- 1The Key Laboratory of Mariculture Ministry of Education, Ocean University of China, Qingdao, China
- 2Fisheries College, Ocean University of China, Qingdao, China
- 3College of Environmental Science and Engineering, Guilin University of Technology, Guilin, China
In recent years, Harpadon nehereus gradually become a dominant species with great potential for exploitation in the East China Sea, and it is worth investigating whether H. nehereus would tolerate cold stress to continue to expand into the colder northern waters. The molecular regulation level is favorable evidence to explore the cold tolerance of H. nehereus, a total of 6,650, 1,936, and 2,772 differentially expressed genes (DEGs) in transcription regulation, and 4,409, 1,250, and 2,303 differential alternative splicing genes (DASGs) in alternative splicing regulation were identified in H. nehereus at 13, 15, and 17°C, respectively, importantly, 47 genes were identified as the key candidate genes for cold tolerance in H. nehereus. In transcription regulation, up-regulated DEGs were enriched in metabolic process terms and ribosome, spliceosome pathway, etc., while down-regulated DEGs were enriched in signal transduction terms, focal adhesion, proteoglycans in cancer pathway, etc., at 13, 15, and 17°C, respectively. In alternative splicing regulation, spliceosome, mRNA surveillance pathway, etc., were significantly enriched in DASGs. In a word, H. nehereus adapts to cold environments mainly through transcription and translation, transmembrane transport, protein modification, etc., while cold stress may also induce some diseases in H. nehereus.
Introduction
Global warming is causing a series of climate changes that threaten the survival and distribution of fish, such as ocean warming and frequent extreme winter cold events (Fossheim et al., 2015; Johnson et al., 2018). Water temperature is the master factor to determine nearly all life activities of most fishes, including growth, development, reproduction, behavior, and geographical distribution (Lopez-Olmeda and Sanchez-Vazquez, 2011; Poloczanska et al., 2016). Like other ectotherms, changes in fish distribution areas may vary with daily and seasonal water temperature variations (Morgan et al., 2013). Currently, the impact of ocean warming on fish is a hot topic of interest for researchers, which often overlooks the survival and distribution pressure on fish caused by extremely winter cold temperatures triggered by climate warming. The research on the cold tolerance of fish was more concerned with cultured economics (Xu et al., 2018; Sun et al., 2019; Jiao et al., 2020) and model fish species (Valdez et al., 2005; Ren et al., 2021), while studies on natural marine fish were relatively scarce. In addition, the study found that fish can adapt to variations in water temperature through physiological plasticity or micro-evolution (Whitehead et al., 2013), and the adaptive evolution of temperature tolerance determines their ecological status and distribution range, so the study of cold tolerance of fish in natural marine areas has significance to estimate the variations in their distribution range.
Lizardfish Harpadon nehereus, also known as Bombay duck, is mostly discontinuously distributed in the benthic along the coasts and estuaries of East Africa, Asia, and Indonesia, e.g., the Ganges Estuary, the Bay of Bengal, and the Cambay Bay (Bapat, 1970). In China, H. nehereus is widely distributed in the south of the Yellow Sea, the East China Sea (Liu and Cheng, 2015), and the South China Sea (Zhu et al., 2014). The morphological characteristics of H. nehereus including small eyes, wide mouth, incomplete skull, membranous skin, and gelatinous body with high moisture (>90%) (Roy et al., 2014) give it the resemblance to deep-sea fishes (Wang et al., 2019), whereas H. nehereus inhabits offshore sandy mud bottom in most of a year. Historically, H. nehereus have been overlooked for its low value and less yield in China. In recent decades, with the marine environmental change and the traditional fisheries decline, the population size of H. nehereus in the East China Sea has rapidly expanded (Lin, 2009). Resource investigations of fish communities indicate that H. nehereus has become dominant species in newly expanded areas of its current range along the southern coast of China (Lin, 2009; Sun et al., 2015). The growth of H. nehereus populations may be accompanied by a lack of survival resources and interspecific competition, and it may have forced them to expand to colder northern waters for survival, deserving detailed studies for more evidence besides biological habits (Firdaus et al., 2018), and nutritional processing (Taqwa et al., 2020). This study not only provides molecular insights into the cold tolerance of H. nehereus, but also enriches the genetic information.
The molecular response mechanism of fish to cold stress is subject to complex regulation at multiple levels, e.g., epigenetic modifications, transcription, translation, alternative splicing, and post-translational modifications. Exploring the cold tolerance of fish at the molecular level is the basis for the study of fish adaptation to low-temperature environments. With the development of high-throughput sequencing technology and bioinformatics, RNA-Seq has developed rapidly into a powerful tool for better understanding the candidate genes and potential biological pathways in responding to different environmental stresses (Sun et al., 2020; Sun et al., 2021), as well as predicting new transcripts, simple sequence repeats (SSR), and alternative splicing (AS) (Lan et al., 2014). Although the flexibility of AS contributes to environmental adaption and phenotypic plasticity in organisms (Mastrangelo et al., 2012), studies on cold tolerance of fish were mostly at the transcriptional level, such as Nibea albiflora (Xu et al., 2018), Epinephelus coioides (Sun et al., 2019), Larimichthys crocea (Mininni et al., 2014), Larimichthys polyactis (Liu F. et al., 2020), and Nibea albiflora (Jiao et al., 2020), and lack of studies on alternative splicing. Therefore, the study of cold tolerance in H. nehereus from both transcription and alternative splicing levels is certainly an innovative way that could search for more evidence from multiple regulation levels.
In this study, we performed the transcription and alternative splicing regulation to explore the regulating strategies of cold tolerance in H. nehereus. This study is the first to reveal the molecular mechanism of cold tolerance in H. nehereus, and fill a partial gap in transcription and alternative splicing research on cold tolerance in fish.
Materials and Methods
Experimental Animals
From November 2020 to January 2021, H. nehereus (average body length: 159.71 ± 24.16 mm, weight: 50.53 ± 12.30 g) were collected from the East China Sea, they were harvested from 13, 15, 17, and 24°C natural marine areas. During the sampling process, we tried to ensure that temperature was the only variable in different sampling areas and other environmental indicators were similar, the location and environment information of sampling sites were presented in Supplementary Table 1. We immediately anesthetized the freshly caught H. nehereus with ice in the sampling process, followed by rapid dissection of muscle (M), and put them into liquid nitrogen for preservation. After disembarkation, they were transferred to −80°C refrigerator for storage and subsequent RNA extraction. Each group had three biological duplications and a total of 12 samples were taken for RNA-Seq.
RNA Extraction, Library Preparation, and Sequencing
Total RNA was isolated from tissues sampled by using TRIZOL Reagent (Invitrogen, United States) and treated with RNase-free DNase I (Takara, China) at 37°C for 1 h to remove residual genomic DNA. RNA purity was checked using NanoPhotometer® spectrophotometer (IMPLEN, CA, United States). RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, United States). Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States) was used to assess the quality of extracted RNA. Only high-quality RNA samples were used to construct cDNA libraries, and the libraries were sequenced by paired-end sequencing on the Illumina Novaseq6000 sequencing platform (Illumina, United States).
