
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 28 April 2022
Sec. Evolutionary and Population Genetics
Volume 10 - 2022 | https://doi.org/10.3389/fevo.2022.869626
The species that inhabit systems highly affected by anthropic activities usually exhibit this external influence in their gene pool. In this study, we investigated the genetic patterns of populations of Heleobia atacamensis, a freshwater microgastropod endemic to the Atacama Saltpan, a system historically exposed to environmental changes, and currently subjected to conditions associated with metallic and non-metallic mining and other anthropic activities. Molecular analyses based on nuclear and mitochondrial sequences indicate that the saltpan populations are highly fragmented and that the genetic structure is mainly explained by historical geographic isolation, with little influence of contemporary factors. The microsatellite results suggest a moderate genetic diversity and sharp differentiation mediated by isolation by distance. Additionally, despite the high environmental heterogeneity detected and the marked historical dynamism of the region, our data reveal no signs of demographic instability. The patterns of contemporary gene flow suggest a change in the current genetic structure, based on the geographic proximity and specific environmental conditions for each population. Our results, highlight the role of fragmentation as a modulator of genetic diversity, but also suggest that the historical persistence of isolated populations in naturally dynamic environments could explain the apparent demographic stability detected.
During centuries, anthropogenic activities have modified natural environments worldwide, and have usually fragmented the habitats of various species, which has impacted the gene pool of populations by reducing gene flow and driving genetic drift (Charlesworth and Willis, 2009; Schlaepfer et al., 2018). Likewise, the increased risk of extinction of some species due to the loss of genetic diversity, inbreeding depression, and other consequences of population isolation, has also been documented (Wilcox and Murphy, 1985; Thomas, 2000; Reed, 2004). A little-studied issue corresponds to determining the effect of environmental degradation, that is, any change or harmful disturbance on the environment and on populations that have historically been exposed to natural fragmentation at different time scales. The answer to this question invites studies that lie at the interface between population genetics and evolutionary biology (Coates et al., 2018; Ralls et al., 2018).
In the context of global change, the persistence of a population requires an adaptive response to rapid changes in the conditions that generate environmental stress in organisms. However, the response to this stressor will largely depend on the conditions to which a population has previously adapted (Fitzpatrick and Reid, 2019). For example, it is expected that habitat fragmentation erodes genetic variability and increases genetic divergence between remaining populations (Reed and Frankham, 2003; Lowe et al., 2005). It is also expected that certain life history traits (e.g., low vagility) could imply an increase in the vulnerability of an organism to fragmentation (Schlaepfer et al., 2018). On the other hand, in the populations that inhabit environments that have historically been subject to cycles of fragmentation and connectivity (e.g., the Andean Highlands), it is very likely that they have developed strategies that have allowed them to persist under these conditions (Chevin and Lande, 2010). In this way, this mixture of strategies (and factors) could have a relevant role in the possibilities of adaptive responses in the current scenario of climate change and ecosystem impoverishment at the global level. Additionally, under certain conditions, the development of these adaptive strategies can come from foreign populations through migrants, but on other occasions, they can be generated in situ, giving rise to adaptations that allow the population to persist in a dynamic environment (Chevin et al., 2010; Furness et al., 2015; Wong and Candolin, 2015).
Therefore, research is needed to broaden our understanding of the response of populations that have historically persisted in systems that present cycles of fragmentation and reconnection and that are currently exposed to abrupt modifications as a result of global change. An interesting system to evaluate these predictions corresponds to the Atacama Saltpan, located in northern Chile. In the hypersaline lagoon systems of this saltpan, different species not only have had to respond to the stress related to the physicochemical changes of an active saltpan (e.g., changes in salinity, temperature, pH) (Alonso and Risacher, 1996; Risacher and Alonso, 1996; Risacher and Fritz, 2009), and to the successive cycles between hyperarid and arid periods that characterized the region during the Pliocene, Pleistocene and Holocene (Wirrmann and Mourguiart, 1995; Betancourt et al., 2000; Bobst et al., 2001; Sáez et al., 2012; González-Silvestre et al., 2013; Ritter et al., 2018), but also to the extraction of water for metal (copper) and non-metal (lithium and potassium) mining, in addition to other anthropogenic activities such as tourism and recreation. A recent study indicated that during the last 20 years, there has been a significant degradation of saltpan ecosystems, and these changes have been reflected in a decline in vegetation cover, an increase in temperature, a decrease in soil moisture and an intensification of drought conditions in these systems (Liu et al., 2019). Another study conducted in Tilopozo, an aquifer located in the southern margin of the Atacama Saltpan, indicates that the changes in vegetation cover would be mediated by changes in soil moisture and aquifer structure (type of soil, its fissures, and its permeability), along with sustained water extraction in the system (Soto et al., 2019). In this way, the possibility of evaluating both historical factors (arid and hyperarid cycles) and contemporary factors (anthropic effects and global change) position the Atacama Saltpan as an interesting system to evaluate the effect of fragmentation and environmental dynamism on the pattern of genetics differentiation of the species that inhabit this ecosystem.
The extreme conditions (high solar radiation and surface water evaporation, high daily thermal oscillation, low oxygen pressure) that predominate in the hydrological systems of the Atacama Saltpan have generated a biotic assembly characterized mainly by bacterial communities (Demergasso et al., 2010; Cubillos et al., 2018; Dorador et al., 2018) and macroinvertebrates (Zúñiga et al., 1991). Among the latter is Heleobia atacamensis (Philippi, 1860), a strictly aquatic microgastropod that inhabits Tilopozo, one of the brackish and saline aquifers of the saltpan. Heleobia atacamensis is categorized as Data Deficient by the International Union for Conservation of Nature (IUCN), however, according to environmental legislation in Chile, the species is classified as Critically Endangered by the Ministry of the Environment (RCE DS52/2014). The classification criteria were based mainly on the decrease in habitat quality and given that the species occupies less than 1 km2 (Ministerio del Medio Ambiente, 2014). Although H. atacamensis was described in Tilopozo, populations of Heleobia have recently been detected in several hydrological systems of the saltpan with varying degrees of connectivity; to date, the identity or relationship and structure of these populations of the saltpan have not been described or evaluated.
Previous phylogenetic studies show that H. atacamensis corresponds to a monophyletic lineage different from other species of the genus (Collado et al., 2013), and that the cladogenesis event between this taxon and its closest lineage would have occurred 1.7 (2.5–1.0) Ma (Collado et al., 2016b), a date much later than the establishment of the hyperarid conditions of the Atacama Desert that occurred during the late Oligocene and early Miocene (Muñoz et al., 1997; Clarke, 2006). Therefore, these antecedents suggest that the population divergence process of H. atacamensis would have occurred in hypersaline and highly fragmented hydrological systems. This condition of isolation in saltpan populations could have changed temporarily during periods of higher rainfall mainly as a consequence of glacial cycles (Ammann et al., 2001; Fornari et al., 2001; Nester et al., 2007; Mujica et al., 2014) and intensification of the South American Summer Monsoon (Quade et al., 2008; Marengo et al., 2012; Baker and Fritz, 2015); therefore, the connectivity between populations of H. atacamensis would present dynamic cycles, although fragmentation would predominate. Based on the above, this study attempts to answer the following questions regarding the evolutionary history and persistence of historically fragmented and currently threatened populations:
1. What is the connectivity, genetic diversity pattern and degree of coincidence of demographic events among the populations of H. atacamensis? It is predicted that the genetic flow will be related to the spatial position of the systems, that the genetic diversity will be similar between the populations, and that there will be demographic stability reflecting resilience to historical events of fragmentation and environmental degradation.
2. To what extent are geographic isolation and environmental heterogeneity related to genetic differentiation between populations? Considering the low vagility of Heleobia species and the sensitivity to environmental conditions (Iannacone and Alvariño, 2002; Cazzaniga et al., 2011; Souza et al., 2013; Magalhães et al., 2014; Collado et al., 2021), it is hypothesized that there will be a pattern of isolation by distance and signs of isolation by environment.
3. Is there a change between the historical and contemporary structuring of populations? It is predicted that there will be a change and that the current structure will have components of the past structure, the physiography of the saltpan and differentiation of the ecosystems.
The Atacama Saltpan is the largest salt deposit in Chile and is located on the eastern margin of the Atacama Desert at 2,300 masl. The hydrological system extends approximately 100 km long by 80 km wide at its maximum, representing a total surface area of approximately 3,000 km2, whose lagoon area reaches 12.6 km2 (Alonso and Risacher, 1996; Lowenstein and Risacher, 2009). The surface water contributions come mainly from permanent courses that drain from the Andes Mountains to the north and east of the saltpan, while the underground contributions come from the Andean slope of the saltpan (Alonso and Risacher, 1996; Marazuela et al., 2019). Precipitation is concentrated during the austral summer and averages 10 mm/year; however, considering the high evaporation potential (2,000 mm/year), there is a negative water balance (Alonso and Risacher, 1996; Risacher and Alonso, 1996). On the eastern margin of the saltpan, there are different hydrological subsystems with different degrees of connectivity, and most of these aquifers drain in the center of the saltpan in layers different from the evaporites (Alonso and Risacher, 1996). In this study, eight hydrological systems located on the eastern and southern fringes of the Atacama Saltpan were analyzed (Figure 1).
