- 1Natural History Museum of Geneva, Geneva, Switzerland
- 2Department of Genetics and Evolution, University of Geneva, Geneva, Switzerland
Introduction
Darwin pointed out that “species of different genera and classes have not changed at the same rate” (Darwin, 1859, chapter X). Besides, he coined the expression “living fossils” for lineages whose “new forms will have been more slowly formed, and old forms more slowly exterminated” (chapter IV), among other characteristics. This expression has become popular, but has sometimes been misunderstood as meaning that some organisms do not evolve. It has also been commonly used by paleontologists and evolutionary biologists to describe a general pattern of relative stasis in morphological evolution in some lineages. Darwin's definition of the concept was imprecise and he considered that “species and groups of species, which are called aberrant, and which may fancifully be called living fossils, will aid us in forming a picture of the ancient forms of life” (Darwin, 1859, Chapter XIV). For more than 200 years, nevertheless, debates have raged on the definition of the concept (e.g., Bennett et al., 2017, 2018; Lidgard and Love, 2018; Turner, 2019), and more generally on the merits of its use in the life sciences (e.g., Casane and Laurenti, 2013; Naville et al., 2015). Although Darwin (1859) cited several taxa of fish as examples of “living fossils,” he did not mention the coelacanths, or actinistians, which were only known as fossils at his time. Huxley, however, soon after (1866) noticed the low anatomical disparity of coelacanths throughout their history. Since that time, and especially after the discovery of the living Latimeria in 1938 (Smith, 1939), the coelacanth has become an iconic symbol of the “living fossil” due to the slow morphological evolution illustrated by the fossil record of the clade, and its supposed affinities with tetrapods. Only the question of evolutionary rate is addressed here, not the question of ancestral status or other “living fossil” characteristics attributed to coelacanths. The low rate of evolution based on a lasting generalist morphological Bauplan has been confirmed by most subsequent authors who have worked on the group (Schaeffer, 1952; Cloutier, 1991; Forey, 1998; Schultze, 2004; Zhu et al., 2012; Cavin and Guinot, 2014), knowing that there are also exceptions to this general Bauplan (e.g., Friedman and Coates, 2006; Wendruff and Wilson, 2012; Cavin et al., 2017). However, part of the community of researchers working on fossil and living coelacanths avoids using this expression.
Most twenty-first century genetic studies have confirmed that the substitution rate of the genome of Latimeria was found to be slower than that of other vertebrate lineages in the mitochondrial (Sudarto et al., 2010; Nikaido et al., 2011; Lampert et al., 2012; Kadarusman et al., 2020) as well as in the nuclear genome (Nikaido et al., 2013) at least for the genes encoding proteins (Amemiya et al., 2010, 2013), when measured in substitution per year, although alternative interpretations remain (Bockmann et al., 2013; Casane and Laurenti, 2013; Grandcolas et al., 2014; Minelli and Baedke, 2014; Naville et al., 2015). It should be noted that although there is evidence for active transposable dynamics in the Latimeria genome (Smith et al., 2012; Chalopin et al., 2014; Naville et al., 2015), these elements have found to be highly conserved (Smith et al., 2012). The fact that at the level of transposable element activity, the coelacanth genome does not reveal slow dynamics (Chalopin et al., 2014) is thus not related to the global substitution rate.