Identification and Enrichment Analysis of Differentially Expressed Genes
Firstly, clean reads from raw data were obtained using Fastp software by removing reads containing adapter, reads containing ploy-N, and low-quality reads (Chen et al., 2018). Subsequently, the clean reads were mapped to the H. nehereus reference genome using STAR software (Dobin et al., 2013). Based on the numbers of clean reads mapped to the genome sequence, the differential expressed level of genes was calculated using featureCounts (Liao et al., 2014). The expression of each transcript was normalized using the transcripts per million (TPM) algorithm, and then read-count data was subjected to differential expressed analysis using the DESeq2 R package (Love et al., 2014). The differentially expressed genes (DEGs) were identified by | log2FC| ≥ 1 (which corresponds to Fold Chang ≥ 2 or ≤ −2) and p-adjusted (p-adj) ≤ 0.01. Moreover, DEGs were divided into up-regulated DEGs and down-regulated DEGs, and Gene Ontology (GO) (Ashburner et al., 2000) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000) pathway enrichment analyses (p-adj < 0.05) were performed for up-and down-regulated DEGs, respectively.
Identification of Differential Alternative Splicing
In addition to transcription regulation, mRNA data was used to study alternative splicing regulation by evaluating exon expression. Differential exon usage was assessed using the DEXSeq R package. The operational steps are described in detail in the DEXSeq manual.1 Threshold as p-adjusted (p-adj) ≤ 0.01 and log2FC ≥ 2 was set for screening DAS.
Ethics Statement
All animal experiments were conducted following the guidelines and approval of the respective Animal Research and Ethics Committees of the Ocean University of China, and the field studies did not involve endangered or protected species. Following the Management Rule of Laboratory Animals (Chinese Order No. 676 of the State Council, revised 1 March 2017).
Results
Base Data Statistics About Transcriptome
In this study, 12 cDNA libraries were constructed and sequenced, and clean Q20 and Q30 were over 95 and 90%, respectively. The range of clean reads mapped to the genome sequences was 84.41–97.39%, among which uniquely mapped reads over 75% (Supplementary Table 2). All the above results proved that the data were highly accurate, and ensured the reliability of subsequent analysis results.
Analysis of Differentially Expressed Genes
A large number of DEGs were mined in H. nehereus under low-temperature environments, 6,650 (974 up- and 5,676 down-regulated), 1,936 (352 up- and 1,584 down-regulated), and 2,772 (426 up- and 2,346 down-regulated) genes were differentially expressed with | log2FC| ≥ 1 and p-adj ≤ 0.01 at 13, 15, and 17°C, respectively (Figure 1). Under the three cold stresses, unique and shared DEGs were shown in Figure 2, among which 734 DEGs were shared.
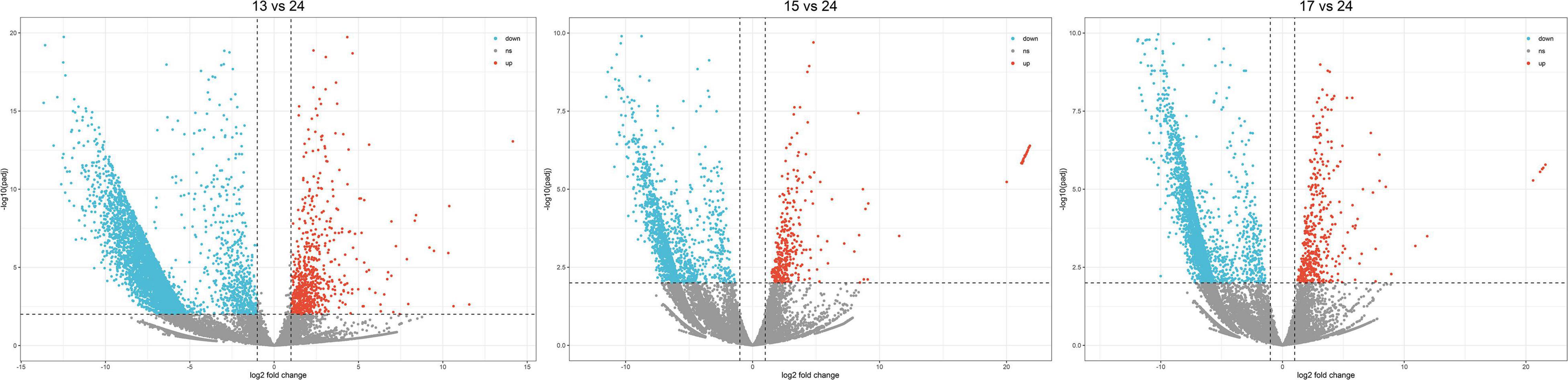
Figure 1. Volcano scatter plots of differentially expressed genes. Red points represent significantly up-regulated differentially expressed genes. Blue points represent significantly down-regulated differentially expressed genes.
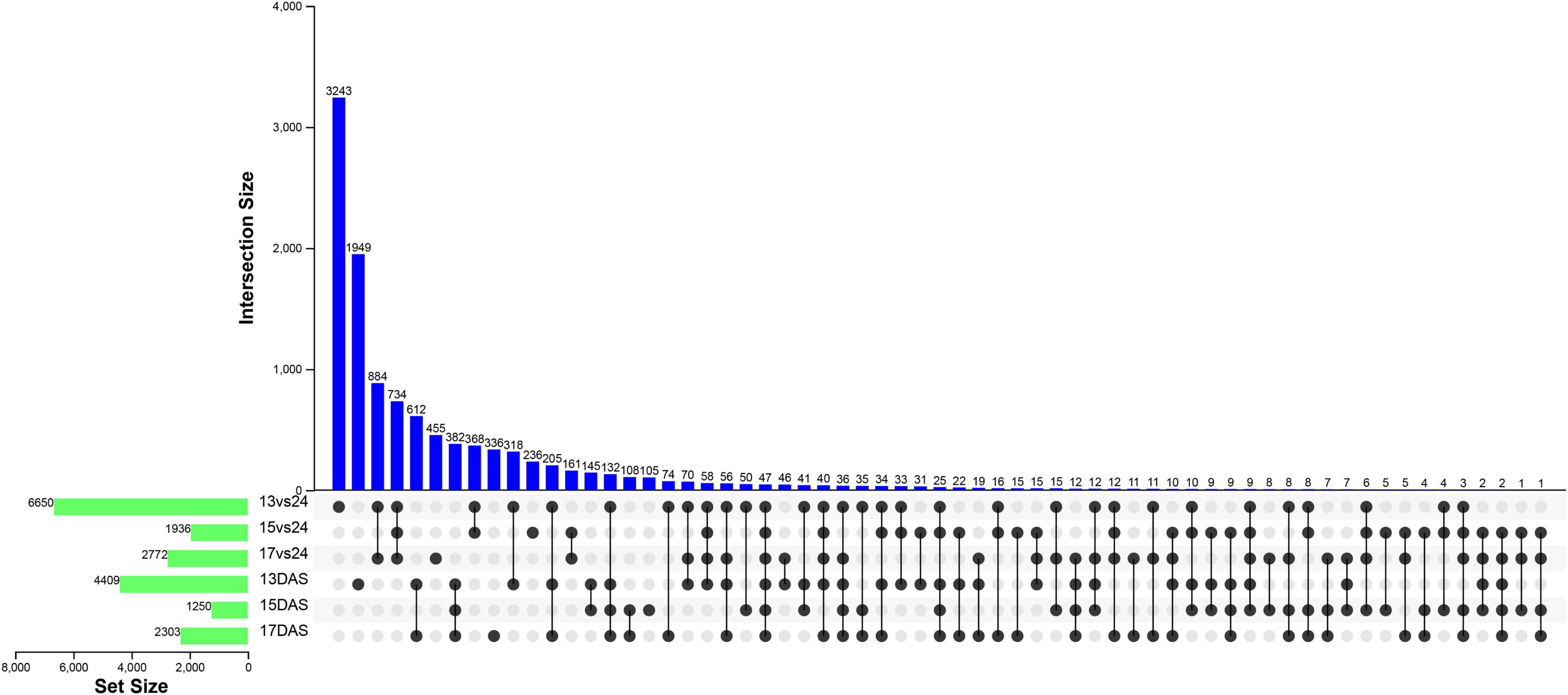
Figure 2. Upset plot of interactions between transcription and alternative splicing regulation of differentially expressed genes. 13vs24, 15vs24, and 17vs24 and 13DAS, 15DAS, and 17DAS represented the number of differentially expressed genes at the transcription and alternative splicing regulation, respectively.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis
Gene ontology enrichment results can be divided into three parts: Biological process (BP), Cell component (CC), and Molecular function (MF). Up-regulated DEGs (UDEGs) were enriched in 75 (BP: 55, CC: 8, and MF: 12), 32 (BP: 15, CC: 10, and MF: 7), and 46 (BP: 22, CC: 18, and MF: 6) GO terms at 13, 15, and 17°C, respectively; Down-regulated DEGs (DDEGs) were enriched in 42 (BP: 25, CC: 8, and MF: 9), 29 (BP: 11, CC: 8, and MF: 10), and 10 (BP: 3, CC: 2, and MF: 5) GO terms at 13, 15, and 17°C, respectively. In addition, biological processes in GO enrichment results of DEGs were highlighted (Figure 3).
