- 1Hubei Engineering Research Center for Protection and Utilization of Special Biological Resources in the Hanjiang River Basin, School of Life Sciences, Jianghan University, Wuhan, China
- 2Hubei Key Laboratory of Environmental and Health Effects of Persistent Toxic Substances, Jianghan University, Wuhan, China
- 3Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Wuhan, China
Dam construction on the upper Yangtze River has dramatically altered riverine ecosystems and caused habitat fragmentation of fishes, which might influence the genetic structure of fish populations. In this study, we examined the possible genetic effects of dam construction on Chinese lizard gudgeon (Saurogobio dabryi) populations in the upper Yangtze River, China. Seven populations were sampled, and genetic structure was analyzed using single nucleotide polymorphism (SNP) markers through the specific locus amplified fragment sequencing (SLAF-seq) method. The numbers of SNPs were lower in the upstream populations than in the downstream populations. Genetic similarity was increased from downstream to upstream. The upstream populations of S. dabryi might be more vulnerable to genetic drift than those downstream. Structure analysis indicated three distinct genetic groups of S. dabryi in the upper Yangtze River, among which the genetic differentiation values (Fst) were at a high level. The genetic differentiation of S. dabryi exhibited a close correlation with spatial distance. We did not detect a significant correlation between isolation time and genetic differentiation, suggesting that impacts of dams on the genetic structure of S. dabryi can be relatively minimal on a short time scale. The results quantify the genetic diversity and population structure patterns of S. dabryi after habitat fragmentation caused by dams, which will provide a reference for resource protection and management of this species in the upper Yangtze River.
Introduction
Dam construction has damaged the continuity of natural rivers and dramatically altered riverine ecosystems, causing habitat fragmentation of fishes. This affects the connectivity and migration of fish species, leading to increased inbreeding probability, enhanced genetic drift, and blocked gene flow, thereby reducing genetic diversity and increasing genetic differentiation (Pavlova et al., 2017; Nakajima et al., 2020; Machado et al., 2021). Low genetic diversity might reduce the viability of individuals and populations in the fragmented habitats (Cook and Sullivan, 2018).
The upper Yangtze River is a hotspot of diverse fish species. However, it has been facing a serious issue of habitat fragmentation due to the construction of a cascade of hydropower dams, such as Three Gorges Dam, Xiangjiaba Dam, Xiluodu Dam, Baihetan Dam, and others (Cheng et al., 2015). Many fishes have been greatly affected in aspects, such as population size decline, spawning ground loss, and reduced reproductive success. If key habitats are lost or are no longer accessible, such destruction may have a serious negative impact on local fish species (Zhang et al., 2021), and it is particularly difficult to conduct fish conservation. Understanding the genetic effects of habitat fragmentation is a basis for conservation and management actions for fishes in the Yangtze River.
At present, studies on the genetic effects of habitat fragmentation caused by dams in the Yangtze River mostly focus on large and migratory fishes (Wang et al., 2007; Song et al., 2016; Liu et al., 2017, 2020; Dong et al., 2019), while research on small-sized species is very limited. Saurogobio dabryi is a small-sized fluvial fish species in the sub-family Gobioninae (Cyprinidae) and prefers to inhabit flowing waters. Its sexual maturation is early and the minimum age of sexual maturity is 1 year (Liu et al., 2019). This species produces drifting eggs, which need flowing water to be hatched (Lin et al., 2018). After dam construction, many river sections are changed from lotic water to close lentic water, which has greatly altered the habitats of S. dabryi and will potentially reduce its reproductive success (Xu et al., 2019). The spawning time of S. dabryi has become shortened and the reproductive rate has decreased (Gao, 2016). Li et al. (2016) reported the genetic structure of three populations from Chishui River, Hejiang and Yibin of the upper Yangtze River based on the mitochondrial Cyt b gene sequence, but the sample regions were near one another and not blocked by dams, thus, genetic effects caused by dam blockage were not involved. Liu (2020) studied the genetic differentiation of four populations in the Jialing River based on the mitochondrial D-loop sequence and 10 microsatellite loci, but the sample regions were limited to a tributary of the upper Yangtze River. The previous studies focused on populations of S. dabryi at relatively small spatial scales and included a small number of genetic markers, which yielded limited genetic information on this species.
Genome-scan approaches, such as specific locus amplified fragment sequencing (SLAF-seq), present large numbers of genome-wide single nucleotide polymorphisms (SNPs) and increase coverage throughout the genome using thousands of SNP markers, and thus can improve the capacity to unravel genetic diversity and genetic divergence within species (Zhang et al., 2020). In this study, we used SNP markers produced by the SLAF-seq method to assess the genetic diversity and genetic differentiation of seven S. dabryi populations in the upper Yangtze River. The aim was to reveal the genetic diversity and differentiation patterns of S. dabryi in fragmented habitats of the upper Yangtze River and to probe possible genetic effects of isolation by dams. The results are expected to provide a reference for natural resource protection and management of this species in the upper Yangtze River.
