
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 10 August 2022
Sec. Evolutionary and Population Genetics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.932004
This article is part of the Research Topic Genetic History of Human Populations Along the Ancient Silk Road View all 18 articles
 Edward Allen1†
Edward Allen1† Yao Yu1†
Yao Yu1† Xiaomin Yang2†
Xiaomin Yang2† Yiran Xu3,4
Yiran Xu3,4 Panxin Du5
Panxin Du5 Jianxue Xiong5
Jianxue Xiong5 Dian Chen6
Dian Chen6 Xiaohong Tian7
Xiaohong Tian7 Yong Wu7
Yong Wu7 Xiaoli Qin1
Xiaoli Qin1 Pengfei Sheng1,3,4*
Pengfei Sheng1,3,4* Chuan-Chao Wang2,8,9,10*
Chuan-Chao Wang2,8,9,10* Shaoqing Wen1,3,4*
Shaoqing Wen1,3,4*Han/non-Han interactions were engrained among the border regions of ancient Imperial China. Yet, little is known about either the genetic origins or the lifeways of these border peoples. Our study applies tools from ancient deoxyribonucleic acid (DNA) and stable isotope analysis to the study of a Han dynasty population at the Shichengzi site in modern-day Xinjiang. Isotopic analysis (δ13C and δ15N) of human (n = 8), animal (n = 26), and crop remains (n = 23) from Shichengzi indicated that dietary patterns among site inhabitants could be split among agro-pastoral and agricultural groups based on differences in the collagen 15N ratios. DNA analysis divided the four Shichengzi samples into two groups, with one group primarily harboring the ancient Northeast Asian (ANA) related ancestry, while the other showed a dominant Late Neolithic Yellow River (YR_LN) related ancestry. Both ancient DNA and stable isotope evidence point to the Northeast Asian origins of pastoralists and East Asian origins of Han agriculturalists, who, nonetheless, shared a single burial space at Shichengzi. This study thus provides clear evidence for the multiple origins and identities of populations across the porous border represented by the Han Empire and surrounding regions and proposes a new model for the interpretation of border culture in early Imperial China.
An increasing number of studies seek to combine ancient deoxyribonucleic acid (DNA) work with multiple forms of scientific and classic archeological analysis (Haak et al., 2008; Frei et al., 2015, 2019; Mittnik et al., 2019; Blank et al., 2021; Ingman et al., 2021; Patterson et al., 2022). Integrating DNA and carbon, nitrogen stable isotope analysis has improved our knowledge of ancient origins and human/non-human paleodietary practices (Wilson et al., 2007; Alkass et al., 2013; Ames et al., 2015). Such a multidisciplinary approach can offer fascinating new insights into ancient population history and subsistence in various contexts but has rarely been applied to the bordering regions of Chinese imperial history (e.g., Ning et al., 2019). Employing the research methods of this emerging field, our study opens new vistas for border research through a combination of DNA data with paleodietary and other archeological evidence at a Han dynasty border site.
From 2014 to 2019, Shichengzi (43°36′59.1″N, 89°45′43.2″E, 1,770 m amsl), located in Qitai County, Xinjiang, on the northern slopes of the Tianshan Mountains, was identified as a major Han dynasty garrison and was scientifically excavated (Figure 1 and Supplementary Figure 1). Laid out over approximately 110,000 m2, C-14 dates suggest a site occupation from the first century BC to the third century AD (Sheng et al., 2020). Multiple architectural features were uncovered and dated to the period of the agricultural garrison. These included the main gate, rammed mud walls, moats, buildings, a wealth of pottery, tiles, weapons, and agricultural implements (Area A, Area C, and Area D). Burials and a single Han-style kiln were excavated in Area B (Tian et al., 2018, 2020). Sheng et al. (2020) subsequently characterized Shichengzi as a “melting pot” of Han and non-Han culture, using archaeobotanical evidence to demonstrate a local adaptation to the Tianshan Mountain settings against relatively amenable climactic conditions circa 2000–1700 BP (Sheng et al., 2021).

Figure 1. (a) Map of northwestern China. Small dots represent Han cities found in Xinjiang. (b–e) Main findings at Areas A–D at the Shichengzi site.
From the first century BC to the third century, interactions between the Han and agro-pastoralist empires and peoples of Central Asia AD were considered a high point in early Silk Road cross-cultural communications (Millward, 2013; Spengler, 2019). In China’s written histories, Han Emperor Wu (141–87 BC) inspired this dramatic shift circa 119 BC, seeking allies and shoring up Han presence in a quest to subjugate the feared Xiongnu confederation to his direct north (Ban, 1962; Fan, 2000). Subsequent Han emperors strengthened the Han military presence in the “Western Regions” (primarily located in modern-day Xinjiang) (Zhang and Tian, 2015; Li, 2017). Chinese scholars have historically documented and voluminously researched the forced migration of ethnic Han Chinese from the Central Plains region to the empire’s western extremities during these decades (see Yang, 1991; Zhu, 2012). Against the diverse ecological backdrop of Central Asia, a stratified Han order was established in agricultural garrisons (tuntian 屯田; lit. “fortified agricultural fields”) (Luo et al., 2018) and other settlement types. This helped stabilize the Han Empire’s political and economic influence on the steppe. In practice, however, inter-migration and exchange between groups are believed to have been widespread. One scholar characterized the “cultural mediation, assimilation, rejection, or integration” between the Han center and its peripheries (Di Cosmo, 2009) around this time. With Shizhengzi, the question of interaction between Han, Xiongnu, and Western Region groups became one of acute interest.
Here, we fuse traditional archeological approaches with C and N isotopes, and ancient DNA data analysis of four individuals at Shichengzi (Figure 1a), previously confirmed as a Han dynasty agricultural garrison (tuntian) (Sheng et al., 2020). Divergent genetic and isotopic profiles, nonetheless, shared similar burial practices at this site. Considered as a combination of diverging dietary practices around a common burial space and possibly site use, we argue that this “mutualism” was negotiated against a backdrop of Han expansion/regional accommodation and long-established, flexible pastoralist subsistence strategies. Our study contributes to current knowledge by demonstrating one way, in which Han migrants and agro-pastoralists of the Western Regions interacted within the agricultural garrison context and explores the possible consequences for our understanding of Han border formation processes at the northwest borders. The methodological approach combines archeological sciences and history and will be useful in re-evaluating border formation in imperial Chinese history in short-term and long-term contexts.
To determine the date of Shichengzi’s occupation, human and animal bone, charcoal, and crop seed samples (n = 14) recovered from the site were sent to Beta Analytic Inc., Miami, Florida, United States, for radiocarbon analysis. All dates were calibrated using the IntCal 20 calibration curve (Reimer et al., 2020) and OxCal v4.3.2.1 Detailed sample information is listed in Supplementary Table 1.
A total of ten burials, one sacrificial horse pit and one pottery kiln, were excavated in the west of the Shichengzi site. The burials were divided into six vertical shaft earth pit tombs, three vertical shaft side-chamber burials, and a vertical shaft burial with a second-tier ledge (ercengtai). All were individually prone burials with the head facing west and a trough-shaped coffin. A mortise-tenon structure was employed for the inner-coffin and outer-coffin discovered in the second-tier ledge burial.
The side-chamber burials M1 and M3 contained Han dynasty wuzhu coins (M3), a bronze plaque, a bronze ring, an iron knife, and various utilitarian potteries (M1). The burial at M2, the second-tier ledge burial, was entombed in a wooden outer coffin, setting this tomb dramatically apart from M1 and M3. M2 was also buried with goat astragalus bones, silk, lacquer, and beads. The burial style and choice of burial goods strongly suggest two different populations, likely a Han population (like M3) and a pastoralist population (like M2), as pointed out in Sheng et al. (2020).
The collagen from human bone samples at Shichengzi (n = 8) was extracted using protocols outlined in Richards and Hedges (1999). Carbonized seeds (n = 2) of Setaria italica and Panicum miliaceum collected from Shichengzi by flotation were also pretreated for stable C and N isotopic determination. We collated published stable carbon and nitrogen isotopic results for multiple varieties of food resources found at Shichengzi, including isotope data (n = 47) of charred wheat/naked barley grains, sheep/goat, cattle, and horse, as well as dog bones from Shichengzi. Additionally, we revisited the existing C and N isotopic evidence for the paleo diet at Shirenzigou (n = 38), dated 2200–1900 BP, and two typical Han cemeteries at Heishuiguo (n = 116) and Huangwan (n = 7), located in the Hexi Corridor during 2100–1700 BP. This would provide comparative data on subsistence strategies. All existing isotopic data are summarized in Supplementary Tables 2, 3.