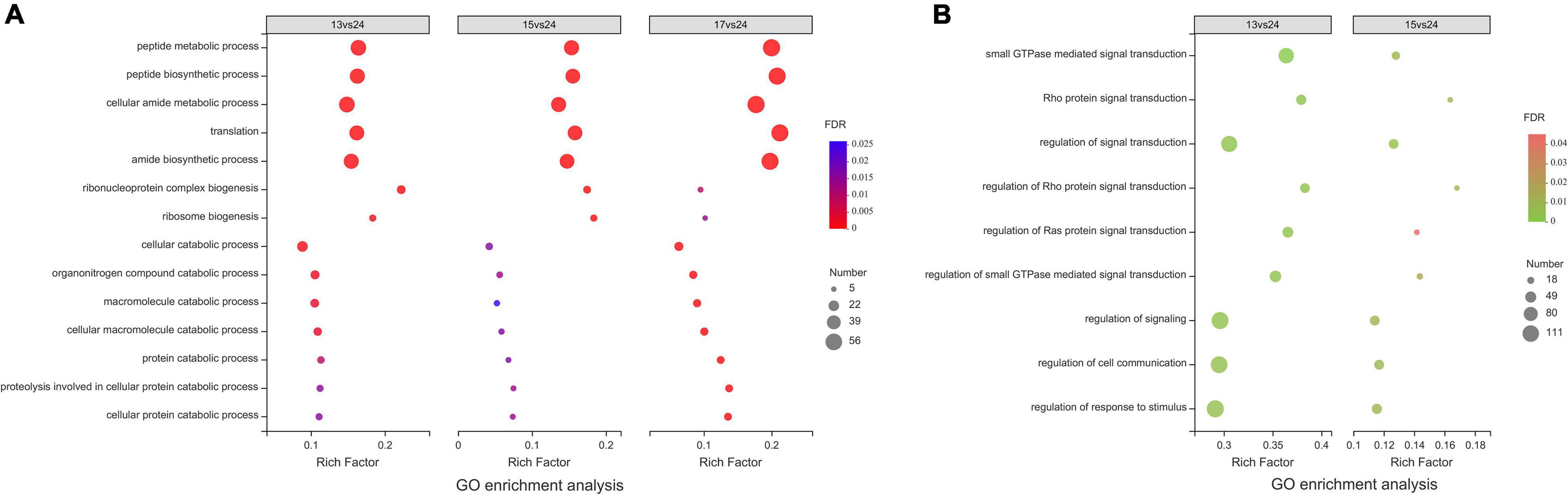
Figure 3. Intersection of biological processes in GO enrichment results of differentially expressed genes at different low temperatures. (A,B) Represented the intersection of biological processes results of up-and down-regulated DEGs, respectively.
Up-regulated differentially expressed genes enrichment results showed that 8, 2, and 5 KEGG pathways were significantly enriched in 13, 15, and 17°C, respectively, among which “ribosome” and “spliceosome” pathways were common in three low-temperature environments (Figure 4); DDEGs enrichment results showed that 56, 32, and 29 KEGG pathways were significantly enriched in 13, 15, and 17°C, respectively, and “focal adhesion,” “proteoglycans in cancer,” and other 12 pathways were common in all three low-temperature environments (Figure 5).
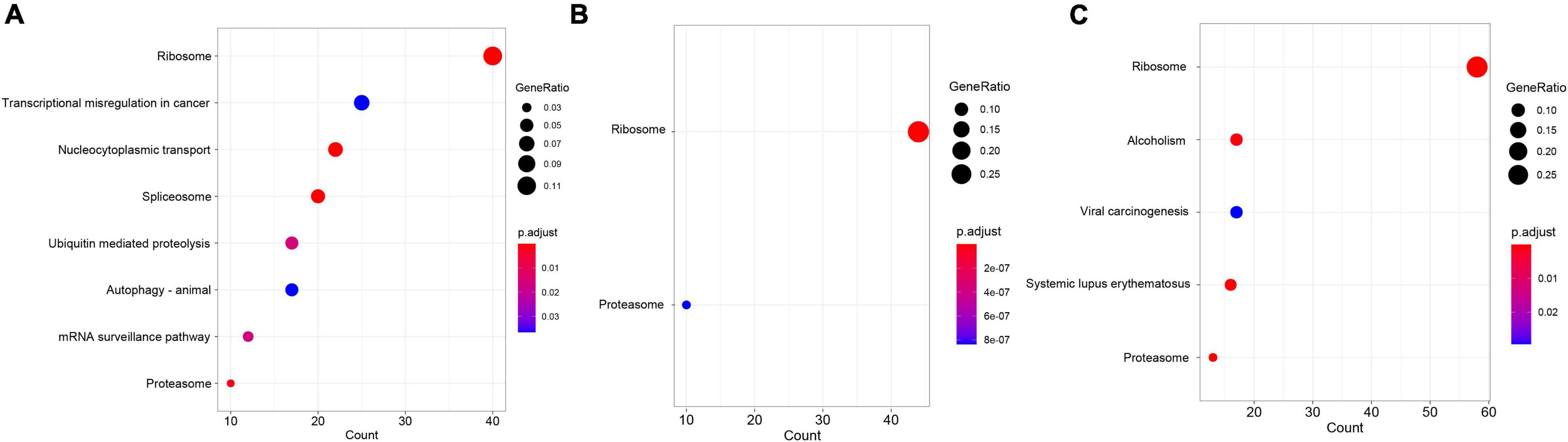
Figure 4. KEGG enrichment results of up-regulated DEGs at different low temperatures. (A–C) Represented KEGG enrichment results at 13, 15, and 17°C, respectively.