Materials and Methods
Study Areas and Sample Collection
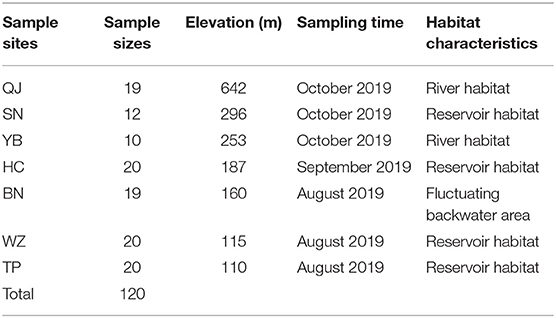
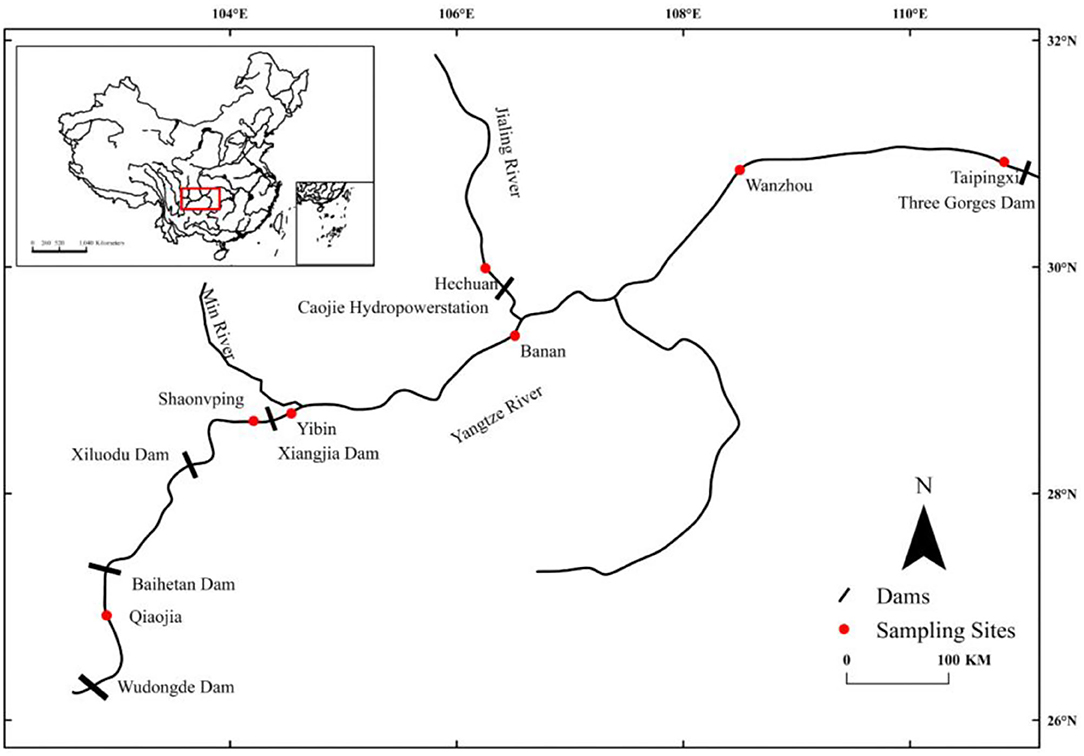
A length of the river of about 1,500 km distance from the Wudongde Dam to the Three Gorges Dam on the upper Yangtze River was selected as the research area. This zone has been fragmented by hydropower dams, such as Wudongde Dam (closure in 2016), Baihetan Dam (2015), Xiluodu Dam (2007), Xiangjiaba Dam (2008), and Three Gorges Dam (2003) from upstream to downstream. These dams are large, impassable, and power-generating dams, which lack fishways (Cheng et al., 2015). Individuals of S. dabryi were sampled at seven sites from August to October 2019 (Table 1). These sites were Qiaojia (QJ), Shaonvping (SN), Yibin (YB), Hechuan (HC), Banan (BN), Wanzhou (WZ), and Taipingxi (TP) (Figure 1). Sampling site QJ is between Wudongde Dam and Baihetan Dam. Sampling site SN is between Xiluodu Dam and Xiangjia Dam. Sampling sites YB, BN, WZ, and TP are in the long river section between Xiangjia Dam and Three Gorges Dam. The length of this river section is about 1,000 km with changes from river habitat (400 km) to reservoir habitat (600 km), and it is a relatively intact habitat for fishes to complete their life histories, especially for fishes producing drifting eggs. Sampling site HC is in the lower reach of the Jialing River, a tributary of the upper Yangtze River. There is Caojie hydropower station (closure in 2005) between HC and the main stream. QJ and YB are river habitats. SN, HC, WZ, and TP are reservoir habitats. BN is in fluctuating backwater area of the Three Gorges Reservoir. When the impounding height of the Three Gorges Reservoir is 145 m from June to September, the water of BN flows. When the impounding height is 175 m from October to May of the next year, the water of BN closes to lentic (Table 1).
Individuals were captured using trammel or gill nets. Fin clips were taken and preserved in 95% ethanol until DNA extraction.
Preparation of SLAF Sample for Sequencing, Data Processing, and SNP Calling
Twenty individuals per population were randomly selected for DNA extraction when the sample size was >20. If the population sample size was <20, all samples were taken for screening.
Total genomic DNA was extracted from fin tissues using a phenol-chloroform protocol (Green and Sambrook, 2012), DNA quality was checked by a NanoDrop One Spectrophotometer (Thermo Scientific) and the samples were used for library construction. A total of 120 samples of S. dabryi were used to generate genome-wide SNP data using the SLAF sequencing methodology (Sun et al., 2013), a new sequencing technology much like RAD-seq, which targets a reduced representation of the genome. As there was no reference genome sequence for S. dabryi, the genome of a related species, Cyprinus carpio (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/951/615/GCF_000951615.1_common_carp_genome) (Xu et al., 2014) was selected to perform in silico digestion prediction. The restriction enzymes RsaI + HaeIII were used for digestion. SLAF tag length was selected between 314 and 364 bp. Based on the prediction, we then proceeded to library construction. Genomic DNA (about 500 ng) was digested to completion with RsaI (New England Biolabs, NEB), T4 DNA ligase (NEB), adenosine triphosphate (ATP), and RsaI adapters at 37°C. Restriction-ligation reactions were heat-inactivated at 65°C and then digested with the second restriction enzyme HaeIII (NEB) at 37°C. Duplex tag-labeled sequencing adapters were ligated to the A-tailed DNA with T4 DNA ligase. PCR reactions were performed using diluted restriction-ligation samples, deoxyribonucleoside triphosphates (dNTPs), high-fidelity DNA polymerase (NEB), and PCR primers. PCR products were purified and then separated by electrophoresis in a 2% agarose gel. Fragments 300–450 bp (with indexes and adaptors) in size were excised and purified using QIAquick Gel Extraction Kit (Qiagen). The gel-purified product was sequenced on the Illumina HiSeq 2500 system.
