
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 15 July 2022
Sec. Evolutionary and Population Genetics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.895056
This article is part of the Research TopicInsights in Evolutionary and Population Genetics: 2022View all 10 articles
Cacao (Theobroma cacao L.) is the basis of the lucrative confectionery industry with “fine or flavour” cocoa attracting higher prices due to desired sensory and quality profiles. The Amazonas Region (north Peru) has a designation of origin, Fine Aroma Cacao, based on sensory quality, productivity and morphological descriptors but its genetic structure and ancestry is underexplored. We genotyped 143 Fine Aroma Cacao trees from northern Peru (Bagua, Condorcanqui, Jaén, Mariscal Cáceres, and Utcubamba; mainly Amazonas Region), using 192 single nucleotide polymorphic markers. Identity, group, principal coordinate, phylogenetic and ancestry analyses were conducted. There were nine pairs of matched trees giving 134 unique samples. The only match within 1,838 reference cacao profiles was to a putative CCN 51 by a Condorcanqui sample. The “Peru Uniques” group was closest to Nacional and Amelonado-Nacional genetic clusters based on FST analysis. The provinces of Bagua and Utcubamba were genetically identical (Dest = 0.001; P = 0.285) but differed from Condorcanqui (Dest = 0.016–0.026; P = 0.001–0.006). Sixty-five (49%) and 39 (29%) of the Peru Uniques were mixed from three and four genetic clusters, respectively. There was a common and strong Nacional background with 104 individuals having at least 30% Nacional ancestry. The fine aroma of cacao from Northern Peru is likely due to the prevalent Nacional background with some contribution from Criollo. A core set of 53 trees was identified. These findings are used to support the continuance of the fine or flavour industry in Peru.
Domestication and use of Theobroma cacao L. (cacao; chocolate tree) dates back to ∼5,000 years from ruins of the Chinchipe culture, Palanda, south-eastern Ecuador and Montegrande, Jaen, Peru (Valdez, 2013; Ochoa, 2017; De la Fuente, 2018; Olivera-Núñez, 2018; Zarrillo et al., 2018). Cacao is used to refer to the plant while cocoa is used for the fermented and dried seeds and their processed products. Cacao is a tropical dicot Malvaceae tree (Alverson et al., 1999; Bayer et al., 1999) native to the Amazon basin of South America (Toxopeus, 1985; Motamayor and Lanaud, 2002; Bartley, 2005). The fruits produce seeds that are used in the pharmaceutical and cosmetic industries but primarily as the raw ingredients for the multibillion dollar chocolate industry (Oddoye et al., 2013; Wickramasuriya and Dunwell, 2018). The consumption of chocolate and its products is estimated to increase by 3% each year (Wickramasuriya and Dunwell, 2018) and acts as the main economic driver of global cacao farming (Tacer-Caba, 2019).
Cacao crops are critical for local economies of about 6 million smallholder farmers in Latin America, Africa, and Asia (Rice and Greenberg, 2000; Beg et al., 2017). Peru produced 160,289 metric tonnes of cocoa in 2020 making it the 9th largest producer of cocoa worldwide (FAO, 2022). In the Amazonas region of Peru, cacao is the second most economically important crop with a cultivated and harvested area of 13,416.83 ha (Instituto Nacional de Estadística e Informática [INEI], 2012). In this region, three provinces are the main producers of cacao: Bagua with the highest cocoa production (75%), followed by the Utcubamba, and Condorcanqui provinces (Torres-Armas and Gonzáles-Castro, 2018). The discovery of high-yielding and disease-resistant varieties is needed to support the growing global cacao industry (Goenaga et al., 2009; Phillips-Mora et al., 2013). The conservation and utilisation of cacao genetic diversity are crucial for the sustainable cultivation of cacao (Zhang and Motilal, 2016; Laliberté et al., 2018).
The cocoa industry recognises “bulk cocoa” and “fine or flavour cocoa” with the latter garnering a higher premium price. While bulk cacao still contributes to more than 80% of worldwide production (Wickramasuriya and Dunwell, 2018), there has been an increase in demand for fine or flavour chocolate, along with consumer appreciation for the traditional histories and origin of native cacao varieties (Mejía et al., 2021). Peru has been designated a 75% producer and exporter of fine flavour cocoa (International Cocoa Organization [ICCO], 2021) and is thus well poised to capitalise on this consumer base. Cacao from the Peruvian Amazonas region currently has a designation of origin, namely Fine Aroma Cacao, based on its peculiar characteristics in terms of its sensory quality (aroma and flavour) (Instituto Nacional de Defensa de la Competencia y de la Protección de la Propiedad Intelectual [INDECOPI], 2016). These qualities have given a high value and demand which strongly improve the competitiveness of Peruvian Amazonas cocoa in the foreign market (Oliva and Maicelo-Quintana, 2020). Five groups of cacao (Bagüinos, Cajas, Indes, Toribianos, Utkus) were identified according to these sensory features, in addition to productivity and morphological descriptors (Oliva-Cruz, 2020). Sensory evaluation of Bagua type cacao determined that this upper Amazon variety differed from the native “Chuncho” cacao found in Quillabamba, Cusco (Céspedes-Del Pozo et al., 2018; Mejía et al., 2021). The Indes and Bagüinos morphotypes had the best floral and fruity sensory characteristics and the highest dry weight and number of seeds (Oliva-Cruz, 2020).
Bulk cocoa traditionally comes from Forastero cacao while “fine or flavour” cocoa can be obtained from Criollo, Nacional and some Trinitario varieties (Pridmore et al., 2000). Cacao was traditionally classed as Criollo, Forastero and Trinitario varieties, based on morphological and agronomical traits with the latter variety being thought to be a hybrid of the former two (Toxopeus, 1985; Pridmore et al., 2000). Forastero encompassed a range of cacao types including the Amelonado variety responsible for the basis of the West African bulk cocoa industry and the Nacional variety from Ecuador known for its fine Arriba flavour. The Refractario variety also from Ecuador arose out of a mass field selection program in the 1920s for witches’ broom disease resistance (Pound, 1938, 1943; Bartley, 2001).
A variety of molecular approaches have enabled better separation and understanding of the true genetic diversity and varietal classification than the traditional names and industry convention. A review of these molecular approaches can be found in Livingstone et al. (2012); Motilal et al. (2017), and Everaert et al. (2020). Genetic diversity is higher when there are unique samples that increase differentiation within and among groups. Accurate identity analysis is, however, dependent on the number of markers, as well as, the composition of the marker set used for both microsatellite markers (Motilal et al., 2009) and single nucleotide polymorphism (SNP) markers (Mahabir et al., 2020).
The use of microsatellite markers is currently being supplanted by SNP markers especially for large genetic diversity studies. Genotyping of cacao germplasm with SNPs has been performed using the novel integrated fluid circuit (IFC) technology (Osorio-Guarín et al., 2017), which increased the throughput per run, simplified setup of reactions, and decreased the running cost (Xu, 2016). Lately, the analysis of the genetic diversity and population structure of cacao have used a set of reduced and informative SNP markers (Singh and Singh, 2015; Cosme et al., 2016; Osorio-Guarín et al., 2017; Mahabir et al., 2020; Wang et al., 2020). The identification and authentication of fine flavour cacao varieties have also employed SNPs (Fang et al., 2014; Arevalo-Gardini et al., 2019).
Genetic clustering of cacao was clarified by Motamayor et al. (2008) who used 106 microsatellite markers to identify ten genetic groups (Amelonado, Contamana, Criollo, Curaray, Guiana, Iquitos, Marañon, Nacional, Nanay, and Purús) in the Amazon basin of South America. The clustering of these groups has been supported and refined by Thomas et al. (2012) and Nieves-Orduña et al. (2021). Five of the 10 genetic clusters (Contamana, Iquitos, Marañon, Nacional, and Nanay) occur in Peru (Motamayor et al., 2008). Nieves-Orduña et al. (2021) identified 23 chloroplast microsatellite haplotypes on a sample of 233 cacao plants with the highest variation being found in western Amazonia; particularly in the north-western Amazon with Peru having seven unique haplotypes. The genetic clustering of cacao is expected to change as more wild natural stands of cacao are explored in the Amazon. North-eastern Peru hosts a wide diversity and genetic variability of cacao that is under-explored (Motamayor et al., 2008; Thomas et al., 2012). Two traditional fine or flavour varieties in Peru are the small-seeded variety known as Chuncho from the Urubamba valley in southern Peru; and the “Piura Porcelana” variety with large pale seeds mainly cultivated in Piura, Amazonas, and Cajamarca provinces of northern Peru (Arevalo-Gardini et al., 2019). Céspedes-Del Pozo et al. (2018), using 96 single nucleotide polymorphism (SNP) markers reported that the native cacao variety “Chuncho” –from La Convencion, Cusco in southern Peru – was distinct but closest to the Contamana population, Beni population (unique population from Beni River in Bolivia, Zhang et al., 2012), and cacao from the Madre de Dios region. “Piura Porcelana” formed an immediate sister clade to the Nacional group (Arevalo-Gardini et al., 2019).
Additionally, Chia-Wong et al. (2018) tried to assess about 80 fine or flavour trees from the five principal cacao regions of Peru (Amazonas, Cusco, San Martin, Piura, and Huánuco) with 18 microsatellites but the amplification was unsuccessful. Zhang et al. (2006a), using 15 microsatellites, demonstrated that cacao in Huallaga and Ucayali Valleys were distinct groups. The Huallaga farmer selections were shown to be mainly hybrids of Trinitario and Upper Amazon Forastero accessions (Zhang et al., 2011).
Saavedra-Arbildo et al. (2018) demonstrated from fruit and seed morphology that the cacao in the Peruvian regions of Amazonas, Cusco and Piura were similar in thickness of fruit wall, fruit length, water content of testa, and seed width but differed in depth of primary furrows, fruit mass, seed mass, fruit width, number of seeds, dry mass of seeds, seed length, and seed thickness. In addition, the northern regions of Amazonas and Piura appeared more similar to each other than Cusco although all three areas were differentiated on the basis of number of seeds and seed length with the Piura region having the greatest proportion of white seeds in fruits that were generally elliptic-obovate with obtuse apices and little to no rugosity (Saavedra-Arbildo et al., 2018).
There is a scarcity of recent work on the phenotypic and genetic diversity of cacao in Peru, far less for northern Peru. In addition, the use of the current SNP marker technology is limited to a few studies. Studies on the genetic diversity of cacao and especially fine aroma cacao in northern Peru are lacking. The goal of this study is to determine the genetic uniqueness, genetic diversity and ancestry of Fine Aroma Cacao from the Peruvian Amazonas region by SNP genotyping. In addition, we examined if three provinces (Bagua, Condorcanqui, and Utcubamba) were genetically distinct and harboured new cacao genetic clusters. The resultant information is expected to be a significant addition to our understanding of the genetic diversity of cacao in Peru and how it can be leveraged to bolster the fine flavour status in Peru.