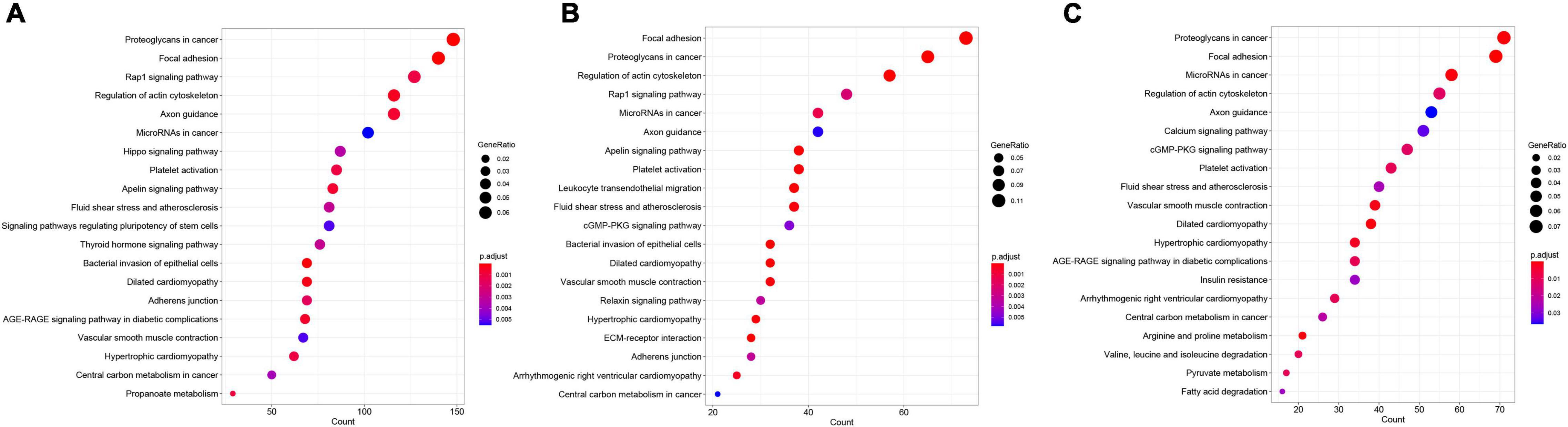
Figure 5. KEGG enrichment results of down-regulated DEGs at different low temperatures. (A–C) Represented KEGG enrichment results at 13, 15, and 17°C, respectively.
Analysis of Differential Alternative Splicing
As a result, 18,062, 2,674, and 6,424 exons in 4,409, 1,250, and 2,303 genes were differential alternative splicing (DAS) at 13, 15, and 17°C, respectively, among which 382 DAS genes (DASGs) were common in three low-temperature environments (Figure 2). Moreover, 47 genes were common to both DEGs and DASGs, among which 19 genes were up-regulated expressed in both transcription and alternative splicing regulation (Table 1). With the combination of GO annotation and manual literature searches, 19 genes were grouped roughly into four categories, including transcription and translation, transmembrane transport, posttranslational modification, and others (Table 1).
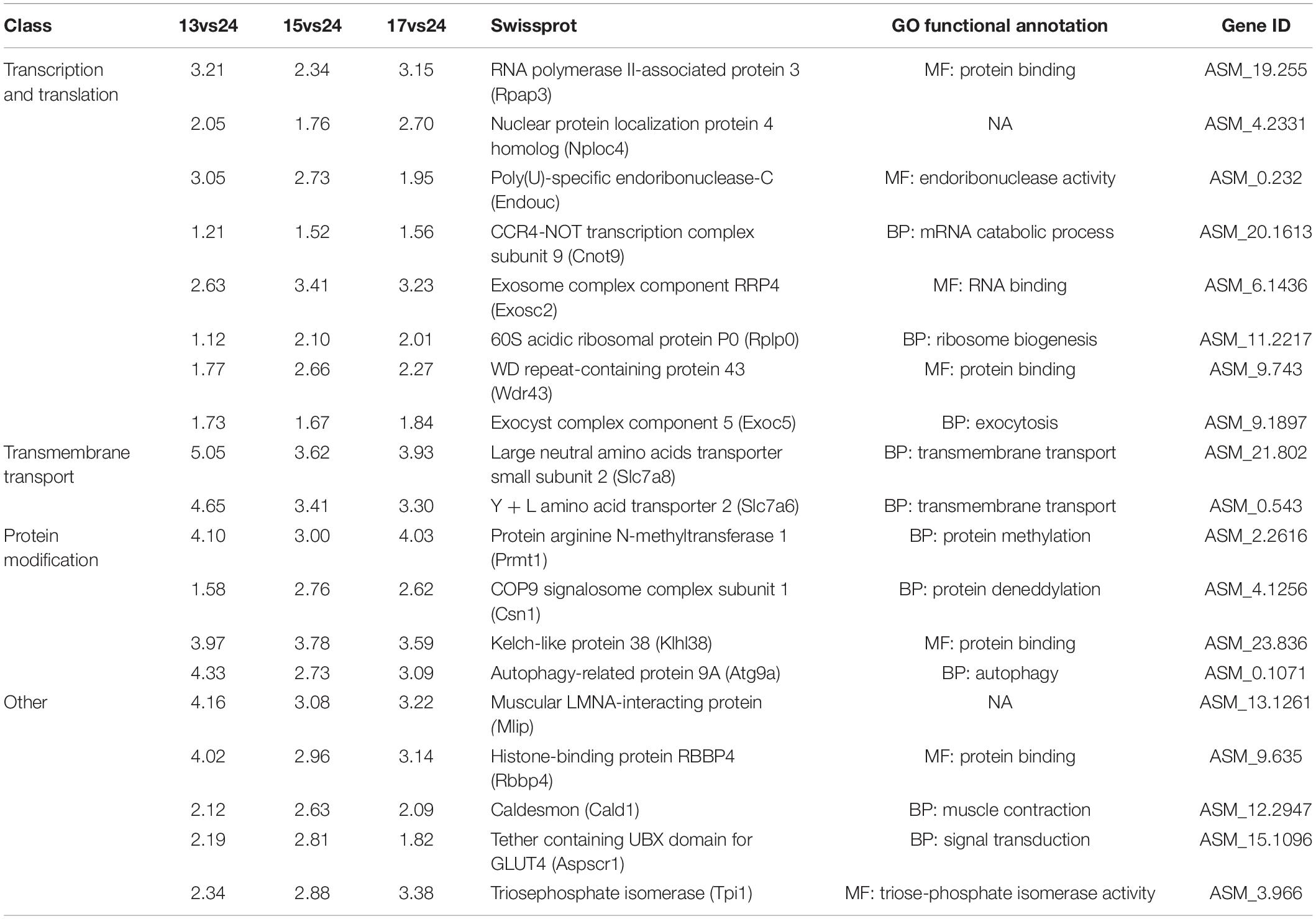
Table 1. Functional annotation and classification of 19 up-regulated key candidate genes in transcriptional and alternative splicing regulation.
Kyoto encyclopedia of genes and genomes enrichment results showed that DASGs were enriched in 34, 12, and 24 pathways at 13, 15, and 17°C, respectively. “Spliceosome,” “mRNA surveillance pathway,” and other five pathways were significantly enriched in three low-temperature environments (Figure 6).