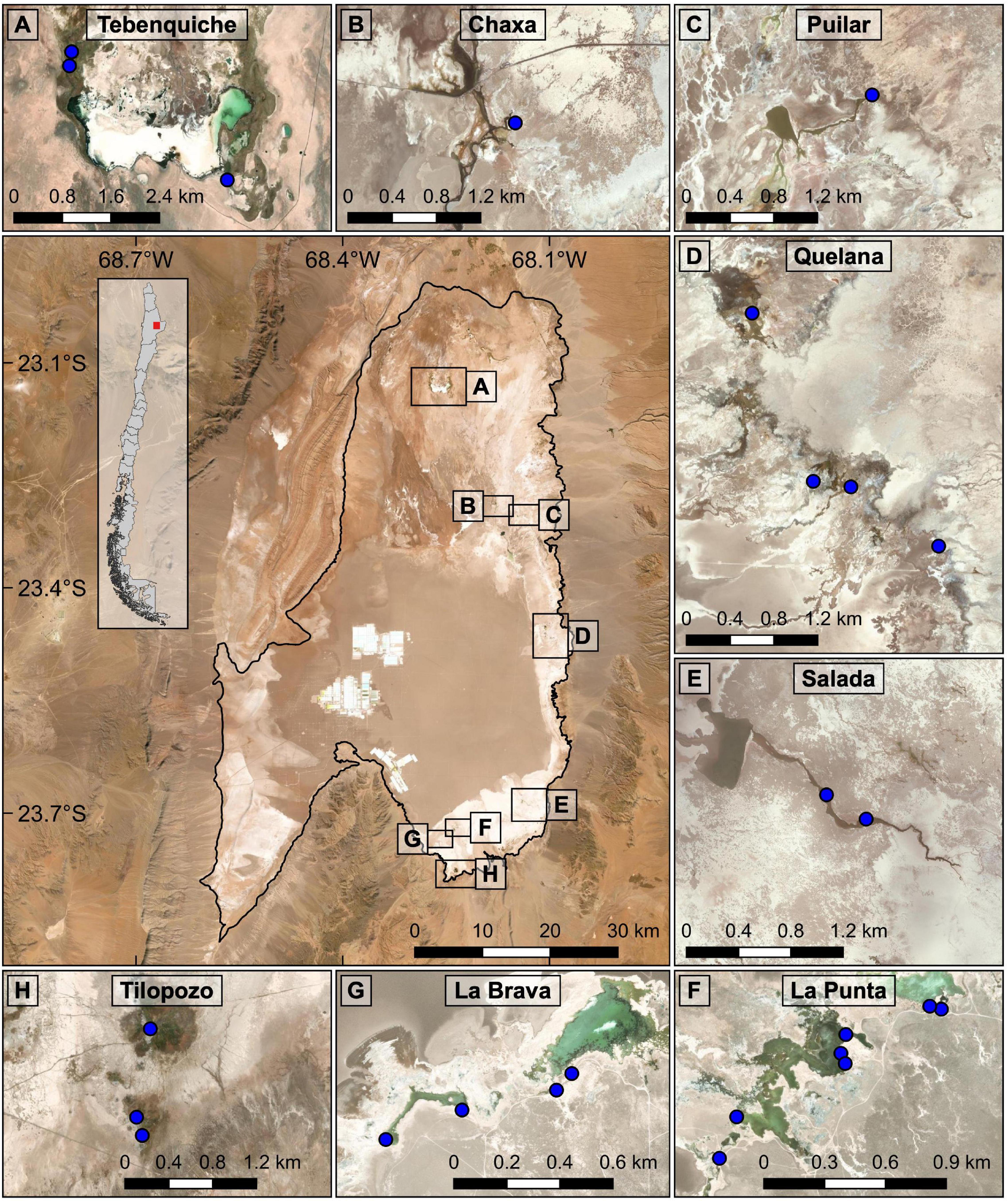
Figure 1. Map showing the Atacama Saltpan. The boundary of the saltpan is depicted in the central figure. The eight systems analyzed and the sampling points considered for each system are indicated (A–H). Maps created in QGIS Geographic Information System v3.4.9 with ESRI world imagery (ESRI, DigitalGlobe, GeoEye, Earthstar Geographics, CNES/Airbus DS, USDA, USGS, AeroGRID, IGN, and the GIS User Community).
The specimens of Heleobia were collected from hydrological systems of the Atacama Saltpan between 2017 and 2018, and even when the sampling periods were in different years, the gene pool and morphological features of the individuals from the same system were shared, and thus, were considered as the same population. The snails were collected by hand or using a sieve of 1 mm mesh opening from the sediment or macrophytes present in the systems. The authors of this study were authorized to collect specimens of the genus Heleobia in the Atacama Saltpan according to Authorization No. 620195 of the National Forest Corporation (CONAF) and Exempt Resolution No. 1253/2018 of the Undersecretary of Fisheries and Aquaculture (SUBPESCA). According to the geography, the sampling points within the saltpan were divided into eight systems: Tebenquiche, Chaxa, Puilar, Quelana, Salada, La Punta, La Brava and Tilopozo (Figures 1A–H; photographs of the systems are shown in Supplementary Figure 1). The conformation of these systems was determined by grouping sampling sites that belong to the same watercourse, that is, that correspond to transects within the same tributary or body of water.
DNA extraction was performed using the Cetyl Trimethyl Ammonium Bromide (CTAB) method (Winnepenninckx et al., 1993). The amplification was performed by polymerase chain reaction (PCR) considering three mitochondrial markers and three nuclear markers. The methodology applied followed the protocols of Collado and Méndez (2012) and Valladares et al. (2018). The details of the markers and primers used in the study are shown in Supplementary Table 1. Both strands of the amplified products were sequenced by Macrogen Inc. (Seoul, South Korea). The sequences were edited and then aligned using the algorithms ClustalW (Larkin et al., 2007) and MUSCLE (Edgar, 2004) in the program CodonCode Aligner v3.6.1 (CodonCode Corporation, Dedham, MA).
Phylogenetic reconstructions were performed under the Maximum Likelihood (ML) and Bayesian Inference (BI) algorithms using the mitochondrial sequences (COI, 12S, and 16S). For ML and BI, the best model of sequence evolution was previously selected using bModelTest (Bouckaert and Drummond, 2017). Reconstruction by ML was performed in RAxML v8.0 (Stamatakis, 2014), and node support was obtained by performing a bootstrap analysis of 1,000 pseudoreplicates. Bayesian reconstruction was performed in MrBayes v3.2.6 (Ronquist et al., 2012) using the Markov Chain Monte Carlo (MCMC) method. The BI analysis was run three times for 100 million generations each time, with a sampling of trees every 10,000 generations. The consensus tree considered a burn-in period of 25%. The reconstructions of ML and BI were performed in the CIPRES cluster of the San Diego Supercomputer Center (Miller et al., 2010).
In the phylogenetic reconstructions, the species Heleobia chimbaensis (Biese, 1944), H. ascotanensis (Courty, 1907) and H. carcotensis Collado, Valladares, and Méndez, (2016) were used as a sister group considering its geographical proximity to the Atacama Saltpan. Together, Heleobia sp. of Parinacota was used as an external group sensu lato to root the phylogenetic tree. Additionally, two H. atacamensis sequences obtained from the National Center for Biotechnology Information (NCBI-GenBank) database were included in the analyses (Supplementary Table 2).
The reconstruction of the species tree and estimation of divergence times were performed using Bayesian inference in the program BEAST v2.5.2 (Bouckaert et al., 2019). For this analysis, the mitochondrial and nuclear genes (complete set; 6 loci) were considered. The analysis was performed using a multispecies coalescent tree (MSCT) reconstruction in the StarBEAST2 v0.14.0 package (Ogilvie et al., 2017). For the MSCT inference, the Yule tree model was implemented with a strict clock model using Heleobia australis (d’Orbigny, 1835) as outgroup. For this analysis, the individuals were assigned to its system of origin. For the StarBEAST2 analysis, a mutation rate of 1.7% per million years proposed for the cytochrome oxidase subunit 1 (COI) gene in Cochliopidae (Wilke et al., 2009) and three independent runs of 100 million generations, sampling every 10,000 generations was used. For the MSCT analyses, five individuals were selected per system, the missing sequences were coded as missing, and the alignment was partitioned by gene. The convergence of the results was analyzed in Tracer v1.7.1 (Rambaut et al., 2014) and the results were summarized in a single ultrametric tree using TreeAnnotator v2.5.1 (Drummond et al., 2012). To assess the phylogenetic congruence between the mitochondrial and nuclear loci, phylogenetic trees were estimated for the datasets separately using the same parameters as the analysis of the complete dataset.
The genetic diversity of each system was characterized from the COI gene due to its greater variability using the following diversity indices estimated in Arlequin v3.0 (Excoffier and Lischer, 2010): Number of polymorphic sites (S), number of haplotypes (KH), haplotype diversity (H) and the average of differences between pairs of sequences (Π). The relationships between the haplotypes were visualized by constructing a haplotype network through the median-joining algorithm (Bandelt et al., 1999) using PopART v1.7 (Leigh and Bryant, 2015). Genetic distances between hydrological systems were estimated in MEGA11 v11.0.8 (Tamura et al., 2021). To determine the number of genetic groups and spatial population structure within the saltpan, an analysis was performed in the GENELAND v4.0.7 package (Guillot et al., 2005) in R v3.6.1 software (R Development Core Team, 2019). The most likely number of clusters (CT) was identified by performing fifteen independent MCMC analyses of 108 iterations and thinning every 10,000 iterations. The range was limited between CT = 1 and CT = 8 (total of systems analyzed), and a burn-in of 25%.
For this study, the specific primers and PCR conditions for H. atacamensis developed by Fabres et al. (2020) were used. The standardization of the PCR was performed using the protocol described by Schuelke (2000). The amplification products were analyzed by the Pontificia Universidad Católica de Chile (Santiago, Chile) sequencing services. Subsequently, fragment analysis was performed using GeneMapper v4.0 software (Applied Biosystems, Waltham, MA). The excess homozygotes or heterozygotes was evaluated with the program MICRO-CHECKER v2.2.3 (van Oosterhout et al., 2004), and for each locus, the presence of stuttering and null alleles were searched.
Genetic diversity indices including number of alleles (A), number of private alleles (AP), percentage of polymorphic loci (% PL), observed heterozygosity (HO), expected heterozygosity (HE) and fixation index (F) were calculated in GenAlEx v6.5 (Peakall and Smouse, 2012). The allelic richness (AR) was estimated in the PopGenReport v3.0.4 package (Adamack and Gruber, 2014), performing a rarefaction based on the smaller sample size (n = 32). To evaluate whether the individuals within each population were significantly more or less related under the assumption of random mating, we calculated the population average of relatedness (R) and the estimated significance considering 10,000 permutations in GenAlEx. The degree of genetic structuring between pairs of populations was calculated using the FST indices in the GENETIX v4.0.5 program (Belkhir et al., 2004).
The number of genetic groups (K) was evaluated using the genotypes of the individuals through a Bayesian analysis implemented in STRUCTURE v2.3.4 (Pritchard et al., 2000; Falush et al., 2003, 2007). From 1 to 8 groups, 600,000 replicates were analyzed by performing an MCMC analysis considering a burn-in period of 300,000 parameters, and the results were obtained based on 15 runs. The structuring parameters were calculated for each K-value under the admixture model, allelic frequencies correlated between groups and using locality of origin information as LocPrior (Pritchard et al., 2000; Falush et al., 2003). The most likely number of populations (K) was evaluated considering the value of the log-likelihood of the observed data [LnP (D)] and the second-order change rate of the log-likelihood of the data in different runs of K (ΔK) described in Evanno et al. (2005); the results were analyzed using the Pophelper v2.3.0 package (Francis, 2017). The consistency between the different STRUCTURE runs was evaluated using CLUMPP v1.1.2 (Jakobsson and Rosenberg, 2007).
