
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 09 June 2022
Sec. Conservation and Restoration Ecology
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.928816
This article is part of the Research TopicEffects of Non-Random Sources of Alteration on Biodiversity and Ecosystem FunctioningView all 19 articles
Cyanobacteria bloom is a global aquatic ecological problem that seriously threatens human health and social development. The outbreak of cyanobacteria bloom is affected by various environmental factors, among which light dose is an essential factor. In this study, the growth changes of cyanobacteria under different amounts of natural light were studied by simulating different depths of Taihu Lake, and we used 16S rRNA and non-targeted metabolomics for sequencing to reveal the effects of light on the diversity of cyanobacteria and coexisting microorganisms, and to analyze the changes of related genes, functional structures and internal metabolism involved in nitrogen cycling. The result shows that excessive and insufficient light could limit the growth, photosynthesis, and EPS secretion of cyanobacteria, resulting in an antioxidant stress response. At the same time, the amount of natural light affects the vertical distribution of cyanobacteria, and under the condition of 1/3 natural light, cyanobacteria first appeared to float. In addition, the amount of natural light affects the diversity, abundance, and metabolites of cyanobacteria and coexisting microorganisms, and the expression of nifH, nirK, and nirS, three nitrogen-fixing genes, is significantly different in different genera. This study provides valuable information on the molecular mechanism of the effects of the amount of natural light on cyanobacteria bloom.
- Insufficient and excessive light affects the growth of algae cells.
- Light dose affects the microbial community structure of symbiosis with algae.
- The amount of light affects the expression of metabolites and related nitrogen-fixing genes of microorganisms.
Cyanobacteria are one of the earliest organisms on earth and are considered significant contributors to the formation and nitrogen fixation of the earth’s atmosphere (Sylvain et al., 2013). Cyanobacteria have a series of unique physiological characteristics, such as nitrogen fixation mechanism, temperature and light adaptation mechanism, and suspension mechanism,
enabling them to occupy a competitive advantage in a relatively harsh environment. Furthermore, cyanobacteria blooms are easy to form in freshwater lakes and reservoirs in summer (Hou et al., 2021). In recent years, the frequent occurrence of cyanobacteria blooms in Taihu Lake in China has seriously threatened ecosystem security and human health, making more research scholars want to uncover the underlying mechanism and explore more effective prevention measures.
The environmental factors affecting cyanobacteria bloom are complex, and light dose is considered to be one of the most critical environmental factors (Figueredo and Giani, 2009; Desortovaa and Pun Ocha, 2011). Early studies consider cyanobacteria to be the dominant species under high turbidity and insufficient light conditions (Havens et al., 2003) and confirm that some cyanobacteria could lie dormant for several months in the absence of light and grow rapidly immediately after light restoration (Montechiaro and Giordano, 2006). At the same time, the light will affect the vertical distribution of cyanobacteria in the water. Cyanobacteria through the continuous assembly and rupture of intracellular air sacs suspend in appropriate water depth to obtain the optical light conditions so that they can reproduce in large numbers (Wallace et al., 2000; Pinilla, 2006). At present, most studies are based on laboratory control of light dose to explore its influence on the growth of cyanobacteria (Kruger et al., 1981; Wiedner et al., 2003; El Semary, 2010; Salvador et al., 2016). However, the difference between the growth of cyanobacteria under natural light and indoor culture is obvious, so it is more practical to discuss the physiological and biochemical characteristics of algae under natural light.
Cyanobacteria is one of the most widespread nitrogen-fixing microorganisms, playing a pivotal role in nitrogen fixation in the water environment, and the energy needed for nitrogen fixation is mainly supplied by photosynthesis (Kuypers et al., 2018). Plants, eukaryotes, prokaryotes, and other organisms coexisting in the water environment where cyanobacteria live are also involved in the nitrogen fixation process. Plants can form chloroplasts for photosynthesis by forming endosymbiosis with cyanobacteria. Eukaryotes have also been reported to achieve nitrogen-fixing symbiosis with cyanobacteria. Cyanobacteria can sit on the cell wall of a kind of single-celled algae, donating fixed nitrogen and receiving fixed carbon in return (Thompson et al., 2012; Martínez-Pérez et al., 2016). It has also been widely reported that prokaryotic microorganisms participate in nitrogen fixation, nitrification, and denitrification in the process of nitrogen cycling, and the related links are mainly driven by microorganisms through regulating the corresponding enzymes encoded by their functional genes (Garnier et al., 2006; Zhong et al., 2017). For example, most ammonia-oxidizing bacteria belonging to Betaproteobacteria and Gammaproteobacteria can encode ammonia monooxygenase (AMO) to oxidize ammonia to hydroxyl amine (Hooper et al., 1997; Arp and Stein, 2003). Proteobacteria and Verrucomicrobia have a variety of methano-oxidizing bacteria that can encode hydroxylamine oxidoreductase (HAO) to convert hydroxyl amine into NO, which can be further oxidized to nitrous acid or reduced to N2O. The diversity of microorganisms involved in the N cycle can be discovered in these relevant reports (Nyerges and Stein, 2009; Maalcke et al., 2014). Meanwhile, the responses of microbial N function genes to environmental changes in different ecosystems are different (Tu et al., 2016; Kou et al., 2019; Yu et al., 2019), which contains oxygen produced by photosynthesis may affect the activity of enzymes involved in nitrogen fixation (Berman-Frank et al., 2003). However, there is a lack of research on the effects of natural light on the expression of nitrogen-fixing genes and metabolites of cyanobacteria and coexisting microorganisms.
The research on cyanobacteria mainly focuses on the effects of nutrient salts and other factors on algal growth and bloom formation, while studies on the relationship between algae and light have mainly focused on experimental simulations, lacking the effects of natural outdoor light on algal growth. In particular, there is a lack of studies on the effects of light on the algae and algae symbiosis microbial diversity distribution, metabolites, and nitrogen fixation gene expression. Therefore, in this paper, we investigated under different amounts of natural light the physiological and biochemical responses of Microcystis aeruginosa at low concentrations as well as the expression changes of the diversity of coexisting microorganisms, metabolic process, and nitrogen-fixing process-related genes. The main research contents are as follows: (1) growth and photosynthesis of algae cells; (2) EPS secretion and protein synthesis of algal cells; (3) Changes in antioxidant enzyme system in algae cells; (4) Diversity analysis of coexisting microorganisms in the growth environment of algae cells; (5) Correlation analysis between metabolites and microorganisms; (6) Expression of genes related to microbial involvement in nitrogen fixation. This study reveals the influence mechanism of natural light on the growth of cyanobacteria and the formation of algal blooms.
Microcystis aeruginosa is a microcystin-producing typical alga. The Microcystis aeruginosa (FACHB-905) used in this study was purchased from Wuhan Institute of Hydrobiology, Chinese Academy of Sciences (Wuhan, China). Plexiglas columns were cleaned and filtered (filter membrane 0.45 μm) Taihu Lake water was added, followed by indoor cultures of Microcystis aeruginosa. The culture conditions were direct cultivation under natural field conditions.
The experimental device is shown in Figure 1. The experiment was carried out in four plexiglass cylinders (height: 200cm, diameter: 20cm), with an injection port set at the bottom of cylinder, and four sampling outlets set at heights of 10, 60, 120, and 180 cm, respectively (the average water depth of Taihu lake is 1.8 m). The culture (100mL) is added to the experimental device in batches and inoculated Microcystis aeruginosa into a sterilized plexiglass water column (60 L) at an initial cell density of 1.0 × 104 cells/mL. Four experiments adopted four different natural light conditions (full light S1, 2/3 amount of light S2/3, 1/3 amount of light S1/3, and no light S0, respectively).