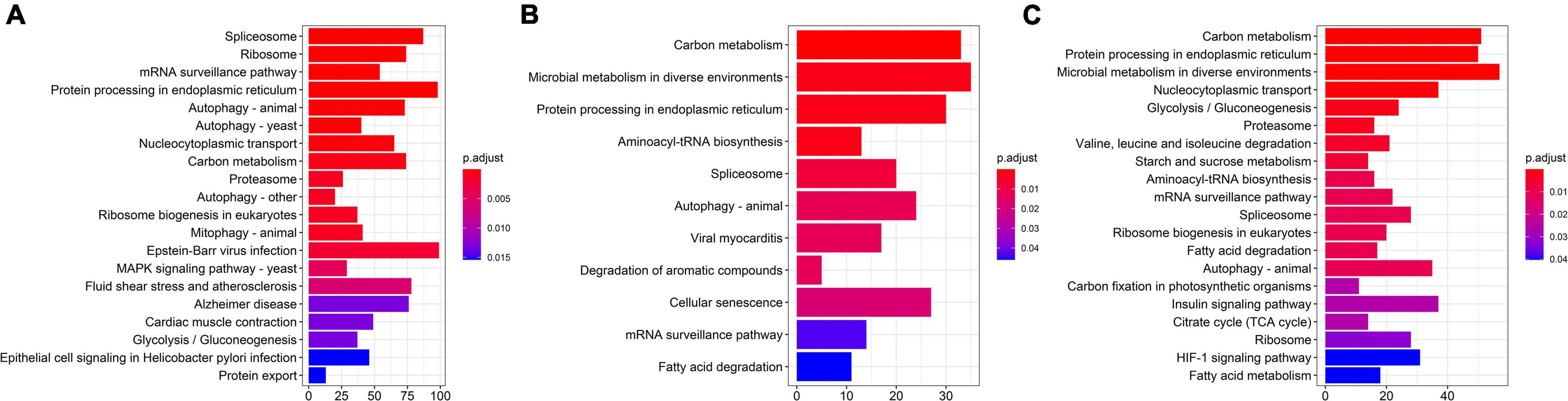
Figure 6. KEGG enrichment results of differential alternative splicing genes (DASGs) at different low temperatures. (A–C) Represented KEGG enrichment results at 13, 15, and 17°C, respectively.
Discussion
Overall, numerous DEGs and their relevant pathways were involved in cold stresses, and the number of down-regulated DEGs was much more than up-regulated DEGs in transcription regulation, this may reduce the energy requirement for body storage in low-temperature environments (Wen et al., 2017). Taking all the results together, H. nehereus adapts to low-temperature environments mainly through transcription and translation, transmembrane transport, and protein modification.
Transcription and Translation
The temperature has a decisive influence on the structure and function of cellular components such as nucleic acids and proteins (Somero and Hochachka, 1971). When fish faces cold stress, biomolecules such as nucleic acids and proteins do not fold and assemble correctly, resulting in reduced activity (Zhang et al., 2018). In this study, UDEGs were involved in many biological processes and pathways related to transcription and translation such as translation, ribosome biogenesis, ribosome, spliceosome, and nucleocytoplasmic transport (Figures 3A, 4), similar to the results of Danio rerio (Long et al., 2013) and Oryzias latipes (Ikeda et al., 2017). The spliceosome pathway played an important role in regulating the adaptation of H. nehereus to cold stress, so alternative splicing analysis was of great significance in the study of cold tolerance of H. nehereus. DASGs enrichment results showed that transcription and translation related pathways such as spliceosome, ribosome, and mRNA surveillance pathway were also significantly enriched. Up-regulated DEGs and DASGs in ribosome biogenesis and pathways may be a compensatory response to the cell’s ability to enhance protein synthesis under low-temperature environments.
In addition, Rpap3, Nploc4, Endouc, Cnot9, Exosc2, Rplp0, Wdr43, and Exoc5 proteins were classified as transcription and translation. Rpap3 functions as co-chaperone for both Hsp70 and Hsp90 (Benbahouche et al., 2014), the increase of Rpap3 appeared to be accompanied by an increase in Hsp70 and Hsp90 proteins, but this did not turn out to be the case, and Hsp70 and Hsp90 were reduced in H. nehereus from cold environments. Rpap3 may play a more role in regulating cell survival and apoptosis (Benbahouche et al., 2014), renewing cells to maintain normal physiological functions of H. nehereus cells under low-temperature environments. Endou proteins belong to the Eukaryotic Endou ribonuclease family of enzymes. Recent studies demonstrated that Endou plays important roles in cellular processes, including regulating ER structure, RNA degradation, and cell survival (Poe et al., 2014; Schwarz and Blower, 2014). The reason for the increase of Endouc might also be the maintenance of cell normal physiological processes. Noteworthy, autophagy, as a cell cleaner (Galluzzi and Green, 2019), was heavily enriched in the results (Figures 4A, 6A–C and Table 1), this result reinforced our speculation that H. nehereus maintained normal cellular physiological functions by accelerating the removal of damaged cell structures by autophagy and the renewal of damaged cell structures by associated proteins, such as Rpap3 and Endou. In addition, studies showed that Exosc2 knockout zebrafish showed larval lethality 13 days post-fertilization, with microcephaly neurons, myelin deficiency, and loss of spinal motor neurons (Yatsuka et al., 2021). Furthermore, Exosc8, which is in the same family as Exosc2, its mutation causes hypomyelination with spinal muscular atrophy (Boczonadi et al., 2014), combined with the result that thecardiac muscle contraction pathway and Cald1 protein (Zheng et al., 2009) were significantly expressed (Figure 6A and Table 1), we suspected that the increase of Excos2 was to avoid excessive muscle contraction in H. nehereus under low-temperature environments.
Transmembrane Transport
Low-temperature stimulation can reduce the fluidity of biofilm (including cell membrane and organelle membrane), change the structure of biofilm, and affect the function of membrane and membrane binding protein (Los and Murata, 2004). The phenomenon that cold stress affects the fluidity and structure of fish biofilms seems to be common, e.g., Takifugu fasciatus (Wen et al., 2019), O. latipes (Ikeda et al., 2017), and Pearl gentian grouper (Miao et al., 2021). Therefore, the cold stress may have affected the fluidity and structure of the biofilm in H. nehereus, which formed a protective mechanism to maintain the normal physiological function of the biofilm through the increase of transmembrane transport-related proteins such as Slc7a8 and Slc7a6 (Table 1). The increase in Slc7a8 and Slc7a6 proteins seems unsurprising, and the same family as them, Slc7a11 (Zhou et al., 2019), Slc25a5 (Ren et al., 2021), Slc16a1 (Long et al., 2015), and Slc2a10 (Scott and Johnston, 2012) were also found to be differentially expressed in other fish under low-temperature environments. Slc7a as an amino acid transporter (Fotiadis et al., 2013), the increase of Slc7a6 and Slc7a8 may also be due to the need for more frequent material exchange of cells inside and outside for H. nehereus in low-temperature environments.
Protein export and endocytosis pathway in KEGG, and protein metabolisms, peptide metabolic process, peptide biosynthetic process, cellular amide metabolic process, and amide biosynthetic process terms in GO that require transmembrane transport were also significantly enriched (Figures 3A, 6A). The above results indicated that a large number of stress proteins were synthesized in H. nehereus cells and secreted to act extracellularly, while endocytosis could break down damaged macromolecules for recycling. The secretion of stress proteins and the endocytosis of macromolecules are inseparable from the transmembrane transport function. Additionally, the fluidity of the membrane was enhanced by altering the content of membrane lipid and the proportion of unsaturated fatty acids (Johnston and Roots, 1964), and the fatty acid metabolism pathway was enriched probably to maintain the fluidity of the biofilms in this study (Figure 6C).