The estimation of recent bottlenecks for each of the eight systems of the Atacama Saltpan was performed independently using BOTTLENECK v1.2 (Piry et al., 1999), based on the graphical analysis of Luikart et al. (1998) and Wilcoxon signed-rank tests (WSRs) used to compare the heterozygosity observed with that expected from the number of alleles observed given the sample size, assuming mutation-drift equilibrium for each locus of each system. The two-phase model (TPM) and the stepwise mutational model (SMM) were used to simulate mutation-drift equilibria. For TPM, two values (10 and 30) were tested to estimate the variance of the geometric distribution with a low probability of single-step mutations (70%). Finally, to estimate the contemporary genetic flow between populations, a Bayesian analysis was performed using the software BAYESASS v3.0.8 (Wilson and Rannala, 2003). This analysis was performed using 50 million iterations and a burn-in period of 25%, and sampling was performed every 5,000 iterations. The mixing parameters for allelic frequencies, migration rates and inbreeding coefficients were defined as 0.35, 0.25, and 0.60, respectively. Ten independent runs with different seed values were performed to examine the consistency in the results.
The existence of a pattern of genetic divergence associated with IBD and IBE phenomena was simultaneously tested by a multiple matrix regression with randomization (MMRR) analysis on genetic distance matrices (dependent variable), environmental distance matrices and geographic distance matrices (independent variables) (Wang, 2013). This analysis allowed us to evaluate whether spatial distances (denoted by coefficient βD) and environmental heterogeneity (denoted by coefficient βE) have an effect on the genetic divergence of the populations in the saltpan. The result of the MMRR analysis is a multiple regression that explicitly indicates whether the response variable changes significantly with respect to the independent variables and the direction of this relationship. In this way, the result indicates the fit of the model (using r2) and the significance of the model coefficients. The spatial distance matrix corresponded to the standardized geographic distance between the systems studied. The FST microsatellite matrix (FST(SSR)) was used as a proxy for genetic distance. The FST matrix was linearized according to the formula [FST/(1-FST)] proposed by Rousset (1997). The environmental distance matrix was constructed based on six physicochemical parameters measured in the study systems. The variables considered were temperature, salinity, conductivity, dissolved oxygen, oxygen saturation and pH. Each parameter was measured at least three times (different seasons when possible) to obtain an adequate description of the systems (Supplementary Table 3). The measurements per system were averaged, and the variance was described by a PCA. Subsequently, the environmental distances matrix was generated using all the main components in the vegan v2.5-5 package (Oksanen et al., 2007). The MMRR analysis was performed using the lgrMMRR function with 10,000 permutations in PopGenReport. In addition, to investigate the relative influence of the independent variables (geographic distance and environmental heterogeneity) on genetic dissimilarity, we used multiple regression on distance matrices (MRM) (Lichstein, 2007) to evaluate the relationship between each explanatory variable and the dependent variable. The values for the MRM models were obtained through 10,000 permutations in the ecodist v2.0.1 package (Goslee and Urban, 2007).
The results of bModelTest indicated that the best model of nucleotide substitution for the mitochondrial genes 12S and 16S was HKY, and for COI TN93; on the other hand, for the nuclear markers H3 and ITS2, the best model was GTR + I + G, and for ITS1, it was HKY + G. Original sequences obtained in the present study were submitted to GenBank (accession numbers in Supplementary Table 1).
The BI analysis produced a consensus tree that recovered the same lineages as the ML analysis (-ln Likelihood = –3232.276) using mitochondrial sequences. The phylogenies obtained using BI and ML were concordant, and in both, the samples from the Atacama Saltpan were recovered in a clade differentiated from the rest of the Altiplano localities (Figure 2A and Supplementary Figure 2). Considering node support and spatial distribution, among the samples from the saltpan, four main lineages were recovered. The first lineage contains individuals from Puilar and Chaxa and some from Tebenquiche (Lineage 1); the second group contains individuals from Tilopozo and the rest of the representatives of Tebenquiche (Lineage 2); a third lineage groups all individuals from Quelana and Salada, in addition to some of the individuals from La Punta and one specimen from Puilar (Lineage 3); and the fourth lineage recovered the remaining individuals from La Punta specimens and all the individuals sampled at La Brava (Lineage 4). Two individuals from Quelana were recovered in a polytomy with lineages 1 and 2.
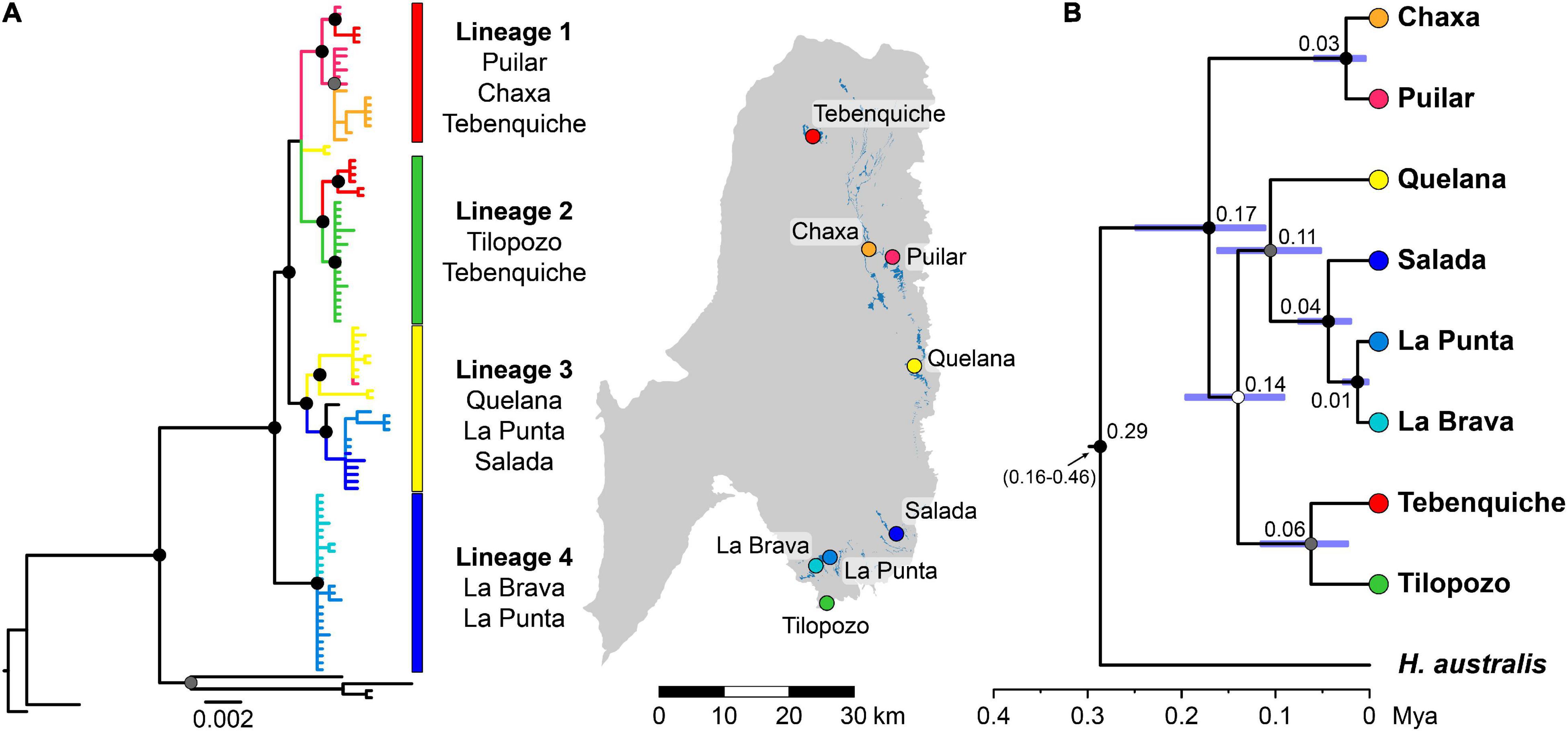
Figure 2. Phylogenetic relationships of Heleobia populations of the Atacama Saltpan (A) Bayesian inference tree of Heleobia using three mitochondrial loci (12S, 16S, and COI). Tree is rooted with different species of Heleobia of the Atacama Desert and southern Altiplano (see full results in Supplementary Figure 1). (B) Maximum clade credibility (MCC) tree of Heleobia obtained from StarBEAST2 analysis based on nuclear and mitochondrial loci (H3, ITS1 and ITS2; 12S, 16S, and COI). Heleobia australis (Buenos Aires, Argentina) was used as outgroup. Mean divergence time estimates are shown with 95% highest posterior density (HPD; blue bars). For both phylogenetic estimations (A,B), black, gray, and white filled circles indicate nodes that were recovered with posterior probability (PP) ≥ 0.95, 0.75– < 0.95, and < 75, respectively. Sampling sites are color-coded (as in other figures) to assist comparisons.
The results of the MSCT estimation using the six markers (mitochondrial and nuclear) were consistent in the different runs performed. The topology of the tree showed a strong spatial component of the systems analyzed (Figure 2B). The reconstruction presented high values of node support, and the lineages agreed with the geographic arrangement of the saltpan, with the exception of the lineage from Tebenquiche (system located in the extreme north of the saltpan), which was recovered with the samples from Tilopozo (type-locality of H. atacamensis). The topology of the MSCT estimated from the nuclear sequences was partially different from the mitochondrial MSCT (Supplementary Figure 3). However, despite this inconsistency, neither dataset disproportionately influenced the reconstruction of the complete set, since features from both trees were recovered in the phylogeny obtained using concatenated sequences. Finally, the divergence times suggest that the diversification process in the saltpan would have begun 170 (110–250) ky ago during the late Pleistocene; however, the wide range covered by the highest posterior density (HPD) region estimated for each node reflects that the estimation of the divergence times requires more information (e.g., fossil calibration or mutation rates for all markers).
The values of the genetic diversity indices estimated from the COI sequences (252 individuals) are shown in Table 1. The results suggest that the lowest genetic variability was found in the La Brava System (southern sector), where a single haplotype was observed for the entire system. In contrast, the greatest variability was found in La Punta (also southern sector), with thirteen haplotypes. Genetic distances between Heleobia individuals grouped by hydrological system ranged from 0.3% (Chaxa-Puilar) to 1.7% (La Brava-Salada) with a mean genetic distance of 1.1% (Supplementary Table 4).