Rate of Evolution
The debate over the concept of “living fossils,” in the sense of slowly evolving organisms, is part of a larger research agenda dealing with the rate of evolution. The rate of morphological evolution is usually measured as a unit of change per generation time (Haldane, 1949; Gingerich, 2001; Evans et al., 2012), while molecular evolution is usually measured as substitution rate per site per year (e.g., Lanfear et al., 2010). Substitution rates are affected by evolutionary and life history traits, such as population size, body size, mutation rate, and generation time (Martin and Palumbi, 1993; Pulquerio and Nichols, 2007; Thomas et al., 2010). Among these factors, generation time imposes a minimum time for the transmission of the genome throughout the generations, in particular when it carries mutations under selection. In order to control for this critically important factor when modeling gene evolution over time, substitution rates are generally given per generation rather than per year (Loewe and Hill, 2010). This allows, for instance, to compare gene dynamics in a population genetics or in a macroevolutionary context assuming that species have similar generation times (Vinciguerra and Burns, 2021). However, when studying the tempo of evolutionary change through time across species within a broader evolutionary framework, molecular evolution is usually given as a function of the number of years or rather million years instead of generations (Folk et al., 2019; Yohe et al., 2020). First discussed in the early 1990s (Ohta, 1993), the relationship between molecular evolution and generation time is actually more complex than expected, notably given that other factors are also at work. Among those is the number of germline cell divisions per generation (Lynch, 2010). For instance, the number of germ line divisions per generation varies greatly among mammalian species: while the human male germline is estimated to have 401 cell generations over an individual's lifetime, the male mouse germline has an average of only 62 (Drost and Lee, 1995; Bromham, 2011)—meaning that the number of copy errors per generation is expected to be more than 6 times higher in humans than in mice. As each cell division in the germline is expected to face the same mutation rate, this difference thus partly buffers the difference in generation time between humans and mice in an attempt to compare their molecular evolution across millions years. In her seminal review, Bromham (2011) points to factors, which in addition to differences in generation time and number of copy errors per generation in the germline, also affect rates of molecular evolution. Among those are the DNA damage level induced by metabolic and environmental energy. For instance, endothermic vertebrates that use metabolism to maintain a constant body temperature (birds and mammals) have higher absolute rates of molecular evolution than ectothermic vertebrates, such as reptiles and ray-finned fish (Martin et al., 1992). Another factor pointed by Bromham is body size and longevity, which might directly trigger selection for DNA repair mechanisms and thus lower levels of molecular evolution (Promislow, 1994). The impact of fecundity on molecular evolution might be driven by a similar selective process in which selection to reduce the rate of harmful mutations would be stronger in species producing smaller number of offspring per year (Welch et al., 2008). Additional factors encompass population sizes, with species characterized by smaller populations experiencing larger molecular evolutionary rates, owing to faster rates of fixation in arising mutations (i.e., substitution rates) along the selection-drift equilibrium (Charlesworth, 2009).
Discussion
Neither the generation time nor the number of copy errors per generation in the germline can be easily identified in non-model species. These life history traits, as well as others such as fecundity or metabolic rates, also cannot be identified when dealing with extinct taxa, in which only a few life history traits are accessible to paleontologists. Some parameters affecting molecular evolution rates can, however, be identified directly on fossil material, such as body size, or can be indirectly inferred, such as generation time. Starting from the importance underlined above of the generation time in the estimation of the nucleotide substitution rate, and evolution rate in general, recent studies shed new light on a probable cause of the slow rate of evolutionary change in the coelacanth lineage.
In a new analysis of the growth lines on a set of scales of L. chalumnae, Mahé et al. (2021) showed that the lifespan in this species is probably around 100 years, the sexual maturity reached about 50 years old at the earliest (49–69), and the gestation time lasts around 5 years. Accordingly, the time before genetic transmission between generations is 54 years at minimum. This is 12.5 times longer that an average time calculated for a set of actinopterygian species (4.4 years when 50% of individuals attain their first reproduction years calculated on 215 individuals from 98 populations of 76 species) and 4.2 times longer that an average time calculated for a set of shark species (13 years calculated for 19 individuals from 11 populations of 9 species) (He and Stewart, 2001) (Figure 1A). In addition to longevity and body size, the slow gestation time might select for DNA repair mechanisms more strongly than in actinopterygians and chondrosteans. So far, no analysis has estimated the number of copy errors per generation in the germline but any other parameter considered equivalent, the evolutionary rate of Latimeria is slower in the same proportion compared to the other two clades, i.e., more than 10 times slower than ray-finned fish and almost 5 times slower than sharks. The discovery by Mahé et al. (2021) is limited to Latimeria among coelacanths, and it has been hypothesized that this life history trait is associated with the stable environments in which these animals live (Cupello et al., 2019). However, Mesozoic coelacanths lived in a variety of marine and freshwater aquatic environments that were very different from the extant mesobenthic habitat, and the stable environmental conditions are not enough to explain a slow rate of evolution.