Protein Modification
The low temperature could induce protein modifications such as phosphorylation and acetylation, which change proteins’ chemical properties and structure and affect their functions. Prmt1, Csn1, Klhl38, Atg9a, and Mlip proteins were classified as protein modification (Table 1), and protein modification related pathways such as proteasome, ubiquitin mediated proteolysis, and protein processing in endoplasmic reticulum pathways were also significantly enriched in low-temperature environments of H. nehereus (Figures 4A, 6A,B). This result was similar to the pathway enriched by other fish under cold stress, like the proteasome pathway in D. rerio (Long et al., 2013) and Pampus argenteus (Zhang et al., 2022), and protein processing in endoplasmic reticulum pathway in D. rerio (Long et al., 2013), Lota lota lota (Yang et al., 2021), and O. latipes (Ikeda et al., 2017). The ubiquitin proteasome system catalyzes the great majority of the protein degradation, including both the rapid degradation of misfolded and regulation proteins (Zhao et al., 2015), therefore, the ubiquitin protease system may clear the misfolded proteins and accelerate the renewal of damaged cells for H. nehereus under low-temperature environments (Han et al., 2020). Although the endoplasmic reticulum is the primary place of protein synthesis, it is also the site of protein degradation (Christianson and Ye, 2014). According to the significant enrichment of proteasome, ubiquitin mediated proteolysis, autophagy, and other protein degradation pathways, we prefer the protein processing in the endoplasmic reticulum pathway to play a role in protein degradation in H. nehereus under low-temperature environments. Many pathways related to protein degradation were significantly enriched, indicating that cold stress may affect the balance of protein synthesis and degradation.
Furthermore, UDEGs were enriched into disease-related pathways, for example, transcriptional misregulation in cancer, systemic lupus erythematosus, alcoholism, viral carcinogenesis pathways, and viral myocarditis (Figures 3A, 4), suggesting that cold stress may induce various diseases in H. nehereus; DDEGs were enriched into signal transduction terms, focal adhesion, regulation of actin cytoskeleton, and other pathways (Figures 3B, 5), indicating that signal transduction, cell adhesion, and cytoskeleton regulation were reduced in H. nehereus under low-temperature environments. Moreover, as stress proteins, the heat shock proteins (HSPs) family does not only play a role in thermal shock but also plays a regulatory role in other stresses, such as heavy metals, osmotic stress, cold stress, etc. (Basu et al., 2002). Strong evidence suggests that HSPs have critical roles in helping fish cope with environmental change (Weber and Bosworth, 2005), but 12 HSP genes were down-regulated differentially expressed in this study, and the differential expression levels of HSP genes seem to be different in different fish coping with cold stress, e.g., the HSP genes were up-regulated expressed in channel catfish, L. lota lota and Puntius tetrazona (Weber and Bosworth, 2005; Liu L. L. et al., 2020; Yang et al., 2021), while down-regulated expressed in O. latipes, D. rerio, and Oratosquilla oratoria (Ikeda et al., 2017; Lou et al., 2019; Han et al., 2020). In a similar situation, the focal adhesion pathway of Gasterosteus aculeatus was down-regulated expressed (Metzger and Schulte, 2018), which was similar to the result of H. nehereus, while L. polyactis was up-regulated expressed (Liu F. et al., 2020). Therefore, the difference in regulation strategies of different fish for adapting to low-temperature environments is worth further exploration.
Conclusion
Cold tolerance of fish is important in determining fish distribution and survival in the changing oceans. In this study, a total of 6,650, 1,936, and 2,772 DEGs in transcription regulation and 4,409, 1,250, and 2,303 DASGs in alternative splicing regulation were identified in H. nehereus at 13, 15, and 17°C, respectively, importantly, 47 genes were identified as the key candidate genes for cold tolerance in H. nehereus. It’s worth noting that ribosome and spliceosome pathways associated with transcription and translation; protein export and endocytosis pathways associated with transmembrane transport; proteasome and protein processing in endoplasmic reticulum pathways associated with protein modification; and transcriptional misregulation in cancer and viral carcinogenesis pathway associated with the disease were significantly enriched in H. nehereus under low-temperature environments. This study provided a certain theoretical basis for future research into the molecular strategies of cold tolerance in fishes. Concurrently, this study is also one of the few papers using transcriptome data to study the regulating strategies of fish under natural low-temperature environments, which can reflect the real molecular mechanism of H. nehereus when coping with cold and other complex natural environmental factors.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA820367), Sequence Read Archive (accession: SRR18494828, SRR18494829, SRR18494830, SRR18494831, SRR18494832, SRR18494833, SRR18494834, SRR18494835, SRR18494836, SRR18494837, SRR18494838, and SRR18494839). Further queries can be directed to the corresponding author(s).
Ethics Statement
The animal study was reviewed and approved by Animal Research and Ethics Committees of the Ocean University of China.
Author Contributions
ZS: methodology, software, formal analysis, data curation, writing−original draft, and visualization. LH: investigation, resources, validation, and funding acquisition. YK: software, formal analysis, and validation. LW: formal analysis, data curation, and validation. BK: conceptualization, validation, writing−review and editing, supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by National Natural Science Foundation of China (Nos. U20A2087 and 41976091).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all members of the laboratory for valuable discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.912113/full#supplementary-material
Footnotes
References
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., and Butler, H. (2000). Gene ontology: tool for the unification of biology. Nat Genet 25, 25–29. doi: 10.1038/75556
Bapat, S. V. (1970). The Bombay duck, Harpodon nehereus (Ham.). Bull. Cent. Mar. Fish. Res. Inst. 21, 1–87.
Basu, N., Todgham, A. E., Ackerman, P. A., Bibeau, M. R., Nakano, K., Schulte, P. M., et al. (2002). Heat shock protein genes and their functional significance in fish. Gene 295, 173–183. doi: 10.1016/s0378-1119(02)00687-x
Benbahouche, N. E., Iliopoulos, I., Torok, I., Marhold, J., Henri, J., Kajava, A. V., et al. (2014). Drosophila Spag Is the Homolog of RNA Polymerase II-associated Protein 3 (RPAP3) and Recruits the Heat Shock Proteins 70 and 90 (Hsp70 and Hsp90) during the Assembly of Cellular Machineries. J. Biol. Chem. 289, 6236–6247. doi: 10.1074/jbc.M113.499608
Boczonadi, V., Muller, J. S., Pyle, A., Munkley, J., Dor, T., Quartararo, J., et al. (2014). EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat. Commun. 5:4287. doi: 10.1038/ncomms5287
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Christianson, J. C., and Ye, Y. H. (2014). Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat. Struct. Mol. Biol. 21, 325–335. doi: 10.1038/nsmb.2793
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Firdaus, M., Lelono, T. D., Saleh, R., Bintoro, G., and Salim, G. (2018). The expression of the body shape in fish species Harpadon nehereus (Hamilton, 1822) in the waters of Juata Laut, Tarakan city, North Kalimantan. AACL Bioflux 11, 613–624. doi: 10.5281/zonodo.1250264
Fossheim, M., Primicerio, R., Johannesen, E., Ingvaldsen, R. B., Aschan, M. M., Dolgov, A. V., et al. (2015). Recent warming leads to a rapid borealization of fish communities in the Arctic. Nat. Clim. Change 5, 673–677.