Sampling was performed on the teeth of 4 individuals at Shichengzi. Ancient DNA extraction and Illumina double-stranded DNA sequencing library preparation were performed at Fudan University and established protocols for working with ancient human DNA were followed (Knapp et al., 2012; Sun et al., 2021). Human remains were surface-cleaned and ground to a fine powder. We used 100 mg of bone powder to extract the DNA using the robotic magnetic beads method. We prepared double-stranded libraries following Meyer’s protocols (Meyer and Kircher, 2010) but with minor modifications and without conducting UDG treatment for all samples. Libraries were amplified with indexing primers in two parallel PCRs using Q5 High-Fidelity DNA Polymerase (NEB). We qualified the clean-up libraries using Qubit 2.0. Finally, we sequenced the libraries on an Illumina HiSeq X10 instrument at the Annoroad Company, China, in the 150-bp paired-end sequencing design.
We clipped the Illumina sequencing adapters using AdapterRemoval v2.2.0 (Schubert et al., 2016). We mapped the merged reads with 30 or more bases to the human reference genome (hs37d5) using BWA v0.7.17 (Li and Durbin, 2010) with parameters “-l 1024 -n.01.” We removed PCR duplicates using DeDup v0.12.3 (Peltzer et al., 2016). We used the trimBam function in bamUtils v1.0.13 (Jun et al., 2015)2 to trim the first and last two base pairs (bp) of each read to remove deamination-based 5′ C > T and 3′ G > A misincorporations and minimize the impact of postmortem DNA damage on genotyping. For the SNPs in the “1240k” panel (Mathieson et al., 2015), we randomly sampled a single high-quality base from a high-quality base (Phred-scaled base quality score 30 or higher) as a pseudo-diploid genotype, using the pileupCaller program.3
The quality of ancient genomic material was assessed through a suite of methods. First, we tabulated patterns of post-mortem chemical modifications expected for ancient DNA using mapDamage v2.0.6 (Jónsson et al., 2013). Second, we estimated mitochondrial contamination rates from modern humans for all individuals using Schmutzi v1.5.1 (Renaud et al., 2015). Third, we measured the nuclear genome contamination rate in males based on X chromosome data as implemented in ANGSD v0.910 (Korneliussen et al., 2014). Since males have only a single copy of the X chromosome, mismatches between bases, aligned to the same polymorphic position, and beyond the level of sequencing error are considered evidence of contamination.
We assigned the biological sex of the ancient samples with the aid of the programs Rx (Mittnik et al., 2016) and Ry (Skoglund et al., 2013).
For mtDNA, we employed the log2fasta program built-in Schmutzi (Renaud et al., 2015) to call mitochondrial consensus sequences from the Schmutzi output. Variations that appeared when checked against rCRS were re-checked in BAM (Binary Alignment Map) files through visual inspection with IGV software (Helga et al., 2013). We then used Haplogrep 2 (Weissensteiner et al., 2016) to assign haplogroups. Y chromosome haplogroups were examined by aligning a set of positions in the ISOGG (International Society of Genetic Genealogy4 and Y-full5 databases, and analysis was performed in the case of a base and mapping quality exceeding 30. Haplogroup determination was performed with the script Yleaf.py in Yleaf software (Ralf et al., 2018), which provides outputs for allele counts of ancestral and derived SNPs along a path of branches of the Y-chromosome tree. Finally, we re-checked SNPs by visual inspection with IGV software (Helga et al., 2013).
We utilized READ (Kuhn et al., 2018) software to detect the degree of genetic kinship between ancient individuals.
We merged our data with two sets of previously published worldwide populations’ genotype datasets using the mergeit program from EIGENSOFT (Patterson et al., 2006), one is based on the Affymetrix Human Origins array dataset containing 597,573 SNPs (“HO” dataset) (Mallick et al., 2016; Jeong et al., 2019), while the other is based on 1240K capture dataset (1,233,013 SNPs, including all the ancient samples and shotgun-sequenced modern samples) (Haak et al., 2015; Damgaard et al., 2018a,b; Narasimhan et al., 2019; Ning et al., 2019, 2020; Wang et al., 2019, Wang et al., 2021; Jeong et al., 2020; Yang et al., 2020; Mao et al., 2021; Zhang et al., 2021).
We performed principal component analysis (PCA) as implemented in the smartpca v16000 (Patterson et al., 2006) on the HO dataset with options “lsqproject: YES” and “shrinkmode: YES” and projected ancient individuals onto the calculated components. We also conducted an unsupervised ADMIXTURE analysis with ADMIXTURE v1.3.0 (Alexander et al., 2009) after pruning for linkage disequilibrium by PLINK v1.90 (Chang et al., 2015) using parameters “– indep-pairwise 200 25 0.4” and generated a total of 271,578 SNPs for ADMIXTURE analysis. We separately labeled individuals with significant differences in clusters and ancestries.
We used outgroup-f3 statistics (Patterson et al., 2012) to measure the genetic relationship between two populations following their divergence from the outgroup. We calculated f4 statistics built-in ADMIXTOOLS (Patterson et al., 2012) with the “f4mode: YES” parameter to further reveal the genetic differences between the two studied subgroups, and their genetic relationships with other ancient and modern eastern Eurasian populations. All f statistics were based on a 1240k dataset.
We further utilized the pairwise qpWave (Patterson et al., 2012; Agranat-Tamir et al., 2020) to test whether pairs of populations form genetic clade related to a set of outgroups.
We applied the qpAdm program (Patterson et al., 2012) based on the 1240k dataset to model ancestry proportions of our ancient Shichengzi population related to one or two different ancestral sources from a set of reference populations, along with parameters “allsnp: YES” and “details: YES.” We also used a model competition approach to find the best model. We took pairs of fitted models, added the source population from one to the reference population set of the other, and assessed whether it continued to fit (Sirak et al., 2021).
All AMS radiocarbon dates for archeological materials recovered from Shichengzi are presented in Supplementary Table 1. Dates ranged from 40 cal BC to 420 cal AD (2σ). Pending more evidence from unearthed artifacts and Chinese historical literature, we believe that Shichengzi was occupied from the first century BC to the third century AD.
As plotted in Figure 2, eight sampled human individuals could be divided into two groups based on differences in δ15N values. The δ15N values of these human samples ranged from 9.6 to 14.3%, suggesting the consumption of different volumes of proteins. The mean δ15N value of individuals in Group A (M1, M2, M4, M5, M6, and M8) was 13.8 ± 0.4%, demonstrating consumption of considerable 15N-enriched foods, such as meat and milk products. In contrast, the two individuals in Group B (M3 and M9) exhibited relatively low δ15N values (9.6% and 10.3%), revealing a consumption of less protein-rich foods than Group A. The δ13C values from the osteological material ranged from -16 to -18.2%, suggesting a similar dietary pattern among individuals partaking in diets harboring C3-based proteins. All raw data are laid out in detail in Supplementary Table 4.
