- 1Key Laboratory of Pollinating Insect Biology, Institute of Apicultural Research, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Apiculture Institute of Jiangxi Province, Nanchang, China
Vespa velutina nigrithorax and Vespa velutina auraria are two subspecies of Vespa velutina Lepeletier. V. velutina preys managed honey bees, other pollinators, and insects. However, the Vespa midgut microbiota of three forms, namely queens, workers, and males have not been reported, thus the objective of this study was to analyze the midgut bacterial diversity of the three forms of V. v. nigrithorax and V. v. auraria. Our results showed that Proteobacteria, Firmicutes, Bacteroidetes, Tenericutes, and Actinobacteria were the most abundant phyla, and Lactobacillus (17.21%) and Sphingomonas (11.39%) were the most abundant genera in the midgut of V. v. nigrithorax and V. v. auraria. We found that the midgut bacterial compositions of the V. velutina males were special, in terms of richness and diversity of bacterial communities, as well as the content of lactic acid bacteria. By comparing the gut bacterial compositions of Vespa from different regions (Japan, South Korea, Italy, and China), it was discovered that the gut bacterial compositions were very similar at the phylum and class level, and Gammaproteobacteria, Bacilli, and Alphaproteobacteria were the most abundant classes of bacteria and consistent in the genus Vespa. Besides, though Vespa from different regions had quite different gut bacterial communities at the genus level, Lactobacillus and other lactic acid bacteria were abundant and played important roles in protection and metabolism in V. velutina collected from different regions. This is the first report of midgut bacterial diversity of three forms queens, workers, and males of V. velutina. Our findings provide insight that Proteobacteria and Firmicutes (especially Lactobacillus and other lactic acid bacteria) are consistent and may play important roles in the genus Vespa. The understanding of the microbiome in the midgut of Vespa and the discovery of the vital bacteria would provide useful information to design pest biological control agents. Thus, the significance of this study is to provide a basis for the study of the relationship between gut microbiota and physiology and health of Vespa, as well as the control of Vespa.
Introduction
Vespa velutina Lepeletier is an Asian hornet that includes three forms, namely queens, workers, and males. The hornet heavily preys Apis mellifera (Tan et al., 2007; Monceau et al., 2013), and Apis cerana (Tan et al., 2007) in both native and invaded areas. Some reviews have summarized the information on V. velutina (Monceau et al., 2014; Kishi and Goka, 2017).
Vespa velutina nigrithorax du Buysson and Vespa velutina auraria Smith are subspecies of V. velutina (Archer, 1993; Nguyen et al., 2006). The yellow-legged hornet, V. v. nigrithorax has established its population in parts of Europe (Villemant et al., 2011) including France (Villemant et al., 2011; Robinet et al., 2017), Spain (Lopez et al., 2011; Goldarazena et al., 2015; Leza et al., 2018), Italy (Bertolino et al., 2016), Germany (Bertolino et al., 2016), the United Kingdom (Budge et al., 2017), and the Netherlands and Switzerland (Robinet et al., 2019). V. v. nigrithorax has also established its population in South Korea (Kim et al., 2006; Choi et al., 2012, 2013). The invasion risk of V. v. nigrithorax has been assessed by Villemant et al. (2011) and Barbet-Massin et al. (2013). The yellow-legged hornet preys on managed honey bees and other insects (Monceau et al., 2013, 2014; Arca et al., 2014, 2015).
Recently, many studies focused on the hornet such as microsatellite variation (Choi et al., 2013), the complete mitochondrial genome (Kim et al., 2017), rapid molecular methods for identification (Stainton et al., 2018), flight capacities (Sauvard et al., 2018), control (Turchi and Derijard, 2018), as well as the gut microbiota. Gut bacteria play an essential role in nutrition, digestion, detoxification, memory variation, and the efficiency of protection against pathogens (Cardoza et al., 2006; Engel et al., 2012), for example, bacteria in honey bee gut (Engel and Moran, 2013; Kwong et al., 2014; Jia et al., 2016, 2017; Li et al., 2021). Suenami et al. (2019) characterized and compared the gut microbiome in two hornet species, Vespa mandarinia and Vespa simillima. The two species have simple gut microbiota, composed of seven or eight “core” operational taxonomic units (OTUs). Seo et al. (2018) briefly compared the intestine bacterial microbiota of Asian hornets (V. v. nigrithorax) and honey bees and found out that the relative ratio of bacterial populations of Asian hornets was different from that of honey bees. However, the intestine bacterial diversity in three forms of hornet has not been investigated. Although the gut microbial composition in different castes of the V. v. nigrithorax was reported recently (Cini et al., 2020), the gut microbial composition of the males, one of the three forms, was not investigated. Besides, the gut microbial community of V. v. auraria has also not been reported. Furthermore, the midgut is the largest digestive organ of bees, and the microbes in it are very important for their health and fitness, while the above studies collected the whole intestine as samples and did not focus on the microbial composition in the midgut of V. velutina. Therefore, the studies of Vespa midgut bacteria are still limited.
In this study, we analyzed the midgut bacterial communities of queens, workers, and males of V. v. nigrithorax and V. v. auraria through sequencing technology. We found that the midgut bacterial compositions of the V. velutina males were special, in terms of richness and diversity of bacterial communities, as well as the content of lactic acid bacteria. When considering the queens, differences in forms did not seem to have an obvious effect on the midgut bacterial communities of V. velutina. Moreover, the midgut bacterial communities of V. v. nigrithorax and V. v. auraria were similar. Besides, the gut bacterial compositions of Vespa from different species or countries were quite different at the genus level. It was remarkable that Lactobacillus and other lactic acid bacteria were abundant and might play important roles in studies related to V. velutina. Here we provide a characterization of the midgut bacterial composition of V. velutina and found the specificity of the midgut microbiome of V. velutina males. The understanding of the microbiome in the midgut of Vespa and the discovery of the vital bacteria would provide useful information to design a more efficient method of control through the development of a set of natural enemies as biological control agents.