The highest depth tag in each SLAF was taken as the reference sequence tag. SLAF paired-end reads with clear index information were clustered and detected using BLAT (Kent, 2002). Sequences with over 80% identity were considered as one SLAF locus. Both GATK (McKenna et al., 2010) and Samtools (Li et al., 2009) were used to call SNPs. VCFtools (Danecek et al., 2011) was used to filter SNPs, which met all four criteria: QUALITY (QUAL) value ≥30, minor allele frequency (MAF) ≥5%, significance level of Hardy-Weinberg equilibrium (HWE) test ≥0.01, and no more than four individuals with missing genotypes at each locus among all individuals.
Genetic Analysis
VCFtools software (Danecek et al., 2011) was used to calculate the nucleotide diversity index (π), Tajima's and Fu's FS D value within populations, and the genetic differentiation coefficient (Fst) between pairs of populations. Perl scripts (Goodswen et al., 2010) were used to calculate the expected heterozygosity (He) and the observed heterozygosity (Ho). The population structure of S. dabryi was analyzed by Admixture software (Alexander et al., 2009), and groups with K values ranging from 1 to 10 were clustered, separately. The clustering results were cross-validated and the optimal number of clusters was determined by the method of minimized cross-validated error rate. A maximum likelihood (ML) tree based on the General Time Reversible model (GTR)+proportion of invariable sites (I)+gamma distributed site-to-site variation (G) model by the Bayesian inference method was constructed using RAxMLv8.1.17 software (Stamatakis, 2014) with 100,000 replicates. A principal coordinate analysis (PCA) plot was generated using Eigen software (Price et al., 2006) to visualize the population differentiation. Identity-by-state (IBS) was calculated by Plink software (Purcell et al., 2007) to assess the genetic similarity of groups.
Genetic Effect of Environmental Factors
Spatial distance between sites (measured along river channels), drainage area, and river length were measured on large-scale catchment topographic maps in ArcGIS10.5. The channel slope was the drop per unit reach length, which was calculated according to reach an elevation of 5 km upstream and 5 km downstream of the sample sites. The isolation time was calculated from the dates of dam closure. The elevation at each sample site was recorded by the Global Positioning System (GPS) (accuracy of 1 m; Table 1). The correlation between the parameters of genetic diversity (SNP numbers, π, and Ho) and the environmental factors (drainage area, river length, channel slope, and elevation) were performed using the cor.test function in the R package. The influence of environmental variables (such as spatial distance, isolation time, and channel slope) on population genetic differentiation was calculated by the Mantel test using the Vegan procedure of the R package (Dixon, 2003). The value of genetic differentiation used linearized Fst values [Fst/(1 – Fst)]. Significance was tested using 1,000 permutations.
Results
Statistics and Evaluation of Sequencing Data
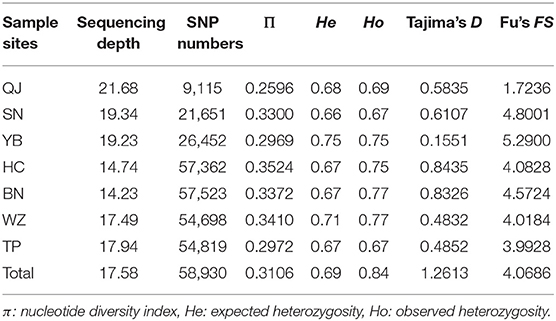
A total of 237.86 Mb of clean reads and 59.87 Gb of clean data were generated for 120 S. dabryi by SLAF sequencing. The average Q30 quality score was 94.98% and the average guanine-cytosine (GC) content was 42.52%. After low-quality reads, adaptors, and barcode sequences were removed, a total of 989,072 SLAF tags were obtained in which 237,400 tags were polymorphic. Population sequencing depth was from 14.23 × to 21.68 ×, with an average sequencing depth of 17.58 ×. A total of 1,300,189 SNPs were obtained, among which there were 58,930 SNPs that passed the SNP filtration criteria (Table 2).
Genetic Diversity
The number of SNPs within the population ranged from 9,115 (QJ population) to 57,523 (BN population), exhibiting great variation among populations in the upper Yangtze River. The number of SNPs in the QJ population was significantly less than in the SN and YB populations (t-test, p < 0.01), The number of SNPs in the SN and YB populations was significantly less than in the HC, BN, WZ, and TP populations (t-test, p < 0.01), showing that the numbers of SNPs were lower in the upstream populations, which suggested that more alleles might have been lost by random genetic drift. There were no significant differences for other metrics of genetic variation (π, He, and Ho) among the seven populations (t-test, p > 0.05). All seven populations had positive values of Tajima's D and Fu's FS, indicating that S. dabryi in the upper Yangtze River had experienced a recent bottleneck (Table 2).
Population Structure
K = 3 had the smallest cross-validation error (Figure 2a), thus supporting three genetic groups. Individuals in QJ belonged to group 1 (in gray), individuals in SN and YB belonged to group 2 (in orange), and individuals in WZ and TP belonged to group 3 (in blue). Groups 2 and 3 both existed within BN and HC populations and exhibited almost equal average probabilities of membership (Figure 2b). This result was also indicated by the PCA, wherein the seven populations formed three distinct groups (QJ; SN\YB; HC\BN\WZ\TP), but there was one individual of the HC population that was clustered into the SN\YB group (Figure 2c). The ML tree also supported this arrangement (Figure 2d). It was clear that the upstream and downstream populations belonged to different genetic groups.