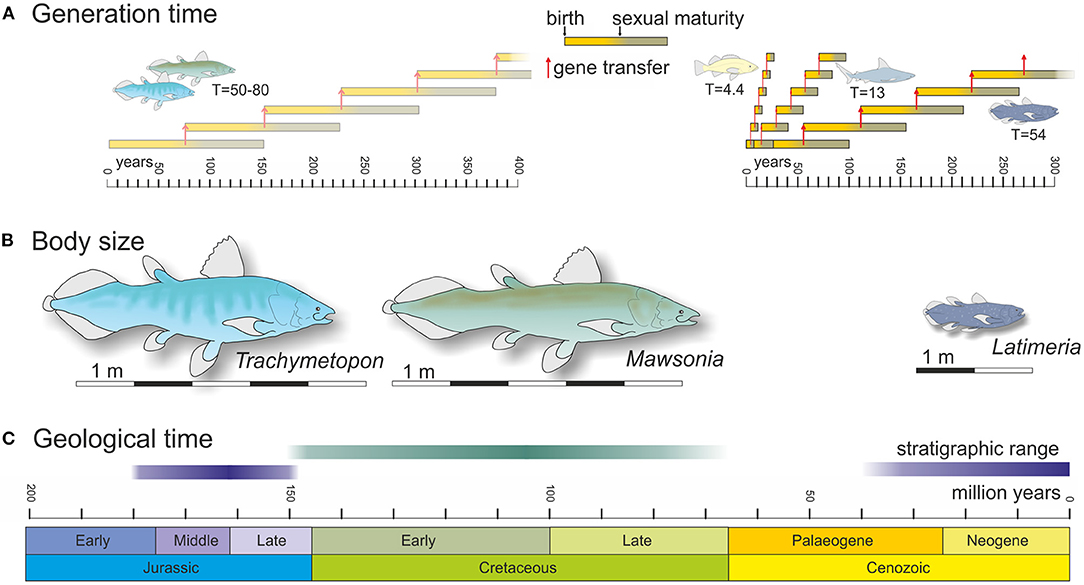
Figure 1. Selected life history traits and stratigraphical range for Latimeria and two Mesozoic mawsoniid coelacanths, Trachymetopon and Mawsonia. (A) Generation time for Latimeria (from Mahé et al., 2021), and for actinopterygians and sharks (from He and Stewart, 2001) (right), and estimated for extinct Trachymetopon and Mawsonia (left). (B) Body size of Latimeria, Trachymetopon and Mawsonia (from Cavin et al., 2021a). (C) Stratigraphical ranges of Latimeria, Trachymetopon and Mawsonia (from Cavin et al., 2021b).
There are exceptions to the slow evolutionary pace observed in Mesozoic coelacanths. For instance, morphological outliers from the generalist coelacanth Bauplan, such as the Middle Triassic Foreyia, may have resulted from heterochronic evolution caused by a change in the expression of developmental patterning genes (Cavin et al., 2017). But overall, new fossil data shows that Jurassic and Cretaceous coelacanths from the mawsoniid family, the sister family to the latimeriids, also evolved morphologically very slowly and probably had a very long generation time, as expressed by their giant body size, and this despite living in coastal marine, brackish and freshwater environments (Cavin et al., 2021a). The marine genus Trachymetopon, which reached about 5 m in length, stretched from the Toarcian to the Kimmeridgian of Europe, during about 30 million years, while the Late Jurassic to Late Cretaceous Mawsonia, which occurred in brackish and fresh waters from western Gondwana and North America, and reached 5.3 m in length, lasted from 50 to 80 million years as revealed by the identification of some specimens from the Late Cretaceous (Cavin et al., 2021a,b) (Figures 1B,C). Although there is no direct way to estimate fecundity and metabolic rates, as well as the age of sexual maturity on these extinct forms, the large body size attained by these coelacanths indicates that the first two of these parameters were likely low and the third likely high. If Latimeria, which barely reached 2 m in length can reach 100 years, the giants Trachymetopon and Mawsonia may have lived centuries to reach 5 meters in body length. Following the comparison, sexual maturity should have been reached even later in these giants than in Latimeria (Figure 1A). These points are partly speculative, but they might explain why these Mesozoic genera have spanned tens of millions of years with almost no morphological change. More generally, long generation and gestation times may explain the slow morphological evolution of coelacanths through deep times, without necessarily introducing other physiological or environmental peculiarities.
Author Contributions
LC and NA contributed equally to the analysis of the data and to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
LC received a Swiss National Science Foundation Grant: Evolutionary pace in the coelacanth clade: New evidence from the Triassic of Switzerland (200021-172700).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jeremy Gauthier (Natural History Museum of Geneva) for the discussion while writing this Opinion, as well as the editor, Michel Laurin, and the reviewer, Patrick Laurenti, for lively scientific exchanges.