The experiment was repeated three times for each group, and the test cycle was 33 days. During the experiment, the average daytime light intensity at the location of the experimental device was 1.01 × 105 lx measured by an illuminometer (9:00–10:00 am, 0.38 × 105lx; 11:00–12:00 am, 1.01 × 105 lx; 2:00–3:00 pm, 1.04 × 105 lx). To ensure the light dose and experimental accuracy, the experimental device was placed on the open field on the shore of Zhushan Bay in the northwest of Taihu Lake (120.087969°E, 31.469109°N). We took 30 mL water samples from each of the four sampling outlets every three days for analysis and evaluated the effects of light on diversity, molecular phylogeny, and metabolic activity of Microcystis aeruginosa.
The total chlorophyll content (chlorophyll-a) was measured by ethanol extraction according to the method of Chao (Chao et al., 2008). Extracellular polymeric substances include the soluble extracellular polysaccharides (sEPS) and the bound extracellular polysaccharides (bEPS). EPS extraction for protein quantification and analysis was performed as follows: 10 mL of sample solution were centrifuged at 2500 g for 15 min (VELOCITY 14R, Dynamica, Australia) and the supernatant was collected to measure sEPS content. Then, the precipitate was suspended in a 0.05% NaCl solution, Ultrasonicated (SB-3200D, SCIENTZ, China) at 150 W for 60 s, and centrifuged at 12000 g for 15min. The resulting supernatant was carefully collected for the measurement of bEPS. The concentrations of the two forms of EPS were analyzed using the phenol-sulfuric acid method.
To establish the concentration and activity of plant enzymes, 20 mL algal cells were collected from each group and centrifuged at 2500 × g for 15 min. Then transferred into PBS (0.01m) and homogenized at 12000 r⋅min-1 for 2 min (FSH-2a, XINRUI INSTRUMENT, 3). Disrupted cells and centrifuged at 12000 g at 4°C for 15 min to obtain supernatants. The activity of the SOD, CAT, and POD was assessed in supernatants using a commercialized chemical assay kit: TP assay kit, SOD assay kit, CAT assay kit, POD assay kit (Nanjing Jiancheng Bioengineering Institute, China).
A sample of 200 mL was collected and filtered through a 0.45 μm fiber membrane, which was collected and used for testing. Total genomic DNA samples were extracted using the OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek, Norcross, GA, United States), following the manufacturer’s instructions, and stored at −20°C prior to further analysis. Then, the extracted genomic DNA was detected by 1% agarose gel electrophoresis. PCR amplicons were purified with Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, United States). After the individual quantification step, amplicons were pooled in equal amounts, and pair-end 2250 bp sequencing was performed using the Illumina NovaSeq platform with NovaSeq 6000 SP Reagent Kit (500 cycles) at Shanghai Personal Biotechnology Co., Ltd (Shanghai, China). Referring to previous studies, the selection of amplification primer sequences was shown in Table 1 (Cantera and Stein, 2007; Zhang et al., 2017).
A sample of 500 mL was collected and filtered through a 0.45 μm fiber membrane, which was collected and used for testing. Take a sample of the filter membrane and put it into a 2 mL EP tube, accurately add 3 mL of acetonitrile: isopropanol: Water (3:3:2, V/V/V) Mixed solution (−20°C), and add 3–4 2 mm zirconium beads; Put it into a high flux tissue grinder, shocked at 30 Hz for 20 s, allowed to stand for 10 s, cycled eight times, and sonicated in an ice water bath for 5 min; After centrifugation for 2 min, 500 μL of the supernatant solution is taken and added into a new 2 mL EP tube. The vacuum concentrator was concentrated to dryness (8-10 h), And the remaining supernatant was placed on a −80°C refrigerator for backup; Add 80 μL of 20 mg/mL MEOX solution for redissolution, vortex vibration for 30 s, and incubate at 60°C for 60 min; Finally, add 100 μL BSTFA-TMCS (99:1) reagent, react at 70°C for 90 min, emulsion at 14 000 RPM for 3 min, And add 90–100 μL of supernatant into the detection bottle; Samples were placed in sealed cuvettes to be tested and processed for GC-TOF upper detection within 24 h. Gas Chromatography was performed on a DB-5ms Capillary column (30 m × 250 μm I.D., 0.25 μm film thickness, Agilent J & W Scientific, Folsom, CA, United States) to separate the effects at a constant flow of 1 mL/min helium. 1 μL of the sample was injected in a split mode in A 1:10 Split ratio by the auto-sampler. The injection temperature was 280°C. The temperature of the transfer line ion source was 320°C and 230°C, respectively. The programs of temperature-rise were followed by an initial temperature of 50°C for 0.5 min, 15°C/min rate up to 320°C, and Staying at 320°C for 9 min. Mass spectrometry was performed using a full scan method with a scan rate of 10 spec/s, electron energy of –70 V, and a solvent delay of 3 min.
GraphPad Prism Software was used to process data, while data was analyzed using SPSS software (Version 22.0, SPSS Inc., Chicago, IL, United States). Comparisons between different treatments (flow and static) were performed by two-way analysis of variance (ANOVA) and subsequent Fisher LSD comparison tests. Both bacterial community structure and abundance in V. natans leaf biofilms were assessed using the online platform Majorbio I-Sanger Cloud Platform.1 Each experiment was performed in triplicate and repeated at least 3 times.
Algal cell density is often used to measure the algae growth. To study the effect of the amount of natural light on the growth of Microcystis aeruginosa, we compared the cell density of Microcystis aeruginosa at different water depths (1.8, 1.2, 0.6, 0.1m) under four natural light conditions (Figure 2): full light (Figure 2A), 2/3 amount of light (Figure 2B), 1/3 amount of light (Figure 2C), and no light (Figure 2D). The results showed that the growth of Microcystis aeruginosa varied significantly with different amounts of light and different water depths. By comparing the average density of algae in the four groups, we found that the algae growth was the best under 2/3 amount of light in each water depth. On the 33rd day, the algae density from top to bottom reached 1.81 × 10 6 cells/mL, 1.16 × 106 cells/mL, 2.09 × 106 cells/mL, and 0.94 × 106 cells/mL, and the average density of algae was the highest among the four groups (1.50 × 106 cells/mL). This indicated that the growth of algal cells is best under 2/3 amount of light, and excessive or insufficient natural light affects the growth of algae cells. Furthermore, we believe that the growth rate of algae cells in the lower layer is lower than that in the upper layer (0.1 m < 0.6 m < 1.2 m < 1.8 m deep water), which may be due to the blocking of natural light by the algae cells in the upper layer, affecting the photosynthesis of the algae cells in the lower layer (Havens et al., 2003). Under the condition of no light, the algae density in each water depth was lower than that in other groups and different from the other three groups, the algae density showed a trend of increasing first and then decreasing, and reached the maximum on the 21st day. This indicates that insufficient light would limit the growth of algae cells and accelerate the decay of algae cells.