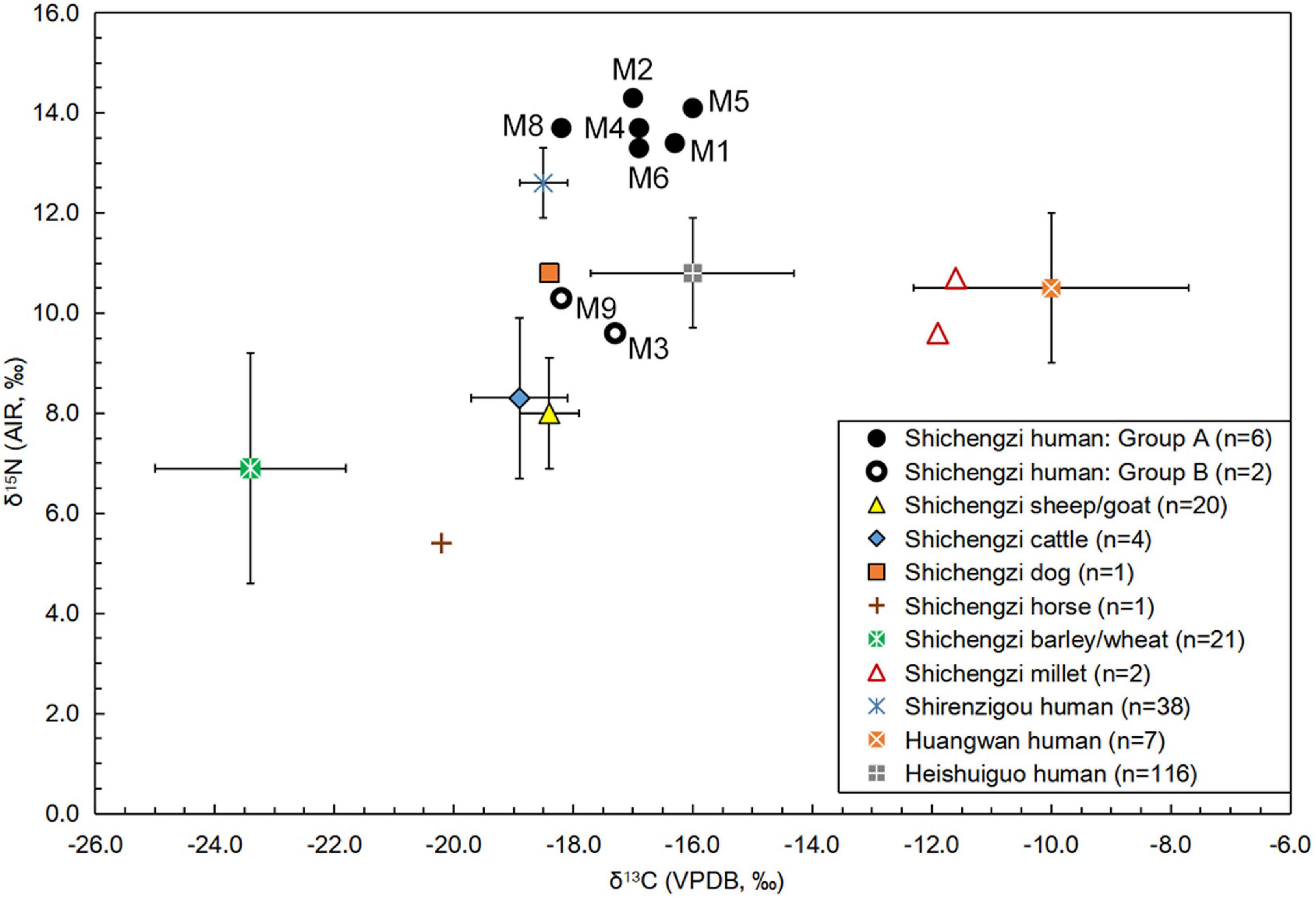
Figure 2. Stable C and N isotopic data of the crop, animal, and human discovered from the Shichengzi and other three relevant sites.
As shown in Figure 2, most negative δ13C and δ15N values were derived from charred wheat and naked barley seed samples. These ranged from -25.8 to -19.8% (mean ± SD: -23.4 ± 1.6%) and 3.1–11.8% (mean ± SD: 6.9 ± 2.3%). Newly acquired stable carbon and nitrogen isotopic results for foxtail millet and common millet stood at -11.9, -11.6%, and 9.6, 10.7%, respectively. This crop data established an isotope baseline for C3 and C4 crop foods at Shichengzi. The δ13C and δ15N values of a horse (Equus caballus) bone recovered from the site emerged at -20.2 and 5.4%, respectively. Since this N isotope value was lower than most wheat and naked barley grains, it suggested that the horse consumed a diet heavily influenced by wild C3 terrestrial grasses and/or shrubs. In addition, the mean δ13C and δ15N values of sheep/goat (n = 20) and cattle (n = 4) stood at -18.4 ± 0.5, -18.9 ± 0.8% and 8 ± 1.1, 8.3 ± 1.6%, revealing that domestic herbivore diets were influenced by C3-based proteins primarily based on Triticoid cereals and wild plants.
Previous isotopic studies have demonstrated that 15N in human collagen is enriched by + 3∼5% relative to diet (Hedges and Reynard, 2007). This provided a baseline for further interpretation of the Shichengzi diet upon the above-mentioned C, N isotopic baseline for potential food resources around the site. As shown in Figure 2, the mean δ15N value of these humans in Group A (13.8%) was 5.8, 5.5, and 6.9 (>3∼5%) higher than values for sheep/goat and cattle bones and crop grains of wheat and naked barley, respectively, suggesting consumption of high degrees of meat and milk products derived from domesticated herbivores found in the site vicinity. In contrast, the mean δ15N value of the Group B population (10%) was 2, 1.7, and 3.1 (<3∼5% or around 3%) higher than the mean 15N values of sheep/goat and cattle bones, as well as wheat and naked barley seeds, respectively. Moreover, both individuals in Group B exhibited a lower δ15N value than a canine sample (10.8%) recovered from Shichengzi, suggesting that this group regularly dined on barley and wheat foods, with minimal consumption of meat from domestic sheep/goat and cattle. If so, it is reasonable to believe that Group A individuals represent pastoral inhabitants, and Group B populations were likely agriculturalists.
Comparative δ13C and δ15N data collected from three sites near Shichengzi have been summarized in Supplementary Table 3 and plotted in Figure 2. The mean δ13C and δ15N values of agro-pastoralists found at Shirenzigou (Figure 1a) were –18.5 ± 0.4 and 12.6 ± 0.7%, respectively, showing that Shirenzigou diets resembled individuals from Group A at Shichengzi and were significantly influenced by 15N enriched C3-based proteins. On the contrary, mean δ13C and δ15N values of Han populations at the Huangwan site and Heishuiguo site in the Hexi Corridor (see Figure 1a) were -10 ± 2.3, 10.5 ± 1.5% and -16 ± 1.7, 10.8 ± 1.1%, respectively. Although individuals at Huangwan yielded a far enriched δ13C value, both Han populations were akin to the Shichengzi B population exhibiting nearly identical and markedly low-level of δ15N levels, indicating that their consumption of animal foods was rather limited and pointing to a basis of inter-group dietary similarity. Thus, despite variability within our assigned categories, when considered within overall food webs, alongside corresponding animal diets, and within the broader northwest China Han context, we feel confident in dividing the Shichengzi population into an agricultural and an agro-pastoral group.
We initially screened four ancient individuals by shallow shotgun sequencing of one Illumina sequencing library per individual, resulting in coverage from 0.023 to 0.064X (Supplementary Table 5). We verified the authenticity of the genome data through a series of methods. All samples showed typical characteristics of ancient DNA with postmodern chemical damage (Supplementary Figure 2). All individuals exhibited negligible contamination from modern populations and were, thus, suitable for subsequent analysis, although we note that M4 showed low mitochondrial coverage that would prove insufficient for estimating the mitochondrial contamination rate. Our kinship analysis confirmed that all pairs of individuals were unrelated. We retained all individuals for subsequent analysis. Mitochondrial haplogroups for all Shichengzi individuals belonged to D4 clades, including D4j15, D4c2b, D4j7, and D4c2c. The two males (M3 and M9) were assigned to Y chromosomal haplogroup O (i.e., O1b*-F435 and O2a2b2a1a-F4110), a typical East Asian haplogroup. Due to low coverage, the other male, M4, was only tentatively assigned to haplogroup N1a1a1a1a-CTS1077, which is prevailing in Northeast Asia, implying the dual paternal origins of the Shichengzi population (Supplementary Table 5).
We observed that the four Shichengzi samples were separated into two groups in the PCA plot (Figure 3). The pair of individuals M1 and M4 clustered in the upper section, together with ancient Neolithic to Iron Age eastern Mongolia Plateau (including Mongolia_N, Ulaanzuukh_SlabGrave, lateXiongnu) and Lake Baikal Eneolithic Hunter-Gatherer (HG) (including Russia_Lokomotiv and Russia_Shamanka populations). The pair of individuals M3 and M9 clustered with ancient agricultural populations from the Yellow River Basin, as well as modern Han Chinese. Our ADMIXTURE analysis revealed a similar pattern (K = 4; Figure 4), with the four Shichengzi individuals divided into two groups with different ancestry compositions. The samples M1 and M4 mainly harbored Ancient Northeast Asian (ANA) related ancestry, which was enriched in ANA HG (represented by AR_EN), while the samples M3 and M9 exhibited a genetic profile akin to millet farmers in the Yellow River Basin from the Late Neolithic Age onward (included YR_LN, YR_LBIA) (Ning et al., 2020).