References
Amemiya, C. T., Alföldi, J., Lee, A. P., Fan, S., Philippe, H., MacCallum, I., et al. (2013). The African coelacanth genome provides insights into tetrapod evolution. Nature 496, 311–316. doi: 10.1038/nature12027
Amemiya, C. T., Powers, T. P., Prohaska, S. J., Grimwood, J., Schmutz, J., Dickson, M., et al. (2010). Complete HOX cluster characterization of the coelacanth provides further evidence for slow evolution of its genome. Proc. Nat. Acad. Sci. USA 107, 3622–3627. doi: 10.1073/pnas.0914312107
Bennett, D. J., Sutton, M. D., and Turvey, S. T. (2017). Evolutionarily distinct “living fossils” require both lower speciation and lower extinction rates. Paleobiology 43, 34–48. doi: 10.1017/pab.2016.36
Bennett, D. J., Sutton, M. D., and Turvey, S. T. (2018). Quantifying the living fossil concept. Palaeontol. Electron. 21, 14A. doi: 10.26879/750
Bockmann, F. A., De Carvalho, M. R., and De Carvalho, M. (2013). The salmon, the lungfish (or the coelacanth) and the cow: a revival? Zootaxa 3750, 265–276. doi: 10.11646/zootaxa.3750.3.6
Bromham, L. (2011). The genome as a life-history character: why rate of molecular evolution varies between mammal species. Philos. Trans. R. Soc. B Biol. Sci. 366, 2503–2513. doi: 10.1098/rstb.2011.0014
Casane, D., and Laurenti, P. (2013). Why coelacanths are not 'living fossils': a review of molecular and morphological data. Bioessays 35, 332–338. doi: 10.1002/bies.201200145
Cavin, L., and Guinot, G. (2014). Coelacanths as “almost living fossils”. Front. Ecol. Evol. 2, 49. doi: 10.3389/fevo.2014.00049
Cavin, L., Mennecart, B., Obrist, C., Costeur, L., and Furrer, H. (2017). Heterochronic evolution explains novel body shape in a Triassic coelacanth from Switzerland. Sci. Rep. 7, 13695. doi: 10.1038/s41598-017-13796-0
Cavin, L., Piuz, A., Ferrante, C., and Guinot, G. (2021a). Giant Mesozoic coelacanths (Osteichthyes, Actinistia) reveal high body size disparity decoupled from taxic diversity. Sci. Rep. 11, 11812. doi: 10.1038/s41598-021-90962-5
Cavin, L., Toriño, P., Van Vranken, N., Carter, B., Polcyn, M. J., and Winkler, D. (2021b). The first late cretaceous mawsoniid coelacanth (Sarcopterygii: Actinistia) from North America: evidence of a lineage of extinct 'living fossils'. PLoS ONE 16, e0259292. doi: 10.1371/journal.pone.0259292
Chalopin, D., Fan, S., Simakov, O., Meyer, A., Schartl, M., and Volff, J. N. (2014). Evolutionary active transposable elements in the genome of the coelacanth. J. Exp. Zool. Part B Mol. Dev. Evol. 322, 322–333. doi: 10.1002/jez.b.22521
Charlesworth, B. (2009). Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10, 195–205. doi: 10.1038/nrg2526
Cloutier, R. (1991). Patterns, trends, and rates of evolution within the Actinistia. Environ. Biol. Fishes 32, 23–58. doi: 10.1007/BF00007444
Cupello, C., Clément, G., Meunier, F. J., Herbin, M., Yabumoto, Y., and Brito, P. M. (2019). The long-time adaptation of coelacanths to moderate deep water: reviewing the evidences. Bull. Kitakyushu Museum Nat. Hist. Human Hist. Series A 17, 29–35.
