats (T. chinensis, T. chinensis-S. salsa, and S. salsa communities) were suffered by short-time flooding and strong evaporation, which should be the major reasons and thus to aggregate the higher EC and lower SMC in these surface soils (Kearney and Fagherazzi, 2016; Zhou et al., 2020). Moreover, soil nutrients of the lower micro-habitats were taken away by tidal fluctuation, meanwhile accumulated in the upper micro-habitats due to litter decompositions (Chomel et al., 2016; Koceja et al., 2021; Yang et al., 2021).
The present study found the bacterial community structures were significantly different in the five micro-habitats on the tidal creek sections, and these differences may be greatly affected by the tidal action and soil properties together. On one hand, the bacterial community structures of lower micro-habitats (PM and BM) had more diversities than these of upper micro-habitats (TC, TSC, and SC). This may be due to that more frequent tidal action contributed to the passive dispersal of bacterial taxa via the marine bacteria. It has been confirmed that several marine bacteria were enrichment in PM and BM, including the dominant bacterial families JTB 255 (γ-Proteobacteria) (Dyksma et al., 2015), Flavobacteriaceae (Flavobacteriia) (Cho and Giovannoni, 2004), and OM1 clade (Actinobacteria) (Morris et al., 2012). In addition, the soil properties may contribute main influence to the differences in bacterial community structures (Figure 6A). Although pH was regarded as the most important environmental factor to affect soil bacterial communities (Fierer, 2017; Xun et al., 2019), the soil nutrient, salinity, and moisture were more important to the bacterial community structures in our study. Many pieces of evidence have supported our result that the soil nutrient availability and salinity were the predominant reasons for bacterial communities in estuary wetland (An et al., 2019; Li et al., 2019; Rath et al., 2019; Zhang et al., 2020). On the other hand, the differences in bacterial community structures were also exhibited in dominant bacterial taxa, in particular with a high variable in the α-, δ-, γ-Proteobacteria, Bacteroidetes, Gemmatimonadetes, Flavobacteriia, Acidobacteria, and Sphingobacteriia, accounting for about 70% (Figure 3C). Indeed, the study found EC, SMC, SOC, and TN may be greatly responsible for them between the lower and upper micro-habitats (Figures 6B,C).
Recent years, SRB and AAPB have got many attentions on sulfur cycling, carbon mineralization, and sequestration in the coastal wetland (Stegman et al., 2014; Jørgensen et al., 2019; Rolando et al., 2022). The present study found SRB and AAPB were characterized as the bacterial biomarkers in tidal creek salt marsh, and may partly result in the differences of structures and functions. SRB were widely distributed in anoxic environments, and the lower micro-habitats (PM and BM) were suffered by long-time seawater flooding and abundance , which were suitable for the SRB groups (Zeleke et al., 2013). Our study found that SRB of 10 genera were great enrichment, which suggests that they may be the potential indicators of the sulfur cycle in lower micro-habitats. Moreover, AAPB were genetically diverse in Proteobacteria (Bryant and Frigaard, 2006; Ritchie and Johnson, 2012), in agreement with our results across α-, β-, and γ-Proteobacteria (Figure 5). Some studies reported that Rhodobacteraceae were high in abundance up to 25% of the total bacterial community in seawater (Buchan et al., 2005; Lenk et al., 2012). In the present study, the Rhodobacteraceae (Actibacterium, Marivita, Roseivivax, Roseovarius, and Seohaeicola) were greatly enriched in the lower micro-habitats (PM and BM), which may be associated with dispersal between the lower micro-habitats and seawater. In addition, the Rhodospirillaceae was abundant in upper micro-habitats (TC, TSC, and SC). Several studies have reported that Rhodospirillaceae was ability to N2 fixation and positive relation of soil nutrient contents and plant successions (Madigan et al., 1984; Ding et al., 2017).
Conclusion
Our study found that the bacterial communities were greatly diverse and heterogeneous in the tidal creek salt marsh, and the diversities were greatly altered by the effects of tidal action, soil salinity, moisture, and nutrients. In particular, the important biomarker taxa of SRB and AAPB groups were close correspondence in different micro-habitats. These results provide fundamental insights into unveiling bacterial community diversities and important bacterial biomarkers of different micro-habitats.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found at NCBI: http://www.ncbi.nlm.nih.gov/bioproject/851955, or with the accession number: PRJNA851955.