A total of 143 trees (15–20 years old) of Fine Aroma Cacao were sampled mainly from farmers’ fields in three provinces of the Amazonas region, in northern Peru (Bagua, Condorcanqui, Utcubamba; Supplementary Table 1 and Figure 1) and were deposited in the herbarium of Universidad Nacional Toribio Rodríguez de Mendoza (KUELAP), Peru (Thiers, 2016). A permit for scientific research on wild flora (RDG N° D000319-2020-MINAGRI-SERFOR-DGGSPFFS, with authorisation code N° AUT-IFL-2020-051) was provided by Servicio Nacional Forestal y de Fauna Silvestre (SERFOR). For each site, the date, time, and GPS coordinates were recorded. The 143 test trees from northern Peru were compared to reference profiles of cacao accessions belonging to the 10 genetic clusters of Motamayor et al. (2008) as well as Trinitario and Refractario accessions. A maximum of 1,838 reference accessions from the International Cocoa Genebank Trinidad were used. Reference profiles are maintained and curated by the Cocoa Research Centre (CRC), The University of the West Indies.
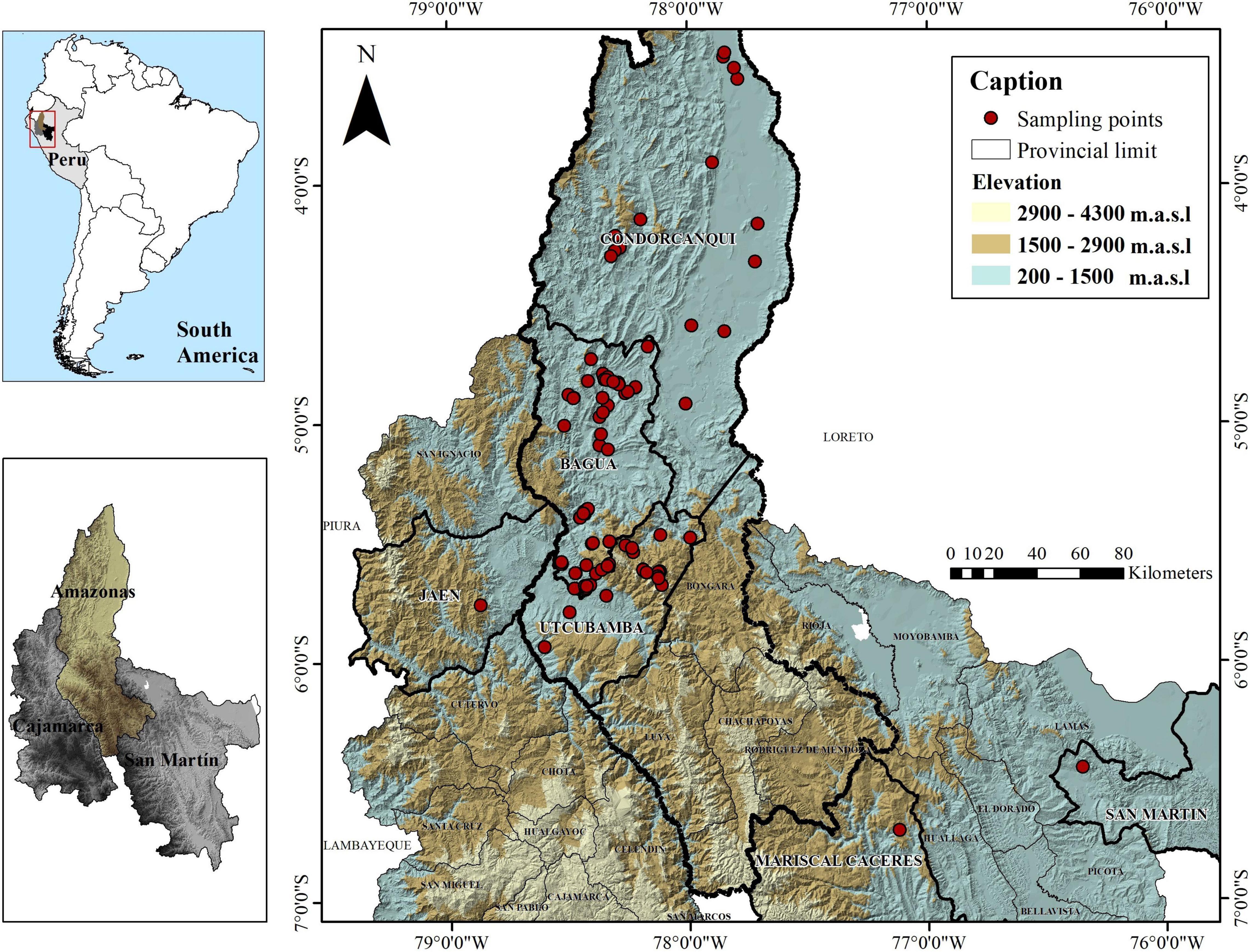
Figure 1. Distribution of collected Fine Aroma Cacao samples from northern Peru. The national, provincial and district boundaries were obtained from the Geoportal of the National Geographic Institute of Peru (IGN) in shapefile format with a DATUM WGS 1984 for illustrative purposes only.
Tissue samples were taken from the distal regions of healthy cacao leaves and stored in pre-labelled 1.5 mL Safelock Eppendorf tubes containing silica gel desiccant. Six leaf discs (6 mm diameter) were prepared from each test plant using the BioArk leaf collection kit from LGC Biosearch Technologies. The plates were shipped to LGC Genomics, United Kingdom for DNA extraction and SNP genotyping using their proprietary KASP chemistry. Genotyping was performed at 192 SNP sites from flanking sequences provided by the Cocoa Research Centre (CRC) of The University of the West Indies (Motilal et al., 2017; Mahabir et al., 2020; Supplementary Table 2). Returned multilocus data from LGC was curated by removing SNPs and samples with more than 7% missing data. This is expected to reduce the impact of missing data on the genetic analyses. Seven of the 192 SNPs (TcSNP 0456, 0701, 1,038, 1,156, 1,229, 1,408, 1,457) had 100% missing data and were removed from subsequent analyses.
Multilocus SNP profiles were analysed using GenAlEx v6.502 (Peakall and Smouse, 2006, 2012). This software was used for frequency analysis, group differentiation tests, principal coordinate analysis (PCoA) and to prepare in files for other programs. Identity analyses were conducted in Cervus v3.0 (Kalinowski et al., 2007), phylogenetic analysis in DARwin v6 (Perrier et al., 2003; Perrier and Jacquemoud-Collet, 2006), ancestry analysis in STRUCTURE v2.3.4 (Pritchard et al., 2000) and core collection identification in PowerCore v1.0 (National Institute of Agricultural Biotechnology, 2006). Statistical tests to determine if there were significant differences in genetic parameters were conducted in MedCalc Statistical Software v12.7.7 (MedCalc Software bvba, 2013).
Identity analyses were conducted in Cervus v3.0 (Kalinowski et al., 2007). A minimum of 170 matching loci with a flexibility mismatch of 5 loci was applied to identify possible groups of matched samples among the data matrix of 185 SNPs/143 Peru test trees (Supplementary Table 3). Missing data occurred at 0–7 SNPs with an average of 0.60% (standard error = 0.05) in the entire dataset. Members within a group are equivalent to each other but not equivalent to members of other groups. One member of each group was retained to obtain a maximal list of unique Peru samples (Peru Uniques). Identity analysis was conducted between the Peru Uniques dataset and 1,838 CRC reference profiles mainly from the International Cocoa Genebank Trinidad. The reference profiles had data at 175 SNPs so identity analysis was conducted using a minimum of 160 matching loci with a flexibility mismatch of 5 loci. In this dataset, missing data was present at 0–42 SNPs per sample (mode = 6) with an average of 7.69% (standard error = 0.16) in the entire dataset. The probability of identity among siblings (PIDSIB), was obtained to estimate the chance of a false match. The PIDSIB is the probability that two siblings drawn at random from a population have identical genotypes (Evett and Weir, 1998; Waits et al., 2001) and was recommended to be used in cacao (Zhang et al., 2006b).
The 134 Peru Uniques had 8 and 26% missing data at TcSNP0230 and TcSNP1350, respectively, over the maximal set of 185 SNPs. In addition, three monomorphic SNPs (TcSNP: 0097, 0383, 1,158) were present. These five SNPs were removed so that missing data was present at 0–14 SNPs per sample (mode = 0) with an average of 0.41% (standard error = 0.05) in the entire dataset. Frequency analysis was conducted in GenAlEx v6.502 (Peakall and Smouse, 2006, 2012) to obtain descriptive genetic measures of number of effective alleles (Nrm e); Shannon’s Information Index (I); observed, expected and unbiased expected heterozygosities (Ho, He, uHe, respectively); the fixation index (F); and individual heterozygosities (Hind) of the sampled trees for each sampled provinces and the Peru Uniques.
Principal coordinate analysis was conducted on the set of Peru Uniques in relation to 390 reference accessions (40 Amelonado, 8 Contamana, 12 Criollo, 17 Curaray, 56 Guiana, 32 Iquitos, 70 Marañon, 40 Nacional, 60 Nanay, 5 Purús, 25 Amelonado/Nacional hybrids, and 25 Amelonado/Criollo hybrids) using 170 SNPs. Population references are from selected accessions with exclusive membership to their respective genetic clusters. Similarly hybrid references were selected based on contributions from only the two required genetic clusters. Accessions and SNPs were chosen to minimise missing data. In this dataset, missing data was present at 0–14 SNPs per sample (mode = 0) with an average of 0.44% (standard error = 0.03) in the entire dataset. The analysis in GenAlEx v6.502 (Peakall and Smouse, 2006, 2012) implemented a standardised linear genetic distance.
Phylogenetic analysis was performed on the same dataset as for the PCoA in DARwin v6 (Perrier et al., 2003; Perrier and Jacquemoud-Collet, 2006). The program accepts allelic data and creates a simple matching dissimilarity index. Missing data was set at 50, 70, or 90% with the default pairwise allele deletion to construct dissimilarity matrices with 1,000 bootstraps. Tree construction employed the weighted Neighbor-Joining algorithm with 1,000 bootstrap replicates. Bootstrap values > = 70% were displayed on the trees.
An analysis of molecular variance (AMOVA) was conducted using the same dataset as for the PCoA using 999 permutations in GenAlEx v6.502 (Peakall and Smouse, 2006, 2012). Group differentiation based on Jost Dest statistic (Jost, 2008, 2009) was conducted on a refined dataset. This dataset involved the same reference groups as for the PCoA but with 136 SNPs to get less than 6% missing data per group and with an average of 0.13% (standard error = 0.01) missing data within the dataset. In addition, the Peru Uniques was decomposed into provincial groups that contained at least five samples. The provinces of Bagua (n = 16), Condorcanqui (n = 24) and Utcubamba (n = 91) were retained. If members of duplicate groups were present, only one sample per group province was retained. In this dataset, there was also less than 6% missing data per group and with an average of 0.12% (standard error = 0.01) missing data. The Dest pairwise calculations were performed in GenAlEx v6.502 (Peakall and Smouse, 2006, 2012) using 999 permutations and 999 bootstraps. Phylogenetic clusters in the collected samples were identified and Jost Dest was used to determine if the clusters were separate groupings. Datasets were examined for private alleles.