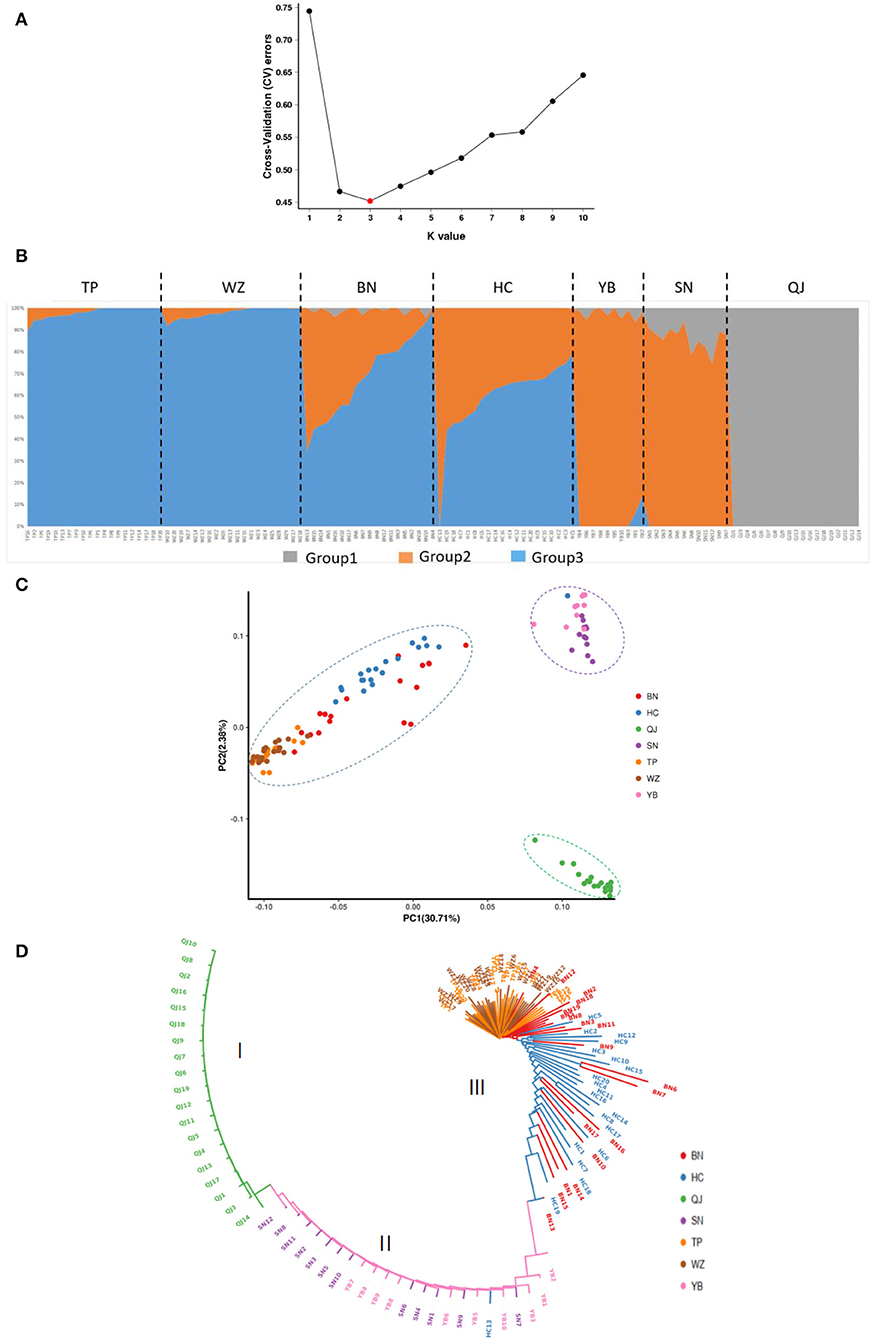
Figure 2. Genetic structure of seven Saurogobio dabryi (S. dabryi) populations based on SNP markers. (A) The optimal number of clusters was determined by minimized cross-validated error rate. (B) Population structure (K = 3) of all individuals. Different colors represent the probability of an individual belonging to different genetic groups. (C) PCA plots of every individual. (D) ML phylogenetic tree. Cluster I included all QJ individuals, cluster II included all SN and YB individuals and one HC individual, and cluster III included all BN, WZ, and TP individuals and the rest of the HC individuals.
Identity-by-state analysis was conducted to assess similarity within three distinct groups. To avoid the interference, we eliminated the BN and HC populations that exchanged genes with both upstream and downstream neighbors. IBS value was the highest in group 1 (the QJ population), followed by group 2 (the SN and YB populations), and the lowest in group 3 (the WZ and TP populations), indicating that the degree of genetic similarity within groups was increased from downstream to upstream (Figure 3).
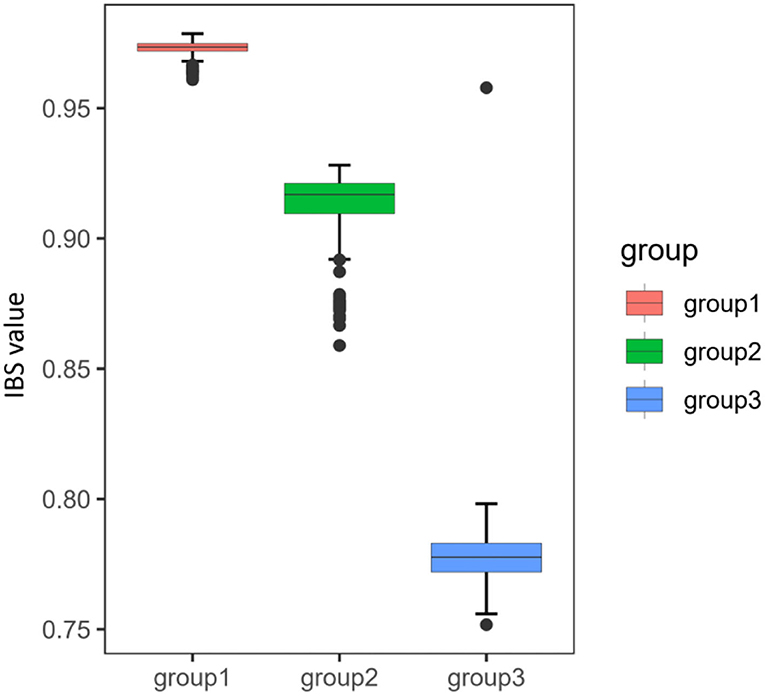
Figure 3. Genetic similarity within three distinct groups by identity-by-state (IBS). The upper and lower ranges of the boxplot, with the upper limit being the upper quartile and the lower limit being the lower quartile. The horizontal line inside the box represents the median. The whiskers were extended to the 95th percentile.