Fotiadis, D., Kanai, Y., and Palacin, M. (2013). The SLC3 and SLC7 families of amino acid transporters. Mol. Aspects Med. 34, 139–158. doi: 10.1016/j.mam.2012.10.007
Galluzzi, L., and Green, D. R. (2019). Autophagy-Independent Functions of the Autophagy Machinery. Cell 177, 1682–1699. doi: 10.1016/j.cell.2019.05.026
Han, B. S., Luo, J. T., Jiang, P. L., Li, Y., Wang, Q., Bai, Y. J., et al. (2020). Inhibition of Embryonic HSP 90 Function Promotes Variation of Cold Tolerance in Zebrafish. Front. Genet. 11:541944. doi: 10.3389/fgene.2020.541944
Ikeda, D., Koyama, H., Mizusawa, N., Kan-No, N., Tan, E., Asakawa, S., et al. (2017). Global gene expression analysis of the muscle tissues of medaka acclimated to low and high environmental temperatures. Compar. Biochem. Physiol. Genom. Proteom. 24, 19–28. doi: 10.1016/j.cbd.2017.07.002
Jiao, S., Nie, M. M., Song, H. B., Xu, D. D., and You, F. (2020). Physiological responses to cold and starvation stresses in the liver of yellow drum (Nibea albiflora) revealed by LC-MS metabolomics. Sci. Total Environ. 715:136940. doi: 10.1016/j.scitotenv.2020.136940
Johnson, N. C., Xie, S. P., Kosaka, Y., and Li, X. C. (2018). Increasing occurrence of cold and warm extremes during the recent global warming slowdown. Nat. Commun. 9:1724. doi: 10.1038/s41467-018-04040-y
Johnston, P. V., and Roots, B. I. (1964). Brain lipid fatty acids and temperature acclimation. Comp. Biochem. Physiol. 11, 303–309. doi: 10.1016/0010-406x(64)90111-2
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Lan, D. L., Xiong, X. R., Wei, Y. L., Xu, T., Zhong, J. C., Zhi, X. D., et al. (2014). RNA-Seq analysis of yak ovary: improving yak gene structure information and mining reproduction-related genes. Sci. Chin. Life Sci. 57, 925–935. doi: 10.1007/s11427-014-4678-2
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lin, L. S. (2009). Spatial distribution and environmental characteristics of Harpodon nehereus in the East China Sea region. J. Shanghai Ocean Univ. 18, 66–71.
Liu, F., Chu, T. Q., Wang, M. J., Zhan, W., Xie, Q. P., Lou, B., et al. (2020). Transcriptome analyses provide the first insight into the molecular basis of cold tolerance in Larimichthys polyactis. J. Compar. Physiol. B-Biochem. Syst. Environ. Physiol. 190, 27–34. doi: 10.1007/s00360-019-01247-3
Liu, L. L., Zhang, R., Wang, X. W., Zhu, H., and Tian, Z. H. (2020). Transcriptome analysis reveals molecular mechanisms responsive to acute cold stress in the tropical stenothermal fish tiger barb (Puntius tetrazona. Bmc Genom. 21:737. doi: 10.1186/s12864-020-07139-z
Liu, Y., and Cheng, J. A. (2015). preliminary analysis of variation characteristics of structure and average trophic level of the main fishery species caught by paired bottom trawl in the East China Sea and the Yellow Sea during the fall season. J. Fisheries Chin. 39, 691–702.
Long, Y., Song, G., Yan, J., He, X., Li, Q., and Cui, Z. (2013). Transcriptomic characterization of cold acclimation in larval zebrafish. BMC Genom. 14:612. doi: 10.1186/1471-2164-14-612
Long, Y., Yan, J. J., Song, G. L., Li, X. H., Li, X. X., Li, Q., et al. (2015). Transcriptional events co-regulated by hypoxia and cold stresses in Zebrafish larvae. BMC Genom. 16:385. doi: 10.1186/s12864-015-1560-y
Lopez-Olmeda, J. F., and Sanchez-Vazquez, F. J. (2011). Thermal biology of zebrafish (Danio rerio). J. Thermal Biol. 36, 91–104.
Los, D. A., and Murata, N. (2004). Membrane fluidity and its roles in the perception of environmental signals. Biochim. Et Biophys. Acta-Biomembr. 1666, 142–157. doi: 10.1016/j.bbamem.2004.08.002
Lou, F. R., Gao, T. X., and Han, Z. Q. (2019). Transcriptome analyses reveal alterations in muscle metabolism, immune responses and reproductive behavior of Japanese mantis shrimp (Oratosquilla oratoria) at different cold temperature. Compar. Biochem. Physiol. D-Genom. Proteom. 32:100615. doi: 10.1016/j.cbd.2019.100615
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom. Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mastrangelo, A. M., Marone, D., Laido, G., De Leonardis, A. M., and De Vita, P. (2012). Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 185, 40–49. doi: 10.1016/j.plantsci.2011.09.006
Metzger, D. C. H., and Schulte, P. M. (2018). Similarities in temperature-dependent gene expression plasticity across timescales in threespine stickleback (Gasterosteus aculeatus). Mol. Ecol. 27, 2381–2396. doi: 10.1111/mec.14591
Miao, B. B., Niu, S. F., Wu, R. X., Liang, Z. B., Tang, B. G., Zhai, Y., et al. (2021). Gene Expression Profile and Co-Expression Network of Pearl Gentian Grouper under Cold Stress by Integrating Illumina and PacBio Sequences. Animals 11:1745. doi: 10.3390/ani11061745
Mininni, A. N., Milan, M., Ferraresso, S., Petochi, T., Di Marco, P., Marino, G., et al. (2014). Liver transcriptome analysis in gilthead sea bream upon exposure to low temperature. Bmc Genom. 15:765. doi: 10.1186/1471-2164-15-765
Morgan, M. J., Garabana, D., Rideout, R. M., Roman, E., Perez-Rodriguez, A., and Saborido-Rey, F. (2013). Changes in distribution of Greenland halibut in a varying environment. Ices J. Mar. Sci. 70, 352–361.
Poe, J. C., Kountikov, E. I., Lykken, J. M., Natarajan, A., Marchuk, D. A., and Tedder, T. F. (2014). EndoU is a novel regulator of AICD during peripheral B cell selection. J. Exp. Med. 211, 57–69. doi: 10.1084/jem.20130648
Poloczanska, E. S., Burrows, M. T., Brown, C. J., Molinos, J. G., Halpern, B. S., Hoegh-Guldberg, O., et al. (2016). Responses of Marine Organisms to Climate Change across Oceans. Front. Mar. Sci. 3:62. doi: 10.3389/fmars.2016.00062
Ren, J., Long, Y., Liu, R., Song, G. L., Li, Q., and Cui, Z. B. (2021). Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish. Int. J. Mol. Sci. 22:3028. doi: 10.3390/ijms22063028
Roy, V. C., Kamal, M., Faridullah, M., Haque, S. A., and Reza, M. S. (2014). Influence of salt and herbal substance on the drying and reconstitution performance of Bombay duck. Harpodon nehereus J. Fisheries 2, 59–63. doi: 10.17017/jfish.v2i1.2014.13
Schwarz, D. S., and Blower, M. D. (2014). The calcium-dependent ribonuclease XendoU promotes ER network formation through local RNA degradation. J. Cell Biol. 207, 41–57. doi: 10.1083/jcb.201406037
Scott, G. R., and Johnston, I. A. (2012). Temperature during embryonic development has persistent effects on thermal acclimation capacity in zebrafish. Proc. Natl. Acad. Sci. U S A 109, 14247–14252. doi: 10.1073/pnas.1205012109
Somero, G. N., and Hochachka, P. W. (1971). Biochemical adaptation to environment. Am. Zool. 11, 159–167.