Materials and Methods
Vespa Samples
The experimental samples were collected in November 2018, V. v. auraria from Shijingshan (N40°13′35.10″, E116°04′15.63″) in Beijing, and V. v. nigrithorax from Nanchang (N28°25′9.78″, E114°41′29.94″) in Jiangxi Province, China.
Dissection and DNA Extraction
The queens, workers, and males of V. v. auraria and V. v. nigrithorax were dissected. Five males, five queens, and five workers were taken from each of the five colonies of V. v. nigrithorax and five colonies of V. v. auraria. All of the queen samples in this study were really gynes. A total of 150 individuals, which included 75 samples of each subspecies, were placed in the refrigerator for 60 s at −20°C, and the midguts were dissected on ice using sterile forceps. Five midguts were pooled in a 2 ml sterile centrifuge tube and treated with liquid nitrogen before being stored in the freezer at −80°C for subsequent DNA extraction. The DNA of midgut bacteria was extracted referring to the previous methods (Dai et al., 2018). Total genomic DNA was quantified by BioSpectrometerR kinetic (Eppendorf, Germany), and their integrity was verified using agarose electrophoresis. The bacterial 16S rRNA gene V3–V4 region was targeted with the primer pair 341f/806r (341F: CCTAYGGGRBGCASCAG, 806R: GGACTACNNGGGTATCTAAT) for the microbial community diversity analysis, and the purified products were sequenced on the Illumina HiSeq 2500 platform at Novogene Bioinformatics Technology Co., Ltd., Beijing, China (Dai et al., 2018). The obtained sequences were normalized to make the samples comparable at the same sequencing depth for the following analysis. To identify specific taxa in the midgut microbiota, we performed a linear discriminant analysis effect size (LEFSe) analysis that revealed changes in the abundance of bacterial taxa accounted for the observed differences in the midgut microbiota.
Statistics
The normalized sequences were classified into OTUs at 97% similarity using UCLUST (Version 1.2.22). The taxonomy of the OTUs was assigned by blasting against SILVA database 132 with default parameters. Alpha diversity observed species (richness) and the Shannon index (diversity indices) were performed with Mothur (Version v.1.30). Beta diversity including unweighted unifrac distances between samples was performed with QIIME (Version 1.8.0). The UPGMA (Unweighted Pair-Group Method with Arithmetic Mean) cluster analysis was performed using the Unweighted Unifrac Distance matrix.
Results
Sequencing Results and Quality Control
Paired-end sequencing of 16S rRNA V3–V4 gene was produced from 30 samples. After trimming the barcodes, primers and filtering chimeras, short and low-quality reads, 75,715 effective sequences were obtained, and the average length of effective sequence reads was 416 bp. In all samples of three forms of V. v. nigrithorax and V. v. auraria, 93.9% of the sequences were assigned to the phylum level, 90.6% to the class level, 84.4% to the order level, 76.6% to the family level, 51.0% to the genus level, and 16.4% to the species level.
Alpha Diversity
Beeswarm plots of observed OTUs and the Shannon index for individual sample groups are shown in Figure 1. There were significant differences between V. v. auraria males and workers (p = 0.006), between males of V. v. auraria and V. v. nigrithorax (p = 0.007) in the observed species (Figure 1A). There were significant differences between V. v. auraria males and workers (p = 0.0141), and between V. v. nigrithorax males and workers (p = 0.0155) in the Shannon index (Figure 1B). It was suggested that in V. v. auraria, males showed significantly higher richness rank and a higher level of diversity compared to the workers. On the contrary, in V. v. nigrithorax, workers showed a significantly higher level of diversity compared to the males. Moreover, when comparing the two subspecies, the richness and diversity of bacterial communities in the males of V. v. auraria were significantly higher than that in the males of V. v. nigrithorax.