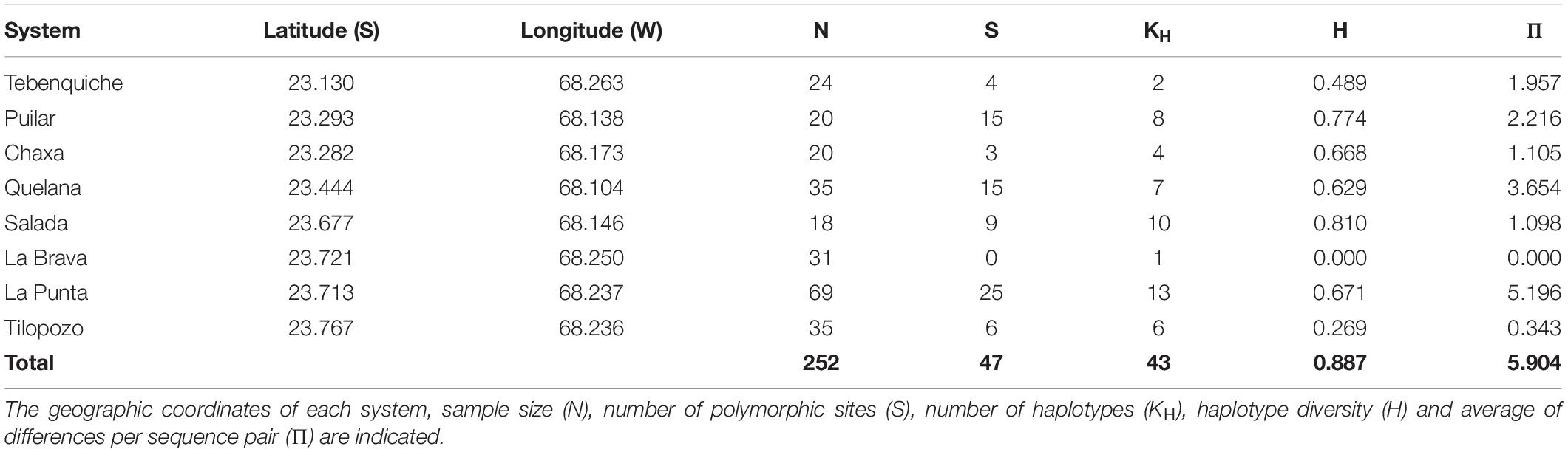
Table 1. Indices of genetic diversity obtained from partial sequences of the COI gene for each sampling system of the Atacama Saltpan.
The haplotype network (Figure 3) showed that most haplotypes were shared between different localities, with the exception of Tilopozo (southern margin of the saltpan), which exhibited exclusive haplotypes. The individuals from La Punta were recovered in two main haplogroups, one associated with the La Brava samples and another separated by ten mutational steps that contained the rest of the La Punta samples and all of the Salada samples.
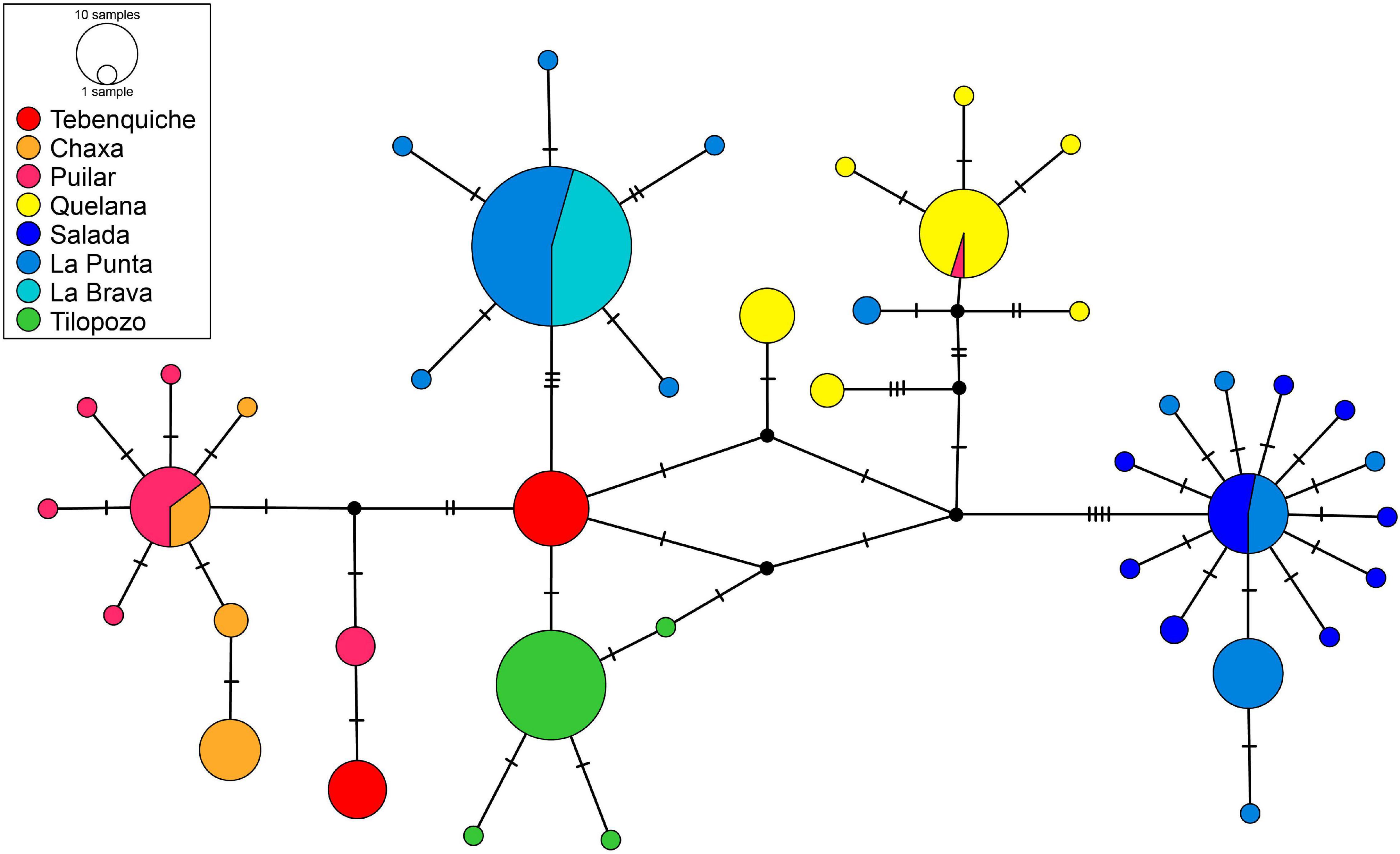
Figure 3. Haplotype network of Heleobia populations of the Atacama Saltpan obtained from partial sequences of the COI gene. The segments on the lines represent the number of mutational steps. Circle sizes are proportional to haplotype frequencies and colors represent the respective sample localities.
The analysis of the number of genetic groups and the spatial population structure obtained with GENELAND indicate the existence of five genetic groups (posterior probability = 0.82). However, the posterior probabilities associated with the definition of the boundaries and spatial belonging between the detected clusters were moderate (on average 0.7), which suggests that the populations would be in a process of divergence (Supplementary Figure 4). The first cluster would be composed of individuals from Tebenquiche and Tilopozo (systems at the geographic edges of the saltpan). The second cluster recovered the individuals collected in Puilar and Chaxa. The third group contains only individuals from Quelana (center of the saltpan). The fourth cluster corresponded to the specimens collected in Salada. The fifth cluster contained the individuals analyzed from La Punta and La Brava.
Eleven microsatellites of 303 individuals from the eight systems of the Atacama Saltpan were analyzed (Supplementary Data Sheet 1). This dataset corresponds to the total number of individuals in the mitochondrial set, except for two individuals of Salada, which did not amplify satisfactorily. In addition, the sample size was increased in most of the systems (except Salada) for the estimation of demographic and population structure parameters. The variability of the populations, the values of heterozygosity (HO, HE), the number of alleles per population and the values of FIS are presented in Table 2. None of the populations deviated from the Hardy-Weinberg equilibrium (no significant FIS values). The results show low values of heterozygosity in all the populations studied. On the other hand, high levels of population average of relatedness (R) were found in most of the populations of the saltpan, with Tilopozo showing the highest value (R = 0.290) and La Brava the lowest (R = 0.038).
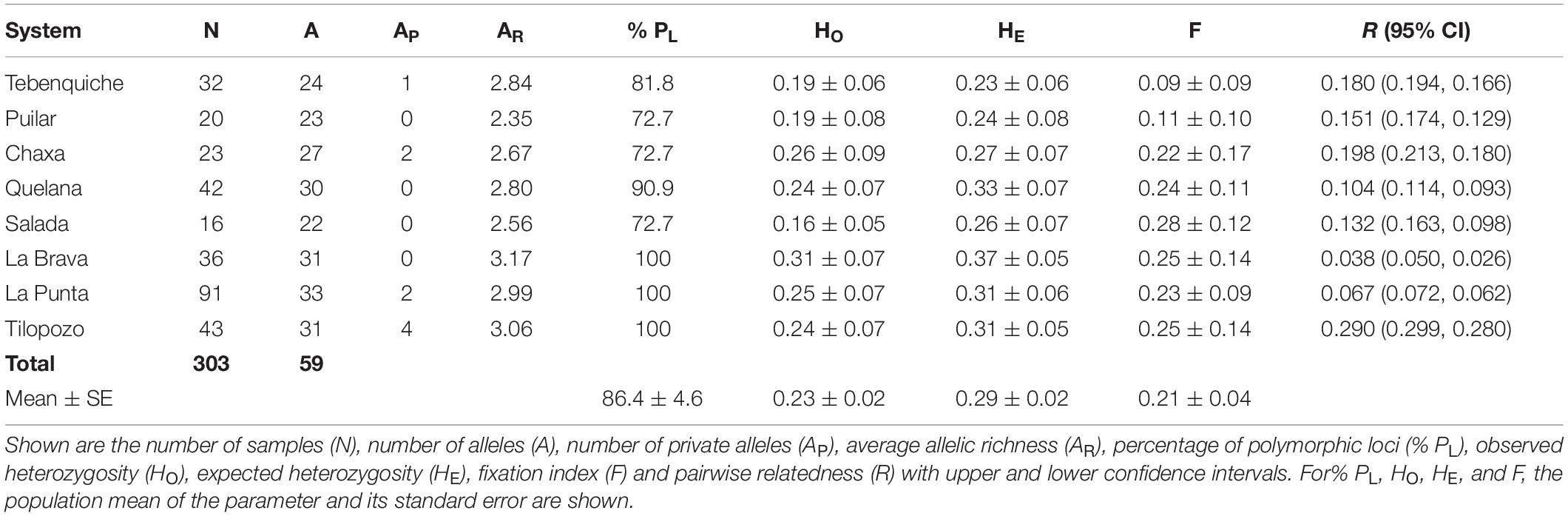
Table 2. Genetic diversity parameters determined from the analysis of the 11 microsatellite loci of eight populations of H. atacamensis.
All the paired comparisons of FST were significant, and the results showed that the closest systems had lower differentiation values (Table 3). Salada presented moderate differentiation values in all comparisons (between 0.224 and 0.490), and Tilopozo showed the highest differentiation values with respect to the rest of the systems (between 0.302 and 0.490).