Population Genetic Differentiation
Fst values among the three clusters (QJ; SN\YB; HC\BN\WZ\TP) were high (Fst = 0.166–0.416), implying considerable genetic differentiation (Table 3). The QJ population most upstream had diverged highly from other populations. The Fst values between the SN and YB populations were very small (0.007), but they were highly differentiated from other populations. The YB, TP, WZ, and BN populations were in the same river section. However, there was high differentiation between the YB population in the upstream river habitat and the TP, WZ, and BN populations in the downstream reservoir habitat.
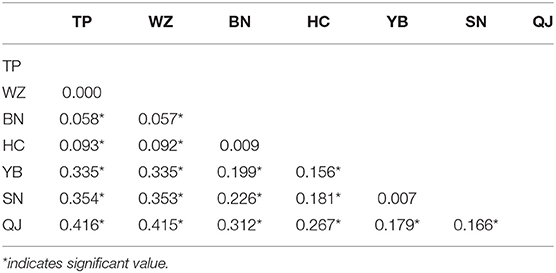
Table 3. Genetic differentiation (Fst) among seven populations of S. dabryi in the upper Yangtze River.
Genetic Effect of Environmental Factors
Correlation analysis indicated a significant negative relationship between the numbers of SNPs and the elevations of the seven populations (r = −0.867, p < 0.05). There were fewer SNPs within upstream populations in the high-elevation region than within downstream populations in the low-elevation region. The numbers of SNPs within populations had no significant correlation with drainage area, river length, and channel slope (Figure 4). In addition, the other genetic diversity metrics (π and Ho) also had no correlation with these environmental factors.
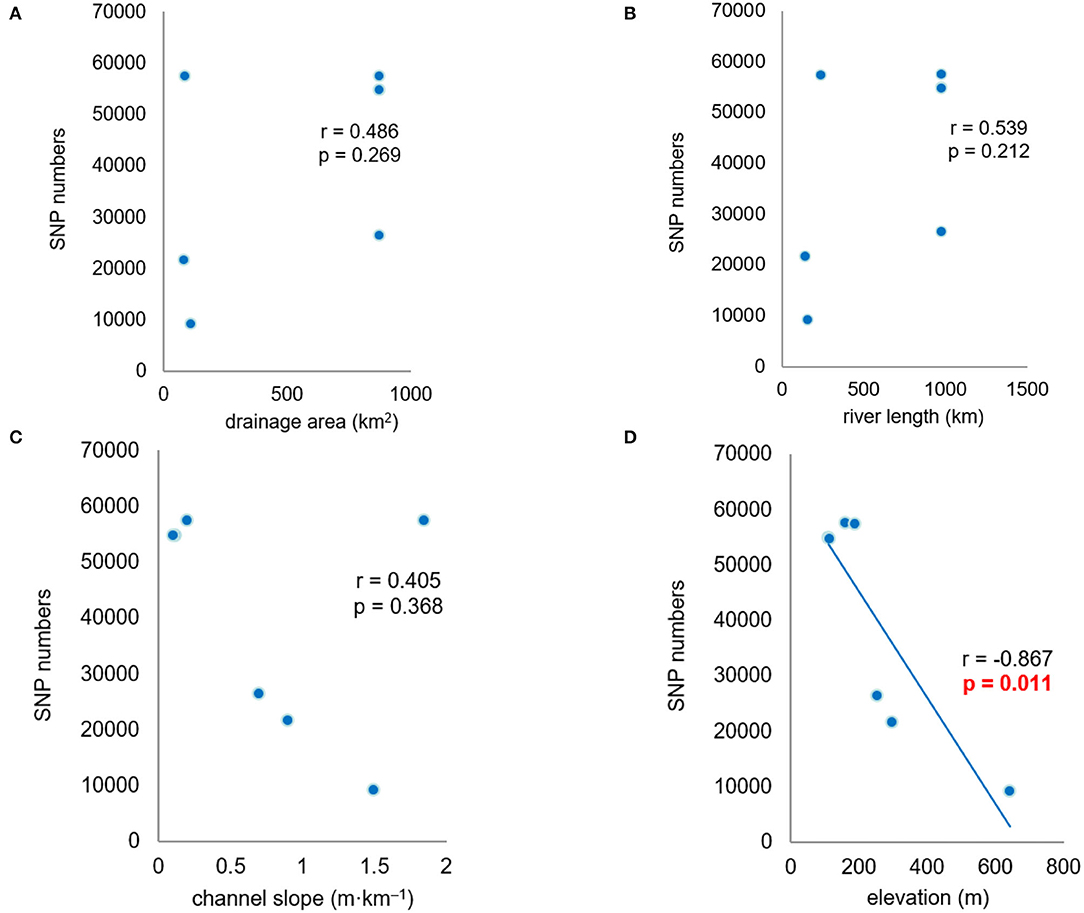
Figure 4. Correlation analysis between the single nucleotide polymorphism (SNP) numbers and environmental factors. (A–D) r is correlation coefficient, p value is used to determine the result of hypothesis testing.
Genetic differentiation exhibited a close correlation with spatial distance among the populations (r = 0.907, p < 0.01). No significant correlation was found between genetic differentiation and isolation times. There was no correlation between genetic differentiation and channel slope (Figure 5).