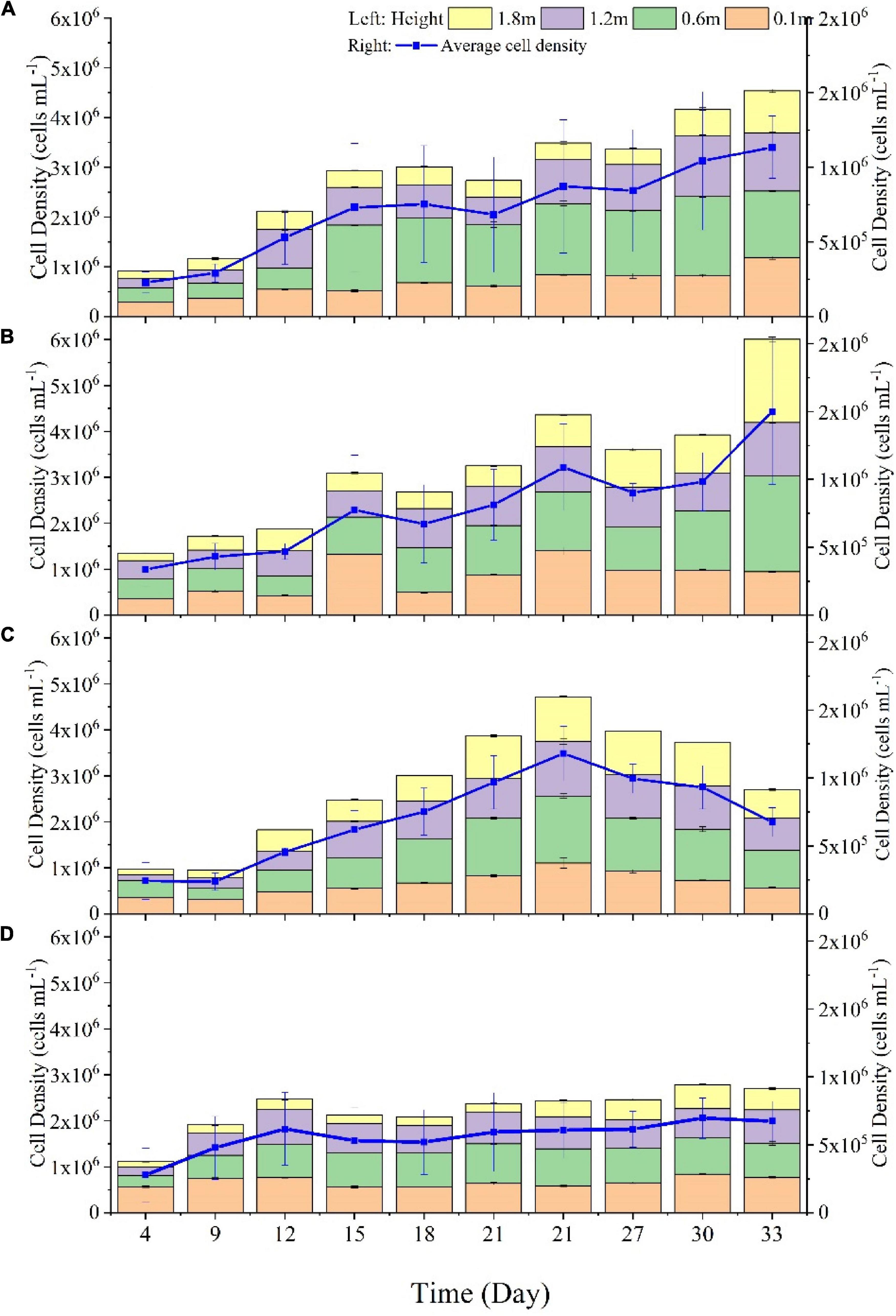
Figure 2. The cell density of Microcystis aeruginosa at different water depths (1.8, 1.2, 0.6, 0.1m) under four natural light conditions: full light (A), 2/3 amount of light (B), 1/3 amount of light (C), and no light (D).
Through culturing Microcystis aeruginosa under different light intensities, found that when the incident light dose was less than 120 lx, the specific growth rate of Microcystis aeruginosa was negative, which means the extinction trend appeared (Chen et al., 2016). In the enclosure of Microcystis blooms in summer, Sun Yang-cai et al. (2007) shaded the water surface to reduce the light dose at 0.1 m depth to 800 lx, which makes the concentration of chlorophyll a in the enclosure decrease from 176.9 μg/L before shading to 21.4 μg/L after 7 days. In conclusion, the growth of algal cells is closely related to the amount of natural light, and both excessive and insufficient light can limit the growth and photosynthesis of algal cells (Havens et al., 2003).
Microcystis aeruginosa can secrete EPS to form a protective film on the cell surface to cope with external stimuli. EPS has strong viscosity and is closely related to the formation of the Microcystis aeruginosa community (Xiao et al., 2018). The main components of EPS in algae cells are polysaccharides and proteins, as well as a small amount of lipids and DNA (Phélippé et al., 2019). To further study the influence mechanism of the amount of natural light on the growth of Microcystis aeruginosa, on the last day, we measured the contents of soluble EPS (s-EPS) in the growth environment and binding EPS (b-EPS) on the surface of algae cells (Figure 3A) and the aggregation proportion of algae cells (Figure 3B). The results showed that the concentration of s-EPS was higher than that of b-EPS under different amounts of natural light, and the concentration of s-EPS was between 19.68 and 41.10 μg/mL, and that of b-EPS was between 3.25 and 10.86 μg/mL. The concentrations of the two EPS were significantly lower than those in other groups under the condition of no light (S0 group). The amount of EPS secreted by algal cells will affect the size of cells aggregation proportion, thus changing the morphology of the cell community. We compared the agglomeration rate of algae cells in each group (Figure 3B) and found that under the condition of 2/3 amount of light (S2/3 Group), the aggregation proportion of algae cells reached the highest, which was positively correlated with EPS concentration. Studies have shown that the acetyl group in polysaccharide components of EPS is an important factor affecting the hydrophobicity and adhesion of algae cells, which can promote the aggregation of algae cells (Chen et al., 2016). The aggregation proportion in the S0 group was significantly lower than that of other groups, indicating that insufficient light could reduce the secretion of EPS, and the reduction of EPS could reduce the aggregation proportion of algae cells.

Figure 3. Changes of soluble EPS (S-EPS) in the growth environment of algae and binding EPS (B-EPS) on the surface of algae cells contents (A) and aggregation proportion (B) in each group. The upper and lower boundaries represent the smallest observed value (lower edge) and the largest observed value (upper edge), the black dot in the middle is the mean, and the red dotted line is the median line.
In order to further explore the effects of different amounts of natural light on the growth of cyanobacteria cells, this study compared the levels of four common antioxidant stress indicators, including total reactive oxygen species (ROS), superoxide dismutase (SOD), peroxidase (POD) and malondialdehyde (MDA), in each group on the 30th day (Figure 4). The results showed that the concentrations of ROS and MDA were lowest under the condition of two-thirds amount of light, and all indexes reached the maximum values under no light condition, indicating that 2/3 amount of light was the most suitable for the growth of algae cells. The stress degree of algae cells is the most severe under no light condition, which can stimulate the activities of SOD and POD in the antioxidant enzyme system to remove ROS in cells and alleviate the oxidative damage of algae cells (Romeo et al., 2000). Other sources of ROS include the oxidation of glycolic acid and the degradation of fatty acids. Alpha Hydroxyl Acid, the final product of chloroplast photorespiration, enters the peroxisome and is oxidized to glycolic acid with the help of O2 to produce H2O2. H2O2 is also produced when fatty acids are degraded by acyl-coA oxidase. ROS produced by peroxisomes are used for seed and pollen germination, fruit ripening, senescence, and stomatal movement (Anuj et al., 2020).