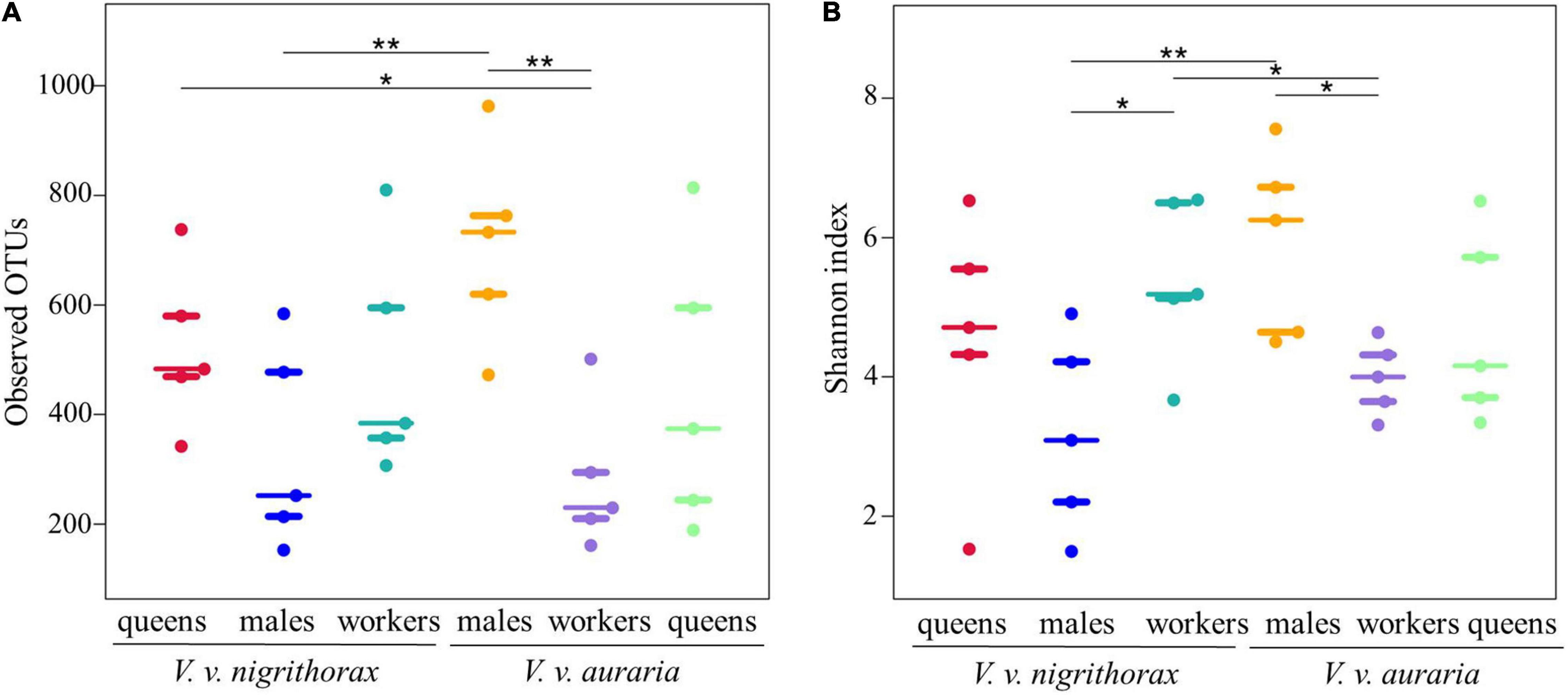
Figure 1. Alpha diversity in Vespa velutina nigrithorax and Vespa velutina auraria midgut bacteria. (A) The amount of bacterial diversity was determined by comparing observed operational taxonomic units (OTUs) and (B) Shannon index. “*” indicates significance at the 0.05 level, and “**” indicates significance at the 0.01 level.
Intestinal Bacterial Communities
Based on the average relative abundance, Proteobacteria (51.13%), Firmicutes (36.35%), Bacteroidetes (5.12%), Tenericutes (3.15%), and Actinobacteria (2.22%) were the most abundant phyla in the midgut of three types of V. v. nigrithorax and V. v. auraria (Figure 2). γ-Proteobacteria (32.51%), Bacilli (28.05%), and α-Proteobacteria (18.46%) were the highest abundant classes. Enterobacteriaceae (23.83%), Streptococcaceae (17.92%), and Sphingomonadaceae (11.46%) were the most abundant family. Major genera (abundant >1%) were Lactobacillus, Sphingomonas, Spiroplasma, Weissella, Serratia, Megamonas, Fructobacillus, Moraxella, and Bombella. Lactobacillus (17.21%) and Sphingomonas (11.39%) were the most abundant genera.
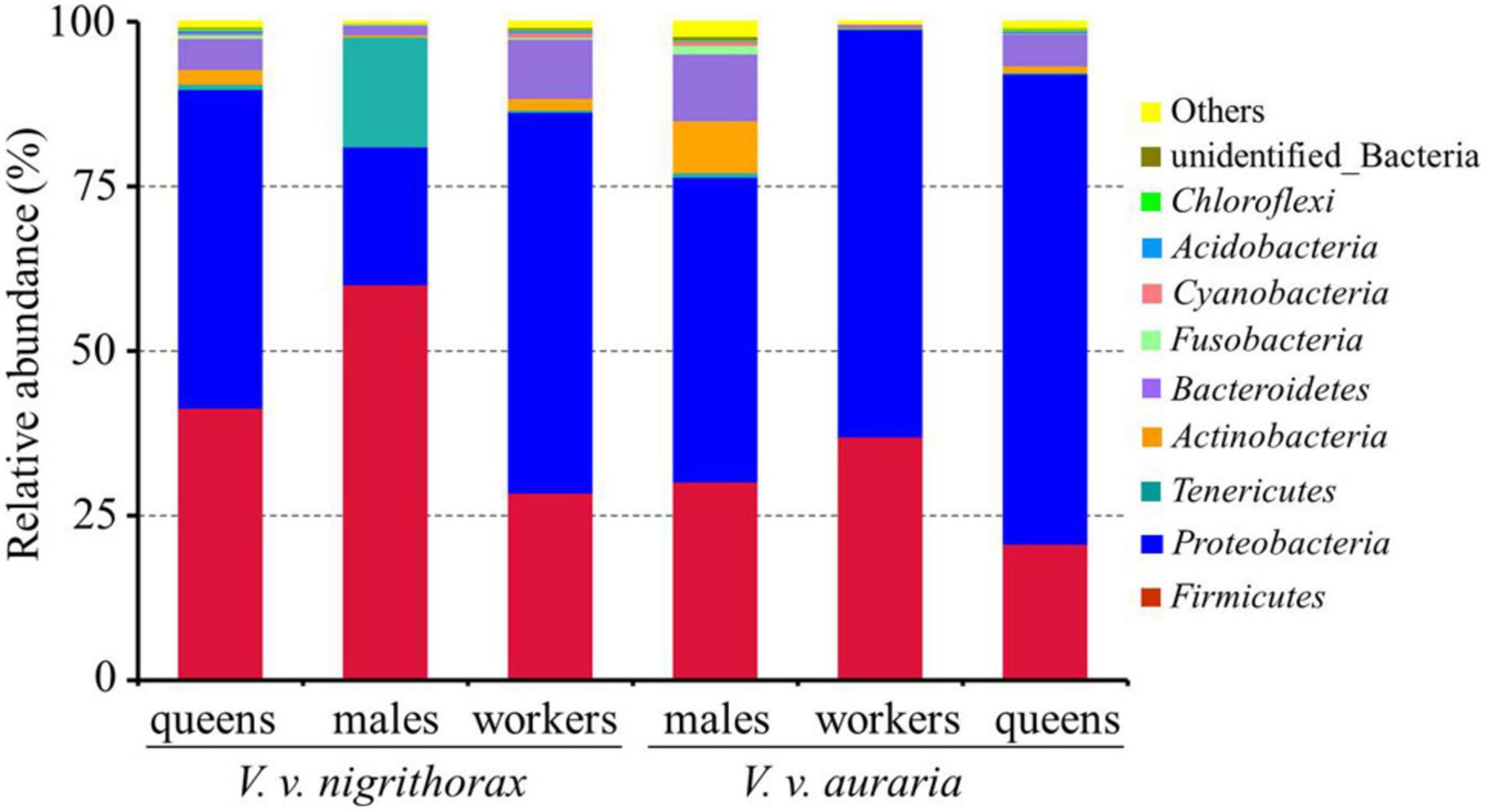
Figure 2. Relative abundance of the dominant midgut bacterial communities in Vespa velutina nigrithorax and Vespa velutina auraria at the phylum level. Each bar represents the average relative abundance of each bacterial taxon within a group.
Linear Discriminant Analysis Effect Size
To detect the classified bacterial taxa with significant abundance differences among the different forms of V. velutina, a biomarker analysis based on the linear discriminant analysis (LDA) effect size (LEfSe) method was used. As shown in Figure 3, 48 bacterial clades presented statistically significant differences with an LDA threshold of 4.0. The largest number of bacterial taxa were presented in the males of V. v. auraria (AD), followed by the workers of V. v. nigrithorax (NW). A total of 13 bacterial taxa were significantly enriched in the males of V. v. auraria, including Clostridiales, Bacteroidales, Actinobacteria, Lachnospiraceae, and Bacillales. Most of the above bacteria are involved in nutrient metabolism, especially for protein degradation, sugar fermentation, and defending insects against harmful microorganisms by producing antimicrobial compounds (Visser et al., 2012; Wang et al., 2020). In addition, 10 bacterial taxa were significantly enriched in the workers of V. v. nigrithorax. It was interesting to note that seven bacterial taxa were significantly enriched in the males of V. v. nigrithorax (ND), and most of them belonged to lactic acid bacteria, including Lactococcus, Streptococcaceae, Lactobacillales, Lactococcus lactis, and Weissella cibaria, which are considered beneficial bacteria and are associated with fermentation.