Population structure of the 143 samples was determined via the model-based clustering method implemented in STRUCTURE v2.3.4 (Pritchard et al., 2000). Reference accessions that represent the 10 genetic clusters identified by Motamayor et al. (2008) were cloned to obtain a sample size of 200 for each population. An initial run using number of populations (K) from nine to 14 [the expected 10 of Motamayor et al. (2008) plus one more than the number of expected clusters in the collected samples] was conducted. A dataset of 154 SNPs was used to obtain minimal missing data. An admixture model with an inferred alpha value, independent allele frequency with 100,000 burnins and 200,000 Markov Chain Monte Carlo (MCMC) repetitions was used with 10 iterations at each K value. The optimal K was selected based on best differentiation of samples to maintain Motamayor et al. (2008) grouping and on the ad hoc method of Evanno et al. (2005). Then with a maximal dataset of 170 SNPs in the Peru samples and cloned population references an admixture model with an inferred alpha value, independent allele frequency with 300,000 burnins and 600,000 MCMC repetitions was used at the chosen K with 10 iterations. The run with the most positive ln P(D) was chosen to represent the ancestral background. A minimum level of 5% was used as evidence of the presence of a genetic group. A minimum level of 95% without a 5% level in any other group was used to establish exclusive membership to a genetic group. The distribution of the predominant ancestral group(s) for the Bagua, Condorcanqui and Utcubamba provinces was tested for equivalence using the comparison of proportions test in MedCalc Statistical Software v12.7.7 (MedCalc Software bvba, 2013).
The Peru Uniques typed at the maximal number of SNPs underwent core selection in PowerCore v1.0 (National Institute of Agricultural Biotechnology, 2006) under its heuristic algorithm. The number of SNPs was reduced to those with less than 6% missing data. The core set was then compared to the entire set of Peru Uniques as well as to the group remaining after the core was removed from the entire set. Comparison was performed at summary statistics (Ne, I, Ho, He, uHe), private alleles and Jost Dest as obtained in GenAlEx v6.502 (Peakall and Smouse, 2006, 2012).
In the dataset of 143 trees/185 SNPs, there were nine pairs of matched samples (Table 1). Eight of these pairs were within the same province with the exception of INDES095 from Bagua being matched at all 185 SNPs to INDES098 from Utcubamba. A PIDSIB of 2.21 × 10–28 was obtained for the dataset of 143 trees/185 SNPs. Removal of one sample from each of the nine pairs gave a set of 134 unique samples (Peru Uniques). The Peru Uniques compared to 1,838 reference accession profiles at 175 common SNPs returned only one possible match of a putative CCN 51 to CCA015 from Condorcanqui with 171 matching loci and one mismatched locus. A PIDSIB of 1.862 × 10–30 was obtained for the dataset of 1,972 samples/175 SNPs. The average minor allele frequency over all the 562 samples is 0.261 (Supplementary Tables 3, 4).
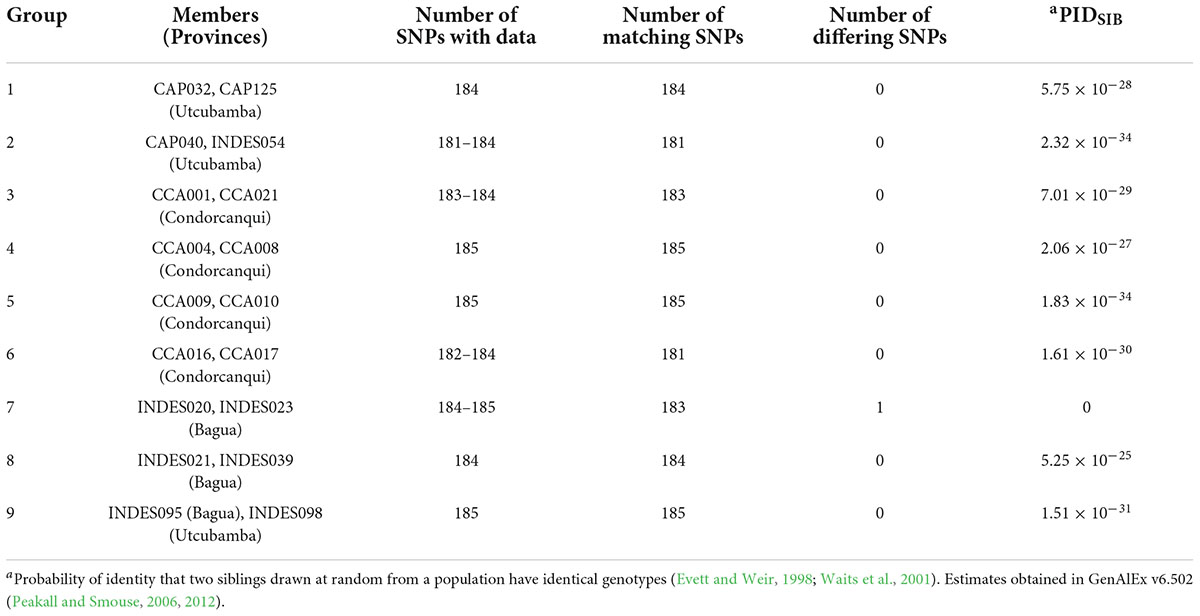
Table 1. Groups of matched samples in 143 cacao trees in northern Peru using 185 single nucleotide polymorphism (SNP) markers.
The resultant frequency analysis showed that Ho was close to He with a very low (0.006) fixation index (Table 2). Using this same set of 180 SNPs, the provinces of Bagua, Condorcanqui, and Utcubamba had a low to zero fixation index (Table 2) with slightly higher He than Ho in Condorcanqui but slightly higher Ho than He in the other two provinces. The Hind in the Peru Uniques ranged from 0.056 to 0.578 with all samples being heterozygous (Supplementary Table 5). The lowest Hind values were observed in the Utcubamba province (INDES032, Hind = 0.056; INDES002, Hind = 0.089). The highest Hind values were observed in the Bagua (INDES070, Hind = 0.578) and the Utcubamba provinces (INDES061, Hind = 0.578). There was an absence of low Hind (0–0.015) in Condorcanqui and Mariscal Cáceres provinces. The single sample from Jaén had a low heterozygosity (Hind = 0.117; Supplementary Table 5).
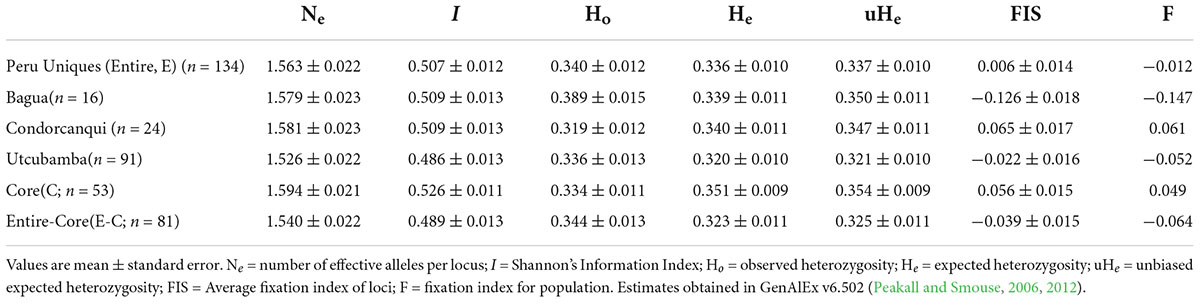
Table 2. Descriptive genetic statistics for set of unique cacao and its core collection in north Peru with 180 SNPs.
The 134 Peru Uniques were distributed across three quadrants in a linear pattern from Amelonado to Nacional but excluding close association with Criollo, Marañon and Guiana groups (Figure 2). There was no apparent sub-clustering of samples. The PCoA explained 23.9, 10.8, and 9.5% on the first three axes, respectively.
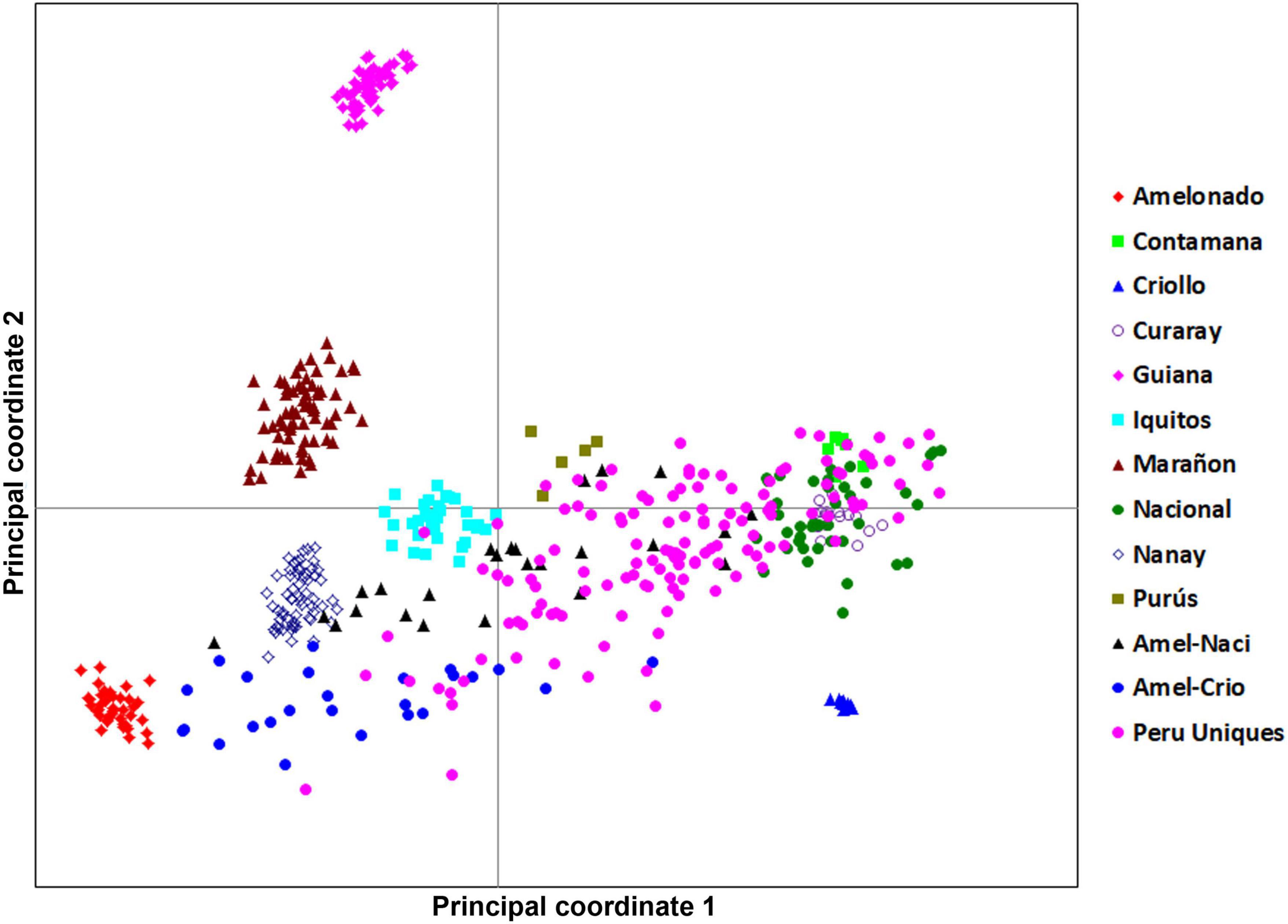
Figure 2. A principal coordinate analysis 2D-scatter plot of 134 Peru Uniques and 390 reference accessions using 170 SNP genetic data. The first and second axes explained 23.86 and 10.80% of the variation, respectively.