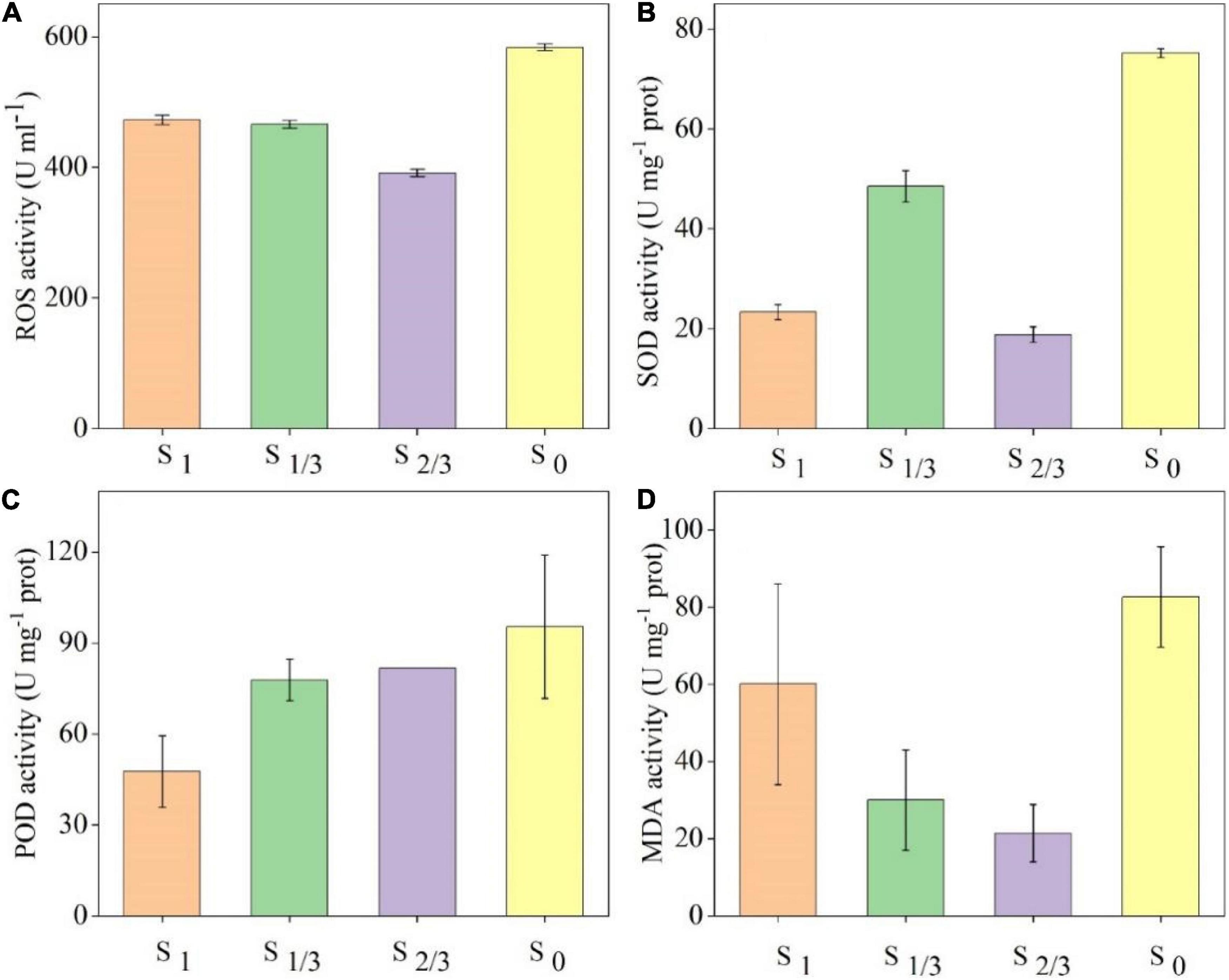
Figure 4. Oxidative stress indexes of algae cells grown under different amounts of light: (A) ROS content, (B) and the concentration changes of superoxide dismutase (SOD), (C) peroxidase (POD), (D) malondialdehyde (MDA).
In addition, when the external stress exceeds the balance capacity of the cellular antioxidant system, the oxygen free radicals generated by the non-enzymatic system in algae cells will trigger lipid peroxidation to form lipid peroxides (Kaur et al., 2006), which can explain why the concentration of MDA in S0 group is the highest.
In this paper, we used 16S rRNA sequencing to investigate the influence of different light levels on microbial community diversity in the growth environment of Microcystis aeruginosa, and the results are shown in Figure 5. The Venn diagram (Figure 5A) shows the distribution of the number of operational taxonomic units (OTUs) for each group of samples, and common and unique OTUs can be used to show composition similarity and overlap of different samples. The number of OTUs for each group was 449 (S1), 370 (S2/3), 1083 (S1/3), and 1934 (S0), indicating that different amounts of natural light had significant effects on microbial diversity in cyanobacteria growing environment. To further explore the richness and species variation of each microflora, we compared the relative abundance of microbial communities in each group at the level of phylum classification (Figure 5B). It turns out that at the phylum level, Proteobacteria, Firmicutes, Bacteroidetes, Cyanobacteria, and Actinobacteria were the dominant bacteria in the growth environment of cyanobacteria under different light conditions. These dominant bacteria have been confirmed to be ubiquitous in polluted water and can promote the stability of EPSs secretion (Lv et al., 2014; Wang et al., 2016; Liu et al., 2017). The relative abundances of these phyla changed significantly under different light conditions. Proteobacteria and Firmicutes reached the maximum relative abundance under 2/3 and 1/3 natural light, respectively. The relative abundances of Bacteroidetes and Cyanobacteria reached the maximum under full light conditions, while that of Actinobacteria reached the maximum under no light conditions. Cyanobacteria and bacteria usually interact in this microenvironment, including mutualism and mutual inhibition (Chen et al., 2007; Sun Yang-cai et al., 2007; Bai et al., 2016; Mehmeti et al., 2016). Among these bacteria interacting with cyanobacteria, many studies have shown that Proteobacteria are the most abundant bacterial group in the water environment of Taihu Lake (Ma et al., 2016; Zhu et al., 2019).
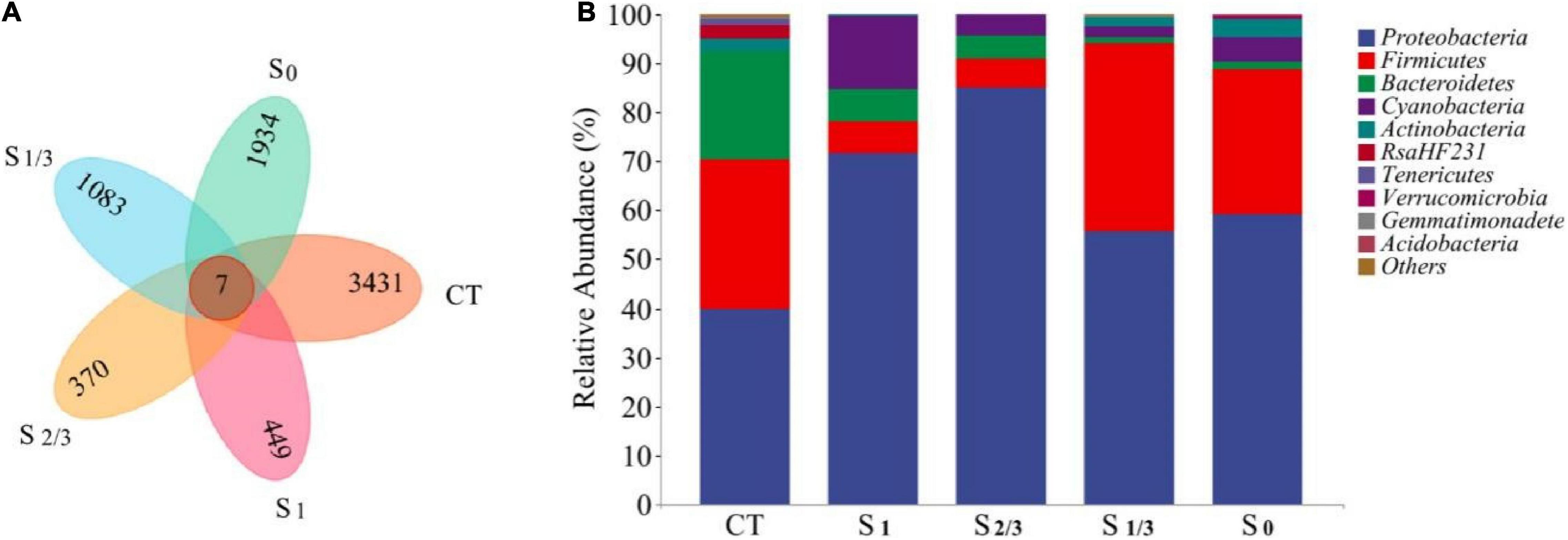
Figure 5. Microbial community analysis of different samples: (A) Venn diagram of the distribution of OTUs. (B) The relative abundance of microbial communities at the phylum level.