Author Contributions
ZW: conceptualization, writing—original draft, and editing. KY: resources and formal analysis. DZ, YL, BG, YY, XW, ZR, WW, and XC: investigation and data curation. JYa: visualization. JYu: supervision and review. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (U1806218, 41871087, 31800011, and U2106214). Moreover, the Coastal Resources and Environment Team for Blue-Yellow Area as follow: The Project of the Cultivation Plan of Superior Discipline Talent Teams of Universities in Shandong Province-The Coastal Resources and Environment Team for Blue-Yellow Area.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.950605/full#supplementary-material
References
An, J. X., Liu, C., Wang, Q., Yao, M. J., Rui, J. P., Zhang, S. H., et al. (2019). Soil bacterial community structure in Chinese wetlands. Geoderma. 15, 290–299. doi: 10.1016/j.geoderma.2018.09.035
Arriola, J. M., and Cable, J. E. (2017). Variations in carbon burial and sediment accretion along a tidal creek in a florida salt marsh. Limnol. Oceanogr.62, S15–S28. doi: 10.1002/lno.10652
Bryant, D. A., and Frigaard, N. U. (2006). Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14, 488–496. doi: 10.1016/j.tim.2006.09.001
Buchan, A., González, J. M., and Moran, M. A. (2005). Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005
Cabezas, A., Mitsch, W. J., Macdonnell, C., Zhang, L., Bydatek, F., and Lasso, A. (2017). Methane emissions from mangrove soils in hydrologically disturbed and reference mangrove tidal creeks in southwest Florida. Ecol. Eng. 114, 57–65. doi: 10.1016/j.ecoleng.2017.08.041
Chang, E. R., Veeneklaas, R. M., and Bakker, J. P. (2010). Seed dynamics linked to variability in movement of tidal water. J. Veg. Sci. 18, 253–262. doi: 10.1111/j.1654-1103.2007.tb02536.x
Chirol, C., Haigh, I. D., Pontee, N., and Thompson, C. E. L. (2018). Parametrizing tidal creek morphology in mature saltmarshes using semi automated extraction from lidar. Remote Sens. Environ. 209, 291–311. doi: 10.1016/j.rse.2017.11.012
Cho, J. C., and Giovannoni, S. J. (2004). Robiginitaleabiformata gen. nov., sp. nov., a novel marine bacterium in the family Flavobacteriaceae with a higher G+C content. Int. J. Syst. Evol. 54, 1101–1106. doi: 10.1099/ijs.0.03023-0
Chomel, M., Guittonny-Larchevêque, M., Fernandez, C., Gallet, C., DesRochers, A., Par,é, D., et al. (2016). Plant secondary metabolites: a key driver of litter decomposition and soil nutrient cycling. J. Ecol. 104, 1527–1541. doi: 10.1111/1365-2745.12644
Coban, O., De Deyn, G. B., and van der Ploeg, M. (2022). Soil microbiota as game-changers in restoration of degraded lands. Science. 375. doi: 10.1126/science.abe0725
Cui, B. S., Cai, Y. Z., Xie, T., Ning, Z. H., and Hua, Y. Y. (2016). Ecological effects of wetland hydrological connectivity: problems and prospects. J. Beijing Normal Univ. 52, 738–746. doi: 10.16360/j.cnki.jbnuns.2016.06.011
Ding, L.j, Su, J. Q., Li, H., Zhu, Y. G., and Cao, Z. H. (2017). Bacterial succession along a long-term chronosequence of paddy soil in the Yangtze River Delta, China. Soil Biol. Biochem. 104, 59–67. doi: 10.1016/j.soilbio.2016.10.013
Dyksma, S., Bischof, K., Fuchs, B. M., Hoffmann, K., Meier, D., Meyerdierks, A., et al. (2015). Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 10, 1939–1953. doi: 10.1038/ismej.2015.257