Phylogenetic trees based on 50, 70, or 90% missing data thresholds to retain sample pairs were similar (Supplementary Figures 1, 2) and the tree based on 70% missing data is provided in Figure 3. The 134 Peru Uniques were mainly distributed between reference clusters rather than within clusters and were closest to, and arrayed along, the Nacional, Contamana, Curaray, Iquitos, Purús, and Nanay genetic groups (Figure 3). Two samples, CCA027 (Condorcanqui) and CAP045 (Utcubamba), were associated with the Amelonado and Criollo clusters with CAP045 being closest to the Criollo group. INDES095 from the Bagua province was an immediate sister clade to the Nanay group. Three phylogenetic clusters (Phylo A, B, C) in the Peru Uniques were found (Figure 3) and each cluster contained a variable number of samples from each of the three main provinces. The PhyloA cluster (represented by CAP086 from Utcubamba) contained 14 individuals (including the three samples from Mariscal Cáceres) and was positioned between the Iquitos and Purús genetic groups. The PhyloB cluster (represented by INDES064 from Utcubamba) contained 33 individuals and formed a sister clade with the Contamana/Curaray clade. The PhyloC cluster (represented by CAP107 from Utcubamba) contained 64 individuals (including the single sample from Jaén) and was positioned between the Nacional and Contamana/Curaray clades.

Figure 3. Phylogram (based on 70% missing data) of unique cacao samples collected from Northern Peru (134 samples) and 390 reference accessions using 170 single nucleotide polymorphisms. Three phylogenetic clusters (PhyloA-C) from the samples from north Peru are indicated together with a representative sample. All three representative samples are from Utcubamba and each cluster also contain samples from Bagua and Condorcanqui. Three other samples from north Peru are indicated – CAP45 (Utcubamba), CCA27 (Condorcanqui) and INDES95 (Bagua). The three samples from Mariscal Cáceres are in PhyloA and the one sample from Jaén is in PhyloC. ≥ 70% bootstrap values are displayed.
The AMOVA that incorporated the 134 Peru Uniques as a unit group partitioned 54.5% within genetic clusters and 45.5% among genetic clusters (Supplementary Table 6). The genetic differentiation (FST = 0.455) was significant (P = 0.001) (Supplementary Table 6). The FST among pairwise groups (Supplementary Table 7) indicated that the set of Peru Uniques was closest to the group of Amelonado/Nacional hybrids (0.126) and then to the Nacional cluster (0.155). The Jost Dest measure of group differentiation among the reference groups were all significant (P = 0.001, 0.002) with maximal values of 0.626 (Amelonado vs. Criollo) and minimal values of 0.089 (Amelonado vs. Amelonado/Criollo) and 0.098 (Amelonado/Nacional vs. Amelonado/Criollo) (Figure 4 and Supplementary Tables 8, 9). Private alleles were only present from two SNPs (TcSNP0097, TcSNP1158) in the Contamana group.
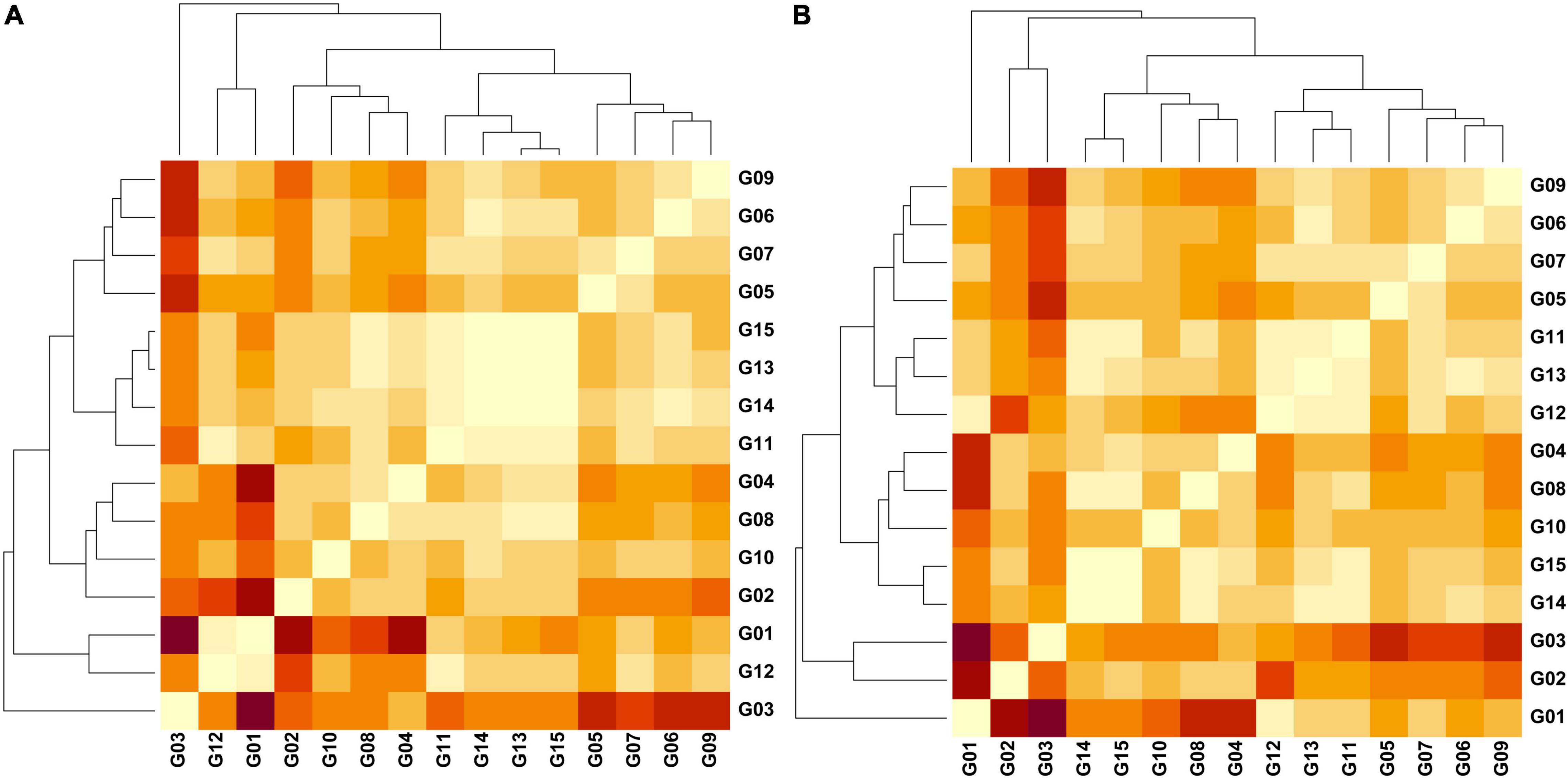
Figure 4. Heatmaps representing pairwise Jost differentiation indices (Dest) (light = 0.00/dark = 0.626) among 15 groups of cacao samples based on the genetic variation of 136 SNPs examining (A) three regions and (B) three phylogenetic clusters in north Peru. Groups – G1 (Amelonado; n = 40), G2 (Contamana; n = 8), G3 (Criollo; n = 12), G4 (Curaray; n = 17), G5 (Guiana; n = 56), G6 (Iquitos; n = 32), G7 (Marañon; n = 70), G8 (Nacional; n = 40), G9 (Nanay; n = 60), G10 (Purús; n = 5), G11 (Amelonado/Nacional; n = 25), G12 (Amelonado/Criollo; n = 25), G13A (Bagua; n = 16), G14A (Condorcanqui; n = 24), G15A (Utcubamba; n = 91), G13B (PhyloA; n = 14), G14B (PhyloB; n = 33), G15B (PhyloC; n = 64). Dest values obtained in GenAlEx v6.502 (Peakall and Smouse, 2006, 2012).
Jost Dest measure was obtained from a dataset of 136 SNPs for which missing data was less than 6% in any of the grouped samples. The Dest values were significant (P = 0.001) for all pairwise comparisons involving the three Peruvian provinces and any of the 12 reference groups with maximal Dest (0.347) recorded for Criollo vs. Condorcanqui (Supplementary Table 8). The reference group Iquitos was closest to Condorcanqui (Dest = 0.098) whereas Nacional was closest to Bagua (0.075) and Utcubamba (0.066). The three provinces were also close to the set of Amelonado/Nacional hybrids (Dest = 0.070 – 0.076). Among the three provinces, significant and low Dest values were obtained for Bagua vs. Condorcanqui (Dest = 0.016; P = 0.006) and Condorcanqui vs. Utcubamba (Dest = 0.016; P = 0.006) but Bagua vs. Utcubamba was low and non-significant (Dest = 0.001; P = 0.285). Phylogenetic clusters of the 10 population groups of Motamayor et al. (2008), two reference mixed groups (Amelonado/Nacional and Amelonado/Criollo) and the three clades (Phylo A, B, C) in the Peruvian dataset were all significantly different (P = 0.001) from each other (Supplementary Table 9). The PhyloB and PhyloC clades were closest to each other (Dest = 0.037) while the PhyloA and PhyloC clades were furthest from other Peruvian samples (Dest = 0.144). The three clades in the Peruvian dataset were all close to the Amelonado/Nacional group (Dest = 0.094–0.097). The PhyloA clade was closest to the Iquitos (Dest = 0.082) and the Amelonado/Criollo (Dest = 0.094) reference groups. The PhyloB clade was closest to the Amelonado/Nacional (Dest = 0.094) reference group. The PhyloC clade was closest to the Nacional (Dest = 0.063) and the Amelonado/Nacional (Dest = 0.095) reference groups.
At K = 10, the reference populations were all resolved into the expected groupings of Motamayor et al. (2008) and the Amelonado/Nacional and Amelonado/Criollo hybrids presented the ancestry profile to match their expected founder populations (Figure 5A). However, the optimal K value by Evanno’s method was for 11 populations. At K = 11, the 10 reference populations were resolved, but one population in each run was split into two distinct groups. The population that was split was inconsistent and occurred in the Amelonado (once), Contamana (twice), Criollo (thrice), Curaray (twice), Guiana (once), and Marañon (once) of the 10 iterations. An example is presented in Figure 5B. Nonetheless, the Peru Uniques at both K = 10 and K = 11 in the initial analysis exhibited a strong and frequent Nacional background (Figures 5A,B) which was supported by the more stringent analysis at K = 10 (Supplementary Tables 10, 11).