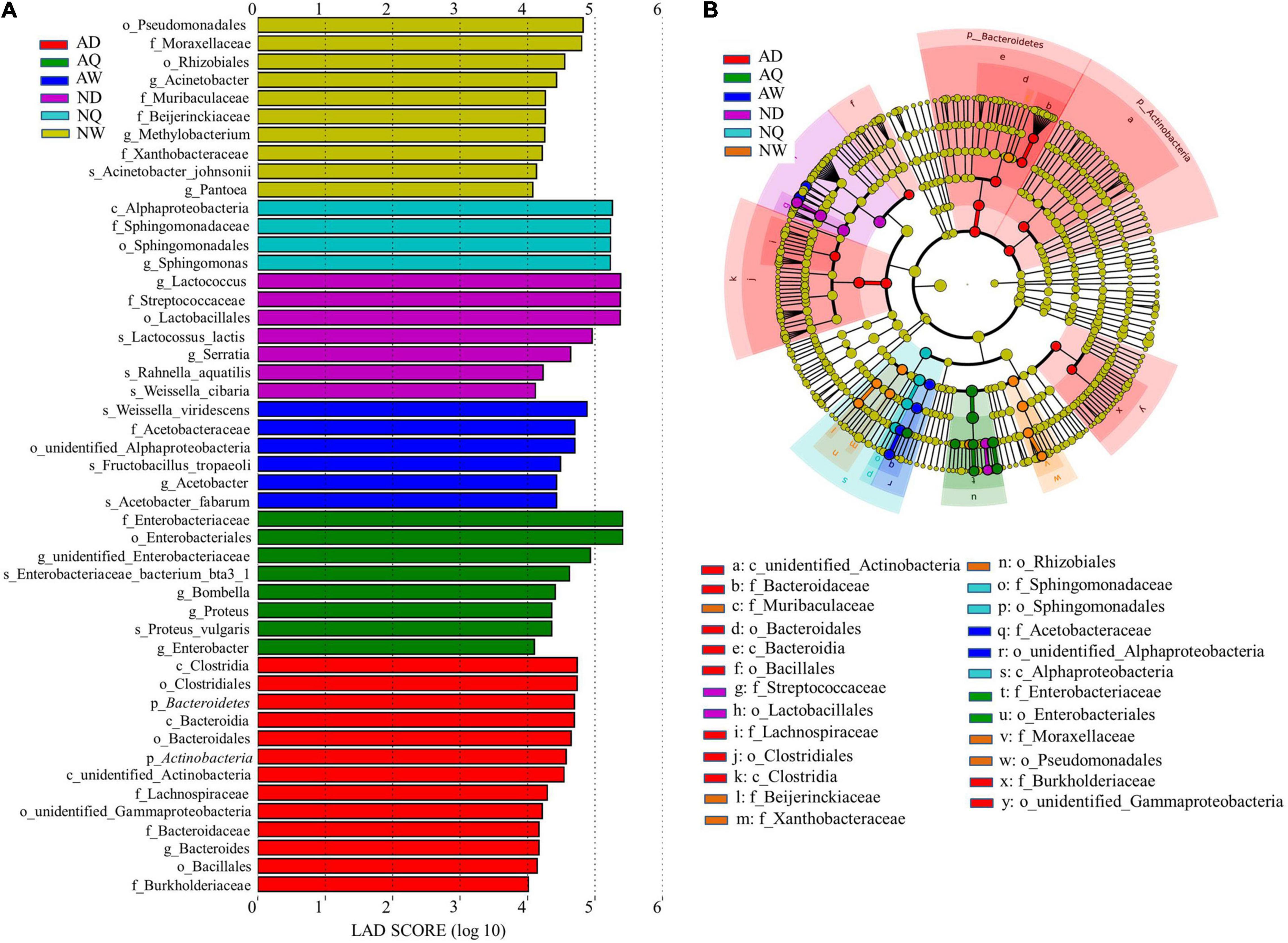
Figure 3. Linear discriminant analysis effect size (LEfSe) analysis of microbial abundance among samples of Vespa velutina nigrithorax and Vespa velutina auraria. (A) Linear discriminant analysis (LDA) score identified the size of differentiation among samples of V. v. nigrithorax and V. v. auraria with a threshold value of 4.0. (B) The cladogram of microbial communities. AD, V. v. auraria males; AQ, V. v. auraria queens; AW, V. v. auraria workers; ND, V. v. nigrithorax males; NQ, V. v. nigrithorax queens; NW, V. v. nigrithorax workers.
Discussion
Vespa velutina is considered a pest of A. cerana in parts of China (Tan et al., 2007), and an invasive species in Europe (Monceau et al., 2014). V. velutina may have adverse impacts on domestic Vespa species or other insect populations (Wilson et al., 2009; Beggs et al., 2011; Choi et al., 2012). This is the first report of midgut bacterial diversity of three forms queens, workers, and males of V. velutina.
The whole gut (midgut and hindgut) microbial composition of V. velutina has been reported by Cini et al. (2020). They found that microbial communities showed differences in composition across reproductive phenotypes (workers and future queens) (Cini et al., 2020). While in this study, the midgut bacterial composition of queens showed no significant differences in richness rank and level of diversity compared to that of the workers and males, so it seems that the differences in forms have no obvious effect on the midgut bacterial communities of V. velutina. Cini et al. (2020) also reported that the worker stage harbored a significantly higher number of bacterial OTUs compared to all other sample types (larva, newly emerged females, gyne, nest, and meconium), while there were no differences displayed by Shannon and Evenness indices among all sample types. However, in the present study, the richness and diversity of midgut bacterial communities of males were significantly higher than that of workers in V. v. auraria. In addition, the lactic acid bacteria were significantly enriched in the males of V. v. nigrithorax. Therefore, in V. velutina, the midgut bacterial communities of males were special and their specific roles in metabolism and health are also worthy of further study. Besides, based on the results of