Fennessy, S. (2014). Wetland ecosystems and global change. Global Environm. Chan. 255–261. doi: 10.1007/978-94-007-5784-4_129
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Glaser, C., Frei, S., Massmann, G., and Gilfedder, B. S. (2021). Tidal creeks as hot-spots for hydrological exchange in a coastal landscape. J. Hydrol. 597, 126158. doi: 10.1016/j.jhydrol.2021.126158
Gong, Z. H., Mou, K. N., Wang, Q. W., Qiu, H. C., Zhang, C., and Zhou, D. M. (2021). Parameterizing the Yellow River Delta tidal creek morphology using automated extraction from remote sensing images. Sci. Total Environ. 769, 144–572. doi: 10.1016/j.scitotenv.2020.144572
Huang, L. B., Bai, J. H., Wen, X. J., Zhang, G. L., Zhang, C. D., Cui, B. S., et al. (2020). Microbial resistance and resilience in response to environmental changes under the higher intensity of human activities than global average level. Global Change Biol. 26, 2377–2389. doi: 10.1111/gcb.14995
Jørgensen, B. B., Findlay, A. J., and Pellerin, A. (2019). The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 10, 849. doi: 10.3389/fmicb.2019.00849
Kearney, W., and Fagherazzi, S. (2016). Salt marsh vegetation promotes efficient tidal channel networks. Nat. Commun. 7, 12287. doi: 10.1038/ncomms12287
Kim, D. (2018). Modeling spatial and temporal dynamics of plant species richness across tidal creeks in a temperate salt marsh. Ecol. Indic. 93, 188–195. doi: 10.1016/j.ecolind.2018.04.080
Koceja, M. E., Bledsoe, R. B., Goodwillie, C., and Peralta, A. L. (2021). Distinct microbial communities alter litter decomposition rates in a fertilized coastal plain wetland. Ecosphere. 12, 1–18. doi: 10.1002/ecs2.3619
Lenk, S., Moraru, C., Hahnke, S., Arnds, J., Richter, M., Kube, M., et al. (2012). Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J. 6, 2178–2187. doi: 10.1038/ismej.2012.66
Li, H., Chi, Z. F., Li, J. L., Wu, H. T., and Yan, B. X. (2019). Bacterial community structure and function in soils from tidal freshwater wetlands in a Chinese delta: Potential impacts of salinity and nutrient. Sci. Total Environ. 696, 134029. doi: 10.1016/j.scitotenv.2019.134029
Li, J. Y., Chen, Q. F., Li, Q., Zhao, C. S., and Feng, Y. (2021). Influence of plants and environmental variables on the diversity of soil microbial communities in the Yellow River Delta wetland, China. Chemosphere. 274, 129967. doi: 10.1016/j.chemosphere.2021.129967
Liu, L. Y., Qu, F. Z., Li, Y. Z., Yu, J. B., Yang, J. S., and An, C. B. (2020). Correlation between creek tidal distribution and vegetation coverage in the Yellow River Delta coastal wetland. Chinese J. Ecol. 39, 1830–1837. doi: 10.13292/j.1000-4890.202006.006
Lv, X. F., Ma, B., Yu, J. B., Chang, S. X., Xu, J. M., Li, Y. Z., et al. (2016). Bacterial community structure and function shift along a successional series of tidal flats in the Yellow River Delta. Sci. Rep. 6, 36550. doi: 10.1038/srep36550
Madigan, M., Cox, S. S., and Stegeman, R. A. (1984). Nitrogen fixation and nitrogenase activities in members of the family Rhodospirillaceae. J. Bacteriol. 157, 73–78. doi: 10.1128/jb.157.1.73-78.1984
Mallin, M. A., and Lewitus, A. J. (2004). The importance of tidal creek ecosystems. J. Exp. Mar. Biol. Ecol. 298, 145–149. doi: 10.1016/S0022-0981(03)00356-3
Mooraki, N., Sari, E. A., Soltani, M., and Valinassab, T. (2009). Spatial distribution and assemblage structure of macrobenthos in a tidal creek in relation to industrial activities. Int. J. Environm. Sci. Technol. 6, 651–662. doi: 10.1007/BF03326106