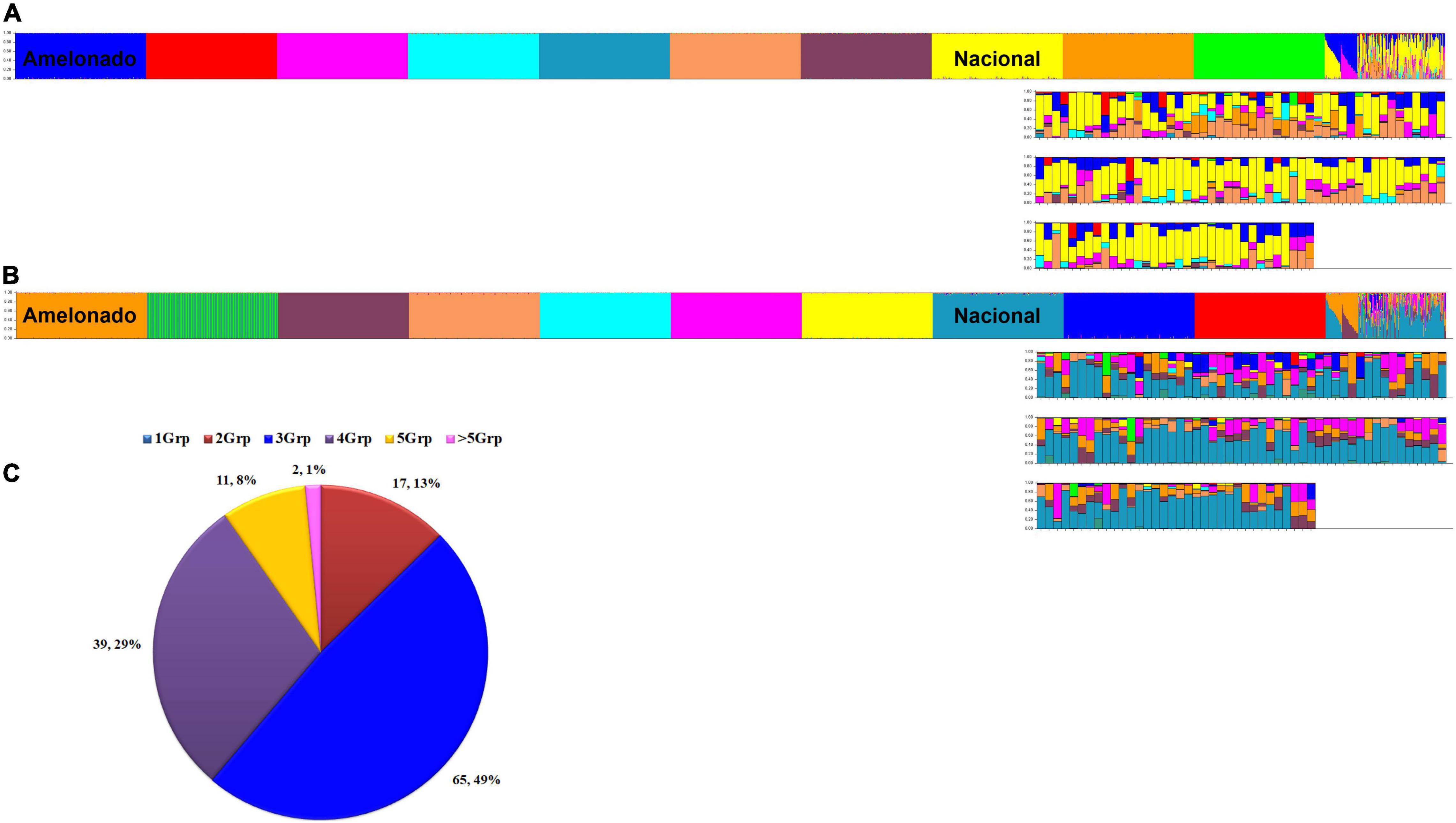
Figure 5. Ancestry of 134 unique cacao samples (Peru Uniques) collected from northern Peru. Ancestry at K = 10 (A) and K = 11 (B) using 154 SNPs obtained from STRUCTURE (Pritchard et al., 2000) output using model based on 100,000 burnins, 200,000 Markov Chain Monte Carlo (MCMC) simulations, admixture ancestry model and independent allele frequencies. Samples are arranged as 10 reference populations (Amelonado, Contamana, Criollo, Curaray, Guiana, Iquitos, Marañon, Nacional, Nanay, Purús), Amelonado/Nacional, Amelonado/Criollo and Peru Uniques. Distribution of admixture classes (C) in Peru Uniques from 1 to more than five genetic groups (Grp) obtained from STRUCTURE (Pritchard et al., 2000) output using model based on 170 SNPs, 300,000 burnins, 600,000 MCMC simulations, admixture ancestry model and independent allele frequencies.
The 134 Peru Uniques were all admixed with contributions from 2 to 7 genetic groups and with a mixture of three groups being the most frequent (Figure 5C). Apart from Nacional, at least 50% ancestry was present from Amelonado (CCA27, Condorcanqui), Contamana (CAP107, Utcubamba; INDES70, Bagua), Criollo (CAP45; Utcubamba) and Iquitos (CAP37, INDES18, INDES62: Utcubamba; CCA16, Condorcanqui). The combination of only Amelonado with Criollo ancestry was only found in CAP45 and CCA27. The single sample collected from Jaén had 85% Nacional, 8% Curaray, and 5% Iquitos ancestral background. The three samples from Mariscal Cáceres had a common background of Amelonado, Criollo, and Iquitos. However, apart from the actual contributions being different, two of these (INDES 101, 106) were higher in Iquitos (41%) ancestry whereas the other (INDES112) had Nanay as the major component (37%). A set of 49 samples combined both Criollo and Nacional ancestry (each at a minimum of 10%) with the majority (36) coming from the Utcubamba province and the remainder from the Bagua (8) and Condorcanqui (3) provinces. Sixteen samples lacked Nacional ancestry and contained instead an Amelonado/Criollo background with other groups except for INDES95 from Bagua which was mixed with Nanay (44%), Iquitos (36%), and Curaray (13%). The proportion of cacao trees with at least 25% Nacional ancestry was highest in Utcubamba (91.1%; 82 of 90), then Bagua (75%; 12 of 16) with the lowest occurrence in Condorcanqui (58.3%, 14 of 24). However, only the comparison of Condorcanqui to Utcubamba was significantly different (P = 0.0003; Supplementary Table 12).
A set of 53 samples were identified as a core collection from the 134 Peru Uniques and 182 SNP loci (Supplementary Table 13). Statistical measures in the core were higher than that of the entire set and the entire set without the core except for Ho which showed the reverse trend (Table 2). However, private alleles were lacking and Jost Dest was non-significant being estimated as 0 (P = 0.993) and 0.001 (P = 0.177) for the aforementioned two comparisons.
Examining the genetic diversity and ancestry of cacao from its centre of diversity is essential to better understand its population structure and the judicious conservation and cultivation of native varieties. In this study 143 cacao trees from north-western Peru (the majority from the Amazonas region) were SNP genotyped via 185 informative SNPs to examine their genetic diversity and ancestry. Overall, the findings indicated that the samples had moderate gene diversity (He = 0.336) and shared ancestry with the Nacional, Amelonado, Iquitos and Criollo groups. The 143 samples had few matching duplicates (nine groups with two members each) and these were usually within a province rather than across provinces. This internal duplicate matching was lower than that recorded for cacao collected in Belize (Motilal et al., 2010) and for farm selections in Dominica (Gopaulchan et al., 2019), Dominican Republic (Boza et al., 2013), Hawaii (Nagai et al., 2009), Nicaragua (Trognitz et al., 2011), the Huallaga and Ucayali valleys in Peru (Zhang et al., 2006a), Puerto Rico (Cosme et al., 2016) but higher than that reported in one farm in Jamaica (Lindo et al., 2018), Vietnam (Everaert et al., 2017) or for the ICS and TRD accessions in Trinidad (Johnson et al., 2009). Furthermore, an absence of duplicates was reported for 164 trees in Bolivia (Zhang et al., 2012), 93 trees in Tumaco, Colombia (Yacenia Morillo et al., 2014), for 53 trees in Sulawesi, Indonesia (Dinarti et al., 2015), for 220 trees in the Juanjui province of the Huallaga valley, Peru (Zhang et al., 2011), and for 109 trees in Uganda (Gopaulchan et al., 2019). A set of 134 Peru Uniques was obtained after removal of duplicate samples and only one sample (CCA015; Condorcanqui) matched to an external reference variety with a very low PIDSIB (1.862 × 10–30) in the identity analysis dataset. The internal and external match analyses indicated that the cacao samples collected in north Peru were generally distinct and unique. This is promising for maintaining relic diversity, identifying genotypes best suited to local conditions and maintaining the distinctiveness of the Peruvian fine aroma cacao industry. The few duplicate trees may represent very closely related varieties that are unable to be resolved with the SNP panel in this study. If not, then these duplicated samples may represent clonal propagated material that was disseminated in earlier years. The presence of a sample similar to a putative CCN 51 supports the latter view and represents a cautionary note for north Peru. However, the ancestry profile of the sample in north Peru (CCA015) was different from that reported in Boza et al. (2014) suggesting that the CCN 51 may have been a mislabelled reference accession. CCA015, while probably not CCN 51, represents an example of a sample without Nacional but with Criollo ancestry which will contribute to the fine aroma designation.
The low or zero fixation indices are indicative of the absence of inbreeding. This supports the use of crosses between trees sampled from the Peruvian Amazon region for genetic improvement. A moderate level of gene diversity was observed (He = 0.32–0.34) for the Peru Uniques as well as in the Bagua, Condorcanqui, and Utcubamba provinces. This was similar to on-farm cacao in Dominica (He = 0.320; Gopaulchan et al., 2020), in Honduras and Nicaragua (He = 0.367; Lukman et al., 2014), and Uganda (He = 0.332; Gopaulchan et al., 2019) but higher than in Colombia (He = 0.28; Yacenia Morillo et al., 2014), Ghana (He = 0.245; Padi et al., 2015), and Chuncho cacao from the La Convención province in south Peru (He = 0.230; Céspedes-Del Pozo et al., 2018). The He of cacao in north Peru was lower than that reported in Bolivia (He = 0.56; Zhang et al., 2012), Cameroon (He = 0.50; Efombagn et al., 2008), of the Juanjui province of San Martin in north Peru (He = 0.741; Zhang et al., 2011) and that of Ecuador (He = 0.496; Loor Solorzano et al., 2009). The moderate He observed in this study is probably reflective of the lack of imported varieties to give rise to differential hybrid material. The higher He reported above may also have been due in part to the use of microsatellites in those studies.