principal component analysis (PCA), principal coordinate analysis (PCoA) of Unweighted Unifrac Distance (Supplementary Figure 1), and cluster analysis (Supplementary Figure 2), it was found that the differences inside groups (queens, males, and workers) were greater than that between groups (V. v. nigrithorax and V. v. auraria), which indicated that the midgut bacterial communities of V. v. nigrithorax and V. v. auraria were similar.
Proteobacteria, Firmicutes, Bacteroidetes, Tenericutes, and Actinobacteria were the most abundant phyla in the midgut of V. v. nigrithorax and V. v. auraria. When comparing the gut bacterial compositions of the genus Vespa from different studies (Supplementary Table 1), it was found that the gut bacterial compositions were very similar at the phylum and class level, even if these samples were collected from different species (or subspecies) and regions (from different countries). In V. mandarinia, V. simillima, V. v. nigrithorax, and V. v. auraria, the most abundant and shared phyla of bacteria were Proteobacteria and Firmicutes, and the most abundant and shared classes of bacteria were Gammaproteobacteria, Bacilli, and Alphaproteobacteria. It was suggested that the above bacteria were consistent in the genus Vespa. However, when comparing the gut bacterial compositions at the low taxonomic levels, there were differences among different species. It was interesting to discover that the differences in gut bacterial compositions at the genus level were obvious. In our study, the gut bacteria compositions at the genus level were quite different from that in V. mandarinia, V. simillima (Suenami et al., 2019), and V. v. nigrithorax (Seo et al., 2018), but similar to that in V. v. nigrithorax (Cini et al., 2020). In fact, Bifidobacterium and Lactobacillus were the main abundant genera in V. v. nigrithorax (Cini et al., 2020), but were rare in the guts of V. mandarinia, V. simillima (Suenami et al., 2019) and V. v. nigrithorax (Seo et al., 2018). In this study, the main abundant genera included Lactobacillus and several other lactic acid bacteria except Bifidobacterium, thus consolidating their presence as one of the leading bacterial communities in the gut of V. velutina. The above results indicated that Vespa from different regions (Japan, South Korea, Italy, and China) have quite different gut bacterial communities at the genus level, thus we speculated that geographic location might affect the compositions of Vespa gut bacteria, but not at the high taxonomic levels, such as the level of phylum and class.