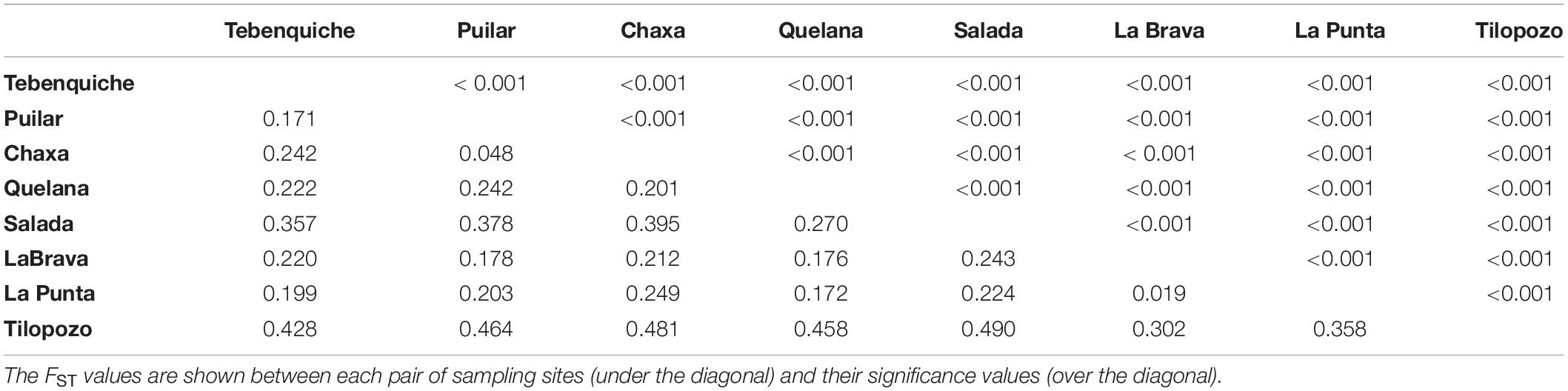
Table 3. Genetic structuring among populations of the Atacama Saltpan obtained using 11 microsatellite loci.
The most likely number of populations in the saltpan, determined using the Evanno method, was K = 4 (Figure 4). The four populations defined were: Population 1 contained samples from the Tebenquiche, Chaxa and Puilar systems (northern systems of the saltpan); Population 2 recovered individuals from Quelana (center of the saltpan); Population 3 recovered the individuals collected in the Salada, La Brava and La Punta systems (south-central of the saltpan); finally, Population 4 recovered all individuals from Tilopozo (type-locality of H. atacamensis).
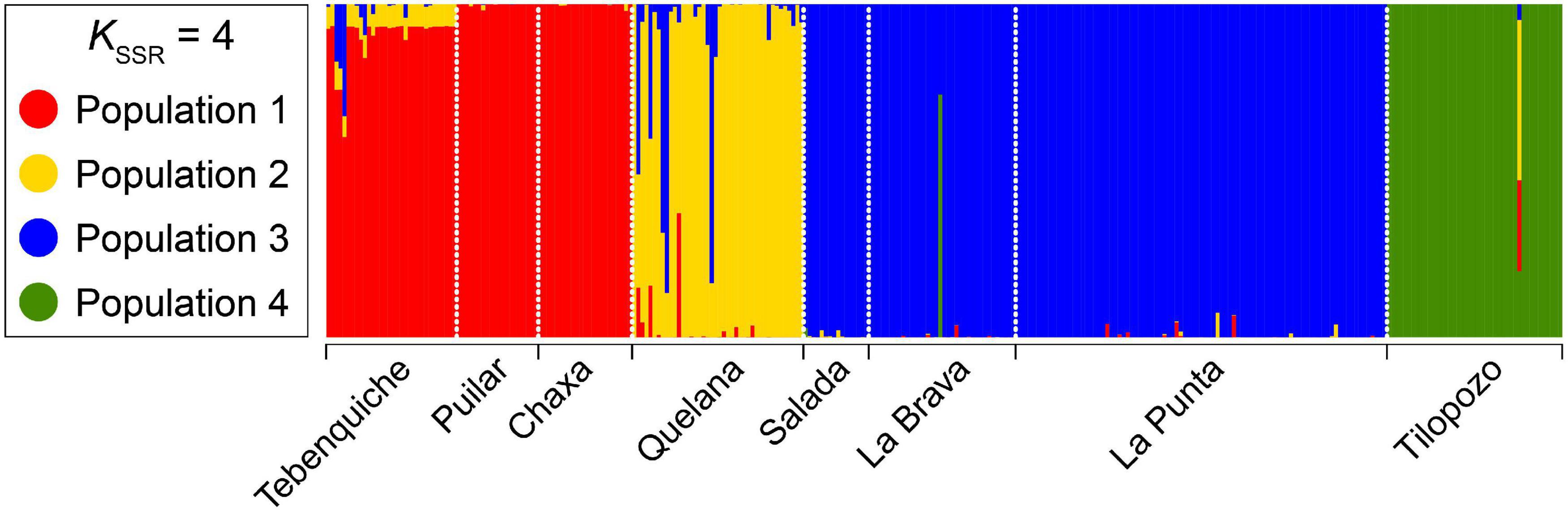
Figure 4. Population structure inferred for gastropods of the genus Heleobia of the Atacama Saltpan obtained from 11 microsatellite loci using STRUCTURE.
No significant signs of genetic bottlenecks were detected in any of the systems analyzed, using both the TPM and SSM models (all p-values > 0.1 from the Wilcoxon tests); in the same way, all systems presented L-shaped normal distributions, indicating the absence of abrupt demographic declines in populations.
The environmental characterization based on the six physicochemical features (temperature, pH, dissolved oxygen, oxygen saturation, salinity and conductivity) showed high environmental heterogeneity between and within systems (Supplementary Figure 5). Tilopozo differed from the rest of the systems mainly by temperature and salinity, Tebenquiche and Chaxa showed similar conditions of high salinity, Salada showed a high environmental heterogeneity mainly due to temperature and oxygen concentrations, La Punta and La Brava showed environmental similarity, as well as Puilar and Quelana. The MMRR analysis for the complete set of variables showed a significant and positive correlation between geographic and genetic distance (βD = 0.23, p < 0.05), indicating a pattern of isolation by distance; however, a significant relationship between environmental heterogeneity and genetic differentiation was not detected (βE = 0.06, p > 0.05), suggesting an absence of isolation by environment. The model considering both predictive variables, was significant but had a low coefficient of determination (r2 = 0.22, p < 0.05). The relative contribution of environmental variables to genetic differentiation determined by the MRM analysis generated a significant model with a high coefficient of determination (r2 = 0.77, p < 0.05), and was consistent with the MMRR analysis regarding that the geographic distance showed a positive and significant correlation with genetic distance (b = 0.05, p < 0.05). With respect to the rest of the predictive variables, both oxygen saturation and pH showed a significant and positive relationship (b = 1.11, p < 0.05; and b = 6.05, p < 0.05, respectively) with genetic differentiation.
The estimated migration rates showed a high value of self-recruitment in the Tebenquiche, Quelana, La Punta and Tilopozo systems, indicating a reduced genetic flow between these systems and the rest of the localities. In contrast, in Chaxa, an evident mixture of genotypes from Puilar was observed. Similarly, Salada presented a unidirectional flow of migrants from La Punta. With respect to the type-locality of H. atacamensis (Tilopozo), this system presents the highest value of self-recruitment of the sample, which indicates that there is a contemporary isolation of this locality compared to the rest of the saltpan (Table 4).
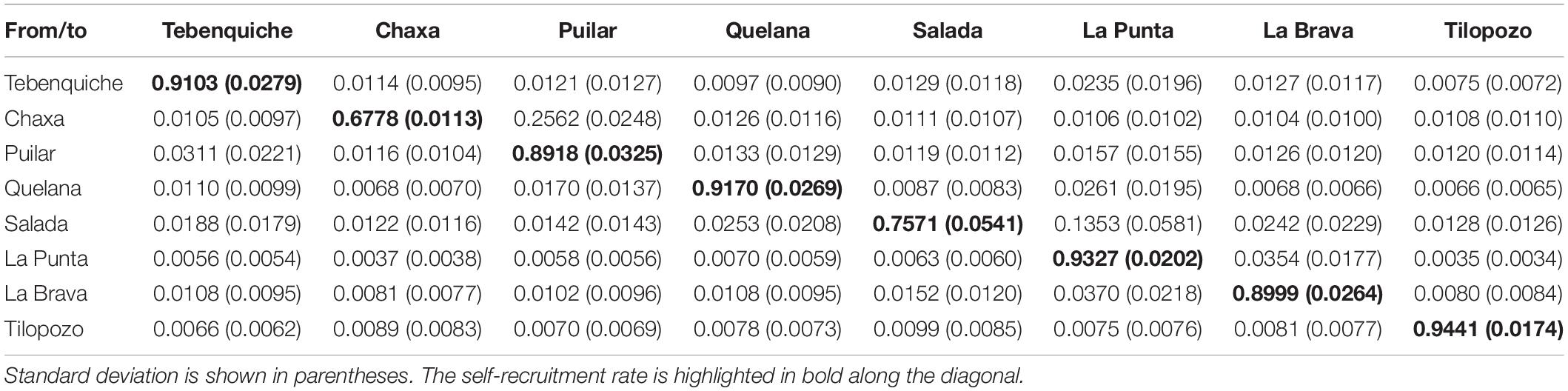
Table 4. Estimated contemporary migration rate for Heleobia populations of Atacama Saltpan obtained using 11 microsatellite loci.
The analysis of nuclear and mitochondrial sequences, in addition to the study of microsatellites of eight isolated systems of the Atacama Saltpan, showed a high level of genetic structuring among Heleobia populations studied. In addition, this genetic structuring would be recent if we consider that there is a significant relationship between the level of genetic divergence and the geographic distance between the systems. It is likely that the combination of genetic drift and reduced gene flow, added to endogenous factors such as adaptation to environmental dynamism, has shaped the underlying complex genetic pattern of Heleobia populations that inhabit the saltpan. In this sense, the results of the study show the existence of previously undetected cryptic diversity in historically fragmented populations of the genus.
The results of the phylogenetic analysis suggest that the populations of Heleobia of the Atacama Saltpan correspond to a genetic and evolutionary unit differentiated from the rest of the species of the Altiplano and the Atacama Desert. Even when the tree was reconstructed based on mitochondrial sequences, the reciprocal monophyly and the high node support values indicate that the diversification process of Heleobia in the Atacama Saltpan would correspond to an in situ process. This would be an expected pattern considering the limited vagility of Heleobia species (Cazzaniga et al., 2011). In this sense, multiple studies have shown that, in general, each species of the genus is restricted to a single hydrological system or basin (Kroll et al., 2012; Collado et al., 2013, 2016a,2016b). In addition, the study revealed a genetic structure within the system congruent with the geographic separation of the sampled sites (latitudinal differentiation), a result similar to that reported in H. ascotanensis, a species that inhabits the Ascotán Saltpan located approximately 200 km north of the Atacama Saltpan (Valladares et al., 2018). The mitochondrial tree also showed that two samples from Quelana (Lineage 3) were recovered in polytomy with Lineage 1 (Puilar, Chaxa and Tebenquiche) and Lineage 2 (Tilopozo and the rest of the individuals from Tebenquiche). Similarly, an individual from Puilar was recovered in Lineage 3 (Quelana, La Punta, and Salada). The branching pattern indicate that individuals with gene pools from other systems persist in these lineages, suggesting a connection between localities. This makes sense considering that Quelana is located in the center of the saltpan, therefore, it is possible that there is (or has been) gene flow between this system and adjoining systems.