Estimates of Hind revealed that the majority of the collected samples were heterozygous with few highly homozygous trees. In contrast, Lerceteau et al. (1997) reported a high level of homozygous trees in two old plantations (80–100 years) in Ecuador. This indicated a low incidence of inbreed individuals in the current study and the presence of good cross-compatibilities among a greater number of founder individuals in the current study. Highly heterozygous samples should be assessed for vigour and productivity. The eleven samples with low heterozygosity should be assessed for self-compatibility toward obtaining pure lines for breeding purposes. Differential phenotypes selected from these two groups may be useful to find QTL (quantitative trait locus) for tree breeding purposes. Although the samples had mainly heterozygous individuals, the Shannon Index of diversity was similar to that reported in Dominica (Gopaulchan et al., 2020) and Uganda (Gopaulchan et al., 2019) but lower than in Honduras and Nicaragua (Ji et al., 2013), and in Indonesia (Lukman et al., 2014). The samples from north Peru were therefore lower in diversity and probably reflects the lower occurrence of introduced germplasm from other countries.
The PCoA revealed an underlying pattern of mixed types between Amelonado and Nacional groups that was supported by the phylogenetic, group differentiation and ancestry analyses. Three distinct phylogenetic clusters were present in the collected germplasm and supported by Dest statistics. AMOVA, FST, and Dest analyses supported the distinction of the Peru Uniques from the reference groups and the provinces of Bagua, Condorcanqui and Utcubamba from the reference groups. However, the provinces of Bagua and Utcubamba were similar to each other. This differed from Oliva-Cruz (2020) who found that these two provinces differed in ecotype composition. Yet, the proportion of Nacional trees from the current study was similar between these two provinces. These results and the identity analyses suggest that the sampled germplasm in north Peru contained unique multilocus profiles with possible inter-provincial differentiation and greater similarity between the Bagua and Utcubamba provinces. The province of Condorcanqui is recommended for further collection to verify its difference. Likewise, the province of Bagua was represented by 16 samples and increasing the sample size would allow for better resolution of inter-provincial differentiation. Bulking of cacao samples for fermentation or marketing purposes could be undertaken for the provinces of Bagua and Utcubamba. A similar recommendation for bulking across regions was obtained for Dominica (Gopaulchan et al., 2020). Further refinement could be achieved by propagating and maintaining the three phylogenetic clusters as distinct units provided that their sensory profiles are different. Additional collection and SNP genotyping to ascertain the frequency and distribution of these three clades in north Peru should be undertaken.
The clade PhyloC with 64 members was a good candidate for a new genetic group based on the phylogram (Figure 3). Initial population modelling in STRUCTURE (Pritchard et al., 2000) and assessed with the method of Evanno et al. (2005) fitted 11 groups. However, this was at the expense of splitting an accepted genetic cluster into two distinct groups instead of identifying PhyloC as a separate group. Furthermore, the two mixed reference groups (Amelonado/Nacional and Amelonado/Criollo) were also differentiated from Dest estimates from all other groups indicative that samples just need to be in groups rather than true populations to have differing estimators of genetic differentiation. None of the three test clades had any private alleles that could have supported the presence of a different genetic cluster. The results were therefore interpreted as PhyloA, PhyloB, and PhyloC being better fitted as clades of germplasm with hybrid ancestry from the genetic grouping of Motamayor et al. (2008). Hence, the collected samples from north Peru were mainly unique admixed cacao trees but did not comprise a novel genetic cluster and did not contain a subset that could be a novel group.
New populations in cacao were reported for Bolivia (Zhang et al., 2012), Colombia (Osorio-Guarín et al., 2017), and Peru (Céspedes-Del Pozo et al., 2018). However, these reports of new populations may be tentative due to limitations in each study. The Beni population in Bolivia was shown to be distinct from the Ucayali population from FST values (Zhang et al., 2012). However, an ancestry plot as well as a phylogenetic tree were not provided and the possibility of a sister clade to the Ucayali population cannot be ruled out. The Ucayali population contains members of the SCA accessions (Zhang et al., 2011) which belong to the Contamana cluster (Motamayor et al., 2008). Hence, the Beni population could be like clade PhyloB which was significantly different by Dest statistic from Contamana but was not a unique group. Furthermore, the PCoA study of Zhang et al. (2012) was limited by the few representative members of the accepted 10 genetic clusters and was probably lacking Purús members which may have resulted in an artificial separation of the Beni germplasm from the reference accessions.
Osorio-Guarín et al. (2017) indicated that two new cacao groups in Colombia were present based primarily on their ancestry result. However, examination of their graph revealed that the Iquitos and Nanay genetic clusters were not resolved from each other and the Curaray population was composed of three groups including Contamana and Amelonado. This suggests that the new groups were at the expense of established populations and further modelling is required to firmly establish whether these are new genetic clusters, subgroups of existing populations or sister clades of related germplasm. Céspedes-Del Pozo et al. (2018) reported that the Chuncho cacao from the La Convención province in Cusco, Peru was a distinct genetic cluster even from Contamana. These authors found very close genetic distances (0.06–0.07) of Chuncho to the Beni, Madre de Dios and Ucayali groups similar to the close Dest values for the Peru Uniques and the clades PhyloA, Phylob and PhyloC to the Nacional and Iquitos genetic groups in the current study. Furthermore, the PCoA plot of Céspedes-Del Pozo et al. (2018) apparently did not employ members from six known genetic groups including Nacional and Curaray thereby compromising the suggested distinct clustering of Chuncho cacao. In addition, some members of the Ucayali/Urubamba which are likely members of the Contamana cluster were dispersed among the Chuncho samples. Examination of the ancestry graphs of Céspedes-Del Pozo et al. (2018) indicated that two genetic groups, likely Iquitos and Nanay, were unresolved as in Osorio-Guarín et al. (2017). The allocation of Chuncho to a new group may therefore need further validation.
The fit to the 10 genetic groups of Motamayor et al. (2008) agreed with the close Dest values to Iquitos, Nacional, Amelonado/Nacional and Amelonado/Criollo groups. As a unit group, the Peru Uniques was closest to the set of Amelonado/Nacional mixed references with the Bagua, Condorcanqui and Utcubamba provinces being closest to Nacional and Amelonado/Nacional groups. A similar result was returned for test clades PhyloB and PhyloC whereas PhyloA was closest to the Iquitos group. Actual ancestry estimates supported the predominance of Nacional ancestry variably mixed mainly with Amelonado and Iquitos with additional contributions from Contamana, Criollo and Nanay groups. Only two Amelonado/Criollo admixed samples were found in north Peru, moderate Criollo (≥30%) ancestry was found in only six samples and only 16 samples lacked Nacional ancestry. This suggests that the fine aroma of cacao in north Peru is likely due to the Nacional background. However, the Condorcanqui province had a lower occurrence of Nacional members and it may be worthwhile to rejuvenate or infill farms with accessions having high Nacional ancestry already present in this region to ensure that the fine aroma status is maintained. The 16 samples lacking Nacional ancestry should be revisited and assessed for disease, productivity and flavour traits. If they prove to have superior traits including valuable or marketable flavour attributes, these samples can be cloned and maintained for breeding purposes. However, if acceptable trait combinations are lacking, these trees should not be clonally propagated or used to obtain open-pollinated seeds for distribution to farmers. This will help to maintain the fine aroma designation. Similarly, the set of 49 samples that contained both Criollo and Nacional ancestry should be examined for their flavour profile. If a distinctive flavour profile is found, this group can be clonally propagated and distributed to farmers. Self- and cross-compatibilities should be ascertained prior to distribution to identify the best possible mix to achieve fruit set on farms.
The genetic diversity of the fine aroma cacao in north Peru could be adequately represented by a set of 53 samples. The 143 sampled trees of this study have been clonally propagated as rooted cuttings and maintained as three different germplasm collections in an altitudinal gradient in the Utcubamba province (420 masl, 779620.6S, 9363856.4W; 480 masl, 792305.8S, 9364081.9W; 950 masl, 801491.0S, 9364914.0W). This collection will be expanded as additional genotyping is obtained on germplasm from future field collections. Furthermore, phenotyping of the three germplasm collections would provide information on phenotypic diversity that can be used to complement the genetic diversity of the set of 53 accessions and hence obtain a best core collection and a working collection. The core collection should be safeguarded by having an internal safety duplication where each accession is represented by at least five clonal copies and by having replicates of the core collection at different sites within the Peruvian Amazonas region. This would facilitate access to budwood for propagation to resupply farms with best local material to maintain the fine aroma status of cacao in north Peru.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
DB, MC, and MO conceived the idea and acquired the funding for the research and collecting expedition. LM and AM curated the data and selected the reference accessions. LM conducted the data analysis. DB, MC, AM, and LM contributed to the first draft of the manuscript. All authors reviewed, edited, and approved the final version of the manuscript.
This study was supported by the Fondo Nacional de Desarrollo Científico, Tecnológico y de Innovación Tecnológica (FONDECYT) funded by the Project through the Contract N 026-2016 “Círculo de Investigación para la Innovación y el fortalecimiento de la cadena de valor del cacao nativo fino de aroma en la zona nor oriental del Perú (CINCACAO)”. This study was also partially funded by the Project through the Contract 030-2018-FONDECYT-BM-IADT-MU and 142-2018-FONDECYT-BM-IADT-MU.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are most grateful to Jani Mendoza, Rosmery Robles, Jhordy Perez, and Daniel Tineo for their technical and logistical assistance. We also thank to Clelia Jima Chamiquit and Stefhany Valdeiglesias Ichillumpa for their translations to Awajun and Quechua languages, respectively.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.895056/full#supplementary-material
Alverson, W. S., Whitlock, W. A., Nyffeler, R., Bayer, C., and Baum, D. (1999). Phylogeny of the core Malvales: evidence from ndhF sequence data. Am. J. Bot. 86, 1474–1486. doi: 10.2307/2656928
Arevalo-Gardini, E., Meinhardt, L. W., Zuñiga, L. C., Arévalo-Gardni, J., Motilal, L., and Zhang, D. (2019). Genetic identity and origin of “Piura Porcelana”—a fine-flavored traditional variety of cacao (Theobroma cacao) from the Peruvian Amazon. Tree Genet. Genomes 15:11. doi: 10.1007/s11295-019-1316-y
Bartley, B. G. D. (2001). Refractario—an explanation of the meaning of the term and its relationship to the introductions from Ecuador in 1937. Ingenic Newslett. 6, 10–15.