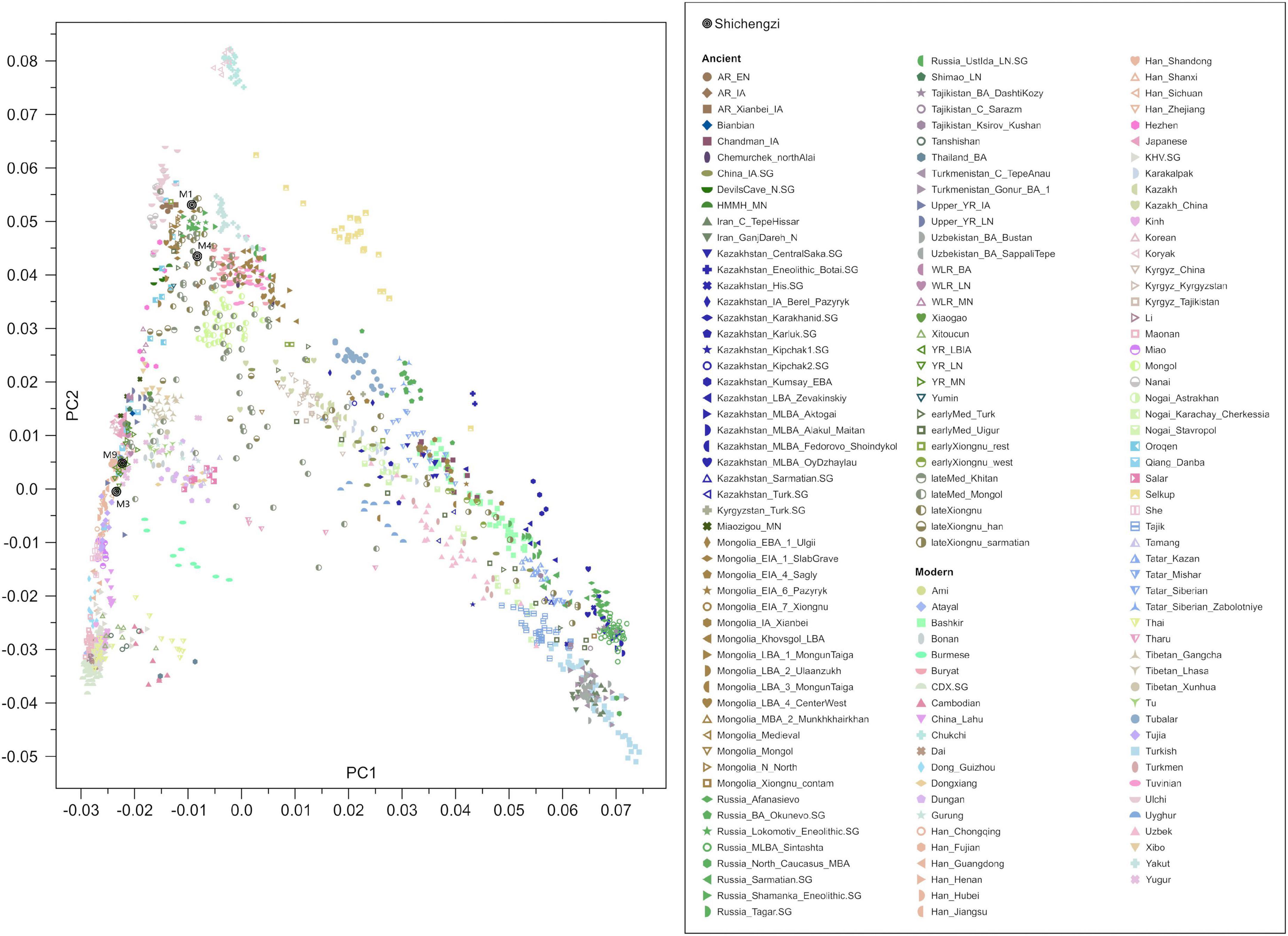
Figure 3. PCA of ancient samples. Projection of ancient samples onto PCA dimensions 1 and 2 defined by East Asian, Central Asian, European, Siberian, and other populations.
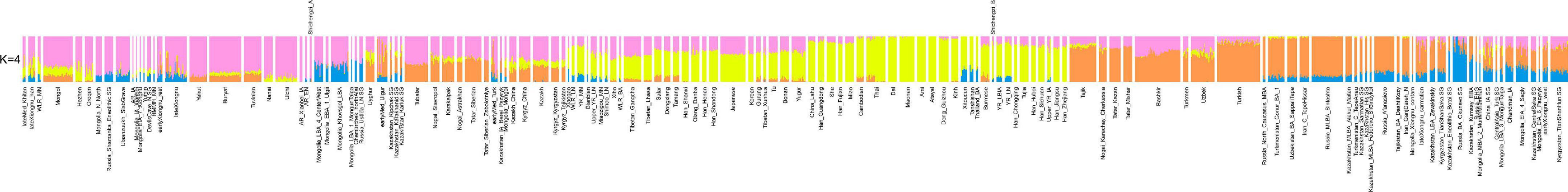
Figure 4. The result of ADMIXTURE analysis (K = 4) among Eurasian groups shows that two Shichengzi groups harbor disparate ancestral composition patterns. Populations mainly from southern China and Southeast Asia (yellow), Mongolia (light magenta), and West Eurasia (Orange and blue).
Next, we utilized outgroup-f3 analysis to further determine the genetic differences among Shichengzi individuals and explore the genetic affinity of Shichengzi to modern/ancient Eurasians on a quantitative basis. Here the high genetic heterogeneity among Shichengzi groups was vividly exemplified: M3 and M9 shared more alleles with Yellow River millet farmers (YR_LN and YR_LBIA) and modern Han groups, while M1 and M4 exhibited a close genetic relationship with Northeast Asian populations who harbored a larger amount of ANA ancestry, including ANA (Neolithic Mongolia_N_ East/Russia_Lokomotiv_Eneolithic/DevilsCave_N/Russia_ Shamanka_Eneolithic, Bronze Age’s Ulaanzuukh_SlabGrave/WLR_BA_o) (Jeong et al., 2020; Ning et al., 2020; Wang et al., 2021) and modern Tungusic-speaking groups (Figure 5 and Supplementary Table 6). We confirmed the above finding by f4 statistics (Supplementary Figure 3). Pairwise qpWave analysis revealed a similar clustering pattern with outgroup-f3 statistics but further demonstrated the genetic difference among Shichengzi individuals (Figure 6). M3 and M9 were genetically homogeneous with Yellow River millet farmers (p > 0.05), while M1 clustered not only with Yellow River millet farmers but also with HG from Mongolia Plateau and Baikal Lake (p > 0.05). Notably, M4 only clustered with HG from Mongolia Plateau and Baikal Lake (p > 0.05). Based on those results from PCA, ADMIXTURE, f-statistics, and pairwise-qpWave homogeneous test, we grouped the four individuals into Shichengzi_A (M1, M4) and Shichengzi_B (M3, M9). Shichengzi_A showed a closer genetic relationship with Neolithic HG in Mongolia Plateau and Baikal Lake and Bronze Age Ulaanzuukh_SlabGrave populations, who derived the majority of the ancestry from ANA, as opposed to Yellow River millet farmers [positive values of f4 (Mbuti, Shichengzi_A; Yellow River, Mongolia Plateau)]. Shichengzi_A also shared more alleles with modern Tungusic and Mongolic speaking populations than Sino-Tibetan speakers as reflected in positive values of f4 (Mbuti, Shichengzi_A; Sino-Tibetan, Tungusic/Mongolic) (Supplementary Figures 4, 5). By contrast, Shichengzi_B showed a closer genetic affinity with Yellow River millet farmers than with ancient populations of Mongolia Plateau, as reflected in negative values of f4 (Mbuti, Shichengzi_B; Yellow River, Mongolia Plateau). Shichengzi_B also shared more alleles with Sino-Tibetan populations than with Altaic speaking groups, as shown in the negative values of f4 (Mbuti, Shichengzi_B; Sino-Tibetan, Altaic) (Supplementary Figures 4, 5).