At the same time, Bacteroidetes is also a common bacterial species in the algal sphere, and a major component of hydrolytic acidification reactions (Ma et al., 2016). The microbial community composition in the study system also differed under different light conditions. In addition, they found that light dose had a significant impact on the microbial community through the study on lake surface water, which is consistent with the conclusions of our study (Paver and Kent, 2017).
The results of PICRUSt (Figure 6) showed that the main microbial functions in the growth environment of Microcystis aeruginosa include biosynthesis, degradation/utilization/assimilation, detoxification, generation of precursor, metabolite, and energy, glycan pathways, macromolecule modification and metabolic Clusters (Figure 6A). Among them, amino acid biosynthesis occupies the most abundant biosynthesis function, which provides essential energy and carbon source for bacteria (Lopez-Gonzalez et al., 2015). Other studies have shown that during amino acid metabolism, bacteria can use ammonia to synthesize amino acids and humus, which is conducive to denitrification (Wu et al., 2017). In addition, the light dose can significantly affect other biosynthesis functions such as cofactor, prosthetic group, electron carrier, and vitamin biosynthesis. nucleoside and nucleotide biosynthesis, fatty acid and lipid biosynthesis, and carbohydrate biosynthesis. Bacteria can synthesize fatty acids using acetyl coenzyme a (Co A) as a precursor, and fatty acids are the main components of the cell membrane. By regulating the type and composition of fatty acids, bacteria can regulate cell membrane fluidity, and maintain membrane stability and normal physiological functions to adapt to adversity stress (Zhang and Rock, 2008).
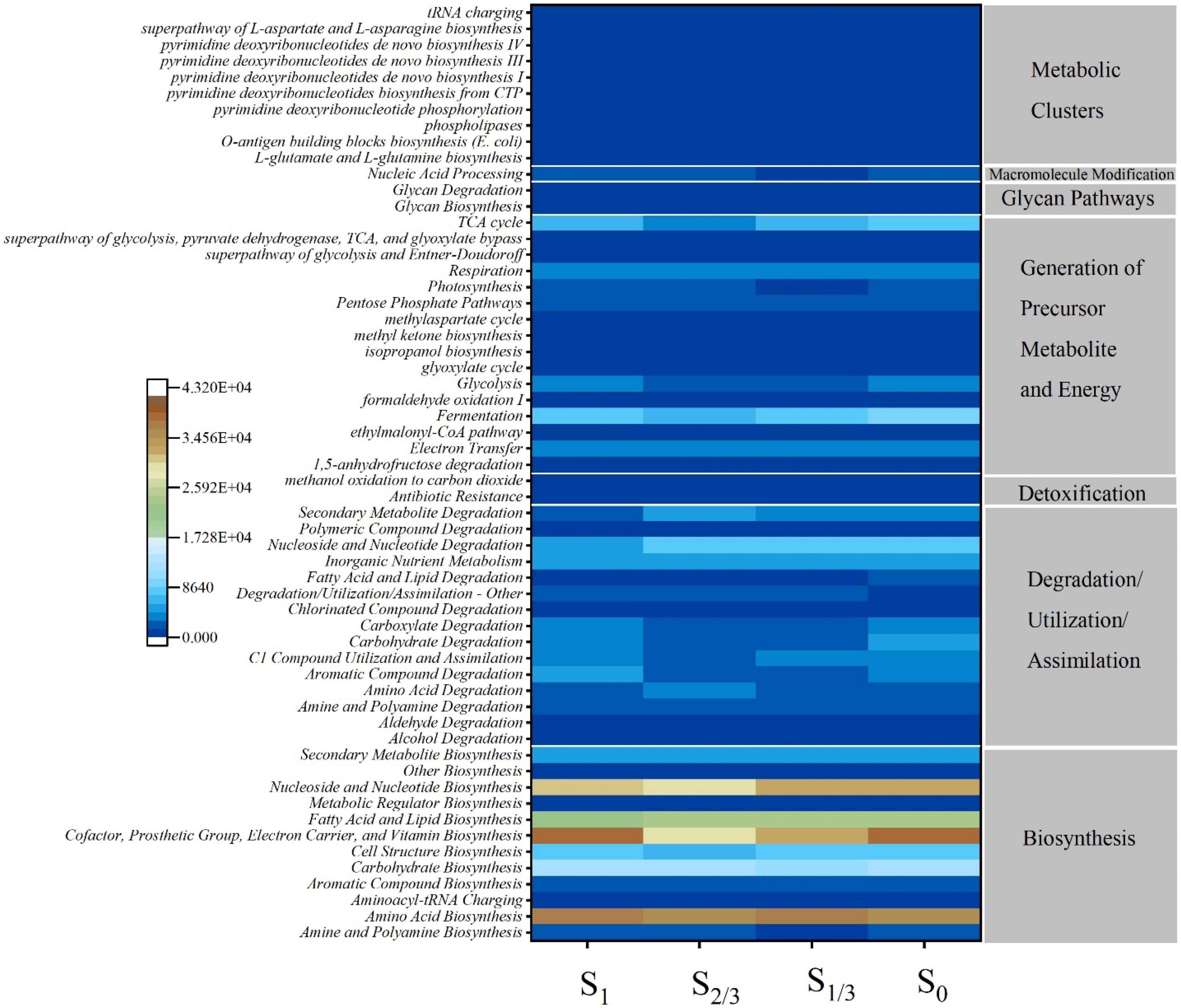
Figure 6. Metabolic pathways of different functional categories predicted by PICRUSt algorithm based on the microbial composition of 16S rRNA. Legends represent the richness of the predicted readings, red for metabolic function with high richness and blue for metabolic function with low richness.
To explore the effect of light on the relationship between cyanobacterial growth and microorganisms, we determined the metabolome of microorganisms in the environment using GC-MS. The thermography about the correlation between microflora and metabolites (Figure 7) showed that 26 of the 50 metabolites have at least one significant correlation with microorganisms. Most of the microorganisms in Bacteroidetes and Cyanobacteria were negatively correlated with the change of metabolites, while most of the microorganisms in Actinomycetes were positively correlated with the change of metabolites. Among Proteobacteria, Acinetobacter, Blastomonas, Gemmobacter, Mesorhizobium, and Porphyrobacter were all negatively correlated with metabolites, while Arenimonas and Hydrogenophaga were positively correlated with metabolites.