The topology of the phylogenetic tree with the complete dataset (mitochondrial and nuclear sequences) was partially discordant with respect to that generated only based on mitochondrial markers. The main difference was in the position of the snails from La Brava and La Punta in comparison with the other lineages; however, in both phylogenetic estimations, the influence of the spatial arrangement of the systems was noticeable. According to the MSCT reconstruction, the diversification process started approximately 170 ky ago (late Pleistocene), which could be related to the wet periods that have been reported for the area (Sáez et al., 2012; Ritter et al., 2018), although the lack of fossil calibration prevents a more robust dating.
The mitochondrial diversity indices obtained for the populations of Heleobia of the Atacama Saltpan were substantially lower than those reported in other Heleobia species that present fragmented populations (Valladares et al., 2018). On the other hand, the genetic distances between systems (COI sequences) were comparable to previous studies in closely related species of Heleobia and that also used mitochondrial information (Collado et al., 2013, 2016b). Although there are no studies based on microsatellites for the genus, the microsatellite diversity indices obtained in the present study are also lower than those obtained in fragmented populations of other taxa of the Altiplano region (Cruz-Jofré et al., 2016; Guerrero-Jiménez et al., 2017). A unique situation occurs in the south-central systems of the saltpan, since in La Punta, two highly differentiated haplogroups were detected, one which also constituted the only haplotype of La Brava and the second was shared with Salada. Considering the poor genetic diversity of La Brava relative to La Punta and the phylogenetic relationships between the populations showing that they are closely related. These results suggest that the individuals of La Brava would have originated from La Punta migrants and that, in turn, a portion of the haplotypes recovered in La Punta would have come from Salada. This hypothesis is compatible with a model of fluctuating connectivity in the saltpan, a pattern proposed for aquatic organisms that inhabit hypersaline systems in the region (Morales et al., 2011; Collado and Méndez, 2013; Cruz-Jofré et al., 2016; Valladares et al., 2018). Something similar occurs between Chaxa and Puilar, although the relationship is more diffuse, since both share a main haplotype.
The fluctuating connectivity model proposed for the Atacama Saltpan mainly considers the topography of the saltpan (see Munk et al., 2018) and the occurrence of extreme rainfall events in the region (Wirrmann and Mourguiart, 1995; Betancourt et al., 2000; Bobst et al., 2001; Sáez et al., 2012; González-Silvestre et al., 2013; Ritter et al., 2018). In the case of Heleobia, these components are even more critical given that it is a strictly aquatic microgastropod, therefore, migration between systems could have occurred exclusively as a result of a passive movement associated with flood events that would connect hydrologically isolated systems.
The results obtained using mitochondrial sequences and nuclear microsatellites indicate that the genetic variability found in the populations of Heleobia of the saltpan can be divided into three levels: first, the systems located in the northern region of the saltpan (Tebenquiche, Puilar and Chaxa) and the systems located in the south (Salada, La Punta and La Brava), which host most of the genetic variability, and in the case of Chaxa-Puilar and Salada-La Punta, are inhabited by individuals with shared haplotypes (suggesting genetic flow). Second, Quelana presents intermediate genetic diversity and mainly haplotypes not shared with other systems, indicating genetic isolation from the rest of the systems, even when it is located in the geographic center of the saltpan. Third, Tilopozo has low gene flow values compared with other populations and reduced genetic variability. The low genetic diversity detected in this system is probably explained by various historical demographic events that affected its population size. A particular case was detected in La Brava, since this system presented the lowest mitochondrial genetic diversity (a single haplotype shared with La Punta), but a high number of alleles and the highest allelic richness estimated from microsatellites. If it is also considered that La Brava presented the lowest level of relatedness (R = 0.038), no private alleles and was recovered in the same population with La Punta and Salada (microsatellite results), it is probable that a recent colonization has occurred from a nearby system to La Brava.
According to the microsatellite analysis, five distinct populations are recognized in the Atacama Saltpan. In Tilopozo (type-locality of H. atacamensis), the population with the lowest levels of contemporary gene flow was recovered, suggesting that this population has remained isolated from the rest (supporting what was obtained in the MSCT reconstruction and COI haplotype network). The individuals of the northern systems of the saltpan (Tebenquiche, Puilar and Chaxa) formed a delimited group, although they present some genotypes associated with the Quelana population. Specimens from the Salada, La Punta and La Brava systems formed a genetic cluster, which would correspond to a population limited to the south-central sector of the saltpan. Gene flow analyses suggest that the movement of individuals mainly occurs between nearby systems (Puilar-Chaxa and Salada-La Punta); however, this also reflects the environmental similarity of the systems that present the highest values of connectivity. Tilopozo presents unique environmental conditions (measured as physicochemical variation), and at the same time, it presents the lowest values of gene flow. This pattern is repeated in the northern systems, which overlap in the environmental space and have high degrees of connectivity.
Similarly, temporal habitat influence in terms of connectivity and fragmentation can be inferred by comparing the multilocus phylogenetic estimation (i.e., historical genetic structure) and the results of microsatellite analyses (i.e., contemporary genetic structure). Although both results show the relationship between genetic structure and physiography, the main difference is the relationship between Tebenquiche (north of the saltpan) and Tilopozo (south of the saltpan). These systems are the most distant (∼70 km), however, in all phylogenetic reconstructions they were recovered in the same lineage. In contrast, the analyses using microsatellites grouped Tebenquiche with Chaxa and Puilar (all are systems located in the north of the saltpan) and Tilopozo was defined as an independent population. In the case of phylogenetic reconstruction, the Tilopozo/Tebenquiche relationship is likely due to polymorphism retention, considering the geographic distance between the systems, which would prevent the connection between them even in extreme rainfall events. On the other hand, the contemporary genetic pattern is consistent with the spatial arrangement of the systems and with the possible migration routes in extreme rainfall events. This if we consider that the differences in altitude between localities (see Munk et al., 2018) would only promote the connection of nearby systems and toward the center of the saltpan.
The results indicate that the genetic differentiation pattern of Heleobia throughout the saltpan is intimately related to the spatial isolation of the populations. This pattern of IBD was significant in all analyses and is consistent with studies in hydrological systems within desert environments (Murphy et al., 2012, 2015; Valladares et al., 2018). Consequently, high levels of population relatedness were detected in most of the systems, which is probably not only related to the geographic isolation but also to the low vagility and reproductive biology reported for Heleobia atacamensis (Collado et al., 2021). In this sense, hydrological systems composed of springs with a certain degree of isolation and in a desert environment, are habitats where endemic species have evolved into a variety of taxa, which apparently maintain species in relatively stable and isolated conditions (Moline et al., 2004; Murphy et al., 2009; Sei et al., 2009; Trajanovski et al., 2010). The present study shows how in an endemic gastropod, the identification of genetically differentiated populations provides information on the hydrological structure underlying a complex system of lagoons and aquifers such as the Atacama Saltpan.
The results show that much of the current genetic diversity of the populations of Heleobia of the Atacama Saltpan is related to different degrees of connectivity and geographic isolation. In addition, the low indices of diversity and high degree of fragmentation indicate that the saltpan populations of Heleobia are highly differentiated. This is crucial when establishing management plans for H. atacamensis, given that much of the genetic diversity detected resides in small isolated populations. Therefore, inappropriate management that does not consider the evolutionary history of these lineages could trigger an event of outbreeding depression or a depletion of the genotypic pool (Cook and Sgrò, 2019). For example, the low diversity and high degree of isolation detected in Quelana is of concern; all the analyses detected it as a highly differentiated population with a low degree of connectivity, and clearly repopulating with other populations does not align with the recommended procedure (Waller, 2015; Ralls et al., 2018). On the other hand, in Tilopozo, in addition to the genetic differentiation detected, the environmental variables (e.g., temperature and salinity) were different from all other systems of the saltpan. This condition turns the type-locality of H. atacamensis into a unique habitat in the system, whose ecological imprint is strongly related to the degree of genetic differentiation of the population. Therefore, an artificial restoration of genetic flow in the Atacama Saltpan should be restricted to populations that present a population decline or an extreme decline in genetic diversity. Finally, based on the present results, there was no evidence of contemporary changes in the saltpan impacting demographic stability of the Heleobia populations. We propose that this may be explained by different strategies that this species has developed in response to the historical environmental dynamism that has occurred in the system. However, it is necessary to note that this fragile equilibrium largely depends on the local conditions for each lineage in the saltpan. Therefore, conservation plans for the species that are distributed in the Atacama Saltpan should be geared toward the preservation and characterization of the conditions (biotic and abiotic) that characterize each system. In addition, it is necessary to implement studies that allow monitoring the temporal change in terms of diversity and genetic connectivity of the of Heleobia populations of the saltpan.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
MV designed the experiments. MV and GC performed the sampling. AF and PS performed the experiments. MV and AF analyzed the data. MM obtained funding for the original project idea and contributed in work proposal. MV wrote the manuscript with input from GC. All authors reviewed the manuscript.
This work was supported by the Compañía Minera Albemarle Ltda., and Centro de Ecología Aplicada Ltda., Chile.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
MV acknowledges a Postdoctoral Fellowship VRIP UBB No. 121859/2266/2021. MM acknowledges to FONDECYT-ANID 1200419. MV thank Ian J. Wang for his help with IBD/IBE analyses. We also thank Elizabeth Chihuailaf, Natalia C. Muñoz-Herrera and Franco Cruz-Jofré for their assistance during samplings. We would like to thank the reviewers for their comments and suggestions that helped us to improve the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.869626/full#supplementary-material
Adamack, A. T., and Gruber, B. (2014). PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol. Evol. 5, 384–387. doi: 10.1111/2041-210X.12158
Alonso, H., and Risacher, F. (1996). Geoquímica del salar de atacama, parte 1: origen de los componentes y balance salino. Rev. Geol. Chile 23, 113–122. doi: 10.5027/andgeoV23n2-a01
Ammann, C., Jenny, B., Kammer, K., and Messerli, B. (2001). Late quaternary glacier response to humidity changes in the arid andes of chile (18–29°S). Palaeogeogr. Palaeoclimatol. Palaeoecol. 172, 313–326. doi: 10.1016/S0031-0182(01)00306-6
Baker, P. A., and Fritz, S. C. (2015). Nature and causes of quaternary climate variation of tropical South America. Quat. Sci. Rev. 124, 31–47. doi: 10.1016/j.quascirev.2015.06.011
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N., and Bonhomme, F. (2004). GENETIX 4.05, Logiciel sous Windows TM Pour la Génétique des Populations. Montpellier: Laboratoire Génome, Populations, Interactions. CNRS Umr 5171, Université de Montpellier r.