The gut microbiota is important for the health status of the host (Engel et al., 2012; Engel and Moran, 2013; Kwong et al., 2014). Gilliamella, Lactobacillus, and Bifidobacterium, which are the core genera in the gut of honey bees (Kwong and Moran, 2016; Ma et al., 2019), are also the main genera of bacteria in Vespa. It has been suggested that V. simillima gut is exposed to honey bee-derived microbial populations and likely selects for specific microbial groups (Suenami et al., 2019). Though Suenami hypothesized that the hornets do not require Lactobacillus and Bifidobacterium because they may already have alternative fermenters, Lactobacillus and Bifidobacterium are the two abundant genera of bacteria in V. v. nigrithorax and V. v. auraria (Cini et al., 2020 and this study). Besides, the results of this study showed that lactic acid bacteria were significantly enriched in the midgut of V. v. nigrithorax males. Recent studies show that Lactobacillus Firm-5 species enhances bees’ memory and honeybee gut Lactobacillus modulates host learning and memory behaviors via regulating tryptophan metabolism (Li et al., 2021; Zhang et al., 2022). Besides, Bifidobacterium spp. significantly contributes to the honey bee’s metabolism and health (Chen et al., 2021). Therefore, we presume that Lactobacillus and Bifidobacterium still play important roles in the protection and metabolism of V. velutina. It is necessary to carry out more research to verify the roles of Lactobacillus and Bifidobacterium in the life of V. velutina in the following study.
This discovery provides insight into the intestinal microbiota of V. velutina. Besides, further studies on the relationship between Vespa and gut microbial communities (especially Proteobacteria and Firmicutes) are necessary to elucidate how gut microbiota influence Vespa physiology and biology. On this basis, it will be feasible and meaningful to design pest biological control agents targeting the key microbes in the gut of Vespa to control them. One shortcoming of this study is that V. v. nigrithorax and V. v. auraria samples were collected from two sites in northern and southern China which were different in geographical environment and climatic conditions. However, it is rare to find two V. velutina subspecies in the same wild site.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: The datasets generated in this study can be found in the sequence read archive (SRA) archive in GenBank under the BioProject: PRJNA833662.
Author Contributions
P-LD and FL conceived this research and designed the experiments. P-LD, LZ, FL, S-LM, and YY participated in the design and interpretation of the data. P-LD, FL, P-HW, S-LM, YY, X-LW, and W-GY performed the experiments and analysis. P-LD, LZ, and FL wrote the manuscript. P-LD, LZ, FL, X-LW, and Q-YD participated in the revisions of the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2021-IAR), the Beijing Natural Science Foundation (6202032), and the Central Public-Interest Scientific Institution Basal Research Fund (IAR-CPSIBRF-2020-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Wujun Jiang, Jiamin Qin, Qun Tu, Qun Luo, and Xijian Xu (Jiangxi Institute of Apicultural Research) for collecting samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.934054/full#supplementary-material
References
Arca, M., Mougel, F., Guillemaud, T., Dupas, S., Rome, Q., Perrard, A., et al. (2015). Reconstructing the invasion and the demographic history of the yellow-legged hornet, Vespa velutina, in Europe. Biol. Invasions 17, 2357–2371.
Arca, M., Papachristoforou, A., Mougel, F., Rortais, A., Monceau, K., Bonnard, O., et al. (2014). Defensive behaviour of Apis mellifera against Vespa velutina in France: testing whether European honeybees can develop an effective collective defence against a new predator. Behav. Process. 106, 122–129. doi: 10.1016/j.beproc.2014.05.002
Archer, M. E. (1993). A phylogenetic study of the species of the genus Vespa (Hymenoptera: Vespinae). Entomol. Scand. 24, 469–478. doi: 10.1163/187631293X00226
Barbet-Massin, M., Rome, Q., Muller, F., Perrard, A., Villemant, C., and Jiguet, F. (2013). Climate change increases the risk of invasion by the Yellow-legged hornet. Biol. Conserv. 157, 4–10. doi: 10.1016/j.biocon.2012.09.015
Beggs, J. R., Brockerhoff, E. G., Corley, J. C., Kenis, M., Masciocchi, M., Muller, F., et al. (2011). Ecological effects and management of invasive alien Vespidae. Biocontrol 56, 505–526. doi: 10.1007/s10526-011-9389-z
Bertolino, S., Lioy, S., Laurino, D., Manino, A., and Porporato, M. (2016). Spread of the invasive yellow-legged hornet Vespa velutina (Hymenoptera: Vespidae) in Italy. Appl. Entomol. Zool. 51, 589–597. doi: 10.1007/s13355-016-0435-2
Budge, G. E., Hodgetts, J., Jones, E. P., Ostoja-Starzewski, J. C., Hall, J., Tomkies, V., et al. (2017). The invasion, provenance and diversity of Vespa velutina Lepeletier (Hymenoptera: Vespidae) in Great Britain. PLoS One 12:e0185172. doi: 10.1371/journal.pone.0185172
Cardoza, Y. J., Klepzig, K. D., and Raffa, K. F. (2006). Bacteria in oral secretions of an endophytic insect inhibit antagonistic fungi. Ecol. Entomol. 31, 636–645. doi: 10.1111/j.1365-2311.2006.00829.x