Morris, R. M., Frazar, C. D., and Carlson, C. A. (2012). Basin-scale patterns in the abundance of SAR11 subclades, marine Actinobacteria (OM1), members of the Roseobacter clade and OCS116 in the South Atlantic. Environ. Microbiol. 14, 1133–1144. doi: 10.1111/j.1462-2920.2011.02694.x
Murray, N. J., Phinn, S. R., DeWitt, M., Ferrari, R., Johnston, R., Lyons, M. B., Clinton, N., Thau, D., and Fuller, R. A. (2019). The global distribution and trajectory of tidal flats. Nature. 565, 222–225. doi: 10.1038/s41586-018-0805-8
Rath, K. M., Fierer, N., Murphy, D. V., and Rousk, J. (2019). Linking bacterial community composition to soil salinity along environmental gradients. ISME J. 13, 836–846. doi: 10.1038/s41396-018-0313-8
Ritchie, A. E., and Johnson, Z. I. (2012). Abundance and genetic diversity of aerobic anoxygenic phototrophic bacteria of coastal regions of the Pacific ocean. Appl. Environ. Microbiol. 78, 2858–2866. doi: 10.1128/AEM.06268-11
Rolando, J. L., Kolton, M., Song, T., and Kostka, J. E. (2022). The core root microbiome of Spartina alterniflora is predominated by sulfur-oxidizing and sulfate-reducing bacteria in Georgia salt marshes, USA. Microbiome. 10, 37. doi: 10.1186/s40168-021-01187-7
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Stegman, M. R., Cottrell, M. T., and Kirchman, D. L. (2014). Leucine incorporation by aerobic anoxygenic phototrophic bacteria in the Delaware estuary. ISME J. 8, 2339–2348. doi: 10.1038/ismej.2014.75
Tan, L. S., Ge, Z. M., Fei, B. L., Xie, L. N., Li, Y. L., Li, S. H., et al. (2020). The roles of vegetation, tide and sediment in the variability of carbon in the salt marsh dominated tidal creeks. Estuar. Coast. Shelf Sci. 239, 106752. doi: 10.1016/j.ecss.2020.106752
Trifunovic, B., Vázquez-Lule, A., Capooci, M., Seyfferth, A. L., Moffat, C., and Vargas, R. (2020). Carbon dioxide and methane emissions from temperate salt marsh tidal creek. Journal of Geophysical Research: Biogeosciences. 125, 1–16. doi: 10.1029/2019JG005558
Vandenbruwaene, W., Meire, P., and Temmerman, S. (2012). Formation and evolution of a tidal channelnetwork within a constructed tidal marsh. Geomorphology, 151-152:–No match found–114–125. doi: 10.1016/j.geomorph.2012.01.022
Wang, Q., Xie, T., Ning, Z. H., Chen, C., Man, Y., and Cui, B. S. (2021). Enhancement of lateral connectivity promotes the establishment of plants in saltmarshes. Sci. Total Environ. 767, 145484. doi: 10.1016/j.scitotenv.2021.145484
Wilson, C. A., Hughesb, Z. J., FitzGeraldb, D. M., Hopkinsonc, C. S., Valentined, V., and Kolkere, A. S. (2014). Saltmarsh pool and tidal creek morphodynamics: dynamic equilibrium of northern latitude saltmarshes? Geomorphology. 213, 99–115. doi: 10.1016/j.geomorph.2014.01.002
Wu, Y. N., Liu, J. K., Yan, G. X., Zhai, J. X., Cong, L., Dai, L. Y., Zhang, Z. M., and Zhang, M. X. (2020). The size and distribution of tidal creeks affects salt marsh restoration. J. Environ. Manage. 259, 110070. doi: 10.1016/j.jenvman.2020.110070
Xiao, D. R., Deng, L., Kim, D. G., Hang, C. B., and Tian, K. (2019). Carbon budgets of wetland ecosystems in china. Global Change Biology. 25. doi: 10.1111/gcb.14621
Xun, W. B., Li, W., Xiong, W., Ren, Y., Liu, Y. P., Miao, Y. Z., et al. (2019). Diversity-triggered deterministic bacterial assembly constrains community functions. Nature Communicat. 10, 3833. doi: 10.1038/s41467-019-11787-5