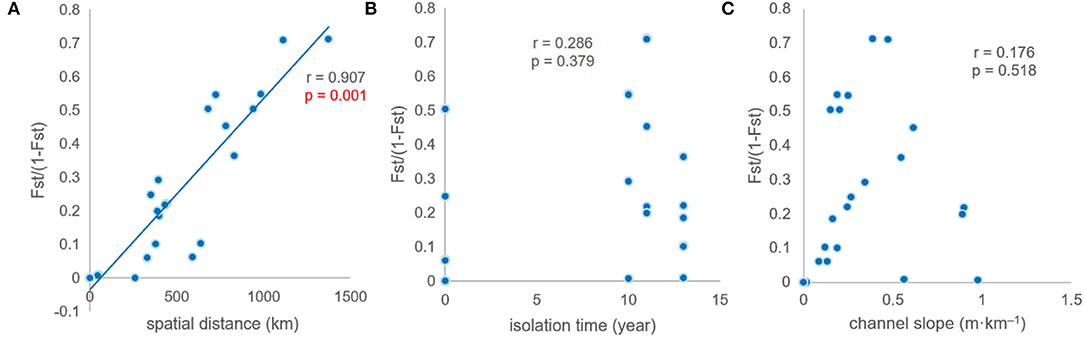
Figure 5. Mantel tests between genetic differentiation and environmental factors. (A–C) r is correlation coefficient, p value is used to determine the result of hypothesis testing.
Discussion
In this study, no significant differences in the values of π, He, and Ho were observed among these populations, but the number of SNPs was lower in the QJ, SN, and YB populations, suggesting that more alleles might have been lost in the upstream populations due to random genetic drift. Compared with other parameters of genetic diversity, variation in the number of SNPs was more apparent. Consistently, the genetic similarity was higher within the upstream group. The two genetic values reflected that genetic diversity was lower within the upstream populations than in the downstream populations. The percentage in the abundance of S. dabryi was about 4.1% of the total catch in the QJ section in 2006–2012 (Tang et al., 2014), which was lower than that in the YB section, 11.3% in 2007–2009 (Xiong et al., 2015), which indicated that S. dabryi was historically less abundant upstream than downstream. Historically smaller populations might account for the lower genetic diversity in the upstream populations. Some other cyprinid fishes in the upper Yangtze River showed a similar trend, lower genetic diversity upstream, such as Coreius heterodon (Yan et al., 2008; Cheng et al., 2013), Ancherythroculter nigrocauda (Zhai et al., 2019), and Megalobrama pellegrini (Wang et al., 2019). In this study, the upstream populations of S. dabryi with lower allelic richness and higher genetic similarity were more vulnerable to genetic drift than downstream populations. Upstream populations should be given more conservation attention to avoid inbreeding and gene pool degradation.
We have detected a recent bottleneck event according to the positive values of Tajima's D and Fu's FS. S. dabryi prefers to inhabit and spawn in flowing water, but dam construction has changed the river into lentic water habitats. It has been found that the spawning time has become shortened and the reproductive rate of S. dabryi has decreased (Gao, 2016). In addition, environmental features in the upper Yangtze River also have changed in response to dam construction (Cheng et al., 2015). Under such conditions, the dam-induced fragmentation might lead to the decline of the fishes (Blanton et al., 2019). The recent bottleneck events may possibly have resulted in the loss of allelic richness of S. dabryi.
Structure analyses indicated that the seven populations of S. dabryi represented three distinct groups. It was clear that upstream and downstream populations presented different genetic structures. The YB, WZ, and TP populations were in the same river section with no dam barrier, but the YB population was in the riverine habitat, and the WZ and TP were in reservoir habitat. The YB population was obviously differentiated from WZ and TP, suggesting that different local flow conditions can lead to genetic differentiation (Landguth et al., 2014).
Genetic differentiation of S. dabryi exhibited a close correlation with spatial distance. Significant genetic differentiation was shown between populations except for YB-SN, WZ-TP, and BN-HC. It was found that the populations of YB-SN and BN-HC without differentiations were distributed very closely regardless of isolation by dams. These populations might have been genetically differentiated before the dam construction. S. dabryi is a small-sized fish species with a finite capacity movement (Liu et al., 2019). It tends to move short distances more frequently than long distances and thus had gene flow over a limited geographic region. Such distance-attenuated movement might give rise to population structure consistent with an isolation-by-distance (IBD) evolutionary model (Roberts et al., 2013). Interestingly, the genetic differentiation between populations WZ and TP was not significant although the distance between them is relatively long. WZ and TP are both reservoir habitats and have similar ecological environments, which might bring the similar ecological adaptation. Therefore, such genetic structure of S. dabryi might be attributed to spatial distance and ecological adaptation.
At present, we did not detect a significant correlation between isolation time and genetic differentiation of S. dabryi. It is generally considered that genetic differentiation on a short time scale is difficult to detect. No genetic change was detected in rainbow trout (Oncorhynchus mykiss) populations 5–10 generations or in galaxiid (Galaxias platei) populations 20–30 generations after dam construction (Deiner et al., 2007; Vera-Escalona et al., 2015). However, dam-induced isolation on a long time scale has had a significant impact on other fish populations. Gouskov et al. (2016) found a dam, which was built over 100 years ago and had a strong effect on population structure of the carp (Squalius cephalus) in the Rhine River Basin. In our study, these dams are around 20 years old or even younger (Cao, 2019) about 20 or less generations for S. dabryi. The effect of isolation has not obviously emerged. Hence, the genetic impacts of dams can be relatively minimal on a short time scale. Environmental changes have occurred in the upper Yangtze River after the construction of these dams. With the increase in isolation time, these changes will possibly influence the genetic structure of S. dabryi, and thus genetic impacts of dams need long-term monitoring.
Conclusion
High-throughput DNA sequencing technology has provided a large number of molecular markers and powerful tools for the study of population genetics. In this study, we identified a large number of SNP loci in populations of S. dabryi in the upper Yangtze River. The results showed that the numbers of SNPs were lower within the upstream populations, and more alleles might have been lost in the upstream populations. It suggests that the upstream populations are more vulnerable to genetic drift and thus should be given more attention in resource conservation. These populations of the upper Yangtze River formed three distinct groups, with significant genetic differentiation among three groups, and should be considered as three different genetic units for conservation management. Our results exemplify the genetic diversity and population structure of S. dabryi after habitat fragmentation caused by dams, which will provide a reference for natural resource protection and management of this species in the upper Yangtze River.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA759087.
Ethics Statement
The animal study was reviewed and approved by Jianghan University.