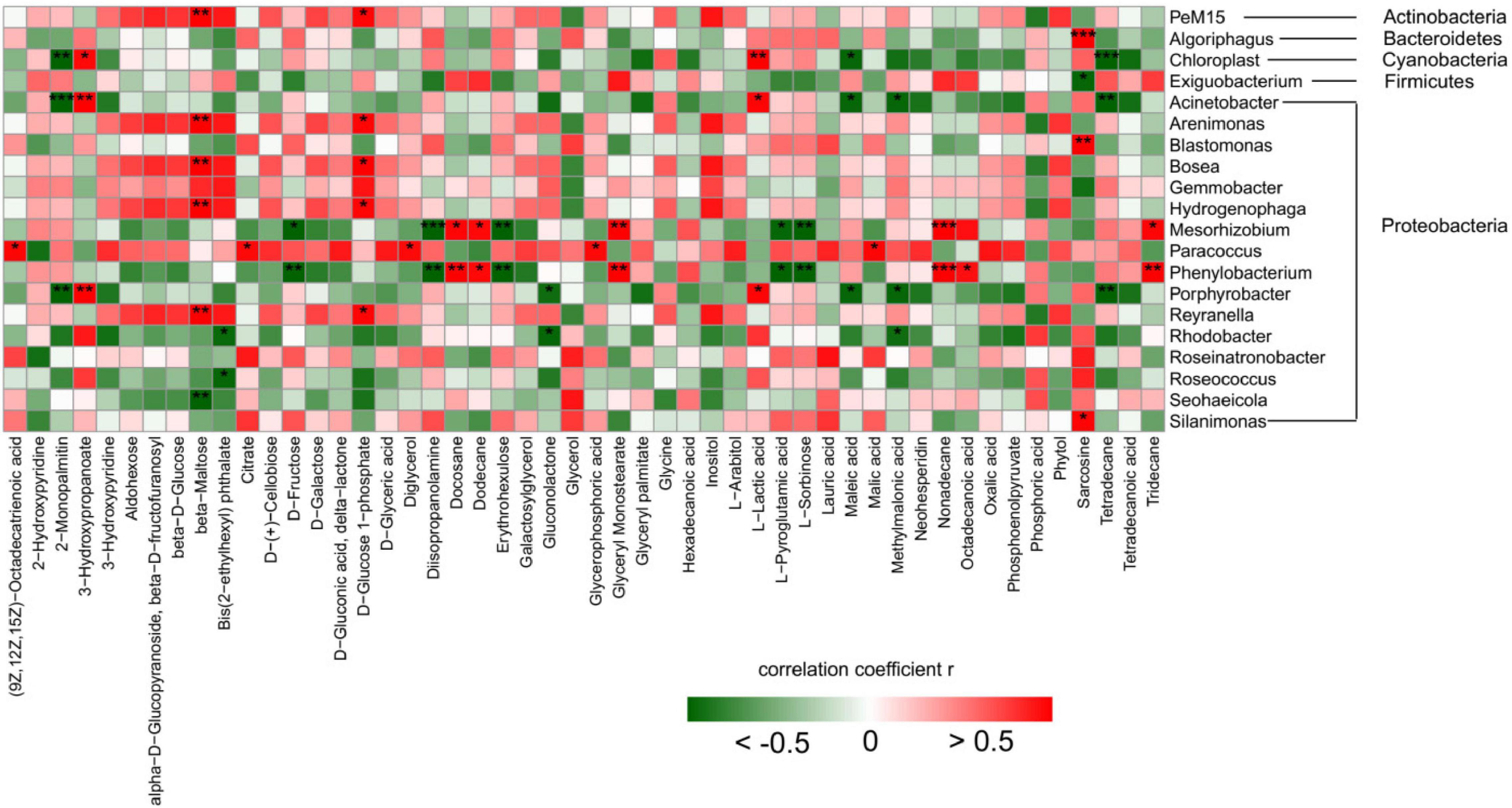
Figure 7. Thermograms of correlation between bacteria and metabolites. The notes on the right are the top 20 bacteria genus; Below are the names of the top 50 metabolites in abundance. Correlation analysis: Pearson correlation coefficient and P value were calculated, and the number of * was marked according to the P value (P < 0.001, *** means extremely significant correlation; P < 0.01, ** means very significant correlation; P < 0.05, * means significant correlation; P > 0.05, no * means insignificant correlation; Red represents positive correlation, green represents negative correlation).
The genus of Cyanobacteria showed a significant negative correlation with 2-Monopalmitin, Maleic acid, and Tetradecane, while a significant positive correlation with L-Lactic acid and 3-Hydroxypropanoate. L-lactic acid is generated from the catalytic reduction of pyruvate by NAD-dependent lactate dehydrogenase (nLDH) (Goffin et al., 2004). This process involves glycolysis, gluconeogenesis, fructose and mannose metabolism, pyruvate metabolism, and other metabolic pathways. Actinomycetes showed a significant positive correlation with beta-Maltose, D-Gluconic acid, and delta-lactone. Bacteroidetes showed a significant positive correlation with Sarcosine, and Firmicutes showed a significant negative correlation with Phytol. In summary, metabolomics analysis showed that cyanobacteria and microorganisms in the environment produced a large number of metabolites under different light conditions, and the relative contents of metabolites in various bacteria were significantly affected by different amounts of light.
Microorganisms participate in nitrogen fixation, nitrification, denitrification, and other processes in the nitrogen cycle by releasing enzymes encoded by their functional genes (Zhong et al., 2017). NifH is a marker gene commonly used to determine nitrogen-fixing microorganisms in the environment, which is involved in the process of transforming N2 into NH3 in the nitrogen-fixing cycle (Zehr et al., 2003). During the nitrogen cycle, nitrite reductase (Nir) encoded by nirK or nirS genes can catalyze NO2– to NO reduction, which is often used as a marker gene to characterize the diversity and abundance of denitrifying bacteria in the environment (Regan et al., 2017; Ma et al., 2019). To further explore the effect of light conditions, we analyzed the expression of genes related to nitrogen-fixing microorganisms in the growth environment of Microcystis aeruginosa at the end of the experiment (Figure 8).
The results showed that there were significant differences in the expressions of nifH(a), nirK(b), and nirS(c) genes in different strains under different light levels.
Among them, the nifH gene is mainly expressed in Flavobacterium, Rhodopseudomonas, Desulfovibrio, and Rhizobium. Several kinds of microbes, including Desulfovibrio, are characterized by an increase in nifH expression with the decrease of light dose (Figure 8A), while the expression of nitrogen-fixing gene nirK in Pseudomonas, Alcaligenes, Rhizobium, and other microorganisms gradually increased with the decrease of light exposure (Figure 8B). The nirS gene was expressed in various microorganisms including Azotobacter, Azospirillum, Bacillus, Rhodobacteria, Bradyrhizobium, Pseudomonas, Dechlorococcus, and Thioalgae (Figure 8C).
For example, Proteobacteria, Anaerobic ammonia-oxidizing Bacteria, and Bacteroidetes can reduce nitrite to nitric oxide (Maia and Moura, 2014). These microorganisms have been found in many hypoxic environments, and nirK and nirS are involved in the expression of nitrite reductase (Bartossek et al., 2010). However, these nitrogen-fixing genes have also been found in other bacteria, such as anaerobic ammonium-oxidizing bacteria, nitrite and methane-oxidizing bacteria, and ammonia-oxidizing bacteria and archaea. These bacteria do not contain nirK and nirS genes but may use other nitrite reductases to reduce nitrite to nitric oxide (Kartal and Keltjens, 2016).
In addition, some researchers have found that the expression of microbial N functional genes is affected by climatic factors (e.g., precipitation or temperature), and most of the environmental variables significantly related to N functional genes (e.g., precipitation, soil organic matter, etc.) are also closely related to basic metabolism of microorganisms (De Vries et al., 2012; Zhou et al., 2021). Consequently, the amount of natural light can be an important factor affecting the expressions of related genes of nitrogen-fixing microorganisms in the nitrogen cycle, which provides a more in-depth discussion and theoretical basis for the important role of coexisting microorganisms in the nitrogen-fixing process in the growth environment of cyanobacteria.
In conclusion, this paper deeply discusses the effects of natural light on the growth state of cyanobacteria individuals and communities, as well as the response of cyanobacteria molecular systems and cyanobacterial coexistence community structure, biosynthetic functions, metabolism, and nitrogen fixation processes. We conclude that both excessive and insufficient light can limit cyanobacterial growth, affect photosynthesis and EPS production of algae, and influence the vertical distribution of cyanobacteria in the water body. At the same time, the amount of light can alter the structure of the microbial communities and their metabolites in the cyanobacterial growth environment, and may also affect the expression of nitrogen-fixing genes associated with microorganisms that coexist with cyanobacteria. This study is of great significance to further study the potential mechanism of cyanobacterial blooms.