Yang, J. S., Zhan, C., Li, Y. Z., Zhou, D., Yu, Y., and Yu, J. B. (2018). Effect of salinity on soil respiration in relation to dissolved organic carbon and microbial characteristics of a wetland in the Liaohe River estuary, Northeast China. Sci. Total Environ. 642, 946–953. doi: 10.1016/j.scitotenv.2018.06.121
Yang, X. L., Wang, X. T., Xiao, S., Liu, Z. Y., Zhou, X. H., Du, G. Z., et al. (2021). Dominant plants affect litter decomposition mainly through modifications of the soil microbial community. Soil Biol. Biochem. 161, 108399. doi: 10.1016/j.soilbio.2021.108399
Yu, X. J., Zhang, Z. S., Xue, Z. S., Song, X. L., Zhang, H. R., and Wu, H. T. (2018). Morphological characteristics and connectivity of tidal channels in the yellow river delta for 7 periods since 1989. Wetland Sci. 16, 517–523. doi: 10.13248/j.cnki.wetlandsci.2018.04.010
Zeleke, J., Sheng, Q., Wang, J. G., Huang, M. Y., Xia, F., Wu, J. H., et al. (2013). Effects of Spartina alterniflora invasion on the communities of methanogens and sulfate-reducing bacteria in estuarine marsh sediments. Front. Microbiol. 4, 243. doi: 10.3389/fmicb.2013.00243
Zhang, G. L., Bai, J. H., Tebbe, C. C., Zhao, Q. Q., Jia, J., Wang, W., et al. (2020). Salinity controls soil microbial community structure and function in coastal estuarine wetlands. Environ. Microbiol. 23, 1020–1037. doi: 10.1111/1462-2920.15281
Zhang, H. X., Zheng, S., Ding, J. W., Wang, O. M., and Liu, F. H. (2017). Spatial variation in bacterial community in natural wetland-river-sea ecosystems. J. Basic Microbiol. 57, 536–546. doi: 10.1002/jobm.201700041
Zhao, X. S., Cui, B. S., Sun, T., and He, Q. (2010). The relationship between the spatial distribution of vegetation and soil environmental factors in the tidal creek areas of the Yellow River Delta. Ecol. Environm. Sci. 19, 1855–1861. doi: 10.16258/j.cnki.1674-5906.2010.08.009
Zhou, T. Z., Xin, P., Li, L., Barry, D. A., and Šimunekb, J. (2020). Effects of large macropores on soil evaporation in salt marshes. J. Hydrol. 584, 124754. doi: 10.1016/j.jhydrol.2020.124754
Zhu, Z. C., Bouma, T. J., Zhu, Q., Cai, Y. P., and Yang, Z. F. (2021). Effects of waves and sediment disturbance on seed bank persistence at tidal flats. Front. Mar. Sci. 8, 728065. doi: 10.3389/fmars.2021.728065
Keywords: tidal creek, micro-habitats, bacterial community, sulfate-reducing bacteria, ecological functions
Citation: Wang Z, Yang K, Yu J, Zhou D, Li Y, Guan B, Yu Y, Wang X, Ren Z, Wang W, Chen X and Yang J (2022) Soil Bacterial Community Structure in Different Micro-Habitats on the Tidal Creek Section in the Yellow River Estuary. Front. Ecol. Evol. 10:950605. doi: 10.3389/fevo.2022.950605
Received: 23 May 2022; Accepted: 22 June 2022;
Published: 01 August 2022.
Edited by:
Chuanyu Gao, Northeast Institute of Geography and Agroecology (CAS), ChinaReviewed by:
Lei Qin, Northeast Institute of Geography and Agroecology (CAS), ChinaXiaofei Lv, China Jiliang University, China
Copyright © 2022 Wang, Yang, Yu, Zhou, Li, Guan, Yu, Wang, Ren, Wang, Chen and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junbao Yu, anVuYmFvLnl1QGdtYWlsLmNvbQ==; Jisong Yang, eWFuZ2ppc29uZ0BsZHUuZWR1LmNu
 Zhikang Wang
Zhikang Wang Kaixin Yang
Kaixin Yang Junbao Yu
Junbao Yu Di Zhou1
Di Zhou1 Yunzhao Li
Yunzhao Li Bo Guan
Bo Guan Yang Yu
Yang Yu Jisong Yang
Jisong Yang