Chen, J., Wang, J., and Zheng, H. (2021). Characterization of Bifidobacterium apousia sp. nov., Bifidobacterium choladohabitans sp. nov., and Bifidobacterium polysaccharolyticum sp. nov., three novel species of the genus Bifidobacterium from honey bee gut. Syst. Appl. Microbiol. 44:126247. doi: 10.1016/j.syapm.2021.126247
Choi, M. B., Lee, S. A., Suk, H. Y., and Lee, J. W. (2013). Microsatellite variation in colonizing populations of yellow-legged Asian hornet, Vespa velutina nigrithorax, in South Korea. Entomol. Res. 43, 208–214. doi: 10.1111/1748-5967.12027
Choi, M. B., Martin, S. J., and Lee, J. W. (2012). Distribution, spread, and impact of the invasive hornet Vespa velutina in South Korea. J. Asia Pac. Entomol. 15, 473–477. doi: 10.1016/j.aspen.2011.11.004
Cini, A., Meriggi, N., Bacci, G., Cappa, F., Vitali, F., Cavalieri, D., et al. (2020). Gut microbial composition in different castes and developmental stages of the invasive hornet Vespa velutina nigrithorax. Sci. Total Environ. 745:140873. doi: 10.1016/j.scitotenv.2020.140873
Dai, P. L., Yan, Z. X., Ma, S. L., Yang, Y., Wang, Q., Hou, C. S., et al. (2018). The herbicide glyphosate negatively affects midgut bacterial communities and survival of honey bee during larvae reared in vitro. J. Agric. Food Chem. 66, 7786–7793. doi: 10.1021/acs.jafc.8b02212
Engel, P., Martinson, V. G., and Moran, N. A. (2012). Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. U. S. A. 109, 11002–11007. doi: 10.1073/pnas.1202970109
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Goldarazena, A., Heredia, I. P. D., Romon, P., Iturrondobeitia, J. C., Gonzalez, M., Lopez, S., et al. (2015). Spread of the yellow-legged hornet Vespa velutina nigrithorax du Buysson (Hymenoptera: Vespidae) across northern Spain. Bull. OEPP EPPO Bull. 45, 133–138. doi: 10.1111/epp.12185
Jia, H. R., Dai, P. L., Geng, L. L., Jack, C. J., Li, Y. H., Wu, Y. Y., et al. (2017). No effect of Bt Cry1Ie toxin on bacterial diversity in the midgut of the Chinese honey bees, Apis cerana cerana (Hymenoptera, Apidae). Sci. Rep. 7:41688. doi: 10.1038/srep41688
Jia, H. R., Geng, L. L., Li, Y. H., Wang, Q., Diao, Q. Y., Zhou, T., et al. (2016). The effects of Bt Cry1Ie toxin on bacterial diversity in the midgut of Apis mellifera ligustica (Hymenoptera: Apidae). Sci. Rep. 6:24664. doi: 10.1038/srep24664
Kim, J. K., Choi, M. B., and Moon, T. Y. (2006). Occurrence of Vespa velutina Lepeletier from Korea, and a revised key for Korean Vespa species (Hymenoptera: Vespidae). Entomol. Res. 36, 112–115. doi: 10.1111/j.1748-5967.2006.00018.x
Kim, J. S., Jeong, J. S., and Kim, I. (2017). Complete mitochondrial genome of the yellow-legged Asian hornet, Vespa velutina nigrithorax (Hymenoptera: Vespidae). Mitochondrial DNA B Resour. 2, 82–84. doi: 10.1080/23802359.2017.1285211
Kishi, S., and Goka, K. (2017). Review of the invasive yellow-legged hornet, Vespa elution migrator (Hymenoptera: Vespidae), in Japan and its possible chemical control. Appl. Entomol. Zool. 52, 361–368. doi: 10.1007/s13355-017-0506-z
Kwong, W. K., Engel, P., Koch, H., and Moran, N. A. (2014). Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. U.S.A. 111, 11509–11514. doi: 10.1073/pnas.1405838111
Kwong, W. K., and Moran, N. A. (2016). Gut microbial communities of social bees. Nat. Rev. Microbiol. 14, 374–384. doi: 10.1038/nrmicro.2016.43
Leza, M., Miranda, M. A., and Colomar, V. (2018). First detection of Vespa velutina nigrithorax (Hymenoptera: Vespidae) in the Balearic Islands (Western Mediterranean): a challenging study case. Biol. Invasions 20, 1643–1649. doi: 10.1007/s10530-017-1658-z
Li, L., Solvi, C., Zhang, F., Qi, Z., Chittka, L., and Zhao, W. (2021). Gut microbiome drives individual memory variation in bumblebees. Nat. Commun. 12:6588. doi: 10.1038/s41467-021-26833-4
Lopez, S., González, M., and Goldarazena, A. (2011). Vespa velutina Lepeletier, 1836 (Hymenoptera: Vespidae): first records in Iberian Peninsula. Bull. OEPP EPPO Bull. 41, 439–441. doi: 10.1111/j.1365-2338.2011.02513.x
Ma, S. L., Yang, Y., Jack, C. J., Diao, Q. Y., Fu, Z. M., and Dai, P. L. (2019). Effects of Tropilaelaps mercedesae on midgut bacterial diversity of Apis mellifera. Exp. Appl. Acarol. 79, 169–186. doi: 10.1007/s10493-019-00424-x
Monceau, K., Bonnard, O., and Thiery, D. (2014). Vespa velutina: a new invasive predator of honeybees in Europe. J. Pest Sci. 87, 1–16. doi: 10.1007/s10340-013-0537-3
Monceau, K., Maher, N., Bonnard, O., and Thiery, D. (2013). Predation pressure dynamics study of the recently introduced honeybee killer Vespa velutina: learning from the enemy. Apidologie 44, 209–221. doi: 10.1007/s13592-012-0172-7
Nguyen, L. T. P., Sait, F., Kojima, J., and Carpenter, J. M. (2006). Vespidae of Viet Nam (Insecta: Hymenoptera) 2. Taxonomic notes on Vespinae. Zool. Sci. 23, 95–104. doi: 10.2108/zsj.23.95
Robinet, C., Darrouzet, E., and Suppo, C. (2019). Spread modelling: a suitable tool to explore the role of human-mediated dispersal in the range expansion of the yellow-legged hornet in Europe. Int. J. Pest Manag. 65, 258–267. doi: 10.1080/09670874.2018.1484529
Robinet, C., Suppo, C., and Darrouzet, E. (2017). Rapid spread of the invasive yellow-legged hornet in France: the role of human-mediated dispersal and the effects of control measures. J. Appl. Ecol. 54, 205–215.