Betancourt, J. L., Latorre, C., Rech, J., Quade, J., and Rylander, K. A. (2000). A 22,000-year record of monsoonal precipitation from northern chile’s atacama desert. Science 289, 1542–1546. doi: 10.1126/science.289.5484.1542
Bobst, A. L., Lowenstein, T. K., Jordan, T. E., Godfrey, L. V., Ku, T. L., and Luo, S. (2001). A 106 ka paleoclimate record from drill core of the salar de atacama, northern Chile. Palaeogeogr. Palaeoclimatol. Palaeoecol. 173, 21–42. doi: 10.1016/S0031-0182(01)00308-X
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., et al. (2019). BEAST 2.5: an advanced software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 15:e1006650. doi: 10.1371/journal.pcbi.1006650
Bouckaert, R. R., and Drummond, A. J. (2017). bModelTest: bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 17:42. doi: 10.1186/s12862-017-0890-6
Cazzaniga, N. J., Matínez, S. A., De Francesco, C. G., Collado, G. A., Ciocco, N. F., Ovando, X. M. C., et al. (2011). “Diversidad, biología y ecología de especies del género Heleobia de la Provincia Malacológica de Cuyo, Argentina,” in El Género Heleobia (Caenogastropoda: Cochliopidae) en América del Sur. Amici Molluscarum, ed. Cazzaniga, N. J. (Santiago: Sociedad Malacológica de Chile (SMACH)), 11–48.
Charlesworth, D., and Willis, J. H. (2009). The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796. doi: 10.1038/nrg2664
Chevin, L.-M., and Lande, R. (2010). When do adaptive plasticity and genetic evolution prevent extinction of a density-regulated population? Evolution 64, 1143–1150. doi: 10.1111/j.1558-5646.2009.00875.x
Chevin, L.-M., Lande, R., and Mace, G. M. (2010). Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol. 8:e1000357. doi: 10.1371/journal.pbio.1000357
Clarke, J. D. A. (2006). Antiquity of aridity in the chilean atacama desert. Geomorphology 73, 101–114. doi: 10.1016/j.geomorph.2005.06.008
Coates, D. J., Byrne, M., and Moritz, C. (2018). Genetic diversity and conservation units: dealing with the species-population continuum in the age of genomics. Front. Ecol. Evol. 6:165. doi: 10.3389/fevo.2018.00165
Collado, G. A., Chihuailaf, E., Muñoz-Herrera, N., Contreras, M., Novoa, F., and Valladares, M. A. (2021). Reproductive aspects of the poorly known and critically endangered freshwater snail Heleobia atacamensis (Gastropoda: Truncatelloidea). PeerJ 9, e11550. doi: 10.7717/peerj.11550
Collado, G. A., and Méndez, M. A. (2012). Phylogenetic relationships and taxonomy of Altiplano populations of Biomphalaria (Gastropoda: Planorbidae): inference from a multilocus approach. Zool. J. Linn. Soc. 165, 795–808. doi: 10.1111/j.1096-3642.2012.00829.x
Collado, G. A., and Méndez, M. A. (2013). Microgeographic differentiation among closely related species of Biomphalaria (Gastropoda: Planorbidae) from the Andean Altiplano. Zool. J. Linn. Soc. 169, 640–652. doi: 10.1111/zoj.12073
Collado, G. A., Valladares, M. A., and Méndez, M. A. (2013). Hidden diversity in spring snails from the Andean Altiplano, the second highest plateau on Earth, and the Atacama Desert, the driest place in the world. Zool. Stud. 52:50. doi: 10.1186/1810-522X-52-50
Collado, G. A., Valladares, M. A., and Méndez, M. A. (2016a). A new species of Heleobia (Caenogastropoda: Cochliopidae) from the Chilean Altiplano. Zootaxa 4137, 277–280. doi: 10.11646/zootaxa.4137.2.8
Collado, G. A., Valladares, M. A., and Méndez, M. A. (2016b). Unravelling cryptic species of freshwater snails (Caenogastropoda, Truncatelloidea) in the Loa River basin, Atacama Desert. Syst. Biodivers. 14, 417–429. doi: 10.1080/14772000.2016.1153526
Cook, C. N., and Sgrò, C. M. (2019). Poor understanding of evolutionary theory is a barrier to effective conservation management. Conserv. Lett. 12, 1–10. doi: 10.1111/conl.12619
Cruz-Jofré, F., Morales, P., Vila, I., Esquer-Garrigos, Y., Hugueny, B., Gaubert, P., et al. (2016). Geographical isolation and genetic differentiation: the case of Orestias ascotanensis (Teleostei: Cyprinodontidae), an andean killifish inhabiting a highland salt pan. Biol. J. Linn. Soc. 117, 747–759. doi: 10.1111/bij.12704
Cubillos, C. F., Aguilar, P., Grágeda, M., and Dorador, C. (2018). Microbial communities from the world’s largest lithium reserve, salar de atacama, chile: life at high licl concentrations. J. Geophys. Res. Biogeosciences 123, 3668–3681. doi: 10.1029/2018JG004621
Demergasso, C., Dorador, C., Meneses, D., Blamey, J., Cabrol, N., Escudero, L., et al. (2010). Prokaryotic diversity pattern in high-altitude ecosystems of the Chilean Altiplano. J. Geophys. Res 115:G00D09. doi: 10.1029/2008JG000836
Dorador, C., Fink, P., Hengst, M., Icaza, G., Villalobos, A. S., Vejar, D., et al. (2018). Microbial community composition and trophic role along a marked salinity gradient in Laguna Puilar, Salar de Atacama, Chile. Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 111, 1361–1374. doi: 10.1007/s10482-018-1091-z
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol 29, 1969–1973. doi: 10.1093/molbev/mss075
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fabres, A. A., Valladares, M. A., Sáez, P. A., Collado, G. A., Pastenes, L., and Méndez, M. A. (2020). Novel microsatellite markers for an endangered freshwater snail, Heleobia atacamensis (Caenogastropoda: Cochliopidae), from the Atacama Saltpan. Molluscan Res. 40, 231–235. doi: 10.1080/13235818.2020.1775367
Falush, D., Stephens, M., and Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587. doi: 10.1111/j.1471-8286.2007.01758.x
Falush, D., Stephens, M., and Pritchard, J. K. (2007). Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 7, 574–578.
Fitzpatrick, S. W., and Reid, B. N. (2019). Does gene flow aggravate or alleviate maladaptation to environmental stress in small populations? Evol. Appl. 12, 1402–1416. doi: 10.1111/eva.12768
Fornari, M., Risacher, F., and Féraud, G. (2001). Dating of paleolakes in the central Altiplano of Bolivia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 172, 269–282. doi: 10.1016/S0031-0182(01)00301-7
Francis, R. M. (2017). pophelper: an R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17, 27–32. doi: 10.1111/1755-0998.12509
Furness, A. I., Lee, K., and Reznick, D. N. (2015). Adaptation in a variable environment: Phenotypic plasticity and bet-hedging during egg diapause and hatching in an annual killifish. Evolution. 69, 1461–1475. doi: 10.1111/evo.12669
González-Silvestre, L., Maldonado, A., Núñez, L., Cartajena, I., Carrasco, C., and de Souza, P. (2013). Condiciones paleovegetacionales y asentamientos humanos durante el formativo. temprano: análisis de polen del sitio tulán-85 (1.530/1.260-460/420 años cal. a.C.), cuenca del salar de atacama. Chungará (Arica) 45, 387–410. doi: 10.4067/S0717-73562013000300003
Goslee, S. C., and Urban, D. L. (2007). The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22, 1–19. doi: 10.18637/jss.v022.i07
Guerrero-Jiménez, C. J., Peña, F., Morales, P., Méndez, M., Sallaberry, M., Vila, I., et al. (2017). Pattern of genetic differentiation of an incipient speciation process: the case of the high Andean killifish Orestias. PLoS One 12:e0170380. doi: 10.1371/journal.pone.0170380
Guillot, G., Mortier, F., and Estoup, A. (2005). Geneland: a computer package for landscape genetics. Mol. Ecol. Notes 5, 712–715. doi: 10.1111/j.1471-8286.2005.01031.x
Iannacone, J., and Alvariño, L. (2002). Efecto del detergente doméstico alquil aril sulfonato de sodio lineal (LAS) sobre la mortalidad de tres caracoles dulceacuícolas en el Perú. Ecol. Apl. 1, 81–87. doi: 10.21704/rea.v1i1-2.234
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Kroll, O., Hershler, R., Albrecht, C., Moreno Terrazas, E., Apaza, R., Fuentealba, C., et al. (2012). The endemic gastropod fauna of Lake Titicaca: correlation between molecular evolution and hydrographic history. Ecol. Evol. 2, 1517–1530. doi: 10.1002/ece3.280
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leigh, J. W., and Bryant, D. (2015). PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Lichstein, J. W. (2007). Multiple regression on distance matrices: a multivariate spatial analysis tool. Plant Ecol. 188, 117–131. doi: 10.1007/s11258-006-9126-3
Liu, W., Agusdinata, D. B., and Myint, S. W. (2019). Spatiotemporal patterns of lithium mining and environmental degradation in the Atacama Salt Flat, Chile. Int. J. Appl. Earth Obs. Geoinf. 80, 145–156. doi: 10.1016/j.jag.2019.04.016
Lowe, A. J., Boshier, D., Ward, M., Bacles, C. F. E., and Navarro, C. (2005). Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity 95, 255–273. doi: 10.1038/sj.hdy.6800725
Lowenstein, T. K., and Risacher, F. (2009). Closed basin brine evolution and the influence of Ca–Cl Inflow Waters: death valley and bristol dry lake California, Qaidam Basin, China, and Salar de Atacama, Chile. Aquat. Geochemistry 15, 71–94. doi: 10.1007/s10498-008-9046-z
Luikart, G., Allendorf, F., Cornuet, J.-M., and Sherwin, W. (1998). Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 89, 238–247. doi: 10.1093/jhered/89.3.238
Magalhães, T. R. F., Neves, R. A. F., Valentin, J. L., and Figueiredo, G. M. (2014). Do the changes in temperature and light affect the functional response of the benthic mud snail Heleobia australis (Mollusca: Gastropoda)? An. Acad. Bras. Cienc. 86, 1197–1206. doi: 10.1590/0001-3765201420130093
Marazuela, M. A., Vázquez-Suñé, E., Ayora, C., García-Gil, A., and Palma, T. (2019). Hydrodynamics of salt flat basins: the Salar de Atacama example. Sci. Total Environ. 651, 668–683. doi: 10.1016/j.scitotenv.2018.09.190
Marengo, J. A., Liebmann, B., Grimm, A. M., Misra, V., Silva Dias, P. L., Cavalcanti, I. F. A., et al. (2012). Recent developments on the South American monsoon system. Int. J. Climatol. 32, 1–21. doi: 10.1002/joc.2254
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). Creating the CIPRES science gateway for inference of large phylogenetic Trees. Proc. Gatew. Comput. Environ. Work. 1, 1–8.