Author Contributions
HL performed the laboratory work and wrote the manuscript. FX designed the research and revised the manuscript. DZ collected the samples and revised the manuscript. XD and DC collected the samples. YC, YW, and MX revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This research was supported by the National Key R & D Program of China (2018YFD0900903), the National Natural Science Foundation of China (nos. 51979123 and 51779105), the Innovative Research Team Foundation of the Department of Education of Hubei Province, China (T2020034), and the Scientific Research Project of Jianghan University (2021kjzx006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to Dr. Jinyun Luo for helping with data analysis, Jin Cai, Dong Wang, and Tianjiao Lei for assisting with the sample collecting.
References
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individual. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Blanton, R. E., Cashner, M. F., Thomas, M. R., Brandt, S. L., and Floyd, M. A. (2019). Increased habitat fragmentation leads to isolation among and low genetic diversity within populations of the imperiled Kentucky Arrow Darter (Etheostoma sagitta spilotum). Conserv. Genet. 20, 1009–1022. doi: 10.1007/s10592-019-01188-y
Cao, W. X (2019). Water ecological restoration in the cascade development of hydropower in the upper reaches of the Yangtze River. Technol. Econ. Changjiang 2, 5–10. doi: 10.19679/j.cnki.cjjsjj.2019.0201
Cheng, F., Li, W., Castello, L., Murphy, B. R., and Xie, S. G. (2015). Potential effects of dam cascade on fish: lessons from the Yangtze River. Rev. Fish Biol. Fish. 25, 569–585. doi: 10.1007/s11160-015-9395-9
Cheng, F., Li, W., Wu, Q., Hallerman, E., and Xie, S. (2013). Microsatellite DNA variation among samples of bronze gudgeon, Coreius heterodon, in the mainstem of the Yangtze River, China. Ichthyol. Res. 60, 165–171. doi: 10.1007/s10228-012-0329-4
Cook, D. R., and Sullivan, S. M. P. (2018). Associations between riffle development and aquatic biota following low head dam removal. Environ. Monit. Assess. 190, 1–14. doi: 10.1007/s10661-018-6716-1
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Deiner, K., Garza, J. C., Coey, R., and Girman, D. J. (2007). Population structure and genetic diversity of trout (Oncorhynchus mykiss) above and below natural and man-made barriers in the Russian River, California. Conserv. Genet. 8, 437–454. doi: 10.1007/s10592-006-9183-0
Dixon, P (2003). Vegan, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1658/1100-9233
Dong, W. W., Wang, D. Q., Tian, H. W., Pu, Y., Yu, L. X., Duan, X. B., et al. (2019). Genetic structure of two sympatric gudgeon fishes (Xenophysogobio boulengeri and X. nudicorpa) in the upper reaches of Yangtze River Basin. Peer J. 7, e7393. doi: 10.7717/peerj.7393
Gao, T. H (2016). Studies on Gobioninae fish resources and habitat selections in the upper Yangtze River (PhD dissertation). Southwest University, Chongqing, 77–78.
Goodswen, S., Gondro, C., Watson-Haigh, N., and Kadarmideen, H. (2010). FunctSNP: an R package to link SNPs to functional knowledge and dbAutoMaker: a suite of Perl scripts to build SNP databases. BMC Bioinform. 11:311. doi: 10.1186/1471-2105-11-311
Gouskov, A., Reyes, M., Wirthner-Bitterlin, L., and Vorburger, C. (2016). Fish population genetic structure shaped by hydroelectric power plants in the upper Rhine catchment. Evol. Appl. 9, 394–408. doi: 10.1111/eva.12339
Green, M. R., and Sambrook, J. (2012). Molecular Cloning: A Laboratory Manual, 4th Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Kent, W. J (2002). BLAT – the BLAST-like alignment tool. Genome Res. 12, 656–664. doi: 10.1101/gr.229202
Landguth, E. L., Muhlfeld, C. C., Waples, R. S., Jones, L., Lowe, W. H., Whited, D., et al. (2014). Combining demographic and genetic factors to assess population vulnerability in stream species. Ecol. Appl. 24, 1505–1524. doi: 10.1890/13-0499.1
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, X. B., Tang, Q. Y., Yu, D., and Liu, H. Z. (2016). Genetic diversity and population history of longnose gudgeon (Saurogobio dabryi) in the upper Yangtze River and Chishui River based on cytochrome b gene sequences. Chin. J. Zool. 51, 833–843.
Lin, P. C., Liu, F., Li, M. Z., Gao, X., and Liu, H. Z. (2018). Spatial pattern of fish assemblages along the river-reservoir gradient caused by the Three Gorge Reservoir (TGR). Acta Hydrobiol. Sin. 46, 1124–1134.
Liu, D., Li, X., and Song, Z. (2020). No decline of genetic diversity in elongate loach (Leptobotia elongata) with a tendency to form population structure in the upper Yangtze River. Glob. Ecol. Conserv. 23, e01072. doi: 10.1016/j.gecco.2020.e01072
Liu, H. Y., Xiong, F., Duan, X. B., Tian, H. W., Liu, S. P., and Chen, D. Q. (2017). Low population differentiation revealed in the highly threatened elongate loach (Leptobotia elongata Bleeker), a species endemic to the fragmented upper reaches of the Yangtze River. Biochem. Syst. Ecol. 70, 22–28. doi: 10.1016/j.bse.2016.10.015
Liu, Y (2020). Population genetic analysis of Saurogobio dabryi and S. punctatus in the Jialing River (Master dissertation). China West Normal University, Nanchong, Sichuan.
Liu, Y. Y., Zeng, Y., Xiong, X. Q., Lv, Z. Y., Hu, Y., Jiang, C. M., et al. (2019). Difference and adaptation of reproductive biological characteristics of Saurogobio dabryi in Jialing River cascade water conservancy project. Freshw. Fish. 49, 3–8.