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
ZW: conducting experiments, data analysis, and writing – original draft preparation. GL: data analysis and writing – review and editing. HH: curation and writing – review and editing. WZ: conceptualization and methodology. JW: investigation and resources. SH and ZZ: supervision, writing – review and editing, and funding acquisition. All authors contributed to the article and approved the submitted version.
This study was supported by ABA Chemicals, Chengdu University of Technology Research Startup Fund (10912-KYQD2020-08431), and National Environmental Protection Key Laboratory of Cooperative Control and Joint Remediation of Water and Soil Pollution Open Fund Project (GHB-2020-010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Anuj, C., Antul, K., and Nirmaljit, K. (2020). Ros and oxidative burst:roots in plant development. Plant Divers. 42, 35–45. doi: 10.1016/j.pld.2019.10.002
Arp, D. J., and Stein, L. Y. (2003). Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 38, 471–495. doi: 10.1080/10409230390267446
Bai, L. L., Xu, H. C., Wang, C. H., Deng, J. C., and Jiang, H. L. (2016). Extracellular polymeric substances facilitate the biosorption of phenanthrene on cyanobacteria Microcystis aeruginosa. Chemosphere 162, 172–180. doi: 10.1016/j.chemosphere.2016.07.063
Bartossek, R., Nicol, G. W., Lanzen, A., Klenk, H. P., and Schleper, C. (2010). Homologues of nitrite reductases in ammonia-oxidizing archaea: diversity and Genomic Context. Environ. Microbiol. 12, 1075–1088. doi: 10.1111/j.1462-2920.2010.02153.x
Berman-Frank, I., Lundgren, P., and Falkowski, P. (2003). Nitrogen fixation and the evolution of photosynthetic characteristics in cyanobacteria. Res. Microbiol. 154, 157–164.
Cantera, J. J. L., and Stein, L. Y. (2007). Molecular diversity of nitrite reductase genes (nirK) in nitrifying bacteria. Environ. Microbiol. 9, 765–776. doi: 10.1111/j.1462-2920.2006.01198.x
Chao, W., Song, H. Z., Pei, F. W., Hou, J., Wei, L., and Wen, J. Z. (2008). Metabolic adaptations to ammonia-induced oxidative stress in leaves of the submerged macrophyte Vallisneria natans (Lour.) Hara. Aquat. Toxicol. 87, 88–98. doi: 10.1016/j.aquatox.2008.01.009
Chen, M., Xie, M., Wu, W., and Yu, F. (2016). Chemical characteristics of capsular polysaccharide and water-soluble released exopolysaccharide from Microcystis. Chin. J. Lake Sci. 28, 609–615.
Chen, X. S. Y.-C., Zeng, X.-W., Li, C.-J., and Kong, H.-N. (2007). The inhibition of low light dose on the growth of Microcystis aeruginosa in raw water. China Environ. Sci. 3, 352–355.
De Vries, F. T., Manning, P., Tallowin, J. R. B., Mortimer, S. R., Pilgrim, E. S., Harrison, K. A., et al. (2012). Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol. Lett. 15, 1230–1239.
Desortovaa, B., and Pun Ocha, P. (2011). Variability of phytoplankton biomass in a Lowland River: response to climate change in China. J. Clim. Change 41, 160–166.
El Semary, N. A. (2010). Investigating factors affecting growth and cellular mcyB transcripts of Microcystis aeruginosa PCC 7806 using Real - time PCR. Ann. Microbiol. 60, 181–188. doi: 10.1007/s13213-010-0024-5
Figueredo, C. C., and Giani, A. (2009). Phytoplankton community in the tropical lake of Lagoa Santa (Brazil): combined effect of Cylindrospermopsis hydroscopicity and hydroscopicity on Cylindrospermopsis hydroscopicity. J. Hydroscopicity 39, 264–272.
Garnier, J., Cebron, A., Tallec, G., Billen, G., Sebilo, M., and Martinez, A. (2006). Nitrogen behaviour and nitrous oxide emission in the tidal Seine River estuary (France) as influenced by human Activities in the upstream watershed. Biogeochemistry 77, 305–326. doi: 10.1007/s10533-005-0544-4
Goffin, P., Lorquet, F., Kleerebezem, M., and Hols, P. (2004). Major role of NAD-dependent lactate dehydrogenases in aerobic lactate utilization in Lactobacillus plantarum during early stationary phase. J. Bacteriol. 186, 6661–6666. doi: 10.1128/JB.186.19.6661-6666.2004
Havens, K. E., James, R. T., East, T. L., and Smith, V. H. (2003). N:P ratios, light limitation, and cyanobacterial dominance in a subtropical lake impacted by non-point source nutrient pollution. Environ. Pollut. 122, 379–390. doi: 10.1016/s0269-7491(02)00304-4
Hooper, A. B., Vannelli, T., Bergmann, D. J., and Arciero, D. M. (1997). Enzymology of the oxidation of Ammonia to nitrite by Bacteria. Antonie Leeuwenhoek 71, 59–67. doi: 10.1023/a:1000133919203
Hou, W., Song, Y., Lu, N., and Zhang, R. (2021). Synergy effect of environmental factors on the growth and toxins production by Microcystis aeruginosa. Nat. Environ. Pollut. Technol. 20, 891–898.
Kartal, B., and Keltjens, J. T. (2016). Anammox Biochemistry: a tale of heme C Proteins. Trends Biochem. Sci. 41, 998–1011. doi: 10.1016/j.tibs.2016.08.015
Kaur, G., Alam, M. S., Jabbar, Z., Javed, K., and Athar, M. (2006). Evaluation of antioxidant activity of Cassia siamea flowers. J. Ethnopharmacol. 108, 340–348. doi: 10.1016/j.jep.2006.05.021
Kou, Y., Li, C., Li, J., Tu, B., Wang, Y., and Li, X. (2019). Climate and soil parameters are more important than denitrifier abundances in controlling potential denitrification rates in Chinese grassland soils. Sci. Total Environ. 669, 62–69. doi: 10.1016/j.scitotenv.2019.03.093
Kruger, G. H. J., Eloff, J. N., and Pretorius, J. A. (1981). Morphological changes in toxic and non-toxic microcystis isolates at different irradiance levels. J. Phycol. 17, 52–56.
Kuypers, M. M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276.
Liu, H. B., Song, X., Guan, Y. N., Pan, D., Li, Y. H., Xu, S. Y., et al. (2017). Role of illumination intensity in microcystin development using Microcystis aeruginosa as the model algae. Environ. Sci. Pollut. Res. 24, 23261–23272. doi: 10.1007/s11356-017-9888-2
Lopez-Gonzalez, J. A., Suarez-Estrella, F., Vargas-Garcia, M. C., Lopez, M. J., Jurado, M. M., and Moreno, J. (2015). Dynamics of bacterial microbiota during lignocellulosic waste composting: studies upon its structure, functionality and biodiversity. Bioresour. Technol. 175, 406–416. doi: 10.1016/j.biortech.2014.10.123
Lv, Y., Wan, C., Lee, D. J., Liu, X., and Tay, J. H. (2014). Microbial communities of aerobic granules: granulation mechanisms. Bioresour. Technol. 169, 344–351. doi: 10.1016/j.biortech.2014.07.005
Ma, J. R., Qin, B. Q., Paerl, H. W., Brookes, J. D., Hall, N. S., Shi, K., et al. (2016). The persistence of cyanobacterial (Microcystis spp.) blooms throughout winter in Lake Taihu, China. Limnol. Oceanogr. 61, 711–722.