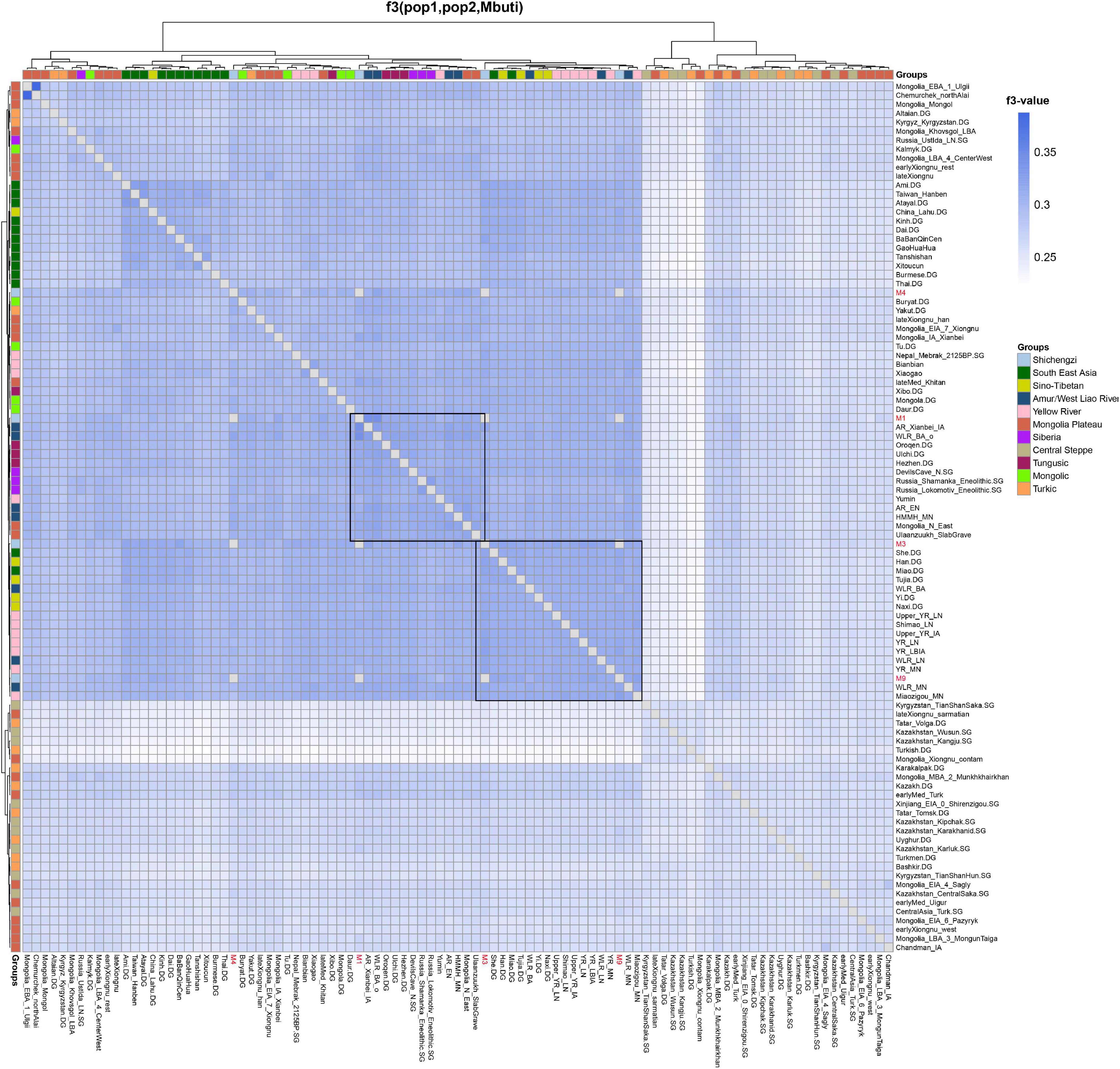
Figure 5. The results of f3(X1, X2; Mbuti) based on the 1240k dataset show the genetic relationship among East Eurasians. Darker colors indicated a larger-f3 value and more shared alleles between a pair of tests.
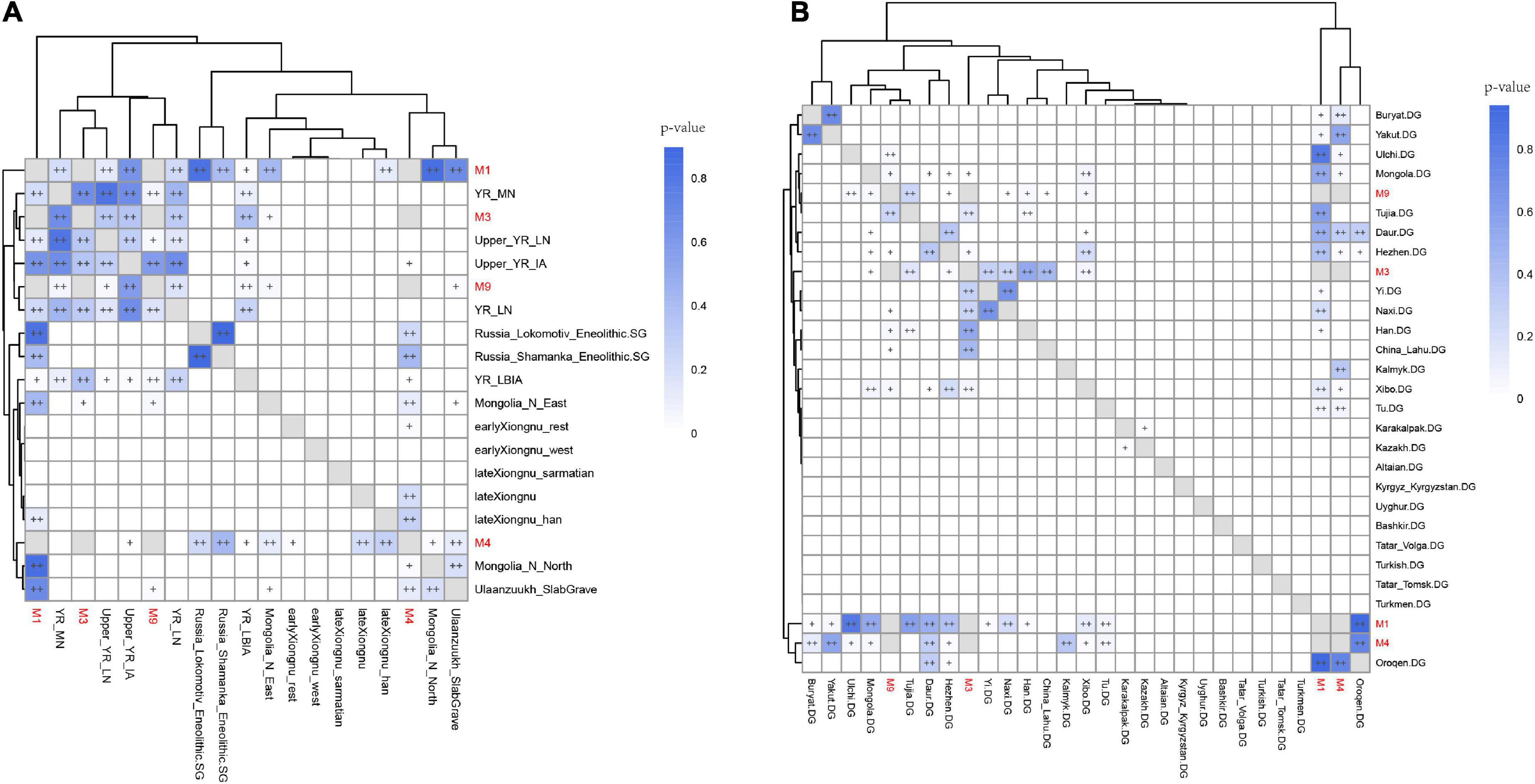
Figure 6. The results of qpWave analysis show the different genetic affinity patterns of four Shichengzi individuals to ancient populations in the Yellow River valley (A) and Mongolia Plateau (B). “++” represented values greater than 0.05, and “+” represented values lower than 0.05 but larger than 0.01. p > 0.05 indicated the supported genetic clade between pairs. Computations were based on the outgroup set (Mbuti.DG, Onge.DG, Yana_UP.SG, Loschbour.DG, Iran_GanjDareh_N, Turkmenistan_Gonur_BA_1, Russia_Afanasievo, Anatolia_N, Kazakhstan_Eneolithic_Botai.SG, AR19K, Liangdao2, Japan_Jomon, Miaozigou_MN).
We further used qpWave and qpAdm to explore the number of ancestry sources and plausible admixture models for the Shichengzi groups (Supplementary Table 7). A minimum of two ancestral streams could indicate the likely origins of both Shichengzi groups. Modeling for Shichengzi_A showed a likely descent from ancient Mongolia Plateau populations, harboring mostly ANA ancestry; while one-way modeling with Ulaanzuukh_SlabGrave as a single source failed when Mongolia_N_North was included in the outgroup set (p = 0.0063), but a fitted two-way model was acquired only with the inclusion of Ami as the other source at an estimated proportion of 25.8 ± 7.9%. The models with YR_LN as a source failed to explain the genetic variation of Shichengzi_A, suggesting a very limited genetic influence from farming groups. Shichengzi_B exhibited a thoroughly different profile in its ancestry composition by deriving ancestry from YR_LN rather than ANA-related populations. When we added Mongolia_North to the outgroup set, the models involving YR_LN as a single source still provided a fit for Shichengzi_B (0.37 < p < 0.81). The admixture modeling results at the individual level of M3 and M9, M1, and M4 were consistent with the Shichengzi_B and Shichengzi_A results at the group level, respectively.