Bartley, B. G. D. (2005). The Genetic Diversity of Cacao and Its Utilization. Cambridge MA: CABI Publishing. doi: 10.1079/9780851996196.0000
Bayer, C., Fay, M. F., de Bruijn, A. Y., Savolainen, V., Morton, C. M., Kubitzki, K., et al. (1999). Support for an expanded family concept of Malvaceae within a recircumscribed order Malvales: a combined analysis of plastid atpB and rbcL DNA sequences. Bot. J. Linn. 129, 267–303. doi: 10.1111/j.1095-8339.1999.tb00505.x
Beg, M. S., Ahmad, S., Jan, K., and Bashir, K. (2017). Status, supply chain and processing of cocoa–A review. Trends Food Sci. Technol. 66, 108–116. doi: 10.1016/j.tifs.2017.06.007
Boza, E. J., Irish, B. M., Meerow, A. W., Tondo, C. L., Rodríguez, O. A., Ventura-López, M., et al. (2013). Genetic diversity, conservation, and utilization of Theobroma cacao L.: genetic resources in the Dominican Republic. Genet. Resour. Crop Evol. 60, 605–619. doi: 10.1007/s10722-012-9860-4
Boza, E. J., Motamayor, J. C., Amores, F. M., Cedeño-Amador, S., Tondo, C. L., Livingstone, D. S., et al. (2014). Genetic characterization of the cacao cultivar CCN 51: its impact and significance on global cacao improvement and production. J. Amer. Soc. Hort. Sci. 139, 219–229. doi: 10.21273/JASHS.139.2.219
Céspedes-Del Pozo, W. H., Blas-Sevillano, R., and Zhang, D., and University students (2018). “Assessing genetic diversity of cacao (Theobroma cacao L.) nativo Chuncho in La Convención, Cusco-Perú,” in Proceedings of the International Symposium on Cocoa Research (ISCR), Lima.
Chia-Wong, J. A., Márquez-Dávila, K. J., Cárdenas-Salazar, H., Hurtado-Gonzales, O. P., Huaman-Camacho, T., Céspedes-Del-Poso, W., et al. (2018). “Avances en el estudio de las bases genéticas y organolépticas del cacao fino o de aroma en el Perú,” in Proceedings of the International Symposium on Cocoa Research (ISCR), Lima.
Cosme, S., Cuevas, H. E., Zhang, D., Oleksyk, T. K., and Irish, B. M. (2016). Genetic diversity of naturalized cacao (Theobroma cacao L.) in Puerto Rico. Tree Genet. Genomes 12:88. doi: 10.1007/s11295-016-1045-4
Dinarti, D., Susilo, A. W., Meinhardt, L. W., Ji, K., Motilal, L. A., Mischke, S., et al. (2015). Genetic diversity and parentage in farmer selections of cacao from Southern Sulawesi, Indonesia revealed by microsatellite markers. Breed. Sci. 65, 438–446. doi: 10.1270/jsbbs.65.438
Efombagn, I. B. M., Motamayor, J. C., Sounigo, O., Eskes, A. B., Nyassé, S., Cilas, C., et al. (2008). Genetic diversity and structure of farm and GenBank accessions of cacao (Theobroma cacao L.) in Cameroon revealed by microsatellite markers. Tree Genet. Genomes 4, 821–831. doi: 10.1007/s11295-008-0155-z
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Everaert, H., De Wever, J., Tang, T. K. H., Vu, T. L. A., Maebe, K., Rottiers, H., et al. (2020). Genetic classification of Vietnamese cacao cultivars assessed by SNP and SSR markers. Tree Genet. Genomes 16:43. doi: 10.1007/s11295-020-01439-x
Everaert, H., Rottiers, H., Pham, P. H. D., Ha, L. T. V., Nguyen, T. P. D., Tran, P. D., et al. (2017). Molecular characterization of Vietnamese cocoa genotypes (Theobroma cacao L.) using microsatellite markers. Tree Genet. Genomes 13:99. doi: 10.1007/s11295-017-1180-6
Evett, I. W., and Weir, B. S. (1998). Interpreting DNA Evidence: Statistical Genetics for Forensic Scientists. Sunderland, MA: Sinauer Associates.
Fang, W., Meinhardt, L. W., Mischke, S., Bellato, C. M., Motilal, L., and Zhang, D. (2014). Accurate determination of genetic identity for a single cacao bean, using molecular markers with a nanofluidic system, ensures cocoa authentication. J. Agric. Food Chem. 62, 481–487. doi: 10.1021/jf404402v
FAO (2022). FAOSTAT Crops and Livestock Products. License: CC BY-NC-SA 3.0 IGO. Available online at: https://www.fao.org/faostat/en/#data/QCL (accessed May 28, 2022).
Goenaga, R., Irizarry, H., and Irish, B. (2009). TARS Series of Cacao Germplasm Selections. HortScience 44, 826–827. doi: 10.21273/HORTSCI.44.3.826
Gopaulchan, D., Motilal, L. A., Bekele, F. L., Clause, S., Ariko, J. O., Ejang, H. P., et al. (2019). Morphological and genetic diversity of cacao (Theobroma cacao L.) in Uganda. Physiol. Mol. Biol. Plants. 25, 361–375. doi: 10.1007/s12298-018-0632-2
Gopaulchan, D., Motilal, L. A., Kalloo, R. K., Mahabir, A., Moses, M., Joseph, F., et al. (2020). Genetic diversity and ancestry of cacao (Theobroma cacao L.) in Dominica revealed by single nucleotide polymorphism markers. Genome 63, 583–595. doi: 10.1139/gen-2019-0214
Instituto Nacional de Defensa de la Competencia y de la Protección de la Propiedad Intelectual [INDECOPI] (2016). Denominación de Origen Cacao Amazonas Perú. Lima: INDECOPI.
Instituto Nacional de Estadística e Informática [INEI] (2012). Características de la Unidad Agropecuaria in IV Censos Nacional Agropecuario. Lima: INEI.
Ji, K., Zhang, D., Motilal, L. A., Boccara, M., Lachenaud, P., and Meinhardt, L. W. (2013). Genetic diversity and parentage in farmer varieties of cacao (Theobroma cacao L.) from Honduras and Nicaragua as revealed by single nucleotide polymorphism (SNP) marker. Genet. Resour. Crop. Evol. 60, 441–453. doi: 10.1007/s10722-012-9847-1
Johnson, E. S., Bekele, F., Brown, S., Song, Q., Zhang, D., Meinhardt, L. W., et al. (2009). Population structure and genetic diversity of the Trinitario cacao (Theobroma cacao L.) from Trinidad and Tobago. Crop Sci. 49, 564–572. doi: 10.2135/cropsci2008.03.0128
Jost, L. (2008). GST and its relatives do not measure differentiation. Mol. Ecol. 17, 4015–4026. doi: 10.1111/j.1365-294X.2008.03887.x
Jost, L. (2009). D vs, GST: response to Heller and Siegismund (2009) and Ryman and Leimar (2009). Mol. Ecol. 18, 2088–2091. doi: 10.1111/j.1365-294X.2009.04186.x
Kalinowski, S. T., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x
Laliberté, B., End, M., Cryer, N., Daymond, A., Engels, J., Eskes, A. B., et al. (2018). “Conserving and exploiting cocoa genetic resources: The key challenges,” in Achieving Sustainable Cultivation of Oil Palm, ed. D. Burleigh (Cambridge: Science Publishing), 19–46.
Lerceteau, E., Quiroz, J., Soria, J., Flipo, S., Pe’tiard, V., and Crouzilat, D. (1997). Genetic differentiation among Ecuadorian Theobroma cacao L. accessions using DNA and morphological analyses. Euphytica 95, 77–87. doi: 10.1023/A:1002993415875
Lindo, A. A., Robinson, D. E., Tennant, P. F., Meinhardt, L. W., and Zhang, D. (2018). Molecular characterization of cacao (Theobroma cacao) germplasm from Jamaica using single nucleotide polymorphism (SNP) markers. Trop. Plant Biol. 11, 93–106. doi: 10.1007/s12042-018-9203-5
Livingstone, D. S., Freeman, B., Motamayor, J. C., Schnell, R. J., Royaert, S., Takrama, J., et al. (2012). Optimization of a SNP assay for genotyping Theobroma cacao under field conditions. Mol. Breeding 30, 33–52. doi: 10.1007/s11032-11011-19596-11034
Loor Solorzano, R. G., Risterucci, A. M., Courtois, B., Fouet, O., Jeanneau, M., Rosenquist, E., et al. (2009). Tracing the native ancestors of modern Theobroma cacao L. population in Ecuador. Tree Genet. Genomes 5, 421–433. doi: 10.1007/s11295-008-0196-3
Lukman Zhang, D., Susilo, A. W., Dinarti, D., Bailey, B., Mischke, S., et al. (2014). Genetic identity, ancestry and parentage in farmer selections of cacao from Aceh, Indonesia revealed by single nucleotide polymorphism (SNP) markers. Trop. Plant Biol. 7, 133–144. doi: 10.1007/s12042-014-9144-6
Mahabir, A., Motilal, L. A., Gopaulchan, D., Ramkissoon, S., Sankar, A., and Umaharan, P. (2020). Development of a core SNP panel for cacao (Theobroma cacao L.) identity analysis. Genome 63, 103–114. doi: 10.1139/gen-2019-0071
MedCalc Software bvba (2013). MedCalc Statistical Software version 12.7.7. Ostend: MedCalc Software bvba.
Mejía, A., Meza, G., Espichan, F., Mogrovejo, J., and Rojas, R. (2021). Chemical and sensory profiles of Peruvian native cocoas and chocolates from the Bagua and Quillabamba regions. Food Sci. Technol. 41, 576–582. doi: 10.1590/fst.08020
Motamayor, J. C., Lachenaud, P., da Silva e Mota, J. W., Loor, R., Kuhn, D. N., Brown, J. S., et al. (2008). Geographic and genetic population differentiation of the Amazonian chocolate tree (Theobroma cacao L). PLoS One 3:e3311. doi: 10.1371/journal.pone.0003311
Motamayor, J. C., and Lanaud, C. (2002). “Molecular analysis of the origin and domestication of Theobroma cacao L,” in Managing Plant Genetic Diversity, eds J. M. M. Engels, V. Ramanatha Rao, A. H. D. Brown, and M. T. Jackson (Oxon: CABI Publishing). doi: 10.1079/9780851995229.0077
Motilal, L. A., Sankar, A., Gopaulchan, D., and Umaharan, P. (2017). “Cocoa,” in Biotechnology of plantation crops, eds P. Chowdappa, A. Karun, M. K. Rajesh, and S. V. Ramesh (New Delhi: Daya Publishing House), 313–354.
Motilal, L. A., Zhang, D., Umaharan, P., Mischke, S., Boccara, M., and Pinney, S. (2009). Increasing accuracy and throughput in large-scale microsatellite fingerprinting of cacao field germplasm collections. Trop. Plant Biol. 2, 23–37. doi: 10.1007/s12042-008-9016-z
Motilal, L. A., Zhang, D., Umaharan, P., Mischke, S., Mooleedhar, V., and Meinhardt, L. W. (2010). The relic Criollo cacao in Belize - Genetic diversity and relationship with Trinitario and other cacao clones held in the International Cocoa Genebank. Trinidad. Plant Genet. Resour. 8, 106–115. doi: 10.1017/S1479262109990232
Nagai, C., Heinig, R., Olano, C. T., Motamayor, J. C., and Schnell, R. J. (2009). “Fingerprinting of cacao germplasm in Hawaii,” Cacao Report No. 1. Waipahu, HI: Hawaii Agriculture Research Center.