Ma, Y., Zilles, J. L., and Kent, A. D. (2019). An evaluation of primers for detecting denitrifiers via their functional genes. Environ. Microbiol. 21, 1196–1210. doi: 10.1111/1462-2920.14555
Maalcke, W. J., Dietl, A., Marritt, S. J., Butt, J. N., Jetten, M. S., Keltjens, J. T., et al. (2014). Structural basis of biological NO generation by octaheme oxidoreductases. J. Biol. Chem. 289, 1228–1242. doi: 10.1074/jbc.M113.525147
Martínez-Pérez, C., Mohr, W., Löscher, C., Dekaezemacker, J., Littmann, S., Yilmaz, P., et al. (2016). The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat. Microbiol. 1:16163. doi: 10.1038/nmicrobiol.2016.163
Mehmeti, E., Stankovic, D. M., Chaiyo, S., Svorc, L., and Kalcher, K. (2016). Manganese dioxide-modified carbon paste electrode for voltammetric determination of riboflavin. Microchim. Acta 183, 1619–1624. doi: 10.1007/s00604-016-1789-4
Montechiaro, F., and Giordano, M. (2006). Effect of prolonged dark incubation on pigments and photosynthesis of the cave-dwelling cyanobacterium Phormidium autumnale (Oscillatoriales, Cyanobacteria). Phycologia 45, 704–710.
Nyerges, G., and Stein, L. Y. (2009). Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol. Lett. 297, 131–136. doi: 10.1111/j.1574-6968.2009.01674.x
Paver, S. F., and Kent, A. D. (2017). Direct and context-dependent effects of light, temperature, and phytoplankton shape bacterial community composition. Ecosphere 8:e01948.
Phélippé, M., Gonçalves, O., Thouand, G., Cogne, G., and Laroche, C. (2019). Characterization of the polysaccharides chemical diversity of the cyanobacteria arthrospira platensis. Algal Res. 38:101426. doi: 10.1016/j.algal.2019.101426
Pinilla, G. A. (2006). Vertical distribution of phytoplankton in a clear water lake of Colombian Amazon. Hydrobiologia 568, 79–90.
Regan, K., Stempfhuber, B., Schloter, M., Rasche, F., Prati, D., Philippot, L., et al. (2017). Spatial and temporal dynamics of nitrogen fixing, nitrifying and denitrifying microbes in an unfertilized grassland soil. Soil Biol. Biochem. 109, 214–226. doi: 10.1016/j.soilbio.2016.11.011
Romeo, M., Bennani, N., Gnassia-Barelli, M., Lafaurie, M., and Girard, J. P. (2000). Cadmium and copper display different responses towards oxidative stress in the kidney of the sea bass Dicentrarchus labrax. Aquat. Toxicol. 48, 185–194. doi: 10.1016/s0166-445x(99)00039-9
Salvador, D., Churro, C., and Valerio, E. (2016). Evaluating the influence of light intensity in MCYA gene expression an microcystin production in toxic strains of Planktothrix agardhii and Microcystis aeruginosa. J Microbiol. Methods 123, 4–12. doi: 10.1016/j.m
Sun Yang-cai, C. X. U., Kong, H., Liang, Y. U., Wu, D. I., and Li, C. (2007). Algae growth potential of Changjiang estuary reservoirs and effectiveness of light shading for algae control. Environ. Pollut. Control 11, 805–808.
Sylvain, M., Snyder, S., Baurès, E., and Thomas, O. (2013). State of knowledge and concerns on cyanobacterial blooms and cyanotoxins. Environ. Int. 59, 303–327. doi: 10.1016/j.envint.2013.06.013
Thompson, A. W., Carter, B. J., Musat, N., Vaulot, D., Musat, N., Vaulot, D., et al. (2012). Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Shows that the ubiquitous Cyanobacterium ucyn-a forms A nitrogen fixing symbiosis with AN Algae. Science 337, 1546–1550.
Tu, Q. C., Deng, Y., Yan, Q. Y., Shen, L. N., Lin, L., He, Z. L., et al. (2016). Biogeographic patterns of soil diazotrophic communities across six forests in the North America. Mol. Ecol. 25, 2937–2948. doi: 10.1111/mec.13651
Wallace, B. B., Bailey, M. C., and Hamilton, D. P. (2000). Simulation of vertical position of bu oyancy regulating microcys tis aeruginosa in a shallow eutrophic lake. Aquat. Sci. 62, 320–333.
Wang, S., Gao, M., Li, Z., She, Z., Wu, J., Zheng, D., et al. (2016). Performance evaluation, microbial enzymatic activity and microbial community of a sequencing batch reactor under long-term exposure to cerium dioxide nanoparticles. Bioresour. Technol. 220, 262–270. doi: 10.1016/j.biortech.2016.08.086
Wiedner, C., Visser, P. M., Fastner, J., Metcalf, J. S., Codd, G. A., and Mur, L. R. (2003). Effects of light on the microcystin content of Microcystis strain PCC 7806. Appl. Environ. Microbiol. Mar. 69, 1475–1481. doi: 10.1128/AEM.69.3.1475-1481.2003
Wu, J., Zhao, Y., Qi, H., Zhao, X., Yang, T., Du, Y., et al. (2017). Identifying the key factors that affect the formation of humic substanc during different materials composting. Bioresour. Technol. 244, 1193–1196. doi: 10.1016/j.biortech.2017.08.100
Xiao, M., Li, M., and Reynolds, C. S. (2018). Colony formation in the cyanobacterium Microcystis[J]. Biol. Rev. 93, 1399–1420. doi: 10.1111/brv.12401
Yu, M., Meng, J., Yu, L., Su, W., Afzal, M., Li, Y., et al. (2019). Changes in nitrogen related functional genes along soil pH, C and nutrient gradients in the charosphere. Sci. Total Environ. 650, 626–632. doi: 10.1016/j.scitotenv.2018.08.372
Zehr, J. P., Jenkins, B. D., Short, S. M., and Steward, G. F. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5, 539–554. doi: 10.1046/j.1462-2920.2003.00451.x
Zhang, M., Bai, S. H., Tang, L., Zhang, Y., Teng, Y., and Xu, Z. (2017). Linking potential nitrification rates, nitrogen cycling genes and soil properties after remediating the agricultural soil contaminated with heavy metal and fungicide. Chemosphere 184, 892–899. doi: 10.1016/j.chemosphere.2017.06.081
Zhang, Y. M., and Rock, C. O. (2008). Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6, 222–233.
Zhong, L., Zhou, X., Wang, Y., Li, F. Y., Zhou, S., Bai, Y., et al. (2017). Mixed grazing and clipping is beneficial to ecosystem recovery but may increase potential N2O emissions in a Semi - arid grassland. Soil Biol. Biochem. 114, 42–51. doi: 10.1016/j.soilbio.2017.07.002
Zhou, S., Xue, K., Zhang, B., Tang, L., Pang, Z., Wang, F., et al. (2021). Spatial patterns of microbial nitrogen-cycling gene abundances along a precipitation gradient in various temperate grasslands at a regional scale. Geoderma 404:115236.
Keywords: visible light, cyanobacteria bloom, metabolic activity, nitrogen-fixing gene, microbial diversity
Citation: Wang Z, Li G, Huang H, Zhang W, Wang J, Huang S and Zheng Z (2022) Effects of Solar Radiation on the Cyanobacteria: Diversity, Molecular Phylogeny, and Metabolic Activity. Front. Ecol. Evol. 10:928816. doi: 10.3389/fevo.2022.928816
Received: 26 April 2022; Accepted: 18 May 2022;
Published: 09 June 2022.
Edited by:
Chao Wang, Chinese Academy of Fishery Sciences, ChinaReviewed by:
Yueming Qu, UK Centre for Ecology and Hydrology (UKCEH), United KingdomCopyright © 2022 Wang, Li, Huang, Zhang, Wang, Huang and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suzhen Huang, aHVhbmdzekBmdWRhbi5lZHUuY24=; Zheng Zheng, enpoZW5naGpAZnVkYW4uZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.