In Han times, the Xiongnu, a pastoralist regime to the dynasty’s north, flourished on the Mongolia Plateau. As stated above, a possible connection between Shichengzi inhabitants and Xiongnu is worth further exploration. In our study, the Xiongnu population was genetically classified into eastern and western groups, with the eastern group (earlyXiongnu_rest, late Xiongnu, and lateXiongnu_Han) exhibiting dominant Northeast Asian or Han-related ancestry (Jeong et al., 2020). We found that sample M4 also formed a genetic clade with early Xiongnu rest as reflected in non-significant values of f4 (X, Mbuti; M4, earlyXiongnu_rest) (Supplementary Figure 3). The pairwise-qpWave analysis also demonstrated that M1 and M4 formed a genetic clade with lateXiongnu_Han, and M4 also exhibited certain similarities with late Xiongnu (Figure 6).
During and after the reign of Emperor Wudi, the Han grand strategy in Western Regions was described historically as “associating with distant countries and attacking those nearby” (yuan jiao jin gong): allying with Western Region states to better take the fight to the feared Xiongnu. The powerful Wusun (乌孙) state was one such contemporary polity subsequently courted by the Han. With this historical information in mind, we also explored the relationship between Shichengzi and Wusun groups. Our f4 analysis in the form of f4 (X, Mbuti; Shichengzi, Wusun) showed a genetically significant difference between Shichengzi and Wusun/Kangju populations in Central Asia, who derived most of their ancestry from West Eurasian groups (Z < -3 when X included West Eurasians, such as Sintashta, Turkmenistan_Gonur_BA_1, and Anatolia_N) (Supplementary Table 8).
We also explored the genetic relationship between Shichengzi and samples from the Shirenzigou site, an early Iron Age agro-pastoralist site on the northern slopes of the eastern Tianshan Mountains, Xinjiang (Ning et al., 2019). We conducted an f4 (X, Mbuti; Shichengzi, Shirenzigou) and observed significant differences between both Shichengzi groups and Shirenzigou people: both Shichengzi groups harbored more East Eurasian ancestry than Shirenzigou (Z > 3 when X included East Eurasians) (Supplementary Figure 6). This refuted the possibility of direct migration between the two locales.
This study has presented a combination of evidence: the paleogenetics of the Shichengzi population; C and N isotope analysis of the Shichengzi paleodiet; and Shichengzi’s excavated burials. Shichengzi inhabitants are divided into two separate genetic groups and paleodietary patterns. This combination of DNA and stable isotope analysis has used methods unavailable to traditional archeology and historiography and conclusively demonstrated the high likelihood of joint agropastoralist/agriculturalist use of the Shichengzi site during the Han (202 BC–AD 8 and AD 25—220) dynasty. Use of site space and burial data (Supplementary Figure 1 and Figures 1b–e), however, points to close interaction between these different groups alongside the expression of differences. Subsequently, we argue that these Shichengzi groups were contemporaries, each group involved in who were both involved in the social and political life of this Han frontier post. Combining our new C and N isotopic evidence of human dietary patterns from the Shichengzi site with isotopic data from three other related sites, Shirenzigou, Huangwan, and Heishuiguo (Figure 1a), strengthens the claim that these pastoralist and agricultural subsistence strategies were maintained within a shared sociopolitical environment around Shichengzi.
The Shichengzi evidence is useful for updating our understanding of lifeways and lifestyles within the Han border system. With a widened array of archeological data, we may also begin to place the archeology of Han borders in a global context. The archeology of Roman border formation processes, for example, has previously addressed the issue of agropastoral/agricultural interaction. Leaning on arguments that stressed the entanglement of plant and animal domestication processes within specific ecosystems – a relationship of “mutualism” as opposed to the artificial and rigid division of agricultural and pastoralist pathways (Langlie and Capriles, 2021; Boucher, 1985), Banning argued for an underlining mutualism in agropastoralist/agriculturalist interactions on the Roman frontier (Banning, 1986). Parker (1987) built on this argument with a critique pointing out that only a “policed” frontier could realistically contain any kind of mutualistic relationship. While Shichengzi undeniably witnessed a degree of “integration” and “accommodation” of Han and non-Han subjects (Di Cosmo, 2009) under the banner of Imperial administrative control and “policing,” we find that such “mutualism” also expressed a localized knowledge and adaptation to new sociopolitical realities. Archeologically speaking, the Han system was adapted by these farming and pastoral peoples. In terms of personal livelihoods and wealth, the impressive grave goods of the likely agro-pastoralist individual M2 may suggest that local elites bought into and indeed perhaps embraced the Han system within their own broader networks. By contrast, the bare burials of Han migrants, such as individual M3, suggesting that they occupied a considerably lowlier status in life, in line with literary evidence suggesting much of the Han population moved to these regions occupied a servile or imprisoned status (Yang, 1991). We can look forward to advances in work on urbanism in the Xinjiang region during and around this time period, work which will highlight the interconnections and regional temporal precedents of local and Han imperial formation processes.
Our DNA and paleodietary analysis has also improved our sense of lifeways and subsistence at Shichengzi and thus other Han frontier garrisons. Recent bioarcheological studies of the Roman frontier, inspired by the social archeological literature of the early 2000s (Meskell and Preucel, 2004), have begun to consider their subjects as “embodied” individuals within this complex liminal space (Gowland, 2017). At Shichengzi, diversity of subsistence pathways and genetic background coalesced into an “embodied” border identity of mutually benefiting pastoralist and agricultural groups. Agro-pastoral groups may have attached themselves to Shichengzi for economic and social benefit and become embodied parts of border reality through the long-term consequences of such decisions. In turn, we can also begin to suggest the possibility that this rapid acclimatization to the agricultural garrison system was equally facilitated by agro-pastoralist adaptations, with engagement running both ways between agricultural groups. This rapid adaptation to Han space further suggests longer-term, synergistic agro-pastoralist/agricultural relationships in the Xinjiang region. The Han system, in a sense, bought and adapted itself to this network.
The divergence in the genetic and paleodietary profile of burials at Shichengzi suggests that Han and Altaic-speaking agriculturalist/agro-pastoralist populations inhabited and used the site extensively. As the agricultural garrison system and evidence in the form of Han-style pottery kilns both suggest, this was a quintessentially imported Han space. Yet, while Han/non-Han burials may be distinguishable by the presence of wuzhu coins in the latter case and the vertical-shaft earth pit burials of agro-pastoralist populations across Xinjiang in this period for the latter (Tian, 2021), the observed customary use of this shared burial chamber and long-term association of both groups with Shichengzi should be considered further as evidence of more long-term, habitual accommodation. Our evidence points to the long-term adaptation of both agriculturalists and non-agriculturalist to the agricultural garrison system, in a process more deeply etched in the region and more flexible than accommodation might suggest. This offers a promising line of analysis as new archeological material for the Western Regions of the Han Empire is unearthed and curated using the methods of scientific archeology.
Our study offers the first ancient DNA data providing coverage of the Han dynasty (202 BC–220 AD) border regions. This is combined with isotopic and burial data and allows us to put forward a new reading of Han border formation and development. Border identity can be understood as an array of plural identities, combining agricultural and agro-pastoral lifeways in highly localized contexts, adapting to new sociopolitical realities, and finding means of mutual benefit and group individuation. Future projects will attempt to extend the study of Han border history through analysis of multiple site types, delving further into short- and long-term border complexity in northwest China, using the tools highlighted in this essay alongside additional traditional and scientific archeological analysis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The permission and oversight were provided by the Ethics Committee of Fudan University of Life Sciences to study their ancient genomes. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
PS, C-CW, SW, and EA conceived the idea for the study. EA, DC, XT, YW, XQ, and PS performed or supervised the archeological work. YX, PD, JX, and SW performed or supervised the wet laboratory work. YY, XY, and C-CW analyzed the data. EA, YY, XY, PS, SW, and C-CW wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the National Key R&D Program of China (2020YFE0201600 and 2020YFC1521607), the National Social Science Fund of China (19VJX074 and 21CKG022), the National Natural Science Foundation of China (32070576, 31801040, and 32111530227), the B&R Joint Laboratory of Eurasian Anthropology (18490750300), the Major Research Program of National Natural Science Foundation of China (91731303), the Major Project of National Social Science Foundation of China granted to C-CW (21&ZD285), Xiaohua Deng (20&ZD248), and SW (20&ZD212), the “Double First-Class University Plan” Key Construction Project of Xiamen University (The Origin and Evolution of East Asian Populations and the Spread of Chinese Civilization, 0310/X2106027), the Nanqiang Outstanding Young Talents Program of Xiamen University (X2123302), the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), the 111 Project (B13016), the Major Project of Marxist Theoretical Research and Construction Project (2021MZD014), and European Research Council (ERC) grant to Dan Xu (ERC-2019-ADG-883700-TRAM).