Darwin, C. (1859). On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray. doi: 10.5962/bhl.title.162283
Drost, J. B., and Lee, W. R. (1995). Biological basis of germline mutation: comparisons of spontaneous germline mutation rates among drosophila, mouse, and human. Environ. Mol. Mutagen. 25, 48–64. doi: 10.1002/em.2850250609
Evans, A. R., Jones, D., Boyer, A. G., Brown, J. H., Costa, D. P., Ernest, S. M., et al. (2012). The maximum rate of mammal evolution. Proc. Nat. Acad. Sci. 109, 4187–4190. doi: 10.1073/pnas.1120774109
Folk, R. A., Stubbs, R. L., Mort, M. E., Cellinese, N., Allen, J. M., Soltis, P. S., et al. (2019). Rates of niche and phenotype evolution lag behind diversification in a temperate radiation. Proc. Nat. Acad. Sci. 116, 10874–10882. doi: 10.1073/pnas.1817999116
Friedman, M., and Coates, M. I. (2006). A new recognized fossil coelacanth highlights the early morphological diversification of the clade. Proc. R. Soc. Ser. B 273, 245–250. doi: 10.1098/rspb.2005.3316
Gingerich, P. D. (2001). Rates of evolution on the time scale of the evolutionary process. Microevol. Rate Pattern Process 2001, 127–144. doi: 10.1007/978-94-010-0585-2_9
Grandcolas, P., Nattier, R., and Trewick, S. (2014). Relict species: a relict concept? Trends Ecol. Evol. 29, 655–663. doi: 10.1016/j.tree.2014.10.002
Haldane, J. B. S. (1949). Suggestions as to quantitative measurement of rates of evolution. Evolution 3, 51–56. doi: 10.1111/j.1558-5646.1949.tb00004.x
He, J. X., and Stewart, D. J. (2001). Age and size at first reproduction of fishes: predictive models based only on growth trajectories. Ecology 82, 784–791. doi: 10.1890/0012-9658(2001)0820784:AASAFR2.0.CO
Kadarusman, H. Y., Pouyaud, L., Hocdé, R., Hismayasari, I. B., Gunaisah, E., et al. (2020). A thirteen-million-year divergence between two lineages of Indonesian coelacanths. Sci. Rep. 10, 192. doi: 10.1038/s41598-019-57042-1
Lampert, K. P., Fricke, H., Hissmann, K., Schauer, J., Blassmann, K., Ngatunga, B. P., et al. (2012). Population divergence in East African coelacanths. Curr. Biol. 22, R439–R440. doi: 10.1016/j.cub.2012.04.053
Lanfear, R., Welch, J. J., and Bromham, L. (2010). Watching the clock: studying variation in rates of molecular evolution between species. Trends Ecol. Evol. 25, 495–503. doi: 10.1016/j.tree.2010.06.007
Lidgard, S., and Love, A. C. (2018). Rethinking living fossils. Bioscience 68, 760–770. doi: 10.1093/biosci/biy084
Loewe, L., and Hill, W. G. (2010). The population genetics of mutations: good, bad and indifferent. Philos. Trans. R. Soc. B. Biol. Sci. 365, 1153–1167. doi: 10.1098/rstb.2009.0317
Lynch, M. (2010). Evolution of the mutation rate. Trends Genet. 26, 345–352. doi: 10.1016/j.tig.2010.05.003
Mahé, K., Ernande, B., and Herbin, M. (2021). New scale analyses reveal centenarian African coelacanths. Curr. Biol. 31, 1–8. doi: 10.1016/j.cub.2021.05.054
Martin, A. P., Naylor, G. J., and Palumbi, S. R. (1992). Rates of mitochondrial DNA evolution in sharks are slow compared with mammals. Nature 357, 153–155. doi: 10.1038/357153a0
Martin, A. P., and Palumbi, S. R. (1993). Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl. Acad. Sci. 90, 4087–4091. doi: 10.1073/pnas.90.9.4087
Minelli, A., and Baedke, J. (2014). Model organisms in evo-devo: promises and pitfalls of the comparative approach. History Philos. Life Sci. 36, 42–59. doi: 10.1007/s40656-014-0004-3
Naville, M., Chalopin, D., Casane, D., Laurenti, P., and Volff, J.-N. (2015). The coelacanth: Can a “living fossil” have active transposable elements in its genome? Mobile Genetic Elem. 5, 55–59. doi: 10.1080/2159256X.2015.1052184
Nikaido, M., Noguchi, H., Nishihara, H., Toyoda, A., Suzuki, Y., Kajitani, R., et al. (2013). Coelacanth genomes reveal signatures for evolutionary transition from water to land. Genome Res. 23, 1740–1748. doi: 10.1101/gr.158105.113
Nikaido, M., Sasaki, T., Emerson, J. J., Aibara, M., Mzighani, S. I., Budeba, Y. L., et al. (2011). Genetically distinct coelacanth population off the northern Tanzanian coast. Proc. Natl. Acad. Sci. 108, 18009–18013. doi: 10.1073/pnas.1115675108
Ohta, T. (1993). An examination of the generation-time effect on molecular evolution. Proc. Natl. Acad. Sci. 90, 10676–10680. doi: 10.1073/pnas.90.22.10676
Promislow, D. E. (1994). DNA repair and the evolution of longevity: a critical analysis. J. Theor. Biol. 170, 291–300. doi: 10.1006/jtbi.1994.1190
Pulquerio, M. J., and Nichols, R. A. (2007). Dates from the molecular clock: how wrong can we be? Trends Ecol. Evol. 22, 180–184. doi: 10.1016/j.tree.2006.11.013
Schaeffer, B. (1952). Rates of evolution in the coelacanth and dipnoan fishes. Evolution 6, 101–111. doi: 10.2307/2405507
Schultze, H.-P. (2004). “Mesozoic sarcopterygians,” in Mesozoic fishes 3 - systematics, paleoenvironments and biodiverity, eds. G. Arratia and A. Tintori (München: Verlag Dr Friedrich Pfeil), 463–492.