Machado, C. B., Braga-Silva, A., Freitas, P. D., and Galetti, P. M. (2021). Damming shapes genetic patterns and may affect the persistence of freshwater fish populations. Freshw. Biol. 67, 603–618. doi: 10.1111/fwb.13866
McKenna, A., Hanna, M., Banks, E., Sivachenk, A. O., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Resour. 20, 1297–1303. doi: 10.1101/gr.107524.110
Nakajima, N., Hirota, S. K., Matsuo, A., Suyama, Y., and Nakamura, F. (2020). Genetic structure and population demography of white-spotted charr in the upstream watershed of a large dam. Water 12, 2406. doi: 10.3390/w12092406
Pavlova, A., Beheregaray, L. B., Coleman, R., Gilligan, D., Harrisson, K. A., Ingram, B. A., et al. (2017). Severe consequences of habitat fragmentation on genetic diversity of an endangered Australian freshwater fish: a call for assisted gene flow. Evol. Appl. 10, 531–550. doi: 10.1111/eva.12484
Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A., and Reich, D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909. doi: 10.1038/ng1847
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Roberts, J. H., Angermeier, P. L., and Hallerman, E. M. (2013). Distance, dams and drift: what structures populations of an endangered, benthic stream fish. Freshw. Biol. 58, 2050–2064. doi: 10.1111/fwb.12190
Song, J., Chang, L. J., and Wang, D. (2016). Multi-locus genomic analysis reveals the genetic diversity and population structure of the rock carp (Procypris rabaudi) in the upper Yangtze River. Biochem. Syst. Ecol. 66, 86–93. doi: 10.1016/j.bse.2016.03.010
Stamatakis, A (2014). RAxML version 8, a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sun, X. W., Liu, D. Y., Zhang, X. F., Li, W. B., Liu, H., Hong, W. G., et al. (2013). SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 8:e58700. doi: 10.1371/journal.pone.0058700
Tang, H., Yang, Z., Gao, S., Wan, L., Zhu, D., Liu, H., et al. (2014). Annual dynamics of fish biodiversity and community structure in the non-impounded reaches of the lower Jinsha River: case study of the Qiaojia section. J. Hydroecol. 6, 7–15.
Vera-Escalona, I., Habit, E., and Ruzzante, D. E. (2015). Echoes of a distant time: effects of historical processes on contemporary genetic patterns in Galaxias platei in Patagonia. Mol. Ecol. 24, 4112–4128. doi: 10.1111/mec.13303
Wang, C. Y., Yu, X. M., and Tong, J. G. (2007). Microsatellite diversity and population genetic structure of redfin culter (Culter erythropterus) in fragmented lakes of the Yangtze River. Hydrobiologia 586, 321–329. doi: 10.1007/s10750-007-0702-x
Wang, H. S., Wang, T., Li, W. J., and Liu, H. Z. (2019). The genetic diversity, individual relatedness and possible mating system of an isolated population of the cyprinid species Megalobrama pellgrini in upper reaches of the Changjiang (Yangtze River), China. J. Oceanol. Limnol. 37, 1042–1050. doi: 10.1007/s00343-019-8152-7
Xiong, F., Liu, H., Duan, X., Liu, S., and Chen, D. (2015). Present status of fishery resources in Yinbin section of the upper Yangtze River. J. Southw. Univ. (Nat. Sci. Ed.) 37, 1–8.
Xu, P., Zhang, X., Wang, X., Li, J. T., Liu, G. M., Kuang, Y. Y., et al. (2014). Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 46, 1212–1219. doi: 10.1038/ng.3098
Xu, W., Yang, Z., Wan, L., Tang, H. Y., and Chen, X. J. (2019). Natural reproduction status of fish species producing pelagic eggs before and after impoundment of Yin-pan Hydropower Station in the lower Wujiang River. J. Hydroecol. 40, 8–15.
Yan, L., Wang, D., Fang, Y., Liu, S., Duan, X., Chang, Y., et al. (2008). Genetic diversity in the bronze gudgeon, Coreius heterodon, from the Yangtze River system based on mtDNA sequences of the control region. Environ. Biol. Fishes 82, 35–40. doi: 10.1007/s10641-007-9248-z
Zhai, D. D., Zhang, Z., Zhang, F. T., Liu, H. Z., Cao, W. X., and Gao, X. (2019). Genetic diversity and population structure of a cyprinid fish (Ancherythroculter nigrocauda) in a highly fragmented river. J. Appl. Ichthyol. 35, 1–8. doi: 10.1111/jai.13897
Zhang, J., Zhang, Y., Feng, Y., Yan, R., Shi, Y., Zhang, Y., et al. (2021). Impact and prospects of water conservation on fish habitat and advances of ecobiology operation in Yangtze River, China: a review. J. Environ. Biol. 42, 1201–1212. doi: 10.22438/jeb/42/5/MRN-1894
Keywords: Saurogobio dabryi, SNP, genetic diversity, population differentiation, habitat fragmentation, the upper Yangtze River
Citation: Liu H, Xiong F, Zhai D, Duan X, Chen D, Chen Y, Wang Y and Xia M (2022) Genetic Diversity and Population Differentiation of Chinese Lizard Gudgeon (Saurogobio dabryi) in the Upper Yangtze River. Front. Ecol. Evol. 10:890475. doi: 10.3389/fevo.2022.890475
Received: 06 March 2022; Accepted: 10 May 2022;
Published: 03 June 2022.
Edited by:
Zhang Min, China Institute of Water Resources and Hydropower Research, ChinaReviewed by:
Zhaobin Song, Sichuan University, ChinaEric M. Hallerman, Virginia Tech, United States
Copyright © 2022 Liu, Xiong, Zhai, Duan, Chen, Chen, Wang and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Xiong, eGY5NjAzQDE2My5jb20=; Xinbin Duan, ZHVhbkB5ZmkuYWMuY24=
 Hongyan Liu
Hongyan Liu Fei Xiong1,2*
Fei Xiong1,2* Dongdong Zhai
Dongdong Zhai Daqing Chen
Daqing Chen Yuanyuan Chen
Yuanyuan Chen Ying Wang
Ying Wang