We are thankful to Prof. Yi Guo at Zhejiang University for his help with the pre-treatment experiment of bone samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.932004/full#supplementary-material
Supplementary Figure 1 | Location of the Shichengzi site and the photograph of the four excavation areas (A–D) at Shichengzi during 2014–2019.
Supplementary Figure 2 | Shared genetic drift estimated via f4-statistics of the form (A) f4(Mbuti, Shichengzi_A; Sino-Tibetan, Altaic) and (B) f4(Mbuti, Shichengzi_B; Sino-Tibetan, Altaic) based on the merged 1240K dataset.
Supplementary Figure 3 | Shared genetic drift estimated via f4-statistics of the form (A) f4(Mbuti, Shichengzi_A; Yellow River, Mongolia Plateau) and (B) f4(Mbuti, Shichengzi_B; Yellow River, Mongolia Plateau) based on the merged 1240K dataset.
Supplementary Figure 4 | Results of f4-statistics performed in the form of f4(X, Mbuti; Shichengzi, Shirezigou) to explore the genetic similarities and differentiation between Shichengzi populations and possible ancestral source populations.
Supplementary Figure 5 | Results of f4-statistics performed in the form of f4(X, Mbuti; Shichengzi_ind, EA) to explore the genetic similarities and differentiation between Shichengzi populations and possible ancestral source populations.
Supplementary Figure 6 | (A) Mapdamage result shows mismatch frequency in different distance from 5’ and 3’ end of sequence read, four colors representing four individuals. (B) Kinship detected between pairs of individuals.
Agranat-Tamir, L., Waldman, S., Martin, M. A. S., Gokhman, D., Mishol, N., Eshel, T., et al. (2020). The genomic history of the bronze age southern levant. Cell 181, 1146–1157.e11. doi: 10.1016/j.cell.2020.04.024
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Alkass, K., Saitoh, H., Buchholz, B. A., Bernard, S., Holmlund, G., Senn, D. R., et al. (2013). Analysis of radiocarbon, stable isotopes and DNA in teeth to facilitate identification of unknown decedents. PLoS One 8:e69597. doi: 10.1371/journal.pone.0069597
Ames, K. M., Richards, M. P., Speller, C. F., Yang, D. Y., Lyman, R. L., and Butler, V. L. (2015). Stable isotope and ancient DNA analysis of dog remains from Cathlapotle (45CL1), a contact-era site on the Lower Columbia River. J. Archaeol. Sci. 57, 268–282. doi: 10.1016/j.jas.2015.02.038
Banning, E. B. (1986). Peasants, pastoralists, and pax romana: mutualism in the highlands of Jordan. Bull. Am. Sch. Orient. Res. 261, 25–50.
Boucher, D. H. (1985). “The idea of mutualism, past, and future,” in The biology of mutualism: ecology and evolution, ed. D. H. Boucher (Oxford: Oxford University Press), 1–28.
Blank, M., Sjögren, K. G., Knipper, C., Frei, K. M., Malmström, H., Fraser, M., et al. (2021). Mobility patterns in inland southwestern Sweden during the Neolithic and Early Bronze Age. Archaeol. Anthropol. Sci. 13:64. doi: 10.1007/s12520-021-01294-4
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4:7. doi: 10.1186/s13742-015-0047-8
Damgaard, P., de, B., Martiniano, R., Kamm, J., Moreno-Mayar, J. V., Kroonen, G., et al. (2018b). The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science 360:eaar7711. doi: 10.1126/science.aar7711
Damgaard, P., de, B., Marchi, N., Rasmussen, S., Peyrot, M., Renaud, G., et al. (2018a). 137 ancient human genomes from across the Eurasian steppes. Nature 557, 369–374. doi: 10.1038/s41586-018-0094-2
Frei, K., Mannering, U., Kristiansen, K., Allentoft, M. E., Wilson, A. S., Skals, I., et al. (2015). Tracing the dynamic life story of a Bronze Age Female. Sci. Rep. 5:10431. doi: 10.1038/srep10431
Frei, K. M., Bergerbrant, S., Sjögren, K. G., Jørkov, M. L., Lynnerup, N., Harvig, L., et al. (2019). Mapping human mobility during the third and second millennia BC in present-day Denmark. PLoS One 14:e0219850. doi: 10.1371/journal.pone.0219850
Gowland, R. L. (2017). Embodied identities in Roman Britain: a bioarchaeological approach. Britannia 48, 175–194.
Haak, W., Brandt, G., Jong, H. N. D., Meyer, C., Ganslmeier, R., Heyd, V., et al. (2008). Ancient DNA, Strontium isotopes, and osteological analyses shed light on social and kinship organization of the Later Stone Age. Proc. Natl. Acad. Sci. U.S.A. 105, 18226–18231. doi: 10.1073/pnas.0807592105
Haak, W., Lazaridis, I., Patterson, N., Rohland, N., Mallick, S., Llamas, B., et al. (2015). Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211. doi: 10.1038/nature14317
Hedges, R., and Reynard, L. (2007). Nitrogen isotopes and the trophic level of humans in archaeology. J. Archaeol. Sci. 34, 1240–1251.
Helga, T., James, T. R., and Jill, P. M. (2013). Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinf. 14, 178–192. doi: 10.1093/bib/bbs017
Ingman, T., Eisenmann, S., Skourtanioti, E., Akar, M., Ilgner, J., Gnecchi Ruscone, G. A., et al. (2021). Human mobility at Tell Atchana (Alalakh), Hatay, Turkey during the 2nd millennium BC: Integration of isotopic and genomic evidence. PLoS One 16:e0241883. doi: 10.1371/journal.pone.0241883
Jeong, C., Balanovsky, O., Lukianova, E., Kahbatkyzy, N., Flegontov, P., Zaporozhchenko, V., et al. (2019). The genetic history of admixture across inner Eurasia. Nat. Ecol. Evol. 3, 966–976. doi: 10.1038/s41559-019-0878-2
Jeong, C., Wang, K., Wilkin, S., Taylor, W. T. T., Miller, B. K., Bemmann, J. H., et al. (2020). A Dynamic 6,000-Year genetic history of eurasia’s eastern steppe. Cell 183, 890–904.e29. doi: 10.1016/j.cell.2020.10.015
Jónsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. F., and Orlando, L. (2013). mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684. doi: 10.1093/bioinformatics/btt193
Jun, G., Wing, M. K., Abecasis, G. R., and Kang, H. M. (2015). An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 25, 918–925. doi: 10.1101/gr.176552.114
Knapp, M., Clarke, A. C., Horsburgh, K. A., and Matisoo-Smith, E. A. (2012). Setting the stage – Building and working in an ancient DNA laboratory. Ann. Anat. 194, 3–6. doi: 10.1016/j.aanat.2011.03.008
Korneliussen, T. S., Albrechtsen, A., and Nielsen, R. (2014). ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15:356. doi: 10.1186/s12859-014-0356-4
Kuhn, J. M. M., Jakobsson, M., and Günther, T. (2018). Estimating genetic kin relationships in prehistoric populations. PLoS One 13:e0195491. doi: 10.1371/journal.pone.0195491
Langlie, B. S., and Capriles, J. M. (2021). Paleoethnobotanical evidence points to agricultural mutualism among early camelid pastoralists of the Andean central Altiplano. Archaeol. Anthropol. Sci. 13:107. doi: 10.1007/s12520-021-01343-y
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Li, N. (2017). The organization and management of the tuntian system in the Western Regions during Han dynasties. Agricult. Archaeol. 1, 124–132.
Luo, L., Wang, X., Lasaponara, R., Xiang, B., Zhen, J., Zhu, L., et al. (2018). Auto-Extraction of linear archaeological traces of tuntian irrigation canals in miran site (China) from Gaofen-1 Satellite Imagery. Remote Sens. 10:718.
Mallick, S., Li, H., Lipson, M., Mathieson, I., Gymrek, M., Racimo, F., et al. (2016). The simons genome diversity project: 300 genomes from 142 diverse populations. Nature 538, 201–206. doi: 10.1038/nature18964
Mao, X., Zhang, H., Qiao, S., Liu, Y., Chang, F., Xie, P., et al. (2021). The deep population history of northern East Asia from the Late Pleistocene to the Holocene. Cell 184, 3256–3266.e13. doi: 10.1016/j.cell.2021.04.040
Mathieson, I., Lazaridis, I., Rohland, N., Mallick, S., Patterson, N., Roodenberg, S. A., et al. (2015). Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499-503. doi: 10.1038/nature16152
Meskell, L., and Preucel, R. W. (2004). Identities. a companion to social archaeology. Oxford: Oxford University Press.