Sun, P., Dai, F., Chen, Y., Shan, X., and Jin, X. (2015). Seasonal variations in structure of fishery resource in the Yangtze River Estuary and its adjacent waters. Progr. Fishery Sci. 36, 8–16.
Sun, Z., Lou, F., Zhang, Y., and Song, N. (2020). Gill Transcriptome Sequencing and De Novo Annotation of Acanthogobius ommaturus in Response to Salinity Stress. Genes 11:631. doi: 10.3390/genes11060631
Sun, Z. C., Lou, F. R., Zhao, X., and Song, N. (2021). Characterization and analysis of transcriptome complexity using SMRT-Seq combined with RNA-Seq for a better understanding of Acanthogobius ommaturus in response to temperature stress. Int. J. Biol. Macromol. 193, 1551–1561. doi: 10.1016/j.ijbiomac.2021.10.218
Sun, Z. Z., Tan, X. H., Xu, M. L., Liu, Q. Y., Ye, H. Q., Zou, C. Y., et al. (2019). Liver transcriptome analysis and de novo annotation of the orange-spotted groupers (Epinephelus coioides) under cold stress. Compar. Biochem. Physiol. D-Genom. Proteom. 29, 264–273. doi: 10.1016/j.cbd.2018.12.008
Taqwa, A., Burhanuddin, A. I., Niartiningsih, A., and Nessa, M. N. (2020). Nomei fish (Harpadon nehereus, Ham. 1822) reproduction biology in Tarakan waters. IOP Confer. Ser. Earth Environ. Sci. 473:012012.
Valdez, D. M., Miyamoto, A., Hara, T., Edashige, K., and Kasai, M. (2005). Sensitivity to chilling of medaka (Oryzias latipes) embryos at various developmental stages. Theriogenology 64, 112–122. doi: 10.1016/j.theriogenology.2004.11.006
Wang, K., Shen, Y., Yang, Y., Gan, X., Liu, G., Hu, K., et al. (2019). Morphology and genome of a snailfish from the Mariana Trench provide insights into deep-sea adaptation. Nat. Ecol. Evol. 3, 823–833. doi: 10.1038/s41559-019-0864-8
Weber, T. E., and Bosworth, B. G. (2005). Effects of 28 day exposure to cold temperature or feed restriction on growth, body composition, and expression of genes related to muscle growth and metabolism in channel catfish. Aquaculture 246, 483–492.
Wen, B., Jin, S. R., Chen, Z. Z., Gao, J. Z., Wang, L., Liu, Y., et al. (2017). Plasticity of energy reserves and metabolic performance of discus fish (Symphysodon aequifasciatus) exposed to low-temperature stress. Aquaculture 481, 169–176.
Wen, X., Hu, Y. D., Zhang, X. Y., Wei, X. Z., Wang, T., and Yin, S. W. (2019). Integrated application of multi-omics provides insights into cold stress responses in pufferfish Takifugu fasciatus. Bmc Genom. 20:563. doi: 10.1186/s12864-019-5915-7
Whitehead, A., Zhang, S. J., Roach, J. L., and Galvez, F. (2013). Common functional targets of adaptive micro- and macro-evolutionary divergence in killifish. Mol. Ecol. 22, 3780–3796. doi: 10.1111/mec.12316
Xu, D. D., You, Q. C., Chi, C. F., Luo, S. Y., Song, H. B., Lou, B., et al. (2018). Transcriptional response to low temperature in the yellow drum (Nibea albiflora) and identification of genes related to cold stress. Compar. Biochem. Physiol. D-Genom. Proteom. 28, 80–89. doi: 10.1016/j.cbd.2018.07.003
Yang, T. Y., Zhang, Y., Meng, W., Zhong, X., Shan, Y., and Gao, T. X. (2021). Comparative transcriptomic analysis brings new insights into the response to acute temperature acclimation in burbot (Lota lota lota). Aquacult. Rep. 20:100657.
Yatsuka, H., Hada, K., Shiraishi, H., Umeda, R., Morisaki, I., Urushibata, H., et al. (2021). Exosc2 deficiency leads to developmental disorders by causing a nucleotide pool imbalance in zebrafish (vol 533, pg 1470, 2020). Biochem. Biophys. Res. Commun. 546, 200–200. doi: 10.1016/j.bbrc.2020.10.044
Zhang, M., Hu, J. B., Zhu, J. J., Wang, Y. J., Zhang, Y. Y., Li, Y. Y., et al. (2022). Transcriptome, antioxidant enzymes and histological analysis reveal molecular mechanisms responsive to long-term cold stress in silver pomfret (Pampus argenteus). Fish Shellfish Immunol. 121, 351–361. doi: 10.1016/j.fsi.2022.01.017
Zhang, Y., Burkhardt, D. H., Rouskin, S., Li, G. W., Weissman, J. S., and Gross, C. A. (2018). A Stress Response that Monitors and Regulates mRNA Structure Is Central to Cold Shock Adaptation. Mol. Cell 70, 274–286. doi: 10.1016/j.molcel.2018.02.035
Zhao, J. H., Zhai, B., Gygi, S. P., and Goldberg, A. L. (2015). mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. U S A 112, 15790–15797. doi: 10.1073/pnas.1521919112
Zheng, P. P., Severijnen, L. A., Willemsen, R., and Kros, J. M. (2009). Caldesmon is essential for cardiac morphogenesis and function: in vivo study using a zebrafish model. Biochem. Biophys. Res. Commun. 378, 37–40. doi: 10.1016/j.bbrc.2008.10.165
Zhou, T., Gui, L., Liu, M. L., Li, W. H., Hu, P., Duarte, D. F. C., et al. (2019). Transcriptomic responses to low temperature stress in the Nile tilapia. Oreochromis niloticus. Fish Shellfish Immunol. 84, 1145–1156. doi: 10.1016/j.fsi.2018.10.023
Keywords: differentially expressed genes, differential alternative splicing, low-temperature environments, marine fish, RNA-seq
Citation: Sun ZC, Huang LL, Kong YF, Wang LL and Kang B (2022) Regulating Strategies of Transcription and Alternative Splicing for Cold Tolerance Harpadon nehereus Fish. Front. Ecol. Evol. 10:912113. doi: 10.3389/fevo.2022.912113
Received: 04 April 2022; Accepted: 16 May 2022;
Published: 07 June 2022.
Edited by:
Hui Zhang, Institute of Oceanology (CAS), ChinaReviewed by:
Fangrui Lou, Yantai University, ChinaShengyong Xu, Zhejiang Ocean University, China
Mir Mohammad Ali, Sher-e-Bangla Agricultural University, Bangladesh
Tianxiang Gao, Zhejiang Ocean University, China
Copyright © 2022 Sun, Huang, Kong, Wang and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Kang, YmthbmdmaXNoQDE2My5jb20=
 Zhicheng Sun
Zhicheng Sun Liangliang Huang3
Liangliang Huang3 Yefu Kong
Yefu Kong Linlong Wang
Linlong Wang Bin Kang
Bin Kang