Smith, J. J., Sumiyama, K., and Amemiya, C. T. (2012). A living fossil in the genome of a living fossil: Harbinger transposons in the coelacanth genome. Mol. Biol. Evol. 29, 985–993. doi: 10.1093/molbev/msr267
Smith, J. L. B. (1939). A living coelacanthid fish from South Africa. R. Soc. South Africa. 143, 748–750. doi: 10.1038/143748a0
Sudarto, Lalu, X. C., Kosen, J. D., Tjakrawidjaja, A. H., Kusumah, R. V., Sadhotomo, B., et al. (2010). Mitochondrial genomic divergence in coelacanths (Latimeria): slow rate of evolution or recent speciation? Mar. Biol. 157, 2253–2262. doi: 10.1007/s00227-010-1492-7
Thomas, J. A., Welch, J. J., Lanfear, R., and Bromham, L. (2010). A generation time effect on the rate of molecular evolution in invertebrates. Mol. Biol. Evol. 27, 1173–1180. doi: 10.1093/molbev/msq009
Turner, D. D. (2019). In defense of living fossils. Biol. Philos. 34, 1–22. doi: 10.1007/s10539-019-9678-y
Vinciguerra, N. T., and Burns, K. J. (2021). Species diversification and ecomorphological evolution in the radiation of tanagers (Passeriformes: Thraupidae). Biol. J. Linnean Soc. 133, 920–930. doi: 10.1093/biolinnean/blab042
Welch, J. J., Bininda-Emonds, O. R., and Bromham, L. (2008). Correlates of substitution rate variation in mammalian protein-coding sequences. BMC Evol. Biol. 8, 1–12. doi: 10.1186/1471-2148-8-53
Wendruff, A. J., and Wilson, M. V. H. (2012). A fork-tailed coelacanth, Rebellatrix divaricerca, gen. et sp. nov. (Actinistia, Rebellatricidae, fam. nov.), from the lower Triassic of Western Gondwana. J. Verteb. Paleontol. 32, 499–511. doi: 10.1080/02724634.2012.657317
Yohe, L. R., Fabbri, M., Hanson, M., and Bhullar, B.-A. S. (2020). Olfactory receptor gene evolution is unusually rapid across Tetrapoda and outpaces chemosensory phenotypic change. Curr. Zool. 66, 505–514. doi: 10.1093/cz/zoaa051
Keywords: Actinistia, generation time, giants, Jurassic, Cretaceous, rate of evolution
Citation: Cavin L and Alvarez N (2022) Why Coelacanths Are Almost “Living Fossils”? Front. Ecol. Evol. 10:896111. doi: 10.3389/fevo.2022.896111
Received: 14 March 2022; Accepted: 09 May 2022;
Published: 23 May 2022.
Edited by:
Michel Laurin, UMR7207 Centre de recherche sur la paléobiodiversité et les paléoenvironnements (CR2P), FranceReviewed by:
Patrick Laurenti, Université de Paris, FranceCopyright © 2022 Cavin and Alvarez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lionel Cavin, bGlvbmVsLmNhdmluQHZpbGxlLWdlLmNo
 Lionel Cavin
Lionel Cavin Nadir Alvarez
Nadir Alvarez