Meyer, M., and Kircher, M. (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010:pdb.prot5448. doi: 10.1101/pdb.prot5448
Mittnik, A., Massy, K., Knipper, C., Wittenborn, F., and Promerová, M. B. (2019). Kinship-based social inequality in Bronze Age Europe. Science 366, 731–734. doi: 10.1126/science.aax6219
Mittnik, A., Wang, C.-C., Svoboda, J., and Krause, J. (2016). A molecular approach to the sexing of the triple burial at the upper paleolithic site of dolní vìstonice. PLoS One 11:e0163019. doi: 10.1371/journal.pone.0163019
Narasimhan, V. M., Patterson, N., Moorjani, P., Rohland, N., Bernardos, R., Mallick, S., et al. (2019). The formation of human populations in South and Central Asia. Science 365:eaat7487. doi: 10.1126/science.aat7487
Ning, C., Li, T., Wang, K., Zhang, F., Li, T., Wu, X., et al. (2020). Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat. Commun. 11:2700. doi: 10.1038/s41467-020-16557-2
Ning, C., Wang, C.-C., Gao, S., Yang, Y., Zhang, X., Wu, X., et al. (2019). Ancient genomes reveal yamnaya-related ancestry and a potential source of Indo-European Speakers in Iron Age Tianshan. Curr. Biol. 29, 2526–2532.e4. doi: 10.1016/j.cub.2019.06.044
Parker, S. T. (1987). Peasants, Pastoralists, and “Pax Romana”: A Different View. Bull. Am. Sch. Orient. Res. 265, 35–51.
Patterson, N., Isakov, M., Booth, T., Büster, L., Fischer, C. E., Olalde, I., et al. (2022). Large-scale migration into Britain during the Middle to Late Bronze Age. Nature 601, 588–594. doi: 10.1038/s41586-021-04287-4
Patterson, N., Moorjani, P., Luo, Y., Mallick, S., Rohland, N., Zhan, Y., et al. (2012). Ancient Admixture in Human History. Genetics 192, 1065–1093. doi: 10.1534/genetics.112.145037
Patterson, N., Price, A. L., and Reich, D. (2006). Population Structure and Eigenanalysis. PLoS Genetics 2:e190. doi: 10.1371/journal.pgen.0020190
Peltzer, A., Jäger, G., Herbig, A., Seitz, A., Kniep, C., Krause, J., et al. (2016). EAGER: efficient ancient genome reconstruction. Genome Biol. 17:60. doi: 10.1186/s13059-016-0918-z
Ralf, A., Montiel González, D., Zhong, K., and Kayser, M. (2018). Yleaf: software for human Y-Chromosomal haplogroup inference from next-generation sequencing data. Mol. Biol. Evol. 35, 1291–1294. doi: 10.1093/molbev/msy032
Reimer, P., Austin, W., Bard, E., Bayliss, A., Blackwell, P. G., Ramsey, C. B., et al. (2020). The IntCal20 Northern Hemisphere radiocarbon age calibration curve (0–55 cal kBP). Radiocarbon 62, 725–757.
Renaud, G., Slon, V., Duggan, A. T., and Kelso, J. (2015). Schmutzi: Estimation of Contamination and Endogenous Mitochondrial Consensus Calling for Ancient DNA. Genome Biol. 16:224. doi: 10.1186/s13059-015-0776-0
Richards, M. P., and Hedges, R. E. M. (1999). Stable isotope evidence for similarities in the types of marine foods used by late mesolithic humans at sites along the atlantic coast of Europe. J. Archaeol. Sci. 26, 717–722.
Schubert, M., Lindgreen, S., and Orlando, L. (2016). AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9:88. doi: 10.1186/s13104-016-1900-2
Sheng, P., Liu, Y., Tian, X., Wu, Y., and Guan, Y. (2021). Paleo-environmental implications of the micro-botanical remains recovered from a military garrison of Han Dynasty in Xinjiang. J. Archaeol. Sci. 39:103176.
Sheng, P., Storozum, M., Tian, X., and Wu, Y. (2020). Foodways on the Han dynasty’s western frontier: archeobotanical and isotopic investigations at Shichengzi, Xinjiang, China. Holocene 30, 1174–1185.
Sirak, K. A., Fernandes, D. M., Lipson, M., Mallick, S., Mah, M., Olalde, I., et al. (2021). Social stratification without genetic differentiation at the site of Kulubnarti in Christian Period Nubia. Nat. Commun. 12:7283. doi: 10.1038/s41467-021-27356-8
Skoglund, P., Storå, J., Götherström, A., and Jakobsson, M. (2013). Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 40, 4477–4482. doi: 10.1016/j.jas.2013.07.004
Spengler, R. N. III (2019). Fruit from the Sands: The Silk Road Origins of the Foods We Eat. Berkeley: University of California Press.
Sun, X., Wen, S., Lu, C., Zhou, B., Curnoe, D., Lu, H., et al. (2021). Ancient DNA and multimethod dating confirm the late arrival of anatomically modern humans in southern China. Proc. Natl. Acad. Sci. U.S.A. 118:e2019158118. doi: 10.1073/pnas.2019158118
Tian, X. (2021). Archaeological excavation and findings from the Shichengzi site in Qitai County, Xinjiang. Cult, Relics World 7, 71–74.
Tian, X., Feng, Z., and Wu, Y. (2020). The 2018 Excavation of the Shichengzi Site in Qitai County. Xinjiang. Archaeology 12, 21–40.
Tian, X., Wu, Y., Duo, S., Zhang, S., and Chen, X. (2018). The 2016 Excavation of the Shichengzi Site in Qitai County, Xinjiang. Cult. Relics 5, 4–25.
Wang, C. C., Reinhold, S., Kalmykov, A., Wissgott, A., Brandt, G., Jeong, C., et al. (2019). Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat. Commun. 10:590. doi: 10.1038/s41467-018-08220-8
Wang, C.-C., Yeh, H.-Y., Popov, A. N., Zhang, H.-Q., Matsumura, H., Sirak, K., et al. (2021). Genomic insights into the formation of human populations in East Asia. Nature 591, 413–419. doi: 10.1038/s41586-021-03336-2
Weissensteiner, H., Pacher, D., Kloss-Brandstätter, A., Forer, L., Specht, G., Bandelt, H.-J., et al. (2016). HaploGrep 2: Mitochondrial Haplogroup Classification in the Era of High-Throughput Sequencing. Nucleic Acids Res. 44, W58–W63. doi: 10.1093/nar/gkw233
Wilson, A. S., Taylor, T., Ceruti, M. C., Chavez, J. A., Reinhard, J., Grimes, V., et al. (2007). Stable isotope and DNA evidence for ritual sequences in Inca child sacrifice. Proc. Natl. Acad. Sci. U.S.A. 104, 16456–16461. doi: 10.1073/pnas.0704276104
Yang, J. P. (1991). Several questions of the tuntian farming during the Western Han dynasty. J. Chin. Soc. Econ. History 4, 11–18.
Yang, M. A., Fan, X., Sun, B., Chen, C., Lang, J., Ko, Y.-C., et al. (2020). Ancient DNA indicates human population shifts and admixture in northern and southern China. Science 369, 282–288. doi: 10.1126/science.aba0909
Zhang, A., and Tian, H. (2015). The remains of the site the layout of the defense cities of the Han dynasty in the Western Regions. J. Chin. Historic. Geogr. 30, 47–55.
Zhang, F., Ning, C., Scott, A., Fu, Q., Bjørn, R., Li, W., et al. (2021). The genomic origins of the Bronze Age Tarim Basin mummies. Nature 599, 256-261. doi: 10.1038/s41586-021-04052-7
Keywords: ancient DNA, isotope analysis, Han population, lifestyle, Silk Road, Western Regions
Citation: Allen E, Yu Y, Yang X, Xu Y, Du P, Xiong J, Chen D, Tian X, Wu Y, Qin X, Sheng P, Wang C-C and Wen S (2022) Multidisciplinary lines of evidence reveal East/Northeast Asian origins of agriculturalist/pastoralist residents at a Han dynasty military outpost in ancient Xinjiang. Front. Ecol. Evol. 10:932004. doi: 10.3389/fevo.2022.932004
Received: 29 April 2022; Accepted: 18 July 2022;
Published: 10 August 2022.
Edited by:
Gyaneshwer Chaubey, Banaras Hindu University, IndiaReviewed by:
Sara V. Good, University of Winnipeg, CanadaCopyright © 2022 Allen, Yu, Yang, Xu, Du, Xiong, Chen, Tian, Wu, Qin, Sheng, Wang and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengfei Sheng, c2hlbmdwZW5nZmVpQGZ1ZGFuLmVkdS5jbg==; Chuan-Chao Wang, d2FuZ0B4bXUuZWR1LmNu; Shaoqing Wen, d2Vuc2hhb3FpbmcxOTgyQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.