Sauvard, D., Imbault, V., and Darrouzet, E. (2018). Flight capacities of yellow-legged hornet (Vespa velutina nigrithorax, Hymenoptera: Vespidae) workers from an invasive population in Europe. PLoS One 13:e0198597.
Seo, J., Yang, S. H., Kim, I. S., and Yeonjong, K. (2018). Intestine bacterial microbiota of Asian hornet (Vespa velutina nigrithorax) and honey bee. Korean J. Environ. Agric. 37, 135–140.
Stainton, K., Hall, J., Budge, G. E., Boonham, N., and Hodgetts, J. (2018). Rapid molecular methods for in-field and laboratory identification of the yellow-legged Asian hornet (Vespa velutina nigrithorax). J. Appl. Entomol. 142, 610–616.
Suenami, S., Nobu, M. K., and Miyazaki, R. (2019). Community analysis of gut microbiota in hornets, the largest eusocial wasps, Vespa mandarinia and V. simillima. Sci. Rep. 9:9830. doi: 10.1038/s41598-019-46388-1
Tan, K., Radloff, S. E., Li, J. J., Hepburn, H. R., Yang, M. X., Zhang, L. J., et al. (2007). Bee hawking by the wasp, Vespa velutina, on the honeybees Apis cerana and A. mellifera. Naturwissenschaften 94, 469–472. doi: 10.1007/s00114-006-0210-2
Turchi, L., and Derijard, B. (2018). Options for the biological and physical control of Vespa velutina nigrithorax (Hym.: Vespidae) in Europe: a review. J. Appl. Entomol. 142, 553–562.
Villemant, C., Barbet-Massin, M., Perrard, A., Muller, F., Gargominy, O., Jiguet, F., et al. (2011). Predicting the invasion risk by the alien bee-hawking Yellow-legged hornet Vespa velutina nigrithorax across Europe and other continents with niche models. Biol. Conserv. 144, 2142–2150.
Visser, A. A., Nobre, T., Currie, C. R., Aanen, D. K., and Poulsen, M. (2012). Exploring the potential for Actinobacteria as defensive symbionts in fungus-growing termites. Microb. Ecol. 63, 975–985. doi: 10.1007/s00248-011-9987-4
Wang, S. C., Wang, L. Y., Fan, X., Yu, C., Feng, L., and Yi, L. (2020). An insight into diversity and functionalities of gut microbiota in insects. Curr. Microbiol. 77, 1976–1986. doi: 10.1007/s00284-020-02084-2
Wilson, E. E., Mullen, L. M., and Holway, D. A. (2009). Life history plasticity magnifies the ecological effects of a social wasp invasion. Proc. Natl. Acad. Sci. U.S.A. 106, 12809–12813. doi: 10.1073/pnas.0902979106
Keywords: Vespa, three forms, midgut bacterial community, Proteobacteria, Firmicutes, Lactobacillus
Citation: Zhang L, Liu F, Wang X-L, Wang P-H, Ma S-L, Yang Y, Ye W-G, Diao Q-Y and Dai P-L (2022) Midgut Bacterial Communities of Vespa velutina Lepeletier (Hymenoptera: Vespidae). Front. Ecol. Evol. 10:934054. doi: 10.3389/fevo.2022.934054
Received: 02 May 2022; Accepted: 25 May 2022;
Published: 21 June 2022.
Edited by:
Ying Wang, Shandong Agricultural University, ChinaReviewed by:
Wang Tianwei, Chinese Academy of Sciences (CAS), ChinaAbebe Jenberie Wubie, International Centre of Insect Physiology and Ecology (ICIPE), Kenya
Copyright © 2022 Zhang, Liu, Wang, Wang, Ma, Yang, Ye, Diao and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing-Yun Diao, ZHF5dW4xQDEyNi5jb20=; Ping-Li Dai, ZGFpcGluZ2xpQGNhYXMuY24=
†These authors have contributed equally to this work and share first authorship
 Li Zhang
Li Zhang Feng Liu
Feng Liu Xin-Ling Wang1
Xin-Ling Wang1 Qing-Yun Diao
Qing-Yun Diao Ping-Li Dai
Ping-Li Dai