National Institute of Agricultural Biotechnology (2006). PowerCore (v. 1.0). A program applying the advanced M strategy using heuristic search for establishing core or allele mining sets. Ranchi: National Institute of Agricultural Biotechnology.
Nieves-Orduña, H. E., Müller, M., Krutovsky, K. V., and Gailing, O. (2021). Geographic patterns of genetic variation among cacao (Theobroma cacao L.) populations based on chloroplast markers. Diversity 13:249. doi: 10.3390/d13060249
Oddoye, E. O. K., Agyente-Badu, C. K., and Gyedu-Akoto, E. (2013). “Cocoa and its by-products: Identification and utilization,” in Chocolate in Health and Nutrition, eds R. R. Watson, V. R. Preedy, and S. Zibadi (Totowa, NJ: Humana Press), 23–37. doi: 10.1007/978-1-61779-803-0_3
Oliva, M., and Maicelo-Quintana, J. L. (2020). Identification and selection of ecotypes of fine native cocoa aroma from the north-eastern zone of Peru. Rev. Investig. Agroproducc. Sustent. 4, 31–39. doi: 10.25127/aps.20202.556
Oliva-Cruz, S. M. (2020). Caracterización socioeconómica de la diversidad biológica de cacao Criollo fino de aroma en comunidades rurales de la región Amazonas. Ph. D. thesis. Chachapoyas-Perú: Universidad Nacional Toribio Rodríguez De Mendoza De Amazonas.
Olivera-Núñez, Q. (2018). Jaén, Arqueología y Turismo Yanapay Andina Consultores. Jaén: Municipalidad Provincial de Jaén.
Osorio-Guarín, J. A., Berdugo-Cely, J., Coronado, R. A., Zapata, Y. P., Quintero, C., Gallego-Sánchez, G., et al. (2017). Colombia a source of cacao genetic diversity as revealed by the population structure analysis of germplasm bank of Theobroma cacao L. Front. Plant Sci. 8:1994. doi: 10.3389/fpls.2017.01994
Padi, F. K., Ofori, A., Takrama, J., Djan, E., Opoku, S. Y., Dadzie, A. M., et al. (2015). The impact of SNP fingerprinting and parentage analysis on the effectiveness of variety recommendations in cacao. Tree Genet. Genomes 11:44. doi: 10.1007/s11295-015-0875-9
Peakall, R., and Smouse, P. E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1111/j.1471-8286
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Perrier, X., Flori, A., and Bonnot, F. (2003). “Data analysis methods,” in Genetic diversity of cultivated tropical plants, eds P. Hamon, M. Seguin, X. Perrier, and J. C. Glaszmann (Montpellier: Enfield Science Publishers), 43–76.
Perrier, X., and Jacquemoud-Collet, J. P. (2006). DARwin software. Available online at: http://darwin.cirad.fr/darwin (accessed July 11, 2014).
Phillips-Mora, W., Arciniegas-Leal, A., Mata-Quirós, A., and Motamayor-Arias, J. C. (2013). Catalogue of Cacao Clones: Selected by CATIE for Commercial Plantings. Turrialba: CATIE.
Pound, F. J. (1938). “Cacao and witches’ broom disease (Marasmius perniciosus) of South America,” in Archives Cacao Research, Vol. 1, ed. H. Toxopeus (Washington DC: American Cacao Research Institute and Brussels), 20–72.
Pound, F. J. (1943). “Cacao and witches’ broom disease (Marasmius perniciosa),” Report on a recent visit to the Amazon territory of Peru, September 1942–February 1943. Trinidad: Yuille’s Printery.
Pridmore, R., Crouzillat, D., Walker, C., Foley, S., Zink, R., Zwahlen, M. C., et al. (2000). Genomics, molecular genetics and the food industry. J. Biotechnol. 78, 251–258. doi: 10.1016/s0168-1656100000202-00209
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1111/j.1471-8286.2007.01758.x
Rice, R. A., and Greenberg, R. (2000). Cacao cultivation and the conservation of biological diversity. Ambio 29, 167–173. doi: 10.1579/0044-7447-29.3.167
Saavedra-Arbildo, R. P., Cárdenas-Salazar, H., Márquez-Dávila, K. J., Beraun-Cruz, Y., Carranza-Cruz, M. S., Hurtado-Gonzalez, O. P., et al. (2018). “Colecta y estudio de las características morfológicas y organolépticas en fruta fresca y licor de arboles de cacao (Theobroma cacao L.) con atributos de poseer caracteristicas de fino y de aroma,” in Proceedings of the International Symposium on Cocoa Research (ISCR), Lima.
Singh, B. D., and Singh, A. K. (2015). “Mapping populations,” in Marker-Assisted Plant Breeding: Principles and Practices, eds B. D. Singh and A. K. Singh (New Delhi: Springer), doi: 10.1007/978-81-322-2316-0_5
Tacer-Caba, Z. (2019). “The concept of superfoods in diet,” in The Role of Alternative and Innovative Food Ingredients and Products in Consumer Wellness, ed. C. M. Galanakis (Amsterdam: Academic Press), doi: 10.1016/B978-0-12-816453-2.00003-6
Thiers, B. (2016). Index Herbariorum. A global directory of public herbaria and associated staff. Bronx, NY: New York Botanical Garden’s Virtual Herbarium.
Thomas, E., van Zonneveld, M., Loo, J., Hodgkin, T., Galluzzi, G., and van Etten, J. (2012). Present spatial diversity patterns of Theobroma cacao L. in the Neotropics reflect genetic differentiation in Pleistocene refugia followed by human-influenced dispersal. PLoS One 7:e47676. doi: 10.1371/journal.pone.0047676
Torres-Armas, E. A., and Gonzáles-Castro, J. B. (2018). Caracterización de productores en la cadena de valor del cacao fino de aroma de Amazonas. Conocimiento para Desarrollo 9, 113–120.
Toxopeus, H. (1985). “Botany, types and populations,” in Cocoa, 4th Edn, eds G. A. R. Wood and R. A. Lass (London: Longman), 11–37.
Trognitz, B., Scheldeman, X., Hansel-Hohl, K., Kuant, A., Grebe, H., and Hermann, M. (2011). Genetic population structure of cacao plantings within a young production area in Nicaragua. PLoS One 6:e16056. doi: 10.1371/journal.pone.0016056
Valdez, F. (2013). “Prefacio,” in Arqueologia Amazonica: las civilizaciones ocultas del bosque tropical, ed. F. Valdez (Quito: IRD Editions).
Waits, L. P., Luikart, G., and Taberlet, P. (2001). Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol. Ecol. 10, 249–256. doi: 10.1046/j.1365-294X.2001.01185.x
Wang, B., Motilal, L. A., Meinhardt, L. W., Yin, J., and Zhang, D. (2020). Molecular characterization of a cacao germplasm collection maintained in Yunnan, China using single nucleotide polymorphism (SNP) markers. Trop. Plant Biol. 13, 359–370. doi: 10.1007/s12042-020-09267-y
Wickramasuriya, A., and Dunwell, J. M. (2018). Cacao biotechnology: current status and future prospects. Plant Biotechnol. J. 16, 4–17. doi: 10.1111/pbi.12848
Xu, W. (2016). Functional Nucleic Acids Detection in Food Safety: Theories and Applications. Singapore: Springer, doi: 10.1007/978-981-10-1618-9
Yacenia Morillo, C., Morillo, A. C., Muñoz, F. J. E., Ballesteros, P. W., and González, A. (2014). Caracterización molecular con microsatélites amplificados al azar (RAMs) de 93 genotipos de cacao (Theobroma cacao L.). Agronomia Colombiana 32, 315–325. doi: 10.15446/agron.colomb.v32n3.46879
Zarrillo, S., Gaikwad, N., Lanaud, C., Powis, T., Viot, C., Lesur, I., et al. (2018). The use and domestication of Theobroma cacao during the mid-Holocene in the upper Amazon. Nat. Ecol. Evol. 2, 1879–1888. doi: 10.1038/s41559-018-0697-x
Zhang, D., Arevalo-Gardini, E., Mischke, S., Zuñiga-Cernandes, L., Barreto-Chavez, A., and Adriazola, et al. (2006a). Genetic diversity and structure of managed and semi-natural populations of cacao (Theobroma cacao) in the Huallaga and Ucayali valleys of Peru. Ann. Bot. 98, 647–655. doi: 10.1093/aob/mcl146
Zhang, D., Mischke, S., Goenaga, R., Hemeida, A. A., and Saunders, J. A. (2006b). Accuracy and reliability of high-throughput microsatellite genotyping for cacao clone identification. Crop Sci. 46, 2084–2092. doi: 10.2135/cropsci2006.01.0004
Zhang, D., Gardini, E. A., Motilal, L. A., Baligar, V., Bailey, B., Zuñiga-Cernades, L., et al. (2011). Dissecting genetic structure in farmer selections of Theobroma cacao in the Peruvian Amazon: implications for on farm conservation and rehabilitation. Trop. Plant Biol. 4, 106–116. doi: 10.1007/s12042-010-9064-z
Zhang, D., Martínez, W. J., Johnson, E. S., Somarriba, E., Phillips-Mora, W., Astorga, C., et al. (2012). Genetic diversity and spatial structure in a new distinct Theobroma cacao L. population in Bolivia. Genet. Resour. Crop Evol. 59, 239–252. doi: 10.1007/s10722-011-9680-y
Keywords: core collection, fine or flavour cocoa, genetic structure, group differentiation, Nacional ancestry, north Peru, phylogeny, Peruvian Amazonas region
Citation: Bustamante DE, Motilal LA, Calderon MS, Mahabir A and Oliva M (2022) Genetic diversity and population structure of fine aroma cacao (Theobroma cacao L.) from north Peru revealed by single nucleotide polymorphism (SNP) markers. Front. Ecol. Evol. 10:895056. doi: 10.3389/fevo.2022.895056
Received: 12 March 2022; Accepted: 29 June 2022;
Published: 15 July 2022.
Edited by:
Alison G. Nazareno, Federal University of Minas Gerais, BrazilReviewed by:
Alessandro Alves-Pereira, State University of Campinas, BrazilCopyright © 2022 Bustamante, Motilal, Calderon, Mahabir and Oliva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danilo E. Bustamante, ZGFuaWxvLmJ1c3RhbWFudGVAdW50cm0uZWR1LnBl
†ORCID: Danilo E. Bustamante, orcid.org/orcid.org/0000-0002-5979-6993; Lambert A. Motilal, orcid.org/orcid.org/0000-0001-7978-5717; Martha S. Calderon, orcid.org/orcid.org/0000-0003-3611-140X; Amrita Mahabir, orcid.org/orcid.org/0000-0003-3611-140X; Manuel Oliva, orcid.org/orcid.org/0000-0002-9670-0970
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.