Ministerio del Medio Ambiente (2014). Inventario Nacional de Especies de Chile: Heleobia Atacamensis (Philippi, 1860). 10o Proceso Clasif. Especies. MMA, Chile. Available online at: http://mma.gob.cl. (Accessed March 8, 2022)
Moline, A. B., Shuster, S. M., Hendrickson, D. A., and Marks, J. C. (2004). Genetic variation in a desert aquatic snail (Nymphophilus minckleyi) from Cuatro Ciénegas. Coahuila, Mexico. Hydrobiologia 522, 179–192. doi: 10.1023/B:HYDR.0000029972.80491.d3
Morales, P., Vila, I., and Poulin, E. (2011). Genetic structure in remnant populations of an endangered cyprinodontid fish. Orestias ascotanensis, endemic to the Ascotán salt pan of the Altiplano. Conserv. Genet. 12, 1639–1643. doi: 10.1007/s10592-011-0245-6
Mujica, M. I., Latorre, C., Maldonado, A., González-Silvestre, L., Pinto, R., de Pol-Holz, R., et al. (2014). Late Quaternary climate change, relict populations and present-day refugia in the northern Atacama Desert: a case study from Quebrada La Higuera (18° S). J. Biogeogr. 42, 76–88. doi: 10.1111/jbi.12383
Munk, L. A., Boutt, D. F., Hynek, S. A., and Moran, B. J. (2018). Hydrogeochemical fluxes and processes contributing to the formation of lithium-enriched brines in a hyper-arid continental basin. Chem. Geol. 493, 37–57. doi: 10.1016/j.chemgeo.2018.05.013
Muñoz, N., Charrier, R., and Reutter, K.-J. (1997). “Evolución de la cuenca del Salar de atacama: inversión tectónica y relleno de una cuenca de antepaís de retroarco,” in Proceedings of the 8th Congreso Geológico Chileno, (Antofagasta), 195–199.
Murphy, A. L., Pavlova, A., Thompson, R., Davis, J., and Sunnucks, P. (2015). Swimming through sand: connectivity of aquatic fauna in deserts. Ecol. Evol. 5, 5252–5264. doi: 10.1002/ece3.1741
Murphy, N. P., Adams, M., and Austin, A. D. (2009). Independent colonization and extensive cryptic speciation of freshwater amphipods in the isolated groundwater springs of Australia’s Great Artesian Basin. Mol. Ecol. 18, 109–122. doi: 10.1111/j.1365-294X.2008.04007.x
Murphy, N. P., Breed, M. F., Guzik, M. T., Cooper, S. J. B., and Austin, A. D. (2012). Trapped in desert springs: phylogeography of Australian desert spring snails. J. Biogeogr. 39, 1573–1582. doi: 10.1111/j.1365-2699.2012.02725.x
Nester, P. L., Gayó, E., Latorre, C., Jordan, T. E., and Blanco, N. (2007). Perennial stream discharge in the hyperarid Atacama Desert of northern chile during the latest Pleistocene. Proc. Natl. Acad. Sci. U.S.A. 104, 19724–19729. doi: 10.1073/pnas.0705373104
Ogilvie, H. A., Bouckaert, R. R., and Drummond, A. J. (2017). StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Mol. Biol. Evol. 34, 2101–2114. doi: 10.1093/molbev/msx126
Oksanen, J., Blanchet, F., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2007). The vegan package. Community Ecol. Packag. 10, 631–637.
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in excel. population genetic software for teaching and research –an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Piry, S., Luikart, G., and Cornuet, J.-M. (1999). BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 90, 502–503. doi: 10.1093/jhered/90.4.502
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Quade, J., Rech, J., Betancourt, J. L., Latorre, C., Quade, B., Rylander, K. A., et al. (2008). Paleowetlands and regional climate change in the central Atacama Desert, northern Chile. Quat. Res. 69, 343–360. doi: 10.1016/j.yqres.2008.01.003
R Development Core Team (2019). R: A Language and Environment for Statistical Computing 3.6.1. Vienna: R Foundation for Statistical Computing.
Ralls, K., Ballou, J. D., Dudash, M. R., Eldridge, M. D. B., Fenster, C. B., Lacy, R. C., et al. (2018). Call for a paradigm shift in the genetic management of fragmented populations. Conserv. Lett. 11:e12412. doi: 10.1111/conl.12412
Rambaut, A., Suchard, M. A., and Drummond, A. J. (2014). Tracer v1.6. Available online at: http//beast.bio.ed.ac.uk/Tracer. (accessed March 8, 2022).
Reed, D. H. (2004). Extinction risk in fragmented habitats. Anim. Conserv. 7, 181–191. doi: 10.1017/S1367943004001313
Reed, D. H., and Frankham, R. (2003). Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237. doi: 10.1046/j.1523-1739.2003.01236.x
Risacher, F., and Alonso, H. (1996). Geoquímica del Salar de Atacama, parte 2: evolución de las aguas. Rev. Geol. Chile 23, 123–134.
Risacher, F., and Fritz, B. (2009). Origin of salts and brine evolution of Bolivian and Chilean salars. Aquat. Geochemistry 15, 123–157. doi: 10.1007/s10498-008-9056-x
Ritter, B., Binnie, S. A., Stuart, F. M., Wennrich, V., and Dunai, T. J. (2018). Evidence for multiple Plio-Pleistocene lake episodes in the hyperarid Atacama Desert. Quat. Geochronol. 44, 1–12. doi: 10.1016/j.quageo.2017.11.002
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rousset, F. (1997). Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145, 1219–1228. doi: 10.1007/BF00341816
Sáez, A., Cabrera, L., Garcés, M., Van den Bogaard, P., Jensen, A., and Gimeno, D. (2012). The stratigraphic record of changing hyperaridity in the Atacama desert over the last 10Ma. Earth Planet. Sci. Lett. 355–356, 32–38. doi: 10.1016/j.epsl.2012.08.029
Schlaepfer, D. R., Braschler, B., Rusterholz, H.-P., and Baur, B. (2018). Genetic effects of anthropogenic habitat fragmentation on remnant animal and plant populations: a meta-analysis. Ecosphere 9:e02488. doi: 10.1002/ecs2.2488
Schuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 18, 233–234. doi: 10.1038/72708
Sei, M., Lang, B. K., and Berg, D. J. (2009). Genetic and community similarities are correlated in endemic-rich springs of the northern Chihuahuan Desert. Glob. Ecol. Biogeogr. 18, 192–201. doi: 10.1111/j.1466-8238.2008.00436.x
Soto, J., Román-Figueroa, C., and Paneque, M. (2019). A model for estimating the vegetation cover in the high-altitude Wetlands of the Andes (HAWA). Land 8, 1–17. doi: 10.3390/land8010020
Souza, F. M., Brauko, K. M., Lana, P. C., Muniz, P., and Camargo, M. G. (2013). The effect of urban sewage on benthic macrofauna: a multiple spatial scale approach. Mar. Pollut. Bull. 67, 234–240. doi: 10.1016/j.marpolbul.2012.10.021
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thomas, C. D. (2000). Dispersal and extinction in fragmented landscapes. Proc. R. Soc. London. Ser. B Biol. Sci. 267, 139–145. doi: 10.1098/rspb.2000.0978
Trajanovski, S., Albrecht, C., Schreiber, K., Schultheiß, R., Stadler, T., Benke, M., et al. (2010). Testing the spatial and temporal framework of speciation in an ancient lake species flock: the leech genus Dina (Hirudinea: Erpobdellidae) in Lake Ohrid. Biogeosciences 7, 3387–3402. doi: 10.5194/bg-7-3387-2010
Valladares, M. A., Méndez, M. A., and Collado, G. A. (2018). Influenced but not determined by historical events: genetic, demographic and morphological differentiation in Heleobia ascotanensis from the Chilean Altiplano. PeerJ 6:e5802. doi: 10.7717/peerj.5802
van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Waller, D. M. (2015). Genetic rescue: a safe or risky bet? Mol. Ecol. 24, 2595–2597. doi: 10.1111/mec.13220
Wang, I. J. (2013). Examining the full effects of landscape heterogeneity on spatial genetic variation: A multiple matrix regression approach for quantifying geographic and ecological isolation. Evolution 67, 3403–3411. doi: 10.1111/evo.12134
Wilcox, B. A., and Murphy, D. D. (1985). Conservation strategy: the effects of fragmentation on extinction. Am. Nat. 125, 879–887. doi: 10.1086/284386
Wilke, T., Schultheiß, R., and Albrecht, C. (2009). As time goes by: a simple fool’s guide to molecular clock approaches in invertebrates*. Am. Malacol. Bull. 27, 25–45. doi: 10.4003/006.027.0203
Wilson, G. A., and Rannala, B. (2003). Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191. doi: 10.1093/genetics/163.3.1177
Winnepenninckx, B., Backeljau, T., and De Wachter, R. (1993). Extraction of high molecular weight DNA from molluscs. Trends Genet. TIG 9:407. doi: 10.1016/0168-9525(93)90102-N
Wirrmann, D., and Mourguiart, P. (1995). Late quaternary spatio-temporal limnological variations in the Altiplano of Bolivia and Peru. Quat. Res. 43, 344–354. doi: 10.1006/qres.1995.1040
Wong, B. B. M., and Candolin, U. (2015). Behavioral responses to changing environments. Behav. Ecol. 26, 665–673. doi: 10.1093/beheco/aru183
Keywords: connectivity, conservation, historic fragmentation, isolation by distance (IBD), isolation by environment (IBE), microgeographic differentiation
Citation: Valladares MA, Fabres AA, Collado GA, Sáez PA and Méndez MA (2022) Coping With Dynamism: Phylogenetics and Phylogeographic Analyses Reveal Cryptic Diversity in Heleobia Snails of Atacama Saltpan, Chile. Front. Ecol. Evol. 10:869626. doi: 10.3389/fevo.2022.869626
Received: 04 February 2022; Accepted: 06 April 2022;
Published: 28 April 2022.
Edited by:
Izeni Pires Farias, Federal University of Amazonas, BrazilReviewed by:
Artur Osikowski, University of Agriculture in Krakow, PolandCopyright © 2022 Valladares, Fabres, Collado, Sáez and Méndez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco A. Méndez, bW1lbmRlekB1Y2hpbGUuY2w=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.